Abstract
Gene expression occurs in two essential steps: transcription and translation. In bacteria, the two processes are tightly coupled in time and space, and highly regulated. Tight regulation of gene expression is crucial. It limits wasteful consumption of resources and energy, prevents accumulation of potentially growth inhibiting reaction intermediates, and sustains the fitness and potential virulence of the organism in a fluctuating, competitive and frequently stressful environment. Since the onset of studies on regulation of enzyme synthesis, numerous distinct regulatory mechanisms modulating transcription and/or translation have been discovered. Mostly, various regulatory mechanisms operating at different levels in the flow of genetic information are used in combination to control and modulate the expression of a single gene or operon. Here, we provide an extensive overview of the very diverse and versatile bacterial gene regulatory mechanisms with major emphasis on their combined occurrence, intricate intertwinement and versatility. Furthermore, we discuss the potential of well-characterized basal expression and regulatory elements in synthetic biology applications, where they may ensure orthogonal, predictable and tunable expression of (heterologous) target genes and pathways, aiming at a minimal burden for the host.
Keywords: transcription factors, sigma factors, attenuation control, regulatory RNA, RNA polymerase, synthetic biology
Bacterial gene regulatory mechanisms are generally complex, multi-layered and tightly interwoven, but once fully unraveled isolated constituents provide valuable cis- and trans-acting elements for applications in synthetic biology requiring orthogonal, predictable, and tunable expression of (heterologous) target genes and pathways.
INTRODUCTION
The study of bacterial gene regulation definitely took off with the pioneering and groundbreaking work on regulation of lac operon encoded enzyme synthesis in Escherichia coli (Jacob and Monod 1959, 1961; Gilbert and Müller-Hill 1966, 1967). Very importantly, it was rapidly realized that induction of the lac operon in the presence of its substrate and induction of a lysogen for bacteriophage lambda on the one hand and repression of amino acid biosynthesis genes and operons in the presence of the end product on the other are two manifestations of the same phenomenon: negative regulation of transcription initiation by a trans-acting DNA-binding transcription regulator (historical accounts by Maas 1991, 2007). Since then, numerous distinct mechanisms of gene regulation have been identified. And most importantly, it has become increasingly clear that the expression of a single bacterial gene or operon is generally controlled not by one but by a combination of diverse mechanisms, potentially operating at different levels (transcription, translation) and stages (initiation, elongation, termination) of gene expression. Furthermore, and to add to the complexity and versatility of bacterial regulatory systems, the effect of a single regulatory molecule, regardless whether it is a DNA- or RNA-binding protein, a cis- or trans-acting small regulatory RNA, may vary depending on the specific target, its interaction with a small effector molecule or associated protein(s) and/or its interference/cooperation with other regulatory processes. This review provides an extensive overview of the various mechanisms modulating bacterial gene expression and illustrates the versatility of bacterial regulation, the intertwinement of regulatory mechanisms and the integration of multiple signals to generate an adapted regulatory response in function of environmental growth conditions and cellular needs. Much of the information provided here is derived from E. coli, still the best-studied microorganism. However, the underlying basic principles of these regulatory mechanisms are more generally valid for both Gram-negative and Gram-positive organisms, even though the target of a specific regulatory molecule may differ and a specific gene or operon may be regulated by a different mechanism in distinct bacteria. This is among others (a. o.) illustrated below for regulation by the alarmone ppGpp and of pyrimidine biosynthesis in the Gram-positive soil bacterium Bacillus subtilis.
As bacterial gene regulation mechanisms are mostly multi-layered and intricate, they are generally too complex to be incorporated as such in the design of synthetic circuits. However, once fully unraveled appealing properties of well-characterized isolated cis- and trans-acting basic gene expression and regulatory elements (parts) can be captured, engineered and further optimized as standardized parts for application in synthetic biology. In this review, we emphasize the interest of various regulatory systems for such SynBio applications. Profound knowledge of transcriptional and post-transcriptional gene regulatory mechanisms is thus not only of fundamental interest, but it may also lead to economically/industrially relevant applications and applications in human and animal health care, such as diagnostics, therapeutics, bioremediation, the development of specific gene silencing antibacterial drugs or the construction of well-characterized artificial gene expression systems for microbial synthesis of diverse (bio)chemicals from renewable resources. The latter is gaining interest as a valuable alternative to the classical (petrochemical) and frequently environmentally less friendly production schemes and methods (Jullesson et al.2015). Synthetic biology for microbial production relies a. o. on the construction of stable, predictable and tunable orthogonal gene expression systems showing maximal independence of and minimal interference with the host metabolism in order to avoid an important burden (Mutalik et al.2013; Singh 2014; Wu et al.2016). Orthogonal gene expression and orthogonality of gene expression and regulatory elements are used in the context of this review, and more broadly in synthetic biology, to indicate the use of components (naturally occurring heterologous or artificial) that are functionally insulated from the action of the endogenous gene expression and regulatory machineries, and ideally only recognize their cognate elements. Thus, heterologous orthogonal sigma factors introduced in a host will only bind to their cognate promoters introduced in this host and not to promoters of the endogenous genes and neither will they cross-react among them. Similarly, orthogonal ribosomes will only bind their cognate ribosome-binding sites (RBS). However, it is important to realize that orthogonal gene expression is never completely independent of the host since transcription and translation will generally still rely on some components of the host's basal gene expression machineries or at least require its metabolic energy providing machinery and other cellular resources. Nevertheless, the use of orthogonal gene regulatory elements provides a means to modulate and optimize as desired the expression of a specific set of genes without considerably affecting the vast majority of the endogenous ones, and contributes to limit metabolic burden. The rational design of such expression systems requires ‘toolboxes’ containing well-characterized ‘parts’ including promoter and ribosome-binding sites of various strengths (libraries), and regulatory elements, which can be assembled in a combinatorial manner (De Mey et al.2007; Lucks et al.2008; Davis, Rubin and Sauer 2011; Brewster, Jones and Phillips 2012; Kahl and Endy 2013; Tripathi, Zhang and Lin 2014; Bradley, Buck and Wang 2016; Rangel-Chavez, Galan-Vasquez and Martinez-Antonio 2017). In this respect, alternative sigma factors, DNA- and RNA-binding regulatory proteins, riboswitches and small regulatory RNAs are among the regulatory elements that hold great promise, as further developed in this review. However, it should be stressed that optimizing gene expression addresses only one aspect of synthetic circuits. Other major bottlenecks in heterologous production may be related to feedback inhibition or allosteric regulation of enzyme activity, protein folding and stability, post-translational modifications, etc., but this is beyond the scope of this review.
Basal bacterial transcription and translation: tightly coupled processes
Gene expression occurs in two steps, transcription and translation, which in bacteria are tightly coupled in space and time (Fig. 1A) (McGary and Nudler 2013). This coupling is an absolute necessity due to the generally very short half-life of bacterial mRNA molecules, which is on average a few minutes only (Rauhut and Klug 1999). Furthermore, various regulatory mechanisms may affect the rate of mRNA degradation (positively or negatively) and the accessibility of mRNAs for the translation machinery, which again has an effect on mRNA stability (see the section Regulation of mRNA stability). Thus, transcription, translation and mRNA degradation are tightly interconnected processes.
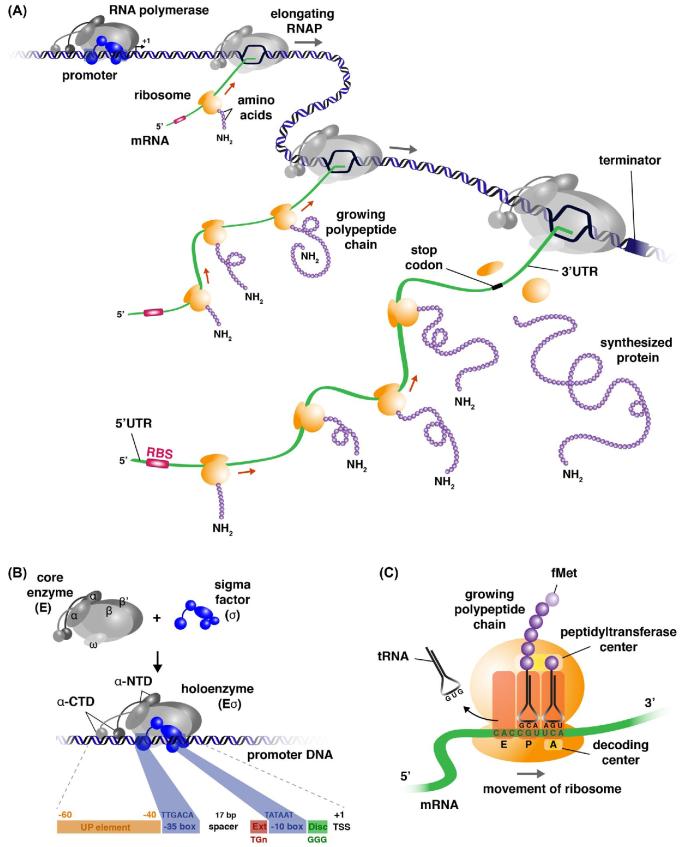
Figure 1.
Bacterial transcription and translation is coupled. (A) Simplified schematic view of mRNA and protein synthesis with several RNA polymerase (RNAP) molecules simultaneously transcribing a single gene, and several ribosomes translating a single monocistronic mRNA. Transcription starts with binding of RNAP holoenzyme to the promoter region, mRNA synthesis proceeds in the direction 5΄ to 3΄ and stops at a Rho-dependent or -independent terminator. In both instances, transcription termination is accompanied by release of the mRNA molecule and dissociation of RNAP from the DNA template. Notice that during transcription elongation σ (blue colored) may (as in the figure) or may not dissociate from the complex. Translation by fully assembled 70S ribosomes (after initial binding of the 30S subunit to the RBS) starts at the initiation codon (mostly AUG, positioned in the P site of the ribosome), proceeds in the direction N-terminus to C-terminus and stops at a nonsense codon. Translation termination is accompanied by the release of the polypeptide chain from the tRNA and recycling of the ribosomes in separate 30S and 50S subunits. The magenta colored segment near the 5΄-end of the mRNA represents the ribosome-binding site (RBS) that comprises the Shine–Dalgarno sequence and the initiation codon. 5΄-UTR and 3΄-UTR correspond to untranslated regions of the mRNA near its 5΄- and 3΄-end, respectively. For simplicity, initiation, elongation and termination factors are not shown (see Box 1 for more information on bacterial transcription and translation). (B) Schematic representation of the RNAP holoenzyme composed of the core enzyme (α2ββ΄ω) and a σ factor that is responsible for promoter recognition. UP is the upstream promoter element that is contacted by α-CTD, whereas the –10 and –35 promoter elements are contacted by different parts of σ (see Fig. 2C for details). Ext represents the extended –10 promoter element and Disc the discriminator site. TSS represents the transcription start, which is mostly a purine. Sequences and distances correspond to the consensus promoter for the housekeeping σ factor (σ70) of E. coli. (C) Schematic view of an elongating fully assembled 70S ribosome with three binding sites for tRNA molecules. A, the aminoacyl site with an incoming aminoacylated tRNA selected on basis of codon–anticodon complementarity; P, the peptidyl site with a tRNA carrying the growing peptide chain; and E, the exit site for binding of an uncharged tRNA after transfer of the growing peptide chain from the P site to the A site-bound tRNA followed by a translocation cycle.
Bacterial transcription is performed by a single RNA polymerase (RNAP), which consists of a catalytic core (E), with subunit composition α2ββ΄ω, that associates with a sigma factor (σ) to form the holoenzyme (Eσ) (Fig. 1B). More information on the composition of the basal transcription apparatus, the different stages in transcription initiation and processes of transcription termination is provided in Box 1. σ factors ensure specific promoter recognition by directly binding to two conserved hexanucleotide boxes centered around the positions –35 and –10 (Pribnow box) with respect to the transcription initiation site (+1) for σ70 family members, and positions –24 and –12 for σ54 family members (Figs 1B and 2) (Saecker, Record and deHaseth 2011; Cho et al.2014). These hexanucleotide stretches constitute the major specificity determinants of the promoter and are connected by a spacer of conserved length but variable sequence, though spacer and surrounding region sequences may also contribute to promoter specificity, as recently demonstrated for extra-cytoplasmic function (ECF) σ factors from B. subtilis (Gaballa et al.2018). Besides a major housekeeping σ factor bacteria generally contain several alternative σ factors (Mittenhuber 2002). They are used to direct the synthesis of specific sets of genes by interacting with specific, conserved promoter elements (Fig. 2). It appears that the reduced capacity of alternative σ factors to melt promoter sequences ensures their more stringent promoter recognition compared to the housekeeping σ (Koo et al.2009). Many promoter regions contain binding sites for more than one σ factor to ensure transcription in different growth conditions. Direct sequence-specific DNA binding by a subunit of RNAP is unique to the bacterial domain of life. It is already a form of gene regulation, and one that holds great promise for exploitation in the design of orthogonal synthetic circuits (see the section Alternative σ factors for orthogonal expression of synthetic circuits). Mutations affecting the conserved hexanucleotide promoter elements generally have drastic effects on the transcription initiation frequency and promoter specificity. In contrast, randomizing the connecting linker sequence allows tuning of the transcription initiation frequency and promoter libraries exhibiting a wide range of expression levels were constructed for various organisms including Gram-negative and Gram-positive bacteria (Solem and Jensen 2002; De Mey et al.2007; Dehli, Solem and Jensen 2012; Rytter et al.2014; Gilman and Love 2016; Guiziou et al.2016). This proved to be possible without loss of specificity/orthogonality (Bervoets et al.2018).
Box 1. The basal bacterial transcription and translation machineries.
The basal bacterial transcription apparatus. In bacteria, all RNA species are synthesized by a single RNA polymerase composed of a core enzyme (α2ββ΄ω) that associates with a sigma factor (σ) (Fig. 1A and B). All subunits of the core enzyme are highly conserved in sequence and structure among bacteria, and homologs of the core subunits are also present in Eukarya and Archaea (Korkhin et al.2009; Werner and Grohmann 2011). In contrast, σs that associate with the core and provide promoter-specific recognition are unique to Bacteria but highly variable in sequence, size and domain structure (Gruber and Gross 2003; Haugen, Ross and Gourse 2008; Feklístov et al.2014) as further detailed in the main body of the text (Fig. 2). Most bacteria possess several σ factors, a primary housekeeping (also called vegetative) σ factor (σ70, also designed σD in E. coli, σA in many other bacteria) that is used for transcription initiation of all unconditionally essential genes and most genes whose product is required in all growth conditions, and a variable number of alternative sigma factors that are used to express specific sets of genes in response to particular stress conditions, the assembly and function of flagella, chemotaxis and certain developmental processes (Paget 2015; Davis et al.2017). All σ factors in a cell compete for binding to the same core enzyme and interact with two conserved hexanucleotide stretches in the promoter, centered at position –10 (also called Pribnow box) and –35 with respect to the transcription initiation site (+1) for the housekeeping σ and other sigma-70 family members (Mauri and Klump 2014; Kandavalli, Tran and Ribeiro 2016) (Fig. 1B and 2). Ideally these hexanucleotides are separated by a 17 bp spacer. Some promoters (generally with a highly degenerated or no –35 element) have an extended –10 sequence, with the conserved TGN dinucleotide immediately preceding the Pribnow box (Fig. 1B). Strong promoters may also contain a UP element (Upstream Promoter), an A+T-rich sequence preceding (–40 to –60) the –35 box, which is contacted by the C-terminal domain of one or both α-subunits (α-CTD) (Fig. 1B) (Ross et al.1993; Gaal et al.1996; Estrem et al.1999). It appears that structural properties of promoter regions correlate with functional features (Meysman et al.2014). The promoter sequence will to a large extent determine the basal transcription initiation frequency but this frequency may be strongly influenced (enhanced, reduced) by various regulatory mechanisms. These mechanisms are the major focus of this review and are extensively discussed in the main body of the text. Initial binding of Eσ to the promoter DNA generates a closed binary complex (RPc) that subsequently isomerizes to the open complex (RPo), which is characterized by local DNA melting over a distance of 13 bp (from position −11 to +3). Incorporation of the first nucleotide generates the ternary initiating complex (RPi), which upon further transcript elongation will generally go through a number of abortive initiation cycles whereby short RNA products (generally ≤12 nt) are released, before evolving into the elongating complex (RPe) that is characterized by promoter escape and productive RNA synthesis. This transition from RPi to RPe is a
rate-limiting step in transcription initiation (generally 1 to 2 s) and hence already a form of gene regulation since as long as the RNAP has not liberated the promoter a new initiation round cannot be started. Though it is generally stated that the σ factor dissociates from the core RNAP upon transcription elongation, and must do so to free the RNA exit channel, it appears now that E. coli σ70 may remain associated with the elongating enzyme (Harden et al.2016). For more detailed information on the mechanism of bacterial transcription initiation, the reader is referred to excellent and recent reviews (Saecker, Record and deHaseth 2011; Grylak-Mielnicka et al.2016; Lloyd-Price et al.2016; Henderson et al.2017).
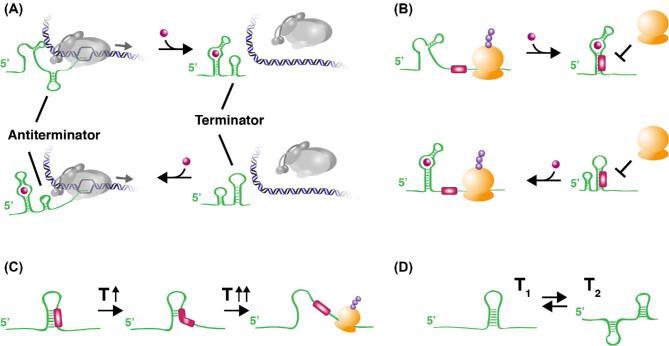
Bacterial genes are frequently grouped in operons and a polycistronic transcription unit may cover up to tens of genes that are coordinately expressed from a common control region. The transcription elongation rate is not uniform but discontinuous and influenced by local pause sites and RNAP-associated factors that play key regulatory roles (Artsimovitch and Landick 2000). Operons may contain internal secondary promoters (generally rather weak and constitutive) to allow adjustment of the expression of downstream genes. Transcription termination can occur in two distinct manners, depending on the termination signal. Intrinsic or Rho-independent termination occurs at G+C-rich stem-loop structures immediately followed by a stretch of U-residues in the mRNA. These secondary RNA structures are also exploited in regulatory systems such as transcriptional attenuation, where they are stabilized and used to prematurely stop the RNA polymerase in particular conditions only (Henkin and Yanofsky 2002). Rho-dependent termination makes use of the Rho protein and its cofactor NusG. Rho is a hexameric ring-shaped molecule with ATPase activity that binds to a conserved rut site in the RNA, moves along the transcript and forces dissociation of the transcription complex. In E. coli, intrinsic and Rho-dependent termination contribute more or less equally to transcription termination (Ray-Soni, Bellecourt and Landick et al.2016). Besides its essential role in generating the 3΄-end of a transcript, Rho is also involved in premature transcription termination as a regulatory process of gene expression. About 27% of all annotated E. coli genes have 5΄-untranslated sequences (5΄-UTR) longer than 80 nucleotides (nt) that are natural substrates for Rho-dependent termination and in many instances this premature termination process is counteracted by the binding of small regulatory RNAs (sRNAs) to the 5΄-UTR (Sedlyarova et al.2016). Rho-dependent termination and targeting of nascent transcripts that are not translated efficiently is also essential to limit antisense transcript accumulation and concommittant R-loop formation (Raghunathan et al.2018). The importance of Rho and its cofactor NusG is underscored by the observation that both rho and nusG are essential genes in E. coli and many other bacteria. For recent reviews on transcription termination and the multifunctional role of Rho, see Peters, Vangeloff and Landick (2011), Grylak-Mielnicka et al. (2016), Kriner, Sevostyanova and Groisman (2016), Porrua, Boudvillain and Libri (2016) and Ray-Soni, Bellecourt and Landick (2016).
The basal bacterial translation apparatus. Bacterial protein synthesis is performed on ribosomes, 70S particles composed of a small (30S) and a large (50S) subunit that assemble on mRNA (Fig. 1A). As transcription, translation consists of three major phases, initiation, elongation and termination, followed by ribosome recycling (Laursen et al.2005). Both subunits consist of ribosomal RNA (16S rRNA for the 30S, 5S and 23S rRNA for the 50S subunit) associated with a large number of ribosomal proteins. Peptide bond formation is catalyzed by the 23S rRNA; hence, it is a ribozyme (RNA molecule with catalytic activity) that uses the energy of the acyl bond on the aminoacylated tRNA (aa-tRNA) to drive the condensation reaction. Ribosomes bear three binding sites for tRNA: (i) the A (aminoacyl) site that accepts the incoming aa-tRNA (except the initiator tRNAi, which frequently carries formylmethionine in bacteria, and immediately enters the P site), a process that is based on codon–anticodon recognition; (ii) the P (peptidyl) site, which holds the tRNA attached to the growing peptide chain; and (iii) the E (exit) site that bears the deacylated tRNA before it exits the ribosome in the next translocation cycle (Fig. 1C). Translation initiation starts with the cooperative binding of mRNA and the translation initiation factors IF1 and GTP-bound IF2 to respectively that part of the A and P sites present on the small subunit, which at this stage is already associated with initiation factor IF3 bound in the E site. GTP-bound IF2 brings the charged tRNAi to the P site. Subsequent GTP hydrolysis is accompanied by the assembly of the full ribosome and dissociation of the initiation factors. Translation generally starts with the first AUG codon (or more rarely and less efficiently with GUG or UUG) following the ribosome-binding site (RBS; alias Shine–Dalgarno [SD]), which is complementary to the 3΄-end of 16S rRNA of the 30S subunit. All subsequent aa-tRNAs for chain elongation are brought to the A site of assembled 70S ribosomes in association with GTP-bound elongation factor EF-Tu, which after GTP hydrolyses is recycled with the use of GTP and EF-Ts. Coordinated translocation along the mRNA is performed with the help of GTP-bound EF-G. Hence, protein synthesis is an energetically very costly process that requires one ATP (for the aminoacylation of tRNA for the corresponding amino-acyl tRNA synthetase) and two GTP molecules for every amino acid that is incorporated. Protein synthesis will terminate when one of three stop codons (UAA, UAG, UGA or a combination) is encountered and occupies the A site. This process requires a termination factor of class-I (RF1 or RF2) and class-II (RF3, a GTP-ase) and hydrolysis of another GTP molecule to release the peptide chain from the peptidyl-tRNA and the dissociation of the release factors. Termination is followed by dissociation of the 70S particles into 30S and 50S constituents (with the help of RRF, the ribosomal release factor, and IF3), a requisite for their recycling and reuse in a next initiation round. In polycistronic mRNAs, each open reading frame (ORF) is generally preceded by an RBS. The efficiency of protein synthesis strongly depends on the complementarity of the SD sequence with the 3΄-end of 16S rRNA, its distance to the initiation codon, the nature of the latter and of the subsequent codons near the N-terminus of the protein and the concentration of aa-tRNAs in the cell. The latter will influence the velocity of ribosome movement and this is exploited in regulatory mechanisms of the attenuation type, which may affect both premature transcription termination and translation initiation (Henkin and Yanofsky 2002). For more information on bacterial translation, see Moreno et al. (2000), Ramakrishnan (2002) and Laursen et al. (2005).
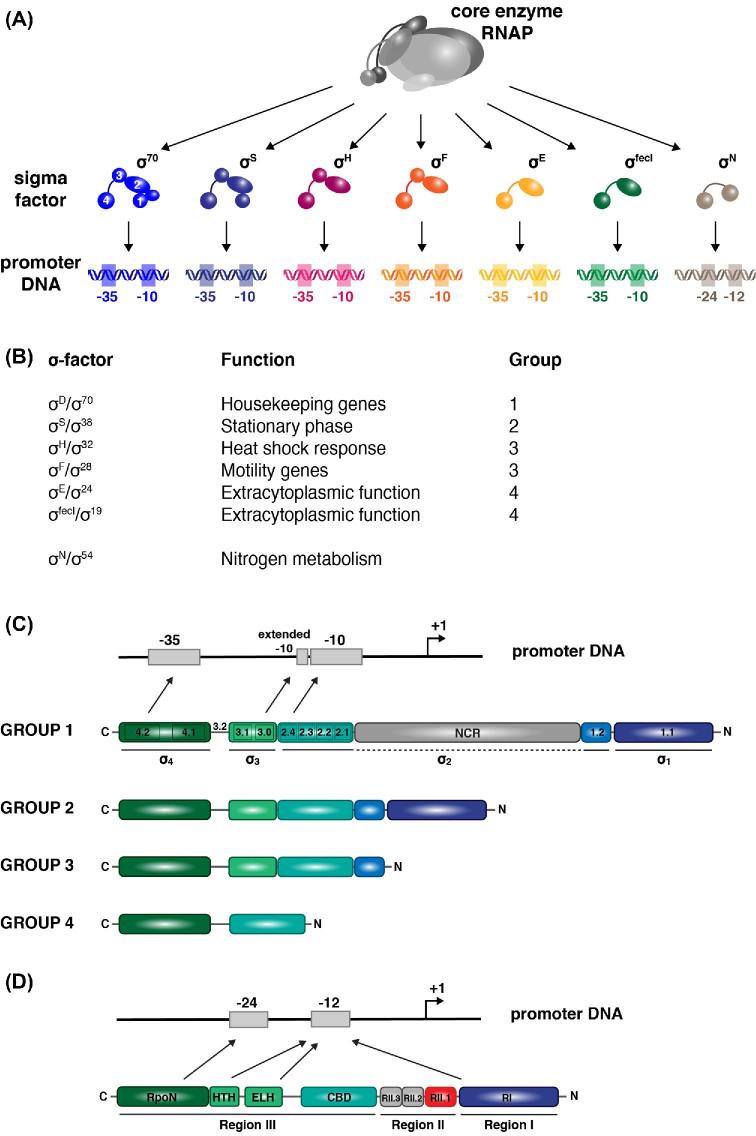
Figure 2.
Domain composition of σ factors and their interaction with cognate promoter sequences. (A) The seven σ factors of E. coli, which all bind competitively to similar regions of the unique core RNAP but interact with specific promoter sequences centered around positions –10 and –35 for the six members of the σ70 family and around –12 and –35 for σN, the sole representative of the σ54 family. Notice that some cross-talk may exist in the binding of alternative sigma factors to non-cognate promoter sequences as observed for the housekeeping σ70 and σS in E. coli and among members of the ECF (extracytoplasmic function) group of σ factors in general. (B) Function and division in four groups of the six σ70 family members of E. coli based on domain composition, and σ54 that forms a distinct family on its own. (C) Promoter structure and domain composition of the four groups of σ70 family members. NCR stands for non-conserved region. Arrows indicate interactions of specific subdomains of σ with promoter sequences. σ1.1 plays an inhibitory role in promoter binding. (D) Promoter structure and domain composition of σ54. RpoN is the region that specifically interacts with the –24 promoter region and is the most conserved domain among σ54 proteins. HTH stands for the helix-turn-helix motif that interacts with the –12 promoter sequence, and ELH for extra long helix. CBD is the RNAP core-binding domain. RI interacts with RIII and plays an inhibitory role, blocking the entry of the DNA template strand. It is also a site of contact for activator proteins. RII penetrates deeply in the DNA-binding channel and also plays an inhibitory role and has to be displaced in the transcribing complex.
