Summary
Ubiquitin-like (Ubl) posttranslational modifications are potential targets of therapeutics. However, the only known mechanism for inhibiting a Ubl activating enzyme is through targeting its ATP-binding site. Here we identify an allosteric inhibitory site in the small ubiquitin-like modifier (SUMO) activating enzyme (E1). This site was unexpected because both it and analogous sites are deeply buried in all previously solved structures of E1s of ubiquitin-like modifiers (Ubl). The inhibitor not only suppresses SUMO E1 activity, but also enhances degradation in vivo, presumably due to a conformational change induced by the compound. In addition, the lead compound increased the expression of miR-34b and reduced c-Myc levels in lymphoma and colorectal cancer cell lines and a colorectal cancer xenograft mouse model. Identification of this first-in-class inhibitor of SUMO E1 is a major advance in modulating Ubl modifications for therapeutic aims.
Keywords: covalent inhibitor, allosteric inhibitor, ubiquitin-like modification, activating enzyme, E1, SUMO, c-Myc
eTOC Blurb
Ubiquitin-like (Ubl) post-translational modifications are potential targets for developing novel therapeutics for life-threatening diseases. Here, we have identified a new allosteric, covalent inhibitory site for inhibiting an activating enzyme of Ubl. This finding could spur innovative drug discovery efforts targeting Ubl modifications.
Graphical Abstract

Introduction
Aberrations in post-translational modifications by ubiquitin or ubiquitin-like proteins (Ubl), such as the small ubiquitin-like modifiers (SUMO), are associated with the pathogenesis of life-threatening diseases, such as cancer (Sarge and Park-Sarge, 2011; Zhu et al., 2010), neurodegenerative disorders (Steffan et al., 2004; Subramaniam et al., 2009), and viral infection (Jaber et al., 2009; Kim et al.). For example, multiple studies indicate that SUMOylation is dysregulated in many types of cancers and that the SUMO activating enzyme (SAE, SUMO E1) could be a potential target to inhibit c-Myc and KRas-dependent oncogenesis (He et al., 2017; Kessler et al., 2011; Luo et al., 2009; Yu et al., 2015) and reduce cancer cell stemness and resistance (Bogachek et al., 2016; Du et al., 2016). The activating enzyme catalyzing ubiquitin-like Atg8 and Atg12 modifications in autophagy, known as Atg7, has been shown as an indirect target for KRas-dependent oncogenesis (Guo et al., 2013; Rosenfeldt et al., 2013). Despite the importance of Ubl modifications in dysregulated signaling pathways in diseased cells, only a handful of FDA-approved drugs targeting this type of post-translational modifications have been developed. This deficiency illustrates knowledge gaps in targeting these enzymes by small molecules and underscores the need to discover novel chemotypes and mechanisms to inhibit Ub1 modifications.
An Ubl modification requires several steps that are catalyzed by three enzymes, referred to as E1 (activating enzyme), E2 (conjugation enzyme) and E3 (ligase). The SUMO E1 is a heterodimer of SAE1 and Uba2 (also known as SAE2). Briefly, an Ubl is first activated by E1 through ATP hydrolysis and forms a thioester conjugate with E1. The Ubl is then transferred to E2, forming a thioester conjugate with E2. Finally, the Ubl is transferred to target proteins, a step usually catalyzed by an E3. Usually, Ubl modifications add new docking sites to target proteins. For example, SUMO modifications enables new protein-protein interactions through the SUMO-interacting motif (SIM) in receptor proteins (Song et al., 2004; Song et al., 2005). At least three members of the SUMO family (SUMO1, 2, and 3) are ubiquitin-like proteins that can conjugate to other cellular proteins by a biochemical mechanism similar to ubiquitylation (Hay, 2005; Sarge and Park-Sarge, 2009; Yeh, 2009). Currently, the only known mechanism to inhibit the E1 enzymes targets their ATP-binding sites (Brownell et al., 2010; Soucy et al., 2009).
Historically, covalent drugs have had great success (e.g., aspirin and penicillin), and covalent drugs have become a focus in anticancer and antiviral drug discovery (Kalgutkar and Dalvie, 2012; Singh et al., 2011). These compounds contain low reactivity warheads that allow covalent adducts to form only when a non-covalent complex forms first. Their duration of action depends on the turnover rate of their protein targets, independent of the drug’s stability in the blood.
In this study, we describe the identification and characterization of a covalent inhibitor chemotype of SUMO E1 that binds to an unexpected allosteric site conserved in E1 enzymes. The site is deeply buried in previously published structures of SUMO E1 and the analogous site is buried in other Ubl E1s. The lead compound not only blocks enzymatic function, but also induces SUMO E1 degradation in cells, presumably by inducing a conformational change in the E1 upon binding. Structure activity relationship (SAR) and time dependent inhibition studies show that the specific covalent inhibitory adduct formation is a result of non-covalent binding. The lead compound also shows efficacy in colorectal cancer xenograft and patient-derived xenograft (PDX) models. The findings described here is a major advance in our knowledge on targeting these enzymes by small molecules to translate information on dysregulation of this type of post-translational modifications in diseases into novel therapies.
Results
Discovery of a Highly-Specific SUMO E1 Inhibitor by HTS.
To identify novel SUMOylation inhibitors, we conducted high throughput screening of 290,921 compounds (PubChem AID 2011) using a fluorescence resonance energy transfer (FRET) based primary assay (PubChem AID 2006) and a chemiluminescence-based secondary assay using AlphaScreen (Amplified Luminescent Proximity Homogeneous Assay Screen) technology to eliminate false positive hits (PubChem AID 2018). Both assays detected the formation of SUMO1-RanGAP1 conjugation in the presence of E1 and E2 enzymes as described previously (Rouleau et al., 2008). Hits were counter-screened by an ubiquitylation assay to eliminate any inhibitors that were not specific to SUMOylation. One hit showed high specificity to SUMOylation (CID 9549553, Figure 1A) - out of 780 assays through which this compound has been documented in PubChem when this manuscript was first prepared, the assays in which it produced an IC50 less than 7 μM for a human protein target are either SUMOylation biochemical assays or cellular assays of RORγ and human tyrosyl-DNA phosphodiesterase 1, both of which are SUMOylation substrates. In addition, the compound did not inhibit ubiquitylation biochemical assays at concentrations up to 100 μM. These previous screening data indicated that this compound has a unique specificity to SUMOylation.
Figure 1. Identification of a selective SUMOylation inhibitor by HTS.
(A) Summary of the assays that the HTS hit was most potent for human proteins based on IC50 values among 780 assays tested as shown by data deposited in PubChem.
(B and C) In vitro SUMOylation assay of SUMO conjugation to RanGAP1 detected by AlphaScreen (B) and ubiquitylation assay detected by time-resolved fluorescence resonance energy transfer (C, see also PubChem AID 2658). One representative curve from four experiments is shown for SUMOylation (B). The results from five experiments are shown for ubiquitylation (C). The structure of COH000 is shown to the right in (B).
Thirty-six analogs were synthesized to gain structure-activity relationship (SAR) information (Table 1). The SAR was observed suggesting that the compound inhibits SUMOylation in a specific manner (Table 1). These studies resulted in an improved SUMOylation inhibitor (referred as COH000) (PubChem CID 46835111, SID 99212997; IUPAC: dimethyl 1-((R)-1-(phenylamino)-2-(p-tolyl)ethyl)7-oxabicyclo[2.2.1]hepta-2,5-diene-2,3-dicarboxylate) (Figure 1B). COH000 inhibited SUMOylation with an average IC50 of approximately 0.2 μM in vitro (Figure 1B) but did not inhibit ubiquitylation in an Ubc13-mediated poly-ubiquitylation assay tested under the same condition at concentrations up to 100 μM (Figure 1C). Therefore, COH000 has over 500-fold selectivity for SUMOylation than ubiquitylation. With regard to SAR specificity, many of the analogs were diastereomers, with R or S configurations around the chiral carbon center indicated in Table 1 and in general the R isomer were more potent than the S isomers.
Discovery of a Covalent Allosteric Inhibitory Site in the SUMO E1.
We conducted a series of enzymatic analysis to understand how COH000 inhibits SUMOylation. The compound inhibits the formation of SUMO-E1 thioester formation, suggesting that it inhibits SUMO E1. An E1 catalyzes two steps in the cascade of reactions: (1) ATP-dependent adenylation of the C-terminus of Ubl and release of pyrophosphate (PPi), and (2) transfer of Ubl to the Cys-domain, forming a thioester bond with the catalytic Cys (Bohnsack and Haas, 2003; Haas and Rose, 1982). We examined the adenylation step using a previously established method (Tatham et al., 2003; Wang et al., 2010; Wang and Chen, 2010; Wang et al., 2009); when 32P-labeled PPi is mixed with unlabeled ATP in the presence of SUMO E1, ATP gradually gains radioactivity as PPi gradually loses radioactivity. Thus, the ATP:PPi isotope exchange rate specifically measures the rate of the adenylation step. The 32PPi:ATP isotope exchange assay showed that COH000 inhibited SUMO adenylation (Figure 2A), and the inhibitory effect was not overcome with increasing concentrations of ATP or SUMO1. This result indicates that COH000 inhibits SUMO adenylation without directly competing with ATP or SUMO1 binding.
