Abstract
Specificity is one of the most important and complex properties that is central to both natural antibody function and therapeutic antibody efficacy. However, it has proven extremely challenging to define robust guidelines for predicting antibody specificity. Here we evaluated the physicochemical determinants of antibody specificity for multiple panels of antibodies, including >100 clinical-stage antibodies. Surprisingly, we find that the theoretical net charge of the complementarity-determining regions (CDRs) is a strong predictor of antibody specificity. Antibodies with positively charged CDRs have a much higher risk of low specificity than antibodies with negatively charged CDRs. Moreover, the charge of the entire set of six CDRs is a much better predictor of antibody specificity than the charge of individual CDRs, variable domains (VH or VL) or the entire variable fragment (Fv). The best indicators of antibody specificity in terms of CDR amino acid composition are reduced levels of arginine and lysine and increased levels of aspartic and glutamic acid. Interestingly, clinical-stage antibodies with negatively charged CDRs also have a lower risk for poor biophysical properties in general, including a reduced risk for high levels of self-association. These findings provide powerful guidelines for predicting antibody specificity and for identifying safe and potent antibody therapeutics.
Keywords: aggregation, CDR, mAb, non-specific binding, polyspecificity
Introduction
The specificity of antibodies – defined as their ability to strongly recognize their targets and weakly recognize off-target molecules – is central to their natural functions in the immune system. Seminal work on understanding the evolution of antibody specificity during B-cell maturation revealed that a majority of immature B-cells are polyspecific, and their specificity is greatly increased during the process of B-cell maturation and production of antigen-specific antibodies (Wardemann et al., 2003; Keenan et al., 2008). Defects in this natural process of antibody specificity maturation are linked to autoimmune disorders (Goodnow, 2007).
The widespread use of antibodies as therapeutics has led to much interest in understanding determinants of antibody specificity. These efforts include new or improved experimental methods for profiling antibody specificity that evaluate antibody interactions with themselves (i.e. self-association) (Jacobs et al., 2010; Sule et al., 2013; Sun et al., 2013; Jayaraman et al., 2014; Liu et al., 2014; Estep et al., 2015; Kelly et al., 2015; Li et al., 2015; Wu et al., 2015; Alam et al., 2018; Avery et al., 2018; Geng et al., 2016a,b) and with diverse types of biomolecules, including proteins, nucleic acids and polysaccharides (Wardemann et al., 2003; Mouquet et al., 2010; Hotzel et al., 2012; Xu et al., 2013; Avery et al., 2018; Datta-Mannan et al., 2015a,b; Kelly et al., 2017a,b). Interestingly, antibody specificity has recently been shown to be a key physicochemical predictor of the relative likelihood of success of antibody drugs in the clinic (Jain et al., 2017).
Nevertheless, it has remained challenging to define the molecular-level determinants of antibody specificity for two main reasons. First, antibody specificity is a relative concept and its definition is based on experimental measurements that are dependent on the type of non-target molecules used for specificity analysis. Therefore, it is not possible to compare antibody specificity measurements from different studies that use different polyspecificity reagents. Second, it is difficult to define the sequence determinants of antibody specificity given that antibody variable regions – and especially the complementarity-determining regions (CDRs) – display significant sequence variation. Therefore, the small portion of maximal chemical (amino acid) diversity that is typically sampled in a given study of antibody specificity is often insufficient to determine how antibody sequence generally impacts antibody specificity.
The goal of this study is to identify key sequence determinants of antibody specificity and address some of the challenges that have previously limited such analysis. Based on prior work (Wardemann et al., 2003; Birtalan et al., 2008, 2010; Sharma et al., 2014; Dobson et al., 2016; Datta-Mannan et al., 2015a,b; Kelly et al., 2017a,b; Tiller et al., 2017a,b), we reasoned that the amino acid composition of antibody CDRs is the primary determinant of antibody specificity. Therefore, we first sought to identify sequence determinants of antibody specificity using common framework antibodies with sequence variation only in a single CDR (heavy chain CDR3, HCDR3). Next, we sought to test the generality of these findings using a larger panel of antibodies with much more sequence variation and whose specificity has been rigorously profiled using several different types of experimental measurements (Jain et al., 2017). Finally, we evaluated the connection between CDR sequence determinants of antibody specificity and antibody biophysical properties in general, including antibody hydrophobicity, self-association and aggregation. Here we report key molecular-level determinants of antibody specificity and demonstrate how specific types of chemical properties of antibody CDRs govern antibody specificity and other biophysical properties for diverse antibodies, including those in clinical development.
Materials and Methods
Cloning of antibody variants
The scFv variants were cloned from an existing pET-17b bacterial expression plasmid containing the parent scFv gene between the HindIII and KpnI restriction sites. The scFv contains an N-terminal pelB sequence and C-terminal 3× FLAG and 7× His tags. scFv variants were created using custom DNA primers (Integrated DNA Technologies) and site-directed mutagenesis with PfuUltra II fusion polymerase (600850, Agilent Technologies). The scFv variants were also subcloned into the pBIOCAM5 mammalian expression vector (39344, Addgene) at the N-terminus of human IgG1 Fc. Each scFv gene was amplified from the bacterial expression plasmids using primers that incorporate NcoI and NotI restriction sites at the 5′ and 3′ ends, respectively. The genes were then ligated into the pBIOCAM5 mammalian expression vector (39344, Addgene). The resulting scFv-Fc fusions contain a 6× His tag and 3× FLAG tag at the C-terminus.
Antibody mammalian expression and purification
The scFv-Fc variants were expressed transiently using an adherent HEK293T cell line (CRL-3216, ATCC). Cultures were seeded with 2 million cells in a 75 cm2 tissue culture (T75) flask (10 062-860, VWR) containing 15 mL of DMEM-GlutaMAX (10 569-044, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (35 010CV, Corning) and 1% penicillin-streptomycin (15140122, Thermo Fisher Scientific). The cultures were grown at 37°C with 5% CO2 until ~70% confluency (2 days). The cells were then transfected with expression plasmids using Lipofectamine 2000 (11668019, Thermo Fisher Scientific). First, 20 μg of lipofectamine and 8 μg of plasmid DNA were added separately to 0.5 mL of Opti-MEM (31 985-062, Thermo Fisher Scientific) and incubated for 10 min. Next, the lipofectamine solution was combined with the DNA solution and incubated for 30 min. Following incubation, the lipofectamine/DNA mixture (1 mL total volume) was added to each flask. Three T75 flasks were used for expression of each scFv-Fc antibody. The expression cultures were grown at 37°C with 5% CO2 for 4 days, and the media containing the secreted antibodies was collected.
The scFv-Fc antibodies were purified by adding 0.5 mL of a 50% slurry of Protein A agarose resin (20333, Thermo Fisher Scientific) to the collected media and incubating overnight at 4°C with rocking. Next, the resin was collected by vacuum into a 10 mL centrifuge column (89898, Thermo Fisher Scientific) and washed with 100–150 mL of PBS (pH 7.4). The antibodies were eluted by incubating the resin in 0.5–1 mL of 0.1 M glycine (pH 2.5) for 15 min. After elution, the antibodies were neutralized by addition of 1 M K2HPO4 (100 μL per mL of elution volume). Aggregate content was analyzed using size-exclusion chromatography (SEC). For expression batches with >90% monomer, the scFv-Fc antibodies were buffer exchanged twice into PBS prior to use (Zeba desalting columns, 89882, Thermo Fisher Scientific). Samples with <90% monomer were further purified by preparative SEC using a Shimadzu high performance liquid chromatography system. The scFv-Fc antibodies were injected (250 μL) onto two YMC-Pack-Diol-200 columns (30 cm × 8 mm; YMC) in series using PBS with 200 mM arginine (pH 7.4) at a flow rate of 0.6 mL/min as the mobile phase. Signal was monitored by UV absorbance at 280 nm, and 0.3 mL fractions were collected. The collected fractions were pooled, concentrated (Amicon Ultra-0.5 mL centrifugal filter, UFC505096, Millipore) and buffer exchanged once into PBS (Zeba desalting columns, 89882, Thermo Fisher Scientific). The resulting antibody concentration was measured using the MicroBCA assay (23235, Thermo Fisher Scientific). The antibodies were re-analyzed by SEC to ensure >90% monomer and evaluated by SDS-PAGE (WG1203BX10, Thermo Fisher Scientific).
