Abstract
Background: This study presents a label-free method of separating macrophages and fibroblasts, cell types critically associated with tumors.
Materials and Methods: Contactless dielectrophoresis (DEP) devices were used to separate fibroblasts from macrophages by selectively trapping one population. An ImageJ macro was developed to determine the percentage of each population moving or stationary at a given point in time in a video.
Results: At 350Vrms, 20 kHz, and 1.25 μL/min, more than 90% of fibroblasts were trapped while less than 20% of macrophages were trapped.
Conclusions: Contactless DEP was used to study macrophage and fibroblast separation as a proof-of-concept study for separating cells in the tumor microenvironment. The associated ImageJ macro could be used in other microfluidic cell separation studies.
Keywords: microfluidics, microenvironment, cell separation
Introduction
Immune, stem, and stromal cells are actively recruited to the tumor microenvironment. Secretory factors can change their phenotype into a tumor-associated cell type that supports tumor cell survival and progression. Resident cells such as endothelial and epithelial cells or stem and progenitor cells are similarly affected. Macrophages and fibroblasts are often found as accessory cells in the tumor microenvironment.1 They have critical functions such as immune suppression, angiogenesis support, and extracellular matrix remodeling (macrophages) or provide structural and metabolic support (fibroblasts).2
Understanding the molecular mechanisms of how the tumor microenvironment modulates the phenotype of resident cells and how recruited cells contribute to tumorigenesis is critical for the development of treatment strategies that could suppress the development of tumor-supportive phenotype of the tumor-associated cells to suppress tumor growth and progression. For this reason, it is important to be able to separate and characterize specific cell types out of biopsies as a processing step.
Here, we demonstrate a method known as dielectrophoresis (DEP) for separating different cell types. DEP is a phenomenon in which particles, in this case cells, exposed to the gradient of an electric field are polarized depending on the characteristics of the cells and the medium that surrounds them. This polarization induces movement of the cells along that gradient.3 This phenomenon can be used to trap cells or divert them from normal streamlines.4,5 Characterization of these cell types using DEP has been previously performed,5 but cell separation has not been achieved before.
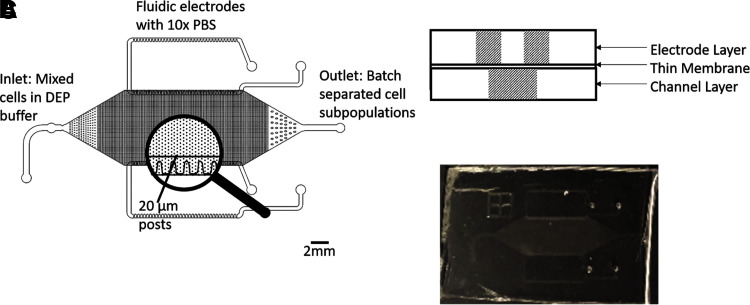
We have previously developed a technique called contactless DEP, which employs a polydimethylsiloxane (PDMS) microfluidic device containing a cell flow chamber. This chamber contains an array of 20 μm posts where cells trap based on the gradient of an applied electric field. Contactless fluidic electrodes are filled with conductive fluid and separated from the main channel by a thin PDMS membrane, as shown in Figure 1. Applying voltage using contactless electrodes filled with 10 × concentrated phosphate-buffered saline (PBS) eliminates problems with cell mortality as is seen in traditional DEP by preventing electrolysis and bubble formation in the chip, as well as avoiding contact between regions of high electric field and cells. In addition to improving cellular viability, utilizing small post structures allows better control of cell selectivity by preventing pearl chaining and cell–cell interactions. Previous works by this group have shown this improved viability and selectivity.6–8
FIG. 1.
Schematic representation of chip design. (A) Top view of multilayer chip and post structure. (B) Side view of three-layer design. (C) Image of DEP chip. DEP, dielectrophoresis.
