Abstract
Cells from all three domains of life, Archaea, Bacteria and Eukarya, produce extracellular vesicles (EVs) which are sometimes associated with filamentous structures known as nanopods or nanotubes. The mechanisms of EV biogenesis in the three domains remain poorly understood, although studies in Bacteria and Eukarya indicate that the regulation of lipid composition plays a major role in initiating membrane curvature. EVs are increasingly recognized as important mediators of intercellular communication via transfer of a wide variety of molecular cargoes. They have been implicated in many aspects of cell physiology such as stress response, intercellular competition, lateral gene transfer (via RNA or DNA), pathogenicity and detoxification. Their role in various human pathologies and aging has aroused much interest in recent years. EVs can be used as decoys against viral attack but virus-infected cells also produce EVs that boost viral infection. Here, we review current knowledge on EVs in the three domains of life and their interactions with the viral world.
Keywords: extracellular vesicles, nanotubes, Archaea, virus, evolution, LUCA
This review discusses the current research on extracellular vesicle (EV) biology and their various roles in the three domains of life: Archaea, Bacteria and Eukarya. The physiological and/or evolutionary relationships between EVs and viruses are also examined.
INTRODUCTION
The release of membrane-bound vesicles is a universally conserved cellular process that occurs in all three domains of life (Deatherage and Cookson 2012; Schatz and Vardi 2018) (Fig. 1). The production of these extracellular vesicles (EVs) has been systematically observed each time researchers have investigated this phenomenon, suggesting that all cells are potentially capable of EV production. These EVs can transport various molecular cargoes and deliver them to recipient cells by fusion with the cytoplasmic membrane and/or by endocytosis in eukaryotes, modifying their physiology.
Figure 1.
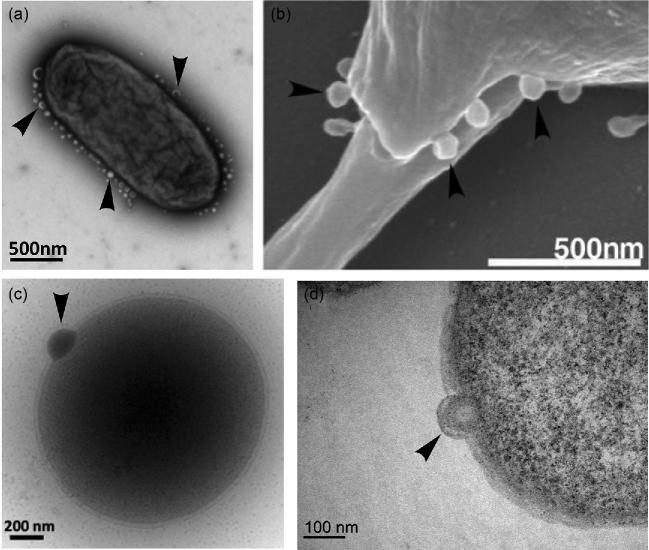
Biogenesis of extracellular vesicles in the three domains of life. Vesicle budding indicated with arrows. (a) TEM showing hypervesiculation in the bacterium S. typhimurium. Image kindly provided by Mario F. Feldman (University of Alberta, Canada). (b) SEM showing microvesicles budding from the eukaryote Leishmania donovani. Image reprinted from Silverman et al. (2008). (c) Cryo-TEM of vesicle budding from the archaeon T. kodakaerensis. The protrusion of the S layer can also be observed clearly. (d) TEM of ultrathin cell sections of vesicle budding from T. kodakaerensis. Figures (c) and (d) provided by the authors.
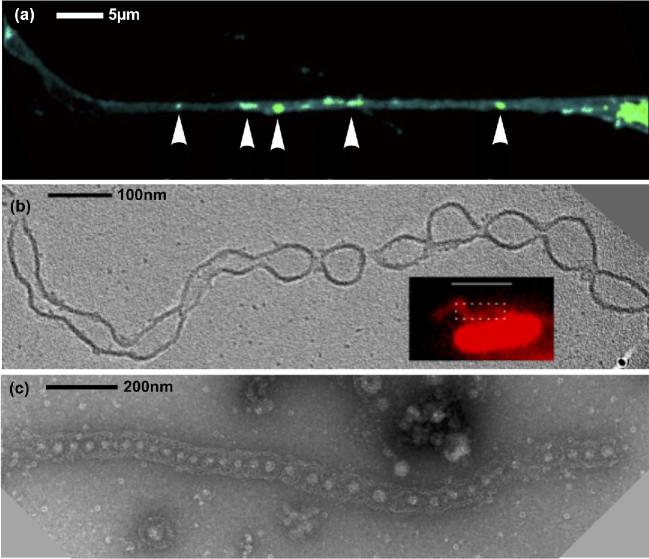
Because of their small size (from 20 to 500 nm for most of them), EVs have been mainly observed by electron microscopy, either as free particles in the culture medium following concentration or as nascent particles budding from the cell membranes (refer to Fig. 1). EVs observed by transmission electron microscopy (TEM) are usually heterogeneous in size with irregular shapes, such as a cup-shaped appearance, possibly due to sample preparation. However, they appear perfectly spherical when observed by cryo-electron microscopy without chemical fixation or contrasting (Koning et al.2013; Raposo and Stoorvogel 2013; Gorlas et al.2015; Pérez-Cruz et al.2015; Milasan et al.2016) (Fig. 1c). EVs are sometimes associated with long filamentous structures connecting cells in the three domains of life, known as nanopods and nanotubes in Archaea and Bacteria and tunneling nanotubes (TNTs) or ‘microvillus-like protrusions’ in Eukaryotes (Fig. 2) (Marguet et al.2013; Koistinen et al.2015; Keller et al.2017; Nawaz and Fatima 2017; Stempler et al.2017). These nanotubes often contain arrays of EVs surrounded by membranes, suggesting that they could be involved in EV production (Soler et al.2008; Dubey and Ben-Yehuda 2011; Shetty et al.2011; Marguet et al.2013). Notably, nanotubes seem to be involved in the production of EVs by some cancerous human cells (Rilla et al.2013).
