Abstract
Local delivery of simvastatin (SIM) has exhibited potential in preventing inflammation and limiting bone loss associated with experimental periodontitis. The primary aim of this study was to analyze transcriptome changes that may contribute to SIM's reduction of periodontal inflammation and bone loss. We evaluate the global genetic profile and signaling mechanisms induced by SIM on experimental periodontitis bone loss and inflammation. Twenty mature female Sprague Dawley rats were subjected to ligature‐induced experimental periodontitis around maxillary second molars (M2) either unilaterally (one side untreated, n = 10) or bilaterally (n = 10). After the ligature removal at day 7, sites were injected with either carrier, pyrophosphate (PPi ×3), 1.5‐mg SIM‐dose equivalent SIM‐pyrophosphate prodrug, or no injection. Three days after ligature removal, animals were euthanized; the M1‐M2 interproximal was evaluated with μCT, histology, and protein expression. M2 palatal gingiva was harvested for RNA sequencing. Although ligature alone caused upregulation of proinflammatory and bone catabolic genes and proteins, seen in human periodontitis, SIM‐PPi upregulated anti‐inflammatory (IL‐10, IL‐1 receptor‐like 1) and bone anabolic (insulin‐like growth factor, osteocrin, fibroblast growth factor, and Wnt/ β‐catenin) genes. The PPi carrier alone did not have these effects. Genetic profile and signaling mechanism data may help identify enhanced pharmacotherapeutic approaches to limit or regenerate periodontitis bone loss. © 2018 The Authors. JBMR Plus Published by Wiley Periodicals, Inc. on behalf of the American Society for Bone and Mineral Research.
Keywords: MOLECULAR PATHWAYS–REMODELING, GH/IGF‐1, WNT/Β‐CATENIN/LRPS, DENTAL BIOLOGY, ANABOLICS
Introduction
Periodontitis is a bacterial‐initiated disease stimulating an inflammatory response between the periodontal sulcus and the alveolar crest, which results in the destruction of connective tissue attachment to the tooth root and crestal bone loss. It is the primary cause of tooth loss in humans.1 Periodontitis affects approximately 47% of the adult population in the United States, the equivalent of 65 million people.2, 3 Though reduction of the bacterial biofilm around teeth is a cornerstone of therapy, the management of the host inflammatory response and the stimulation of bone regeneration are required in more advanced cases. The bone regeneration in periodontitis is difficult because it must be attempted in a highly inflamed microenvironment; therefore, it requires both the stimulation of bone growth and the resolution of local inflammation. The conventional therapy entails a surgical procedure exposing and debriding the bony defect, then treating the bone surface with synthetic bone matrix and growth factors.4 However, recent clinical evidence suggests that nonsurgical debridement of the tooth root and injection of statin formulations, particularly simvastatin (SIM), can regenerate a significant portion of the bone lost to periodontitis,5, 6 thereby reducing patient morbidity and cost.
Although SIM was originally designed to suppress cholesterol synthesis by inhibiting 3‐hyroxy‐3‐methylglutaryl‐coenzyme A reductase, it also has been shown to have bone anabolic and anti‐inflammatory properties.7 However, the mechanisms for periodontal bone regeneration and inflammation control remain poorly understood.
Naturally occurring periodontitis causes short episodes of bone resorption on a highly variable timetable over decades, so experimental periodontitis (ExP) in the rat model is often used to cause measureable periodontal bone loss over a 1‐week period.8 Ligature‐induced periodontitis used in the current study has been shown to initiate an inflammatory infiltrate in the connective tissue between the sulcular epithelium and bone, composed primarily of the lymphocytes lineage.9 This pattern is similar to histology associated with episodes of human periodontal attachment and bone loss.10, 11 Local SIM injections have been shown in our previous studies to prevent this bone loss or to regenerate bone around teeth.9 The purpose of the current study was to explore the genes and signaling pathways associated with bone turnover and inflammation that are either upregulated or downregulated by the creation of ExP, then altered by local injection of a SIM prodrug. Because the cellular mechanisms maintained and regulated by SIM‐induced signaling have not been extensively explored to enable an understanding of those genes functioning in the repression of periodontitis, our objective in this study was to analyze transcriptome changes that may contribute to reducing periodontal inflammation and bone loss. We analyzed the transcriptome responding to SIM prodrug locally delivered in high doses in a periodontitis inflammatory lesion.
We hypothesized that EXP enhances gene activation (and protein production) of known proinflammatory and bone catabolic mediators, and that locally applied SIM would decrease inflammatory mediators and stimulate bone anabolic genes and protein production.
Materials and Methods
Animal proceduresTwenty mature (10‐ to 12‐month‐old) female retired breeder Sprague Dawley rats were housed and treated in the University of Nebraska Medical Center (UNMC) College of Dentistry Animal Facility under the approval of the UNMC Institutional Animal Care and Use Committee (IACUC #13‐006‐03). Rats were allowed to acclimate 1 week before the first procedure. An induction chamber connected to an isoflurane anesthetic vaporizer initiated anesthesia with 1% to 4% isoflurane/100% O2 (1 to 3 L/min), which was subsequently maintained by the application of a nose cone with 0.5% to 2% isoflurane/100% O2 (0.5 to 1 L/min) in the experimental procedures. Animals’ weight was recorded after anesthesia induction and before all procedures to monitor weight gain/loss. Weights of all rats averaged >300 gm at the conclusion of the study. Rats were randomly divided into two groups of 10 rats each (Table 1). The maxillary second molars had 4‐0 silk ligatures placed subgingivally to induce ExP except for the untreated left side in group 1. All ligatures were removed 1 week later and three local injections of 1.5‐mg SIM‐pyrophosphate prodrug (SIM‐PPi; The details of the SIM‐PPi prodrug—preparation, structure, half‐life, release, efficacy, cleavage in vivo, pharmacokinetics, and biodistribution—are discussed in our US provisional patent application 62/487,692.) were delivered into the palatal gingiva at the mesial, midpalatal, and distal aspects of the second molars to the depth of 2 mm using 26G × ½‐inch sterile syringes. To place SIM in a periodontitis pocket, it must be dissolved or suspended in a liquid carrier for injection. PPi was chosen because it targets bone, but also was inert to gene activation in rat, ExP. Rats were euthanized 3 days later by CO2 asphyxiation. Animals were weighed after euthanasia. Palatal gingiva tissues of the second molars on both sides were collected from rats using sterile #15 blades and evenly pooled into three samples within each group for subsequent RNA extraction. The palates with intact interproximal gingiva were fixed in 10% buffered formalin solution for subsequent μCT and histological evaluation.
Table 1.
Experimental Groups
| Group | N | Day 1 | Day 8 | Day 11 |
|---|---|---|---|---|
| 1 | 10 | Left: Unmanipulated (no ligature or injections) | Left: Unmanipulated (no ligature or injections) | Euthanize |
| Right: ligatures | Remove ligatures | |||
| 2 | 10 | Left: ligatures | Remove ligatures Inject PPi | Euthanize |
| Right: ligatures | Remove ligatures Inject SIM/PPi |
RNA extraction, construction of small RNA libraries, and RNA‐seq
Total RNA was isolated using the RNeasy Protect Mini Kit (Cat # 74124; Qiagen, Valencia, CA, USA) following the manufacturer's instructions and measured for purity and concentration by ultraviolet spectroscopy (NanoDrop 2000c; Thermo Fisher Scientific, Waltham, MA, USA). RNA integrity evaluation, libraries construction, and validation were performed as previously reported.12 RNA sequencing was performed using Illumina HiSeq 2500 (Illumina, San Diego, CA, USA) at the UNMC Bioinformatics and Systems Biology Core.