Figure 2. COH000 is an allosteric covalent inhibitor of the SUMO E1.
(A) ATP:PPi exchange assay to show that COH000 inhibits the SUMO adenylation step of SUMO E1 catalysis. The assay was conducted with various concentrations of the compound at increasing concentrations of ATP (upper panel) or SUMO-1 (lower panel).
(B) Fragment ion assignments for the MS/MS spectrum of the 2+ charge state of peptide COH000 conjugated peptide (residues 14–35 of SAE2, AVAGGRVLVVGAGGIGCELLKN, m/z 849.47). The potential conjugation sites of COH000 with Cys30 are shown to the right.
(C) SUMO E1 catalyzed Ubc9•SUMO thioester formation is inhibited by COH000 in a dose-dependent manner for WT enzyme but not C30S mutant. 0.25 μM E1 WT or C30S was incubated with 2 μM Ubc9, 5 μM SUMO-1, 100 μM ATP, 5 mM MgCl2, 50 mM NaCl, 0.05% Tween-20, 20 mM HEPES pH 7.6 for 15 minutes at 37oC. The figure shows a Coomassie stained SDS-PAGE gel.
(D) Analysis of KI and kinact of COH000 inhibition of SUMO E1. Natural logarithm of the rate of Ubc9·SUMO-1 thioester is plotted against the pre-incubation time of SUMO E1 with the indicated concentrations of COH000 (left panel). After incubation, homogeneous time-resolved fluorescence readings were taken. The negative slope in the graph from the upper panel was used to determine the inactivation rate kobs (right panel) that was used for non-linear regression analysis that obtained KI and kinact. Data are presented as means ± SD, in three independent experiments.
Inhibition of SUMO E1 by COH000 is irreversible. SUMO E1 with or without the addition of COH000 was added to individual dialysis cassettes and dialyzed separately against buffer without COH000. The samples were withdrawn from the cassettes at various time points to examine SUMO E1 activity by SUMOylation assays. Inhibition of SUMO E1 activity was long lasting, and no enzyme activity in samples treated with COH000 was observed up to 8 hours of dialysis (Figure S1A). This result suggests that COH000 has an extremely slow off-rate and is consistent with it being covalently bound to the enzyme.
To verify covalency and determine the possible covalent modification site of COH000 on SUMO E1, mass spectrometry (MS) analysis was conducted on SUMO E1 samples with or without full inhibition by COH000 (>10 fold excess for > 1 h) as determined by enzymatic assays. Each sample was subjected to on-line pepsin digestion then subjected to liquid chromatography tandem MS (LC-MS/MS) analyses. The resulting MS data were searched using the program Sequest to assign spectra to the sequence of SUMO E1 and summarized using the program Scaffold. To identify covalent adducts with COH000, the search was performed using a potential modification of Cys corresponding to the simple addition of COH000 (419.1733 u). The Sequest search yielded 92% sequence coverage for SAE1 and 81% sequence coverage for SAE2 (Data S1). Only one identified peptide from SAE2 (residues 14–35, AVAGGRVLVVGAGGIGCELLKN) had a modification corresponding to the simple adduct of COH000 (+419.17 u) on Cys30 at 100% labeling by intact mass. Based on the molecular weight, the proposed covalent modification is shown in Fig. 2B. The corresponding unmodified peptide was not found in the SAE2 sample without the addition of COH000. However, the peptide AVAGGRVLVVGAGGIGCEL (residues 14–32) was found, which is expected if modification of Cys30 interferes with pepsin cleavage after Leu32. The fragment ion (MS/MS) spectra for both peptides are of sufficient quality to assign their sequences correctly with high confidence (Figure 2B). Two independently inhibited SUMO E1 samples followed by LC-MS/MS analysis yielded the same result.
We specifically analyzed the catalytic Cys residue of SAE2 (Cys173) for any modifications, because modification of this Cys could also eliminate SUMO E1 activity. A large peptide (residues 131–185) was identified in the extracted ion chromatograms (EIC) of the MS scans at the same retention time and nearly equal intensity in four independent runs, two for SUMO E1 without the inhibitor and two for SUMO E1 with full inhibition by the compound (Figure S1B). Thus, COH000 did not modify Cys173. Taken together, these data indicate that COH000 specifically modifies Cys30 and does not modify other Cys residues, including the catalytic Cys173 which is solvent exposed in previously determined crystal structure (Lois and Lima, 2005). The lack of modification of the catalytic Cys is consistent with COH000 not inhibiting the homologous ubiquitin E1, which also contains a solvent exposed catalytic Cys (Figure 1C, lower panel).
To confirm that covalent attachment to Cys30 is the mechanism for inhibiting SUMO E1, we conducted enzymatic assays to determine whether mutation of Cys30 could remove the inhibitory effect. We mutated Cys30 to Ser, and the C30S mutant was active but with activity of less than 50% of that of the wild type SAE2. Although WT enzyme was inhibited by COH000 in a dose-dependent manner, the C30S mutant was not (Figure 2C). The IC50 in this biochemical assay is higher than that in AlphaScreen assay (Figure 1C), due to the more than 10-fold higher SUMO E1 concentrations. These finding supports the covalent adduct formation in inhibiting SUMO E1 by COH000. Cys30 is not involved in binding ATP or SUMO, and thus the identification of this site is consistent with that COH000 does not compete with ATP or SUMO1 binding (Figure 2A). The traditional Michaelis–Menten kinetics could not be obtained for the SUMO E1, nor for the E1s of other ubiquitin-like modifiers, because E1 has two substrates (ATP and SUMO) and the product, AMP-SUMO conjugate, is not released from the enzyme, but initiates covalent modifications of the enzyme as a thioester.
The selectivity of COH000 for modification of Cys30 suggests that non-covalent interactions of COH000 with SUMO E1 may enable covalent adduct formation. The inhibitory effect of COH000 depends both on its concentration and incubation time (Figure 2D, left panel). The time-dependent inhibitory effects were used to calculate the approximate inactivation rate kobs (Figure 2E, right panel), which is defined below:
The hyperbolic curve of kobs versus inhibitor concentration is consistent with the formation of a non-covalent interaction before covalent adduct formation. Non-linear regression analysis was used to fit KI and kinact (Figure 2E, right panel) (Ito et al., 1998).
COH000 Binding Requires a Change of SUMO E1 Conformation.
The covalent attachment to Cys30 is unexpected, because Cys30 is deeply buried Cys residues in all previously determined crystal structures of SUMO E1s, including the adenylation conformation (Figure 3A, upper panel) and thioester formation conformation (Figure 3A, lower panel) (Lois and Lima, 2005; Olsen et al., 2010). Among all eighteen undisulfide Cys residues in SUMO E1, nine are solvent accessible including the catalytic Cys173, but none of them were modified by COH000 in multiple MS/MS analysis of fully inhibited SUMO E1 by COH000. Additionally, inspection of other published Ubl E1 crystal strictures, the analogous sites, although conserved (Figure S1C), are all deeply buried; thus there is no indication that this site could be accessible for covalent inhibition. Therefore, binding of COH000 to the SUMO E1 requires a conformational state of the enzyme not previously observed. SUMO E1 with and without fully inhibition by COH000 had different thermal denaturation profiles (Figure 3B), with inhibitor bound state has higher melting temperature than unbound state. This result indicates that the inhibitor-bound conformation is significantly different from the apo E1 conformation.
Figure 3. COH000 enhances SAE2 degradation in cells.
(A) The COH000 covalent attachment site Cys30 is shown in spheres colored according to the atom types (green, carbon; yellow, sulfur), and the bound ATP or ATP analog (red) and SUMO (magenta) are shown according to published crystal structures in the adenylation active (upper panel, pdb id: 1Y8R) and thioester formation active (lower panel, pdb id: 3KYD) conformations that differ on both the domain orientations and active site structures.
(B) Differential scanning fluorimetry of SUMO E1 with and without fully inhibited by COH000. The derivative of the raw data is shown.
(C) COH000 increased degradation of SAE2 in cells as indicated by pulse-chase experiment. Upper panel, the gel image. Lower panel, the gel band intensity normalized by total input protein detected by Coomassie staining relative to time zero.