Figure 2.
Nanotube production in the three domains of life. (a) TNT connecting eukaryotic (human) cells, with labeled vesicles indicated by arrows. Adapted with permission from Keller et al. (2017): image cropped and arrow style altered. (b) 'Nanotubes' produced by the bacteria S. oneidensis form outer membrane extensions with regular constrictions forming vesicles. Adapted with permission from Subramanian et al. (2018). Image courtesy of Poorna Subramanian (California Institute of Technology, USA). (c) 'Nanopods' produced by the archaeon T. prieurii. Discrete vesicles are surrounded by the cellular S-layer forming a tubular structure. Image kindly provided by Aurore Gorlas (Institute for Integrative Biology of the Cell, Université Paris-Saclay, France).
The importance of EV production as a major phenomenon in the living world was for a long time underestimated, with EVs being initially dismissed as platelets or cellular ‘dust’ (Wolf 1967; Cocucci, Racchetti and Meldolesi 2009) and ignored in most microbiology textbooks. However, EV-focused research over the past two decades has begun to reveal their significance in cell physiology and their diverse biological functions have been extensively documented. It is now well recognized that EVs and related nanotubes can transport a variety of cargoes, including proteins, lipids, sugars and nucleic acids, and play important roles in all types of cell-to-cell interactions. The concentration of cargoes within membrane-bound EVs offers protection against extracellular enzymes and the aqueous environment and allows the secretion of both lipophilic and hydrophobic compounds. In particular, EVs are the only secretion system, proposed to be named secretion system type zero (Guerrero-Mandujano et al.2017), allowing cells to secrete and share with other cells lipids, hydrophobic, leaderless or denatured proteins, or hydrophobic signaling molecules (for recent reviews, see Jurkoshek et al.2016; Penfornis et al.2016; Tkach and Théry 2016; Domingues and Nielsen 2017; Kouwaki et al.2017; Takahashi et al.2017; Toyofuku et al. 2017a). Additionally, the transfer of nucleic acids between cells via EVs is being increasingly investigated. In Archaea and Bacteria, fusion of EVs carrying DNA between cells has been proposed as a novel mechanism for horizontal gene transfer (HGT), in addition to the well-known mechanisms of transformation, transduction and conjugation (Domingues and Nielsen 2017; Erdmann et al.2017). In Eukaryotes and Bacteria, EVs seem to be involved in the long-distance transfer of genetic information via the shuttling of regulatory or mRNA between cells (Tsatsaronis et al.2018).
The impact of EV production on pathogenesis is becoming an increasingly active research field (El Andaloussi et al.2013). It is now well established that many pathogenic bacteria use their EVs to deliver toxic compounds to infected cells (Bitto et al.2017) whereas eukaryotic EVs are involved in many important human pathologies, from cancer to cardiovascular and neurodegenerative diseases with many laboratories exploring their potential to be used as biomarkers or delivery vehicles for therapeutic action (Yáñez-Mó et al.2015; Robbins, Dorronsoro and Booker 2016; Thompson et al.2016; Liu et al.2017; Mateescu et al.2017). EVs are also major players in aging (Takasugi 2018). Remarkably, the composition of EVs changes with age in humans, and a pioneering experiment demonstrated that administration of EVs isolated from young cells ameliorates age-related functional decline in older mice (Zhang et al.2017).
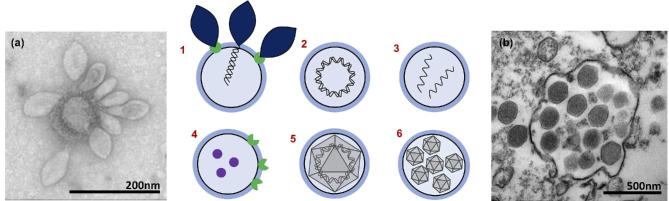
A research area which has become increasingly important in recent years is that of the interactions between EVs and viruses. Strikingly, EVs resemble the virions of enveloped viruses when observed by electron microscopy. Furthermore, EVs can attach to virions (if EVs harbor virus receptors at their surface), engulf viral particles or mimic viral particles by carrying viral proteins, RNA and DNA (Fig. 3). Some EVs containing viral genome or plasmids have been described as viral or plasmid vesicles (plasmidions) and could facilitate the propagation of these mobile elements (Forterre, Da Cunha and Catchpole 2017). Whereas EVs can sometimes act as decoys to limit viral infection, virus themselves can manipulate the production of EVs from infected cells (the virocell, sensu Forterre 2013) to their own benefit (Altan-Bonnet 2016). These observations have fueled speculation on the physiological and/or evolutionary relationships between EVs and viruses, suggesting that studying EVs could be helpful in understanding the origin of viruses themselves (Jalasvuori and Bamford 2008; Forterre and Krupovic 2012).