RNA‐seq analysis
RNA‐seq data were obtained for unmanipulated controls, samples with ligature only (ExP), samples treated with the carrier only following ligature removal (ExP + PPi), and samples treated with SIM and the carrier following ligature removal (ExP + SIM‐PPi). Each group was represented by three biological replicates resulting in 12 samples used for RNA‐seq, which was performed in 75‐bp single‐end mode. Raw reads were analyzed with FASTQC (v. 0.11.5) for quality control.13 Overrepresented (eg, adapter and similar technical) sequences remaining in the raw reads were assessed and subsequently removed using Trimmomatic (v 0.36) in the palindrome mode based on default alignment detection and scoring parameters.14 Trimmomatic also was used for low‐quality base filtering. Maximum information quality filtering was employed with a minimum average read quality threshold of 25. Following technical sequence and low‐quality base removal, reads that were shorter than 36 bp were filtered out. Transcript quantification was done based on the Rnor_6.0 reference genome using Salmon (v. 0.8.2) with the default parameter.15 Salmon uses sample‐specific models such as a correction for GC‐content bias that improves the accuracy of transcription abundance estimates. Transcripts per million (TPM) in Salmon's output was used as the relative abundance measure employed in our downstream analysis. Differential gene expression analysis was done using DESeq2.16
μCT Measurements
The μCT measurements were performed on intact molar areas using a high‐resolution Skyscan 1172 μCT system (Bruker, Kontich, Belgium) as described in our previous study.9 Briefly, maxillae were scanned by the μCT, followed by 3D reconstructions using internal software and reorientation of 3D models to standard position for future evaluation. The distance from the molar cemento–enamel junction (CEJ) to the alveolar bone crest (ABC) was measured in sagittal interproximal views by one masked examiner to confirm ExP bone loss compared to the control.
Histology and immunofluorescence
The palates fixed in 10% buffered formalin solution were decalcified in 5% formic acid solution for 2 weeks, and processed and embedded in paraffin. Eight‐μm‐thick serial sagittal sections were collected and stained with hematoxylin and eosin to show the crestal interproximal bone and gingival landmarks between the first and second molars (Fig. 1 A–C). Immunofluorescence technique as described previously17 was used to analyze the distribution of selected bone anabolic and inflammatory proteins suggested by RNA‐seq data. Briefly, the above‐mentioned sections were blocked with 10% normal goat serum (NGS) in PBS for 1 hour at room temperature in a humidity chamber and incubated with primary antibodies (see below) in PBS containing 10% NGS at 4°C overnight, followed by incubation with 5% NGS‐diluted secondary antibody for 1 hour at room temperature. Normal rabbit serum (ab166640; Abcam, Cambridge, MA, USA) at the same concentrations was used in corresponding negative controls. The antibodies used for the protein assay included rabbit anti‐MMP‐9 antibody (ab76003, 1:200; Abcam), rabbit anti‐IGF‐1 antibody (orb10886, 1: 200; Biorbyt, Cambridge, UK), rabbit anti‐TNF‐α antibody (GTX110520, 1:100; GeneTex, Irvine, CA, USA), goat anti‐rabbit IgG (H+L) cross‐adsorbed secondary antibody, and Alexa Fluor 488 (A‐11011, 1:500; Thermo Fisher Scientific, Waltham, MA). The fluorescence was observed with a fluorescence confocal laser scanning microscope (Nikon A1 CLSM; Nikon, Tokyo, Japan). Measurement of staining intensity was performed in Adobe Photoshop CC 2017 (Adobe, San Jose, CA, USA). Three representative areas were selected from each image for measurement. Staining intensity was determined as the gray value (mean).
Figure 1.
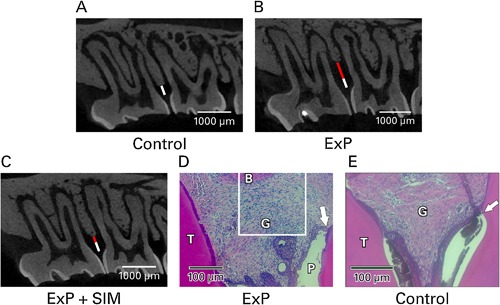
μCT and histology of experimental periodontitis. (A) Normal interproximal bone height between maxillary first and second molars (white bar, control). (B) Experimental periodontitis caused bone loss (ExP; red bar), whereas (C) ExP followed by local simvastatin injections has been shown to cause bone preservation/regeneration after 28 days9 (shorter red bar, ExP + simvastatin [SIM]). (D) Furthermore, histology of ExP showed increased lymphocytic infiltrate between the periodontal pocket (P) and alveolar bone (B), and apical migration of the epithelial attachment along the root surface (arrow). White box indicates the area of interest for the immunofluorescence evaluations in the current study. (E) Untreated control histology showed epithelial attachment at the cementoenamel junction (arrow where enamel has been decalcified away) and minimal inflammatory infiltrate.
Confirmation of differentially expressed genes with PCR
To confirm differentially expressed genes in rat maxillary second‐molar samples, real‐time qPCR was performed as previously described.18 Primary human gingival fibroblast cells (ATCC Cat #PCS‐201‐018; American Type Culture Collection, Manassas, VA, USA) were cultured in a six‐well plate in DMEM (Gibco, Grand Island, NY, USA), adding 5% fetal calf serum (Hyclone; GE Healthcare Life Sciences, Marlborough, MA, USA) plus 100 U/mL penicillin and 100 µg/mL streptomycin (Hyclone; GE Healthcare Life Sciences). Once cells reached 80% confluence: (1) a group of cells were treated with fresh medium with 1 μg/mL LPS (1:1000 dilution, ultrapure lipopolysaccharide from P. gingivalis; Cat # tlrl‐ppglps, Invitrogen, Carlsbad, CA, USA, as used previously19) for 24 hours and 48 hours; (2) a group of cells remained untreated for 24 hours and 48 hours; (3) a group of cells was treated with LPS (1:1000 dilution) for 24 hours followed by an additional 24 hours of 100nM SIM (1:50 dilution of the 5 μM) as previously used by us20; and (4) a group of cells was treated with 100nM SIM (1:50 dilution of the 5 μM) for 24 hours and 48 hours. Each experiment was performed in duplicate. The quantification of biologically active LPS was measured by using HEK‐Blue LPS Detection Kit (Cat # rep‐lps2, Invitrogen). RNA extraction was conducted as previously described using the Arcturus PicoPure RNA Isolation Kit (Thermo Fisher Scientific) to consistently extract high‐quality RNA from a very few cells. RNA (500 ng) was converted to cDNA using Invitrogen Superscript IV VILO Master Mix Invitrogen (Carlsbad, CA) providing a highly efficient and thermostable reverse transcriptase, which promotes a significant cDNA yield at high temperatures in less time. An additional preamplification step was conducted using TaqMan PreAmp Master Mix (Thermo Fisher Scientific) with a custom preamplification pool of genes of interest to amplify small amounts of cDNA without introducing amplification bias—specifically for small, precious mRNA samples—and allow more qPCR reactions. Samples were preamplified for 14 cycles with thermal cycling conditions of 95°C for 15 s and 60°C for 4 min, followed by immediate placement on ice. Finally, samples were diluted with TE buffer (ph 8.0) to 1:20 and placed in 96‐well custom array plates in technical triplicate; qPCR was executed with TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific) reagents. PCR conditions were 40 cycles at 95°C for 15 s, and at 60°C for 60 s. Each technical repeat's gene‐specific ΔCtCt value was subtracted from the housekeeping gene's ΔCtCt value. Then, data were analyzed using ANOVA on the repeat‐normalized ΔΔCtCt values, including the control group; translate effects were estimated from the ANOVA onto the multiplicative scale. The values of nine genes tested were normalized by adjusting for the concentration of an abundant known housekeeping gene, like 18S rRNA, glyceraldehyde 3‐phosphate dehydrogenase (GAPDH), and β‐actin and the ΔCt values of a naive/vehicle group. PCR assays detecting the reference genes, 18S rRNA, GAPDH, and β‐actin, were calculated alongside those for the group of interest. The resulting data set was analyzed.