Translation of bacterial mRNAs by ribosomes, with the help of translation initiation factors, starts with binding of the 30S subunit to the RBS, a short A+G-rich sequence, also called Shine–Dalgarno (SD) sequence, preceding the translation initiation codon (generally AUG) that is (partially) complementary to the 3΄-end of 16S rRNA (Ramakrishnan 2002). More details on the basal bacterial translation machinery are provided in Box 1. Translation starts as soon as the 5΄-end of the mRNA molecule has been synthesized, and several ribosomes may simultaneously translate a single mRNA molecule (polyribosomes) (Fig. 1A and C). In general, each open reading frame (ORF) of a polycistronic mRNA is preceded by a RBS. Variations in the accessibility of the RBS by RNA-binding proteins or reversible trapping in double-stranded RNA structures may greatly affect the translation initiation frequency and hence constitute a form of gene regulation (Duval et al.2015). As with promoter sequences, variations in the RBS allow to tune the translation initiation frequency. RBS libraries have been constructed, algorithms have been developed and tools such as the RBS calculator allow the design of synthetic RBSs and to predict the rate of translation initiation on basis of the 5΄-mRNA sequence (Salis, Mirsky and Voigt 2009; Salis 2011; Egbert and Klavins 2012; Guiziou et al.2016). Furthermore, an orthogonal ribosome–mRNA translation system (O-ribosome) was developed in which an orthogonal 16S rRNA that recognizes an altered SD sequence only translates its cognate O-mRNAs (Rackham and Chin 2005; An and Chin 2009; Liu, Kim and Jewett 2017).
Need for gene regulatory mechanisms
Regulation of gene expression is pivotal for optimal energy management and the generation of a swift and adapted metabolic response to fluctuating environmental conditions and stresses. In combination with regulation of enzyme activity (enzymotropic regulation by a. o. feedback inhibition, allosteric regulation and (reversible) chemical modifications) that operates at a different time scale, regulation of gene expression at transcriptional and post-transcriptional level avoids the accumulation of pathway intermediates and wasteful consumption of resources and energy. Hence, bacteria that have a more varied lifestyle and exhibit a considerable nutrient and metabolic versatility, such as the Gram-positive soil bacterium B. subtilis and the Gram-negative opportunistic pathogen Pseudomonas aeruginosa, bear a much larger number and variety of genes encoding regulatory elements including transcriptional regulators, two-component systems and alternative σ factors than organisms such as intracellular pathogens that thrive in more stable biotopes (Pérez-Rueda, Janga and Martínez-Antonio 2009; Pérez-Rueda and Martínez-Nuñez 2012; Freyre-González et al.2013; Pérez-Rueda et al.2018). At the level of protein production, regulation of transcription initiation has the advantage that it affects the very first step in the process and hence is most interesting in terms of energy saving. On the other hand, regulation of translation initiation, the most regulated step in translation (Duval et al.2015), has the advantage that it has an immediate impact on protein synthesis. Various quantitative proteomic and RNA-Seq studies performed with diverse bacteria have shown that a large part of gene regulation occurs at the post-transcriptional level, including translation initiation and elongation, but also proteolysis and sequestration, especially in response to different stress conditions (Nie, Wu and Zhang 2006; Lu et al.2007; de Sousa et al.2009; Dressaire et al.2010; Picard et al.2012, 2013).
Regulation of gene expression by σ factor competition
Besides their housekeeping σ factor, which is used for the synthesis of most products that are needed in all growth conditions, bacteria possess a variable number of alternative σ factors. As stated above, these recognize specific sets of promoter sequences but compete for binding to the limited pool of core RNAP (about 1300 molecules involved in transcription and another 700 free molecules in an E. coli cell) (Fig. 2A) (Ishihama 2000; Gruber and Gross 2003, Grigorova et al.2006; Feklístov et al.2014; Mauri and Klumpp 2014; Davis et al.2017). This competition already constitutes a form of gene regulation and ensures a mechanism for cross-talk between different classes of genes. Furthermore, functional modules of genes transcribed with the same σ factor guarantee adaptability and evolvability (Binder et al.2016). σ factors generally control global switches in the gene expression profile, mainly in response to stress conditions, and cascades of alternative σ factors are used to coordinate gene expression in time and space (cellular compartmentalization). They steer complex cellular processes including sporulation in B. subtilis, photosynthesis and circadian rhythms in cyanobacteria, aerial hyphae production by Streptomyces coelicolor and lytic propagation of bacteriophages (Burbulys, Trach and Hoch 1991; Hilbert and Piggot 2004; Hinton et al.2005; Hinton 2010). It is in the latter context that bacteriophage T4-encoded alternative σ factors, regulation of their activity by anti-σ factors, and cooperation with transcriptional activator proteins for the temporal and successive expression of viral genes have been discovered (Stevens 1973; Minakhin and Severinov 2005; Tagami et al.2014).
Escherichia coli has seven σ factors (Fig. 2A and B) (Maeda, Fujita and Ishihama 2000), but some other organisms have many more. Thus, the Gram-positive soil bacterium B. subtilis has 19 characterized/predicted σ factors, many of which are involved in sporulation, S. coelicolor encodes over 60 σ factors and the Gram-negative soil dwelling myxobacterium Sorangium cellulosum So0157–2 more than 100 (Mittenhuber 2002; Kill et al.2005; Han et al.2013). Most alternative σ factors belong to the large σ70 family (Lonetto, Gribskov and Gross 1992; Paget 2015), with the exception of σ54 (σN) that forms a distinct family on its own (Fig. 2C and D) (Merrick 1993). Whereas bacteria have in general multiple σ70 type factors, they commonly bear only one σ54 family member. σ70 family members are modular proteins and are divided into four groups based on sequence conservation and domain structure and composition (Fig. 2C) (Campbell et al.2002; Paget and Helmann 2003). Much information was retrieved from the crystal and co-crystal structures of σA, the housekeeping σ factor from the extreme thermophilic Gram-negative bacterium Thermus aquaticus (Campbell et al.2002). Housekeeping σ factors generally belong to group 1 and are composed of four conserved domains connected by flexible linkers (Paget 2015). Each domain is predicted to bind both core RNAP and DNA. Domain 1 (σ1) is unique to group 1 members and its N-terminal sequence (σ1.1) inhibits the binding of free σ factor to the promoter, unless the σ factor has bound core RNAP. This mechanism avoids promoter binding by an isolated σ subunit, which would result in frequent inhibitory and non-productive promoter occupation. Autoinhibition of DNA binding exerted by σ1.1 relies on its high negative charge, which allows it to act as a DNA mimic (Young, Gruber and Gross 2002; Schwartz et al.2008). In addition, region 1.1 plays an important role in the interaction of the holoenzyme with DNA, whereby it facilitates open promoter complex formation (Wilson and Dombroski 1997; Ruff, Record and Artsimovitch 2015). Domain 2 (σ2) contains an exposed α-helix 2.2 predicted to form a primary interface with the core, whereas the region 2.3–2.4 helix interacts with the –10 promoter sequence. It contains aromatic residues important for DNA melting and interaction with the non-template strand of the –10 promoter sequence. Both σ3 and σ4 contain three α helices. One helix in σ3 interacts with the conserved TG dinucleotide of extended –10 promoters. Two helices in σ4 specifically interact with the –35 promoter element. σ factors belonging to group 2 are closely related to group 1 members but are generally dispensable for growth (at least in laboratory conditions). One of its best-studied members is E. coli σS (RpoS), responsible for the general stress and starvation response, and specific gene expression in the stationary growth phase (Landini et al.2014). Its concentration in the cell is highly regulated and varies depending on the nature of the stress and the growth conditions (see below for details). About 23% of all E. coli genes are regulated in response to changes in the level of σS, though to a different degree (Wong et al.2017). A core regulon of 63 genes associated with EσS upon transition from exponential to stationary phase growth was identified by chromatin immunoprecipitation-sequencing (ChIP-seq) (Peano et al.2015). A study performed with Salmonella further revealed that a large number of σS concentration-dependent genes are regulated at the protein level only, indicating an important role for post-transcriptional regulation (Lago et al.2017). In contrast to the housekeeping σ0 factor, σS was recently shown to adopt an open conformation in solution in which the folded σ2 and σ4 domains are interspersed by domains with a high degree of disorder (Cavaliere et al.2018). This open configuration of σS also provides insight into a possible mechanism for regulation of its activity by the chaperone Crl (see below). σ factors belonging to group 3 only contain the domains 2, 3 and 4 (Fig. 2C). They are a. o. responsible for induction of gene expression upon heat shock, sporulation and flagellar biosynthesis. The σ factors from group 4 are among the smallest, bearing only domains σ3 and σ4. They constitute the ECF family, the largest and most divergent class of alternative σ factors (Helmann 2002; Souza et al.2014; Campagne, Allain and Vorholt 2015). They are generally involved in regulation of cell surface and transport systems, and are frequently co-transcribed with a trans-membrane anti-σ factor that interacts with and controls the activity of the cognate σ factor.
σ54 factors were initially identified in the context of nitrogen assimilation, but are also involved in a. o. the utilization of alternative carbon sources, the assembly of motility organs and the production of extracellular alginate. They differ considerably from the σ70 family members in sequence, promoter recognition and transcription initiation mechanism (Fig. 2D) (Österberg, del Peso-Santos and Shingler 2011; Shingler 2011; Yang et al.2015; Glyde et al.2017). Yet, σ54 still binds overlapping sites on the core RNAP and competes with all other σ factors for core binding. Eσ54 holoenzymes interact with two nucleotide stretches centered around –12 and –24 (Doucleff et al.2007; Yang et al.2015). The latter is contacted in the major groove of the DNA by the recognition helix of a helix-turn-helix (HTH) motif that is present in the C-terminal domain of all σ54 proteins. This recognition helix contains nine highly conserved amino acids and is also referred to as the RpoN box. Eσ54 polymerases require an activator protein (also called bacterial enhancer binding protein) of the AAA+ family (ATPases associated with various cellular activities), many of which are part of two-component systems, and the energy of ATP hydrolysis to overcome the high energy barrier for open complex formation (Buck et al.2000; Reitzer and Schneider 2001; Shingler 2011; Bush and Dixon 2012; Yang et al.2015; Zhang et al.2016; Glyde et al.2017). σ54-associated activators frequently act in conjunction with a DNA-bending protein such as IHF (integration host factor) or CRP/CAP (cyclic-AMP receptor protein/catabolite activator protein) that facilitate DNA looping between the upstream bound activator and the RNAP.
As indicated above, all σ factors present in a cell compete for binding to the unique core RNAP. Competition will depend on the affinity for the core and the concentration of free alternative σ factor in the cell. In general, the housekeeping σ factor has the highest affinity, and is the most abundant one in most growth conditions. However, in response to a certain stimulus the concentration of available, active alternative σ factor will increase and it will then successfully compete with and in part displace the housekeeping σ factor. Different strategies are used in nature to regulate the concentration and activity of alternative σ factors in the cell and their sequential production when they are involved in complex cascades of temporal gene expression. Such strategies operate at different levels and include the rate of synthesis (both transcription and translation) and proteolytic turnover, subcellular localization, sequestration and covalent modification (including limited proteolysis) (Fig. 3A and B) (Campbell, Westblade and Darst 2008; Österberg, del Peso-Santos and Shingler 2011; Micevski and Dougan 2013; Treviño-Quintanilla, Freyre-González and Martínez-Flores 2013; Paget 2015).
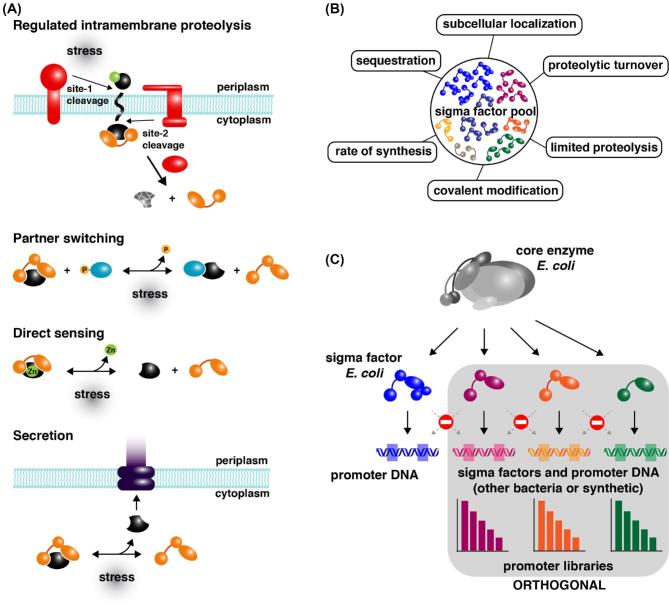
Figure 3.
Various mechanisms for regulation of σ factor activity. (A) Schematic representation of four mechanisms for regulation of σ-anti-σ factor activity involving stress-induced release of the σ factor from the inhibitory σ-anti-σ complex: regulated intramembrane proteolysis (RIP) of the anti-σ factor, partner switching, direct sensing and reduction of anti-σ concentration by secretion through the flagellar export system. Red colored symbols represent proteases. Orange colored symbols represent σ factors belonging to different groups (group 3 represented with three domains and group 4 (ECF) represented with two domains). A black colored symbol represents the cognate anti-σ factor. A blue colored ellipse with a P represents a phosphorylated anti-anti-σ factor. Zn surrounded by a green colored sphere represents a zinc atom. (B) Scheme summarizing different mechanisms used for regulation of σ factor activity in various bacteria. (C) Synthetic orthogonal gene expression system for E. coli based on the introduction of heterologous or artificial (hybrid) σ factors and their cognate specific promoters exhibiting no cross-talk, and fine-tuning of gene expression by use of promoter libraries with a broad range of transcription initiation frequencies without loss of orthogonality.
Some ‘pro-σ’ factors are activated by controlled proteolysis, whereby an autoinhibitory N-terminal extension is removed. Examples hereof are the activation of pro-σE and pro-σK in the mother cell compartment during starvation-induced sporulation of B. subtilis (Hofmeister et al. 1995; Kroos et al.1999; Zhou and Kroos 2004; Imamura et al.2008). However, post-translational control of the activity of alternative σ factors by anti-σ factors appears to be the most widespread mechanism for the regulation of σ factor activity. An anti-σ factor interacts with and stabilizes the cognate σ factor in a form that is incompatible with RNAP binding by concealing the key RNAP-binding determinants in domains 2 and 4 of the σ factor. Anti-σ factors are σ-specific and they frequently form an operon, sometimes also comprising a third gene encoding an anti-anti-σ factor. Release of the σ factor from the σ-anti-σ complex occurs in response to a signal recognized by the anti-σ itself or by additional components. Anti-σ factors are modular proteins, consisting of a σ-binding domain and a sensor/signaling domain that may respond to signals from within or outside the cell. Mechanisms known so far for the release of cytoplasmically located σ factors in response to signals are regulated proteolysis, partner switching and direct sensing (Fig. 3A) (Österberg, del Peso-Santos and Shingler 2011; Paget 2015). Regulated intramembrane proteolysis (RIP) is the mechanism that is commonly used for signal transduction across membranes for the control of ECF σ factors in response to extracytoplasmic stimuli (Heinrich and Wiegert 2009). RIP involves the sequential cleavage of a membrane-traversing anti-σ factor by two membrane-associated proteases (a ‘site-1’ and ‘site-2’ protease) resulting in the release of the cytoplasmic domain, which is bound to the σ factor. Release of the latter from this complex requires further degradation of the anti-σ factor by a cytoplasmic protease. In E. coli, RIP is used to control the release of σ24 (σE) from RseA (the cognate anti-σ factor) in response to outer membrane dysfunction (e.g. upon heat shock), while in B. subtilis σW is released analogously from its trans-membrane anti-σ factor RsiW in response to cell envelope stress induced by antimicrobial peptides or agents (Ades 2004; Heinrich and Wiegert 2009).
‘Partner switching’ is a widespread mechanism that is used by anti-σ factors whose activity is countered by an anti-anti-σ factor that sequesters the anti-σ factor in response to a particular trigger (Fig. 3A) (Herrou et al.2012). Some of these anti-anti-σ factors share structural similarities with their cognate σ factor, suggesting that they have evolved to be mimics (Francez-Charlot et al.2009). The B. subtilis group 3 σ factors σB and σF are two among the best-studied examples of partner switching. These systems consist of four key components: σ factor, anti-σ factor (a protein kinase), anti-anti-σ factor and a phosphatase complex. In absence of the trigger signal, the σ factor is kept in an inactive state by complex formation with the cognate anti-σ factor. However, the latter can alternatively bind to the non-phosphorylated anti-anti-σ factor, but anti-σ itself may phosphorylate the anti-anti-σ factor to antagonize this interaction. The phosphorylation status of the anti-anti-σ factor is thus crucial to the activity of the σ factor. In response to a trigger signal, a phosphatase will dephosphorylate the anti-anti-σ factor, thereby allowing it to bind anti-σ, which consequently results in the release of active σ (Hecker, Pané-Farré and Völker 2007). In other instances, the anti-σ factor directly senses the signal, which induces a conformational change in the protein that results in release of the σ factor (Fig. 3A). An example hereof is the σR/RsrA system of S. coelicolor (Kang et al.1999). RsrA is a cysteine-rich protein, a member of the zinc-binding anti-σ domain (ZASD) family. It lacks membrane-spanning or extracytoplasmic domains. In response to oxidative stress, an intramolecular disulfide bond is formed that triggers the release of the zinc atom and the stabilization of an RsrA conformation that is unable to bind σR (Li et al.2003).
Regulation of anti-σ activity by secretion out of the cell is for instance used in control of the flagellum biogenesis in Salmonella Typhimurium (Smith and Hoover 2009; Fitzgerald, Bonocora and Wade 2014) (Fig. 3A). Transcription of the late flagellar genes (encoding flagellin and chemotaxis functions) requires the alternative σ factor σ28 (σF, FliA), which is kept in an inactive state by complex formation with its anti-σ factor FlgM (Aldridge et al.2006). Expression of the late flagellar genes by EσF will therefore only occur when the concentration of FlgM has been sufficiently lowered by secretion through the flagellar export system. That is when the hook and basal body have already been assembled (Sainsi et al.2011).
Hereunder, we describe to some extent details of the regulation of E. coli σS synthesis and activity, and its competition for core RNAP binding with the housekeeping σ factor, as an example of the multi-layered character of bacterial regulatory mechanisms and their strong intertwinement. The concentration of σS in the cell is tightly regulated at the level of its synthesis (mainly but not exclusively post-transcriptionally) and degradation. Translation of the RpoS mRNA that is naturally repressed by the sequestration of the RBS in an extensive secondary structure present in the 5΄-untranslated region (5΄-UTR) is tightly regulated by the small non-coding RNAs (sRNAs) DsrA (osmotic shock), ArcZ (aerobic/anaerobic growth), RprA (low temperature stress) and OxyS (oxidative stress), each of which is expressed in response to a different stress condition (Repoila, Majdalani and Gottesman 2003; Mika and Hengge 2014). DsrA, RprA and ArcZ activate translation of the rpoS mRNA, whereas OxyS downregulates it. All four regulatory RNAs interact with the sRNA binding and chaperone protein Hfq (originally identified as a host factor required for replication of bacteriophage Qβ) that changes their structure. This conformational change has an impact on the interaction of DsrA, RprA and ArcZ with the 5΄-UTR and their susceptibility to degradation by RNase E, whereas OxyS appears to act through competition in the binding with Hfq (Moon and Gottesman 2011; Henderson et al.2013). Furthermore, DsrA, ArcZ and RprA are also involved in transcriptional control of rpoS, where they suppress premature Rho-dependent transcription termination by inhibiting the binding of the transcription terminator protein and thus stimulate rpoS transcription during the transition to the stationary phase of growth (Sedlyarova et al.2016). See below for more information on mechanisms of regulation of mRNA stability, RNA-binding proteins and the action of small regulatory RNAs. RpoS transcription is also indirectly activated by the alarmone ppGpp. ppGpp in conjunction with DksA stimulates transcription of both the small regulatory RNA DsrA and the anti-adapter protein IraP (see below), which have a positive effect on RpoS activity (Girard et al.2017).
Ιn exponentially growing cells, σS is rapidly degraded by the ATP-dependent protease ClpXP in a RssB (regulator of σS B, alias SprE and MviA) dependent manner (Zhou and Gottesman 1998; Zhou et al.2001). In contrast, its degradation is strongly reduced in the presence of three proteins (anti-adaptors) that act as inhibitors of RssB in response to (i) phosphate starvation (IraP), (ii) magnesium starvation (IraM) and (iii) DNA damage (IraD) (Bougdour, Wickner and Gottesman 2006; Bougdour et al.2008; Battesti, Majdalani and Gottesman 2011, 2015; Battesti et al.2012, 2013; Park et al.2017). Finally, at the transition from the exponential to the stationary growth phase the competition between the housekeeping and the stationary phase sigma factors for core RNAP binding is biased by sequestration of σ70 (sigma D) via two mechanisms: interaction with the Rsd protein (regulator of sigma D) (Patikoglou et al.2007; Hofmann, Wurm and Wagner 2011; Park et al.2013) and binding to the small RNA molecule 6S that is abundantly produced in stationary phase and mimics the structure of an open promoter complex (Wilkomm and Hartmann 2005; Wassarman 2007; Lee et al.2013). This competition is further affected by the small protein Crl of E. coli and other γ-proteobacteria that exerts a positive effect on the assembly of the EσS holoenzyme upon transition from the exponential to the stationary phase and in other stressful growth conditions (Banta et al.2013; Dudin, Lacour and Geiselmann 2013).
Alternative σ factors for orthogonal expression of synthetic circuits
The high degree of sequence conservation of the bacterial core RNAP, the specificity of σ factor-promoter recognition, the abundance and variety of naturally occurring alternative σ factors plus the possibility of generating artificial chimeric ones, combined with the observation that their activity can be modulated, turn alternative and heterologous sigma factors into outstanding candidates for applications in synthetic biology (Fig. 3C). Furthermore, their small size, especially of the ECF family of σ factors, may contribute to limit the burden for the host cell to a minimum. Synthetic biology approaches have recently been applied for the improvement of industrial microbial strains for the production of both native and new-to-the-host products (Jullesson et al.2015; Cheon et al.2016; Pandey et al.2016). One major complication in such approaches is the occurrence of imbalances in the synthesis of pathway intermediates due to lack of adequate gene expression and regulation. This leads inevitably to high metabolic burden for the host organism, which, as a consequence, results in low production efficiencies/yields (Wu et al.2016). Dynamic pathway control and orthogonal transcription to maximally uncouple pathway expression from the host's metabolism may significantly contribute to restrict these negative effects. Rhodius et al. (2013) analyzed σ factors of the ECF family originating from various bacteria for orthogonality towards each other and towards the E. coli host, and demonstrated their potential for use in complex genetic circuits. Pinto et al. (2018) assembled ECF σ factors into regulatory cascades in E. coli and B. subtilis and demonstrated that such ‘autonomous timer circuits’ introduce a tunable time delay between inducer supply and target gene activation. Bervoets et al. (2018) showed orthogonality of various σ factors and their cognate naturally occurring promoters of different strengths, all originating from B. subtilis but belonging to different groups (groups 3 and 4), towards each other and towards the host (E. coli). Furthermore, they generated promoter libraries for three of these σ factors, σB (group 3, general stress response), σF (group 3, sporulation) and σW (group 4, ECF), by randomizing the spacer sequence (or part of it) connecting the –10 and –35 promoter elements. These promoter libraries cover a wide range of transcription initiation frequencies without loss of orthogonality. Together, these elements must allow the design of complex pathways, divided into modules driven by different σ factor–promoter pairs, in which the expression of each individual gene can be fine-tuned (Fig. 3C). σW from B. subtilis and its cognate anti-σ factor (rsiW) were also used to demonstrate that ultrasensitivity from sequestration combined with positive feedback is sufficient to build a bistable switch (Chen and Arkin 2012; see Box 2 for definition of bistable switch). Such bistable switches constitute the canonical solution for building memory units (Burrill and Silver 2010). Similarly, Annunziata et al. (2017) used a σ factor originating from P. aeruginosa and its corresponding anti-σ factor in E. coli to generate an orthogonal multi-input system to control gene expression in a predictable manner. In addition to naturally occurring σ factor–promoter pairs, the modular architecture of σ factors with folded domains connected by less structured linkers allows the generation of chimeric σ factors with distinct promoter selectivity (Rhodius et al.2013). This observation further increases the capability of using multiple combinations of orthogonal expression systems in a single cell.
Box 2. Logic gates and circuits.
In analogy to electronics, logic gates in synthetic biology make use of physical elements and/or signals (e.g. gene regulatory elements, small effector molecules) to perform a logical operation dependent on inputs to produce a single output. Logic circuits are more complex as they contain combinations of several logic gates. Gates may be of different kinds and be used in different combinations (Fig. Box 2). A simple NOT gate has only one input and one output and the latter will only be produced in the absence of the former. In an AND gate configuration, the production of the output (X) relies on the simultaneous presence of at least two inputs (A and B). If only one of both inputs is present, no output will be generated. As a simple example, transcription of a gene generating the output (X) requires the presence of an activator protein (A) and an inducer molecule (B) that combines with the activator protein and enhances its DNA-binding affinity. Either one of them is not sufficient. In more complex AND gate configurations, the production of a single output may rely on more than two inputs. Inversely, a NAND gate (NOT-AND) is a logic gate that always leads to the production of output, except when all inputs are present. A NAND gate is thus complementary to the AND gate. In an OR gate configuration, the presence of either input A or B is sufficient to generate the output. Inversely, in a NOR gate configuration the generation of output is only ensured in the absence of both input signals, but inhibited by either one or both.
Toggle switch. In the context of synthetic biology and gene expression, a toggle switch corresponds to a bistable biological circuit that switches from one stable equilibrium state to another in a reversible manner depending on the input. It exhibits two exclusive states of gene expression (ON and OFF), but without other stable intermediate states.
Oscillator. In an oscillator system the level of output exhibits regular cycles around an unstable steady state. A crucial element in the generation of the oscillator behavior is the presence of an inhibitory feedback loop in gene expression in which an increase in the activity of one gene activates other genes of the circuit that ultimately will inhibit its synthesis. A second requisite of an oscillator is the presence of an in-built time delay that enables the activities of the respective genes in the circuit to fluctuate in regular cycles.