In our device, cells with different bioelectrical phenotypes will begin trapping in the main channel at different applied electric field frequencies. By modulating the applied frequency, the device will selectively trap some cells while allowing others to pass through the device. This selectivity allows separation of highly similar cell types in a label-free manner while maintaining high cellular viability such that they can be cultured or further characterized downstream. Compared with other DEP techniques, this device provides more selective and higher viability separation of cells, which allows more closely related and physically similar cells to be separated, while allowing less similar cells to be separated at a much higher efficiency.
Batch separation can be performed by trapping some of the cells while allowing other cells to flow through and be collected in an output tube. After turning off the voltage, trapped cells release from their posts and can be collected in another output tube.
This study presents the separation of macrophages from fibroblasts using contactless DEP, a label-free microfluidic technique. While the bioelectric properties of these cells have been previously characterized,9 this is the first time separation of these cells has been accomplished using DEP. This article presents a new ImageJ macro that provides a method of using videos obtained during microfluidic DEP experiments. The macro extracts the number of cells moving versus stationary in each video frame, from which can be calculated what percentage of each cell type is trapped. As an example, we are using a mixture of macrophages and fibroblasts, representing different cell types that are found in the tumor microenvironment and contribute to a permissive microenvironment. Other applications, such as microfluidic chemotaxis assays, devices for fluid dynamics studies using fluorescent beads as flow markers, and circulating tumor cell sorting applications, could also benefit from this software.10,11
Theory
The DEP force balanced with the drag force governs the behavior of cells flowing through the contactless DEP device. When an AC electric field is applied, cells are polarized. Depending on the sign of the DEP force, which depends on the sign of the Clausius–Mossotti factor, cells are pushed up or down the electric field gradient. The Clausius–Mossotti factor is written as3:
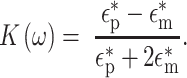 |
In this equation, 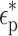 is the complex permittivity of the particle or cell and
is the complex permittivity of the particle or cell and 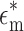 is the complex permittivity of the medium. The complex permittivity is defined as
is the complex permittivity of the medium. The complex permittivity is defined as 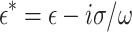 . Where ε is the permittivity, σ is conductivity and ω is angular frequency. The DEP equation for a dipole is then written as3:
. Where ε is the permittivity, σ is conductivity and ω is angular frequency. The DEP equation for a dipole is then written as3:
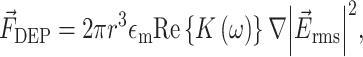 |
where r is the cell radius, 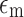 is the conductivity of the medium, Re{K(ω)} is the real part of the Clausius–Mossotti factor as a function of angular frequency (ω), and
is the conductivity of the medium, Re{K(ω)} is the real part of the Clausius–Mossotti factor as a function of angular frequency (ω), and 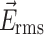 is the root mean squared of the electric field.
is the root mean squared of the electric field.
Materials and Methods
DEP chip and buffer
As discussed in previous works,6–9 the DEP chip design consists of three layers, as shown in Figure 1. It has two fluidic electrodes into which 10 × PBS, a highly conductive solution, is added. The chip also has a channel layer with 20 μm diameter insulating posts made from PDMS (Dow-Corning). The channel and electrode layers are separated from each other by a 14 μm membrane made from PDMS spun on a silanized silicon wafer (University Wafers) at 4000 rpm and then plasma bonded to the other layers.6,7 Wires are inserted into pipette tips full of 10 × PBS that are inserted into the fluidic electrodes, which are connected to a signal generator (Agilent 33500B Series), amplifier (Trek Model 2205), and oscilloscope for monitoring. When an AC signal is applied, a voltage drop occurs across the chip, causing electric field gradients to form around the 20 μm PDMS posts.