Antibody bacterial expression and purification
Bacterial expression plasmids were transformed into BL21(DE3) pLysS cells (200 132, Agilent Technologies). Transformed colonies were inoculated into 200 mL of auto-induction media (Studier, 2005) supplemented with ampicillin (100 μg/mL) and chloramphenicol (35 μg/mL). The expression cultures were incubated for 48 h at 30°C with shaking at 225 rpm. The cells were pelleted, and the scFvs were purified from the supernatant via their polyhistidine tag. The supernatant was incubated with 3 mL of Ni-NTA resin (30230, Qiagen) overnight at 4°C and 80 rpm. The Ni-NTA resin was collected by vacuum and washed with 250 mL of PBS. An additional wash was performed using 3 mL of PBS (pH 7.4) with 50 mM imidazole for 15 min. The scFvs were eluted by incubating the resin with 3 mL of PBS (pH 7.4) supplemented with 500 mM imidazole and 6 M GuHCl (15 min). After elution, the scFvs were allowed to unfold overnight at 4°C and refolded by buffer exchanging twice into PBS (89894, Thermo Fisher Scientific). Aggregates were removed by centrifugation at 21 000×g for 5 min and filtering through a 0.22 μm filter (SLGV004SL, Millipore). The antibody concentration was measured by UV absorbance at 280 nm, and SDS-PAGE (WG1203BX10, Thermo Fisher Scientific) was used to assess purity.
Non-specific binding analysis
For non-specific binding analysis using milk, transparent MaxiSorp 384 well plates (P6366, Sigma) were blocked with 100 μL of 10% (w/v) milk (instant non-fat dry milk, Kroger) in PBS overnight at 4°C and subsequently washed six times. All washes were performed with 100 μL of PBS supplemented with 0.1% Tween 20 (PBST). The scFv-Fc fusions and mAbs were diluted in PBS to a range of concentrations (0–300 nM). After dilution, 25 μL of scFv-Fc or mAb was added to the wells. Binding occurred overnight at room temperature. After three washes, 25 μL of a 1000× dilution of goat anti-human IgG Fc-HRP (A18817, Thermo Fisher Scientific) or a 1000× dilution of goat anti-mouse IgG-HRP (32430, Thermo Fisher Scientific) in PBST was added to the wells for the scFv-Fc fusions and mAbs, respectively. The plate were incubated for 1 h at room temperature and subsequently washed three times. Next, 25 μL of 1-Step Ultra TMB-ELISA substrate (34028, Thermo Fisher Scientific) was added to each well and developed for 5–6 min. The reaction was quenched with 25 μL of 2 M H2SO4. Signals were quantified by measuring the absorbance at 450 nm (SpectraMax Plus384, Molecular Devices). Non-specific binding signals were reported as the signal divided by the background. The background was evaluated without scFv-Fc or mAb but with all other detection reagents.
For the non-specific binding analysis using a panel of non-antigen proteins, transparent MaxiSorp 384 well plates (P6366, Sigma) were coated with 100 μL of 0.2 mg/mL proteins in PBS. The panel of proteins included ovalbumin (pI = 4.6; S25132, Thermo Fisher Scientific), bovine serum albumin (BSA; pI = 4.7; BP9706 Thermo Fisher Scientific), insulin (pI = 5.4; I9278, Sigma), keyhole limpet hemocyanin (KLH; pI = 4.6; H8283, Sigma), ribonuclease A (pI = 9.3; R4875, Sigma), avidin (pI = 10; A9275, Sigma) and lysozyme (pI = 11, L6876, Sigma). The wells were coated at 4°C overnight and subsequently washed six times. All washing steps were performed with 100 μL of PBST. The scFv-Fc fusions were diluted to 100 nM in PBS, added to the wells (25 μL), and allowed to bind overnight at room temperature. Subsequent steps were performed as described for the milk non-specific binding experiments.
Circular dichroism
Antibody (scFv) thermal stability was measured via circular dichroism (CD) using a Jasco 815 spectrophotometer. Ellipticity was monitored at 235 nm while heating from 25 to 95°C. Stock scFv solutions were diluted in PBS to 0.1 mg/mL and heated at a rate of 0.1°C/min. After collecting ellipticity data every 0.5°C, the scFv solutions were cooled back to 25°C and left for 10 min. A second melt was then performed to examine the efficiency of refolding after thermal denaturation. Ellipticity data was normalized from 0 to 1 using the maximum and minimum ellipticity values from the first melt. The melting temperatures were calculated from the first thermal melts. Both the folded and unfolded ellipticities were fit as linear functions of temperature. The fraction of folded scFv at a given temperature in the transition region (ΘT) was then calculated from (ΘT–ΘU)/(ΘF–ΘU), where (ΘU) and (ΘF) represent the predicted ellipticities if the proteins were unfolded or folded, respectively. Linear regression of the fraction folded versus temperature within the transition region was used to determine the apparent melting temperature (temperature at which the fraction folded is equal to 0.5).
Non-native antibody solubility
Non-native scFv solubility was analyzed by heating the antibody solutions in PBS at a concentration of ~0.4 mg/mL at 65°C for 4 h. Following the heat stress, the antibody solutions were cooled to 4°C overnight, centrifuged at 21 000×g for 5 min and the top 50% of the supernatant was removed for concentration analysis. Soluble antibody concentration was then measured using the BCA assay (23225, Thermo Fisher Scientific).
Size-exclusion chromatography
For analysis of the scFv-Fc antibodies, the samples were injected (100 μL at 60 μg/mL) onto an analytical YMC-Pack-Diol-200 column (30 cm × 8 mm; YMC) using a Shimadzu high performance liquid chromatography system. The mobile phase consisted of PBS containing 200 mM arginine (pH 7.4) at a flow rate of 0.7 mg/mL. Monomer content was quantified using the Lab Solutions software. A similar method was performed to assess monomer content following preparative SEC using an scFv-Fc concentration of 10 μg/mL (100 μL injection volume).
For the scFv antibodies, samples were analyzed using the same column described above on an Agilent 1260 Infinity II high-performance liquid chromatography system. First, the scFvs were diluted to 0.40 mg/mL in PBS (pH 7.4) and heated at 55°C for 1 h to reduce the amount of insoluble aggregate. The samples were then centrifuged at 21 000×g for 30 min, and the top 80% of the supernatant was recovered. Next, half of the sample was normalized to 0.25 mg/mL (PBS, pH 7.4) for analysis, and the other half was heated to 65°C for 4 h, incubated overnight at 4°C, and centrifuged at 21 000×g for 30 min to remove aggregates. The antibody solutions were then injected (100 μL) into the column (mobile phase was PBS with 0.2 M arginine, pH 7.4). The column flow rate was 0.7 mL/min, and scFv elution was monitored via UV absorbance at 280 nm.
Analysis of sequence and biophysical data for clinical-stage antibodies
The amino acid sequences and biophysical measurements for the 137 clinical-stage antibodies were obtained from a previous publication (Jain et al., 2017). The CDRs of all antibodies were identified using Kabat numbering. The theoretical net charges of the different antibody regions at pH 7.4 were calculated by summing the charges of Glu (−1), Asp (−1), Arg (+1), Lys (+1) and His (+0.1).
Logistic regression analysis was performed using limits for each biophysical property and assigning each mAb as being either above or below the various limits. Next, the mAbs were binned based on increments of their net charge, and the percentages of mAbs in each bin exceeding the cutoff values were calculated. Finally, a logistic function [y = 1/(1 + exp(–A–Bx))] was fit to the data and the P-value of the independent coefficient (B) was calculated to evaluate statistical significance. This was performed in Microsoft Excel using the ‘Binary Logistic and Probit Regression’ option in the Real Statistics resource pack add-in. First, each mAb was assigned a value of 1 if it exceeded the biophysical limit or a value of 0 if it was below the limit. This binary data along with the binned net charge values were used as the inputs for the logistic regression. Newton’s method was selected for the analysis (20 iterations). The regression analysis was performed using an alpha value of 0.05 and a classification cutoff of 0.5. This analysis was repeated without binning antibodies based on increments of their net charge.
The statistical significance of each correlation was also evaluated using receiver operating characteristic curve (ROC) analysis. The ROC curve was generated from the true positive rate and false positive rate outputs of the logistic regression performed with the Microsoft Excel add-in. The area under the curve was evaluated using the trapezoidal rule.