Figure 3.
EVs and viruses interact in multiple ways. 1 and (a): Virus receptors on vesicles could act as decoys protecting the host from infection. (a) TEM showing several Sulfolobus spindle-shaped virus 1 (SSV1), from the Fuselloviridae family, attached to a membrane vesicle. 2 and 3: Encapsulated DNA/ RNA can be infectious as in pleolipoviruses or plasmidions. 4: Virus receptors and effectors can transfer between cells, promoting infection of non-susceptible hosts. 5: Membrane-bound viruses resist human attack. 6 and (b): VPVs allow for high MOI and 'Trojan horse’-style infection. Image (a) kindly provided by Virginija Krupovic, Institut Pasteur, France. Image (b) kindly provided by Jônatas Santos Abrahão, Institute of Biological Sciences, Universidade Federal de Minas Gerais, Brazil and obtained by the Center of Microscopy of UFMG, Brazil.
Finally, the ubiquity of EVs suggests that their production could have already existed at the time of the last universal common ancestor (LUCA) (Gill and Forterre 2016). However, it remains to be seen if any of the modern mechanisms of EV production are homologous in the three domains of life, testifying for their antiquity, or if different mechanisms of EV production have originated independently in different domains. Unfortunately, our knowledge concerning the mechanisms of EV biogenesis is still very limited, and as yet it has not been possible to draw clear-cut evolutionary connections between their modes of production in different domains. Genetic and biochemical analyses have only begun to elucidate mechanistic aspects of EV production in Bacteria (Wessel et al.2013; Devos et al.2015; Kulp et al.2015; Roier et al.2015, 2016a,b; Resch et al.2016; Turnbull et al.2016; Ojima et al.2017) and Eukaryotes (Muralidharan-Chari et al.2009; Ostrowski et al.2010; Oliveira et al.2010b; Gross et al.2012; Rilla et al. 2013, 2014; Tricarico, Clancy and D'Souza-Schorey 2016). These analyses suggest that membrane protein/lipid composition play a crucial role in EV production (Bonnington and Kuehn 2016; Elhenawy et al.2016; Skotland, Sandvig and Llorente 2017).
All these fascinating new discoveries and hypotheses explain why, in recent years, specialized journals focusing on EVs have been launched, such as the Journal of Extracellular Vesicles, and regular international meetings dedicated to EVs have been established as well as an academic society, the International Society for Extracellular Vesicles (ISEV). The data from various EV studies have been listed in three databases dedicated to EVs, namely Exocarta (lipids, RNA and proteins identified in exosomes), Vesiclepedia (data from different types of EVs) and EVpedia (high-throughput analyses and data on proteins, nucleic acids and lipids EVs) (Mathivanan and Simpson 2009; Kalra et al.2012; Kim et al. 2013, 2015).
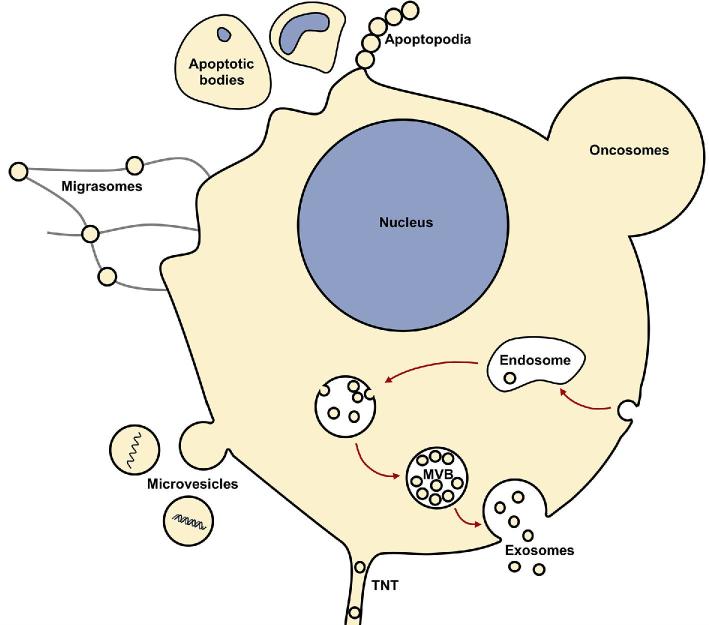
EVs are diverse in origin and nature, and there is little consensus on their classification (Gould and Raposo 2013). They are known under a variety of names such as membrane vesicles, extracellular membrane vesicles, microvesicles, microparticles, exosomes and ectosomes (in Eukaryotes), as well as more specialized names for particles arising from specific cell types e.g. oncosomes (produced by cancer cells), migrasomes (produced by amoeboid cells), apoptotic bodies (produced by cells during apoptosis), etc. (Colombo, Raposo and Théry 2014; Minciacchi et al.2015) (Fig. 4). In recent years, the term ‘extracellular vesicles’ (EV) has been regularly used in most reviews on this topic. This term has been adopted by ISEV and journals devoted to their studies (Mateescu et al.2017). Throughout this review, the term ‘extracellular vesicles’ (EVs) will be used a priori to refer to all types of membrane vesicles in the three domains of life, except when the identification of a specific subset of EVs is well documented, such as the well-known outer membrane vesicles (OMV) produced by Bacteria.