Cellular experiments
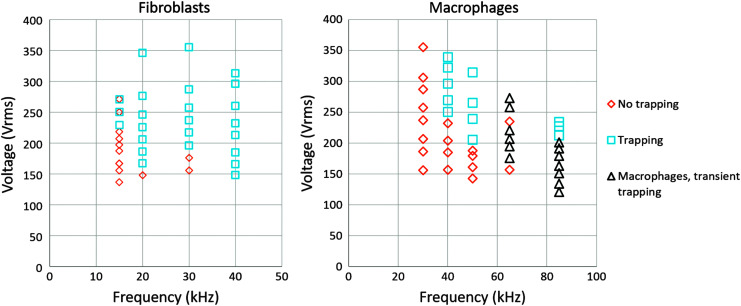
As a proof-of-concept study, we conducted first DEP trapping experiments on separate murine fibroblast (OP9; ATCC) adherent cells and murine peritoneal macrophage (PMJ2-PC1; ATCC) suspensions to determine frequency/voltage/flow rate pairings at which these cells would trap. OP9 cells were grown on adherent dishes within a media of minimum essential medium alpha (Sigma–Aldrich) with 20% (v/v) fetal bovine serum (FBS; Atlanta Biologicals), 2.2 g/L sodium bicarbonate (Fisher), and 1% (v/v) penicillin–streptavidin (ATCC). PMJ2-PC1 mouse macrophage cells were grown in suspension in Dulbecco's modified Eagle medium (Life Technologies) with 5% FBS, 3.7 g/L sodium bicarbonate, and 1% penicillin–streptavidin. Cells were transferred to DEP buffer through repeated centrifugation until a conductivity of less than 120 μS/cm was reached. First, each cell population was run through the device. For a set of frequencies and voltages, videos were obtained of cells flowing through the device. For each voltage/frequency combination, we recorded whether cells were able to trap. A plot of these results is shown in Figure 3. We then performed a separation experiment using a mixture of fibroblasts stained with calcein green (5 μg/mL; Life Technologies) and macrophages stained with calcein red (1.7 μg/mL; Life Technologies) for 15 min. Based on the results from individual population analysis as shown in Figure 3, we chose a flow rate of 1.25 μL/min, 20 kHz, and 346Vrms to separate mixed cells in the device. We turned on the voltage, allowing cells to trap, and then turned it off to test if trapping was reversible. Occasionally, we sped up the flow rate briefly to wash cells through the device. The software OBS (Open Broadcaster Software) was used to record the live view from the Leica X software, and notes were taken when the voltage was turned on/off, or cells were washed. Using VLC media player scene filter, the video was converted to two stacks of images for further analysis. An ImageJ script was written to analyze these video results. Further experimental details on contactless DEP have been discussed at length in our previous publications.6,7
FIG. 3.
Fibroblasts and macrophage trapping regions at 1.25 μL/min. Each point corresponds with a voltage–frequency pairing.
ImageJ script
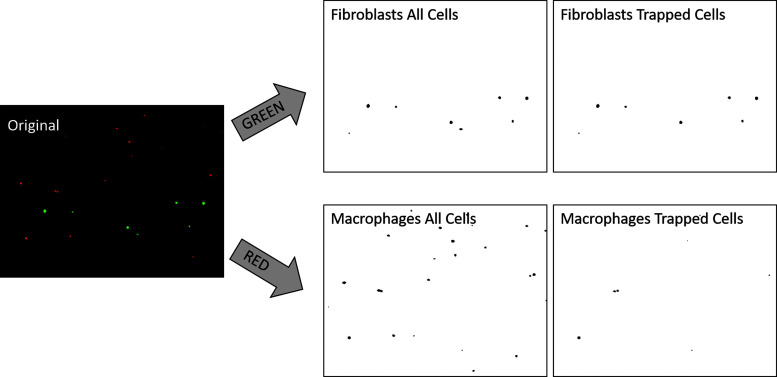
The basic parameters for the code were developed using the macro record function “Plugins>Macros>Record.” A basic outline of the code was developed following existing ImageJ protocols.12,* The macro uses several packages from ImageJ including the Dialog package for providing a window for user commands. First, the command “getString(text question, text answer)” is applied. This command opens a quick dialog box with a single line of question and input without having to use the full Dialog command. Then, the image sequence of interest is opened. The commands File.getParent(source) and File.getName(source) are used to split the full path into the file name and the path without the filename. This is used in multiple places in the code to query the user. The code can be defined as functioning in eight distinct steps, with example outputs shown in Figure 2.
FIG. 2.