The sets of biophysical flags that were most strongly linked to CDR net charge for the clinical-stage antibodies were evaluated using the following procedure. First, the 10 biophysical assays (Jain et al., 2017) were clustered into all possible combinations of groups of 3–10 assays. For each biophysical group size (e.g. 3 assays), a given combination of assays (e.g. BVP, CSI and AC-SINS) was selected and the number of mAbs with <2 biophysical flags was calculated as a function of CDR net charge. Next, the strongest relationships between CDR net charge and the percentage of mAbs with <2 biophysical flags were calculated via the multinomial logistic regression (mnrfit) MATLAB function with the objective of achieving the best (lowest) P-value.
Results
Antibody specificity analysis for common framework antibodies
Toward our goal of understanding how antibody CDR sequence impacts antibody specificity, we first investigated antibody non-specific binding for three single-chain antibody fragments fused to human Fc (scFv-Fc; A10, B2 and AF1) that recognize the Alzheimer’s Aβ42 peptide (Tiller et al., 2017a,b). These antibodies have common frameworks and only differ in their HCDR3 sequences (Fig. 1). The wild-type (WT) antibody is based on the variable regions of 4D5 (Carter et al., 1992). We expressed these antibodies in HEK293T cells, purified them using Protein A chromatography (Fig. S1), and analyzed them using size exclusion chromatography (Fig. S2).
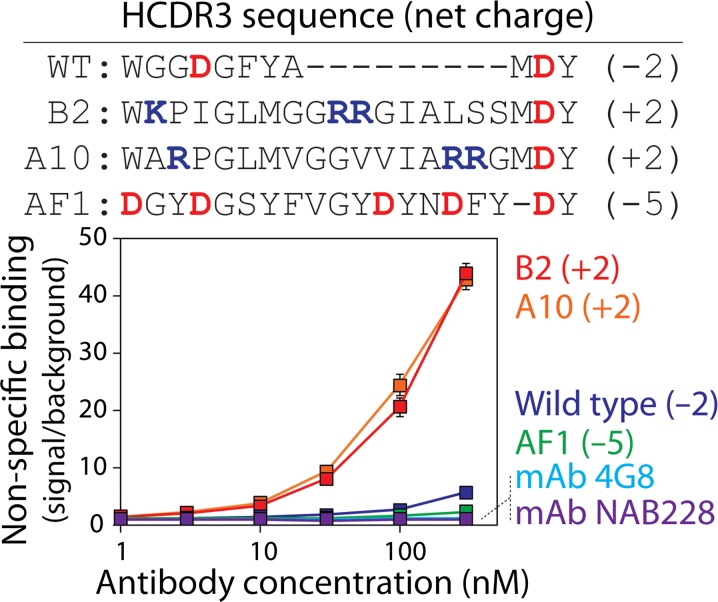
Fig. 1.
Non-specific binding analysis for a panel of antibodies. The levels of non-specific binding for scFv-Fc antibodies (WT, B2, A10 and AF1) and mAbs (4G8 and NAB228) were evaluated using well plates coated with milk proteins. The scFv-Fc antibodies have common frameworks and sequence variation only in heavy chain CDR3 (HCDR3). The amino acid sequences and theoretical net charges of HCDR3 at pH 7.4 are shown for the scFv-Fc antibodies. The levels of non-specific binding for two mAbs generated via immunization are also reported. The reported non-specific binding values are the signals for each antibody divided by the background signal. The background values were evaluated without scFv-Fc antibody or mAb but with all other detection reagents. The average values for three or four independent experiments are shown, and the error bars are standard errors.
We next evaluated the non-specific binding of the antibodies to milk proteins (and other types of molecules in milk) immobilized in well plates (Fig. 1). Increasing concentrations of the A10 and B2 antibodies resulted in large increases in non-specific binding that were much higher than those for the wild-type antibody. Conversely, the AF1 antibody displayed unusually low levels of non-specific binding. These levels were even lower than those for the wild-type antibody and similar to those for multiple Aβ-specific monoclonal antibodies (mAbs; 4G8 and NAB228) generated via immunization. It is notable that the scFv-Fc antibodies with high specificity (wild-type and AF1) had negatively charged HCDR3s, while those with low specificity (A10 and B2) had positively charged HCDR3s.
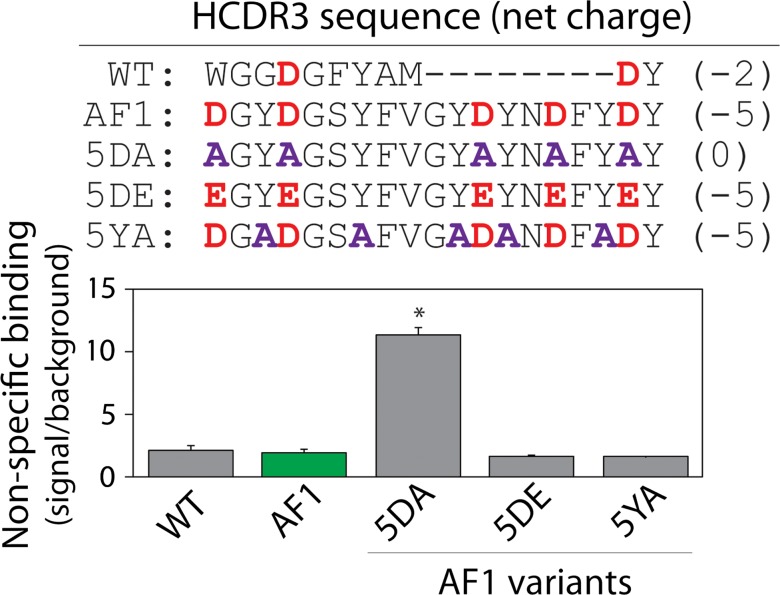
These findings suggest that the presence of negatively charged residues in HCDR3 may play a key role in mediating low levels of non-specific binding. Interestingly, the AF1 antibody has five negatively charged (aspartic acid) residues in HCDR3. To evaluate the importance of these negatively charged residues in promoting high specificity, we generated an alanine mutant in which five aspartic acids were mutated to alanine (5DA; Figs 2 and S1). Removal of aspartic acid residues dramatically increased non-specific binding to levels that approached those for antibodies with positively charged CDRs (Fig. 1). The favorable impact of the negatively charged HCDR3 residues on AF1 specificity was indistinguishable for glutamic and aspartic acid residues (5DE; Fig. 2). Finally, mutation of five tyrosine residues to alanine in HCDR3 of the AF1 antibody (5YA) resulted in similar levels of non-specific binding as AF1. These results suggest that the negatively charged residues in AF1 are important for its high specificity.
Fig. 2.
Analysis of the impact of negatively charged and tyrosine CDR residues on antibody specificity using alanine-scanning mutagenesis. Non-specific binding of the AF1-Fc mutants to milk proteins was evaluated as described in Fig. 1 (100 nM antibody). The impact of replacing aspartic acid with glutamic acid was also evaluated. The theoretical HCDR3 net charges were calculated at pH 7.4. A two-tailed Student’s t-test was used to evaluate statistical significance for each mutant relative to the parental AF1-Fc antibody [P-values <0.01 (*)]. The average values are for three independent experiments, and the error bars are standard errors.