Resource allocator. A frequently encountered problem in synthetic biology and heterologous gene expression is the exhaustion of cellular resources (building blocks such as amino acids, nucleotides and energy) and basal gene expression elements (e.g. core RNAP, ribosomes). This will result in a metabolic burden and unexpected interferences between the host and circuit gene expression as core RNAP and free ribosome numbers may limit gene expression. One manner to avoid or at least reduce these risks consists in the uncoupling of heterologous gene expression from the host machinery via orthogonal transcription and/or translation and the introduction of a resource allocator that allows to control the allocation of resources between the host and the circuit genes. A resource allocator separates the regulation of the expression system (controllers) from the genes encoding the desired cellular functions (actuators). As an example, a controller that ensures the synthesis of a synthetic orthogonal 16S rRNA (that will assemble with other host-encoded ribosomal proteins and rRNA molecules into orthogonal ribosomes in competition with the endogeneous host 16S rRNA) may be combined with a transcriptional repressor that allows to adapt the amount of the orthogonal 16S rRNA produced in response to demands via a negative feedback loop (Darlington et al.2018). If the repressor is constitutively expressed at transcriptional level and translated by the orthogonal ribosome pool itself, then it will act as both a sensor and regulator of the free orthogonal ribosome pool. At low expression of the circuit genes, the ratio of orthogonal ribosomes/repressor mRNA is high and the repressor will be produced in high concentrations. As a consequence, the production of orthogonal 16S rRNA is low and only few ribosomal constituents are used to assemble orthogonal ribosomes. In contrast, when circuit genes are expressed at a higher level, the number of orthogonal ribosomes per regulator mRNA molecule decreases due to the competition with the circuit mRNAs and the repressor concentration in the cell will drop. As a consequence, the synthesis of orthogonal 16S rRNA will increase, as will the pool of cognate orthogonal ribosomes and circuit expression. Similarly the use of split T7 RNAPs in which each output of the controller (various RNAP fragments recognizing a cognate promoter sequence) combines with a common core RNAP fragment to express various gene circuits (each with a different promoter sequence) ensures that the total amount of heterologous RNAP will not cross a certain threshold imposed by the limited amount of available core fragment (the allocator) (Segall-Shapiro et al.2014).
Figure Box 2.
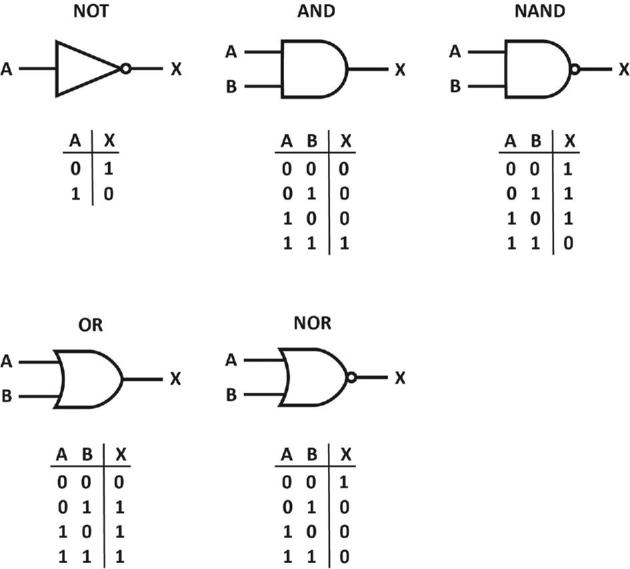
Various types of logic gates. A and B represent inputs (with absence of input indicated with 0 and presence with 1), and X the potential output.
A condition that should absolutely be guaranteed in the application of σ factors (and all other strategies based on single subunit RNAPs or transcription factors, as explained below) for orthogonal (heterologous) gene expression is that their expression in the host organism does not lead to an important burden or growth inhibitory effect (Lynch and Marinov 2015). Even though most tested sigma factors in the above cited examples meet this criterion, this was not always the case (Bervoets et al.2018). To reduce the expression level of the heterologous σ factor(s) (and the accompanying burden), and for reasons of stability of recombinant strain, it is advisable to integrate the corresponding gene into the host genome by generating a single copy knock-in. Depending on the application, σ factor expression may then be driven by a constitutive promoter of well-defined strength (naturally occurring or synthetic), an inducible promoter that can be activated by administering an external stimulus or be linked to an intracellularly produced signal, or be co-transcribed as a polycistronic mRNA with well-defined expression profile (De Paepe et al.2017; Bervoets et al.2018). In such a setup, alternative σ factor–promoter pairs used to drive heterologous gene expression in an orthogonal manner may be combined with well-characterized classical allosteric DNA-binding transcription regulators (or other regulatory mechanisms, see below) to modulate σ factor expression.
In the general context of burden, it is worth mentioning that Ceroni et al. (2018) identified the major transcriptional changes occurring in E. coli host cells upon induction of heterologous gene expression. Irrespective of the nature of the heterologously expressed protein, these appeared to consist essentially of heat-shock response-activated genes. Based on this observation, the authors developed a burden-driven feedback control mechanism that adjusts the expression of the heterologous construct in response to burden. This system relies on the knowledge of the heat-shock response and a CRISPR/dCas9-based (clustered regularly interspaced short palindromic repeats and Crispr-associated proteins) feedback regulation system to specifically target the heterologous gene construct. dCas9 is a ‘dead’ variant of the Cas9 nuclease, which in association with a small guide RNA (sgRNA) still selectively binds specific DNA sequences but no longer cleaves the DNA (Sander and Joung 2014).
Another strategy to generate orthogonal gene expression by attracting the RNAP to specific promoters relies on the exploitation of the strong promoter selectivity exhibited by single subunit RNAPs, generally of viral origin, of which T7 polymerase is certainly the best-characterized and most widely applied example. The system is renowned for its simplicity and transcriptional activity: the polymerase is very compact and the T7 promoter that is orthogonal in many organisms consists of 17 bp only, and the enzyme is highly processive and functions in a wide variety of hosts (reviewed in Borkotoky and Murali 2018). This high processivity has however a drawback since in uncontrolled conditions expression from a T7 promoter may rapidly exhaust the cellular resources resulting in a severe burden and even cell death. Recently applied strategies to reduce this strong activity are the introduction of a stop codon in the T7 RNAP that is then translated under the control of a nonsense suppressor (Angius et al.2018), the expression of an antisense gene cassette and modulation of translation of the T7 RNAP encoding mRNA with a synthetic RBS (Liang et al.2018), or the construction of a self-limiting T7 RNAP expression system (Kar and Ellington 2018).
In the most classical and commercially available application, T7 RNAP is used for the isopropyl-β-d-1-thiogalactopyranoside (IPTG)-induced overproduction of recombinant proteins from a T7-specific orthogonal promoter under control of the lac operator/repressor in E. coli. However, the system was further engineered to allow the combination of multiple orthogonal expression systems in a single cell. Thus, mutants of the polymerase with a different DNA sequence specificity have been produced to generate orthogonal polymerase–promoter pairs that allow for modular control of multiple pathways (Temme et al.2012; Meyer, Ellefson and Ellington 2015). Orthogonality was achieved by swapping the DNA-binding loop of T7 RNAP with homologous sequences mined from sequence databases. Furthermore, split variants of the T7 RNAP consisting of two, three or four parts have been used to create transcriptional AND gates for integration in genetic circuitry as well as a ‘resource allocator’ (Shis and Bennett 2013; Segall-Shapiro et al.2014) (see Box 2 for definition of gates and resource allocator). In these applications, the generation of a functional enzyme relies on the in vivo assembly of different polymerase fragments in various combinations. These fragments may be produced from different host-specific promoters under the control of distinct regulatory proteins and influenced (induced/repressed) by different effector molecules for the construction of logic gates (Iyer et al.2013). To a certain extent, such an assembly strategy mimicks the association of the bacterial RNAP core with different σ factors to generate promoter specificity.
Transcriptional regulation by DNA-binding transcription factors: repression and activation
The transcription initiation frequency of the vast majority of bacterial genes and operons is at least in part regulated (negatively and/or positively) by accessory trans-acting proteins that bind to specific DNA sequences, generally near to or even overlapping the binding site for RNAP (Fig. 4). The ensemble of all genes regulated by the same transcription factor (TF) and dispersed over the chromosome is called a regulon, as coined by Maas and Clark (1964). Bacterial DNA-binding TFs mostly use an HTH or less frequently a β-ribbon motif (ribbon-helix-helix) to bind specific DNA sequences (Nelson 1995; Schreiter and Drennan 2007). The typical bacterial HTH motif is about 20 amino acids (aa) long and comprises two α-helices (called orientation and recognition helix) separated by a short (3–4 aa) turn (Aravind et al.2005). The orientation helix interacts essentially with the sugar-phosphate backbone, whereas the recognition helix establishes sequence-specific interactions. These consist mainly of hydrogen bonds with base-specific groups at the bottom of the major groove and hydrophobic interactions with the methyl group of thymine to unambiguously distinguish the four possible Watson–Crick bps and short specific target sequences within the millions of bps of a bacterial genome (Choo and Klug 1997; Nadassy, Wodak and Janin 1999; Cheng et al.2003; Pan et al.2009). Bacterial HTH proteins generally bind as dimers (or higher oligomeric forms) to imperfect palindromes and interact with two or more major groove segments aligned on one face of the helix. Frequently they also bear an additional DNA-binding element, such as an N-terminal extension (Feng, Johnson and Dickerson 1994), a wing (wHTH) (Brennan 1993) or a hinge helix, as in the LacI/PurR family (Schumacher et al.1994) that further enhances target selectivity and binding affinity. Wings, consisting of antiparallel β-strands, and hinge helices generally interact with the minor groove segment located in between and on the same face as the major groove segments contacted by the recognition helices, whereas N-terminal extensions may interact with the opposite face of the DNA.
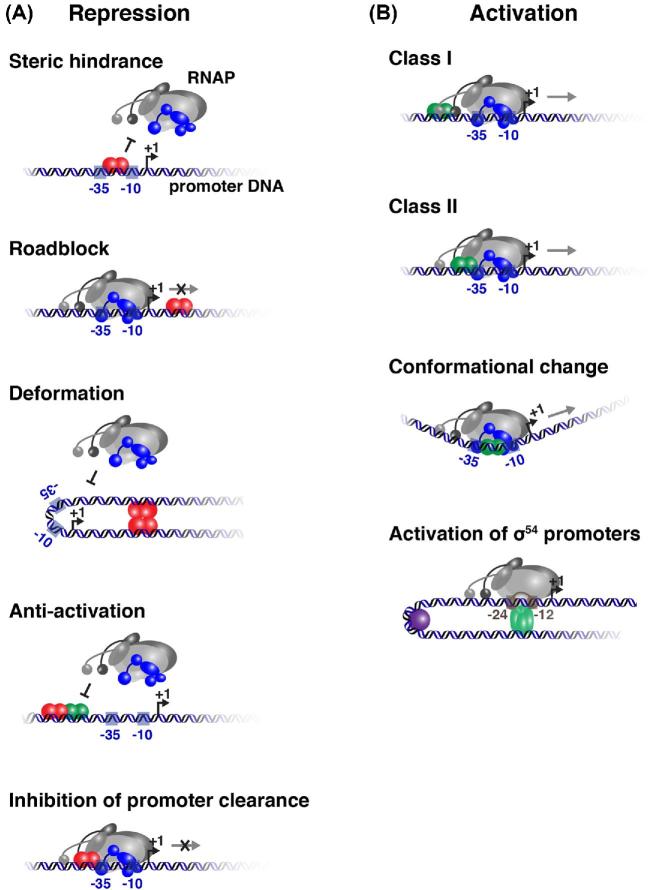
Figure 4.
Regulation of promoter activity by DNA-binding transcription factors. (A) Negative regulation by repressors (red colored symbols). Various possibilities are depicted. In repression by steric hindrance, binding of the repressor in overlap with the RNAP-binding site generates a direct competition in the binding between the regulator and the RNAP. In the roadblock model, binding of the repressor downstream of the promoter element physically inhibits the further progression of the RNAP. Pronounced DNA deformation may result from the binding of repressor proteins upstream and downstream of the promoter making the latter unsuitable for RNAP binding. Negative regulation by anti-activation may result from the mutually exclusive binding of a repressor and an activator to overlapping sites (not shown) or, as shown, from the interference of the repressor with the stimulating effect of the activator on RNAP recruitment. Finally, promoter clearance may be inhibited by a repressor protein that makes strong contacts with both DNA and RNAP thus inhibiting RNAP movement. (B) Stimulation of transcription initiation by activator proteins (green colored symbols). In class I activation, the activator binds at variable distances upstream of the –35 promoter element and makes protein–protein contacts with one or two σ-CTDs, thereby stimulating RNAP recruitment. In class II activation, the regulator binds in overlap with the promoter and makes contacts with α-NTD or σ, or with both. Some activators bind in between the –10 and –35 sequences of promoters with a suboptimal spacing and stimulate promoter activity by untwisting the DNA helix, thus generating a better alignment of the promoter elements for RNAP binding. σ54-dependent promoters require activators of the AAA+ family and the energy of ATP hydrolysis. They generally bind upstream of the promoter elements and act in conjunction with a DNA-bending protein (purple sphere) to facilitate their contact with the RNAP.
A TF that exerts a negative effect on the transcription initiation frequency of a target gene is called a repressor. Repressors can operate through many different mechanisms or combinations thereof, including steric hindrance of RNAP binding or inhibition of later stages of transcription initiation and elongation, local DNA structure alterations (bending, wrapping, looping, twisting) and counteraction of another activating TF (anti-activation) (Fig. 4A). Some TFs bind to operator sequences partially overlapping the conserved –10 or –35 promoter element, thereby sterically interfering with RNAP binding. This is how E. coli ArgR (arginine repressor) represses the P2 promoter of the carAB operon (encoding carbamoylphosphate synthase) (Charlier et al.1988). Others function as a downstream bound ‘roadblock’ for transcription elongation, as does PurR (purine repressor) in regulation of the purB promoter (encoding adenylosuccinate lyase) (He and Zalkin 1992). Repressors may also inhibit transcription by competing with α-CTD for binding a UP element (Quinones et al.2006) or bind at an even larger distance from the promoter. For example, GalR (galactose repressor) binding to two distally located operators of the galETK operon induces DNA looping, which prevents binding of RNAP (Swint-Kruse and Matthews 2009). DNA looping may be facilitated by a DNA-bending protein such as IHF (Moitoso de Vargas, Kim and Landy 1989). DNA wrapping around hexameric PepA (aminopeptidase A, a trigger enzyme) generates a positive toroidal supercoil and represses transcription initiation at the P1 promoter of the E. coli carAB operon (Nguyen Le Minh, Nadal and Charlier 2016). Repression can also be accomplished by a TF acting as ‘anti-activator’, either by directly competing with the activator for binding to overlapping targets or by blocking the activating signal. An example of the latter is CytR (cytidine repressor) that interacts simultaneously with its operator DNA and the activator protein CRP/CAP, thereby counteracting the activity of the latter (Valentin-Hansen, Søgaard-Andersen and Pedersen 1996). Finally, repression may result from direct interaction of the TF with RNAP, such as protein p4 of bacteriophage Φ29 that binds both α-CTD and the DNA upstream of the RNAP-binding site, hence preventing promoter clearance (Monsalve et al.1998). Similarly, lysine-bound ArgP stimulates RNAP binding to the E. coli argO (arginine outward) promoter but sequesters the polymerase in a non-productive complex, whereas arginine-bound ArgP makes slightly different DNA contacts and activates argO transcription (Laishram and Gowrishankar 2007; Nguyen Le Minh et al.2018). Finally, protein Rv1222 of Mycobacterium tuberculosis that interacts with both RNAP and DNA inhibits transcription by anchoring the RNAP onto DNA (Rudra et al.2015). The positively charged C-terminal tail of the Rv1222 interacts with DNA and slows down the RNAP movement during transcription elongation. As this interaction with DNA is electrostatic and not sequence dependent, Rv1222 could in principle inhibit transcription from any promoter.
TFs that stimulate transcription are called activators. Activation of the E. coli arabinose operon by AraC and activation of its lac operon by CRP/CAP were the very first reported cases of bacterial transcription activation (Englesberg et al.1965; Zubay, Schwartz and Beckwith 1970). Activators generally stimulate transcription of promoters that have suboptimal promoter sequences and require an activator protein to stimulate RNAP recruitment. They can act by direct activation through different mechanisms: class I and class II activation, or activation by remodeling of the promoter DNA (Fig. 4B) (Lee, Minchin and Busby 2012). In class I activation, the activator protein binds upstream of the promoter and recruits RNAP by interacting directly with the α-CTD of the polymerase. This is how CRP, the very first transcription activator protein to have been purified, works at some promoters such as the lac operon (Benoff et al.2002; Lawson et al.2004; Shimada et al.2011). Thanks to the length and flexibility of the linker connecting α-CTD to α-NTD of the core polymerase, class I activators may bind at different distances upstream of the promoter. In class II activation, the TF binds to a site overlapping the –35 promoter element and forms direct interactions with domain 4 of σ, with α-NTD or with both (Lee, Minchin and Busby 2012; Feng, Zhang and Ebright 2016). For class II activators, there is very little flexibility in their position on the DNA. An example is autoactivation of bacteriophage λ cI transcription initiation from the λ PRM promoter for repression maintenance (Li, Moyle and Susskind 1994). It is worth noticing that class I and class II activators may function together on the same promoter, and that such an arrangement allows for promoter stimulation that is co-dependent on two distinct signals. Furthermore, some activators such as CRP may function as both class I and class II activators, dependent on the architecture of the promoter-operator region (Lawson et al.2004). A third activation mechanism consists of promoter remodeling, usually of promoters with a suboptimal spacing between the –10 and –35 promoter elements, resulting in their misalignment on the helical surface with respect to the interacting domains of σ. The best-studied examples involve members of the MerR family of metal-binding transcription regulators (Brown et al.2003). In absence of the effector molecule MerR (mercury resistance regulator) binds to a region in between the –10 and –35 promoter elements and bends the DNA away from the polymerase; hence, unliganded MerR acts as a repressor. However, from the same position Hg++-bound MerR untwists the 19 bp long linker region, resulting in a better alignment of the promoter elements, hence facilitating RNAP binding. MerR is thus both a repressor (unliganded) and an activator (liganded) (Heltzel et al.1990; Ansari, Bradner and O’Halloran 1995).
The affinity of DNA-binding TFs (repressors and activators) for particular target sequences is frequently modulated by reversible interaction with one or more small molecules or chemical modifications. They are allosteric proteins that bind inducers and/or co-repressors, or undergo reversible covalent modifications including phosphorylation-dephosphorylation (two-component and phospho-relay systems) or oxidation-reduction (disulfide bond formation) that affect their DNA-binding potential. As an example, binding of allolactose or the artificial inducer IPTG to the C-terminal domain of tetrameric E. coli LacI (lactose operon regulator) or of uracil to dimeric RutR (regulator of pyrimidine utilization) triggers a conformational change that lowers the DNA-binding affinity of the N-terminal domain of the repressor (Matthews and Nichols 1998; Nguyen Ple et al.2010; Nguyen Le Minh et al.2015). In contrast, binding of c-AMP (inducer) to CRP/CAP or of arginine (co-repressor) to hexameric E. coli ArgR strongly enhances the binding affinity and sequence specificity of these regulators for their targets (Charlier et al.1992; Mukhopadhyay, Sur and Parrack 1999; Szwajkajzer et al.2001; Lawson et al.2004). Interestingly, arginine binding to hexameric ArgR shows negative cooperativity, whereby binding of the first arginine molecule results in a 100-fold reduction in the affinity of the five remaining sites (Jin et al.2004; Strawn et al.2010; Pandley et al.2014). Positive cooperativity in effector binding lowers the concentration of effector able to act as a switch for the conversion of apo- to holoregulator. In contrast, negative cooperativity ensures buffering capacity against changes in effector concentration. Some allosteric regulators bind more than one effector molecule and these may exert opposite effects on the regulatory outcome (transforming a repressor into an activator and vice versa) on the same or distinct promoters, as observed for instance for E. coli TyrR (tyrosine repressor) and ArgP (regulator of arginine export and other genes) (Pittard, Camakaris and Yang 2005; Laishram and Gowrishankar 2007; Nguyen Le Minh et al.2018). Some TFs are therefore very versatile (see also below for more details on TyrR action).
Some DNA-binding TFs need a partner signal-sensing protein to exert a regulatory effect. This is particularly so for regulators whose activity depends on their phosphorylation status. Two-component systems consist of a sensor kinase, generally an inner membrane-bound histidine kinase that upon sensing a particular signal (mostly a physicochemical stress) performs autophosphorylation of a specific histidine residue situated on the cytoplasmic surface of the protein, and subsequently transmits the phosphoryl group to a conserved aspartate residue of the cognate response regulator (Egger, Park and Inouye 1997). The vast majority of activating response regulators is active only when phosphorylated, but there are exceptions (Gao, Mack and Stock 2007). A typical example of transcriptional regulation by a two-component system is regulation of the outer membrane proteins OmpC and OmpF by EnvZ (senor kinase) and OmpR (response regulator) in response to the osmolarity of the environment (Yoshida et al.2006). Phosphorylated OmpR (OmpR-P) functions as an activator for ompC but as a repressor of ompF. OmpR-P is also an activator of micF, encoding a small regulatory antisense RNA that negatively affects translation of ompF mRNA by sequestering the RBS site in a double-stranded RNA structure (Coyer et al.1990). This again illustrates the combined use of different regulatory mechanisms (TF and sRNA) and regulation at different stages (initiation of transcription and translation) in the flow of genetic information.
Phopho-relay systems are more complex versions of two-component systems as they contain more interacting partners. As in two-component systems, the initially autophosphorylated sensor kinase transfers the phosphoryl group from a phosphorylated histidine residue to an aspartate containing protein (a response regulator lacking an output domain). Then, the phosphoryl group is transferred to a histidine containing phosphotransfer protein, which in turn interacts with an aspartate containing protein. This transfer may occur a variable number of times, depending on the system, till the phosphoryl group is eventually transmitted to a terminal response regulator that directly affects the transcription initiation frequency. The advantages of multiple phospho-transfers are that (i) they allow the integration of different signals in a regulatory cascade with the use of different kinases and so-called connector proteins that may block phosphorylation reactions or dephosphorylate particular phosphotransfer proteins of the cascade, and (ii) allow interactions between different two-component systems (Bijlsma and Groisman 2003; Mitrophanov and Groisman 2008). A well-characterized example of phospho-relay is the initiation of spore formation with B. subtilis. Sporulation is initiated with the phosphorylation of Spo0F by one of five related kinases (KinA, KinB, KinC, KinD, KinE) that respond to different signals. Then, Spo0F-P passes on the phosphoryl group to Spo0B, which in turn transfers it to Spo0A, the key activator of sporulation (Burbulys, Trach and Hoch 1991). Two-component systems and phospho-relay cascades are abundantly present in Gram-negative (E. coli has about 30, P. aeruginosa 64, the highest number of all sequenced bacterial genomes) and Gram-positive bacteria (B. subtilis has 34) and are involved in the regulation of a variety of critical physiological functions including sporulation, natural competence, antibiotic and heavy metal resistance, transition to stationary phase, osmoregulation, etc. (Fabret, Feher and Hoch 1999; Rodrigue et al.2000). For more details on two-component systems, their signaling mechanism and partner specificity, the reader is referred to Mitrophanov and Groisman (2008), Casino, Rubio and Marina (2009, 2010), Olivera, Ugalde and Martínez-Antonio (2010). Recently it became clear that bacteria also encode many orphan regulators that lack cognate kinases, and new roles have been identified for unphosphorylated response regulators (Desai and Kenney 2017).
Another signaling mechanism consists in the oxidation/reduction of regulatory proteins (Sevilla et al.2018). As an example, modulation of the regulatory properties of OxyR, the H2O2-responsive sensor of oxidative stress and member of the LysR-type family of transcriptional regulators (LTTR), occurs through oxidation of two conserved cysteine residues and the formation of an intramolecular disulfide (Zheng, Aslund and Storz 1998; Kim et al.2002). Oxidation of the regulator alters its DNA-binding affinity and local DNA contacts according to the sliding dimer mechanism (Toledano et al.1994; Teramoto, Inui and Yukawa 2013) that is frequently observed for members of the very large LTTR family (Maddocks and Oyston 2008). Noteworthy, OxyR functions as an activator of defensive genes such as catalase in E. coli and P. aeruginosa, but as a repressor in Corynebacterium glutamicum (Zheng et al.2001; Wei et al.2012; Teramoto, Inui and Yukawa 2013; Pedre et al.2018).
From the above description, it is already evident that TFs may use very different molecular strategies to influence the transcription initiation frequency. Furthermore, the versatility of bacterial TFs is underscored by the observation that a single regulatory protein may bind different effector molecules and have a different effect on the same or different target genes, dependent on the ligand. This is illustrated here with E. coli Lrp (leucine responsive regulatory protein), the prototype of a superfamily of global regulators, also called feast/famine regulators, which are abundantly present in bacteria and archaea as well (Calvo and Matthews 1994; Newman and Lin 1995; Brinkman et al.2003; Kawashima et al.2008; Peeters and Charlier 2010; Song et al.2013). Escherichia coli Lrp, the best-studied member of the family, frequently exerts a regulatory effect on top of a more specific regulator (enhancing or counteracting its effect) or acts in conjunction with other global gene regulators and genome organizers, such as IHF, H-NS (histone-like nucleoid structuring protein), HU (heat unstable nucleoid protein), Fis (factor for inversion stimulation), CRP and ArgP (alias IciA, inhibitor of oriC initiation) to coordinate the global cellular response (about 10% of all genes) upon transitions between rich and lean nutritional conditions (Azam and Ishihama 1999). The ensemble of genes/operons submitted to regulation by such a global regulator is coined a modulon. Besides leucine, its major effector molecule, Lrp is also responsive to a varying extent to methionine, isoleucine, histidine and threonine (Hart and Blumenthal 2011). The molecular strategies employed by Lrp are diverse (van der Woude, Braaten and Low 1992; Kaltenbach, Braaten and Low 1995; Zhi, Mathew and Freundlich 1999; Pul et al.2005; Stoebel, Free and Dorman 2008; Peeters et al.2009). Depending on the specific target, Lrp may act as a repressor or an activator, and in both instances the regulatory outcome may be insensitive, enhanced or reduced in presence of the effector (Hung, Baldi and Hatfield 2002; Tani et al.2002; Cho et al.2008a). These combinations have been described as the concerted (the effect of the regulator is potentiated by the effector) and reciprocal (effector and regulator have opposite effects) manner. Effector binding influences the oligomeric state of the protein that frequently binds (cooperatively) to multiple degenerated binding sites in an operator (Chen, Rosner and Calvo 2001; Chen and Calvo 2002; Chen, Iannolo and Calvo 2005) (frequently overlapping the binding site of another TF) and induces DNA bending (Wang and Calvo 1993).
Escherichia coli TyrR is another example of a versatile TF that may act as both repressor and activator and binds several effector molecules (tyrosine, phenylalanine, ATP) that affect the oligomeric state of the protein and the affinity for different target operator sites that are generally composite and consist of strong (high affinity) and weak (low affinity) binding sites (boxes). Unliganded dimeric TyrR binds to strong boxes only, whereas hexameric TyrR formed by self-association upon binding of tyrosine and ATP binds simultaneously to strong and weak boxes. The regulatory outcome will thus depend on the nature of the effector present, composition of the target site and position of the boxes with respect to the promoter elements. As a result, all TyrR regulon members are regulated in a different manner and a single promoter may be regulated differently depending on the ligand availability (reviewed in Pittard, Camakaris and Yang 2005). As an example, tyrP transcription is slightly repressed by dimeric apo-TyrR, activated by dimeric phenylalanine-bound TyrR, and strongly repressed by hexameric tyrosine-bound TyrR (Yang et al.2004).