(D) MG132 (+) or vehicle (−) was added to cells for 2 hours prior to COH000 treatment at indicated concentrations. After 2 hours of COH000 treatment, the cells were harvested and directly lysed in SDS-sample buffer. SAE2 protein was then analyzed by Western blot. Bracket indicates modified SAE2, likely including ubiquitylation. GAPDH, shown on the same membrane, was used for loading control.
COH000 increased the degradation of SAE2, as shown by pulse-chase assays, likely because the inactive conformation enhanced ubiquitin-proteasome mediated degradation (Figure 3C). Indeed, COH000 caused reduction of SAE2 and increased ubiquitylation of SAE2 in cells (Figure 3D), suggesting that the reduced SAE2 level depends, at least in part, on increased proteasome-dependent degradation.
COH000 Inhibits SUMOylation in Cells.
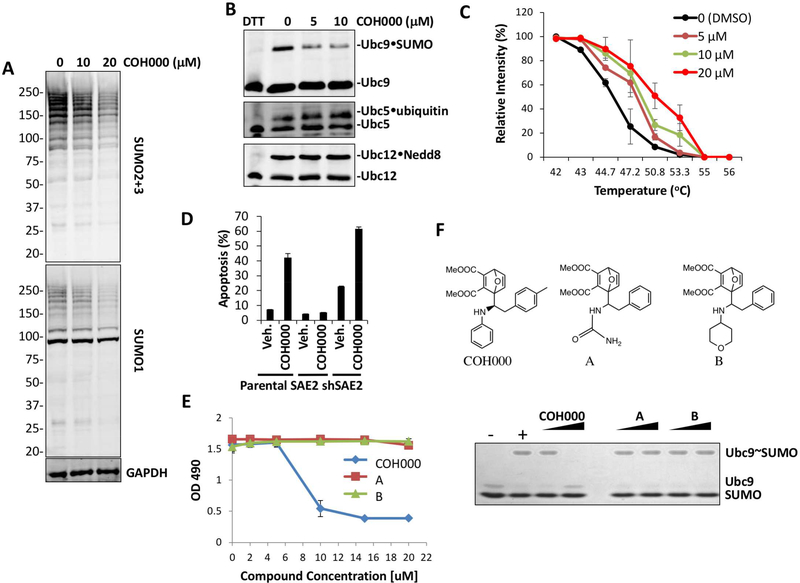
We added COH000 to cell culture media for 18 hours and detected SUMOylated proteins using anti-SUMO-1 and anti-SUMO-2/3 antibodies (Figure 4A). COH000 inhibited SUMOylation in a dose-dependent manner (Figure 4A). To investigate whether COH000 specifically inhibits SUMO E1 in cells, we probed for the formation of activating enzyme-catalyzed intermediates, including SUMO•Ubc9, as well as ubiquitin•Ubc5 and NEDD8•Ubc12 thioesters (Figure 4B). COH000 inhibited only SUMO•Ubc9 thioester formation in a dose-dependent manner and did not inhibit the homologous enzymes catalyzing ubiquitylation or NEDDylation, consistent with its selectivity for SUMOylation in in vitro assays (Figure 1C). The engagement of COH000 with SAE2 was investigated by cellular thermal-shift assay as described previously (Figure 4C and Figure S2A) (Jafari et al., 2014; Martinez Molina et al., 2013). The compound increased the thermal stability of the enzyme in a dose dependent manner (Figure 4C and Figure S2A).
Figure 4. COH000 inhibits SUMOylation in cells.
(A) COH000 inhibits SUMOylation in HCT-116 cells in a dose-dependent manner upon 18 hours treatment.
(B) COH000 selectively inhibits the SUMO E1 in cells. Inhibition of SUMO, Nedd8, and ubiquitin activating enzymes in cells was assessed by Western blot analysis of Ubc9•SUMO, ubiquitin•Ubc5, and Nedd8•Ubc12 thioester formation in HCT-116 cell lysates.
(C) Quantification of thermo-stable SAE2 Western blots from cellular thermal-shift assay (CETSA) to determine COH000 target engagement in cells as previously described (Jafari et al., 2014). Blot intensity was measured by Li-Cor Odyssey software and normalized to intensity of the 42 °C samples for each concentration. Data are presented as means ± STDEV, n = 3, in independent experiments.
(D) Parental, SAE2 -overexpressed or –knockdown HCT116 cells were treated with vehicle (DMSO) or 10 μM COH000 for 20 h. Percentile of apoptotic HCT116 cells were shown in the bar graph. SAE2 overexpression reduced the percentage of apoptotic cells. In contrast, knockdown of SAE2 increased the percentage of apoptotic cells. The data shown are the means of three independent experiments ±STDEV.
(E and F) Correlation of in vitro biochemical activity with cell proliferation. Biochemical assays of SUMO-Ubc9 thioester formation after 10 min reactions with the indicated compounds at 10 and 20 μM as described for Figure 2C (F). Compound A and B are close analogs of COH000 as shown by the structures. “-” and “+” indicate reactions without and with ATP, respectively, as negative and positive controls. COH000, 54 and 55 were added to cell culture media at the indicated concentrations and MTS assays were performed 72 hours later (E). Data shown are the means of three independent experiments ±STDEV.
Previous studies showed that inhibition of SUMOylation by knockdown of SAE2 or Ubc9 induces apoptosis (Kessler et al., 2012). COH000 treatment also consistently induced apoptosis in HCT-116 cells (Figure 4D, Figure S2B) as shown previously (He et al., 2015). The level of apoptosis was reduced when SAE2 was overexpressed by transfection and was increased when SAE2 was knocked down in HCT116 cells (Figure 4D, Figure S2B). The SAE2-dependent effect on COH000-induced apoptosis suggests that apoptosis occurs through COH000 inhibition of SUMO E1. In contrast, COH000 analogs A and B that did not inhibit SUMO E1 in biochemical assays did not inhibit cell proliferation (Figures 4E and 4F). This result suggests that the effect of COH000 on cell proliferation and apoptosis is not due to the covalent reactivity of the molecule.
To further investigate the on- and off-target effects of COH000, we transduced lentiviral vectors expressing WT or a C30S mutant of green-fluorescence-fusion SAE2 (GFP-SAE2) into HEK293 cells, which have high transfection efficiency, to establish cells expressing the exogenous proteins. However, C30S SAE2 does not appear to recapitulate the activity of WT-SAE2 well in cells. We could not obtain stable cell lines expressing similar levels of C30S and WT SAE2. We sorted the cells based on GFP expression to obtain populations of cells that expressed the same levels of WT SAE2 and C30S SAE2 (both SAE2 variants were expressed as GFP fusion proteins). However, within two passages after cell sorting, the C30S SAE2 levels again dropped to approximately 1/3 of those of WT SAE2 (Figure S3). Similar results were obtained using both HCT116 and HEK293 cells. We also attempted to mutate the endogenous SAE2 by CRISPR. After several attempts, including double CRISPR, and screened nearly 400 colonies, we found many heterozygous clones, suggesting that the methods worked, but we did not find a single homozygous clone. Consistent with the much lower enzymatic activity of C30S that of WT SAE2 (Figure 2C), these results suggest that C30S-expressing cells are not an appropriate control. Preliminary profiling of COH000 targets in cells by pull-down with a close alkyne-containing analog of COH000 as a probe suggests that COH000 does not have a large number of other targets (Figure S4).
COH000 Induces miR-34b Expression and Reduces c-Myc expression.
SUMOylation is important to c-Myc-dependent oncogenesis (Hoellein et al., 2014; Kessler et al., 2012). We have shown that inhibition of SUMOylation leads to reduced c-Myc levels through a miRNA-dependent mechanism (Li et al., 2018). The miR-34 family of microRNAs suppresses the expression of proteins, including c-Myc, involved in pluripotency and oncogenesis. miR-34 expression is frequently reduced in cancers. We showed that knockdown of SAE2 increased miR-34b/c expression, which reduced the expression of c-Myc (Li et al., 2018). Treatment of lymphoma and colorectal cancer cells with COH000 also increased the expression of miR-34b, and reduced c-Myc mRNA and protein levels, suggesting that COH000 phenocopies SAE2 knockdown in these experiments (Figure 5A). It appears that any potential COH000 metabolites may not inhibit SUMO E1 in cells as indicated by cell proliferation; cells treated by COH000 for 24 hours followed by culturing for 48 more hours with media containing no COH000 showed nearly identical proliferation as cells treated with COH000 continuously for 72 hours (Figure S5A).
Figure 5. COH000 increases miR-34b expression and reduces c-Myc mRNA and protein levels.