Figure 4.
EV production in Eukaryotes. Multiple types of EVs originate through many complex and varied pathways. Eukaryotic EV functions include protein sorting/trafficking, intercellular communication, host adaptation during infection, metastatic niche adaptation, immune evasion and pathogenesis.
The number of excellent reviews discussing recent and past studies on EVs has exploded of late. Many of them have focused on particular area of EV studies such as HGT (Domingues and Nielsen 2017), immune response regulation (Kouwaki et al.2017), aging (Takasugi 2018), RNA content (Tsatsaronis et al.2018), EVs and cancer (Xu et al.2018) or interactions between EVs and viruses (Altan-Bonnet 2016; Nolte-‘t Hoen et al.2016). Some have covered EVs produced by Bacteria, especially dealing with OMVs (Schwechheimer and Kuehn 2015; O’Donoghue and Krachler 2016; Jan 2017; O’Donoghue et al.2017; Toyofuku et al. 2017a) but most of them are devoted to eukaryotic EVs, with an emphasis on exosomes (Yáñez-Mó et al.2015; Kalra, Drummen and Mathivanan 2016; Guo et al.2017; Rashed et al.2017; De la Torre Gomez et al.2018). In a review sponsored by the European COST action initiative ‘Microvesicles and Exosomes in Disease and Health’, the authors describe in great detail EVs in ‘higher’ and ‘lower’ organisms (Eukaryotes) and devote only a tenth of this review to EVs from pathogenic bacteria, without a single word on EVs produced by Archaea (Yáñez-Mó et al.2015).
The traditional divide between prokaryotes and eukaryotes has profoundly influenced biologists; bacterial EVs have been studied independently of eukaryotic ones, and archaeal EVs for a long time have been mostly ignored. Among eukaryotes, most studies on EVs have focused on human EVs, often without realizing that this was not a specific phenomenon restricted to multicellular ‘higher’ organisms, and that studying this process in other model organisms, including microorganisms, could provide new insights that could be useful to grasp its generality and specificity. Here, we review EV biology and discuss various roles EVs play in the three domains of life, with some emphasis on archaeal EVs (to compensate for their absence in many other reviews) and on the interactions between EVs and the viral world, a research area in which connections between discoveries made in different domains of life is especially striking. We also briefly discuss the possible role of EVs at different steps of cellular evolution, in particular regarding the role of EVs in recent hypotheses on the origin of Eukaryotes (Baum and Baum 2014; Gould, Garg and Martin 2016). We hope that the comparative approach used in this review will help to make the study of EVs a common goal shared by all biologists.
EVs in Bacteria
The domain Bacteria contains very diverse prokaryotic microorganisms unified by common informational machineries for DNA replication, transcription and translation that are strikingly different from those of Eukaryotes and Archaea (Woese, Kandler and Wheelis 1990). Bacteria are also characterized by the presence of peptidoglycan in their cell wall, a rigid biopolymer that creates conditions for EV production quite distinct from those in the other two domains. Peptidoglycan was probably already present in the last bacterial common ancestor and is only lacking today in a few bacterial groups (Mycoplasmatales and some Planctomycetales). Bacteria exhibit various types of cell envelopes that will impact on the nature of their EVs. The vast majority of Bacteria have an outer lipopolysaccharide (LPS)-containing membrane and a rather thin layer of peptidoglycan located in the periplasmic space, i.e. between the outer and inner membrane. They are usually referred to as Gram-negative or diderm bacteria. In contrast, most bacteria of the phylum Firmicutes stain Gram positive and are sometimes referred as monoderm bacteria because they have a single membrane covered by a thick layer of peptidoglycan. Bacteria of the phylum Actinobacteria, including such important species as Streptomyces and Mycobacteria, are rather distinct from both the classical monoderms and diderms. They stain Gram positive because their thin peptidoglycan is directly covered by a thick polysaccharide layer. Many bacteria also contain a proteineous S-layer that plays an important role in the interaction between bacteria and their environment (Fagan and Fairweather 2014). However, this S-layer has been lost in many species that have been studied for a long time in the laboratory, especially those that have been studied for EV production.