Statistical analysis
Rat weight changes and bone loss from CEJ (μCT) among groups were compared by t test. DESeq2 uses a negative binomial model to assess differential expression and employs the Benjamini‐Hochberg procedure21 for multiple hypotheses testing correction. When comparing the transcription abundance between two groups of samples, we used the adjusted p‐value cut‐off of 0.05 to define statistically significant differential expression.
Clustering of samples and/or genes was done using the unweighted pair group method with arithmetic‐mean method utilizing Pearson's correlation as the distance measure.22 The expression data matrix was row‐normalized prior to the application of average linkage clustering. The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.723 was used for functional analysis of the gene lists interrogating Biological Process (BP), Molecular Function, and Cellular Component (CC) Gene Ontology (GO) categories24 and the Kyoto Encyclopedia of Genes and Genomes pathways.25 Biologically relevant categories that were overrepresented in the gene set, but which may be of further interest, were assessed using the Expression Analysis Systematic Explorer (EASE) score in the DAVID tool. The EASE score is the upper bound of the distribution of Jackknife iterative resampling of Fisher exact probabilities with Bonferroni multiple testing correction. Categories containing low numbers of genes were underweighted so that the EASE score would be more robust than the Fisher exact test. The EASE score is a significance level, with smaller EASE scores indicating increasing confidence in overrepresentation. GO categories that had EASE scores of 0.05 or lower were picked as significantly overrepresented. The differentially expressed gene lists were further analyzed using the Ingenuity Pathway Analysis (IPA; Ingenuity Systems, www.ingenuity.com; QIAGEN Silicon Valley, Redwood City, CA, USA) software. IPA is based on the manual curation of scientific literature to identify pathways, networks, and functional categories that are significantly represented in the input gene list. Raw RNA‐seq data are available at the NCBI‐SRA (The National Center for Biotechnology Information–Sequence Read Archive) database under BioProject PRJNA417128.
One‐way ANOVA analysis was performed in Graphpad Prism 7 (GraphPad Software, La Jolla, CA, USA) to compare protein expression among different groups in immunohistochemistry. Means ± 95% CI were shown in a quantification bar graph. qRT‐PCR data analysis was done using ANOVA by built‐in software that estimates statements used to contrast the group effect, which yields ΔΔCt (−0.6848), as well as its SE (0.1185) and 95% CI (−0.9262 to −0.4435). The output data with both confidence level and p value were also generated.
Results
Animal response to ExP reproduces and simulates experimental periodontists in rat model
No rat groups showed significant change in weight during the experimental period. ExP showed significantly more periodontal bone distance from the CEJ to ABC (1.1 ± 0.28 mm) compared to unmanipulated controls (0.67 ± 0.22 mm, p < 0.0005) and increased lymphocytic infiltrate between the periodontal pocket and bone (Fig. 1), which is consistent with our ExP model characterized previously.9 No teeth became hypermobile and all teeth remained fully functional.
RNA‐seq data reveal differentially expressed genes in ExP and genes induced by SIM that are important for periodontal regeneration
The raw RNA‐seq average read count was approximately19.0M single‐end reads per sample, which decreased to approximately 18.6M, following trimming and filtering. Similarly, the average read‐length decreased to 70.0 bp in the processed reads from 74.5 bp in the raw reads. The quality preprocessing increased the average read quality to 34.08 from 32.28 and the percentage of high‐quality bases (with a quality score >20) per sample increased to 95.85% from 92.48%. These results indicate that the read quality was increased significantly at a marginal cost of decreased total number of reads and average read length (see Supplementary Fig. 1). RNA‐seq analysis generated expression data for 31,202 transcripts. Transcripts that showed a TPM value of more than 1 in at least two‐thirds of biological replicates in at least one sample group (control, ExP, ExP + PPi, or ExP + SIM‐PPi) were carried on for downstream analysis rendering 17,093 transcripts. In Fig. 2 A, the genes that are significantly differentially expressed between each pair of sample groups are shown. Experimental periodontitis in the rat model upregulated 1,743 genes and downregulated 1,133 genes compared to unmanipulated contralateral controls (Fig. 2 A). Both the SIM‐PPi and PPi altered a similar number of genes, yet the PPi (carrier) alone had almost no effect beyond the ligature‐induced periodontitis (no upregulated genes, one downregulated gene). Therefore, PPi was considered an inert carrier and all subsequent comparisons were conducted among ExP, ExP + SIM‐PPi, and unmanipulated controls. For the remaining three groups, unsupervised hierarchical clustering analysis was performed using all the transcripts (Fig. 2 B). The clustering results showed clear separation of all three groups from each other, further separating the unmanipulated controls from the ExP and the ExP + SIM‐PPi groups. This blind clustering shows that from a global transcriptional profiling perspective, the application of SIM‐PPi perturbs the ExP group enough to form a distinct group, but not merged with the control, possibly because of the dominant effect of the inflammatory response.
Figure 2.
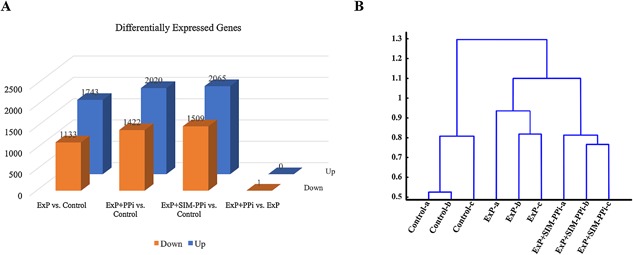
Differentially expressed genes by RNA‐seq in ExP and ExP + SIM‐PPi models. (A) Statistically significantly differentially expressed (adjusted p value <0.05) genes between each pair of sample groups. (B) Unsupervised hierarchical clustering analysis of the three sample groups (control, ExP, ExP + SIM‐PPi) using all the transcripts (B). ExP = experimental periodontitis; SIM‐PPI = simvastatin‐pyrophosphate.
Genes upregulated at least twofold by ExP compared to the control included many proinflammatory markers, including matrix metalloproteinases (MMP 2, MMP 8, MMP 9), IL‐1β, IL‐17, TNF superfamily and receptor, complement components C1q and C5a receptor, LPS‐binding protein, and toll‐like receptor 2 (Fig. 2). Bone turnover factors also were upregulated, including collagen type 1. ExP showed twofold downregulated odontogenic ameloblast associated protein (mediates junctional epithelial attachment to teeth) and IGF‐binding protein 6 (that enhances IGF signaling).
To focus on the unique effect of SIM on gene expression, the overlap between genes dysregulated because of ExP and ExP + SIM‐PPi models was calculated (Fig. 3). Although the two dysregulated gene lists showed a high overlap, indicating the shared effects of the ligature model in both groups, there were a significant number of genes that were dysregulated uniquely because of SIM. Genes upregulated twofold by ExP + SIM‐PPi versus the control, but not ExP alone versus the control, included anti‐inflammatory IL‐10 and IL‐1 receptor‐like 1 (IL1rl1) and IGF‐1 (Fig. 3). The figure highlights significantly enriched functional/pathway categories. In the supplement, we provide the full list of up‐/downregulated genes along with genes uniquely dysregulated in the ExP + SIM‐PPi model and the functional/pathway categories that are significantly enriched in this unique gene list.
Figure 3.