Examples from processed image stack. Videos are available in the Supplementary Data. The original video slice with fluorescent cells is shown, followed by the split and analyzed images of red and green stationary and total cells for the corresponding median image. Accompanying Excel sheets show the cell counts for each slice in the stack.
In the first step, the user is asked for the pathname of an image stack to be imported and whether the image is single or multichannel. Second, the color channels (if more than one) are split into distinct image stacks. The user is given the option to choose a thresholding method from standard ImageJ thresholding methods.13–28 The program then automatically thresholds all images into black and white. The program then calls another standard ImageJ procedure, watershed, to separate clumps of cells into distinct entities before further processing.29 Next, the program asks the user how many images will be used for the median process and uses this to run a loop that calls the process Z Project. In this process, for example, if the user inputs five images, then the program will compare images 1–5, assigning each pixel as black if that pixel is black in the majority of the first five images and white if the pixel is white in the first five images. This “median processed” image containing pixels that correspond with the majority of the pixels in the original stack will be exported into a new stack. The program will then do the same analysis with images 2–6, 3–7, and so on until the end of the stack is reached. At the end, for each image stack, there will be a corresponding median processed stack containing only stationary cells. Next, the program calls the preinstalled ImageJ plugin Analyze Cells. The program asks for four parameters: minimum and maximum cell size in pixels, and minimum and maximum cell circularity. Using these parameters, the program calls Analyze Cells to count and measure each cell, produce new image stacks with outlines for each of the cells, and Excel spreadsheets with cell counts and sizes. Finally, the program saves all these files. An example of outputs is shown in Figure 2.
Results
Previously, we have shown that high flow rate of cells through our contactless dielectrophoresis chip can enhance separation of highly similar cell types.6 The current study was performed in a second mode at much lower flow rates, where bulk separation of more bioelectrically distinct cell populations was possible. This could be used to study a variety of cell combinations. In this study, we found a wide range of frequencies and voltages for which fibroblasts were able to trap but macrophages were not (as shown in Fig. 3), including several frequency/voltage pairings for which the fibroblast population reached nearly complete trapping.
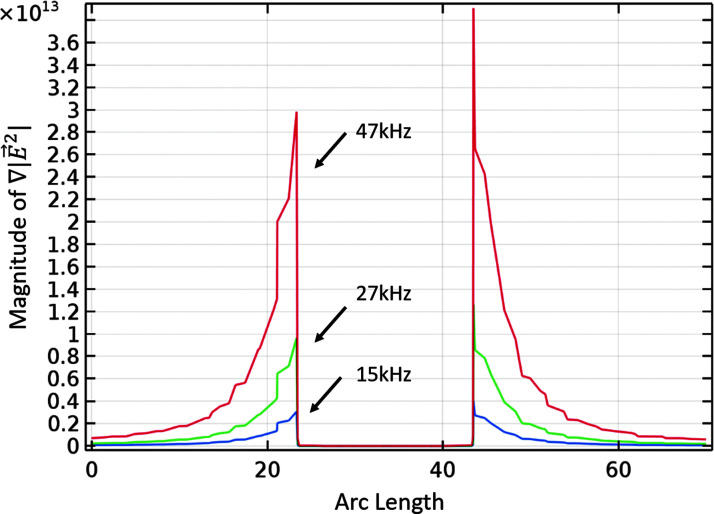
It is important to note that as shown in Figure 3, the voltage required to trap cells decreased with increasing frequency. There are two reasons for this observation. First, this slope corresponds with the fact that the DEP force must be higher than the drag force on a cell to trap in the chip, and the DEP force per volt as a function of frequency increases with increasing Clausius–Mossotti factor. Within this frequency range, the Clausius–Mossotti factor is in the upward slope regimen between crossover frequency and plateau,30 leading to a stronger DEP force per volt applied. Second, in contactless DEP, as the voltage drops across a membrane in the device, the field inside the chip increases per volt applied as a function of frequency. This is shown in Figure 4.
FIG. 4.
Magnitude of the gradient of the electric field squared, across a post in the direction of flow.