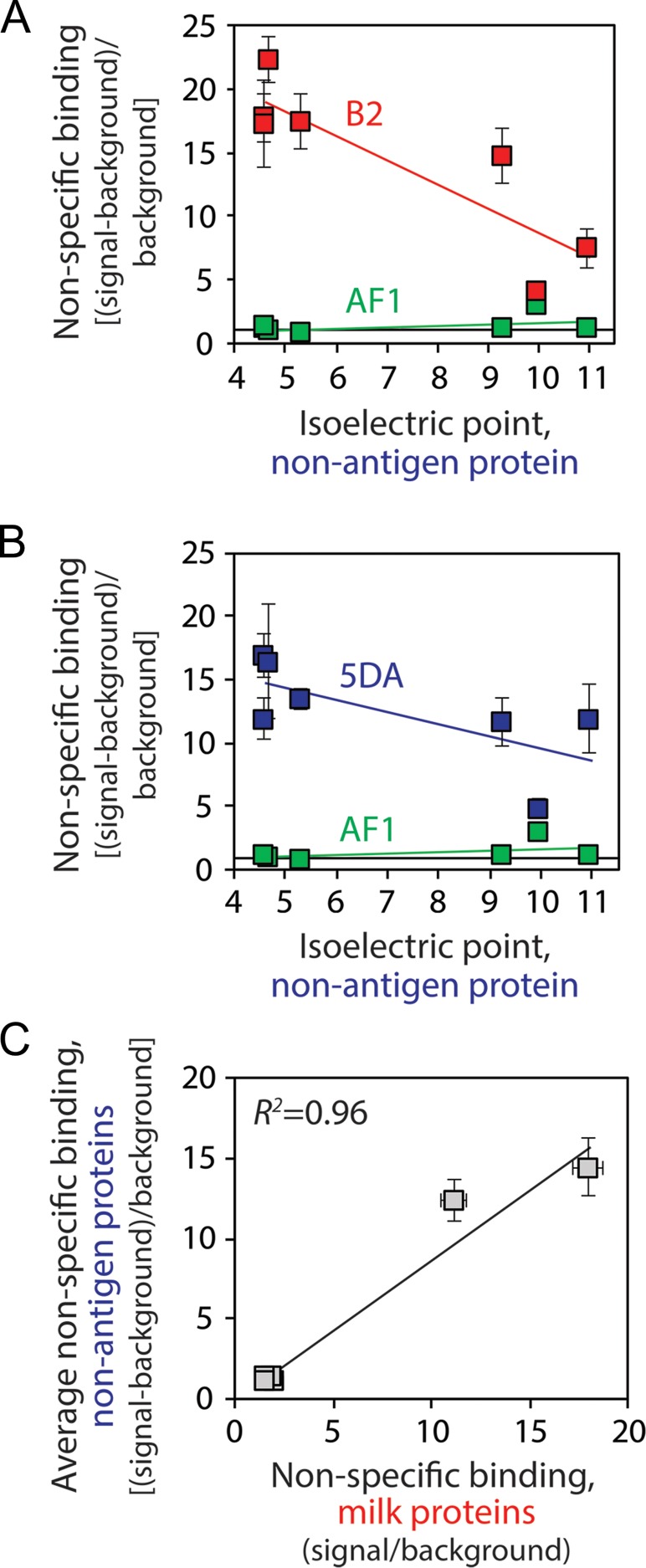
We next evaluated to what extent our findings were dependent on the type of molecules (milk proteins) that we used for measuring antibody non-specific interactions (Fig. 3). Milk is a complex mixture of casein and other molecules, and these milk components are generally expected to be negatively charged at neutral pH. Therefore, our findings that negatively charged CDRs reduce non-specific interactions may simply be due to the use of negatively charged milk proteins. Therefore, we evaluated the non-specific binding of AF1 and its mutants to a panel of proteins with both acidic (ovalbumin, pI 4.6; BSA, pI 4.7; insulin, pI 5.5; keyhole limpet hemocyanin, pI 4.6) and basic (ribonuclease A, pI 9.3; avidin, pI 10; lysozyme, pI 11) isoelectric points. The theoretical isoelectric points of the scFv-Fc antibodies were relatively similar (range of pIs of 8.1 for AF1 to 8.9 for B2). We found that AF1 generally displayed low levels of non-specific binding to the panel of proteins tested, with modestly higher levels of binding to proteins with high isoelectric points. In contrast, the B2 antibody displayed high levels of non-specific binding not only to proteins with low isoelectric points but to proteins with high isoelectric points as well (Fig. 3A). Similar trends were also seen with the AF1 alanine mutant (5DA) without charged residues in HCDR3 (Fig. 3B). Moreover, we find a strong correlation between antibody non-specific binding to milk proteins and the average non-specific binding values for the panel of seven proteins with acidic and basic isoelectric points (R2 of 0.96; Fig. 3C). Therefore, our findings for a small panel of scFv-Fc antibodies suggest that negatively charged residues in antibody CDRs are linked to high specificity.
Fig. 3.
Effect of the isoelectric point of non-specific protein reagents on antibody non-specific binding. (A, B) Non-specific binding (100 nM scFv-Fc) was measured using a panel of non-antigen proteins (ovalbumin, BSA, insulin, KLH, Rnase A, avidin and lysozyme) with a wide range of isoelectric points (pIs of ~4.5–11). The normalized non-specific binding signals are reported as the signal minus background (no scFv-Fc antibody) divided by the background subtracted signal for the wild-type antibody. (C) Comparison of antibody non-specific binding to milk proteins versus the panel of non-antigen proteins reported in (A) and (B). The average values of non-specific binding for six scFv-Fc antibodies (WT, 5YA, 5DE, AF1, 5DA, B2; in order of increasing levels of non-specific interactions) to seven non-antigen proteins are shown in (C). In (A) and (B), the average values are for four to six independent experiments, and the error bars are standard errors. In (C), the average values are for three to four independent experiments, and the error bars are standard errors. The error bar for the average non-specific binding value for wild-type was determined by normalizing each replicate within a single experiment by the mean and calculating the average standard error.
CDR chemical composition is a key determinant of specificity for clinical-stage antibodies
To test the generality of our findings, we next evaluated whether the net charge of HCDR3 or the entire set of six CDRs are key predictors of specificity for clinical-stage antibodies (approved antibody drugs or antibodies in Phase 2/3 clinical trials). We evaluated published antibody non-specific binding data that was obtained using the variable (VH and VL) regions of 137 clinical-stage antibodies and the same antibody constant regions (IgG1 isotype, allele *01 for heavy chain, alleles IGKC*01 and IGLC2*01 for light chain) regardless of the actual antibody isotype (Jain et al., 2017). The non-specific (ELISA) binding data were previously obtained by evaluating antibody interactions with six biomolecules (cardiolipin, keyhole limpet hemocyanin, lipopolysaccharide, single stranded DNA, double stranded DNA and insulin). Analysis of the relationship between non-specific binding and the net charge of HCDR3 or all six CDRs revealed Spearman’s correlation coefficients that were modest (0.20 for HCDR3 and 0.33 for all CDRs) yet highly significant in the case of overall CDR net charge (P-value of 0.00010) and also significant for HCDR3 net charge (P-value of 0.017).
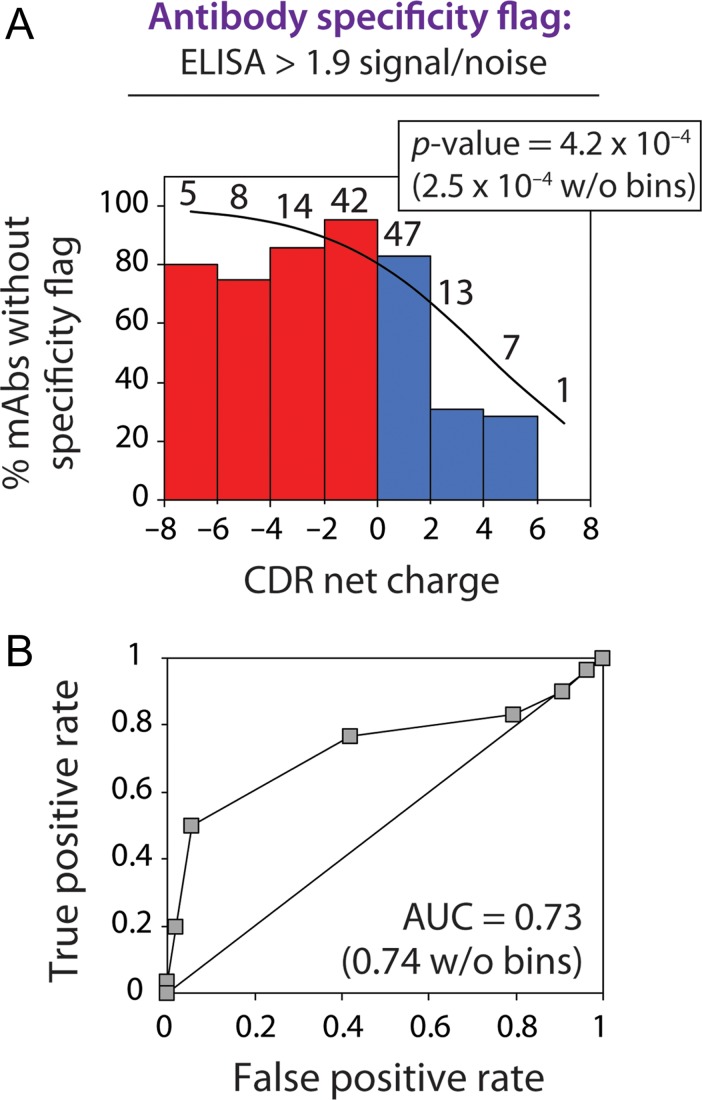
We also noticed that those clinical-stage antibodies with high levels of non-specific binding [values >1.9 signal/background, as reported previously (Jain et al., 2017)] had a higher likelihood of having positively charged CDRs. Importantly, logistic regression reveals that increased positive CDR charge is strongly correlated with increased probability of high levels of antibody non-specific interactions (P-value of 0.00042; Fig. 4A). We also confirmed this correlation using receiver operating characteristic (ROC) curve analysis [area under the curve (AUC) of 0.73; Fig. 4B]. Although these results were generated by grouping antibodies in terms of increments of net charge for visualization purposes, the statistical significance is even higher without grouping antibodies (P-value of 0.00025 and AUC of 0.74).