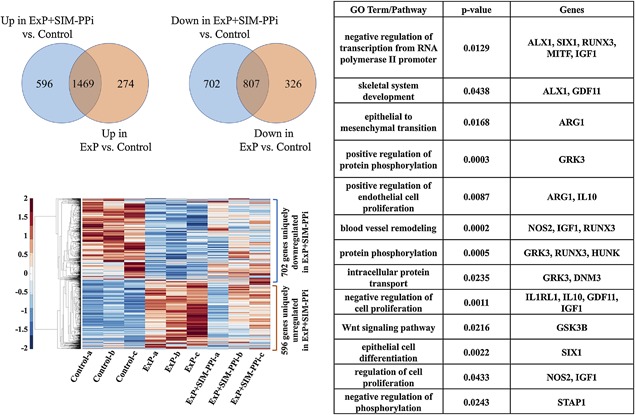
Overlap of genes that are dysregulated in ExP and ExP + SIM‐PPi models compared to the unmanipulated controls by RNA‐seq. Hierarchical clustering of and functional groups and pathways over‐represented by the 596 + 702 genes uniquely dysregulated in the ExP + SIM‐PPi model. ExP = experimental periodontitis; SIM‐PPI = simvastatin‐pyrophosphate.
The data showed that SIM directly activated the IGF‐1 ligand; subsequently, it also increased the expression of several other downstream molecules such Ras and AKT and decreased the expression of SOS, SOCS, PDK1, PKA, and 14‐3‐3. These fine balances between up‐/downregulated genes are necessary to trigger: (1) activation of periodontal ligament growth, (2) induction of alveolar bone regeneration, and (3) repression of periodontitis (Supplementary Figs. 2, 3, and 4). In addition to IGF‐1, SIM also activates the FGF7 ligand, which upregulates AKT and HGF genes, and downregulates SOS and CREB genes, which in tandem, causes periodontal fibroblast growth and maintains periodontal fibers homeostasis (Supplementary Figs. 5 and 6). SIM also induced activation of IL1rl1, which is necessary for immune response and repression of periodontitis by upregulating PKD, MAPK, JNK, ACVRL1, PDGF, CXCL3, SCL, GORASP2, and KLF2 and downregulating IL1, MGEA5, GSK3B, ERBB2, AP1, and ATXN3 (Supplementary Fig. 10). We also observed that SIM treatment caused repression of Wnt/β‐catenin signaling. Loss of Wnt/β‐catenin activates ILK, AKT, TCF4/LEF1, and Frizzled, but also simultaneously represses TGF‐βr, PP2A, Groucho, CBP, CX43, and Dsh genes. Data suggest that simultaneous activation and suppression of these genes (by repression of Wnt/β‐catenin signaling) are necessary for pro‐osteogenesis by enhancing osteoblast differentiation as well as activation of periodontal fibroblast proliferation and morphogenesis. By regulating these genes, Wnt/β‐catenin signaling can induce both bone and fibroblast differentiation and morphology to maintain pro‐osteogenic and fibrogenic homeostasis (Supplementary Figs. 7, 8, and 9). In addition, we also show several other genes (GRK3, ARG1, SIX1, IL‐10, MITF, and OSTN), which are indirectly regulated by IGF‐1, FGF7, Wnt/β‐catenin, and IL1rl1, and play decisive roles in: (1) activating periodontal ligament/fibroblast growth and homeostasis; (2) the induction of alveolar bone repair and regeneration; and (3) immune response and repression of periodontitis. These genes, induced by SIM, are important for periodontal regeneration and in tandem with IGF‐1, FGF7, Wnt/β‐catenin, and IL1rl1, play a vital role in improving and treating periodontitis.
Figure 5.
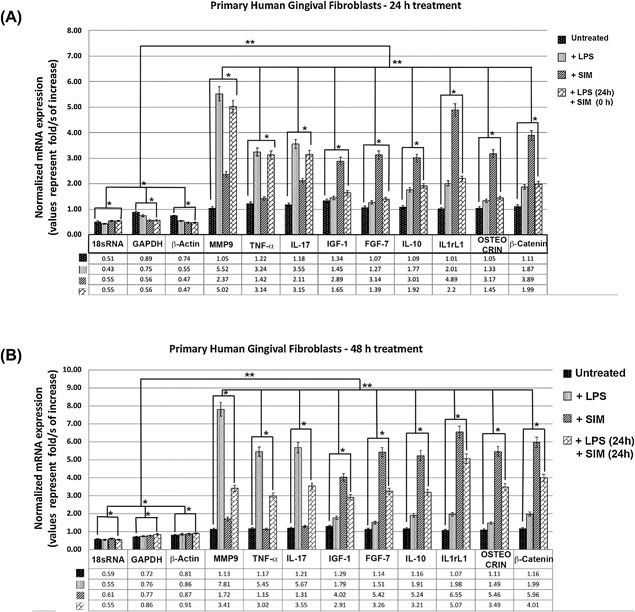
mRNA expression by real‐time qPCR in primary human gingival fibroblasts. (A) Fold change (log2) expression of mRNA relative to reference control genes (18rSRNA, GAPDH, and β‐actin) in primary human gingival fibroblasts treated with LPS and simvastatin for 24 hours. Bar heights indicate mean expression of the genes in samples. Error bars indicate 95% CI estimates of the mean expressions. One asterisk indicates statistically significant difference between the means of a sample set compared to the mean of the control sample set to 5% (correspond to a p value < 0.05); two asterisks indicate statistically significant difference to 1% (correspond to a p value < 0.01). (B) Fold‐change (log2) expression of mRNA relative to reference control genes (18rSRNA, GAPDH, and β‐actin) in primary human gingival fibroblasts treated with LPS and simvastatin for 48 hours. Bar heights indicate mean expression of the genes in samples. Error bars indicate 95% CI estimates of the mean expressions.
SIM treatment altered protein expressions of IGF‐1, MMP‐9, and TNF‐α in ExP
To investigate how SIM activation of inflammatory genes MMP‐9 and TNF‐α and bone regeneration gene IGF‐1 translate into in situ protein levels, the immunofluorescence of these proteins in the periodontium was analyzed. The protein expression of IGF‐1 was substantially upregulated after SIM treatment and mainly localized along the surface of the alveolar bone. In contrast, the expression of inflammatory markers, MMP‐9 and TNF‐α, were predominately within the gingiva. MMP‐9 was reduced after injection of SIM. Similarly, TNF‐α gained noticeable upregulation in periodontitis samples compared to unmanipulated controls, but was reduced in response to SIM treatment (Fig. 4).
Figure 4.
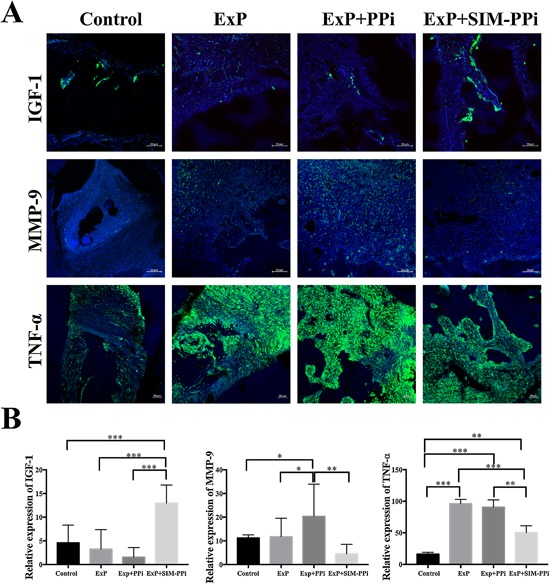
Immunostaining analysis of protein expression of IGF‐1, MMP‐9, and TNF‐α in different sample groups. (A) IGF‐1 showed significantly elevated expression in response to simvastatin treatment(s) compared to experimental periodontitis (ExP and ExP + PPi) and unmanipulated controls. Unmanipulated controls have limited expression of MMP‐9 and TNF‐α, whereas in ExP groups, MMP‐9 and TNF‐α expression was significantly upregulated. However, SIM treatment remarkably reduced the expression of MMP‐9 and TNF‐α. Each bar value, as stated, represents the size and magnification of the image. (B) Quantification of staining intensity of IGF‐1, MMP‐9, and TNF‐α in different sample groups. Statistical analysis was performed by one‐way ANOVA. Error bars represent 95% CI. ExP = experimental periodontitis; SIM‐PPI = simvastatin‐pyrophosphate. *p < 0.05; **p < 0.01; ***p < 0.001.