There exists a range of frequencies and voltages for which fibroblasts will trap but macrophages will not. Based on the results from Figure 3, 350Vrms and 20 kHz were selected to maximize trapping of the fibroblast population while remaining below the onset of trapping for the macrophage population. The macrophages and fibroblasts were stained, mixed, and separated at 20 kHz, 346Vrms, and 1.25 μL/min. The cells were then recorded over a period of time with the voltage being turned on and off, and occasional washes. Output videos, the original data set, and the macro code are included in the Supplementary Data, along with a user guide.
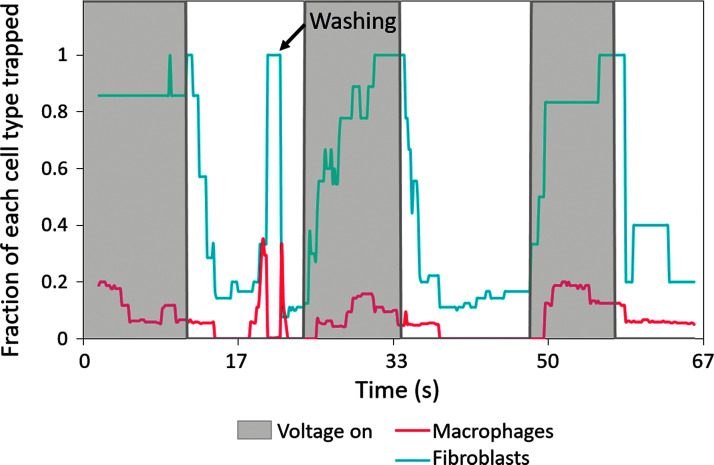
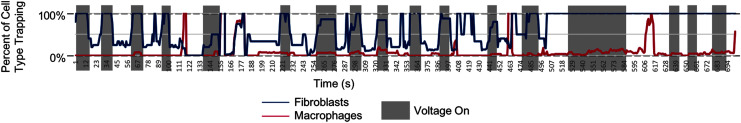
Figures 5 and 6 show the results of the cell separation analysis. The gray boxes represent areas with the voltage on, while white is area with the voltage off. The y-axis represents the fraction of cells trapping at each point in time. When the voltage was on, the majority (more than 90%) of the fibroblasts trapped while very few (less than 20%) to no macrophages trapped. In the instances of fibroblasts not trapping, they were a lighter shade of green, indicating leaking calcein and lowered viability. Instances of macrophages trapping were generally doublets. However, this imaging technique only analyzes a small region of the device, so it is possible that cells not able to trap here would eventually trap upon contact with another post off camera. The voltage was turned off periodically to ensure that any trapping was reversible dependent on voltage, an indicator that the trapping was due to DEP and not cell adherence due to other factors. Periodic washes were performed to ensure fresh batches of cells are flowing through the device and being trapped. This method of recording videos of cells moving and analyzing populations, while providing fewer data points than a downstream batch analysis, allows the researcher to rapidly screen parameters and gives information on the average lag time between turning the voltage on and cells trapping and other important data.
FIG. 5.
Analyzed data for image stack with 6 frames per second, 21 frames per median process, Otsu filtering,22 particle size 10-Infinity pixels, particle circularity 0.1–1. Graph shows the percentage of each cell type trapping at each point in time. As noted, a wash step was performed.
FIG. 6.
Analyzed data for image stack with 1 frame per second, 7 frames per median process, Otsu filtering,22 particle size 10-Infinity pixels, particle circularity 0.1–1. Graph shows the percentage of each cell type trapping at each point in time. Multiple wash steps were performed, brightfield was turned on, and stage was moved to represent different kinds of noise levels.
Figures 5 and 6 show moments with brightfield on to emphasize that this technique is optimized for fluorescence only videos. Similarly, when moving the stage or washing at high flow rate, the rate at which cells move with respect to the frame is very high. To correctly count moving versus trapped cells in this case, a much lower number of median frames are required, and aliasing would likely be present. One problem with this median processing system is that for a given slide, the number of cells counted as trapped can occasionally exceed the total number of cells, as an artifact of the fact that one slide for total counts must be compared to a set of n slides for the processing. However, this should average out over time.