Many control regions bear binding sites for several repressors and/or activators allowing the integration of different signals, resulting in a coordinated, adapted and fine-tuned response. The various regulators may then act independent of each other, in a concerted manner or, in contrast, in a competitive manner. An example is the regulation of the E. coli carAB operon, encoding the sole carbamoylphosphate synthase that ensures the synthesis of all carbamoylphosphate in the cell, a precursor common to the de novo synthesis of arginine and pyrimidine nucleotides. In E. coli and S. Typhimurium, the carAB operon is transcribed from two promoters in tandem (Piette et al.1984; Kilstrup et al.1988). Initiation at the downstream promoter P2 is essentially repressed by arginine-bound hexameric ArgR that sterically interferes with binding of RNAP at P2 but is unable to block an RNAP molecule initiated at promoter P1, 67 nt more upstream (Charlier et al.1988; Wang, Glansdorff and Charlier 1998). Regulation of P1 is complex and involves an interplay between the DNA-binding TFs PurR, RutR and IHF, and the hexameric trigger enzymes (proteins with catalytic activity also exerting a gene regulatory function; Commichau and Stülke 2008) PepA (Aminopeptidase A) and PyrH (UMP-kinase). IHF-induced DNA bending and PepA-induced wrapping of the P1 control region play a major role in the elaboration of higher order nucleo-protein complexes (Minh et al.2009). IHF stimulates P1 activity in minimal medium but potentiates pyrimidine-dependent repression and affects the methylation status of the control region (Charlier et al.1993, 1994, 1995a). RutR stimulates P1 activity in a uracil-sensitive manner (Shimada et al.2007; Nguyen Ple et al.2010; Nguyen Le Minh et al.2015). PepA acts as a strong repressor of P1 activity (Charlier et al.1995b; Nguyen Le Minh, Nadal and Charlier 2016). It is required for the PurR-mediated purine-specific repression (Devroede et al.2004) but abolishes the stimulatory effect of unliganded RutR (Nguyen Le Minh, Nadal and Charlier 2016). Finally, PepA also appears to be involved in the recruitment of PyrH to the P1 control region by protein–protein contacts (Kholti et al.1998; Charlier et al.2000, Marco-Marín, Escamilla-Honrubia and Rubio 2005). Besides this complex regulation by multiple bona fide TFs and trigger enzymes, P1 is subject to stringent control (ppGpp) (Bouvier, Patte and Stragier 1984) and UTP-dependent RNAP stuttering (Han and Turnbough 1998) (see below for the mechanism of RNAP stuttering). For a complete overview of carAB regulation, see Charlier, Nguyen Le Minh and Roovers (2018).
In this context of regulation of pyrimidine nucleotide biosynthesis, it is worth noticing that different bacteria may exhibit a completely different genomic organization of the pathway and use different regulatory strategies for its control. Whereas in E. coli the pyrimidine biosynthesis genes are dispersed all over the chromosome and regulated by distinct mechanisms (or combinations thereof) including activation and repression by TFs, stringent control, UTP-dependent stuttering, transcriptional and translational attenuation, and regulation of mRNA decay, they are grouped in an operon in B. subtilis and mainly regulated by the RNA-binding protein PyrR (reviewed in Turnbough and Switzer 2008). Interestingly, B. subtilis pyrR is the result of a gene duplication (upp) followed by functional divergence, but the regulator still exhibits residual uracil phosphoribosyltransferase activity.
Besides regulation by specific TFs, bacterial gene expression is also influenced on a genome wide scale by nucleoid-associated proteins (NAPs) that are not only involved in genome structuring but also display gene silencing and anti-gene silencing activities (Dillon and Dorman 2010; Seshasayee 2014; Dame and Tark-Dame 2016). Thus, DNA bridges generated upon binding of H-NS (Dame, Wyman and Goosen 2001; Amit, Oppenheim and Stavans 2003) have the potential to trap or exclude RNAP and silence gene expression (Dame et al.2002; Dame, Noom and Wuite 2006). Consequently, H-NS that is present at a constant level per chromosome throughout the cell cycle appears to be a universal repressor (Free and Dorman 1995). This negative action of H-NS may be locally counteracted by other NAPs with DNA bending or wrapping activity, thus interfering with DNA bridge formation. This is the case for HU that has DNA-bending activity and is considered as a regulator of DNA flexibility (Luijsterburg et al.2008). HU binds DNA non-specifically but preferentially interacts with distorted DNA segments such as prebend DNA and four-way junctions (Castaing et al.1995; Dey, Nagaraja and Ramakumar 2017). HU also directly interacts with topoisomerase I, leading to differences in DNA superhelicity, which in turn affect gene expression (Broyles and Pettijohn 1986; Oberto et al.2009; Dorman 2013). Furthermore, some NAPs such as IHF, Fis, Lrp and ArgP that equally bend DNA and may counteract H-NS-mediated silencing exhibit moderate sequence specificity and may act as specific regulators (Charlier et al.1993; Rice et al.1996; Arfin et al.2000; Hung, Baldi and Hatfield 2002; Swinger and Rice 2004; Bradley et al.2007; Peeters et al.2009; Singh and Seshasayee 2017; Skoko et al.2006; Cho et al.2008b; Dey, Nagaraja and Ramakumar 2017; Nguyen Le Minh et al.2018). Hence, the boundary between conventional transcriptional activators and NAPs is becoming increasingly blurred.
Potential of allosteric DNA-binding TFs and two-component systems in synthetic biology
The regulatory potential of well-characterized allosteric DNA-binding TFs and two-component systems, and their sequence-specific interaction with particular target sequences on the DNA are appealing properties for their exploitation in synthetic biology as they may be further rewired to create novel regulatory topologies. Bacteria encode a multitude of transcriptional regulators but only few have been exploited up to date in synthetic biology. Among these, LacI, AraC and TetR are the most frequently used ones for the fine-tuned regulation and induction of gene expression and the assembly of bistable switches, oscillators and logic gates (see Box 2 for definitions) (Fig. 5A) (Gardner, Cantor and Colins 2000; Bertram and Hillen 2008; Khalil and Collins 2010; Shis et al.2014; Cheng et al.2017; Chen et al.2018). In an attempt to increase the possibility of generating more complex circuits, Stanton et al. (2014) generated an orthogonal set of TetR-family repressor/operator combinations by part mining from bacterial genomes. Zeng et al. (2018) constructed a transcriptional AND gate based on split-ligand-inducible TetR proteins and adapted it for use as a protein aggregation sensor in which the larger TetR fragment serves as a detector whereas the smaller fragment is fused to an aggregation-prone protein that serves as a sensor of the aggregation status. In this system, the expression of a split-TetR-repressible fluorescent reporter gene is proportional to protein aggregation. Two-component systems have been used for the generation of biosensors sensing heavy metals and organic pollutants, or were further engineered into chimeric systems for the detection of novel compounds (Ravikumar et al.2017). To be applicable in synthetic biology, gene expression driven and controlled by TFs must exhibit the correct ON and OFF state characteristics and present a sufficiently large dynamic range. These conditions are not necessarily fulfilled with naturally occurring regulatory systems as they may be leaky and not attain sufficiently high expression levels upon derepression or activation required for specific applications. Tuning of this dynamic range in an attempt to obtain desired ON and OFF states was performed by Chen et al. (2018), who constructed a library of promoters covering a spectrum of dynamic ranges by assembling sets of AraC- and LasR-regulated promoters containing the –10 and –35 elements from various promoters, naturally occurring in E. coli or synthetic ones. Besides chemically inducible systems genetically engineered light-sensitive sensors might be advantageously used for the control of gene expression (reviewed in Camsund, Lindblad and Jaramillo 2011). Frangipane et al. (2018) have genetically engineered E. coli with proteorhodopsin and were able to influence the movement of these bacteria in a light-controlled manner allowing the generation of complex shapes starting from a homogeneous population of freely swimming bacteria. Similarly, they successfully reproduced grayscale density images. Light induction has a number of advantages as no chemicals have to be used, its application from outside the reactor is transient and non-invasive, light of different wavelengths is easy to produce and cheap, and as there exist biological sensors responding to different parts of the visible spectrum this opens the possibility of simultaneous multichromatic control of several transcription units. Furthermore, light may serve as an interface between biological systems and computing.
Figure 5.
Theoretical examples of synthetic circuit building for orthogonal gene expression. (A) Hypothetical example of a three-level circuit for orthogonal gene expression combining various gates and integrating multiple signals. The modules on the left represent two AND gates in which inputs I1 and I2 allow the synthesis of a TF and its chaperone (green and salmon colored symbols) that will activate the production of the output O1 (magenta colored), whereas inputs I3 and I4 ensure the synthesis of the two parts of a split TF (or alternatively of a split single subunit RNAP) (purple and light blue colored symbols), which will result in the production of output O2 (taupe colored). O1 and O2 form a hetero-oligomeric TF that will allow the production O3, an orthogonal alternative σ factor (orange colored) that eventually will ensure the synthesis of the final outcome O4. However, this will only occur in the absence of the inputs I5 and I6. In the presence of I5, the formation of the hetero-oligomeric activator (O1-O2) will be inhibited by sequestration of O2 upon binding with an alternative binding partner (dark blue colored), whereas in the presence of I6 the orthogonal σ factor will be sequestered by its cognate anti-σ factor (black colored). (B) Orthogonal gene expression based on toehold switches with co-localized RNA sensing and output modules. In toehold switches, the RBS is not accessible for ribosome binding, unless the RNA secondary structure is disrupted by interaction of the mRNA with a complementary synthetic sRNA. Importantly, and in contrast to other regulatory mechanisms operating at translational level (see Figs 6–8), in toehold switches the RBS is not part of the double-stranded RNA structure, which allows more flexibility in the design of the switch and its cognate synthetic sRNA (Green et al.2014). In the example shown here, synthesis of one or more synthetic sRNA in the cell will allow ribosome binding and translation of the cognate ORFs (here represented by red, yellow, green and blue fluorescent proteins).
Regulation by covalent DNA modifications and DNA rearrangements
Other mechanisms by which the activity of bacterial promoters may be regulated involve the (reversible) chemical modification of bases or changes to the DNA sequence. These modifications may affect the affinity of the DNA for a TF or for RNAP (Nou et al.1993; Blomfield 2001). The most common bacterial chemical modification is DNA methylation, especially of adenine. It is not only used in transcription regulation, pathogenesis and virulence (van der Woude, Braaten and Low 1992; Low, Weyand and Mahan 2001; Heusipp, Fälker and Schmidt 2007), but methylated and hemi-methylated DNA are also involved in regulation of replication initiation, DNA repair, transposition and restriction/modification systems, and may be regarded as a form of bacterial epigenetic regulation (Adhikari and Curtis 2016; Casadesús 2016). Another form of local DNA sequence variation consists of tracts of variable length that repeat a single nucleotide or dinucleotide, often in the vicinity of the –35 promoter element. Here individual cells in a population carry a different number of repeats in the variable region, with each tract length corresponding to a different expression level. As an example, variation in length of a C-tract influences the interaction between the activator protein BvgA (response regulator of virulence genes) and RNAP in the promoter region for fimbrial subunits in Bordetella pertussis (Chen et al.2010). Similarly, variations in length of a short sequence repeat in the control region of the nadA adhesion gene of Neisseria meningitides affects the binding of the repressor NadR (Metruccio et al.2009). Thus, at any moment, variation in short sequence repeats ensures that a subset of cells in the population will have the optimal level of transcription activity for the given condition (van der Woude 2011). This regulation ‘by lottery’, which is driven by repetitive sequences that differ in number from one generation to another, contrasts sharply with regulation by classical TFs for which the effect can be balanced/fine-tuned to the intensity of the input signal.
Another regulatory system involving sequence variation consists in the inversion by site-specific DNA recombination of a DNA segment containing a promoter in response to a particular signal (note that this process may be affected by DNA methylation). Such invertible promoters will switch the expression either ‘ON’ or ‘OFF’, depending on their orientation with respect to the cognate ORFs. Such a DNA switching mechanism is used in the control of fimbrial gene expression in E. coli (Corcoran and Dorman, 2009). Again, such an all-or-none mechanism is in contrast with the action of TFs.
Finally, it is worth noting that some mobile insertion sequences (IS) such as IS3 carry an outward oriented promoter that may reactivate a silent gene upon transposition in its vicinity. Hence, such elements may be considered as mobile promoters (Charlier, Piette and Glansdorff 1982).
Differential regulation of RNAP activity by small molecules and substrates
Besides the binding of alternative σ factors, small ligands that directly interact with the bacterial RNAP provide a mechanism by which the enzyme can respond swiftly and efficiently to environmental changes. The best studied of such small ligands is likely the alarmone ppGpp (guanosine 3΄-diphosphate, 5΄-diphosphate) that binds near the active site of E. coli RNAP and interacts with the β and β΄ subunits (Artsimovitch et al.2004; Ross et al.2013). In E. coli and other Gram-negative bacteria, but not in Gram-positives (see below), ppGpp exerts its effects in cooperation with DksA (DnaK suppressor), a protein that binds in the secondary (dNTP substrate binding) channel of the polymerase and amplifies the regulatory impact of ppGpp by reducing the longevity of the RNAP–promoter complex (Paul et al.2004; Perederina et al.2004; Vrentas et al.2005; Lennon et al.2012). ppGpp (and pppGpp that is converted into ppGpp) is synthesized by ribosome-associated RelA (relaxed) in response to metabolic stress, when amino acid availability is restricted to the extent that translation is limited and uncharged tRNAs accumulate in the A site of ribosomes, and SpoT (a hydrolase that can also function as a synthetase). ppGpp-dependent inhibition of transcription generally occurs at promoters that form unstable open complexes (Barker, Gaal and Gourse 2001; Barker et al.2001; Magnusson, Farewell and Nyström 2005; Dalebroux and Swanson 2012; Mechold et al.2013). These so-called stringent promoters typically have a short G+C-rich sequence stretch (discriminator box) near the start of transcription that forms a high-energy barrier for the isomerization from the closed to the open complex and is non-optimally contacted by σ700 region 1.2 (Haugen et al.2006). Stringent promoters are generally associated with genes encoding products involved in translation (e.g. ribosomal RNAs and proteins), DNA replication and pyrimidine nucleotide synthesis such as the E. coli carAB and pyrBI (aspartate transcarbamylase) operons (Turnbough 1983; Bouvier, Patte and Stragier 1984; Burgos et al.2017). Stringent promoters generally also perform poorly at low concentrations of the initiating nucleotide, mostly ATP. It has been proposed that ppGpp controls expression of the translation machinery in response to sudden starvation, whereas ATP availability controls expression in response to growth rate (Paul et al.2004). Besides these negative effects, ppGpp also positively influences the expression of numerous genes and pathways, including amino acid biosynthesis and uptake, universal stress proteins and rpoS, encoding the alternative sigma factor σS that itself affects transcription initiation at σS-dependent promoters (Magnusson, Farewell and Nyström 2005). It has also been proposed that ppGpp redistributes the available RNAP between stringent (characterized by unstable open complexes) and non-stringent (characterized by lower RNAP-binding affinity and/or the requirement for an alternative σ factor) promoters (Carmona et al.2000; Kvint, Farewell and Nyström 2000; Barker, Gaal and Gourse 2001; Laurie et al.2003). ppGpp-dependent regulation is complex and not fully unraveled, and although the stringent response is rather well conserved among bacterial species, it also exhibits differences related to the particular lifestyle of the organism in both the players involved in sensing and responding to stimuli, and the downstream targets (Boutte and Crosson 2009, 2011; Corrigan et al.2016; Pulschen et al.2017). In Gram-positive bacteria, the effect of (p)ppGpp is rather indirect and mediated by lowering the intracellular levels of GTP, which leads to a decrease in transcription of mRNAs with a GTP-initiating nucleotide, including most rRNA promoters (Krásny and Gourse 2004; Srivatsan and Wang 2008; Wolz, Geiger and Goerke 2010).
The bacterial RNAP holoenzyme incorporates its four nucleoside triphosphate (NTP) substrates with different efficiency (CTP > ATP > GTP > UTP) and its activity is affected by fluctuations in the levels of these NTPs, with the concentration of the initiating NTP (iNTP) being the most crucial. Some promoters, including those of rRNA, some tRNAs and fis, require even higher concentrations of the iNTP than promoters on average. As a consequence, depletion of NTPs in extended stationary growth phase particularly affects these promoters (Murray, Schneider and Gourse 2003; Sojka et al.2011).
At some promoters such as the pyrC (dihydroorotase) and pyrD (dihydroorotate dehydrogenase) genes of E. coli and S. Typhimurium, the ratio of different NTPs will determine the choice of the starting nucleotide and this will in turn affect the potential of the resulting mRNA to form a more or less stable secondary structure at its 5΄-end (Kelln and Neuhard 1982; Frick, Neuhard and Kelln 1990; Sørensen et al.1993; Liu and Turnbough 1994; Turnbough and Switzer 2008). At high intracellular CTP/GTP ratios pyrC and pyrD, transcription will preferentially start with CTP, 6 to 7 nt downstream of the Pribnow box. This transcript has two extra C-residues at the 5΄-end that are not present in the transcript that is made at low CTP/GTP ratios and starts with a guanosine residue. The presence of two extra C-residues allows the formation of a stable secondary structure at the 5΄-end of the mRNA that sequesters the RBS and hence inhibits translation initiation. Thus, even though the initial signal is sensed by the initiating RNAP, the final regulatory outcome is situated at the translational level. However, further emphasizing the intertwinement of regulatory processes, the reduced translation initiation of the longer pyrC transcript also affects its degradation rate (Wilson, Chan and Turnbough 1987; Wilson et al.1992).
High UTP concentrations and local nucleotide sequence near the transcription start site may also result in reiterative transcription or ‘stuttering’ of the RNAP, which is the repeated addition of the same nucleotide at the 3΄-end of the growing transcript (Turnbough 2011). Stuttering is due to slippage between the growing transcript and the DNA template and typically occurs at stretches of at least three consecutive identical nucleotides in the template (Xiong and Reznikoff 1993; Cheng, Dylla and Turnbough 2001). It results in the addition of a homopolymeric RNA stretch and inhibits further downstream elongation. After the addition of a number of identical residues, the transcript can either dissociate from the initiating transcription complex or proceed to the normal addition of nucleotides and productive elongation. In E. coli, several genes and operons of the pyrimidine nucleotide metabolism (biosynthesis, uptake and interconversion, together >30 genes) are at least in part regulated by reiterative transcription (Turnbough 2011). All what is required appears to be a stretch of at least three consecutive thymidine residues in the non-template strand shortly after the start of transcription. As an example, the 5΄-end of the pyrBI transcript has the sequence 5΄-AATTTG. High intracellular UTP concentrations result in the abundant synthesis of short transcripts with the sequence AAUUUn (with n 1 to >30), and this inhibits the production of normal productive transcripts by approximately 7-fold (Liu, Heath and Turnbough 1994).
Regulation of mRNA stability
mRNA degradation plays a key role in controlling gene expression in bacteria, where decay rates can differ from less than a minute to more than an hour (Rauhut and Klug 1999). RNA stability is important for setting both the number of mRNA molecules that may be translated into functional proteins and the number of sRNAs that can exert a regulatory function (see the section Regulation of gene expression by small RNAs). mRNAs that are not actively translated are prone to Rho-dependent transcription termination and mRNA degradation by the membrane-bound degradosome, a large macromolecular complex that in E. coli comprises RNase E (endonuclease), PNPase (polynucleotide phosphorylase, exonuclease) and RNA helicase (Carpousis 2007). Degradation generally starts with internal cleavage of the mRNA by RNase E, which preferentially cuts in A+U-rich single-stranded regions, but may also be initiated from the 5΄-end (Nillson and Uhlin 1991; Mackie 1992, 2013; Luciano et al.2017). Access of the degradosome to these sites can be blocked by initiating or elongating ribosomes and, as a consequence, transcripts that have reduced translation may be subject to more rapid degradation (Deana and Belasco 2005). Additionally, though less frequently, RNA decay in E. coli starts with cleavage by other endonucleases, such as RNase III, RNase G (a paralog of RNase E), RNase P (a ribozyme involved in maturation of the 5΄-end of tRNA molecules) or RNases from toxin–antitoxin modules (Kaga et al.2002; Deutcher 2006; Richards and Belasco 2016; Masuda and Inouye 2017). RNase E can bind monophosphorylated but not di- and triphosphorylated RNA ends, and there is now growing evidence suggesting that the triphosphate structure at the 5΄-end of bacterial mRNA plays a protective role against degradation, similar to the role of the eukaryotic 5΄-cap site (Luciano et al.2017). Its conversion into a 5΄-monophosphate end would then initiate a cascade of endo- and exoribonuclease catalyzed cleavages. The RNA pyrophosphohydrolase RppH plays a crucial role in this process but even though the enzyme is capable of removing pyrophosphate from the 5΄-end of a transcript it is much more active on dephosphorylated ends. In E. coli, the conversion of the triphosphate to the monophosphate 5΄-end appears to start with the removal of the γ phosphate by an as yet unidentified enzyme, followed by the RppH-catalyzed removal of the β phosphate (Luciano et al.2017). RppH is conserved in diverse bacterial species and is involved in the degradation of many but not all mRNAs as it shows some sequence preference (two unpaired nucleotides at the end and preference for a purine in second position). Its activity is modulated by the DapF protein (diaminopimelate isomerase) that forms a complex with RppH (Lee et al.2014; Gao et al.2018). RppH is involved in various crucial cellular processes, including virulence, ribosome biogenesis and resistance to osmotic shock. Its association with an enzyme (DapF) involved in the biosynthesis of lysine and peptidoglycan is therefore intriguing, especially since the catalytic activity of the enzyme is not required for its moonlighting stimulatory action on RppH-dependent mRNA decay (Lee et al.2014). Remarkably, RNase E itself also associates with a metabolic enzyme, the glycolytic enzyme enolase, but little is known about this association (Chandran and Luisi 2006).
RNase E-dependent cleavage in intercistronic regions is also used to differentially regulate the expression of distinct genes of an operon. As an example, RNase E-dependent processing in the intercistronic papB-papA region of the bicistronic papBA mRNA involved in Pap pili synthesis in uropathogenic E. coli results in the accumulation of the papA-specific mRNA (encoding the major structural protein of the pilus shaft) over the initially produced bicistronic papBA and papB mRNA (encoding the PapB transcriptional regulator), the latter being rapidly degraded (Nilsson and Uhlin 1991; Nilsson, Naureckiene and Uhlin 1996). Additional differential expression of more downstream located genes in the papBAHCDJKEFG operon is generated by partial transcription termination at a terminator, situated between papA and papH, that considerably reduces the amount of downstream transcripts (Båga et al.1985).
Evidently and inevitably, mRNA decay rates will be directly linked to other gene regulatory processes affecting the translation initiation frequency such as translational attenuation and translation elongation rates, to the binding of small antisense RNAs and proteins, and to environmental effects affecting mRNA structure such as temperature, pH, salt concentration and the binding of small effector molecules, mainly to the 5΄-UTR (see the sections Regulation of gene expression by RNA-binding proteins, Riboswitches: RNA sensors and RNA thermometers, and Regulation of gene expression by small RNAs). Furthermore, the degradation of some mRNA species in E. coli is regulated in a growth rate-dependent manner (Nilsson et al.1984).
Regulation of gene expression by RNA-binding proteins
Regulatory proteins that bind specific RNA sequences or structures (RBPs) may modulate gene expression through different mechanisms: (i) directly affect the susceptibility of the target RNA to degradation, (ii) modify the accessibility of the RBS for ribosome binding (either positively or negatively), (iii) act as a chaperone for interaction of the target with other effector molecules (including sRNAs) and (iv) alter the formation of transcription terminator/anti-terminator structures (Fig. 6) (Duval et al.2015). Just as ribosomes, some RBPs including Hfq may exert their effect by directly shielding the recognition sites of RNases in mRNAs or small regulatory RNAs, thus protecting them from degradation (Moll et al.2003). Hfq thus not only promotes or facilitates the interaction between small regulatory RNAs (sRNA) and their target mRNA, as illustrated above (regulation of σS), but may also directly protect mRNAs and sRNAs from RNase E cleavage, as demonstrated for the OmpA mRNA and the sRNAs DsrA and RyhB (Moll et al.2003). Other RBPs affect the secondary structure of their cognate mRNA targets upon binding, resulting in RNase recognition sites that become buried or more exposed, thus positively or negatively affecting the stability of these molecules (Fig. 6A). Furthermore, RNA-modifying enzymes or RBPs that actively recruit RNases may influence the mRNA turnover (see the section Regulation of mRNA stability) (Fig. 6C).
Figure 6.
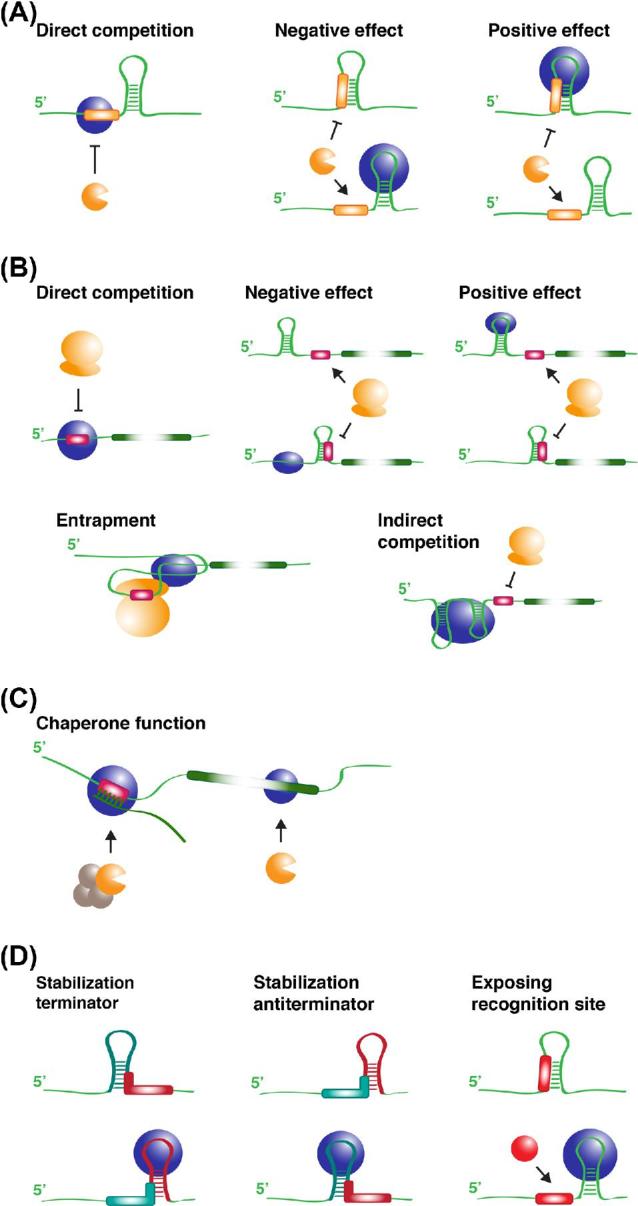
Regulation of mRNA stability and regulation of bacterial translation and transcription with RNA-binding proteins (RBP). (A) Regulated access to ribonucleases. A RBP (blue colored) may directly compete with the binding of a nuclease (yellow colored) to an overlapping site on the mRNA, liberate a recognition site for a ribonuclease on the mRNA resulting in a negative effect on gene expression or favor the formation of a secondary structure in which the target site (yellow colored box) for the ribonuclease is not readily accessible. The latter results in a positive effect on gene expression. (B) Regulated translation initiation. A RBP may (i) directly compete with the ribosome (30S subunit) for binding to the RBS (magenta box), (ii) favor the formation of structure in which the RBS is trapped in a double-stranded secondary structure, (iii) favor the formation of a secondary structure that liberates the RBS, hence facilitating ribosome binding and stimulating translation, (iv) inhibit translation initiation by stabilizing a complex in which the 30S subunit of the ribosome is trapped in an incompetent state by the RNA and (v) indirectly inhibit translation initiation by generating a steric clash with ribosome binding to the RBS. Green colored rectangles represent ORFs. (C) Improved access to proteases by the chaperone function of the RBP that may act in conjunction with a small regulatory RNA (green line) complementary to the mRNA. (D) Transcriptional attenuation. A RBP may positively or negatively affect premature transcription termination by stabilizing a terminator structure (dark red colored hairpin), or, inversely, by stabilizing an anti-terminator structure (sea-green colored hairpin), or still by exposing a recognition site for a transcription terminator protein such as Rho (bright red colored) (or alternatively a target site for a ribonuclease).