Induction of SIM in primary human gingival fibroblasts showed activation of anti‐inflammatory and probone anabolic marker mRNA expression by real‐time qPCR
We also further assessed the expression levels of the nine differentially expressed genes listed in Table 2 using real‐time qPCR in primary human gingival fibroblasts treated with LPS, followed by the additional treatment with SIM. Our qPCR results (see Fig. 5) were in agreement with our RNA‐seq and protein expression results, which demonstrates that LPS treatment induces the mRNA expression of several proinflammatory markers, such as MMP‐9, TNF‐α, and IL‐17 [folds of increase: 5.52, 3.24, and 3.55, respectively, in 24 hours (Fig. 5 A); 7.81, 5.45, and 5.67 in 48 hours (Fig. 5 B)] without having any significant effect on anti‐inflammatory and/or probone anabolic factors markers such as IGF‐1, FGF‐7, IL‐10, IL1rL1, osteocrin (Ostn), and β‐catenin. On the contrary, SIM treatment alone did not have a significant effect on proinflammatory molecules; however, it induced the expression of probone anabolic factors markers such as IGF‐1, FGF‐7, IL‐10, IL1rL1, Ostn, and β‐catenin [folds of increase: 2.89, 3.14, 3.01, 4.89, 3.17, and 3.89, respectively, in 24 hours (Fig. 5 A); 4.02, 5.42, 5.24, 6.55, 5.46, and 5.96 in 48 hours (Fig. 5 B)]. However, an additional 24 hours of LPS treatment with SIM treatment (24 hours) showed significantly decreased proinflammatory molecules, MMP‐9, TNF‐α, and IL‐17 [folds of decreased to 3.41, 3.02, and 3.55, respectively (Fig. 5 B)]. When we analyzed the mRNA expression patterns of the genes in the RNA‐seq data set, we obtained concordant profiles for the genes that were uniquely differentially expressed in ExP versus the control (FC| > 2.0) and SIM‐PPi versus the control (FC| > 2.0), instead of expanding our analysis to all genes identified in the literature because of qRT‐PCR technical limitations.
Table 2.
Genes That Are Uniquely Significantly Differentially Expressed (Adjusted p Value <0.05, |FC| >2.0) in ExP Versus Control
| ENSEMBLE Gene ID | Gene name | Adjusted p value | Fold change (ExP/control) | Function |
|---|---|---|---|---|
| ENSRNOG00000004649 | IL‐1β | 0.032582311 | 1.946 | |
| ENSRNOG00000013820 | TNF‐α | 6.52E‐07 | 3.81 | |
| ENSRNOG00000012467 | IL‐17(IL‐17a) | 9.30E‐07 | 3.854 | |
| ||||
| ENSRNOG00000017539 | MMP9 | 1.57E‐46 | 7.726 | |
| ENSRNOG00000047800 | C5a (C5ar1) | 7.58E‐11 | 3.513 |
|
| ENSRNOG00000009822 | TLR‐2 | 1.05E‐08 | 2.625 |
|
ExP = experimental periodontitis.
Discussion
Placement of the ligature around the rat maxillary second molar is a standard model for inducing inflammation and bone loss (ExP) and testing pharmacotherapeutic interventions.9, 26 Mature retired breeders were chosen for this study because growth of the mandible had ceased, and though multiple reproductive cycles may weaken trabecular microarchitecture, cortical bone (focus of the current study) actually becomes more robust.27 Within 11 days of ligature placement, a large number of proinflammatory genes associated with both rat and human periodontitis were upregulated. These included two cytokines most often associated with periodontitis, IL‐1β and TNF.28, 29 In addition, IL‐17 is a potent pro‐osteoclast activator linked to periodontitis pathogenesis.30 Matrix metalloproteinases, specifically MMP 2, MMP 8, and MMP 9, are key drivers of collagen and bone destruction in periodontitis, as well as regulators of periodontal inflammation.31 Complement, particularly the C5a receptor, has been increasingly implicated in periodontitis etiology because blocking this receptor in rats inhibits periodontal breakdown.32 The major initiator of periodontitis is the LPS component of the bacterial biofilm accumulated in the gingival crevice; increased LPS‐binding protein (LBP) is correlated to the amount of ligature‐induced periodontitis in baboons.33 The expression of toll‐like receptor 2 has been shown to increase in periodontal disease.34 Therefore, the upregulation of these genes is consistent with the induction of periodontitis inflammation.
Bone loss associated with ExP was radiographically evident after 1 week of ligature placement in the current and previous studies.9, 26 Therefore, regulation of genes involved in bone turnover should be evident when the surrounding gingiva was sampled 3 days after ligature removal. The upregulation of collagen type 1 gene corresponds to the synthesis and release of collagen type 1 as the primary bone metabolism event during periodontitis bone turnover, and has promoted its use as a marker of disease activity.35 The downregulation of genes responsible for periodontal homeostasis and bone growth also occurred with periodontitis bone loss. Odontogenic ameloblast‐associated protein has been shown to mediate junctional epithelial attachment to teeth, is present in a healthy periodontium, and is absent in the pathologic periodontitis pocket36 as was created in ExP. Decreasing insulin‐like growth factor 1 (IGF‐1) binding proteins are associated with variable tissue outcomes, ranging from periodontal attachment loss to decreased IGF‐1.37 IGF‐1 has been shown to be significantly underexpressed in periodontitis lesions compared to healthy periodontal sites.38
MMP‐9 is a well‐established inflammatory marker in periodontitis; elevated expression of MMP‐9 was demonstrated in close relationship to chronic periodontal disease.39, 40 MMPs are critical in the degradation of the extracellular matrix composing the connective tissue attachment apparatus to roots, resulting in attachment loss in periodontitis.41 MMP‐9 in the gingival crevice and surrounding gingiva has been shown to be a primary diagnostic biomarker of human periodontitis,42 and SIM reduced MMP‐9 in gingiva of a rat ExP model similar to the current study.43 TNF‐α is a known potent inflammation‐induced osteoclastogenic cytokine.44 TNF‐α strongly promotes inflammatory osteolysis by stimulating activation of macrophages and osteoclast precursors and enhancing expression of osteoclastogenic cytokines.44 TNF‐α promotes bone loss in periodontitis by activation of the RANK‐L expression pathways.45 Blocking TNF is the principal pharmacotherapeutic in reducing progressive joint inflammation and bone loss.46 Immunoassay findings in the current study, in agreement with previous studies,47 revealed that MMP‐9 showed higher expression in periodontitis with carrier alone, but was significantly reduced when SIM was included. We also observed that SIM treatment substantially repressed activity of TNF‐α. Together, our data clearly suggest that although ligature induces upregulation of inflammation agents and causes bone resorption, SIM exerts both anti‐inflammatory and antiosteoclastogenic effects by antagonizing expression of MMP‐9 and TNF‐α. These results are consistent with the gene expression pattern.