Some cells irreversibly trap, such that even when the voltage is removed, they remain trapped, as can be seen in the baseline fibroblast trapping percentages.
Discussion
We hypothesize that the excellent separability between the two populations relates to their size as well as the structure of the cells, with fibroblasts being highly elongated in culture and the macrophages being spherical and nonadherent. Changes in the cellular morphology have been shown to correlate with changes in the DEP force and in the strength of multipolar behavior.31,32 Even though the cells were trypsinized, they could have retained some degree of cytoskeletal organization reminiscent of their in vitro shape, which could lead to differences in DEP trapping. Cell size also likely contributed to the difference in trapping. We observed that of the macrophages trapping in the fibroblast range, they appeared to be doublets or two cells stuck together. Similarly, fibroblasts that did not trap appeared to be dying, as they did not contain as much calcein green and appeared less bright in the images. We observed an interesting phenomenon at high frequencies, where cells would redirect from their streamlines to the posts, indicating dielectrophoretic trapping but would release before the voltage was turned off. In Figure 3, we called this phenomenon transient trapping. To our awareness, this phenomenon has not been previously observed in other cell types. This phenomenon could be electroporation,33 although if so, higher voltages at the same frequencies would not cause DEP trapping. It is also possible that some kind of mechanism related to multipolar behavior or electrorotation of the trapped fibroblasts could contribute to this behavior.3,30,31 Another interesting aspect of this phenomenon is that in order for trapping to occur, DEP forces must exceed drag forces. Once a cell is trapped on a post, it is generally more stable than when it has not fully trapped. Having DEP forces strong enough to divert cells but then not strong enough to hold the cell onto the posts could be indicative of some kind of cytoskeletal reorganization. This would explain why it has only been observed in our chip in macrophages, a cell line that is highly plastic and never adherent, and is only seen at higher frequencies where there would be stronger correlation with internal polarization of the cells.
One drawback to the ImageJ algorithm for tracking and analyzing cells is the slight blurring of voltage on–off boundaries by using the median method. When determining whether a cell is moving, the program needs to reference a sequential subset of images. For example, if 15 images are used to determine if the cell is moving, the first image in the median processed (moving vs. stationary) plot is made from taking the majority of the pixels from images 1–15. If, for example, the voltage was turned on between images 3–4, then the first median processed image will show that the cells were trapped. This can be somewhat compensated for by correlating the set with an image in the center of that set, for example, assigning the median image set 1–15 with original image 7, 2–16 with original image 8, and so on. One way of analyzing, which was performed in Figures 5 and 6, was to divide the number of cells counted in each median processed image (stationary cells) by the total number of cells in the central image from that median set. However, leaving these variables open to user input makes the algorithm flexible for a variety of imaging setups, with different sizes, shapes, and densities of cells.
The development of an ImageJ macro for analyzing videos of moving versus stationary cells can help to speed and automate the analysis of multiple microfluidic device configurations. It can be very helpful for applications such as ours in the area of DEP, where it is necessary to determine the percentage of cells trapped by an electric field. In using this macro, it is necessary to optimize the run parameters before analyzing a full data set, to prevent over- or undersampling of data. The standard thresholding methods in ImageJ can be used to test different threshold methods before running the program.13–28 To optimize the parameters for the median processing and subsequent cell analysis, it may be useful for the researcher to create a file containing a subset of the total number of images for iterative testing, to speed the optimization process. If the number of images chosen is too large, the averaging mechanism may erase some trapped cells. If the number of images chosen is too small, then slow moving particles will show up as trapped. In this case, it is also common to see significant levels of noise, where pieces of particles are visible in one frame of the median processing and absent in the next frame. In the future, this macro could be developed into a full ImageJ plugin or integrated into video processing software so that cell trapping percentages could be analyzed in real time, allowing researchers to obtain data while running experiments. Such developments could help to speed up the optimization of dielectrophoretic setups where optimizing frequency–voltage pairings is necessary to optimize device functionality.