The majority of EV studies in Bacteria have been carried out on Proteobacteria, the most abundant and well-studied phylum of diderm bacteria. Early studies focused on model organisms and/or pathogenic species of Proteobacteria such as Escherichia coli, Neisseria meningitidis, Pseudomonas aeruginosa, Shigella flexneri, Helicobacter pylori, Legionella pheumophila and Shewanella livingstonensis (for early publications, see reviews by Beveridge 1999; Mashburn-Warren and Whiteley 2006; Kulp and Kuehn 2010 and references therein). The study of bacterial EVs was mainly pioneered by studies in the laboratory of Terry Beveridge at a time when this phenomenon was still largely underestimated by microbiologists (Kadurugamuwa and Beveridge 1995; Beveridge 1999). Several types of EVs have been described in Bacteria, with the most studied being the so-called OMVs produced by diderm bacteria. These OMVs are formed by budding of the LPS-containing outer membrane (OM) and mainly contain periplasmic components (Figs 5 and 6). However, EVs containing both the outer and inner membranes of diderm bacteria have been recently identified in several species, and termed outer-inner membrane vesicles (O-IMV) (Pérez-Cruz et al.2013, 2015) (Fig. 6). These EVs contain both periplasmic and cytoplasmic components and originate from the cytoplasmic membrane. EVs have also been observed from monoderm bacteria with thick cell walls, such as Firmicutes and Actinobacteria (Prados-Rosales et al.2014). This was surprising, since it was expected that EVs could not escape such large barriers. How these EVs traverse the wall is, as yet, unknown (Brown et al.2015). Finally, it is increasingly appreciated that bacterial EVs are heterogeneous populations of EVs with various size, density and cargo content, whose production and relative distribution change with the physiological state (Dauros Singorenko et al.2017). The current challenge is to develop methods allowing to reproducibly analyze specific types of EVs.
Figure 5.
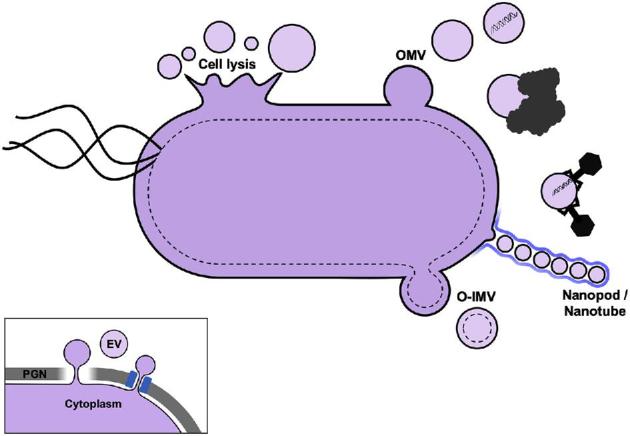
EV production in Bacteria. Two main types of EVs originate from diderm bacteria (OMVs and O-IMVs); however, cell lysis and nanotubes also produce EVs. Functions include intercellular communication, HGT, biofilm formation/maintenance, biomineralization, pathogenesis, viral defense, disposal/detoxification and relief of envelope stress. Inset: EVs in Firmicutes are produced from the single cytoplasmic membrane and must cross the thick PGN layer either by degradation of PGN or through pores.
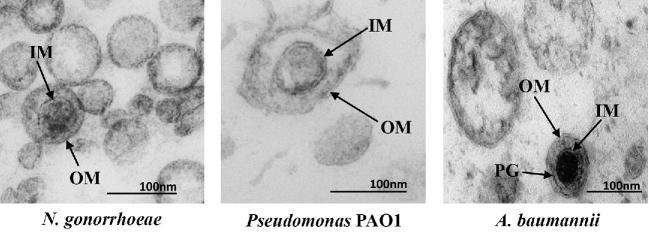
Figure 6.
TEM of ultrathin sections of EVs from three bacterial species. Both OMVs and O-IMVs are observed in these EV preparations, with features of O-IMVs indicated. O-IMVs are surrounded by an external bilayer, probably corresponding to the outer membrane (OM) of the cell, and an inner membrane (IM), probably corresponding to the cytoplasmic membrane, which entraps electron dense material. In the image of O-IMVs from A. baumannii, the putative peptidoglycan layer (PG) can be seen. Images kindly provided by Elena Mercade and Carla Pérez-Cruz (Universitat de Barcelona, Spain).
The production of EVs by Bacteria is not an artifact of laboratory culture conditions. Indeed, Bacteria have been shown to produce EVs in biofilms and during infections (Schooling and Beveridge 2006; Marsollier et al.2007; Deatherage and Cookson 2012; Schwechheimer and Kuehn 2015). EVs in biofilms interact with extracellular DNA to reinforce structural integrity and to also serve as decoys against antibiotics (Schooling, Hubley and Beveridge 2009). For a long time, the presence of EVs in natural environments was largely ignored. A seminal study by Biller and co-workers highlighted the abundance of bacterial vesicles, from the marine phototrophic bacteria Prochlorococcus, in marine ecosystems as well as in the laboratory (Biller et al.2014). Importantly, they showed that Prochlorococcus vesicles can be used as food, supporting the growth of heterotrophic bacterial cultures, suggesting that EVs also impact the marine carbon flux. The authors succeeded in isolating EVs from two very different ocean samples, with concentrations ranging from 105 to 106 vesicles per ml of sea water.