We report for the first time a novel association between SIM treatment upregulation IGF‐1 in experimental periodontitis in rat molars. IGF‐1 has long been thought to be one of the major anabolic factors responsible for limiting or reversing periodontal bone destruction.48 IGF‐1 has been shown to enhance bone regeneration in human periodontal defects49 and to stimulate regeneration of periodontal ligament.50 To further investigate the SIM‐delivery and activation of IGF‐1 in ExP, protein expression confirmed that IGF‐1 expression was low in ExP samples and SIM‐PPi injected into the periodontitis lesion quickly increased IGF‐1 protein expression (Fig. 4). When compared to reparative and regenerative roles of SIM as shown in Fig. 1, it is understandable that SIM‐induced activation of IGF‐1 may play a crucial role in reversing rat periodontitis. It has been shown that IGF‐1 is capable of stimulating PDL fiber bundles, cell proliferation, and local osteoblast precursor proliferation, differentiation, and mineralization of new bone.51 And the major signaling pathways that IGF‐1 uses are both MAPK and PI3K pathways to induce osteogenesis.52 Also, IGF‐1 modulates AKT/GSK3β pathways to induce its anti‐inflammatory effect in rats.53 Activation of FGF7 and IL1rl1 and suppressing Wnt/β‐catenin signaling also may be involved in: (1) activating periodontal ligament/fibroblast growth and homeostasis; (2) induction of alveolar bone repair and regeneration; and (3) immune response and repression of periodontitis.
Induction of IL1rl1, also known as the receptor suppression of tumorigenicity (ST) 2 gene, was noted in response to SIM treatment. ST2 is a crucial binding receptor for IL‐33.54 The cytokine IL‐33 has been recently linked in physiological bone remodeling and proinflammatory cell signaling inhibition.55 Current data, in relation to periodontitis, showed that SIM upregulates ST2, implying that activated IL‐33/ST2 signaling may induce lymphocytes to produce anti‐inflammatory and periodontal repair.56
Our data show that SIM induces an increased expression of the FGF7 gene (Supplementary Figs. 5 and 6) that can potentially augment mineralization. The local delivery of FGF7 increases the expression of osteogenic markers, mineralization with enhanced osteogenesis, and chemoattraction in mandibular bone formation.57 Although the exact mechanism by which SIM‐induced FGF7 facilitates osteogenesis is largely unknown, it has been demonstrated that FGF7 activates dexamethasone, ascorbic acid, and β‐glycerophosphate‐ (DAG‐) induced increases in bone‐like nodule formation and calcium accumulation.58 FGF7‐augmented mRNA expression of RUNX, osterix, bone sialoprotein, and osteocalcin in the presence of DAG, which suggests that FGF7 stimulates osteogenic differentiation.58 Similarly, our data also showed that SIM can increase the expression of RUNX3. Therefore, it is postulated that SIM‐induced increased expression of RUNX3 might be involved in rat alveolar bone formation, regeneration, and mineralization.59
Current data reveal that IL‐10 is upregulated in SIM‐treated rat molar periodontal tissues. IL‐10 is a potent anti‐inflammatory cytokine that also inhibits osteoclastic bone resorption and regulates osteoblastic bone formation.60 Subgingivally delivered SIM in humans has been shown to stimulate IL‐10 in the fluid around periodontitis pockets and to improve periodontal attachment.61 IL‐10 (‐/‐) mice are highly susceptible to bone loss induced by the periodontal pathogen, Porphyromonas gingivalis.62 Therefore, it is suggested that SIM‐induced increased expression of IL‐10 probably facilitates alveolar bone regeneration and limits periodontitis.
SIM also causes upregulation of the Ostn gene that has been recently discovered as a secreted protein produced by cells of the osteoblast lineage, and plays an important role in modulating bone formation and growth.63 Ostn is a soluble osteoblast regulator64 and is expressed in osteoblasts in developing bone.65 It has not previously been associated with SIM application and periodontitis. It has been shown that Ostn expression has an intense immunoreactivity in osteoblasts on bone‐forming surfaces and also in newly incorporated osteocytes.65 Therefore, the current model predicts that increased expression of Ostn in response to SIM treatment induces osteogenesis and modulates alveolar bone formation and growth.
We performed real‐time qPCR and have validated nine genes (MMP‐9, TNF‐α, IL‐17, TGF‐1, FGF‐7, IL‐10, IL1rL1, Ostn, and β‐catenin) in primary human periodontal fibroblasts that were uniquely differentially expressed in ExP versus the control (FC| > 2.0) and SIM‐PPi versus the control (FC| > 2.0), and all agreeing with the RNA‐seq and protein expression results. In addition, our mRNA expression data confirm our RNA‐seq and protein expression data that ExP induces proinflammatory and bone catabolic markers, and subsequent SIM‐PPi treatment causes anti‐inflammatory and bone anabolic genes to be activated.
In summary, gene activation during ExP in the rat reflects many of the key proinflammatory components seen in human periodontitis, including Il‐1β, TNF‐α, IL‐17, MMP‐9, and complement C5a. Local injection of SIM into ExP caused downregulation of MMP‐9 and TNF‐α, upregulation of anti‐inflammatory genes IL‐10 and IL1rl1 in ExP lesions, as well as the potent bone anabolic modulator IGF‐1. The signaling pathways involved in the process are IGF‐1 and FGF7, whereas the Wnt/β‐catenin pathway is repressed. Moreover, our data identify GRK3, ARG1, SIX1, IL‐10, MITF, and OSTN as target genes regulated by IGF‐1, FGF7, Wnt/β‐catenin, and IL1rl1, in response to SIM that are important for periodontal regeneration and may play a vital role in improving and treating periodontitis (Fig. 6). A better understanding of these signaling mechanisms will help to identify enhanced pharmacotherapeutic approaches to limit or regenerate periodontitis bone loss.
Figure 6.
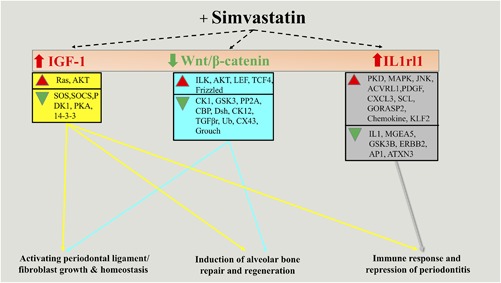
Schematic diagram of simvastatin induced pathways and gene network during rat periodontitis model. Simvastatin directly regulates by IGF‐1, FGF7, Wnt/β‐catenin, and IL‐1 receptor‐like (1IL1rl1) that may play decisive roles in (1) activating periodontal ligament/fibroblast growth and homeostasis, (2) induction of alveolar bone repair and regeneration, and (3) immune response and repression of periodontitis.
Disclosures
ZJ, RAR, and DW are listed as coinventors on a patent application related to the SIM‐PPi prodrug. The other authors report no conflict of interest related to this study.
Supporting information
Supporting Figures S1.
Acknowledgments
This study was financially supported in part by a Mary and Dick Holland Regenerative Medicine Program seed grant, a NIAMS/NIH grant (R01 AR062680) to DW, and a NIDCR, NIH grant (R01DE017986) to AN. JL was supported by a China Scholar Council scholarship (Beijing, China). The authors thank Ms. Marian J. Schmid of the College of Dentistry, University of Nebraska Medical Center (UNMC), Lincoln, for assistance with animal handling, housing, and maintenance in the facility; Yosif AlMoshari, PhD candidate, Department of Pharmaceutical Sciences, UNMC, for assistance with SIM treatment on cells; and Dr. Christian Elowsky of the Morrison Microscopy Core Research Facility, University of Nebraska‐Lincoln, for training on confocal microscopy.
Authors’ roles: Study design: AN, DW, RAR, and HHO; Study conduct: JL, SKC, ZK, MA, XW, ZJ, and RAR; Data collection: JL, SKC, ZK, XW, and ZJ; Data analysis: JL, SKC, ZK, ZJ, and AA; Data interpretation: JL, SKC, AN, DW, RAR, and HHO; Drafting manuscript: JL, SKC, AN, RAR, and HHO; Revising manuscript content: JL, SKC, AN, RR, DW, and HHO; Approving final version of manuscript: AN, RAR, and HHO. JL, SKC, AN, RAR, and HHO take responsibility for the integrity of the data analysis.