(A) Lymphoma (Raji) and colorectal cancer (HCT116) cells were treated COH000 at the indicated concentrations. miR-34b and c-Myc mRNA expression levels were measured by RT-qPCR after 2 days of inhibitor treatments. c-Myc protein levels were detected by Western blots.
(B) HCT116 xenografted Es1e/SCID mice were treated with s.c. peritumoral injection of vehicle or 10 mg/kg COH000. Tumor growth was monitored (left panel). Statistics of apoptosis were detected by TUNEL staining of in tumor tissue from vehicle and COH000-treated mice (right panel). miR-34b levels in tumor cells of the mouse model were also measured. Two-tail Student’s t-test was used to calculate p value. *, p < 0.05, ** p< 0.01.
(C) Representative IHC staining images for SAE2, c-Myc, and TUNEL of xenograft tumor tissues in Es1e/SCID mice from COH000 treatments. Red arrows indicate TUNEL positive staining cells and scale bar presents 50μm.
We used COH000 to evaluate for anti-tumor activity against an HCT-116 cell xenograft mouse model. Because COH000 contains two ester bonds (Figure 1A), a xenograft model of HCT-116 cells was created in a plasma esterase-deficient SCID mouse strain (Es1e/SCID) (Morton et al., 2005) to reduce ester hydrolysis in vivo. Once the tumor became palpable, the tumor bearing Es1e/SCID mice were treated with COH000 or vehicle for 14 days. COH000 significantly inhibited tumor growth (Figure 5B). In addition, COH000 significantly reduced SAE2 levels in tumor tissues (Figure 5C, upper panel), as in cells (Figure 3C). The c-Myc protein level was reduced in inhibitor treated mice tumor (Figure 5C, lower panel) as observed in cells (Figure 5A). In addition, the inhibitor increased apoptosis (Figure 5B, middle panel; Figure 5C middle panel). These observations phenocopy that of SAE2 knockdown using HCT-116 cells carrying Dox-inducible SAE2-shRNA were inoculated by subcutaneous injection into NSG mice (Li et al., 2018). Furthermore, miR-34b level is higher in tumor tissues in COH000-treated mice than vehicle-treated mice.
We also tested the effect of COH000 on primary colorectal tumor cells from patient-derived xenograft tissues (PDX). The primary culture contained colorectal cancer cells as well as stromal and fibroblast cells, and the colorectal cancer cells express an epithelial cell adhesion molecule (EpCAM) marker (Figure S5B and S5C). The sample from Patient #1 had the lowest c-Myc level among the three samples as indicated by the c-Myc staining intensity relative to the EpCAM band, and this sample did not show much further reduction of c-Myc level by COH000 treatment unlike the other two samples. Samples from Patient #2 and #3 expressed higher c-Myc levels and showed reduced c-Myc levels by treatment with COH000. These two samples were more sensitive to COH000 (Figure S5B and S5C). Consistent with the results in colorectal cancer cell line HCT116, COH000 induced apoptosis and decreased cancer cell viability in primary colorectal PDX samples (Figure S5B and S5C).
Discussion
In this study, we have discovered an allosteric inhibitor chemotype that inhibits SUMO E1 through a covalent mechanism. Our further studies demonstrated covalent adduct formation of COH000 to Cys30 of SUMO E1, a cysteine which is not involved in binding ATP or SUMO. This is consistent with the results that COH000 does not compete with ATP or SUMO1 binding (Figure 2A). Cys30 is fully buried in previously determined crystal structures of SUMO E1s (Lois and Lima, 2005; Olsen et al., 2010), and the analogous sites are buried in previously published Ubl E1 crystal structures (Lee and Schindelin, 2008; Noda et al., 2011; Walden et al., 2003). Therefore, binding of COH000 to the SUMO E1 requires a conformational state of the enzyme not previously observed. Sequence comparison of E1 enzymes revealed that the covalent modification site and the amino acid sequence surrounding it is conserved among the human Uba1 (ubiquitin E1), Uba2 (SUMO E1), Uba3 (Nedd8 E1), Uba4 (Urm1 E1), Uba7 (ISG15 E1), and Atg7 (E1 for Atg8 and Atg12) (Figure S1C). Thus, the covalent allosteric inhibition mechanism exemplified by COH000 could be applicable to other E1 enzymes. Findings described here also raise a question of whether E1 enzymes can be allosterically regulated by small molecule metabolites that bind to the same site as COH000.
Covalent inhibitors are well suited for targeting the E1 enzymes of Ubl modifications. Because the E1 enzymes in Ubl modifications, such as the SUMO E1 and Atg7, have a slow turnover rate (Boggio et al., 2004), prolonged inhibition can be achieved without requiring compounds to be stable in circulation, reducing the pharmacokinetic hurdle associated with conventional drug discovery process. In addition, cancers and viruses are less likely to develop resistance to covalent than to non-covalent inhibitors as shown by existing covalent drugs (Kalgutkar and Dalvie, 2012; Singh et al., 2011).
Our finding of a covalent inhibitory mechanism of the E1 enzymes is a major advancement in targeting the Ubl modifications. As we have previously discussed, the lack of drugs targeting Ubl modifications highlights the need to identify new approaches to inhibit this class of enzymes. Members of E1 enzymes are potential targets for inhibiting c-Myc and KRas oncogenes involved in the development of the majority of human cancers and contribute to cancer cell stemness and resistance (Bogachek et al., 2016; Du et al., 2016; He et al., 2017; Kessler et al., 2011; Luo et al., 2009; Yu et al., 2015). Therefore, identification of a mechanism to inhibit the E1 enzymes not only provides a complementary approach to overcome resistance developed through mutations for compounds that target the ATP-binding sites (Xu et al., 2014) but also provides new directions for developing potent and selective inhibitors to potentially treat cancers and other life-threatening diseases.
STAR METHODS and KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal antibody anti- c-Myc | Cell Signaling Technology | Cat# 5605;clone D84C12; RRID:AB_1903938 |
| Rabbit polyclonal anti-UbcH5/UBE2D | Boston BioChem | Cat#A-615; RRID:AB_10699720 |
| Rabbit polyclonal anti-UbcH12/UBE2M | Boston BioChem | Cat# A-655; RRID:AB_10693705 |
| Goat polyclonal anti-UBE2I/UBC9 | Abcam | Cat# ab21193; RRID:AB_2210477 |
| Rabbit polyclonalanti-SAE2/UBA2 | Abcam | Cat# ab185955 RRID:N/A |
| Rabbit polyclonal anti-SAE2/UBA2 | Abcam | Cat# ab58451; RRID:AB_882430 |
| Goat polyclonal anti-AOS1/SAE1 | Santa Cruz Biotech | Cat# sc-46766; RRID:AB_2182926 |
| Rabbit polyclonal anti-c-Myc | Abcam | Cat# ab32072; RRID:AB_731658 |
| Goat polyclonal anti-GAPDH | Santa Cruz Biotech | Cat#sc-48166; RRID:AB_783595 |
| Mouse monoclonal anti-Actin | Sigma Aldrich | Cat#A2228; RRID:AB_476697 |
| Rabbit polyclonal anti-SUMO-1 | Cell Signaling Technology | Cat#4940; Clone: C9H1; RRID:AB_2302825 |
| Mouse monoclonal anti-SUMO2/3 | MBL international | Cat# M114–3; Clone: 1E7; RRID:AB_592769 |
| Mouse monoclonal anti-EpCAM | Cell Signaling Technology | Cat# 2929; Clone: VU1D9; RRID:AB_2098657 |
| Mouse monoclonal anti-Ki67 | Agilent Dako | Cat# GA62661–2; RRID:AB_2687921 |
| Bacterial and Virus Strains | ||
| Lentivirus | This study | N/A |
| Biological Samples | ||
| human colorectal cancer tissue for PDX mouse model generation | City of Hope National Cancer Center-Animal Tumor Model Core | IRB13389 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TAMRA-biotin-azide | Click Chemistry Tools LLC | Cat# 1048–1, CAS: 1797415–74-7 |
| [35S]-Methionine | PerkinElmer Inc | Cat# NEG709A500UC |
| [P32]-labeled sodium pyrophosphate | PerkinElmer Inc | Cat# NEX019001MC |
| Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) | Sigma Aldrich | Cat# C4706; CAS:51805–45-9 |
| Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA) | Sigma Aldrich | Cat# 678937; CAS: 510758–28-8 |
| Critical Commercial Assays | ||
| Nickel Chelate acceptor beads | PerkinElmer | Cat # 6760140 |
| Glutathione donor beads | PerkinElmer | Cat # 6765300 |