Fig. 4.
CDR net charge is inversely correlated with the specificity of clinical-stage mAbs. (A) Logistic regression analysis of the specificity of 137 clinical-stage mAbs (approved antibody drugs or antibodies in clinical trials) as a function of CDR net charge at pH 7.4. Non-specific binding was evaluated using a panel of six biomolecules (cardiolipin, keyhole limpet hemocyanin, lipopolysaccharide, single stranded DNA, double stranded DNA and insulin). Clinical-stage mAbs were flagged if their ELISA non-specific binding signals exceed a value of 1.9 signal/background (Jain et al., 2017). The mAbs were binned based on their CDR net charge values, and the number of antibodies in each bin is reported above each bar. (B) Receiver operating characteristic curve analysis. The corresponding area under curve (AUC) value is also reported.
We also sought to test if the overall net charge of all six antibody CDRs is the best predictor of the risk of high antibody non-specific interactions or if the charge of individual CDRs (such as HCDR3) or other antibody regions [such as individual CDRs or variable (VH or VL) domains] are better predictors of specificity (Table I). Notably, the net charge of individual CDRs or subsets of the CDRs from either the heavy or light chain displayed weaker correlations (P-values of 0.0049–0.57) relative to the net charge of the entire set of six CDRs (P-value of 0.00025). The net charges of individual variable domains (VH and VL) and the variable framework (Fv excluding the CDRs) also displayed much weaker correlations (P-values of 0.011–0.41).
Table I.
Relationship between the net charge of different antibody regions and antibody specificity for clinical-stage mAbs
| Antibody regions | P-value | Area under curve (AUC) | Direction of relationship, net charge vs. specificity |
|---|---|---|---|
| Fv | 0.0112 | 0.66 | − |
| VH | 0.011 | 0.66 | − |
| VL | 0.41 | 0.57 | − |
| Variable framework | 0.12 | 0.59 | + |
| CDRs | 0.00025 | 0.74 | − |
| Heavy CDRs | 0.010 | 0.67 | − |
| H1 | 0.22 | 0.54 | + |
| H2 | 0.024 | 0.64 | − |
| H3 | 0.046 | 0.62 | − |
| Light CDRs | 0.0049 | 0.70 | − |
| L1 | 0.041 | 0.62 | − |
| L2 | 0.0069 | 0.64 | − |
| L3 | 0.57 | 0.60 | − |
Logistic regression analysis of the specificity of 137 clinical mAbs as a function of the theoretical net charge of different antibody regions at pH 7.4. Low antibody specificity was defined as non-specific binding (ELISA) values >1.9 signal/background (Jain et al., 2017). The P-value for the coefficient of the independent variable of the logistic regression function and the area under the curve (AUC) for the receiver operating characteristic curves are reported. A negative relationship between net charge and specificity means that specificity is reduced as net charge is increased, and vice versa for the positive relationship.
The fact that the net charge of the CDRs is correlated with specificity for clinical-stage antibodies suggests that the number of negatively and/or positively charged CDR residues may also be correlated with specificity. Therefore, we performed logistic regression analysis of correlations between the number of certain amino acids in the CDRs of clinical-stage antibodies and the probability of high levels of non-specific binding (Table II). Interestingly, there is a strong positive correlation for increasing numbers of arginine and lysine CDR residues and low specificity (P-value of 0.00023), while there is a strong negative correlation for increasing numbers of aspartic and glutamic acid residues and low specificity (P-value of 0.028). These correlations are more significant than observed for most individual hydrophobic residues or combinations thereof. The one notable exception is leucine, as increasing number of leucine residues in the CDRs is positively correlated with low specificity (P-value is 0.019). Nevertheless, these findings generally demonstrate that CDR net charge (and the number of charged CDR residues) are key factors that determine the specificity of diverse types of antibodies, including those currently in the clinic.
Table II.
Effect of specific types of CDR residues on the specificity of clinical-stage antibodies.
| Amino acids | P-value | Area under curve (AUC) | Direction of relationship, # of amino acids vs. specificity |
|---|---|---|---|
| A | 0.50 | 0.53 | − |
| C | 0.75 | 0.51 | + |
| D | 0.027 | 0.63 | + |
| E | 0.56 | 0.56 | + |
| F | 0.66 | 0.52 | + |
| G | 0.66 | 0.52 | + |
| H | 0.045 | 0.58 | − |
| I | 0.84 | 0.51 | + |
| K | 0.17 | 0.58 | − |
| L | 0.019 | 0.63 | − |
| M | 0.85 | 0.50 | − |
| N | 0.35 | 0.56 | + |
| P | 0.67 | 0.51 | − |
| Q | 0.082 | 0.57 | − |
| R | 0.00039 | 0.71 | − |
| S | 0.45 | 0.56 | + |
| T | 0.91 | 0.50 | − |
| V | 0.52 | 0.53 | + |
| W | 0.077 | 0.63 | + |
| Y | 0.86 | 0.50 | + |
| R, K | 0.00023 | 0.72 | − |
| R, K, H | 0.0010 | 0.69 | − |
| D, E | 0.028 | 0.66 | + |
| F, W | 0.15 | 0.58 | + |
| F, W, Y | 0.73 | 0.53 | + |
| I, M | 0.96 | 0.50 | + |
| I, M, L | 0.065 | 0.59 | − |
Logistic regression analysis of the relationship between the number of each amino acid (or combinations thereof) in the CDRs and the percentage of clinical-stage mAbs with low specificity was performed as described in Table I. A negative relationship between the number of residues and specificity means that specificity is reduced as the number of residues is increased, and vice versa for the positive relationship.
Positively charged CDRs are linked to poor antibody biophysical properties
The primary role of CDR net charge in governing antibody specificity led us to evaluate if it is also important in determining antibody biophysical properties such as self-association and aggregation (Fig. 5). This led us to test the biophysical properties of single-chain antibodies (scFv) with common antibody frameworks (Figs 5 and S3). We reasoned that antibody variants with low specificity would also have increased levels of self-association and aggregation.
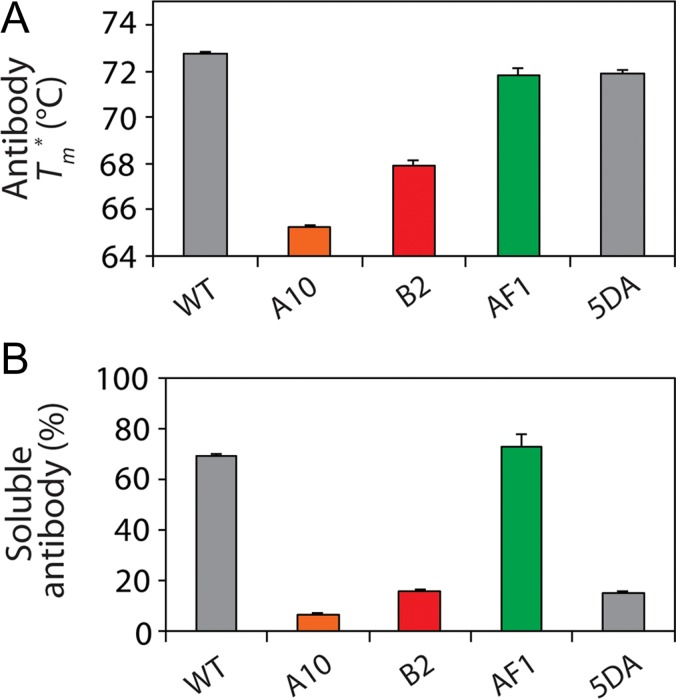
Fig. 5.