Outer membrane vesicles produced by diderm bacteria
Most EVs produced by diderm bacteria derive from the OM and are referred to as OMVs (for recent reviews, see Schwechheimer and Kuehn 2015, Orench-Rivera and Kuehn 2016; Jan 2017; Toyofuku et al. 2017b). During growth, the OM ‘blebs’ outwards and pinches off, forming spherical vesicles (20–250 nm) derived from the OM and trapped periplasmic components (Fig. 5). OMVs thus contain components of the OM and periplasm, such as bacterial lipids, OM proteins, soluble proteins, integral membrane proteins, lipoproteins and glycolipids. In fact, the identification of LPS and OM proteins was used to confirm their OM origin. Mass spectrometry-based proteomic studies were used to characterize the protein components of OMVs (Lee et al.2008; Choi et al.2011; Altindis, Fu and Mekalanos 2014; Jang et al.2014; Kieselbach et al.2015; Lee, Kim and Gho 2016; Yun et al.2017). Although cytoplasmic proteins were believed to be depleted in OMVs (Kulp and Kuehn 2010), some proteomic studies demonstrated their presence, despite following stringent purification protocols (Pérez-Cruz et al.2013; Berleman et al.2014). Further studies are necessary to understand why these cytoplasmic proteins would be associated with OMVs and what role they play (Schwechheimer and Kuehn 2015).
Increased vesiculation may be a response to stress and aid the removal of toxic by-products after exposure to stressful conditions (McBroom and Kuehn 2007; Maredia et al.2012; Macdonald and Kuehn 2013). For example, exposure of Stenotrophomonas maltophilia to the antibiotic ciprofloxacin resulted in an increased release of heterogeneous OMVs (Devos et al.2017).
OMVs can deliver their cargoes to recipient bacteria and have been implicated in inter- and intracellular communication, biofilm formation (Liao et al.2014; Turnbull et al.2016), antibiotic resistance (Rumbo et al.2011), stress response (Maredia et al.2012), toxin delivery (Rompikuntal et al.2012) and the transfer of nucleic acids (Biller et al.2014; Ghosal et al.2015; Sjöström et al.2015; Blenkiron et al.2016; Koeppen et al.2016; Resch et al.2016; Bitto et al.2017; Domingues and Nielsen 2017; Tsatsaronis et al.2018; Dauros-Singorenko et al.2018) (Fig. 5). In addition, OMVs can also deliver their cargoes to eukaryotic cells and have been implicated in pathogenesis (delivery of toxins and virulence factors) and homeostasis of the immune system (Nakao et al.2011; Lee et al.2012; Elhenawy, Debelyy and Feldman 2014; Rakoff-Nahoum, Coyne and Comstock 2014; Hickey et al.2015; Muraca et al.2015; Celluzzi and Masotti 2016; Jan 2017). For instance, Bacteroides thetaiotaomicron OMVs cross can be detected within gut mucosal macrophages suggesting that they can cross the mucosal barrier and initiate intestinal inflammation (Hickey et al.2015; Pathirana and Kaparakis‐Liaskos 2016).
Besides phagocytosis, four routes of OMV uptake have been implicated in host cells. These include macropinocytosis; clathrin-mediated endocytosis; caveolin-mediated endocytosis or non-caveolin, non-clathrin mediated endocytosis (Rewatkar et al.2015; O’Donoghue and Krachler 2016). For more details, refer to the reviews by Villarroya-Beltri et al.2014; Kaparakis-Liaskos and Ferrero 2015; O’Donoghue and Krachler 2016; Bitto et al.2017). A recent study using a genetically encoded OMV probe and cell permeable dye showed that entry and efficiency of uptake are influenced by the bacterial cell wall composition (O’Donoghue et al.2017).
OMVs are resistant to attack by degradative enzymes thus enabling long distance delivery, specificity in host-cell targeting and evasion of the host immune system (Bonnington and Kuehn 2014). As an example, OMVs of Moraxella catarrhalis help cells evade the immune system by bearing antigens which serve as decoys (Tan et al.2007; Perez Vidakovics et al.2010). OMVs can also help in bacterial colonization by selectively killing or promoting the growth of other bacteria species (Kadurugamuwa and Beveridge 1996; Ellis and Kuehn 2010; Hickey et al.2015).
OMVs are a powerful and versatile tool for alternative vaccination strategies due to their immunogenic properties, natural adjuvanticity, uptake by mammalian cells and potential for genetic engineering (van der Pol, Stork and van der Ley 2015). These properties have already been exploited to develop two meningitis vaccines with components from the OM and periplasm of N. meningitidis (Holst et al.2009; Granoff 2010). OMVs can be glycoengineered (geOMVs) to display surface glycans of the pathogen of interest. These geOMVs have been shown to be efficient against Streptococcus pneumoniae in mice and against Campylobacter jejuni in chickens (Price et al.2016). One of the main drawbacks of vesicle-based vaccines is the presence of LPS lipid A, which elicits a strong inflammatory response in the host. Using the glycoengineering approach it may be possible to modify LPS lipid A, thereby reducing its toxicity. To fully exploit OMV-based vaccines, novel genetic tools are needed to load the desired recombinant antigens onto the OMVs. For a recent review on the therapeutic potential of bacterial vesicles, refer to Bitto and Kaparakis-Liaskos (2017).