References
- 1. Albandar J, Brunelle J, Kingman A. Destructive periodontal disease in adults 30 years of age and older in the United States, 1988–1994. J Periodontol. 1999; 70(1):13–29. [DOI] [PubMed] [Google Scholar]
- 2. Eke PI, Dye BA, Wei L, Thornton‐Evans GO, Genco RJ. CDC Periodontal Disease Surveillance Workgroup: James Beck GDRP. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012; 91(10):914–20. [DOI] [PubMed] [Google Scholar]
- 3. Eke PI, Dye B, Wei L, Thornton‐Evans G, Genco R. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012; 91(10):914–20. [DOI] [PubMed] [Google Scholar]
- 4. Hoffmann T, Al‐Machot E, Meyle J, Jervoe‐Storm PM, Jepsen S. Three‐year results following regenerative periodontal surgery of advanced intrabony defects with enamel matrix derivative alone or combined with a synthetic bone graft. Clin Oral Investig. 2016; 20 (2):357–64. [DOI] [PubMed] [Google Scholar]
- 5. Pradeep AR, Priyanka N, Kalra N, Naik SB, Singh SP, Martande S. Clinical efficacy of subgingivally delivered 1.2‐mg simvastatin in the treatment of individuals with Class II furcation defects: a randomized controlled clinical trial. J Periodontol. 2012; 83 (12):1472–9. [DOI] [PubMed] [Google Scholar]
- 6. Pradeep AR, Thorat MS. Clinical effect of subgingivally delivered simvastatin in the treatment of patients with chronic periodontitis: a randomized clinical trial. J Periodontol. 2010; 81 (2):214–22. [DOI] [PubMed] [Google Scholar]
- 7. Mundy GR. Statins and their potential for osteoporosis. Bone. 2001; 29(6):495–7. [DOI] [PubMed] [Google Scholar]
- 8. Vargas‐Sanchez PK, Moro MG, Santos FAD, et al. Agreement, correlation, and kinetics of the alveolar bone‐loss measurement methodologies in a ligature‐induced periodontitis animal model. J Appl Oral Sci. 2017; 25(5):490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bradley AD, Zhang Y, Jia Z, et al. Effect of simvastatin prodrug on experimental periodontitis. J Periodontol. 2016; 87 (5):577–82. [DOI] [PubMed] [Google Scholar]
- 10. Reinhardt RA, Bolton RW, McDonald TL, DuBois LM, Kaidahl WB. In situ lymphocyte subpopulations from active versus stable periodontal sites. J Periodontol. 1988; 59 (10):656–70. [DOI] [PubMed] [Google Scholar]
- 11. Zappa U, Reinking‐Zappa M, Graf H, Espeland M. Cell populations and episodic periodontal attachment loss in humans. J Clin Periodontol. 1991; 18 (7):508–15. [DOI] [PubMed] [Google Scholar]
- 12. Ozturk F, Li Y, Zhu X, Guda C, Nawshad A. Systematic analysis of palatal transcriptome to identify cleft palate genes within TGFbeta3‐knockout mice alleles: RNA‐Seq analysis of TGFbeta3 Mice. BMC Genomics. 2013; 14:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Andrews S. FastQC: a quality control tool for high throughput sequence data. Canbrige, UK: Babraham Institute; 2010. Available from: http://wwwbioinformaticsbabrahamacuk/projects/fastqc/.
- 14. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30 (15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias‐aware quantification of transcript expression. Nat Methods. Apr 2017; 14 (4):417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 2014; 15 (12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu L, Liu J, Li Z, et al. TGFβ3 regulates periderm removal through ΔNp63 in the developing palate. J Cell Physiol. 2015; 230(6):1212–25. [DOI] [PubMed] [Google Scholar]
- 18. Zhu X, Ozturk F, Pandey S, Guda CB, Nawshad A. Implications of TGFbeta on transcriptome and cellular biofunctions of palatal mesenchyme. Front Physiol. 2012; 3:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mesia R, Gholami F, Huang H, et al. Systemic inflammatory responses in patients with type 2 diabetes with chronic periodontitis. BMJ Open Diabetes Res Care. 2016; 4 (1):e000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X, Jia Z, Almoshari Y, Lele SM, Reinhardt RA, Wang D. Local application of pyrophosphorylated simvastatin prevents experimental periodontitis. Pharm Res. 2018; 35 (8):164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995; 57:289–300. [Google Scholar]
- 22. Sneath P. Numerical taxonomy; the principles and practice of numerical classification. San Francisco, CA: WH Freeman; 1973. 573 p.
- 23. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 24. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000; 25 (1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008; 36(Database issue):D480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Struillou X, Boutigny H, Soueidan A, Layrolle P. Experimental animal models in periodontology: a review. Open Dentistry J. 2010; 4:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Bakker CM, Altman‐Singles AR, Li Y, Tseng WJ, Li C, Liu XS. Adaptations in the microarchitecture and load distribution of maternal cortical and trabecular bone in response to multiple reproductive cycles in rats. J Bone Miner Res. 2017; 32 (5):1014–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Spolidorio LC, Ramalho Lucas PD, Steffens JP, et al. Influence of parstatin on experimental periodontal disease and repair in rats. J Periodontol. 2014; 85(9):1266–74. [DOI] [PubMed] [Google Scholar]
- 29. Preshaw PM, Taylor JJ. How has research into cytokine interactions and their role in driving immune responses impacted our understanding of periodontitis? J Clin Periodontol. 2011; 38(s11):60–84. [DOI] [PubMed] [Google Scholar]
- 30. Zenobia C, Hajishengallis G. Basic biology and role of interleukin‐17 in immunity and inflammation. Periodontol 2000. 2015; 69 (1):142–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Franco C, Patricia H‐R, Timo S, Claudia B, Marcela H. Matrix metalloproteinases as regulators of periodontal inflammation. Int J Mol Sci. 2017; 18 (2):440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Damgaard C, Holmstrup P, Van Dyke TE, Nielsen CH. The complement system and its role in the pathogenesis of periodontitis: current concepts. J Periodontol Res. 2015; 50 (3):283–93. [DOI] [PubMed] [Google Scholar]
- 33. Ebersole J, Steffen M, Holt S, Kesavalu L, Chu L, Cappelli D. Systemic inflammatory responses in progressing periodontitis during pregnancy in a baboon model. Clin Exp Immunol. 2010; 162 (3):550–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sumedha S, Kotrashetti V, Nayak R, Nayak A, Raikar A. Immunohistochemical localization of TLR2 and CD14 in gingival tissue of healthy individuals and patients with chronic periodontitis. Biotech Histochem. 2017; 92 (7):487–97. [DOI] [PubMed] [Google Scholar]
- 35. Giannobile WV, Al Shammari KF, Sarment DP. Matrix molecules and growth factors as indicators of periodontal disease activity. Periodontol 2000. 2003; 31(1):125–34. [DOI] [PubMed] [Google Scholar]
- 36. Lee H‐K, Ji S, Park S‐J, et al. Odontogenic ameloblast‐associated protein (ODAM) mediates junctional epithelium attachment to teeth via integrin‐ODAM‐Rho guanine nucleotide exchange factor 5 (ARHGEF5)‐RhoA signaling. J Biolog Chem. 2015; 290 (23):14740–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harb AN, Holtfreter B, Friedrich N, et al. Association between the insulin‐like growth factor axis in serum and periodontitis in the Study of Health in Pomerania: an exploratory study. J Clin Periodontol. 2012; 39 (10):931–9. [DOI] [PubMed] [Google Scholar]