Biophysical properties of single-chain antibodies with high and low specificity. (A) Apparent melting temperature (Tm*) and (B) non-native solubility analysis of scFv antibodies with different HCDR3 net charges and different levels of specificity. In (B), the concentration of soluble antibody was measured after heating at 65°C for 4 h and cooling to 4°C overnight (initial antibody concentration of ~0.4 mg/mL). Average values are reported for (A) two or (B) three independent experiments, and the error bars are standard errors.
Therefore, we first evaluated the conformational stability of each antibody (scFv) variant to determine if simple stability differences may explain any differences in their solubility and aggregation (Figs 5A and S4). Some of the antibodies displayed lower apparent melting temperatures (Tm* values of 65.3 ± 0.1°C and 67.9 ± 0.3°C for A10 and B2, respectively, relative to 72.7 ± 0.1°C for wild-type). However, the AF1 variant (5DA) had a similar stability (71.9 ± 0.2°C) as wild-type (72.7 ± 0.1°C) and the parent AF1 antibody (71.8 ± 0.3°C). We also evaluated the refolding efficiency after thermal denaturation for each antibody and found that the results were generally correlated with their specificities (Fig. S4). The high specificity antibodies (AF1 and wild-type) displayed the highest levels of refolding after thermal denaturation, while the low specificity antibodies (A10 and 5DA) displayed the lowest levels of refolding.
The high levels of refolding after thermal denaturation for the antibodies with high specificity may be due to their low propensity to aggregate when unfolded at high temperature. To test this, we evaluated the non-native solubility of the panel of scFv antibodies after heating them to 65°C for 4 h and cooling them to 4°C overnight (Figs 5B and S3). Notably, the antibodies with low specificity (A10, B2 and 5DA) displayed the lowest solubilities, while those with high specificity (AF1 and wild-type) displayed the highest solubilities.
We next evaluated to what extent CDR net charge is linked to the biophysical properties of clinical-stage antibodies. To accomplish this, we used previously reported measurements of aggregation [accelerated stability (AS)], self-association [affinity-capture self-interaction nanoparticle spectroscopy (AC-SINS), clone self-interaction by bio-layer interferometry (CSI)], hydrophobicity [hydrophobic interaction chromatography (HIC), standup monolayer adsorption chromatography (SMAC) and salt-gradient affinity-capture self-interaction nanoparticle spectroscopy (SGAC-SINS)], and specificity [polyspecificity reagent (PSR), baculovirus particle (BVP), cross-interaction chromatography (CIC) and ELISA assays] (Jain et al., 2017). We used published limits for high levels of hydrophobicity, self-association, non-specific interactions and aggregation for the 10 assays (Fig. 6A) to evaluate the total number of biophysical flags for each mAb (maximum of 10 flags).
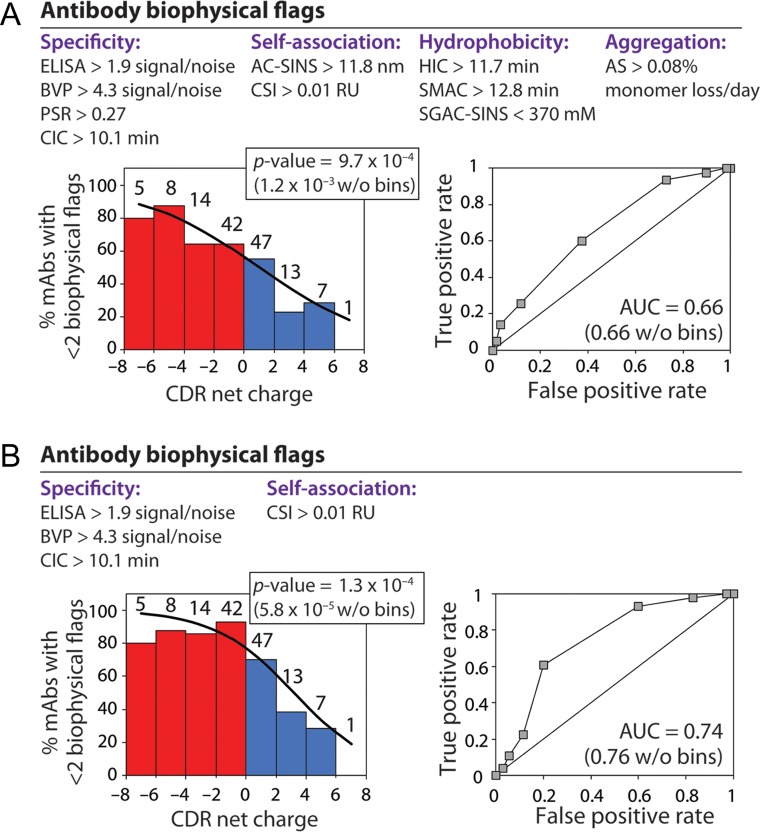
Fig. 6.
Clinical-stage antibodies with positively charged CDRs have increased risk for poor biophysical properties. Logistic regression analysis of the relationship between CDR net charge (pH 7.4) and the percentage of clinical-stage mAbs with <2 biophysical flags. Each mAb was assigned up to 10 biophysical flags based on previously reported measurements of antibody specificity, self-association, hydrophobicity and aggregation (Jain et al., 2017). (A,B) Regression analysis between CDR net charge and the percentage of clinical-stage mAbs with <2 biophysical flags using (A) ten or (B) four biophysical flags. The number of mAbs in each bin is reported above each bar. Receiver operating characteristic curve analysis is also reported in terms of the area under the curve (AUC) values.
We first performed logistic regression analysis to evaluate if increased overall CDR charge is linked to deficits in each antibody biophysical property (Table S1). Importantly, we find that CDR charge is strongly linked to two other measures of non-specific interactions in addition to the ELISA measurements. Increased CDR charge is strongly linked to increased non-specific antibody interactions with baculovirus particles (BVP assay, P-value of 4.3 × 10−4) and soluble membrane proteins (PSR assay, P-value of 3.2 × 10−4). This is notable because mAbs with high levels of non-specific binding to baculovirus particles and soluble membrane proteins have a much higher risk of fast antibody clearance in vivo (Hotzel et al., 2012; Kelly et al., 2015; Dobson et al., 2016).
We also find that CDR net charge is linked to antibody self-association (Table S1). Increased CDR net charge is associated with increased levels of antibody self-association measured by AC-SINS (P-value of 0.0095) and CSI (P-value of 0.010). This is significant because increased antibody self-association is linked to poor solubility (Bethea et al., 2012; Geng et al., 2016a,b; Sule et al., 2012, 2013; Wu et al., 2015) and abnormally high viscosity (Connolly et al., 2012; Yadav et al., 2012; Lilyestrom et al., 2013; Sharma et al., 2014; Tessier et al., 2014; Binabaji et al., 2015; Buck et al., 2015; Nichols et al., 2015).
To evaluate how CDR charge more generally impacts antibody biophysical properties, we performed logistic regression analysis to evaluate the relationship between CDR net charge and the percentage of mAbs with <2 biophysical flags (Fig. 6 and Table S2). We found a significant relationship between the percentage of mAbs with <2 biophysical flags (out of 10 possible flags) and CDR net charge (P-value of 1.2 × 10−3; Fig. 6A). This correlation is even more significant for a subset of four of these flags (P-value of 5.8 × 10−5 for BVP, CIC, CSI and ELISA; Fig. 6B). We also evaluated the role of positively and negatively charged residues on antibody biophysical properties using the same sets of biophysical flags (Table S3). Notably, the number of negatively charged residues (Asp and Glu) was positively correlated with the likelihood of favorable antibody biophysical properties (P-values of 0.0026–0.0031), while the number of positively charged residues (Arg, Lys and His) was negatively correlated with the likelihood for favorable biophysical properties (P-values of 0.0013–0.028). These results collectively demonstrate that CDR net charge and the number of charged CDR residues are key determinants of the biophysical properties of antibodies, including those in clinical development.