Outer-inner bacterial EVs
Bacterial EVs containing both the outer and cytoplasmic membrane were recently described for the Antarctic bacterium S. vesiculosa M7T. These double-membrane vesicles are known as ‘outer-inner membrane vesicles’ (O-IMVs) (Pérez-Cruz et al.2013). Similar EVs were later observed in cultures of diderm pathogenic bacteria such as N. gonorrhoeae, P. aeruginosa PAO1 and Acinetobacter baumannii AB41 (Fig. 6) (Pérez-Cruz et al.2015). As in the case of OMVs, it was suggested that these O-IMVs may be involved in lateral gene transfer and the transfer of cytoplasmic proteins. As O-IMVs are formed by the protrusion of both the outer and cytoplasmic membranes (Fig. 5), they should be enriched in cytoplasmic components, such as DNA and ATP, relative to OMVs. Very little is known about the production mechanism of these O-IMVs, especially with regard to differences between these and classical OMVs. Recently, it has been observed that the diderm bacterium S. maltophilia produced both OMV and O-IMVs following ciprofloxacin induction (Devos et al.2017). In fact, it is possible that most bacteria-producing OMVs also produce O-IMVs to some extent and that O-IMVs have been understudied for methodological reasons and also due to the fact that attention was focused on OMVs.
EVs in Firmicutes
Despite their thick peptidoglycan layer, monoderm Firmicutes also produce EVs. The first hint of the presence of EVs in Firmicutes came from studies with Bacillus subtilis and B. cereus (Dorward and Garon 1990). EVs from B. subtilis are heterogeneous and their diameter correlates with electron density, suggesting that cargo selection and vesicle size may be linked (Brown et al.2015). Following their first observation, several studies have purified EVs from the supernatants of cultured Firmicutes (Klieve et al.2005; Lee et al.2009; Rivera et al.2010; Jiang et al.2014; Olaya-Abril et al.2014; Prados-Rosales et al.2014; Brown et al.2015; Resch et al.2016). In most Firmicutes, the size range of EVs falls within 20 nm–150 nm in diameter. However, some species such as Bacillus spp. and Clostridium perfringens produce larger EVs (up to 400 nm) (Brown et al.2015).
Two non-exclusive hypotheses have been proposed to explain how EVs can cross the large barrier of the peptidoglycan. It was suggested that EVs may be forced through pores in the cell wall by turgor pressure after budding from the cell membrane and/or that the peptidoglycan was locally destroyed by enzymes associated with EVs and/or released with EVs (Brown et al.2015). This second hypothesis was supported by the presence of peptidoglycan-degrading enzymes in EVs isolated from Staphylococcus aureus (Lee et al.2009). It has also been reported that a subpopulation of B. subtilis expresses a prophage-encoded endolysin that degrades peptidoglycan allowing EV release (Toyofuku et al.2017b). More recently, it has been demonstrated that S. aureus produce enzymes, such as phenol-soluble modulins and autolysins, that are implicated in facilitating EV formation (Wang et al.2018).
The membrane and the lumen of EVs from Firmicutes are thought to be derived from the cytoplasmic membrane and the cytoplasm, respectively (MacDonald and Kuehn 2012). These EVs can thus transport a variety of cargoes, including cytoplasmic material, such as RNA and DNA (see below). The protein cargo identified in EVs from Firmicutes includes enzymes involved in peptidoglycan degradation, antibiotic degradation, virulence factors (e.g. anthrolysin, anthrax toxin components, coagulases, hemolysins and lipases) and immunologically active compounds (Marsollier et al.2007; Lee et al.2009; Rivera et al.2010; Gurung et al.2011; Prados-Rosales et al.2011; Thay, Wai and Oscarsson 2013; Brown et al.2014; Resch et al.2016; Vdovikova et al.2017). Proteomic approaches have been used to characterize EVs produced by Firmicutes (Lee et al.2009, 2013; Resch et al.2016). For example, proteomic analysis of Listeria monocytogenes EVs revealed that they were enriched in proteins important for survival and virulence, including the hemolysin listeriolysin O (LLO) (Lee et al.2013), whereas protein analysis of EVs produced by Group A Streptococcus revealed both unique proteins and proteins specifically enriched in EVs among the 195 proteins identified in the EV proteome (Resch et al.2016).
A recent study demonstrated that EVs carrying LLO offered a protective effect against autophagy and cell death (Vdovikova et al.2017). Many Firmicutes are important and well-studied pathogenic species, and many lines of evidence suggest that the EVs produced by Firmicutes are also involved in pathogenesis (Lee et al.2009; Rivera et al.2010; Gurung et al.2011; Prados-Rosales et al.2011; Thay, Wai and Oscarsson 2013; Pathirana and Kaparakis‐Liaskos 2016; Resch et al.2016; Vdovikova et al.2017).
Bacterial EVs as DNA transfer agents
Many studies have now described the association of DNA with bacterial EVs, including OMV, O-IMVs and EVs produced by Firmicutes (Domingues and Nielsen 2017). The possibility that these EVs could be involved in lateral gene transfer has important implications for the transfer of antibiotic resistance and virulence genes but also more generally for bacterial evolution.