| EasysepEpCAM positive selection kit | StemCellTech | Cat # 18356 |
| TUNEL apoptosis detection kit | EMD Millipore | Cat #17–141 |
| Streptavidin Dynabeads | Thermo Scientific | Cat# 11205D |
| Taqman miRNA assay- hsa-miR-34b-5p | Thermo Scientific | Cat # A25576 |
| Taqman Gene Expression Assay-MYC | Thermo Scientific | Cat # 4331182 |
| CellTiter96 AQueous One Solution Cell Proliferation Assay (MTS) | Promega | Cat# G3582 |
| miRNeasy RNA isolation kit | Qiagen | Cat# 217004 |
| TaqMan MicroRNA Reverse Transcription Kit | Thermo Scientific | Cat# 4366596 |
| Deposited Data | ||
| HTS screening for SUMOylation inhibitors | PubChemBioAssay | AID 2011 |
| FRET assay for SUMOylation inhibitor screening | PubChemBioAssay | AID 2006 |
| AlphaScreen for SUMOylation inhibitors | PubChemBioAssay | AID 2018 |
| Experimental Models: Cell Lines | ||
| HCT116 (human, colorectal carcinoma) Gender: male | ATCC | ATCC CCL-247; RRID:CVCL_0291 |
| HT29 (human, colorectal adenocarcinoma) Gender: female | ATCC | ATCC HTB-38; RRID:CVCL_0320 |
| Raji (human, Burkitt’s lymphoma) Gender: male | ATCC | ATCC CCL-86; RRID:CVCL_0511 |
| HEK293T (human, embryonic kidney) Gender: unknown | ATCC | ATCC CRL-3216; RRID:CVCL_0063 |
| HEK293 (human,, embryonic kidney) Gender: unknown | ATCC | ATCC PTA-4488; RRID:CVCL_0045 |
| OVCAR3 (human, ovary adenocarcinoma) Gender: female | ATCC | ATCC HTB-161; RRID:CVCL_0465 |
| Experimental Models: Organisms/Strains | ||
| Mouse: NSG mice, NOD.(SCID)-Il2RG(ko) Sex: both male and female | City of Hope Animal Core facility | N/A |
| Mouse: plasma esterase-deficient SCID mouse strain (Es1e/SCID) Sex: both male and female | Dr. Karen Aboody lab City of Hope | N/A |
| PDX tissues Gender: unknown | City of Hope Animal Core facility and Dr. Marwan Fakih | N/A |
| Oligonucleotides: Site-directed mutagenesis | ||
| C30S-F:5’-GTGGGTGCGGGCGGTATCGGTAGCGAACTGCTGAAAAACC-3’ | This study | N/A |
| C30S-R:5’-GGTTTTTCAGCAGTTCGCTACCGATACCGCCCGCACCCAC-3’ | This Study | N/A |
| Recombinant DNA | ||
| pCMV-VSVG | (Stewart et al., 2003) | Addgene # 8454 |
| pCMV-dR8.2-dvpr | (Stewart et al., 2003) | Addgene # 8455 |
| pCMV-SAE2 | Origene | Cat# SC319458 |
| pLenti-SAE2-mGFP | Origene | Cat# RC201288L4 |
| pLenti-C30S-SAE2-mGFP | This study | N/A |
| GIPZ Human UBA2 shRNA | GE Dharmacon | Clone ID# V2LHS_68112 |
| Software and Algorithms | ||
| proteome software: Scaffold | Proteome Software, Inc. |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to the lead contact, Dr. Yuan Chen (YChen@coh.org)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
For additional information, please see STAR METHODS and KEY RESOURCES TABLE.
Cell lines and lentivirus production
HCT116, HT29, HEK293, HEK293T and OVCAR3 cell lines were grown in Dulbecco’s Modification of Eagle’s Medium (DMEM). Raji cell line was kept in RPMI1640 media. Media was supplemented with 10% heat inactivated fetal calf serum (FBS) (Omega Scientific, Inc.), 2 mM L-glutamine, 100 U ml−1 penicillin, and 100 μg ml−1 streptomycin. All cell lines were kept in a 37°C incubator and supplied with 5% CO2.
Patient-Derived-Xenograft Primary Culture
Patient-derived xenograft model was generated by subcutaneously implant of human colorectal cancer tissue into NSG mice. Human colorectal cancer tissue was obtained with patient consent, as approved by the Research Ethics Board at the City of Hope (IRB13389). Xenograft tumor tissue was washed in PBS, minced and incubated with collagenase (235 U/ml), hyaluronidase (850 U/ml) (Sigma-Aldrich) for 90–120 min at 37°C. DMEM with 10% FBS was added to stop enzymatic digestion. The sample was serially filtered through 70-μm then 40-μm cell strainer. Cells were spun down and resuspended with 1× ice cold red blood cell lysis buffer (Cat# sc-296258, Santa Cruz Tech) and incubated for 2 min to lyse red blood cells. Cells were then used for downstream applications. For further purification to remove stromal or fibroblasts, magnetic isolation was carried out using EpCAM positive selection kit (Cat# 18356, StemCell Tech).
Xenograft Colorectal Cancer Models
All experiments with mice were approved by Beckman Research Institute Animal Care and Use Committee (IACUC #10026) and complied with all relevant federal guidelines and institutional policies. Mice were housed in a controlled environment (12-h light/12-h dark cycle) with access to waterand fed a standard diet generally from 6 to 8 weeks of age. Body weight was measured weekly and food intake was monitored. For colorectal cancer xenograft with COH000 treatment, HCT116 cells were s.c. injected into a plasma esterase-deficient SCID mouse strain (Es1e/SCID)(Morton et al., 2005), male, 8–10 weeks. When the tumors became palpable, COH000 was administered subcutaneously peritumoral injection once a day at a dose of 10 mg per kilogram of body weight. Mice in the control group received equal volumes of vehicle (5% DMSO, 30% solutol in PBS). Tumor volume was measured until the endpoint was reached. Mice were euthanized using CO2 inhalation and tumors were excised.
METHOD DETAILS
Lentivirus production
For lentivirus generation, the envelope plasmid pCMV-VSVG and the packaging plasmid pCMV-dR8.2-dvpr were obtained from Addgene (8454 and 8455, provided by Dr. Bob Weinberg). The DNA plasmid of constitutively expressed SAE2 shRNA was purchased from GE Dharmacon(V2LHS_68112). The lentiviral SAE2 expression plasmid pLenti-SAE2-mGFP was purchased from Origene (Cat# RC201288L4). HEK293T producer cells were transfected with these DNA plasmids, and supernatant containing lentiviral particles was harvested 24 to 48 h after transfection. Then HCT116 or HT29 cells were transduced with the SAE2 shRNA lentivirus. The cells were harvested after 5 daysof virus transduction. For COH000 treatment, HCT116 or HT29 cells were treated with the indicated concentrations for either 20 or 48 h before cell harvest. The lentiviral SAE2 expression plasmid pLenti-SAE2-mGFP was purchased from Origene (Cat# RC201288L4). C30S mutation of SAE2 was obtained by using site-directed mutagenesis and following the standard instructions. The lentivirus carried wild type orC30S mutant SAE2-GFP cDNA was generated as described above. HEK293 or HCT116 cells were then transduced with the lentivirus to express wild type or C30S mutant SAE2-GFP. The transformed cells were first screened by puromycin and sorted by flow cytometry based on GFP intensity.
Transient Transfection
Transient transfection of plasmid DNA (pCMV-SAE2) was performed using Lipofectamine LTX (Cat# 15338100, Thermo Scientific) following the manufacturer’s instructions. After 2 days of DNA transfection, the cells were harvested for Western blot or further COH000 orDMSO (vehicle) incubation.For Western Blot, cells were lysed directly into buffer containing 190 mMTris, pH 6.8, 30% glycerin, and 4% SDS. After protein quantification by the bicinchoninic acid (BCA) method, 100 mMof β-mercaptoethanol was added to the protein sample which was boiled at 95°C.
Antibodies for Immunoblotting
A rabbit monoclonal antibody (mAb) of c-Myc (D84C12; Cell Signaling Technology) was used for immunoblotting (1:1000). For Ubiquitin and Nedd-8 conjugation assay, rabbit anti-UbcH5/UBE2D (A-615) and anti-UbcH12/UBE2M (A-655) were obtained from Boston BioChem. The following rabbit antibodies were obtained from Abcam for immunoblotting or IHC staining in mouse xenograft: goat anti-UBE2I/UBC9 (1:1000) (ab21193), rabbit anti-SAE2/UBA2 (1:1000) (ab185955) and rabbit anti-c-Myc (ab32072). Goat anti-GAPDH polyclonal antibody (pAb) (Santa Cruz Biotechnology, Inc.) and mouse anti-Actin mAb (Sigma Aldrich) were used for immunoblotting as loading controls. For SUMOylation detection, Rabbit anti-SUMO-1 (C9H1; #4940) and mouse anti-SUMO2/3 (1E7; M114–3) antibodies were from Cell Signaling and MBL international. The following antibodies were used for immunoblotting were obtained from Cell Signaling: rabbit anti-c-Myc (D48C12; #5605) and mouse anti-EpCAM (VU1D9; #2929).