Translation initiation requires binding of ribosomes to the RBS. However, accessibility of the RBS is not only important for translation initiation but also influences the extent to which the mRNA may be degraded or even completely transcribed. Therefore, bacteria have evolved a variety of distinct mechanisms to regulate gene expression by limiting the accessibility of the RBS for ribosome binding (Fig. 6B). One such mechanism concerns negative autoregulation, where a RBP binds to the 5΄-end of its own transcript, thus shielding the RBS and inhibiting translation initiation. Examples of this strategy are ribosomal proteins in E. coli and Vibrio cholera (Zengel and Lindahl 1994; Allen et al.2004) and the thrS operon (encoding threonyl-tRNA synthetase) of E. coli, where molecular mimicry (5΄-leader mRNA structure resembles the tRNA substrate) is used for binding (Springer et al.1985, 1986). It is thus a widespread mechanism that extends beyond the Enterobacteriaceae. There are however several ways by which RBPs can inhibit translation. The most common is a ‘competitive’ mechanism, where the RBP and the 30S ribosomal subunit compete for binding to the RBS (Fig. 6B). In another mechanism called ‘entrapment’, the 30S subunit is trapped at the RBS during initiation to prevent further steps in the translation process (Fig. 6B). Here, a ternary complex is formed between the mRNA, the 30S subunit and the regulatory RBP, in which the 30S subunit is trapped in an incompetent state by the mRNA and this state is stabilized by the RBP (Springer et al.1985; Philippe et al.1993). The trapping mechanism appears to be very efficient to inhibit translation when the repressor concentration and/or its affinity is low. Lastly, an RBP may change the secondary structure of the region surrounding the RBS and influence ribosome binding at a distant region (Fig. 6B).
Besides autoregulatory RBPs, bacteria may contain a number of dedicated regulatory RNA-binding proteins such as BpuR, CspA, CsrA, PyrR and TRAP. Likely the best-characterized example is E. coli CsrA (carbon storage regulator A) and its othologs RsmA/RsmE (repressor of secondary metabolites) in Erwinia carotovora and Pseudomonas. CsrA affects the simultaneous expression of various mRNAs involved in multiple processes, including carbon metabolism, peptide transport, biofilm formation, motility, quorum sensing and virulence (reviewed in Romeo, Vakulskas and Babitzke 2013). CsrA may either inhibit or stimulate gene expression (Romeo, Vakulskas and Babitzke 2013; Duval et al.2015; Vakulskas et al.2015; Potts et al.2017). CspA (cold-shock protein A) plays a major role in the control of cold-shock genes (Gualerzi, Giuliodori and Pon 2003). The activity of some RBPs is modulated by interaction with a small effector molecule. They act as allosteric regulators. Thus, binding to and stabilization of particular secondary structures (anti-anti-attenuators) in the 5΄-leader by PyrR (pyrimidine regulator) and TRAP (tryptophan-regulated attenuation protein) from B. subtilis is modulated by the concentration of PRPP (phosphorybosylpyrophosphate) and UMP/UTP, and tryptophan, respectively (Babitzke and Yanofsky 1993; Lu and Switzer 1996; Babitzke 2004; Turnbough and Switzer 2008). PyrR and TRAP affect premature transcription termination (attenuation type of control) in pyrimidine and tryptophan biosynthesis, respectively (Fig. 6D). In addition, TRAP binding also represses translation initiation of the first ORF of the polycistronic trp mRNA of B. subtilis. Furthermore, the activity of TRAP is regulated by anti-TRAP that is induced upon accumulation of uncharged tRNATrp and forms a complex with TRAP, in competition with mRNA binding to the RNA-binding domain of TRAP (Valbuzzi and Yanofsky 2001; Valbuzzi et al.2002).
Some other RBPs constitute a platform for the binding of other molecules (small RNA or protein) that will affect RNA stability or translation efficiency (Fig. 6C). Thus, Hfq binds the sRNAs DsrA, RprA, ArcZ and the mRNA in the context of post-transcriptional regulation of σS production (Battesti, Majdalani and Gottesman 2011; see regulation of σS in the section Regulation of gene expression by sigma factor competition), whereas other regulatory RBPs bind proteins facilitating mRNA degradation, such as PapI (pap operon regulatory protein) or the degradosome (Carpousis 2007; De Lay, Schu and Gottesman 2013).
Another mechanism by which RBPs affect gene expression is by modulating transcription elongation (Santangelo and Artsimovitch 2011). Transcription by the RNAP proceeds until a terminator is reached. At intrinsic terminators, dissociation of the elongation complex is dependent on the mRNA sequence and structure, while factor-dependent terminators need the action of a protein factor such as Rho. Typically, these terminators are present at the end of an operon. However, some exist conditionally within the 5΄-leader region of the transcript. In case of intrinsic termination, RBPs can either favorize the formation of a terminator structure (also called attenuator) by binding to an anti-anti-terminator (and thus inhibiting the formation of the anti-terminator) or the formation of an alternative anti-terminator structure, which prevents the formation of the terminator (Fig. 6D). Generally, the terminator and anti-terminator are mutually exclusive structures. As an example, the ribosomal protein L4 of E. coli interacts with a small hairpin structure in the polycistronic S10 mRNA encoding 11 small ribosomal proteins and in conjunction with NusA causes premature transcription termination at a Rho-independent termination site (Stelzl et al.2003). The activity of this type of RBPs is frequently modulated by binding of a small effector molecule, or controlled via their phosphorylation status. Examples are PyrR/PRPP-UTP and TRAP-trp of B. subtilis, as indicated above. A well-characterized family of widespread bacterial RNA binding anti-terminator proteins with members in Gram-negative and Gram-positive bacteria whose activity is modulated by phosphorylation/dephosphosphorylation is BglG. BglG family regulators control the expression of enzyme II (EII) carbohydrate transporters of the phosphotransferase system (PTS). Escherichia coli BglG, the prototype of the family, regulates expression of the bglGFBH operon that besides the regulator itself encodes the aryl-β-glucoside transporter BglF that belongs to the phosphoenolpyruvate-dependent PTS, and other proteins involved in the utilization of aryl β-glucoside sugars. Only dimeric BglG positively regulates the expression of the bgl operon by binding to sites that partially overlap two Rho-independent transcription terminators, thus stabilizing an alternative anti-terminator conformation (Mahadevan and Wright 1987; Houman, Diaz-Torres and Wright 1990). BglG consists of an RNA-binding domain and two homologous PTS regulation domains, PRD1 and 2, which contain two conserved phosphorylatable histidine residues that are important for the anti-terminator activity of BglG (van Tilbeurgh and Declerck 2001). To be able to dimerize, the PDR1 domain of BglG has to be reversibly dephosphorylated by BglF, which only occurs when BglF is engaged in sugar transport (Amster-Choder and Wright 1992; Görke 2003). In the absence of β-glucoside substrates, BglG is recruited to the cell membrane by BglF that is phosphorylated by enzyme I of PTS and HPr (histidine containing phosphocarrier protein) and phosphorylated BglF and then passes its phosphoryl group to a histidine residue present in the PDR1 domain of BglG, thus inactivating the regulator. In the presence of β-glucosides, however, BglF will rather transfer its phosphoryl group to the sugar molecules and BglG gets dephosphorylated at this site. In addition to this dephosphorylation reaction, activation of BglG requires phosphorylation at histidine residue 208 in its PDR2 domain, a reaction that is catalyzed by HPr (or its paralog FruB) (Rothe et al.2012). The latter appears to act as a carbon catabolite mechanism that downregulates BglG activity when other PTS sugars are available in addition to β-glucosides. In these conditions, sugar transport will drain phosphoryl groups from HPr, leading to less phosphorylated BglG (Görke and Rak 1999). Furthermore, RBPs can also play a role in Rho-dependent termination by inducing a change in the secondary structure of the mRNA, thus exposing a Rho-utilizing site (rut) that is otherwise inaccessible (Fig. 6D).
Finally, RNA-binding proteins may also function as anti-terminators by interacting directly with the elongating RNAP complex and acting at a distance (Greenblatt, Nodwell and Mason 1993; Santangelo and Artsimovitch 2011). The first reported and best-characterized example hereof is the bacteriophage λ protein N that allows the transition between the transcription of early and middle genes of the bacteriophage (Gottesman, Adhya and Das 1980). N binds to the ‘nut’ sites (N-utilization) in the early mRNA molecules initiated from the promoters PR and PL and functions as an anti-terminator, allowing the host RNAP to proceed beyond the terminators tR1 and tL1 (Barik et al.1987; Nodwell and Greenblatt 1991; Rees et al.1997). N binds to a 15 nt stem loop structure near the 5΄-end of the PL and PR transcripts, moves along with the elongating RNAP and acts in conjunction with the host encoded Nus proteins (N-utilizing substance) NusA, B, G and ribosomal protein S10 to override the downstream terminator sites (Schauer et al.1987; Mason and Greenblatt 1991; Mogridge, Mah and Greenblatt 1995; Gusarov and Nudler 2001; Conant et al.2008). Remarkably, NusA is itself part of the E. coli termination machinery but in conjunction with N it participates in anti-termination.
Riboswitches: RNA sensors and RNA thermometers
Some mRNA molecules have the capacity to fold into complex and very specific tertiary structures, generally in their 5΄-UTR, that can act as sensors for different types of chemicals and physical parameters such as small ligands and temperature or pH (Serganov and Nudler 2013) (Fig. 7). Aptamers or natural mRNA sensors that specifically recognize small molecules are better known as riboswitches as they have the ability to ‘switch’ between alternative mRNA conformations that alter gene expression. As riboswitches are an integral part of the mRNA molecule they exclusively act in cis. Riboswitches consist of two distinct but overlapping domains: an aptamer domain that contains the selective binding site and an expression platform that influences gene expression. They act as genetic switches in response to various metabolites including amino acids, vitamins, S-adenosyl methionine, guanine, FMN, thiamine pyrophosphate, ions and second messenger molecules such as c-di-GMP [Bis-(3΄-5΄)-cyclic dimeric guanosine monophosphate] that play an important role as signaling molecule in various processes in both Gram-negative and Gram-positive bacteria, including the control of community behavior, motility, biofilm formation, cell morphogenesis and virulence (Winkler, Cohen-Chalamish and Breaker 2002; Winkler, Nahvi and Breaker 2002; Smith et al.2011; Bordeleau et al.2015; Furukawa et al.2015; Peltier and Soutourina 2017). Riboswitches may affect both translation initiation (by sequestering or freeing a RBS) and transcription termination (attenuation control, by favorizing the formation of either terminator or anti-terminator structures) (Fig. 7A and B). Furthermore, changes in RNA conformation due to ligand binding can directly regulate access to Rho- or RNase E-binding sites, or still may affect mRNA stability via self-cleavage through ligand-induced ribozyme activity (Winkler, Cohen-Chalamish and Breaker 2002; Winkler, Nahvi and Breaker 2002; Serganov and Nudler 2013). Many riboswitches even combine multiple mechanisms that act in parallel and a single transcript may contain more than one sensor domain, each one binding a specific ligand. In such an organization, they have the potential to act as a two-input logic gate where both cognate compounds are required for modulation of gene expression to occur (Sharma, Nomura and Yokobayashi 2008).
Figure 7.
Riboswitches: RNA sensors and RNA thermometers. (A) Ligand (magenta sphere) induced formation of transcriptional terminator or anti-terminator RNA structures in the mRNA (green colored) that result in premature transcription termination of the RNAP (gray colored) or allow downstream transcription elongation, respectively. (B) Translational attenuation mechanisms whereby ligand binding traps the RBS (magenta box) in a RNA secondary structure and is not accessible for ribosome (yellow colored) binding or, inversely, stabilizes an alternative structure and frees the RBS, thus allowing initiation of translation. (C) RNA thermometer. Here the RBS is trapped in a secondary structure and not available for ribosome binding. Melting of this structure in a zipper-like manner at increasing temperatures liberates the RBS, thus allowing translation initiation. (D) Reversible and temperature-dependent formation of alternative secondary RNA structures that may affect premature transcription termination or translation initiation.
In addition to small molecules, different RNA folds, which depend on thermodynamic stabilities, are strongly affected by temperature (Fig. 7C and D). Different thermoresponsive elements, also called RNA thermometers, have been described that allow translation at both elevated or depressed temperatures, and regulate genes associated with cold- or heat-shock response, and virulence genes (Johansson et al.2002; Gualerzi, Giuliodori and Pon 2003; Kortmann and Narberhaus 2012). Examples are the rpoH (σH), agsA and hsp17 genes mediating heat-shock response in respectively E. coli, Salmonella enterica and Synechocystic sp. PCC 6803 and the cIII gene of bacteriophage lambda that favors lysogeny (Altuvia et al.1989; Morita et al.1999; Waldminghaus et al.2007; Kortmann et al.2011). RNA thermometers tend to be relatively simple structures compared to riboswitches that bind small ligands. As a consequence, their presence is difficult to identify/predict in genomic sequences. As riboswitches and RNA thermometers do not require any other regulatory element besides the mRNA itself, and as they exist in the three domains of life, they might represent the oldest forms of gene regulation, already present in the hypothesized RNA world. All currently identified naturally occurring RNA thermometers regulate gene expression at the level of translation initiation. However, recently, the temperature-dependent destabilization of RNA thermometer structures (combined with a U-stretch) was exploited to construct a temperature-responsive transcription terminator (Roßmanith, Weskamp and Narberhaus 2018).
Finally, riboswitches may also sense and interact with uncharged tRNA molecules. In Gram-positive bacteria such as B. subtilis, the expression of many aminoacyl-tRNA synthetase genes and genes involved in amino acid biosynthesis and uptake are regulated by an anti-termination mechanism, in which a specific uncharged tRNA interacts with a cognate ‘T box’ sequence in the 5΄-leader region of the transcripts (Condon, Grunberg-Manago and Putzer 1996; Putzer et al.2002; Green, Grundy and Henkin 2010). This interaction stabilizes an anti-terminator element that competes with the terminator, and hence promotes expression of downstream coding sequences.
Regulation of gene expression by small RNAs
mRNAs can also adopt structures that act as sensors for small regulatory RNAs that affect gene expression via RNA–RNA interactions (Saberi et al.2016). Anti-sense regulatory RNAs can be divided into cis-acting and trans-acting regulatory molecules. Cis-acting RNAs are antisense RNAs, transcribed from the opposite DNA strand and show perfect bp complementarity to the mRNA target. They can vary in size from a few tens to thousands of nucleotides and their abundance in different bacteria varies extensively (Thomason and Storz 2010). Anti-sense RNAs can affect transcription in essentially two manners: transcription interference or transcription attenuation (Fig. 8A). Transcription interference occurs when transcription initiated from one promoter is restrained by the action of a second promoter in cis. Attenuation is a mechanism that causes premature termination of transcription. Base pairing of antisense RNA to the cognate mRNA may induce the formation of a Rho-independent terminator, resulting in an aborted non-functional transcript, or in contrast, induce the formation of an anti-terminator structure thus allowing production of a full-length functional transcript. Another potential mode of regulation by antisense RNAs is to either promote or inhibit degradation of the target mRNA by endo- or exoribonucleases. In bacteria, the endoribonucleases RNase III and RNase E have for instance been linked to RyhB (sRNA regulator of iron acquisition and utilization) antisense RNA-induced target cleavage of sodB mRNA, encoding superoxide dismutase B (Afonyushkin et al.2005).
Figure 8.
Regulation by small RNAs (sRNAs). (A) Regulation of mRNA translation (top part) and transcription attenuation (bottom part) by cis-acting antisense sRNAs. A cis-acting sRNA (blue colored) encoded from the opposite strand as its target gene (green colored) inhibits gene expression at the post-transcriptional level by sequestering the ribosome-binding site (magenta colored rectangle) in a double-stranded RNA secondary structure. Transcriptional attenuation results from sRNA induced formation of a terminator (attenuator; red colored hairpin) structure in the mRNA at the expense of the anti-terminator. (B) Three methods illustrating gene regulation by trans-acting sRNAs. Trans-acting sRNAs are only partially complementary to the target RNA and frequently require the help of a chaperone protein (blue ellipse) such as Hfq in E. coli for stabilization, folding and target binding. Due to the limited bp complementarity with the target, trans-acting sRNAs may bind several distinct mRNAs and either inhibit translation by sequestering the RBS, which may also lead to accelerated mRNA degradation (top part), or inversely free the RBS and hence stimulate translation (middle part). Finally, trans-acting sRNAs bearing multiple binding sites for an inhibitory RNA-binding protein (RBP, blue colored ellipse) indirectly stimulate translation of one or more mRNAs by titration/sequestration of the RBP (bottom part).
Some cis-encoded antisense RNA molecules are produced from plasmids as part of copy-number control systems (RNAI/RNAII of ColE1) (Lacatena and Cesarini 1981), toxin/antitoxin systems (Afonyushkin et al.2005; Thomason and Storz 2010), the production of potentially toxic proteins such as SymE (Fozo, Hemm and Storz 2008) or are involved in the control of transposition frequencies via regulation of transposase synthesis, as observed for IS10 and Tn10 (Kleckner et al.1984). The SymR antisense RNA overlaps the 5΄-end of symE mRNA thus trapping the RBS in a double-stranded RNA structure, which results in repression of symE mRNA translation and enhanced mRNA degradation. Additionally, base pairing between sense and antisense RNA may impact translation of the target mRNA at a RBS distant from the region of interaction by altering the mRNA structure. In most instances, the utilization of antisense RNA as a regulation strategy adds an extra layer of regulation to systems whose expression is already extensively controlled by other mechanisms.
Trans-acting small regulatory RNAs are encoded from loci other than their target genes (Fig. 8B). They are generally about 100 nucleotides long and share limited sequence complementarity with their target and may therefore act on multiple mRNAs in response to environmental cues as pH, temperature and nutrient depletion (Thomason and Storz 2010). Regions of complementarity to the target consist of a seed region of 6–8 contiguous bp, which is the most conserved part of the sRNA. In E. coli, the RNA-binding protein Hfq is generally required to mediate sRNA–mRNA hybrid formation with trans-acting small regulatory RNAs. In this role, Hfq acts as a RNA chaperone to facilitate bp formation and stabilizes the sRNA against degradation (Peer and Margalit 2011; Desnoyer and Massé 2012). sRNAs make use of a variety of mechanisms such as direct binding to the RBS, enhancement of translation by interaction with a transcript that inhibits formation of mRNA secondary structures, guiding Hfq to a site for direct inhibition of translation or titration/sequestration of a RBP by binding to multiple binding sites on the sRNA (Fig. 8B). The latter is how several trans-acting sRNAs including RsmW, RsmY, RsmZ and RsmV operate to titrate the RNA-binding proteins RsmA and/or RsmF that affect mRNA translation of genes involved in the inverse control of acute and chronic virulence in P. aeruginosa (Janssen et al.2018a, b). As with other forms of regulation, repression of translation by sRNAs may lead to mRNA decay. A single transcript may be targeted by several sRNAs as it occurs for the rpoS transcript, involved in stress response and stationary phase growth, that is targeted by DsrA, RprA and ArcZ that all bind the same region of the rpoS transcript to alter its secondary structure, resulting in enhanced translation efficiency and increased rpoS mRNA stability in a highly additive manner (Battesti, Majdalani and Gottesman 2011) (see also above regulation of σS).
Regulatory RNA and synthetic biology
Riboswitches may be advantageously exploited to control gene expression in synthetic pathways as they show (or may be engineered to) high selectivity and dose-dependent control of gene expression and may be used in different hosts. Indeed, as riboswitches do not involve the action of proteins they are supposed to be better transferable to different hosts, to be less cost intensive and faster than protein mediated gene regulation. Furthermore, as they show a modular composition of sensor and response elements they may be combined in a ‘plug-and-play-like’ mode (Etzel and Mörl 2017). However, as naturally occurring riboswitches generally bind cellular metabolites or molecules taken up by the cell such as amino acids and ions, they do not work in an orthogonal manner and their artificial induction may interfere with the host metabolism and vice versa variations in the host metabolism may compromise the use of the riboswitch as a regulatory element for heterologous gene expression. Therefore, engineering is necessary if riboswitches have to be exploited in synthetic biology. This is, however, not evident as there is a tight link between the riboswitch itself and the activity of the RBS that is generally affected by the riboswitch. Nevertheless, riboswitches have considerable potential since synthetic aptamers binding a small molecule of interest can be selected from randomized RNA libraries and the modular design of riboswitches allows the construction of chimeric ones by exchange of aptamer domains and expression platforms (Muranaka et al.2009; Garst, Edwards and Batey 2011; Vinkenborg, Karnowski and Famulok 2011; Ceres et al.2013; Robinson et al.2014; Folliard et al.2017; Harbaugh et al.2018). However, selective ligand binding alone is not sufficient. In order to function as a regulatory switch, the RNA has to undergo a clear ligand induced conformational change with appropriate kinetic and thermodynamic parameters that is translated in a regulatory outcome. Such artificial riboswitches may be selected by a combination of library screening for ligand binding by SELEX (Systematic Evolution of Ligands by Exponential Enrichment) and screening for expression using a dual-reporter gene system (Findeiß et al.2017; Groher et al.2018; Harbaugh et al.2018; Lotz and Suess 2018). Artificial riboswitches may also be generated by dedicated engineering starting from naturally occurring riboswitches and exploiting structural data. Thus, starting from an adenine responsive riboswitch Robinson et al. (2014) created novel orthogonal and chimeric riboswitch–synthetic ligand pairs with superior in vivo gene induction properties that modulate either transcription or translation and are transferable in Gram-negative and Gram-positive bacteria. As a proof of concept, they were shown to regulate bacterial motility genes of E. coli and morphology genes in B. subtilis. In combination, and embedded in the 5΄-UTR of a single mRNA, artificial riboswitches could also be used to engineer logic gates such as AND and NAND (see Box 2 for definitions) (Sharma, Nomura and Yokobayashi 2008). Limitations in the large-scale application of riboswitches as regulatory elements may a. o. be due to potential sensitivity to the genetic context (some riboswitches also affect the structure of the downstream located ORF), low tunability and variability in performance. Furthermore, to be really useful in synthetic biology for a wide range of applications, riboswitches must be modular. In an attempt to overcome these shortcomings, Folliard et al. (2017) used the activating addA riboswitch (binds 2-aminopurine) from Vibrio vulnificus and the repressing btuB riboswitch (binds adenosylcobalamin) from E. coli to develop ribo-attenuators. Riboattenuators consist of a short sequence, bearing a second RBS that is sequestered by a local hairpin, that is inserted between the primary RBS (sequestered by the riboswitch) and the gene of interest. This local hairpin can be opened by a ribosome initiated from the upstream located riboswitch RBS, thus allowing translation of the gene of interest upon freeing the RBS by ligand binding. Advantages of such a system are (i) that it insulates the downstream gene from conformational changes induced by ligand-induced modifications of the riboswitch structure and (ii) reduces variations in expression levels. Similar to metabolite-sensing riboswitches, RNA thermometers can be exploited for the efficient and specific control of gene expression and libraries of RNA thermometers with diverse sensitivity and threshold temperatures have been designed based on thermodynamic computations and experimental assays (Sen et al.2017). The modularity of riboswitches and RNA thermometers was exploited by Roßmanith and Narberhaus (2016) to generate combined regulatory devices with novel functionalities depending on both ligand binding and temperature sensing, which confer dual regulation at the level of transcription and translation.
The application of regulatory strategies based on synthetic trans-acting sRNA has a number of advantages. Their trans-acting character allows the simultaneous targeting of several expression units, target site selection is based on RNA-RNA hybridization according to a simple code compared to protein-RNA or protein–DNA recognition and its strength can be modulated by the creation of libraries. Furthermore, their small size generates little burden for the host cell allowing the simultaneous introduction and expression of multiple sRNAs in the same cell. Libraries of rationally designed synthetic trans-acting sRNAs composed of a scaffold sequence for the recruitment of Hfq and a target-binding sequence were for instance developed to modulate gene expression in the context of metabolic engineering for the overproduction of tyrosine and cadaverine in E. coli (Na et al.2013). Starting from the hypothesis that transcriptional attenuators are modular (in analogy with modularity of riboswitches) and consist of a domain responsible for antisense RNA binding and another for actuating the transcriptional regulation, Takahashi and Lucks (2013) developed orthogonal chimeric trans-acting RNA transcription regulators by fusing and mutagenizing sequences from five different translational attenuators. Toehold switches, de novo designed artificial regulators of gene expression, were developed to enable orthogonal post-transcriptional activation of translation in response to cognate trans-acting sRNAs (Green et al.2014). In toehold switches, translation of an output gene is only possible when a cognate trigger RNA is expressed, and the RNA-RNA interactions are mediated through linear–linear interactions, rather than loop-mediated interactions as in most other naturally occurring and synthetic systems (Fig. 5B). This has the advantage of faster kinetics and stronger thermodynamics (due to the length of the interacting stretches). Furthermore, toehold switches do not rely on direct binding to the RBS for repression but rather on the enclosure of the RBS in a loop formed with secondary structures immediately preceding and following the start codon. This allows the engineering of trigger-switch RNA complexes with low secondary structure in proximity of the start codon and ensures efficient translation in the ON state of the switch. Based on these toehold switch systems, Green et al. (2017) further developed ribocomputing systems composed exclusively of de novo designed RNA nanodevices based on predictable and designable base-pairing rules that regulate gene expression at the post-transcriptional level in E. coli. In this setup, a single-extended RNA transcript is used to co-localize the circuit sensing, computation, signal transduction and output elements of the nanodevices. This has the advantage that it reduces signal loss, lowers the metabolic cost and improves circuit reliability. Although originally developed for E. coli, it should in principle be possible to implement these devices in other organisms. Finally, it is worth mentioning that bacterial CRISPR-Cas defense mechanisms of acquired immunity, naturally used to destroy foreign incoming DNA in the cell (Bhaya, Davidson and Barrangou 2011) and massively applied for gene and genome editing purposes in all domains of life, have also been exploited to generate a non-destructive gene regulatory system in which the target selectivity is based on RNA-DNA hybridization. In these systems, a ‘dead’ or nuclease-null variant of Cas9 that still binds DNA but no longer cleaves it is brought to selected targets on the DNA in association with a single small trans-acting guide RNA (sgRNA) and acts as a transcriptional DNA-binding transcription regulator (Esvelt et al.2013). Furthermore, the CRISPR-Cas interference RNA was also used to expand the dynamic range of gene expression from T7 promoters (McCutcheon et al.2018).