Discussion
Our findings highlight non-intuitive differences between the impacts of positively and negatively charged residues on antibody specificity. Importantly, we found that high levels of negatively charged CDR residues can promote non-specific interactions with positively charged molecules, but these interactions are relatively modest (Fig. 3). However, positively charged CDRs mediate high levels of non-specific interactions to both negatively and positively charged molecules.
We posit that these large differences in antibody specificity are due to key molecular differences between the side chains of positively and negatively charged residues. Our results reveal that arginine is much more important in mediating non-specific interactions than lysine (Table II). Arginine is a large amino acid that can participate in hydrophobic, pseudo-aromatic and hydrogen bonding interactions in addition to electrostatic interactions. The delocalized charge of the guanidinium group – which is much different than the more localized charge of the α-amino group for lysine – is likely part of why arginine displays complex properties that are not explainable based simply on its charge. It is also notable that negatively charged amino acids appear to be better hydrated than positively charged amino acids (Kuntz, 1971), and the guanidinium group is one of the most poorly hydrated ions (Mason et al., 2003, 2004). These factors collectively suggest that positively charged amino acids – and arginine in particular – mediate non-specific antibody interactions via both electrostatic and non-electrostatic interactions.
One of the most surprising aspects of our findings is the key role of negatively charged CDR residues in mediating high antibody specificity. Several previous reports identified positively charged residues in antibody CDRs – and arginine in particular – as a key risk factor for high levels of non-specific interactions (Wardemann et al., 2003; Birtalan et al., 2008, 2010; Kelly et al., 2018; Datta-Mannan et al., 2015a,b; Tiller et al., 2017a,b). However, our results demonstrate that removal of negatively charged CDR residues – without the addition or the presence of any positively charged residues – is sufficient to induce non-specific interactions. This suggests that the favorable contribution of negatively charged residues to antibody specificity cannot be explained simply based on their charge. Instead, we reason that favorable interactions of the acidic side chains with water render negatively charged residues particularly hydrophilic (Kuntz, 1971) and able to prevent hydrophobic interactions that are favored by neighboring aromatic and hydrophobic residues in antibody CDRs. A particularly interesting finding is that aspartic acid is more important as compared to glutamic acid in mediating high specificity (Table II). Aspartic acid also appears more frequently than glutamic acid in clinical antibodies (average fraction of 0.061 ± 0.026 and 0.018 ± 0.020 in the CDRs, respectively; Table S3). This suggests that aspartic acid could be more important for the favorable antibody properties required for clinical use, with specificity being one of the most important.
Another surprising aspect of our work is the finding that CDR charge is not only linked to antibody specificity but also to antibody biophysical properties in general. We have previously shown that negatively charged mutations in antibody CDRs are more effective at preventing antibody aggregation than positively charged mutations (Perchiacca et al., 2014). This is also consistent with other observations that aggregation-resistant antibodies are enriched in negatively charged CDR mutations (Jespers et al., 2004; Arbabi-Ghahroudi et al., 2009; Perchiacca et al., 2011, 2012; Dudgeon et al., 2012; Perchiacca and Tessier, 2012; Lee et al., 2016). However, our findings reveal that positively charged CDRs are linked to increased antibody self-association (Alam et al., 2018) in addition to increased aggregation and reduced specificity. The fact that increased CDR charge is linked to increased self-association is notable because antibodies with high levels of self-association have a much higher risk of abnormally high viscosity (Connolly et al., 2012; Yadav et al., 2012; Lilyestrom et al., 2013; Sharma et al., 2014; Tessier et al., 2014; Binabaji et al., 2015; Buck et al., 2015; Nichols et al., 2015) and fast antibody clearance (Kelly et al., 2015; Dobson et al., 2016). Indeed, several reports have established that positively charged antibody variable regions and even positively charged CDRs can lead to fast antibody clearance (Igawa et al., 2010; Li et al., 2014; Sharma et al., 2014; Bumbaca Yadav et al., 2015; Datta-Mannan et al., 2015a,b). The latter behavior is likely due to antibody non-specific interactions with biomolecules and cells that reduce antibody bioavailability.
Our findings also provide important guidelines for improving therapeutic antibody development. The fact that antibody specificity is the best biophysical predictor of antibody success in the clinic (Jain et al., 2017) suggests that selecting or engineering antibodies with negatively charged or weakly positively charged CDRs is important for generating antibodies with drug-like properties. It will also be important in the future to consider other aspects of CDR chemical composition (e.g. number of hydrophobic and hydrophilic residues) as well as CDR structure (e.g. solvent exposure and spatial positioning of amino acids) in addition to the net charge to improve predictions of antibody specificity. We expect that these efforts – which are currently underway in our laboratory – will greatly improve the identification and engineering of antibody candidates to improve their specificity and likelihood of success in the clinic.
Supplementary Material
Acknowledgements
We thank members of the Tessier lab for their helpful suggestions.
Conflict of interest
P.M.T. has received consulting fees and/or honorariums for presentations of this and/or related research findings at MedImmune, Eli Lilly, Bristol-Myers Squibb, Janssen, Merck, Genentech, Amgen, Pfizer, Adimab, Abbvie, Roche, Boehringer Ingelheim, Bayer, Abbott, DuPont, Schrödinger and Novo Nordisk.
Funding
This work was supported by the National Institutes of Health [R01GM104130 and R01AG050598 to P.M.T.], National Science Foundation [CBET 1159943, 1605266 and 1813963 to P.M.T., Graduate Research Fellowships to L.A.R. and M.C.J.], and the Albert M. Mattocks Chair (to P.M.T).
Authors' contributions
L.A.R, Y.Z., S.D.L, M.C.J. and P.M.T. designed the research, L.A.R. and M.C.J. performed experiments, Y.Z. and S.D.L performed computational analysis, and L.A.R. and P.M.T. wrote the paper with input from the co-authors.
References
- Alam M.E., Geng S.B., Bender C., Ludwig S.D., Linden L., Hoet R. and Tessier P.M. (2018) Mol. Pharm., 15, 150–163. [DOI] [PubMed] [Google Scholar]
- Arbabi-Ghahroudi M., To R., Gaudette N., Hirama T., Ding W., MacKenzie R. and Tanha J. (2009) Protein Eng. Des. Sel., 22, 59–66. [DOI] [PubMed] [Google Scholar]
- Avery L.B., Wade J., Wang M., Tam A., King A., Piche-Nicholas N., Kavosi M.S., Penn S., Cirelli D., Kurz J.C. et al. (2018) mAbs, 10, 244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethea D., Wu S.J., Luo J., Hyun L., Lacy E.R., Teplyakov A., Jacobs S.A., O’Neil K.T., Gilliland G.L. and Feng Y. (2012) Protein Eng. Des. Sel., 25, 531–537. [DOI] [PubMed] [Google Scholar]
- Binabaji E., Ma J. and Zydney A.L. (2015) Pharm. Res., 32, 3102–3109. [DOI] [PubMed] [Google Scholar]
- Birtalan S., Fisher R.D. and Sidhu S.S. (2010) Mol. Biosyst., 6, 1186–1194. [DOI] [PubMed] [Google Scholar]