Cell proliferation Assay
Cell proliferation was measured using a CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS-based) (Cat# G3582, Promega) after COH000 or its analog (54 or 55) treatment at the indicated concentrations and the time points. Briefly, cells were incubated with 20 μL of CellTiter 96 AQueous reagentafter the treatment and incubated at 37oC until color development. Absorbance measurements were performed using a SpectraMax M5 reader (Molecular Device). For assays measuring anti-proliferation effects, all values were normalized to the vehicle treatment. All values are represented graphically as mean ± standard deviation (STDEV) from three independent samples (n=3).
Flow Cytometry Analysis
To determine the level of apoptosis, SAE2 knockdown cells were labeled using the Annexin VAPC Apoptosis Kit (Cat# 550475, BD Bioscience) according to the manufacturer’s instructions. To verify apoptotic effect of COH000 in primary culture cells from patient-derived xenograft tissue, the primary cells were treated with COH000 for 48 h. After cells were harvested, the cells were first stained with anti-EpCAM conjugated with Allophycocyanin (APC) and followed with Annexin V-FITC (Cat# 556419, BD Bioscience) and DAPI staining. The cells were then washed with 1X PBS and fixed in 2% formaldehyde before FACS analysis.
Pulse-Chase Assay
HCT116 were plated at 1 × 105 cells/mL and incubated overnight at 37°C in the presence of 5% CO2. The cells were depleted of methionine for 15 minutes and pulsed labeled with [35S]-Methionine for 2 hours. After the cells were treated with 0.1% DMSO or COH000, they were chased for various times. Immunoprecipitation of SAE2 was followed the standard protocols and samples were separated by SDS-PAGE. The gels were stained by SimplyBlue SafeStain (Cat# LC6060, Thermo Scientific) to ensure even loading. The gel was dried in a gel dryer at 80°C for 2 hours. The radioactive gels were exposed to film overnight at room temperature.
RNA Extraction, Reverse Transcription and Real-Time PCR Quantification Total RNA and microRNA were extracted from cells with a cell density of 75% confluent using miRNeasy RNA isolation kit (Cat# 217004, Qiagen) according to manufacturer’s protocol. MicroRNA cDNA was synthesized from total RNA by using TaqMan MicroRNA Reverse Transcription Kit (Cat# 4366596, Thermo Scientific) and 5X miR-34b specific RT primer according to manufacturer’s protocol. Briefly, reverse transcriptase reactions contained 10 ng of RNA samples, 50 nM RT primer, 1 × RT buffer, 0.25 mM each of dNTPs, 3.33 U/μlMultiScribe reverse transcriptase and 0.25 U/μl RNase Inhibitor (all purchased from cDNA Archive kit of Applied Biosystems). The 15 μl reactions were incubated in an iCycler (Bio-Rad) in a 96-well plate for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C and then held at 4°C. Total RNAs were converted to cDNA by using Omniscript RT kit (Cat# 205113, Qiagen) and oligo d (T) primer according to manufacturer’s protocol. Real-time PCR was performed using an Applied Biosystems 7900HT real-time PCR system. For miRNA cDNA real-time PCR, the 20 μl PCR included 1.33 μl RT product, 1× TaqMan Universal PCR master mix (no UNG) (Cat# 4324020, Thermo Scientific) and 1 μl of Taqmanmir-34b Small RNA Assay mix (Cat # A25576, Thermo Scientific). For real-time PCR of gene expression, the 20 μl PCR included 1 μl RT product, 1× TaqMan Gene Expression Master Mix (Cat# 4369016, Thermo Scientific) and 1 μl of TaqMan Gene Expression Assay primer mix (MYC, Cat#4331182, Thermo Scientific). The reactions were incubated in a 386-well optical plate at 95°C for 1 0 min, followed by 40 to 45 cycles of 95°C for 15s and 60° for 10 min. The threshold cycle (CT) da ta was determinate using default threshold settings. The CT is defined as the fractional cycle number at which the fluorescence passes the fixed threshold. The relative fold change was measured by the ΔΔ(CT) method between each treatment and the control samples in expression: Δ(CT) = CT (miR34b or MYC) – CT (RNU6B or GAPDH); ΔΔ(CT) = Δ (CT; COH000 treatment) − Δ(CT;control samples); fold change = 2^-ΔΔ (CT).
Immunohistochemistry (IHC)
Tumor tissues were fixed in 4% paraformaldehyde and wash with PBS and transfer to 70% ethanol and then embedded in paraffin in accordance with the standard procedure. Sections were stained for hematoxylin and eosin (H&E), Ki67 (MIB-1, Dako), SAE2 (Cat# ab58451, Abcam), c-Myc (Cat# ab32072, Abcam) and TUNEL (TUNEL apoptosis detection kit, EMD Millipore 17–141). For statistic quantification of apoptotic cells, TUNEL staining was performed for 3 mice in each group and 10 fields of each slide were counted.
Biochemical Assays of SUMOylation and High Throughput Screening Assays
Time-resolved fluorescence resonance energy transfer (HTRF) assay was described previously (Alontaga et al., 2012). The overall probe project is described under PubChem BioAssay identifier Summary AID 2011, while the actual HTS screen of 290,921 compounds of the NIH Molecular Libraries Small Molecules Repository (MLSMR) is described under PubChem BioAssay identifier AID 2006. AlphaScreen assay protocol was deposited into PubChem under BioAssay identifier AID 2018. Briefly, SAE, Ubc9, GST-SUMO and His6-RanGap-1 proteins were expressed and purified as described previously (Wang et al., 2010; Wang and Chen, 2010; Wang et al., 2007; Wang et al., 2009).Assay buffer contained 50 mMTris-HCl pH 7.4, 0.3 mM DTT, 10 mM MgCl2, and 0.005% Tween-20. Nickel Chelate acceptor beads (Cat # 6760140) and Glutathione donor beads (Cat # 6765300) were purchased from PerkinElmer. The assays were conducted using Corning 1536-well, white plates (Cat #3725). 2 μL of Mixture 1, containing 12.5 nM SAE and 100 nM His6-RanGap-1 in the assay buffer was mixed with 70 nl of 2 mM compounds dissolved in DMSO. Then 2 μL of Mixture 2, containing 20 mM ATP, 12.5 nM E2 and 30 nM GST-SUMO in assay buffer, was added. After incubation for 90 min at room temperature, 1 μl of pre-mixed Ni acceptor beads and Glutathione Donor beads, at 10 μg/ml each, was added. After incubation for 60 min at room temperature, readings were made on a BMG LabtechPheraStar in an AlphaScreen mode (Ex: 680 nm; Em: 570 nm).
ATP:PPi Exchange Assay to Examine the Adenylation Step of SAE Catalysis
The assay was conducted as previously described (Wang and Chen, 2010). Briefly, the compound COH000 at 0–2 μM was pre-incubated with SAE for 15 minutes at room temperature. Then SUMO-1 protein was added to SAE-compound mixture with final concentrations of 0.1 to 800 μM, and the reaction was initiated by addition of 1 mM ATP containing 50 mMHepes, pH 7.6, 10 mM MgCl2, 0.5 mM DTT, and 1 mM [P32]-labeled sodium pyrophosphate. The mixture was incubated at 37 °C for 20 minutes, and 0.5 ml of 5% TCA containing 10 mM pyrophosphate was added to terminate the reaction. To count for radioactivity in ATP converted from [P32]-labeled pyrophosphate, 0.3 ml of activated charcoal resin (10% w/v in 2% TCA with 10 mM pyrophosphate) was added to the sample and incubated at room temperature for 2 hours with rotating to absorb ATP in the solution. The resin was separated from the supernatant by centrifuging at 15,000 rpm for 5 minutes and washed three times with 1 ml of 2% TCA with 10 mM pyrophosphate, each for 15 minutes with rotating, and was counted by a scintillation counting instrument. All the assays were repeated three times for uncertainty analysis.