Conclusions
We have shown separability of macrophages and fibroblasts using our DEP device. To rapidly analyze these results, we developed an ImageJ installable macro that can be used to count moving versus stationary objects. This code can be used in the future to make processing of DEP data much faster and quantitative. The current version is optimized to be used with moving versus stationary, fluorescently labeled cells or particles. However, it is easily modifiable for novel applications and also comes with user-friendly dialog boxes for easy programming-free input. The code has the DOI 10.5281/zenodo.1204436 at Zenodo. The code, user guide, and video examples are also found in the Supplementary Data for this article.
Acknowledgments
This work was supported by NIH 5R21 CA173092-01 and in part by an unrestricted research grant from CytoRecovery. We also acknowledge Virginia Biosciences Health Research Corporation (VBHRC) and NSF IGERT DGE-0966125 MultiSTEPS.
Footnotes
https://imagej.nih.gov/ij/plugins/index.html
Disclaimer
This article has not been published and is not in press or currently submitted elsewhere. A portion of this article has been posted to the bioRxiv preprint server at https://doi.org/10.1101/292417.
Authors' Contributions
T.A.D., N.B., and N.A. performed experiments. T.A.D. wrote the ImageJ code and performed the analysis. T.A.D., R.V.D., E.M.S., and N.A. contributed to the manuscript writing. E.M.S. prepared the macrophage and fibroblast cells. All co-authors have reviewed and approved this article.
Author Disclosure Statement
R.V.D. has patents in the field of dielectrophoresis. E.M.S. and T.A.D. have patents pending in the field of dielectrophoresis. R.V.D. and E.M.S. have been invited to serve on a scientific advisory board for a company in the area of dielectrophoresis. N.A. and N.B. have nothing to disclose.
Supplementary Material
Supplementary Data
References
- 1.Hanahan D, Weinberg R. The next generation. Cell 2011;144:646–674 [DOI] [PubMed] [Google Scholar]
- 2.Spaw M, Anant S, Thomas SM. Stromal contributions to the carcinogenic process. Mol Carcinog 2017;56:1100–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pohl H. Dielectrophoresis: The Behavior of Neutral Matter in Nonuniform Electric Fields. Cambridge University Press in Cambridge, United Kingdom: Cambridge Monographs on Physics. 1978 [Google Scholar]
- 4.Shafiee H, Caldwell JL, Sano MB, et al. Contactless dielectrophoresis: a new technique for cell manipulation. Biomed Microdevices 2009;11:997–1006 [DOI] [PubMed] [Google Scholar]
- 5.Sano MB, Gallo-Villanueva RC, Lapizco-Encinas BH, et al. Simultaneous electrokinetic flow and dielectrophoretic trapping using perpendicular static and dynamic electric field. Microfluid Nanofluidics 2013;15:599–609 [Google Scholar]
- 6.Douglas TA, Cemazar J, Balani N, et al. A feasibility study for enrichment of highly aggressive cancer subpopulations by their biophysical properties via dielectrophoresis. Electrophoresis 2017;38:1507–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cemazar J, Douglas TA, Schmelz EM, et al. Enhanced contactless dielectrophoresis enrichment and isolation platform via cell-scale microstructures. Biomicrofluidics 2016;10:014109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sano MB, Gallo-Villanueva RC, Lapizco-Encinas BH, et al. Simultaneous electrokinetic flow and dielectrophoretic trapping using perpendicular static and dynamic electric fields. Microfluid Nanofluidics 2013;15:599–609 [Google Scholar]