- Birtalan S., Zhang Y., Fellouse F.A., Shao L., Schaefer G. and Sidhu S.S. (2008) J. Mol. Biol., 377, 1518–1528. [DOI] [PubMed] [Google Scholar]
- Buck P.M., Chaudhri A., Kumar S. and Singh S.K. (2015) Mol. Pharm., 12, 127–139. [DOI] [PubMed] [Google Scholar]
- Bumbaca Yadav D., Sharma V.K., Boswell C.A., Hotzel I., Tesar D., Shang Y., Ying Y., Fischer S.K., Grogan J.L., Chiang E.Y. et al. (2015) J. Biol. Chem., 290, 29732–29741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P., Presta L., Gorman C.M., Ridgway J.B., Henner D., Wong W.L., Rowland A.M., Kotts C., Carver M.E. and Shepard H.M. (1992) Proc. Natl. Acad. Sci. USA, 89, 4285–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly B.D., Petry C., Yadav S., Demeule B., Ciaccio N., Moore J.M., Shire S.J. and Gokarn Y.R. (2012) Biophys. J., 103, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta-Mannan A., Lu J., Witcher D.R., Leung D., Tang Y. and Wroblewski V.J. (2015. a) mAbs, 7, 1084–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta-Mannan A., Thangaraju A., Leung D., Tang Y., Witcher D.R., Lu J. and Wroblewski V.J. (2015. b) mAbs, 7, 483–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson C.L., Devine P.W., Phillips J.J., Higazi D.R., Lloyd C., Popovic B., Arnold J., Buchanan A., Lewis A., Goodman J. et al. (2016) Sci Rep, 6, 38644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudgeon K., Rouet R., Kokmeijer I., Schofield P., Stolp J., Langley D., Stock D. and Christ D. (2012) Proc. Natl. Acad. Sci. USA, 109, 10879–10884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estep P., Caffry I., Yu Y., Sun T., Cao Y., Lynaugh H., Jain T., Vasquez M., Tessier P.M. and Xu Y. (2015) mAbs, 7, 553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng S.B., Wittekind M., Vigil A. and Tessier P.M. (2016. a) Mol. Pharm., 13, 1636–1645. [DOI] [PubMed] [Google Scholar]
- Geng S.B., Wu J., Alam M.E., Schultz J.S., Dickinson C.D., Seminer C.R. and Tessier P.M. (2016. b) Bioconjug. Chem., 27, 2287–2300. [DOI] [PubMed] [Google Scholar]
- Goodnow C.C. (2007) Cell, 130, 25–35. [DOI] [PubMed] [Google Scholar]
- Hotzel I., Theil F.P., Bernstein L.J., Prabhu S., Deng R., Quintana L., Lutman J., Sibia R., Chan P., Bumbaca D. et al. (2012) mAbs, 4, 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa T., Tsunoda H., Tachibana T., Maeda A., Mimoto F., Moriyama C., Nanami M., Sekimori Y., Nabuchi Y., Aso Y. et al. (2010) Protein Eng. Des. Sel., 23, 385–392. [DOI] [PubMed] [Google Scholar]
- Jacobs S.A., Wu S.J., Feng Y., Bethea D. and O’Neil K.T. (2010) Pharm. Res., 27, 65–71. [DOI] [PubMed] [Google Scholar]
- Jain T., Sun T., Durand S., Hall A., Houston N.R., Nett J.H., Sharkey B., Bobrowicz B., Caffry I., Yu Y. et al. (2017) Proc. Natl. Acad. Sci. USA, 114, 944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman J., Wu J., Brunelle M.C., Cruz A.M., Goldberg D.S., Lobo B., Shah A. and Tessier P.M. (2014) Biotechnol. Bioeng., 111, 1513–1520. [DOI] [PubMed] [Google Scholar]
- Jespers L., Schon O., Famm K. and Winter G. (2004) Nat. Biotechnol., 22, 1161–1165. [DOI] [PubMed] [Google Scholar]
- Keenan R.A., De Riva A., Corleis B., Hepburn L., Licence S., Winkler T.H. and Martensson I.L. (2008) Science, 321, 696–699. [DOI] [PubMed] [Google Scholar]
- Kelly R.L., Geoghegan J.C., Feldman J., Jain T., Kauke M., Le D., Zhao J. and Wittrup K.D. (2017. a) mAbs, 9, 1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R.L., Le D., Zhao J. and Wittrup K.D. (2018) J. Mol. Biol., 430, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R.L., Sun T., Jain T., Caffry I., Yu Y., Cao Y., Lynaugh H., Brown M., Vasquez M., Wittrup K.D. et al. (2015) mAbs, 7, 770–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R.L., Zhao J., Le D. and Wittrup K.D. (2017. b) mAbs, 9, 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz I.D. (1971) J. Am. Chem. Soc., 93, 514–516. [DOI] [PubMed] [Google Scholar]
- Lee C.C., Julian M.C., Tiller K.E., Meng F., DuConge S.E., Akter R., Raleigh D.P. and Tessier P.M. (2016) J. Biol. Chem., 291, 2858–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Geng S.B., Chiu M.L., Saro D. and Tessier P.M. (2015) Bioconjug. Chem., 26, 520–528. [DOI] [PubMed] [Google Scholar]
- Li B., Tesar D., Boswell C.A., Cahaya H.S., Wong A., Zhang J., Meng Y.G., Eigenbrot C., Pantua H., Diao J. et al. (2014) mAbs, 6, 1255–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilyestrom W.G., Yadav S., Shire S.J. and Scherer T.M. (2013) J. Phys. Chem. B, 117, 6373–6384. [DOI] [PubMed] [Google Scholar]
- Liu Y., Caffry I., Wu J., Geng S.B., Jain T., Sun T., Reid F., Cao Y., Estep P., Yu Y. et al. (2014) mAbs, 6, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P.E., Neilson G.W., Dempsey C.E., Barnes A.C. and Cruickshank J.M. (2003) Proc. Natl. Acad. Sci. USA, 100, 4557–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P.E., Neilson G.W., Enderby J.E., Saboungi M.L., Dempsey C.E., MacKerell A.D. Jr. and Brady J.W. (2004) J. Am. Chem. Soc., 126, 11462–11470. [DOI] [PubMed] [Google Scholar]
- Mouquet H., Scheid J.F., Zoller M.J., Krogsgaard M., Ott R.G., Shukair S., Artyomov M.N., Pietzsch J., Connors M., Pereyra F. et al. (2010) Nature, 467, 591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols P., Li L., Kumar S., Buck P.M., Singh S.K., Goswami S., Balthazor B., Conley T.R., Sek D. and Allen M.J. (2015) mAbs, 7, 212–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perchiacca J.M., Bhattacharya M. and Tessier P.M. (2011) Proteins, 79, 2637–2647. [DOI] [PubMed] [Google Scholar]
- Perchiacca J.M., Ladiwala A.R., Bhattacharya M. and Tessier P.M. (2012) Protein Eng. Des. Sel., 25, 591–601. [DOI] [PubMed] [Google Scholar]
- Perchiacca J.M., Lee C.C. and Tessier P.M. (2014) Protein Eng. Des. Sel., 27, 29–39. [DOI] [PubMed] [Google Scholar]
- Perchiacca J.M. and Tessier P.M. (2012) Annu. Rev. Chem. Biomol. Eng., 3, 263–286. [DOI] [PubMed] [Google Scholar]
- Sharma V.K., Patapoff T.W., Kabakoff B., Pai S., Hilario E., Zhang B., Li C., Borisov O., Kelley R.F., Chorny I. et al. (2014) Proc. Natl. Acad. Sci. USA, 111, 18601–18606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W. (2005) Protein Expr. Purif., 41, 207–234. [DOI] [PubMed] [Google Scholar]
- Sule S.V., Cheung J.K., Antochshuk V., Bhalla A.S., Narasimhan C., Blaisdell S., Shameem M. and Tessier P.M. (2012) Mol. Pharm., 9, 744–751. [DOI] [PubMed] [Google Scholar]
- Sule S.V., Dickinson C.D., Lu J., Chow C.K. and Tessier P.M. (2013) Mol. Pharm., 10, 1322–1331. [DOI] [PubMed] [Google Scholar]
- Sun T., Reid F., Liu Y., Cao Y., Estep P., Nauman C. and Xu Y. (2013) mAbs, 5, 838–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier P.M., Wu J. and Dickinson C.D. (2014) Expert Opin. Drug Deliv., 11, 461–465. [DOI] [PubMed] [Google Scholar]
- Tiller K.E., Chowdhury R., Li T., Ludwig S.D., Sen S., Maranas C.D. and Tessier P.M. (2017. a) Front. Immunol., 8, 986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller K.E., Li L., Kumar S., Julian M.C., Garde S. and Tessier P.M. (2017. b) J. Biol. Chem., 292, 16638–16652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann H., Yurasov S., Schaefer A., Young J.W., Meffre E. and Nussenzweig M.C. (2003) Science, 301, 1374–1377. [DOI] [PubMed] [Google Scholar]
- Wu J., Schultz J.S., Weldon C.L., Sule S.V., Chai Q., Geng S.B., Dickinson C.D. and Tessier P.M. (2015) Protein Eng. Des. Sel., 28, 403–414. [DOI] [PubMed] [Google Scholar]
- Xu Y., Roach W., Sun T., Jain T., Prinz B., Yu T.Y., Torrey J., Thomas J., Bobrowicz P., Vasquez M. et al. (2013) Protein Eng. Des. Sel., 26, 663–670. [DOI] [PubMed] [Google Scholar]
- Yadav S., Shire S.J. and Kalonia D.S. (2012) J. Pharm. Sci., 101, 998–1011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.