LC-MS/MS Analysis of the Pepsin Digestion of SAE and SAE Inhibited by COH000
One μL of sample was combined with 3 μL of pH 7 buffer (final conc. 5 μM) and injected into the 2 μL exchange loop of our automated MS system. The loop was switched on-line with a flow (2 μL/min) of 10 mMTris-HCl buffer (pH 6.8) and combined with a flow (3 μL/min) of 2M Guanidine-HCl& 100 mM TCEP in 0.8% aqueous formic acid. The combined flow passed through an immobilized pepsin column and the resulting peptides were captured on the enrichment column of an Agilent standard capacity C18 reverse phase LC-MS chip and washed thoroughly with 0.05% aqueous TFA to remove salts. The enrichment column was then switched on-line with the C18 analytical column of the LC-MS chip and peptides were gradient eluted (5 – 44% B in 10 min, 44– 95% B in 1 min, hold 95% B 2.5 min) (Solvent A = 0.05% TFA in water, B = 0.01 % TFA in 80/20 water/acetonitrile) into an Agilent 6520 QTOF mass spectrometer. Each sample was analyzed twice with the same results. Mass spectral data were acquired using a data dependent acquisition to provide fragment ion spectra (MS/MS) for the major ions in each MS spectrum. Although not identical, the base peak chromatograms for the two samples are similar both in intensity and the pattern of the peaks. The MS data were searched with Sequest to assign spectra to the sequence of SAE and the results summarized using Scaffold. The search was also performed using a potential modification of Cys corresponding the simple addition of COH000 (419.1733 u). The Sequest search yielded 92% sequence coverage for the SAE1 and 81% sequence coverage of the SAE2
Cellular Thermal Shift Assay (CETSA)
3×107 HCT116 cells were incubated with COH000 at different concentrations for 2 hours at 37°C prior to CETSA assay. Same volume of DMSO was added as vehicle control. At the end of incubation, 3×106 cells from the same treatment were dispended to each PCR tubes and thermostable SAE2 was examined at designated temperature (42–56°C) by a Veriti 96-well thermal cycler (Bio-Rad). The tubes are kept at room temperature before the heat treatment step. Western blotting was used to detect thermo-stable SAE2 following CETSA.
Click chemistry
1 × 106 of live HCT116 cells were incubated with either DMSO (same volume as COH000) or COH000 at 2 μM at 37°C for 1 h before incubating with 2 μM of probe compound for another 2 h. After that, the cells were harvested, and the cell lysate was prepared to proceed with the click chemistry to conjugate TAMRA-biotin-azide (0.1 mM TAMARA-biotin-azide, 1% SDS, 1 mM TCEP, 0.1 mM TBTA and 1 mM CuSO4) for 1.5 h at room temperature. The proteins pulled down by Streptavidin Dynabeads were visualized by SDS-PAGE and fluorescence imaging by Typhoon scanner (Amersham, GE Healthcare, Chicago, IL). Total cellular protein from the samples was stained with Coomassie blue after SDS-PAGE. For protein identification, 5 × 106 of live HCT116 cells were incubated with either DMSO or COH000 at 2 μM at 37°C for 1 h before incubating with 2 μM of the probe compound for another 2 h. Then, cells were harvested, and the cell lysate was prepared for click chemistry as described above. The proteins were first pull-down by Streptavidin Dynabeads overnight at 4 °C and then separated by SDS-PAGE. The proteins with molecular weight from 32 to 15 kD were isolated from SDS-PAGE, in-gel digestion by trypsin and then identified by LC-MS/MS. The results were analyzed by a proteome software Scaffold (Proteome Software, Inc. Portland, OR).
QUANTIFICATION AND STATISTICAL ANALYSIS
Each experiment was conducted at least three times, as indicated n=3 in Figure Legends. Figures shown are representative. All analyses werecarried out in Microsoft Excel and GraphPad Prism 7. Data shown in bar graphs are mean ± STDEV. P values were evaluated using the analysis of variance (one way ANOVA) or Student’s t-test as indicated in the figures and legends. A P-value <0.05 was considered statistically significant.
Supplementary Material
Table 1.
SUMOylation Potency IC50 (μM)
 |
(Geometric Mean ± S.E. (n=replicates) [stereochemistry at the indicated location]) | |||||
|---|---|---|---|---|---|---|
| R2 |  |
|||||
| -C6H5 | -p-C6H4Cl | -p-C6H4Me | -p-C6H4OMe | |||
| R1 | -CH2C6H5 | 0.69 ± 1.04 (n=8) [R] 4.87 ± 0.947 (n=3) [S] |
0.71 ± 0.04 (n=3) [R] 2.40 ± 0.66 (n=3) [S] |
2.75 ± 0.75 (n=3) [R] 7.18 ± 0.34 (n=3) [S] |
4.79 ± 0.58 (n=3) [R] 12.7 ± 1.77 (n=3) [S] |
|
|
COH000
[R isomer]
-CH2(p-C6H4Me) |
<0.23 ± 0.03 (n=3)
[R]
2.14 ± 0.34 (n=3) [S] |
|||||
| -CH2(p-C6H4OMe) | 0.53 ± 0.05 (n=6) [R] | |||||
| -CH2(p-C6H4OCF3) | 0.92 ± 0.41 (n=10) [?] |
|||||
| -CH2(p-C6H4F) | 1.01 ± 0.37 (n=7) [?] | |||||
| -n-Propyl | 0.84 ± 1.83 (n=11) [R] 2.87 ± 0.09 (n=3) [S] |
|||||
| -Allyl | 1.69 ± 0.46 (n=3) [R] 5.77 ± 0.98 (n=3) [S] 1.33 ± 0.20 (n=3) [?] |
3.82 ± 0.25 (n=3) [R] 4.15 ± 0.34 (n=3) [S] |
12.4 ± 0.71 (n=3) [R] 11.7 ± 0.31 (n=3) [S] |
22.1 ± 2.02 (n=3) [R] 17.6 ± 1.64 (n=3) [S] |
21.0 ± 2.31 (n=3) [R] 15.0 ± 0.59 (n=3) [S] |
|
| -Vinyl | 7.18 ± 0.58 (n=3) [R] 10.4 ± 0.32 (n=3) [S] |
|||||
| -Me | 7.67 ± 1.00 (n=3) [R] 23.7 ± 2.28 (n=3) [S] |
18.2 ± 0.34 (n=3) [R] 11.3 ± 1.51 (n=3) [S] |
34.7 ± 3.53 (n=3) [R] | 55.2 ± 4.69 (n=3) [R] | ||
| -CONH2 | >20* | |||||
| -C5H9O | >20* | |||||
IC50 were determined by SDS-PAGE-based assay. The IC50 of all other compounds were determined by AlphaScreen-based assay.
Highlights.
We identified an allosteric inhibitor chemotype that inhibits an activating enzyme.
The lead compound inhibits the ATP-dependent step of SUMO E1 catalysis.
The compound has specificity to 1 out of 18 non-disulfide bonded Cys residues.
The compound increased miR-34b and reduced c-Myc in cellular and xenograft models.
Significance.
Ubl post-translational modifications are potential targets for developing novel therapeutics for life-threatening diseases, such as c-Myc and KRas driven cancers that lacks targeted therapy. However, only a few drugs are available to target these modifications. This lack of drugs highlights our knowledge deficiency in regulating Ubl enzymes by small molecules. Here, we identified an allosteric, covalent inhibitor chemotype that inhibits the SUMO activating enzyme (E1) by targeting an unexpected buried site. Because the only previously known mechanism to inhibit E1 enzymes is through targeting the ATP-binding site, identification of an allosteric inhibitory mechanism of the activating enzymes could spur innovative drug discovery efforts targeting Ubl modifications. This finding also raises the possibility of allosteric E1 regulation by small molecule metabolites.
Acknowledgments
We thank the City of Hope core facilities, including the Animal Model Core, Bioinformatic Core, NMR Core, Flow Cytometry Core, and Florescence Microscopy Core for excellent technical support, and NIH grants R01GM086171, R01GM102538, and R01CA212119, R01CA216987, and R03DA026556 to Yuan Chen for funding and R44CA189499 to S.X.O. E.S. is a fellowship recipient of the California Institute of Regenerative Medicine. The Conrad Prebys Center for Chemical Genomics wishes to acknowledge the NIH Roadmap Grant U54 HG005033 for providing funds to during its participation as a comprehensive screening center of the Molecular Libraries Probe Production Centers Network (MLPCN). We thank Dr. Sumeet Salaniwal for assistance in preparing Table 1. Dr. Gregory P. Roth, who led the medicinal chemistry studies described here, passed away. Gregory is remembered for his many scientific contributions including those described here.
Footnotes
Declaration of Interests
S.X.O. is an employee and shareholder of SUMO Biosciences, Inc. Y.C. is a founder of SUMO Biosciences, Inc. and a member of its advisory board. Other authors declare no competing interests.