- 38. Choi YS, Kim YC, Ji S, Choi Y. Increased bacterial invasion and differential expression of tight‐junction proteins, growth factors, and growth factor receptors in periodontal lesions. J Periodontol. 2014; 85 (8):e313–e22. [DOI] [PubMed] [Google Scholar]
- 39. Makela M, Salo T, Uitto V‐J, Larjava H. Matrix metalloproteinases (MMP‐2 and MMP‐9) of the oral cavity: cellular origin and relationship to periodontal status. J Dent Res. 1994; 73 (8):1397–406. [DOI] [PubMed] [Google Scholar]
- 40. Marcaccini AM, Novaes AB, Meschiari CA, et al. Circulating matrix metalloproteinase‐8 (MMP‐8) and MMP‐9 are increased in chronic periodontal disease and decrease after non‐surgical periodontal therapy. Clin Chim Acta. 2009; 409 (1):117–22. [DOI] [PubMed] [Google Scholar]
- 41. Desarda H, Gaikwad S. Matrix metalloproteinases & implication in periodontitis–a short review. J Dent Allied Sci. 2013; 2 (2):66–70. [Google Scholar]
- 42. Baeza M, Garrido M, Hernández‐Ríos P, et al. Diagnostic accuracy for apical and chronic periodontitis biomarkers in gingival crevicular fluid: an exploratory study. J Clin Periodontol. 2016; 43 (1):34–45. [DOI] [PubMed] [Google Scholar]
- 43. Mouchrek Júnior JCE, Macedo CG, Abdalla HB, et al. Simvastatin modulates gingival cytokine and MMP production in a rat model of ligature‐induced periodontitis. Clin Cosmet Investig Dentis. 2017; 9:33–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lam J, Takeshita S, Barker JE, Kanagawa O, Ross FP, Teitelbaum SL. TNF‐α induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. J Clin Investig. 2000; 106 (12):1481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hienz SA, Paliwal S, Ivanovski S. Mechanisms of bone resorption in periodontitis. J Immunol Res. 2015; 2015:615486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dimitroulas T, Nikas SN, Trontzas P, Kitas GD. Biologic therapies and systemic bone loss in rheumatoid arthritis. Autoimmun Rev. 2013; 12 (10):958–66. [DOI] [PubMed] [Google Scholar]
- 47. Lazăr L, Loghin A, Bud E‐S, Cerghizan D, Horváth E, Nagy EE. Cyclooxygenase‐2 and matrix metalloproteinase‐9 expressions correlate with tissue inflammation degree in periodontal disease. Rom J Morphol Embryol. 2015; 56 (4):1441–6. [PubMed] [Google Scholar]
- 48. Okada H, Murakami S. Cytokine expression in periodontal health and disease. Crit Rev Oral Biol Med. 1998; 9 (3):248–66. [DOI] [PubMed] [Google Scholar]
- 49. Devi R, Dixit J. Clinical evaluation of insulin like growth factor‐I and vascular endothelial growth factor with alloplastic bone graft material in the management of human two wall intra‐osseous defects. J Clin Diagn Res. 2016; 10 (9):ZC41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Halper J. Advances in the use of growth factors for treatment of disorders of soft tissues In: Halper J, editor. Progress in heritable soft connective tissue diseases. Dordrecht, Germany: Springer; 2014. p. 59–76. [DOI] [PubMed] [Google Scholar]
- 51. Han X, Amar S. IGF‐1 signaling enhances cell survival in periodontal ligament fibroblasts vs. gingival fibroblasts. J Dent Res. 2003; 82 (6):454–9. [DOI] [PubMed] [Google Scholar]
- 52. Wang Y, Bikle DD, Chang W. Autocrine and paracrine actions of IGF‐I signaling in skeletal development. Bone Res. 2013; 1(3):249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang CY, Li XD, Hao ZH, Xu D. Insulin‐like growth factor‐1 improves diabetic cardiomyopathy through antioxidative and anti‐inflammatory processes along with modulation of Akt/GSK‐3β signaling in rats. Korean J Physiol Pharmacol. 2016; 20 (6):613–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Raggatt LJ, Partridge NC. Cellular and molecular mechanisms of bone remodeling. J Biolog Chem. 2010; 285(33):25103–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miller AM. Role of IL‐33 in inflammation and disease. J Inflam. 2011; 8 (1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Beklen A, Tsaous Memet G. Interleukin‐1 superfamily member, interleukin‐33, in periodontal diseases. Biotechn Histochem. 2014; 89(3):209–14. [DOI] [PubMed] [Google Scholar]
- 57. Poudel SB, Bhattarai G, Kim J‐H, et al. Local delivery of recombinant human FGF7 enhances bone formation in rat mandible defects. J Bone Miner Metab. 2017; 35(5):485–96. [DOI] [PubMed] [Google Scholar]
- 58. Jeon Y‐M, Kook S‐H, Rho S‐J, et al. Fibroblast growth factor‐7 facilitates osteogenic differentiation of embryonic stem cells through the activation of ERK/Runx2 signaling. Mol Cell Biochem. 2013; 382(1–2):37–45. [DOI] [PubMed] [Google Scholar]
- 59. Bauer O, Sharir A, Kimura A, Hantisteanu S, Takeda S, Groner Y. Loss of osteoblast Runx3 produces severe congenital osteopenia. Mol Cell Biol. 2015; 35 (7):1097–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang Q, Chen B, Yan F, et al. Interleukin‐10 inhibits bone resorption: a potential therapeutic strategy in periodontitis and other bone loss diseases. BioMed Res Int. 2014; 2014:284836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Grover HS, Kapoor S, Singh A. Effect of topical simvastatin (1.2 mg) on gingival crevicular fluid interleukin‐6, interleukin‐8 and interleukin‐10 levels in chronic periodontitis–a clinicobiochemical study. J Oral Biol CraniofacRes. 2016; 6 (2):85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sasaki H, Okamatsu Y, Kawai T, Kent R, Taubman M, Stashenko P. The interleukin‐10 knockout mouse is highly susceptible to Porphyromonas gingivalis‐induced alveolar bone loss. J Periodon Res. 2004; 39 (6):432–41. [DOI] [PubMed] [Google Scholar]
- 63. Moffatt P, Thomas G, Sellin K, et al. Osteocrin is a specific ligand of the natriuretic peptide clearance receptor that modulates bone growth. J Biol Chem. 2007; 282 (50):36454–62. [DOI] [PubMed] [Google Scholar]
- 64. Thomas G, Moffatt P, Salois P, et al. Osteocrin, a novel bone‐specific secreted protein that modulates the osteoblast phenotype. J Biolog Chem. 2003; 278(50):50563–71. [DOI] [PubMed] [Google Scholar]
- 65. Bord S, Ireland DC, Moffatt P, Thomas GP, Compston JE. Characterization of osteocrin expression in human bone. J Histochem Cytochem. 2005; 53 (10):1181–7. [DOI] [PubMed] [Google Scholar]
- 66. Beklen A, Ainola M, Hukkanen M, Gürgan C, Sorsa T, Konttinen Y. MMPs, IL‐1, and TNF are regulated by IL‐17 in periodontitis. J Dent Res. 2007; 86(4):347–51. [DOI] [PubMed] [Google Scholar]
- 67. Johnson R, Wood N, Serio F. Interleukin‐11 and IL‐17 and the pathogenesis of periodontal disease. J Periodontol. 2004; 75 (1):37–43. [DOI] [PubMed] [Google Scholar]
- 68. Cassatella MA, Meda L, Bonora S, Ceska M, Constantin G. Interleukin 10 (IL‐10) inhibits the release of proinflammatory cytokines from human polymorphonuclear leukocytes. Evidence for an autocrine role of tumor necrosis factor and IL‐1 beta in mediating the production of IL‐8 triggered by lipopolysaccharide. J Exp Med. 1993; 178 (6):2207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figures S1.