Concluding remarks
Bacterial gene regulatory mechanisms are diverse and may operate at different levels and stages in the flow of genetic information. As abundantly documented in this review, gene regulation is generally multi-layered and relies on a combination of different regulatory strategies integrating multiple signals. As these mechanisms are intrinsically intertwined, they are difficult to unravel. Generating a comprehensive picture of a regulatory circuit requires both specific and global, genome-wide in vivo expression studies at mRNA and protein level, and in-depth in vitro biochemical and biophysical analyses to unravel the underlying molecular basis of the mechanisms. Over the years the basal transcription and translation machineries have been dissected in great detail and many regulatory mechanisms have been identified. But, unraveling their interplay in specific pathways and regulatory networks is much more challenging. Even today and after decennia of investigations, we still ignore how the expression of about 50% of the genes is regulated in E. coli, the best-studied organism, and the situation is even worse for other Gram-negative and Gram-positive model organisms and platforms for microbial production such as P. aeruginosa, C. glutamicum, B. subtilis and Streptomyces species to cite only a few. Even in spite of the recent development of genome-wide high-throughput techniques including RNA-seq and Chip-seq for genome-wide transcript quantification and identification of TF-binding sites, respectively, there is still need for new approaches for the systematic large-scale and in-depth unraveling of bacterial gene regulation (Belliveau et al.2018; Hör, Gorski and Vogel 2018).
To exploit bacteria as cell factories in an economically competitive manner for the biological and sustainable production of various and complex compounds, it will be necessary to dispose of robust recombinant strains with high production yields. Synthetic biology facilitates the engineering of such organisms in a rational manner. The import of synthetic circuits of increasing complexity in a host requires the coordinated and fine-tuned expression of several genes without major interference with the host metabolism. Therefore, there is need for libraries of basic cis-acting gene expression elements including promoter and ribosome-binding sites spanning a wide range of activities, various orthogonal gene expression systems at transcriptional and translational level that may be used simultaneously in the same cell, and trans-acting regulatory elements allowing their fine-tuning. To allow the selection of the most productive and robust producing cells, it is imperative that all these elements may be combined efficiently in a combinatorial manner and analyzed in high-throughput screens. This requires a wide variety of standardized parts, toolboxes and seamless assembly techniques. Presently, synthetic biology applications are still in need of more and varied, well-defined parts and tools and will still profit from better characterization of basal and regulatory gene expression elements, including σ factors, TFs, sRNAs, riboswitches and RNA thermometers, and their incorporation in toolboxes to control gene expression at both transcriptional and translational levels in a predictable and tunable manner. Hence, studies unraveling the functioning of naturally occurring bacterial gene regulatory systems and the analysis of the behavior of their constituent parts in artificial systems may be reciprocally beneficial and have an impact at societal level. The fishing pond for such orthogonal elements could be considerably extended by in-depth analyses of less well-studied bacteria and might even be extended to archaeal genomes since these organisms possess regulatory transcription factors that are similar to their bacterial homologs. Furthermore, even eukaryotic DNA-binding motives such as the Zinc-finger with its small size and relatively simple mode of DNA binding and sequence selectivity (MacPherson, Larochelle and Turcotte 2006) could be advantageously exploited to attract chimeric orthogonal regulatory molecules or (single subunit) RNAPs to selected cognate targets. Genome and metagenome mining, bioinformatics and computation to predict the behavior of artificial circuits are therefore expected to play a major role in future developments. It would also be worth to invest in the better characterization of regulatory networks of extremophilic bacteria as they might be better suited as chassis for sustainable microbial production, especially starting from renewable biomass such as low value agricultural waste products including stems and leaved that require pretreatment in harsh conditions.
Acknowledgements
The authors wish to thank Eveline Peeters, Martine Roovers and Louis Droogmans for valuable comments and suggestions.
FUNDING
This work was supported by the Fonds voor Wetenschappelijk Onderzoek Vlaanderen [grant number G.0321.13 N].
REFERENCES
- Ades SE. Control of the alternative sigma factor σE in Escherichia coli. Curr Opin Microbiol 2004;7:157–62. [DOI] [PubMed] [Google Scholar]
- Adhikari S, Curtis PD. DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol Rev 2016;40:575–91. [DOI] [PubMed] [Google Scholar]
- Afonyushkin T, Vecerek B, Moll I et al.. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res 2005;33:1678–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge P, Karlinsey JE, Becker E et al.. Flk prevents premature secretion of the anti-sigma factor FlgM into the periplasm. Mol Microbiol 2006;60:630–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TD, Watkins T, Lindahl L et al.. Regulation of ribosomal protein synthesis in Vibrio cholerae. J Bacteriol 2004;186:5933–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuvia S, Kornitzer D, Teff D et al.. Alternative mRNA structures of the cIII gene of bacteriophage λ determine the rate of its translation initiation. J Mol Biol 1989;210:265–80. [DOI] [PubMed] [Google Scholar]
- Amit R, Oppenheim AB, Stavans J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys J 2003;84:2467–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amster-Choder O, Wright A. Modulation of the dimerization of a transcriptional antiterminator protein by phosphorylation. Science 1992;257:1395–8. [DOI] [PubMed] [Google Scholar]
- An W, Chin JW. Synthesis of orthogonal transcription-translation networks. Proc Natl Acad Sci USA 2009;106:8477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angius F, Ilioaia O, Amrani S et al.. A novel regulation mechanism of the T7 RNA polymerase based expression system improves overproduction and folding of membrane proteins. Sci Rep 2018;8:8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziata F, Matyjaszkiewicz A, Fiore G et al.. An orthogonal multi-input integration system to control gene expression in Escherichia coli. ACS Synth Biol 2017;6:1816–24. [DOI] [PubMed] [Google Scholar]
- Ansari AZ, Bradner JE, O’Halloran TV. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature 1995;374:371–5. [DOI] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Balaji S et al.. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 2005;29:231–62. [DOI] [PubMed] [Google Scholar]
- Arfin SM, Long AD, Ito ET et al.. Global gene expression profiling in Escherichia coli K12. The effects of integration host factor. J Biol Chem 2000;275:29672–84. [DOI] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci USA 2000;97:7090–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Patlan V, Sekine S et al.. Structural basis for transcription regulation by alarmone ppGpp. Cell 2004;117:299–310. [DOI] [PubMed] [Google Scholar]
- Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J Biol Chem 1999;274:33105–13. [DOI] [PubMed] [Google Scholar]
- Babitzke P. Regulation of transcription attenuation and translation initiation by allosteric control of an RNA-binding protein: the Bacillus subtilis TRAP protein. Curr Opin Microbiol 2004;7:132–9. [DOI] [PubMed] [Google Scholar]
- Babitzke P, Yanofsky C. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc Natl Acad Sci USA 1993;90:133–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Båga M, Göransson M, Normark S et al.. Transcriptional activation of a pap pilus virulence operon from uropathogenic Escherichia coli. EM J 1985;4:3887–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta AB, Chumanov RS, Yuan AH et al.. Key features of σS required for specific recognition by Crl, a transcription factor promoting assembly of RNA polymerase holoenzyme. Proc Natl Acad Sci USA 2013;110:15955–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S, Ghosh B, Whalen W et al.. An antitermination protein engages the elongating transcription apparatus at a promoter-proximal recognition site. Cell 1987;50:885–99. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol 2001;305:689–702. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Josaitis CA et al.. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol 2001;305:673–88. [DOI] [PubMed] [Google Scholar]
- Battesti A, Hoskins JR, Tong S et al.. Anti-adaptors provide multiple modes for regulation of the RssB adaptor protein. Genes Dev 2013;27:2722–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 2011;65:189–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. Stress sigma factor RpoS degradation and translation are sensitive to the state of central metabolism. Proc Natl acad Sci USA 2015;112:5159–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Tsegaye YM, Packer DG et al.. H-NS regulation of IraD and IraM antiadaptors for control of RpoS degradation. J Bacteriol 2012;194:2470–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belliveau NM, Barnes SL, Ireland WT et al.. Systematic approach for dissecting the molecular mechanisms of transcriptional regulation in bacteria. Proc Natl Acad Sci USA 2018;115:E4796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoff B, Yang H, Lawson CL et al.. Structural basis of transcription activation: the CAP-αCTD-DNA complex. Science 2002;297:1562–6. [DOI] [PubMed] [Google Scholar]
- Bertram R, Hillen W. The application of Tet repressor in prokaryotic gene regulation and expression. Microb Biotechnol 2008;1:2–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bervoets I, Van Bremt M, Van Nerom K et al.. A sigma factor toolbox for orthogonal gene expression in Escherichia coli. Nucleic Acids Res 2018;46:2133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D, Davidson M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Ann Rev Genet 2011;45:273–97. [DOI] [PubMed] [Google Scholar]
- Bijlsma JJ, Groisman EA. Making informed decisions: regulatory interactions between two-component systems. Trends Microbiol 2003;11:359–66. [DOI] [PubMed] [Google Scholar]
- Binder SC, Eckweiler D, Schulz S et al.. Functional modules of sigma factor regulons guarantee adaptability and evolvability. Sci Rep 2016;6:22212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomfield IC. The regulation of pap and type 1 fimbriation in Escherichia coli. Adv Microb Physiol 2001;45:1–49. [DOI] [PubMed] [Google Scholar]
- Bordeleau E, Purcell EB, Lafontaine DA et al.. Cyclic di-GMP riboswitch-regulated Type IV pili contribute to aggregation of Clostridium difficile. J Bacteriol 2015;197:819–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkotoky S, Murali A. The highly efficient T7 RNA polymerase: a wonder macromolecule in biological realm. Int J Biol Macromol 2018;118:49–56. [DOI] [PubMed] [Google Scholar]
- Bougdour A, Cunning C, Baptiste PJ et al.. Multiple pathways for regulation of σS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol Microbiol 2008;68:298–313. [DOI] [PubMed] [Google Scholar]
- Bougdour A, Wickner S, Gottesman S. Modulating RssB activity: IraP, a novel regulator of σS stability in Escherichia coli. Genes Dev 2006;20:884–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutte CC, Crosson S. Bacterial lifestyle shapes stringent response activation. Trends Microbiol 2009;21:174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutte CC, Crosson S. The complex logic of stringent response regulation in Caulobacter crescentus: starvation signaling in an oligotrophic environment. Mol Microbiol 2011;80:695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier J, Patte JC, Stragier P. Multiple regulatory signals in the control region of the Escherichia coli carAB operon. Proc Natl Acad Sci USA 1984;81: 4139–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley MD, Beach MB, de Koning AP et al.. Effects of Fis on Escherichia coli gene expression during different growth stages. Microbiology 2007;153:2922–40. [DOI] [PubMed] [Google Scholar]
- Bradley RW, Buck M, Wang B. Tools and principles for microbial gene circuit engineering. J Mol Biol 2016;428:862–88. [DOI] [PubMed] [Google Scholar]
- Brennan RG. The winged-helix DNA-binding motif: another helix-turn-helix takeoff. Cell 1993;74:773–6. [DOI] [PubMed] [Google Scholar]
- Brewster RC, Jones DL, Phillips R. Tuning promoter strength through RNA polymerase binding site design in Escherichia coli. PLoS Comput Biol 2012;8:e1002811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman AB, Ettema TJ, de Vos WM et al.. The Lrp family of transcriptional regulators. Mol Microbiol 2003;48:287–94. [DOI] [PubMed] [Google Scholar]
- Brown NL, Stoyanov JV, Kidd SP et al.. The MerR family of transcriptional regulators. FEMS Microbiol Rev 2003;27:145–63. [DOI] [PubMed] [Google Scholar]
- Broyles SS, Pettijohn DE. Interaction of the Escherichia coli HU protein with DNA. Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J Mol Biol 1986;187:47–60. [DOI] [PubMed] [Google Scholar]
- Buck M, Gallegos M-T, Studholme DJ et al.. The bacterial enhancer-dependent σ54 (σN) transcription factor. J Bacteriol 2000;182:4129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 1991;64:545–52. [DOI] [PubMed] [Google Scholar]
- Burgos HL, O’Connor K, Sanchez-Vazquez P et al.. Roles of transcriptional and translational control mechanisms in regulation of ribosomal protein synthesis in Escherichia coli. J Bacteriol 2017;199:e00407–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrill DR, Silver PA. Making cellular memories. Cell 2010;140:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush M, Dixon R. The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol Mol Biol R 2012;76:497–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo JM, Matthews RG. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Rev 1994;58:466–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagne S, Allain FHT, Vorholt JA. Extra cytoplasmic function sigma factors, recent structural insights into promoter recognition and regulation. Curr Opin Struct Biol 2015;30:71–78. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Muzzin O, Chlenov M et al.. Structure of the bacterial RNA polymerase promoter specificity subunit. Mol Cell 2002;9:527–39. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Westblade LF, Darst SA. Regulation of bacterial RNA polymerase σ factor activity: a structural perspective. Cur Opin Microbiol 2008;11:121–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camsund D, Lindblad P, Jaramillo A. Genetically engineered light sensors for control of bacterial gene expression. Biotechnol J 2011;6:826–36. [DOI] [PubMed] [Google Scholar]
- Carmona M, Rodríguez MJ, Martínez-Costa O et al.. In vivo and in vitro effects of (p)ppGpp on the σ54 promoter Pu of the TOL plasmid of Pseudomonas putida. J Bacteriol 2000;182:4711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis AJ. The RNA degradasome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Ann Rev Microbiol 2007;61:71–87. [DOI] [PubMed] [Google Scholar]
- Casadesús J. Bacterial DNA methylation and methylomes. Adv Exp Med Biol 2016;945:35–61. [DOI] [PubMed] [Google Scholar]
- Casino P, Rubio V, Marina A. Structural insight into partner specificity of phosphoryl transfer in two-component signal transduction. Cell 2009;139:325–36. [DOI] [PubMed] [Google Scholar]
- Casino P, Rubio V, Marina A. The mechanism of signal transduction by two-component systems. Curr Opin Struct Biol 2010;20:763–71. [DOI] [PubMed] [Google Scholar]
- Castaing B, Zelwer C, Laval J et al.. HU protein of Escherichia coli binds specifically to DNA that contains single-stranded breaks or gaps. J Biol Chem 1995;270:10291–6. [DOI] [PubMed] [Google Scholar]
- Cavaliere P, Brier S, Filipenko P et al.. The stress sigma factor of RNA polymerase RpoS/σS is a solvent-exposed open molecule in solution. Biochem J 2018;475:341–54. [DOI] [PubMed] [Google Scholar]
- Ceres P, Garst AD, Marcano-Velázquez JG et al.. Modularity of select riboswitch expression platforms enables facile engineering of novel genetic regulatory devices. ACS Synth Biol 2013;2:463–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceroni F, Boo A, Furini S et al.. Burden-driven feedback control of gene expression. Nat Methods 2018;15:387–93. [DOI] [PubMed] [Google Scholar]
- Chandran V, Luisi BF. Recognition of enolase in the Escherichia coli RNA degradasome. J Mol Biol 2006;358:8–15. [DOI] [PubMed] [Google Scholar]
- Charlier D, Gigot D, Huysveld N et al.. Pyrimidine regulation of the Escherichia coli and Salmonella typhimurium carAB operons: CarP and integration host factor (IHF) modulate the methylation status of a GATC site present in the control region. J Mol Biol 1995a;250:383–91. [DOI] [PubMed] [Google Scholar]
- Charlier D, Hassanzadeh G, Kholti A et al.. CarP, involved in pyrimidine regulation of the Escherichia coli carbamoylphosphate synthetase operon encodes a sequence specific DNA-binding protein identical to XerB and PepA, also required for resolution of ColEI multimers. J Mol Biol 1995b;250:392–406. [DOI] [PubMed] [Google Scholar]
- Charlier D, Huysveld N, Roovers M et al.. On the role of the Escherichia coli integration host factor (IHF) in repression at a distance of the pyrimidine specific promoter P1 of the carAB operon. Biochimie 1994;76:1041–51. [DOI] [PubMed] [Google Scholar]
- Charlier D, Kholti A, Huysveld N et al.. Mutational analysis of Escherichia coli PepA, a multifunctional DNA-binding aminopeptidase. J Mol Biol 2000;302:411–26. [DOI] [PubMed] [Google Scholar]
- Charlier D, Nguyen Le Minh P, Roovers M. Regulation of carbamoylphosphate synthesis in Escherichia coli: an amazing metabolite at the crossroad of arginine and pyrimidine biosynthesis. Amino Acids 2018;50:1647–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier D, Piette J, Glansdorff N. IS3 can function as a mobile promoter in E. coli. Nucleic Acids Res 1982;10:5935–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier D, Roovers M, Gigot D et al.. Integration host factor (IHF) modulates the expression of the pyrimidine-specific promoter of the carAB operons of Escherichia coli and Salmonella typhimurium LT2. Mol Gen Genet 1993;237:273–86. [DOI] [PubMed] [Google Scholar]
- Charlier D, Roovers M, Van Vliet F et al.. Arginine regulon of Escherichia coli K-12: a study of repressor-operator interactions and of in vitro binding affinities versus in vivo repression. J Mol Biol 1992;226:367–86. [DOI] [PubMed] [Google Scholar]
- Charlier D, Weyens G, Roovers M et al.. Molecular interactions in the control region of the carAB operon encoding Escherichia coli carbamoylphospahte synthetase. J Mol Biol 1988;204:867–77. [DOI] [PubMed] [Google Scholar]
- Chen D, Arkin AP. Sequestration-based bistability enables tuning of the switching boundaries and design of a latch. Mol Syst Biol 2012;8:620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Decker KB, Boucher PE et al.. Novel architectural features of Bordetella pertussis fimbrial subunit promoters and their activation by the global virulence regulator BvgA. Mol Microbiol 2010;77:1326–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Calvo JM. Leucine-induced dissociation of Escherichia coli Lrp hexadecamers to octamers. J Mol Biol 2002;318:1031–42. [DOI] [PubMed] [Google Scholar]
- Chen S, Iannolo M, Calvo JM. Cooperative binding of the leucine-responsive regulatory protein (Lrp) to DNA. J Mol Biol 2005;345:251–64. [DOI] [PubMed] [Google Scholar]
- Chen S, Rosner MH, Calvo JM. Leucine-regulated self-association of leucine-responsive regulatory protein (Lrp) from Escherichia coli. J Mol Biol 2001;312:625–35. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ho JML, Shis DL et al.. Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors. Nat Commun 2018;9:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AC, Chen WW, Fuhrmann CN et al.. Recognition of nucleic acid bases and base-pairs by hydrogen bonding to amino acid side-chains. J Mol Biol 2003;327:781–96. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dylla SM, Turnbough CL Jr. A long T·A tract in the upp initially transcribed region is required for regulation of upp expression by UTP-dependent reiterative transcription in Escherichia coli. J Bacteriol 2001;183:221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YY, Hirning AJ, Josic K et al.. The timing of transcriptional regulation in synthetic gene circuits. ACS Synth Biol 2017;6:1996–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon S, Kim HM, Gustavsson M et al.. Recent trends in metabolic engineering of microorganisms for the production of advanced biofuels. Curr Opin Chem Biol 2016;35:10–21. [DOI] [PubMed] [Google Scholar]
- Cho BK, Barrett CL, Knight EM et al.. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc Natl Acad Sci USA 2008a;105:19462–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BK, Kim D, Knight EM et al.. Genome-scale reconstruction of the sigma factor network in Escherichia coli: topology and functional states. BMC Biol 2014;12:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BK, Knight EM, Barrett CL et al.. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT tracts. Genome Res 2008b;18:900–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Y, Klug A. Physical basis of a protein-DNA recognition code. Curr Opin Struct Biol 1997;7:117–25. [DOI] [PubMed] [Google Scholar]
- Commichau FM, Stülke J. Trigger enzymes: bifunctional proteins active in metabolism and in controlling gene expression. Mol Microbiol 2008;67:692–702. [DOI] [PubMed] [Google Scholar]
- Conant CR, Goodarzi JP, Weitzel SE et al.. The antitermination activity of bacteriophage lambda N protein is controlled by the kinetics of an RNA-looping-facilitated interaction with the transcription complex. J Mol Biol 2008;384:87–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, Grunberg-Manago M, Putzer H. Aminoacyl-tRNA synthetase gene regulation in Bacillus subtilis. Biochimie 1996;78:381–9. [DOI] [PubMed] [Google Scholar]
- Corcoran CP, Dorman CJ. DNA relaxation-dependent phase biasing of the fim genetic switch in Escherichia coli depends on the interplay of H-NS, IHF and LRP. Mol Microbiol 2009;74:1071–82. [DOI] [PubMed] [Google Scholar]
- Corrigan RM, Bellows LE, Wood A et al.. ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram-positive bacteria. Proc Natl Acad Sci USA 2016;113:E1710–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyer J, Andersen J, Forst SA et al.. micF RNA in ompB mutants of Escherichia coli: different pathways regulate micF RNA levels in response to osmolarity and temperature change. J Bacteriol 1990;172:4143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux ZD, Swanson MS. ppGpp: magic spot beyond RNA polymerase. Nat Rev Microbiol 2012;10:203–12. [DOI] [PubMed] [Google Scholar]
- Dame RT, Noom MC, Wuite GJ. Bacterial chromatin organization by H-NS protein unraveled using dual DNA manipulation. Nature 2006;444:387–90. [DOI] [PubMed] [Google Scholar]
- Dame RT, Tark-Dame M. Bacterial chromatin: converging views at different scales. Curr Opin Cell Biol 2016;40:60–65. [DOI] [PubMed] [Google Scholar]
- Dame RT, Wyman C, Goosen N. Structural basis for preferential binding of H-NS to curved DNA. Biochimie 2001;83:231–4. [DOI] [PubMed] [Google Scholar]
- Dame RT, Wyman C, Wurm R et al.. Structural basis for H-NS mediated trapping of RNA polymerase in the open initiation complex at rrnB P1. J Biol Chem 2002;277:2146–50. [DOI] [PubMed] [Google Scholar]
- Darlington APS, Kim J, Jiménez JI et al.. Design of a translation resource allocation controller to manage cellular resource limitations. Engineering translational resource allocation controllers: mechanistic models, design guidelines, and potential biological implementations. ACS Synth Biol 2018;7:2485–96. [DOI] [PubMed] [Google Scholar]
- Davis JH, Rubin AJ, Sauer RT. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res 2011;39:1131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MC, Kesthely CA, Franklin EA et al.. The essential activities of the bacterial sigma factor. Can J Microbiol 2017;63:89–99. [DOI] [PubMed] [Google Scholar]
- Deana A, Belasco JG. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev 2005;19:2526–33. [DOI] [PubMed] [Google Scholar]
- Dehli T, Solem C, Jensen PR. Tunable promoters in synthetic and systems biology. Subcell Biochem 2012;64:181–201. [DOI] [PubMed] [Google Scholar]
- De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem 2013;288:7996–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mey M, Maertens J, Lequeux GJ et al.. Construction and model-based analysis of a promoter library for E. coli: an indispensable tool for metabolic engineering. BMC Biotechnol 2007;7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe B, Peters G, Coussement P et al.. Tailor-made transcriptional biosensors for optimizing microbial cell factories. J Ind Microbiol Biot 2017;44:623–45. [DOI] [PubMed] [Google Scholar]
- Desai SK, Kenney LJ. To ∼P or not to ∼P? Non-canonical activation by two-component response regulators. Mol Microbiol 2017;103:203–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers G, Massé E. Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Gene Dev 2012;26:726–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sousa AR, Penalva LO, Marcotte EM et al.. Global signatures of protein and mRNA expression levels. Mol Biosyst 2009;5:1512–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutcher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res 2006;34:659–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devroede N, Thia-Toong TL, Gigot D et al.. Purine and pyrimidine-specific repression of the Escherichia coli carAB operon are functionally and structurally coupled. J Mol Biol 2004;336:25–42. [DOI] [PubMed] [Google Scholar]
- Dey D, Nagaraja V, Ramakumar S. Structural and evolutionary analyses reveal determinants of DNA binding specificities of nucleoid-associated proteins HU and IHF. Mol Phylogenet Evol 2017;107:356–66. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol 2010;8:185–95. [DOI] [PubMed] [Google Scholar]
- Dorman CJ. Co-operative roles for DNA supercoiling and nucleoid-associated proteins in the regulation of bacterial transcription. Biochem Soc Trans 2013;42:542–7. [DOI] [PubMed] [Google Scholar]
- Doucleff M, Pelton JG, Lee PS et al.. Structural basis of DNA recognition by the alternative sigma-factor, σ54. J Mol Biol 2007;369:1070–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressaire C, Laurent B, Loubière P et al.. Linear covariance models to examine the determinants of protein levels in Lactococcus lactis. Mol Biosyst 2010;6:1255–64. [DOI] [PubMed] [Google Scholar]
- Dudin O, Lacour S, Geiselmann J. Expression dynamics of RpoS/Crl-dependent genes in Escherichia coli. Res Microbiol 2013;164:838–47. [DOI] [PubMed] [Google Scholar]
- Duval M, Simonetti A, Caldelari I et al.. Multiple ways to regulate translation initiation in bacteria: mechanisms, regulatory circuits, dynamics. Biochimie 2015;114:18–29. [DOI] [PubMed] [Google Scholar]
- Egbert RG, Klavins E. Fine-tuning gene networks using simple sequence repeats. Proc Natl Acad Sci USA 2012;109:16817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger LA, Park H, Inouye M. Signal transduction via histidyl-aspartyl phosphorelay. Genes Cells 1997;2:167–84. [DOI] [PubMed] [Google Scholar]
- Englesberg E, Irr J, Power J et al.. Positive control of enzyme synthesis by gene C in the L-arabinose system. J Bacteriol 1965;90:946–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrem ST, Ross W, Gaal T et al.. Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase alpha subunit. Gene Dev 1999;13:2134–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL et al.. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 2013;10;1116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzel M, Mörl M. Synthetic riboswitches: from plug and pray toward plug and play. Biochemistry 2017;56:1181–98. [DOI] [PubMed] [Google Scholar]
- Fabret C, Feher VA, Hoch JA. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J Bacteriol 1999;181:1975–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feklístov A, Sharon BD, Darst SA et al.. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol 2014;68:357–76. [DOI] [PubMed] [Google Scholar]
- Feng J-A, Johnson RC, Dickerson RE. Hin recombinase bound to DNA: the origin of specificity in major and minor groove interactions. Science 1994;263:348–55. [DOI] [PubMed] [Google Scholar]