DATA AVAILABILITY
The results of high through put screening of SUMOylation inhibitors were deposited to PubChem and can be reached online. PubChem AID 2011, for the high throughput screening of 290,921 compounds. PubChem AID 2006, for the fluorescence resonance energy transfer (FRET) based primary assay. PubChem AID 2018, for thechemiluminescence-based secondary assay using AlphaScreen technology to eliminate false positive hits.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alontaga AY, Bobkova E, and Chen Y (2012). Biochemical analysis of protein SUMOylation. Current protocols in molecular biology / edited by Ausubel Frederick M [et al. ] Chapter 10, Unit10 29. [DOI] [PMC free article] [PubMed]
- Bogachek MV, Park JM, De Andrade JP, Lorenzen AW, Kulak MV, White JR, Gu VW, Wu VT, and Weigel RJ (2016). Inhibiting the SUMO Pathway Represses the Cancer Stem Cell Population in Breast and Colorectal Carcinomas. Stem cell reports 7, 1140–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggio R, Colombo R, Hay RT, Draetta GF, and Chiocca S (2004). A mechanism for inhibiting the SUMO pathway. Molecular cell 16, 549–561. [DOI] [PubMed] [Google Scholar]
- Bohnsack RN, and Haas AL (2003). Conservation in the mechanism of Nedd8 activation by the human AppBp1-Uba3 heterodimer. J Biol Chem 278, 26823–26830. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, et al. (2010). Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Molecular cell 37, 102–111. [DOI] [PubMed] [Google Scholar]
- Du L, Li YJ, Fakih M, Wiatrek RL, Duldulao M, Chen Z, Chu P, Garcia-Aguilar J, and Chen Y (2016). Role of SUMO activating enzyme in cancer stem cell maintenance and self-renewal. Nature communications 7, 12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, et al. (2013). Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes & development 27, 1447–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AL, and Rose IA (1982). The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. The Journal of biological chemistry 257, 10329–10337. [PubMed] [Google Scholar]
- Hay RT (2005). SUMO: a history of modification. Molecular cell 18, 1–12. [DOI] [PubMed] [Google Scholar]
- He X, Riceberg J, Pulukuri SM, Grossman S, Shinde V, Shah P, Brownell JE, Dick L, Newcomb J, and Bence N (2015). Characterization of the loss of SUMO pathway function on cancer cells and tumor proliferation. PloS one 10, e0123882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Riceberg J, Soucy T, Koenig E, Minissale J, Gallery M, Bernard H, Yang X, Liao H, Rabino C, et al. (2017). Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nature chemical biology 13, 1164–1171. [DOI] [PubMed] [Google Scholar]
- Hoellein A, Fallahi M, Schoeffmann S, Steidle S, Schaub FX, Rudelius M, Laitinen I, Nilsson L, Goga A, Peschel C, et al. (2014). Myc-induced SUMOylation is a therapeutic vulnerability for B-cell lymphoma. Blood 124, 2081–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Iwatsubo T, Kanamitsu S, Ueda K, Suzuki H, and Sugiyama Y (1998). Prediction of pharmacokinetic alterations caused by drug-drug interactions: metabolic interaction in the liver. Pharmacological reviews 50, 387–412. [PubMed] [Google Scholar]
- Jaber T, Bohl CR, Lewis GL, Wood C, West JT Jr., and Weldon RA Jr. (2009). Human Ubc9 contributes to production of fully infectious human immunodeficiency virus type 1 virions. Journal of virology 83, 10448–10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari R, Almqvist H, Axelsson H, Ignatushchenko M, Lundback T, Nordlund P, and Martinez Molina D (2014). The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc 9, 2100–2122. [DOI] [PubMed] [Google Scholar]
- Kalgutkar AS, and Dalvie DK (2012). Drug discovery for a new generation of covalent drugs. Expert opinion on drug discovery 7, 561–581. [DOI] [PubMed] [Google Scholar]
- Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et al. (2011). A SUMOylation-Dependent Transcriptional Subprogram Is Required for Myc-Driven Tumorigenesis. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et al. (2012). A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 335, 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ET, Kim YE, Huh YH, and Ahn JH Role of noncovalent SUMO binding by the human cytomegalovirus IE2 transactivator in lytic growth. Journal of virology 84, 8111–8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, and Schindelin H (2008). Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell 134, 268–278. [DOI] [PubMed] [Google Scholar]
- Li YJ, Du L, Aldana-Masangkay G, Wang X, Urak R, Forman SJ, Rosen ST, and Chen Y (2018). Regulation of miR-34b/c-targeted gene expression program by SUMOylation. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois LM, and Lima CD (2005). Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. Embo J 24, 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, and Elledge SJ (2009). A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137, 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Molina D, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, and Nordlund P (2013). Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87. [DOI] [PubMed] [Google Scholar]
- Morton CL, Iacono L, Hyatt JL, Taylor KR, Cheshire PJ, Houghton PJ, Danks MK, Stewart CF, and Potter PM (2005). Activation and antitumor activity of CPT-11 in plasma esterase-deficient mice. Cancer chemotherapy and pharmacology 56, 629–636. [DOI] [PubMed] [Google Scholar]
- Noda NN, Satoo K, Fujioka Y, Kumeta H, Ogura K, Nakatogawa H, Ohsumi Y, and Inagaki F (2011). Structural basis of Atg8 activation by a homodimeric E1, Atg7. Molecular cell 44, 462–475. [DOI] [PubMed] [Google Scholar]
- Olsen SK, Capili AD, Lu X, Tan DS, and Lima CD (2010). Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature 463, 906–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, Au A, Rai TS, Zheng L, Ridgway R, et al. (2013). p53 status determines the role of autophagy in pancreatic tumour development. Nature 504, 296–300. [DOI] [PubMed] [Google Scholar]
- Rouleau N, Wang J, Karras L, Andrews E, Bielefeld-Sevigny M, and Chen Y (2008). Highly sensitive assays for SUMOylation and small ubiquitin-like modifier-dependent protein-protein interactions. Analytical biochemistry 375, 364–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, and Park-Sarge OK (2009). Sumoylation and human disease pathogenesis. Trends in biochemical sciences 34, 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, and Park-Sarge OK (2011). SUMO and its role in human diseases. Int Rev Cell Mol Biol 288, 167–183. [DOI] [PubMed] [Google Scholar]
- Singh J, Petter RC, Baillie TA, and Whitty A (2011). The resurgence of covalent drugs. Nature reviews 10, 307–317. [DOI] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, and Chen Y (2004). Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proceedings of the National Academy of Sciences of the United States of America 101, 14373–14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhang Z, Hu W, and Chen Y (2005). Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. The Journal of biological chemistry 280, 40122–40129. [DOI] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. (2009). An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E, et al. (2004). SUMO modification of Huntingtin and Huntington’s disease pathology. Science (New York, NY 304, 100–104. [DOI] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al. (2003). Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S, Sixt KM, Barrow R, and Snyder SH (2009). Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science (New York, NY 324, 1327–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Chen Y, and Hay RT (2003). Role of two residues proximal to the active site of Ubc9 in substrate recognition by the Ubc9.SUMO-1 thiolester complex. Biochemistry 42, 3168–3179. [DOI] [PubMed] [Google Scholar]
- Walden H, Podgorski MS, and Schulman BA (2003). Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature 422, 330–334. [DOI] [PubMed] [Google Scholar]
- Wang J, Cai S, and Chen Y (2010). Mechanism of E1-E2 interaction for the inhibition of Ubl adenylation. The Journal of biological chemistry 285, 33457–33462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, and Chen Y (2010). Role of the Zn(2+) motif of E1 in SUMO adenylation. The Journal of biological chemistry 285, 23732–23738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hu W, Cai S, Lee B, Song J, and Chen Y (2007). The intrinsic affinity between E2 and the Cys domain of E1 in ubiquitin-like modifications. Molecular cell 27, 228–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lee B, Cai S, Fukui L, Hu W, and Chen Y (2009). Conformational transition associated with E1-E2 interaction in small ubiquitin-like modifications. The Journal of biological chemistry 284, 20340–20348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GW, Toth JI, da Silva SR, Paiva SL, Lukkarila JL, Hurren R, Maclean N, Sukhai MA, Bhattacharjee RN, Goard CA, et al. (2014). Mutations in UBA3 confer resistance to the NEDD8-activating enzyme inhibitor MLN4924 in human leukemic cells. PloS one 9, e93530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh ET (2009). SUMOylation and De-SUMOylation: wrestling with life’s processes. The Journal of biological chemistry 284, 8223–8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Swatkoski S, Holly A, Lee LC, Giroux V, Lee CS, Hsu D, Smith JL, Yuen G, Yue J, et al. (2015). Oncogenesis driven by the Ras/Raf pathway requires the SUMO E2 ligase Ubc9. Proceedings of the National Academy of Sciences of the United States of America 112, E1724–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Sachdeva M, Wu F, Lu Z, and Mo YY (2010). Ubc9 promotes breast cell invasion and metastasis in a sumoylation-independent manner. Oncogene 29, 1763–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.