- 9.Salmanzadeh A, Kittur H, Sano MB, et al. Dielectrophoretic differentiation of mouse ovarian surface epithelial cells, macrophages, and fibroblasts using contactless dielectrophoresis. Biomicrofluidics 2012;6:24104–2410413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nilsson J, Evander M, Hammarström B, et al. Review of cell and particle trapping in microfluidic systems. Anal Chim Acta 2009;649:141–157 [DOI] [PubMed] [Google Scholar]
- 11.Wlodkowic D, Faley S, Zagnoni M, et al. Microfluidic single cell array cytometry for the analysis of tumour apoptosis. Anal Chem 2009;81:5517–5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mutterer J, Rasband W.ImageJ Macro Language Programmer's Reference Guide v1.46d. RSB Homepage 2012: 1. –45 [Google Scholar]
- 13.Sezgin M, Sankur B. Survey over image thresholding techniques and quantitative performance evaluation. J Electron Imaging 2004;13:146–165 [Google Scholar]
- 14.Huang L-K, Wang M-JJ. Image thresholding by minimizing the measures of fuzziness. Pattern Recognit 1995;28:41–51 [Google Scholar]
- 15.Prewitt JMS, Mendelsohn ML. The analysis of cell images. Ann N Y Acad Sci 1966;128:1035–1053 [DOI] [PubMed] [Google Scholar]
- 16.Ridler T, Calvard S. Picture thresholding using an iterative selection method. IEEE Trans Syst Man Cybern 1978;8:630–632 [Google Scholar]
- 17.Li CH, Lee CK. Minimum cross entropy thresholding. Pattern Recognit 1993;26:617–625 [Google Scholar]
- 18.Pun T.A new method for grey-level picture thresholding using the entropy of the histogram. Signal Processing 1980;2:223–237 [Google Scholar]
- 19.Glasbey C.An analysis of histogram-based thresholding algorithms. Graph Models Image Process 1993;55:532–537 [Google Scholar]
- 20.Kittler J, Illingworth J. Minimum error thresholding. Pattern Recognit 1986;19:41–47 [Google Scholar]
- 21.Tsai W-H. Momentpreserving thresholding: A new approach. In: Lawrence O'Gorman and Rangachar Kasturi, eds. Document Image Analysis. Los Alamitos, CA: IEEE Computer Society Press, 1995: 44–60 [Google Scholar]
- 22.Otsu N.A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 1979;9:62–66 [Google Scholar]
- 23.Yen J-C, Chang F-J, Chang S. A new criterion for automatic multilevel thresholding. IEEE Trans Image Process 1995;4:370–378 [DOI] [PubMed] [Google Scholar]
- 24.Zack G, Rogers W, Latt S. Automatic measurement of sister chromatid exchange frequency. J Histochem Cytochem 1977;25:741–753 [DOI] [PubMed] [Google Scholar]
- 25.Shanbhag A.Utilization of information measure as a means of image thresholding. Graph Models Image Process 1994;56:414–419 [Google Scholar]
- 26.Doyle W.Operations useful for similarity-invariant pattern recognition. ACM 1962;9:259–267 [Google Scholar]
- 27.Kapur J, Sahoo P, Wong A. A new method for gray-level picture thresholding using the entropy of the histogram. Computer Vis Graph Image Process 1985;29:273–285 [Google Scholar]
- 28.Sezgin M, Sankur B. Survey over image thresholding techniques and quantitative performance evaluation. J Electron Imaging 2004;13:146–165 [Google Scholar]
- 29.Vincent L, Soille P. Watersheds in digital spaces: An efficient algorithm based on immersion simulations. IEEE Transactions on Pattern Analysis and Machine Intelligence 1991;13:583–598 [Google Scholar]
- 30.Pethig RR.Dielectrophoresis: Theory, Methodology and Biological Applications, 1st edition. Hoboken, New Jersey: Wiley, 2017 [Google Scholar]
- 31.Green NG, Jones TB. Numerical determination of the effective moments of non-spherical particles. J Phys D Appl Phys 2007;40:78–85 [Google Scholar]
- 32.Nili H, Green NG. Higher-order dielectrophoresis of nonspherical particles. Phys Rev E Stat Nonlin Soft Matter Phys 2014;89:1–11 [DOI] [PubMed] [Google Scholar]
- 33.Bonakdar M, Graybill PM, Davalos RV. A microfluidic model of the blood-brain barrier to study permeabilization by pulsed electric field. RSC Adv 2017;7:42811–42818 [DOI] [PMC free article] [PubMed] [Google Scholar]