- Feng Y, Zhang Y, Ebright RH. Structural basis of transcription activation. Science 2016;352:1330–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findeiß S, Etzel M, Will S et al.. Design of artificial riboswitches as biosensors. Sensors 2017;17:pii:E1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald DM, Bonocora RP, Wade JT. Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet 2014;10:e1004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folliard T, Mertins B, Steel H et al.. Ribo-attenuators: novel elements for reliable and modular riboswitch engineering. Sci Rep 2017;7:4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol Mol Biol R 2008;72:579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francez-Charlot A, Frunzke J, Reichen C et al.. Sigma factor mimicry involved in regulation of general stress response. Proc Natl Acad Sci USA 2009;106:3467–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangipane G, Dell’Arciprete D, Petracchini S et al.. Dynamic density shaping of photokinetic E. coli. Elife 2018;7pii:e36608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Free A, Dorman CJ. Coupling of Escherichia coli hns mRNA levels to DNA synthesis by autoregulation: implications for growth phase control. Mol Microbiol 1995;18:101–13. [DOI] [PubMed] [Google Scholar]
- Freyre-González JA, Manjarrez-Casas AM, Merino E et al.. Lessons from the modular organization of the transcriptional regulatory network of Bacillus subtilis. BMC Syst Biol 2013;7:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick MM, Neuhard J, Kelln RA. Cloning, nucleotide sequence and regulation of the Salmonella typhimurium pyrD gene encoding dihydroorotate dehydrogenase. Eur J Biochem 1990;194:573–8. [DOI] [PubMed] [Google Scholar]
- Furukawa K, Ramesh A, Zhou Z et al.. Bacterial riboswitches cooperatively bind Ni2+ or Co2+ ions and control expression of heavy metal transporters. Mol Cell 2015;57:1088–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal T, Ross W, Blatter EE et al.. DNA-binding determinants of the alpha subunit of RNA polymerase: a novel DNA-binding architecture. Gene Dev 1996;10:16–26. [DOI] [PubMed] [Google Scholar]
- Gaballa A, Guariglia-Oropeza V, Dürr F et al.. Modulation of extracytoplasmic function (ECF) sigma factor promoter selectivity by spacer region sequence. Nucleic Acids Res 2018;46:134–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao A, Vasilyev N, Luciano DJ et al.. Structural and kinetic insights into stimulation of RppH-dependent RNA degradation by the metabolic enzyme DapF. Nucleic Acids Res 2018;46:6841–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Mack TR, Stock AM. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci 2007;32:225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Colins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature 2000;403:339–42. [DOI] [PubMed] [Google Scholar]
- Garst AD, Edwards AL, Batey RT. Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol 2011;3 pii:a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W, Müller-Hill B. Isolation of the lac repressor. Proc Natl Acad Sci USA 1966;56:1891–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W, Müller-Hill B. The lac operator is DNA. Proc Natl Acad Sci USA 1967;58:2415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman J, Love J. Synthetic promoter design for new microbial chassis. Biochem Soc Trans 2016;44:731–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard ME, Gopalkrishnan S, Grace ED et al.. DksA and ppGpp regulate the σS stress response by activating promoters for the small RNA DsrA and the anti-adapter protein IraP. J Bacteriol 2017. DOI: 10.1128/JB.00463-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glyde R, Ye F, Darbari VC et al.. Structures of RNA polymerase closed and intermediate complexes reveal mechanisms of DNA opening and transcription initiation. Mol Cell 2017;67:106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görke B. Regulation of the Escherichia coli antiterminator protein BglG by phosphorylation at multiple sites and evidence for transfer of phosphoryl groups between monomers. J Biol Chem 2003;278:46219–29. [DOI] [PubMed] [Google Scholar]
- Görke B, Rak B. Catabolite control of Escherichia coli regulatory protein BglG activity by antagonistically acting phosphorylations. EMBO J 1999;18:3370–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman ME, Adhya S, Das A. Transcription antitermination by bacteriophage lambda N gene product. J Mol Biol 1980;140:57–75. [DOI] [PubMed] [Google Scholar]
- Green AA, Kim J, Ma D et al.. Complex cellular logic computation using ribocomputing devices. Nature 2017;548:117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AA, Silver PA, Collins JJ et al.. Toehold switches: de-novo-designed regulators of gene expression. Cell 2014;159:925–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett 2010;584:318–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J, Nodwell JR, Mason SW. Transcriptional antitermination. Nature 1993;364:401–6. [DOI] [PubMed] [Google Scholar]
- Grigorova IL, Phleger NJ, Mutalik VK et al.. Insights into transcriptional regulation and sigma competition from an equilibrium model of RNA polymerase binding to DNA. Proc Natl Acad Sci USA 2006;103:5332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groher F, Bofill-Bosch C, Schneider C et al.. Riboswitching with ciprofloxacin-development and characterization of a novel RNA regulator. Nucleic Acids Res 2018;46:2121–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol 2003;57:441–66. [DOI] [PubMed] [Google Scholar]
- Grylak-Mielnicka A, Bidnenko V, Bardowski J et al.. Transcription termination factor Rho: a hub linking diverse physiological processes in bacteria. Microbiology 2016;162:433–47. [DOI] [PubMed] [Google Scholar]
- Gualerzi CO, Giuliodori AM, Pon CL. Transcriptional and post-transcriptional control of cold-shock genes. J Mol Biol 2003;331:527–39. [DOI] [PubMed] [Google Scholar]
- Guiziou S, Sauveplane V, Chang HJ et al.. A part toolbox to tune genetic expression in Bacillus subtilis. Nucleic Acids Res 2016;44:7495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I, Nudler E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell 2001;107:437–49. [DOI] [PubMed] [Google Scholar]
- Han K, Li ZF, Zhu LP et al.. Extraordinary expansion of a Sorangium cellulosum genome from an alkaline milieu. Sci Rep 2013;3:2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Turnbough CL Jr. Regulation of carAB expression in Escherichia coli occurs in part through UTP-sensitive reiterative transcription. J Bacteriol 1998;180:705–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbaugh SV, Martin JA, Weinstein J et al.. Screening and selection of artificial riboswitches. Methods 2018;143:77–89. [DOI] [PubMed] [Google Scholar]
- Harden TT, Wells CD, Friedman LJ et al.. Bacterial RNA polymerase can retain σ70 throughout transcription. Proc Natl Acad Sci USA 2016;113:602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart BR, Blumenthal RM. Unexpected coregulator range for the global regulator Lrp of Escherichia coli and Proteus mirabilis. J Bacteriol 2011;193:1054–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen SP, Berkmen MB, Ross W et al.. rRNA promoter regulation by nonoptimal binding of σ region 1.2: an additional recognition element for RNA polymerase. Cell 2006;125:1069–82. [DOI] [PubMed] [Google Scholar]
- Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol 2008;6:507–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Zalkin H. Repression of Escherichia coli purB is by a transcriptional roadblock mechanism. J Bacteriol 1992;174:7121–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker M, Pané-Farré J, Völker U. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol 2007;61:215–36. [DOI] [PubMed] [Google Scholar]
- Heinrich J, Wiegert T. Regulated intramembrane proteolysis in the control of extracytoplasmic function sigma factors. Res Microbiol 2009;160:696–703. [DOI] [PubMed] [Google Scholar]
- Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol 2002;46:47–110 [DOI] [PubMed] [Google Scholar]
- Heltzel A, Lee IW, Totis PA et al.. Activator-dependent preinduction binding of sigma-70 RNA polymerase at the metal-regulated mer promoter. Biochemistry 1990;29:9572–84. [DOI] [PubMed] [Google Scholar]
- Henderson CA, Vincent HA, Casamento A et al.. Hfq binding changes the structure of Escherichia coli small noncoding RNAs OxyS and RprA, which are involved in the riboregulation of rpoS. RNA 2013;19:1089–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KL, Felth LC, Molzahn CM et al.. Mechanism of transcription initiation and promoter escape by E. coli RNA polymerase. Proc Natl Acad Sci USA 2017;114:E3032–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays 2002;24:700–7. [DOI] [PubMed] [Google Scholar]
- Herrou J, Rotskoff G, Luo Y et al.. Structural basis of a protein partner switch that regulates the general stress response of α-proteobacteria. Proc Natl Acad Sci USA 2012;109:E1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusipp G, Fälker S, Schmidt MA. DNA adenine methylation and bacterial pathogenesis. Int J Med Microbiol 2007;297:1–7. [DOI] [PubMed] [Google Scholar]
- Hilbert DW, Piggot PJ. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol R 2004;68:234–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton DM. Transcriptional control in the prereplicative phase of T4 development. Virol J 2010;7:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton DM, Pande S, Wais N et al.. Transcriptional takeover by sigma appropriation: remodeling of the σ70 subunit of Escherichia coli RNA polymerase by the bacteriophage T4 activator MotA and co-activator AsiA. Microbiology 2005;151:1729–40. [DOI] [PubMed] [Google Scholar]
- Hofmann N, Wurm R, Wagner R. The E. coli anti-sigma factor Rsd: studies on the specificity and regulation of its expression. PLoS One 2011;6:e19235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeister AE, Londoño-Vallejo A, Harry E et al.. Extracellular signal protein triggering the proteolytic activation of a developmental transcription factor in B. subtilis. Cell 1995;83:219–26. [DOI] [PubMed] [Google Scholar]
- Hör J, Gorski SA, Vogel J. Bacterial RNA biology on a genome scale. Mol Cell 2018;70:785–99. [DOI] [PubMed] [Google Scholar]
- Houman F, Diaz-Torres MR, Wright A. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell 1990;62:1153–63. [DOI] [PubMed] [Google Scholar]
- Hung SP, Baldi P, Hatfield GW. Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J Biol Chem 2002;277:40309–23. [DOI] [PubMed] [Google Scholar]
- Imamura D, Zhou R, Feig M et al.. Evidence that the Bacillus subtilis SpoIIGA protein is a novel type of signal-transducing aspartic protease. J Biol Chem 2008;283:15287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol 2000;54:499–518. [DOI] [PubMed] [Google Scholar]
- Iyer S, Karig DK, Norred SE et al.. Multi-input regulation and logic with T7 promoters in cells and cell-free systems. PLoS One 2013;8:e78442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Monod J. Genes of structure and genes of regulation in the biosynthesis of proteins. C R Hebd Seances Acad Sci 1959;249:1282–4. [PubMed] [Google Scholar]
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol 1961;3:318–56. [DOI] [PubMed] [Google Scholar]
- Janssen KH, Diaz MR, Gode CJ et al.. RsmV, a small noncoding regulatory RNA in Pseudomonas aeruginosa that sequesters RsmA and RsmF from target mRNAs. J Bacteriol 2018a;200:e00277–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen KH, Diaz MR, Golden M et al.. functional analyses of the RsmY and RsmZ small noncoding regulatory RNAs in Pseudomonas aeruginosa. J Bacteriol 2018b;200:e00736–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Xue W-F, Fukayama JW et al.. Assymetric allosteric activation of the symmetric ArgR hexamer. J Mol Biol 2004;346:43–56. [DOI] [PubMed] [Google Scholar]
- Johansson J, Mandin P, Renzoni A et al.. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 2002;110:551–61. [DOI] [PubMed] [Google Scholar]
- Jullesson D, David F, Pfleger B et al.. Impact of synthetic biology and metabolic engineering on industrial production of fine chemicals. Biotechnol Adv 2015;33:1395–13402. [DOI] [PubMed] [Google Scholar]
- Kaga N, Umitsuki G, Nagai K et al.. RNase G-dependent degradation of the eno mRNA encoding a glycolysis enzyme enolase in Escherichia coli. Biosci Biotech Bioch 2002;66:2216–20. [DOI] [PubMed] [Google Scholar]
- Kahl LJ, Endy D. A survey of enabling technologies in synthetic biology. J Biol Eng 2013;7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach LS, Braaten BA, Low DA. Specific binding of PapI to Lrp-pap DNA complexes. J Bacteriol 1995;177:6449–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandavalli VK, Tran H, Ribeiro AS. Effects of σ factor competition are promoter initiation kinetics dependent. Biochim Biophys Acta 2016;1859:1281–8. [DOI] [PubMed] [Google Scholar]
- Kang JG, Paget MS, Seok YJ et al.. RsrA, an anti-sigma factor regulated by redox change. EMBO J 1999;18:4292–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar S, Ellington AD. Construction of synthetic T7 RNA polymerase expression systems. Methods 2018;143:110–20. [DOI] [PubMed] [Google Scholar]
- Kawashima T, Aramaki H, Oyamada T et al.. Transcription regulation by feast/famine regulatory proteins, FFRPs, in archaea and eubacteria. Biol Pharm Bull 2008;31:173–86. [DOI] [PubMed] [Google Scholar]
- Kelln RA, Neuhard J. Regulation of pyrC expression in Salmonella typhimurium: identification of a regulatory region. Mol Gen Genet 1982;212:287–94. [DOI] [PubMed] [Google Scholar]
- Khalil AS, Collins JJ. Synthetic biology: applications come at age. Nat Rev Genet 2010;11:367–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholti A, Charlier D, Gigot D et al.. pyrH-encoded UMP-kinase directly participates in pyrimidine-specific modulation of promoter activity in Escherichia coli. J Mol Biol 1998;280:571–82. [DOI] [PubMed] [Google Scholar]
- Kill K, Binnewies TT, Sicheritz-Ponten T et al.. Genome update: sigma factors in 240 bacterial genomes. Microbiology 2005;151:3147–50. [DOI] [PubMed] [Google Scholar]
- Kilstrup M, Lu CD, Abdelal A et al.. Nucleotide sequence of the carA gene and regulation of the carAB operon in Salmonella typhimurium. Eur J Biochem 1988;176:421–9. [DOI] [PubMed] [Google Scholar]
- Kim SO, Merchant K, Nudelman R et al.. OxyR: a molecular code for redox-related signaling. Cell 2002;109:383–96. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Morisato D, Roberts D et al.. Mechanism and regulation of Tn10 transposition. Cold Spring Harb Symp Quant Biol 1984;49:235–44. [DOI] [PubMed] [Google Scholar]
- Koo BM, Rhodius VA, Nonaka G et al.. Reduced capacity of alternative sigmas to melt promoters ensures stringent promoter recognition. Gene Dev 2009;23:2426–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkhin Y, Unligil UM, Littlefield O et al.. Evolution of complex RNA polymerases: the complete archaeal RNA polymerase structure. PLoS Biol 2009;7:e1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann J, Narberhaus F. Bacterial RNA thermometers: molecular zippers and switches. Nat Rev Microbiol 2012;10:255–65. [DOI] [PubMed] [Google Scholar]
- Kortmann J, Sczodrok S, Rinnenthal J et al.. Translation on demand by a simple RNA-based thermosensor. Nucleic Acids Res 2011;39:2855–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krásny L, Gourse RL. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J 2004;23:4473–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriner MA, Sevostyanova A, Groisman EA. Learning from the leaders: gene regulation by the transcription termination factor Rho. Trends Biochem Sci 2016;41:690–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L, Zhang B, Ichikawa H et al.. Control of σ factor activity during Bacillus subtilis sporulation. Mol Microbiol 1999;31:1285–94. [DOI] [PubMed] [Google Scholar]
- Kvint K, Farewell A, Nyström T. RpoS-dependent promoters require guanosine tetraphosphate for induction even in presence of high levels of σS. J Biol Chem 2000;275:14795–8. [DOI] [PubMed] [Google Scholar]
- Lacatena RM, Cesareni G. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature 1981;294:623–6. [DOI] [PubMed] [Google Scholar]
- Lago M, Monteil V, Douche T et al.. Proteome remodelling by the stress sigma factor RpoS/σS in Salmonella: identification of small proteins and evidence for post-transcriptional regulation. Sci Rep 2017;7:2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laishram RS, Gowrishankar J. Environmental regulation operating at the promoter clearance step of bacterial transcription. Gene Dev 2007;21:1258–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landini P, Egli T, Wolf J et al.. σS, a major player in the response to environmental stresses in Escherichia coli: role, regulation and mechanisms of promoter recognition. Environ Microbiol Rep 2014;6:1–13. [DOI] [PubMed] [Google Scholar]
- Laurie AD, Bernardo LM, Sze CC et al.. The role of alarmone (p)ppGpp in σN competition for core RNA polymerase. J Biol Chem 2003;278:1494–503. [DOI] [PubMed] [Google Scholar]
- Laursen BS, Sørensen HP, Mortensen KK et al.. Initiation of protein synthesis in bacteria. Microbiol Mol Biol R 2005;69:101–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CL, Swigon D, Murakami KS et al.. Catabolite activator protein: DNA binding and transcription activation. Curr Opin Struct Biol 2004;14:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CR, Kim M, Park YH et al.. RppH-dependent pyrophosphohydrolysis of mRNA is regulated by direct interaction with DapF in Escherichia coli. Nucleic Acids Res 2014;42:12764–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DJ, Minchin SD, Busby SJ. Activating transcription in bacteria. Annu Rev Microbiol 2012;66:125–52. [DOI] [PubMed] [Google Scholar]
- Lee JY, Park H, Bak G et al.. Regulation of transcription from two ssrS promoters in 6S RNA biogenesis. Mol Cells 2013;36:227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon CW, Ross W, Martin-Timasz S et al.. Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Gene Dev 2012;26:2634–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Moyle H, Susskind MM. Target of the transcriptional activation function of phage lambda cI protein. Science 1994;263:75–77. [DOI] [PubMed] [Google Scholar]
- Li W, Bottrill AR, Bibb MJ et al.. The role of zinc in the disulphide stress-regulated anti-sigma factor RsrA from Streptomyces coelicolor. J Mol Biol 2003;333:461–72. [DOI] [PubMed] [Google Scholar]
- Liang X, Li C, Wang W et al.. Integrating T7 RNA polymerase and its cognate transcriptional units for a host-independent and stable expression system in single plasmid. ACS Synth Biol 2018;7:1424–35. [DOI] [PubMed] [Google Scholar]
- Liu C, Heath LS, Turnbough CL Jr. Regulation of pyrBI operon expression in Escherichia coli by UTP-sensitive reiterative RNA synthesis during transcriptional initiation. Gene Dev 1994;8:2904–12. [DOI] [PubMed] [Google Scholar]
- Liu J, Turnbough CL Jr. Effects of transcriptional start site sequence and position on nucleotide-sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli. J Bacteriol 1994;176:2038–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kim DS, Jewett MC. Repurposing ribosomes for synthetic biology. Curr Opin Chem Biol 2017;40:87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J, Startceva S, Kandavalli V et al.. Dissecting the stochastic transcription initiation process in live Escherichia coli. DNA Res 2016;23:203–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetto M, Gribskov M, Gross CA. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol 1992;174:3843–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz TS, Suess B. Small-molecule-binding riboswitches. Microbiol Spectr 2018;6 DOI: 10.1128/microbiolspec.RWR-0025-2018. [DOI] [PubMed] [Google Scholar]
- Low DA, Weyand NJ, Mahan MJ. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect Immun 2001;69:7197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Vogel C, Wang R et al.. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol 2007;25:117–24. [DOI] [PubMed] [Google Scholar]
- Lu Y, Switzer RL. Transcriptional attenuation of the Bacillus subtilis pyr operon by the PyrR regulatory protein and uridine nucleotides in vitro. J Bacteriol 1996;178:7206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano DJ, Vasilyev N, Richards S et al.. A novel RNA phosphorylation state enables 5’end-dependent degradation in Escherichia coli. Mol Cell 2017;67:44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucks JB, Qi L, Whitaker WR et al.. Towards scalable parts families for predictable design of biological circuits. Curr Opin Microbiol 2008;11:567–73. [DOI] [PubMed] [Google Scholar]
- Luijsterburg MS, White MF, van Driel R et al.. The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit Rev Biochem Mol 2008;43:393–418. [DOI] [PubMed] [Google Scholar]
- Lynch M, Marinov GK. The bioenergetics cost of a gene. Proc Natl Acad Sci USA 2015;112:15690–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas WK. The regulation of arginine biosynthesis: its contribution to understanding the control of gene expression. Genetics 1991;128:489–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas WK. The potential for the formation of the arginine biosynthetic enzymes and its masking during evolution. Bioessays 2007;29:484–8. [DOI] [PubMed] [Google Scholar]
- Maas WK, Clark AJ. Studies on the mechanism of repression of arginine biosynthesis in Escherichia coli. II: dominance of repressibility in diploids. J Mol Biol 1964;8:365–70. [DOI] [PubMed] [Google Scholar]
- McCutcheon SR, Chiu KL, Lewis DD et al.. CRISPR-Cas expands dynamic range of gene expression from T7 RNAP promoters. Biotechnol J 2018;13:e1700167. [DOI] [PubMed] [Google Scholar]
- McGary K, Nudler E. RNA polymerase and the ribosome: the close relationship. Curr Opin Microbiol 2013;16:112–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie GA. Secondary structure of the mRNA for ribosomal protein S20. Implications for cleavage by ribonuclease E. J Biol Chem 1992;267:1054–61. [PubMed] [Google Scholar]
- Mackie GA. RNase E: at the interface of bacterial RNA processing and decay. Nat Rev Microbiol 2013;11:45–57. [DOI] [PubMed] [Google Scholar]
- MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol R 2006;70:583–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks SE, Oyston PCF. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 2008;154:3609–23. [DOI] [PubMed] [Google Scholar]
- Maeda H, Fujita N, Ishihama A. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res 2000;28:3497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson LU, Farewell A, Nyström T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol 2005;13:236–42. [DOI] [PubMed] [Google Scholar]
- Mahadevan S, Wright A. A bacterial gene involved in transcription antitermination: regulation at a Rho-independent terminator in the bgl operon of E. coli. Cell 1987;50:485–94. [DOI] [PubMed] [Google Scholar]
- Marco-Marín C, Escamilla-Honrubia JM, Rubio V. First-time crystallization and preliminary X-ray crystallographic analysis of a bacterial-archaeal type UMP-kinase, a key enzyme in microbial pyrimidine biosynthesis. Biochim Biophys Acta 2005;1747:271–5. [DOI] [PubMed] [Google Scholar]
- Mason SW, Greenblatt J. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG and S10. Genes Dev 1991;5:1504–12. [DOI] [PubMed] [Google Scholar]
- Masuda H, Inouye M. Toxins of prokaryotic toxin-antitoxin systems with sequence-specific endoribonuclease activity. Toxins (Basel) 2017;9:E140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews KS, Nichols JC. Lactose repressor protein: functional properties and structure. Prog Nucleic Acid Re 1998;58:127–64. [DOI] [PubMed] [Google Scholar]
- Mauri M, Klumpp S. A model for sigma factor competition in bacterial cells. PLoS Comput Biol 2014;10:e1003845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechold U, Potrykus K, Murphy H et al.. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res 2013;41:6175–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick MJ. In a class of its own—the RNA polymerase sigma factor 54 (sigma N). Mol Microbiol 1993;10:903–9. [DOI] [PubMed] [Google Scholar]
- Metruccio MM, Pigozzi E, Roncarati D et al.. A novel phase variation mechanism in the meningococcus driven by a ligand-responsive repressor and differential spacing of distal promoter elements. PLoS Pathog 2009;5:e1000710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AJ, Ellefson JW, Ellington AD. Directed evolution of a panel of orthogonal T7 RNA polymerase variants for in vivo or in vitro synthetic circuitry. ACS Synth Biol 2015;4:1070–6. [DOI] [PubMed] [Google Scholar]
- Meysman P, Collado-Vides J, Morett E et al.. Structural properties of prokaryotic promoter regions correlate with functional features. PLoS One 2014;9:e88717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micevski D, Dougan DA. Proteolytic regulation of stress response pathways in Escherichia coli. Subcell Biochem 2013;66:105–28. [DOI] [PubMed] [Google Scholar]
- Mika F, Hengge R. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol 2014;11:494–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakhin L, Severinov K. Transcription regulation by bacteriophage T4 AsiA. Protein Expr Purif 2005;41:1–8. [DOI] [PubMed] [Google Scholar]
- Minh PN, Devroede N, Massant J et al.. Insights into the architecture and stoichiometry of Escherichia coli PepA•DNA complexes involved in transcriptional control and site-specific DNA recombination by atomic force microscopy. Nucleic Acids Res 2009;37:1463–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanov AY, Groisman EA. Signal integration in bacterial two-component regulatory systems. Gene Dev 2008;22:2601–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittenhuber G. An inventory of genes encoding RNA polymerase sigma factors in 31 completely sequenced eubacterial genomes. J Mol Microb Biotech 2002;4:77–91. [PubMed] [Google Scholar]
- Mogridge J, Mah TF, Greenblatt J. A protein-RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the lambda N protein. Gene Dev 1995;9:2831–45. [DOI] [PubMed] [Google Scholar]
- Moitoso de Vargas M, Kim S, Landy A. DNA looping generated by DNA bending protein IHF and the two domains of lambda integrase. Science 1989;244:1457–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll I, Afonyushkin T, Vytvytska O et al.. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 2003;9:1308–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve M, Calles B, Mencía M et al.. Binding of phage phi29 protein p4 to the early A2c promoter: recruitment of a repressor by the RNA polymerase. J Mol Biol 1998;283:559–69. [DOI] [PubMed] [Google Scholar]
- Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol 2011;82:1545–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JM, Sørensen HP, Mortensen KK et al.. Macromolecular mimicry in translation initiation: a model for the initiation factor IF2 on the ribosome. IUBMB Life 2000;50:347–54. [DOI] [PubMed] [Google Scholar]
- Morita MT, Tanaka Y, Kodama TS et al.. Translational induction of heat shock transcription factor σ32: evidence for a built-in RNA thermosensor. Gene Dev 1999;13:655–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay J, Sur R, Parrack P. Functional roles of the two cyclic AMP-dependent forms of cyclic AMP receptor protein from Escherichia coli. FEBS Lett 1999;453:215–8. [DOI] [PubMed] [Google Scholar]
- Muranaka N, Sharma V, Nomura Y et al.. An efficient platform for genetic selection and screening of gene switches in Escherichia coli. Nucleic Acids Res 2009;37:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray HD, Schneider DA, Gourse RL. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell 2003;12:125–34. [DOI] [PubMed] [Google Scholar]
- Mutalik VK, Guimaraes JC, Cambray G et al.. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat Methods 2013;10:354–60. [DOI] [PubMed] [Google Scholar]
- Na D, Yoo SM, Chung H et al.. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat Biotechnol 2013;31:170–4. [DOI] [PubMed] [Google Scholar]
- Nadassy K, Wodak SJ, Janin J. Structural features of protein-nucleic acid recognition sites. Biochemistry 1999;38:1999–2017. [DOI] [PubMed] [Google Scholar]
- Nelson HC. Structure and function of DNA-binding proteins. Curr Opin Genet Dev 1995;5:180–9. [DOI] [PubMed] [Google Scholar]
- Newman EB, Lin R. Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli. Annu Rev Microbiol 1995;49:747–75. [DOI] [PubMed] [Google Scholar]
- Nguyen Le Minh P, de Cima S, Bervoets I et al.. Ligand binding specificity of RutR, a member of the TetR family of transcription regulators in Escherichia coli. FEBS Open Biol 2015;5:76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Le Minh P, Nadal M, Charlier D. The trigger enzyme PepA (aminopeptidase A) of Escherichia coli, a transcriptional repressor that generates positive supercoiling. FEBS Lett 2016;590:1816–25. [DOI] [PubMed] [Google Scholar]
- Nguyen Le Minh P, Velázquez-Ruiz C, Vandermeeren S et al.. Differential protein-DNA contacts for activation and repression by ArgP, a LysR-type (LTTR) transcriptional regulator in Escherichia coli. Microbiol Res 2018;206:141–58. [DOI] [PubMed] [Google Scholar]
- Nguyen Ple M, Bervoets I, Maes D et al.. The protein-DNA contacts in RutR•carAB operator complexes. Nucleic Acids Res 2010;38:6286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie L, Wu G, Zhang W. Correlation of mRNA expression and protein abundance affected by multiple sequence features related to translation efficiency in Desulfovibrio vulgaris: a quantitative analysis. Genetics 2006;174:2229–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson G, Belasco JG, Cohen SN et al.. Growth-rate dependent regulation of mRNA stability in Escherichia coli. Nature 1984;312:75–77. [DOI] [PubMed] [Google Scholar]
- Nilsson P, Naureckiene S, Uhlin BE. Mutations affecting mRNA processing and fimbrial biogenesis in the Escherichia coli pap operon. J Bacteriol 1996;178:683–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson P, Uhlin BE. Differential decay of a polycistronic Escherichia coli transcript is initiated by RNase E-dependent endonucleotic processing. Mol Microbiol 1991;5:1791–9. [DOI] [PubMed] [Google Scholar]
- Nodwell JR, Greenblatt J. The nut site of bacteriophage lambda is made of RNA and is bound by transcription antitermination factors on the surface of RNA polymerase. Gene Dev 1991;5:2141–51. [DOI] [PubMed] [Google Scholar]
- Nou X, Skinner B, Braaten B et al.. Regulation of pyelonephritis-associated pili phase-variation in Escherichia coli: binding of the PapI and the Lrp regulatory proteins is controlled by DNA methylation. Mol Microbiol 1993;7:545–53. [DOI] [PubMed] [Google Scholar]
- Oberto J, Nabti S, Jooste V et al.. The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS One 2009;4:e4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera BC, Ugalde E, Martínez-Antonio A. Regulatory dynamics of standard two-component systems in bacteria. J Theor Biol 2010;264:560–9. [DOI] [PubMed] [Google Scholar]
- Österberg S, del Peso-Santos T, Shingler V. Regulation of alternative sigma factor use. Annu Rev Microbiol 2011;65:37–55. [DOI] [PubMed] [Google Scholar]
- Paget MS. Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomolecules 2015;5:1245–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Helmann JD. The σ70 family of sigma factors. Genome Biol 2003;4:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Tsai C-J, Ma B et al.. How do transcription factors select specific binding sites in the genome? Nat Struct Mol Biol 2009;16:1118–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RP, Parajuli P, Koffas MAG et al.. Microbial production of natural and non-natural flavonoids: pathway engineering, directed evolution and systems/synthetic biology. Biotechnol Adv 2016;34:634–62. [DOI] [PubMed] [Google Scholar]
- Pandley SK, Reha D, Zayats V et al.. Binding-competent states for L-arginine in E. coli arginine repressor apoprotein. J Mol Model 2014;20:2330. [DOI] [PubMed] [Google Scholar]
- Park H, McGibbon LC, Potts AH et al.. Translational repression of the RpoS antiadaptor IraD by CsrA is mediated via translational coupling to a short upstream open reading frame. mBio 2017;8:e01355–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YH, Lee CR, Choe M et al.. HPr antagonizes the anti-σ70 activity of Rsd in Escherichia coli. Proc Natl Acad Sci USA 2013;110:21142–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patikoglou GA, Westblade LF, Campbell EA et al.. Crystal structure of Escherichia coli regulator of σ70, Rsd, in complex with σ70 domain 4. J Mol Biol 2007;372:649–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W et al.. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 2004;118:311–22. [DOI] [PubMed] [Google Scholar]
- Peano C, Wolf J, Demol J et al.. Characterization of the Escherichia coli σS core regulon by chromatin immunoprecipitation-sequencing (ChIP-seq) analysis. Sci Rep 2015;5:10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedre B, Young D, Charlier D et al.. Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2 sensing. Proc Natl Acad Sci USA 2018;115:E11623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer A, Margalit H. Accessibility and evolutionary conservation mark bacterial small-RNA target-binding regions. J Bacteriol 2011;193:1690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters E, Charlier D. The Lrp family of transcription regulators in archaea. Archaea 2010;2010:750457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters E, Nguyen Le Minh P, Foulquié-Moreno M et al.. Competitive activation of the Escherichia coli argO gene coding for an arginine exporter by the transcriptional regulators Lrp and ArgP. Mol Microbiol 2009;74:1513–26. [DOI] [PubMed] [Google Scholar]
- Peltier J, Soutourina O. Identification of c-di-GMP-responsive riboswitches. Methods Mol Biol 2017;1657:377–402. [DOI] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN et al.. Regulation through the secondary channel–structural framework for ppGpp-DksA synergism during transcription. Cell 2004;118:297–309. [DOI] [PubMed] [Google Scholar]
- Pérez-Rueda E, Hernandez-Guerrero R, Martinez-Nuñez MA et al.. Abundance, diversity and domain architecture variability in prokaryotic DNA-binding transcription factors. PLoS One 2018;13:e0195332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Rueda E, Janga SC, Martínez-Antonio A. Scaling relationship in the gene content of transcriptional machinery in bacteria. Mol Biosyst 2009;5:1495–501. [DOI] [PubMed] [Google Scholar]
- Pérez-Rueda E, Martínez-Nuñez MA. The repertoire of DNA-binding transcription factors in prokaryotes: functional and evolutionary lessons. Sci Prog 2012;95:315–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Vangeloff AD, Landick R. Bacterial transcription terminators: the RNA 3’-end chronicles. J Mol Biol 2011;412:793–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe C, Eyermann F, Bénard L et al.. Ribosomal protein S15 from Escherichia coli modulates its own translation by trapping the ribosome on the mRNA initiation loading site. Proc Natl Acad Sci USA 1993;90:4394–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Loubière P, Giral L et al.. The significance of translation regulation in the stress response. BMC Genomics 2013;14:588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Milhem H, Loubière P et al.. Bacterial translational regulations: high diversity between all mRNAs and major role in gene expression. BMC Genomics 2012;13:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette J, Nyunoya H, Lusty CJ et al.. DNA sequence of the carA gene and the control region of carAB: tandem promoters, respectively controlled by arginine and the pyrimidines, regulate the synthesis of carbamoyl-phosphate synthetase in Escherichia coli K-12. Proc Natl Acad Sci USA 1984;81:4134–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Vecchione S, Wu H et al.. Engineering orthogonal synthetic timer circuits based on extracytoplasmic function σ factors. Nucleic Acids Res 2018;46:7450–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittard J, Camakaris H, Yang J. The TyrR regulon. Mol Microbiol 2005;55:16–26. [DOI] [PubMed] [Google Scholar]
- Porrua O, Boudvillain M, Libri D. Transcription termination: variations on common themes. Trends Genet 2016;32:508–22. [DOI] [PubMed] [Google Scholar]
- Potts AH, Vakulskas CA, Pannuri A et al.. Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nat Commun 2017;8:1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pul U, Wurm R, Lux B et al.. LRP and H-NS cooperative partners for transcripttion regulation at Escherichia coli rRNA promoters. Mol Microbiol 2005;58:864–76. [DOI] [PubMed] [Google Scholar]
- Pulschen AA, Sastre DE, Machinandiarena F et al.. The stringent response plays a key role in Bacillus subtilis survival of fatty acid starvation. Mol Microbiol 2017;103:698–712. [DOI] [PubMed] [Google Scholar]
- Putzer H, Condon C, Brechemier-Baey D et al.. Transfer RNA-mediated antitermination in vitro. Nucleic Acids Res 2002; 30: 3026–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones M, Kimsey HH, Ross W et al.. LexA represses CTXΦ transcription by blocking access of the α C-terminal domain of RNA polymerase to promoter DNA. J Biol Chem 2006;281:39407–12. [DOI] [PubMed] [Google Scholar]
- Rackham O, Chin JW. A network of orthogonal ribosome x mRNA pairs. Nat Chem Biol 2005;1:159–66. [DOI] [PubMed] [Google Scholar]
- Raghunathan N, Kapshikar RM, Leela JK et al.. Genome-wide relationship between R-loop formation and antisense transcription in Escherichia coli. Nucleic Acids Res 2018;46:3400–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell 2002;108:557–72. [DOI] [PubMed] [Google Scholar]
- Rangel-Chavez C, Galan-Vasquez E, Martinez-Antonio A. Consensus architecture of promoters and transcription units in Escherichia coli: design principles for synthetic biology. Mol Biosyst 2017;13:665–76. [DOI] [PubMed] [Google Scholar]
- Rauhut M, Klug G. mRNA degradation in bacteria. FEMS Microbiol Rev 1999;23:353–70. [DOI] [PubMed] [Google Scholar]
- Ravikumar S, Baylon MG, Park SJ et al.. Engineered microbial biosensors based on bacterial two-component systems and synthetic biotechnology platforms in bioremediation and biorefinery. Microb Cell Fact 2017;16:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Soni A, Bellecourt MJ, Landick R. Mechanisms of bacterial transcription termination: all good things must end. Annu Rev Biochem 2016;85:319–47. [DOI] [PubMed] [Google Scholar]
- Rees WA, Weitzel SE, Das A et al.. Regulation of the elongation-termination decision at intrinsic terminators by antitermination protein N of phage lambda. J Mol Biol 1997;273:797–813. [DOI] [PubMed] [Google Scholar]
- Reitzer L, Schneider BL. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol Mol Biol R 2001;65:422–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repoila F, Majdalani N, Gottesman S. Small non-coding RNAs, co-ordinators of adaptive processes in Escherichia coli: the RpoS paradigm. Mol Microbiol 2003;48:855–61. [DOI] [PubMed] [Google Scholar]
- Rhodius VA, Segall-Shapiro TH, Sharon BD et al.. Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Mol Syst Biol 2013;9:702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice PA, Yang S, Mizuuchi K et al.. Crystal structure of IHF-DNA complex: a protein induced DNA U-turn. Cell 1996;87:1295–306. [DOI] [PubMed] [Google Scholar]
- Richards J, Belasco JG. Distinct requirements for 5’-monophosphate-assisted RNA cleavage by Escherichia coli RNase E and RNase G. J Biol Chem 2016;291:5038–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CJ, Vincent HA, Wu M-C et al.. Modular riboswitch toolsets for synthetic genetic control in diverse bacterial species. J Am Chem Soc 2014;136:10615–24. [DOI] [PubMed] [Google Scholar]
- Roßmanith J, Narberhaus F. Exploring the modular nature of riboswitches and RNA thermometers. Nucleic Acids Res 2016;44:5410–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roßmanith J, Weskamp M, Narberhaus F. Design of a temperature-responsive transcription terminator. ACS Synth Biol 2018;7:613–21. [DOI] [PubMed] [Google Scholar]
- Rodrigue A, Quentin Y, Lazdunski A et al.. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol 2000;8:498–504. [DOI] [PubMed] [Google Scholar]
- Romeo T, Vakulskas CA, Babitzke P. Post-transcriptional regulation on a global scale: form and function of Crs/Rsm systems. Environ Microbiol 2013;15:313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Gosink KK, Salomon J et al.. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science 1993;262:1407–13. [DOI] [PubMed] [Google Scholar]
- Ross W, Vrentas CE, Sanchez-Vazquez P et al.. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell 2013;50:420–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe FM, Bahr T, Stülke J et al.. Activation of Escherichia coli antiterminator BglG requires its phosphorylation. Proc Natl Acad Sci USA 2012;109:15906–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra P, Prajapati RK, Banerjee R et al.. Novel mechanism of gene regulation: the protein Rv1222 of Mycobacterium tuberculosis inhibits transcription by anchoring the RNA polymerase onto DNA. Nucleic Acids Res 2015;43:5855–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff EF, Record MT Jr, Artsimovitch I. E. coli RNA polymerase determinants of open complex lifetime and structure. J Mol Biol 2015;427:2435–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytter JV, Helmark S, Chen J et al.. Synthetic promoter libraries for Corynebacterium glutamicum. Appl Microbiol Biot 2014;98:2617–23. [DOI] [PubMed] [Google Scholar]
- Saberi F, Kamali M, Najafi A et al.. Natural antisense RNAs as mRNA regulatory elements in bacteria: a review on function and applications. Cell Mol Biol Lett 2016;21:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saecker RM, Record MT Jr, deHaseth PL. Mechanism of bacterial transcription initiation: RNA polymerase-promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J Mol Biol 2011;412:754–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsi S, Floess E, Aldridge C et al.. Continuous control of flagellar gene expression by the σ28-FlgM regulatory circuit in Salmonella enterica. Mol Microbiol 2011;79:264–78. [DOI] [PubMed] [Google Scholar]
- Salis HM. The ribosome binding site calculator. Methods Enzymol 2011;498:19–42. [DOI] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol 2009;27:946–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotech 2014;32:347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo TJ, Artsimovitch I. Termination and antitermination: RNA polymerase runs a stop sign. Nat Rev Microbiol 2011;9:319–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer AT, Carver DL, Bigelow B et al.. Lambda N antitermination system: functional analysis of phage interactions with the host NusA protein. J Mol Biol 1987;194:679–90. [DOI] [PubMed] [Google Scholar]
- Schreiter ER, Drennan CL. Ribbon-helix-helix transcription factors: variations on a theme. Nat Rev Microbiol 2007;5:710–20. [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Choi KY, Zalkin H et al.. Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices. Science 1994;266:763–70. [DOI] [PubMed] [Google Scholar]
- Schwartz EC, Shekhtman A, Dutta K et al.. A full-length group 1 bacterial sigma factor adopts a compact structure incompatible with DNA binding. Chem Biol 2008;15:1091–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlyarova N, Shamovsky I, Bharati BK et al.. sRNA-mediated control of transcription termination in E. coli. Cell 2016;167:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall-Shapiro TH, Meyer AJ, Ellington AD et al.. A ‘resource allocator’ for transcription based on a highly fragmented T7 RNA polymerase. Mol Syst Biol 2014;10:742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Apurva D, Satija R et al.. Design of a toolbox of RNA thermometers. ACS Synth Biol 2017;6:1461–70. [DOI] [PubMed] [Google Scholar]
- Serganov A, Nudler E. A decade of riboswitches. Cell 2013;15:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshasayee AS. Gene expression homeostasis and chromosome architecture. Bioarchitecture 2014;4:221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla E, Bes MT, González A et al.. Redox-based transcriptional regulation in prokaryotes: revisiting model mechanisms. Antiox Redox Signal 2018. DOI: 10.1089/ars.2017.7442. [DOI] [PubMed] [Google Scholar]
- Sharma V, Nomura Y, Yokobayashi Y. Engineering complex riboswitch regulation by dual genetic selection. J Am Chem Soc 2008;130:16310–5. [DOI] [PubMed] [Google Scholar]
- Shimada T, Fujita N, Yamamoto K et al.. Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS One 2011;6:e20081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Hirao K, Kori A et al.. RutR is the uracil/thymine-sensing master regulator of a set of genes for synthesis and degradation of pyrimidines. Mol Microbiol 2007;66:744–57. [DOI] [PubMed] [Google Scholar]
- Shingler V. Signal sensory systems that impact σ54-dependent transcription. FEMS Microbiol Rev 2011;35:425–40. [DOI] [PubMed] [Google Scholar]
- Shis DL, Bennett MR. Library of synthetic transcriptional AND gates built with split T7 RNA polymerase mutants. Proc Natl Acad Sci USA 2013;110:5028–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shis DL, Hussain F, Meinhardt S et al.. Modular, multi-input transcriptional logic gating with orthogonal LacI/GalR family chimeras. ACS Synth Biol 2014;3:645–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Seshasayee ASN. Nucleoid-associated proteins: genome level occupancy and expression analysis. Methods Mol Biol 2017;1624:85–97. [DOI] [PubMed] [Google Scholar]
- Singh V. Recent advancements in synthetic biology: current status and challenges. Gene 2014;535:1–11. [DOI] [PubMed] [Google Scholar]
- Skoko D, Yoo D, Bai H et al.. Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein Fis. J Mol Biol 2006;364:777–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Shanahan CA, Moore EL et al.. Structural basis of differential ligand recognition by two classes of bis-(3΄-5΄)-cyclic dimeric guanosine monophosphate-binding riboswitches. Proc Natl Acad Sci USA 2011;108:7757–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TG, Hoover TR. Deciphering bacterial flagellar gene regulatory networks in the genomic era. Adv Appl Microbiol 2009;67:257–95. [DOI] [PubMed] [Google Scholar]
- Sojka L, Kouba T, Barvik I et al.. Rapid changes in gene expression: DNA determinants of promoter regulation by the concentration of the transcription initiating NTP in Bacillus subtilis. Nucleic Acids Res 2011;39:4598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solem C, Jensen PR. Modulation of gene expression made easy. App Environ Microb 2002;68:2397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N, Nguyen Duc T, van Oeffelen L et al.. Expanded target and cofactor repertoire for the transcriptional activator LysM from Sulfolobus. Nucleic Acids Res 2013;41:2932–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen KI, Baker KE, Kelln RA et al.. Nucleotide pool-sensitive selection of the transcriptional start site in vivo at the Salmonella typhimurium pyrC and pyrD promoters. J Bacteriol 1993;175:4137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza BM, Castro TL, Carvalho RD et al.. σ(ECF) factors of gram-positive bacteria: a focus on Bacillus subtilis and the CMNR group. Virulence 2014;5:587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M, Graffe M, Butler JS et al.. Genetic definition of the translational operator of the threonine-tRNA ligase gene in Escherichia coli. Proc Natl Acad Sci USA 1986;83:4384–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M, Plumbridge JA, Butler JS et al.. Autogenous control of Escherichia coli threonyl-tRNA synthetase expression in vivo. J Mol Biol 1985;185:93–104. [DOI] [PubMed] [Google Scholar]
- Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol 2008;11:100–5. [DOI] [PubMed] [Google Scholar]
- Stanton BC, Nielsen AAK, Tamsir A et al.. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat Chem Biol 2014;10:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U, Zengel JM, Tovbina M et al.. RNA-structural mimicry in Escherichia coli ribosomal protein L4-dependent regulation of the S10 operon. J Biol Chem 2003;278:28237–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A. An inhibitor of host sigma-stimulated core enzyme activity that purifies with DNA-dependent RNA polymerase of E. coli following T4 phage infection. Biochem Bioph Res Co 1973;54:488–93. [DOI] [PubMed] [Google Scholar]
- Stoebel DM, Free A, Dorman CJ. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 2008;154:2533–45. [DOI] [PubMed] [Google Scholar]
- Strawn R, Melichercik M, Green M et al.. Symmetric allosteric mechanism of hexameric Escherichia coli arginine repressor exploits competition between L-arginine ligands and resident arginine residues. PLoS Comput Biol 2010;6:e1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr Opin Struct Biol 2004;14:28–35. [DOI] [PubMed] [Google Scholar]
- Swint-Kruse L, Matthews KS. Allostery in the LacI/GalR family: variations on a theme. Curr Opin Microbiol 2009;12:129–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwajkajzer D, Dai L, Fukayama JW et al.. Quantitative analysis of DNA binding by the Escherichia coli arginine repressor. J Mol Biol 2001;312:949–62. [DOI] [PubMed] [Google Scholar]
- Tagami S, Sekine S, Minakhin L et al.. Structural basis for promoter specificity switching of RNA polymerase by a phage factor. Gene Dev 2014;28:521–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi MK, Lucks JB. A modular strategy for engineering orthogonal chimeric RNA transcription regulators. Nucleic Acids Res 2013;41:7577–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani TH, Khodursky A, Blumenthal RM et al.. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc Natl Acad Sci USA 2002;99:13471–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme K, Hill R, Segall-Shapiro TH et al.. Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res 2012;40:8773–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramoto H, Inui M, Yukawa H. OxyR acts as a transcriptional repressor of hydrogen peroxide-inducible antioxidant genes in Corynebacterium glutamicum R. FEBS J 2013;280:3298–312. [DOI] [PubMed] [Google Scholar]
- Thomason MK, Storz G. Bacterial antisense RNAs: how many are there, and what are they doing? Ann Rev Genet 2010;44:167–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano MB, Kullik I, Trinh F et al.. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 1994;78:897–909. [DOI] [PubMed] [Google Scholar]
- Treviño-Quintanilla LG, Freyre-González JA, Martínez-Flores I. Anti-sigma factors in E. coli: common regulatory mechanisms controlling sigma factors availability. Curr Genomics 2013;14:378–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi L, Zhang Y, Lin Z. Bacterial sigma factors as targets for engineered or synthetic transcriptional control. Front Bioeng Biotechnol 2014;2:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbough CL., Jr Regulation of the Escherichia coli aspartate transcarbamylase synthesis by guanosine tetraphosphate and pyrimidine ribonucleoside triphosphates. J Bacteriol 1983;153:998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbough CL., Jr Regulation of gene expression by reiterative transcription. Curr Opin Microbiol 2011;14:142–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbough CL Jr, Switzer RL. Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors. Microbiol Mol Biol R 2008;72:266–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakulskas CA, Potts AH, Babitzke P et al.. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol R 2015;79:193–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valbuzzi A, Gollnick P, Babitzke P et al.. The anti-trp RNA-binding attenuation protein (Anti-TRAP), AT, recognizes the tryptophan-activated RNA binding domain of the TRAP regulatory protein. J Biol Chem 2002;277:10608–13. [DOI] [PubMed] [Google Scholar]
- Valbuzzi A, Yanofsky C. Inhibition of the B. subtilis regulatory protein TRAP by the TRAP-inhibitory protein, AT. Science 2001;293:2057–9. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P, Søgaard-Andersen L, Pedersen H. A flexible partnership: the CytR anti-activator and the cAMP-CRP activator protein, comrades in transcription control. Mol Microbiol 1996;20:461–6. [DOI] [PubMed] [Google Scholar]
- van der Woude MW. Phase variation: how to create and coordinate population diversity. Curr Opin Microbiol 2011;14:205–2011. [DOI] [PubMed] [Google Scholar]
- van der Woude MW, Braaten BA, Low DA. Evidence for global regulatory control of pilus expression in Escherichia coli by Lrp and DNA methylation: model building based on analysis of pap. Mol Microbiol 1992;6:2429–35. [DOI] [PubMed] [Google Scholar]
- van Tilbeurgh H, Declerck N. Structural insights into the regulation of bacterial signaling proteins containing PRDs. Curr Opin Struct Biol 2001;11:685–93. [DOI] [PubMed] [Google Scholar]
- Vinkenborg JL, Karnowski N, Famulok M. Aptamers for allosteric regulation. Nat Chem Biol 2011;7:519–27. [DOI] [PubMed] [Google Scholar]
- Vrentas CE, Gaal T, Ross W et al.. Response of RNA polymerase to ppGpp: requirement for the ω subunit and relief of this requirement by DksA. Gene Dev 2005;19:2378–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldminghaus T, Heidrich N, Brantl S et al.. FourU: a novel type of RNA thermometer in Salmonella. Mol Microbiol 2007;65:413–24. [DOI] [PubMed] [Google Scholar]
- Wang H, Glansdorff N, Charlier D. The arginine repressor of Escherichia coli K-12 makes direct contacts to minor and major groove determinants of the operators. J Mol Biol 1998;277:805–24. [DOI] [PubMed] [Google Scholar]
- Wang Q, Calvo JM. Lrp, a major regulatory protein in Escherichia coli, bends DNA and can organize the assembly of a higher-order nucleoprotein structure. EMBO J 1993;12:2495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM. 6S RNA: a regulator of transcription. Mol Microbiol 2007;65:1425–31. [DOI] [PubMed] [Google Scholar]
- Wei Q, Minh PN, Dötch A et al.. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res 2012;40:4320–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol 2011;9:85–98. [DOI] [PubMed] [Google Scholar]
- Wilkomm DK, Hartmann RK. 6S RNA - an ancient regulator of bacterial RNA polymerase rediscovered. Biol Chem 2005;386:1273–7. [DOI] [PubMed] [Google Scholar]
- Wilson C, Dombroski AJ. Region 1 of σ70 is required for efficient isomerization and initiation of transcription by Escherichia coli RNA polymerase. J Mol Biol 1997;267:60–74. [DOI] [PubMed] [Google Scholar]
- Wilson HR, Archer CD, Liu JK et al.. Translational control of pyrC expression mediated by nucleotide-sensitive selection of transcriptional start sites in Escherichia coli. J Bacteriol 1992;174:514–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson HR, Chan PT, Turnbough CL Jr. Nucleotide sequence and expression of the pyrC gene of Escherichia coli K-12. J Bacteriol 1987;169:3051–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA 2002;99:15908–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 2002;419:952–6. [DOI] [PubMed] [Google Scholar]
- Wolz C, Geiger T, Goerke C. The synthesis and fuction of the alarmone (p)ppGpp in firmicutes. Int J Med Microbiol 2010;300:142–7. [DOI] [PubMed] [Google Scholar]
- Wong GT, Bonocora RP, Schep AN et al.. Genome-wide transcriptional response to varying RpoS levels in Escherichia coli K-12. J Bacteriol 2017;199:pii:e00755–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Yan Q, Jones JA et al.. Metabolic burden: cornerstones in metabolic engineering applications. Trends Biotechnol 2016;34:652–64. [DOI] [PubMed] [Google Scholar]
- Xiong XF, Reznikoff WS. Transcriptional slippage during the transcription initiation process at a mutant lac promoter in vivo. J Mol Biol 1993;231:569–80. [DOI] [PubMed] [Google Scholar]
- Yang J, Hwang JS, Camakaris H et al.. Mode of action of the TyrR protein: repression and activation of the tyrP promoter of Escherichia coli. Mol Microbiol 2004;52:243–56. [DOI] [PubMed] [Google Scholar]
- Yang Y, Darbari VC, Zhang N et al.. Transcription. Structures of the RNA polymerase-σ54 reveal new and conserved regulatory strategies. Science 2015;349:882–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Qin L, Egger LA et al.. Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J Biol Chem 2006;281:17114–23. [DOI] [PubMed] [Google Scholar]
- Young BA, Gruber TM, Gross CA. Views on transcription initiation. Cell 2002;109:417–20. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Jones AM, Thomas EE et al.. A split transcriptional repressor that links protein solubility to an orthogonal genetic circuit. ACS Synth Biol 2018;7:pp 2126–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel JM, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog Nucleic Acid Re 1994;47:331–70. [DOI] [PubMed] [Google Scholar]
- Zhang N, Darbari VC, Glyde R et al.. The bacterial enhancer-dependent RNA polymerase. Biochem J 2016;473:3741–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 1998;279:1718–21. [DOI] [PubMed] [Google Scholar]
- Zheng M, Wang X, Templeton LJ et al.. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol 2001;183:4562–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi J, Mathew E, Freundlich M. Lrp binds to two regions in the dadAX promoter region of Escherichia coli to repress and activate transcription directly. Mol Microbiol 1999;32:29–40. [DOI] [PubMed] [Google Scholar]
- Zhou R, Kroos L. BofA protein inhibits intramembrane proteolysis of pro-σK in an intercompartmental signaling pathway during Bacillus subtilis sporulation. Proc Natl Acad Sci USA 2004;101:6385–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Gottesman S. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J Bacteriol 1998;180:1154–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Gottesman S, Hoskins JR et al.. The RssB response regulator directly targets σS for degradation by ClpXP. Gene Dev 2001;15:627–7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubay G, Schwartz D, Beckwith J. Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc Natl Acad Sci USA 1970;66:104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]