The first description of DNA associated with bacterial EVs was published in 1983 by Nobel laureate Hamilton Smith and colleagues, who described EVs produced by Haemophilus influenzae as ‘specialized membranous structures that protect DNA during Haemophilus transformation’ (Kahn, Barany and Smith 1983). The authors noticed that these so-called transformasomes protected DNA from DNase or restriction enzymes and can therefore constitute a new tool for HGT. Microbiologists remained rather skeptical towards this possibility for a long time. Later, Yaron et al. (2000) reported that EVs produced by the pathogenic strain E. coli O157:H7 can indeed mediate the transfer of virulence genes to other Enterobacteria. It is still unclear how DNA can be packaged into OMVs, which a priori only contain periplasmic components. It is often assumed that DNA is packaged in a subpopulation of ‘secondary’ OMVs formed by aggregation of cell wall fragments after cell lysis. In fact, the packaging of DNA within OMVs has not always been conclusively demonstrated (Renelli et al.2004; Mashburn-Warren and Whiteley 2006; Kulp and Kuehn 2010; Pérez-Cruz et al.2013; Liao et al.2014) and it is often not clear whether the nucleic acids are inside the vesicles or simply associated with the vesicles in such a manner where they resist enzymatic degradation (Domingues and Nielsen 2017). For instance, Pérez-Cruz et al. (2015) did not detect DNA in OMVs from N. gonorrhoeae, whereas they found DNA in O-IMVs produced by this bacterium (see below).
Biller and co-workers recently examined the quantity and distribution of DNA associated with OMVs from different bacteria. Their results demonstrated that the size and quantity of DNA varied amongst the different bacteria and that only a small proportion of EVs contain DNA (Biller et al.2017). Ferrero and colleagues show that most of the DNA associated with OMVs produced by P. aeruginosa is present on the external surface, with a smaller amount located inside OMVs (Bitto et al.2017). This DNA is mostly present in small OMVs of around 20 nm. The authors found that the external DNA includes fragments from the bacterial chromosome, whereas internal DNA was mainly composed of short fragments (1–4 kb) enriched in specific regions encoding proteins involved in virulence, stress response and antibiotic resistance.
Interestingly, studying OMVs from Thermus species, Blesa and Berenguer (2015) have suggested that EVs could function as reservoir of genetic material resistant to heat denaturation for transformation in high temperature environments, as previously proposed for hyperthermophilic archaea (Soler et al.2008). Transfer of DNA mediated by OMVs produced by Thermus species is dependent on the competency of the recipient cells, suggesting that DNA is not delivered by fusion of OMV with host cells (Blesa and Berenguer 2015).
Remarkably, Ferrero and colleagues could demonstrate that P. aeruginosa OMVs can transfer their DNA into the nucleus of eukaryotic cells (Bitto et al.2017). They suggest that the internal DNA plays a role in cell-to-cell communication whereas DNA present at the surface of OMVs performs a different role, being important for biofilm formation and protection. The latter observation is supported by the upregulation of DNA associated with OMVs during biofilm formation in S. mutans (Liao et al.2014).
The presence of DNA in EVs from monoderm organisms was anticipated, considering that their EVs were expected to contain cytoplasmic components. The presence of DNA associated with EVs from Firmicutes and the capacity of these EVs to transfer genetic markers to recipient cells was first observed in Ruminococcus species (Klieve et al.2005). This DNA was described as short fragments of chromosomal DNA ranging from 23 to 90 kb. Interestingly, the authors reported that, unlike chromosomal DNA, EV-associated DNA was resistant to digestion with EcoRI, suggesting differences in the restriction/modification pattern of these DNAs.
Bacterial EVs as RNA transfer agents
The discovery in the first decade of this century that eukaryotic EVs can deliver RNA cargoes to recipient cells, promoting phenotypic changes (see below), prompted several authors to search more recently for RNA associated with bacterial EVs (for review, see Dauros-Singorenko et al.2018; Tsatsaronis et al.2018). RNA was first detected in preparations of EVs produced by Prochlorococcus, and covered a remarkable 95% of the genome (Biller et al.2014). RNA was also detected associated with OMVs of Vibrio cholerae (Sjöström et al.2015), E. coli (Ghosal et al.2015; Blenkiron et al.2016), P. aeruginosa (Koeppen et al.2016) and in EVs of Group A Streptococcus (Resch et al.2016). The majority of RNA in bacterial EVs are rather short (less than 250 nucleotides) and resistant to RNase treatment. However, Sjöström and colleagues reported that RNA associated with V. cholera OMVs was sensitive to RNase, suggesting that it could be located at the surface of the OMVs. Alternatively, the authors suggested that RNAse could have entered these OMVs, impairing their integrity (Sjöström et al.2015). It is likely that, similarly to DNA, RNA could be located both inside and outside of the EVs.
In several studies, RNAs associated with EVs were analyzed using deep RNA-sequencing, revealing a large diversity of RNA molecules (Ghosal et al.2015; Resch et al.2016), including rRNA, tRNA, mRNA and a variety of small RNA species, including CRISPR RNAs guides (Resch et al.2016). Importantly, RNA associated with bacterial EVs can be delivered into the cytoplasm and nuclei of the host cell (Blenkiron et al.2016). Notably, RNA associated with P. aeruginosa OMVs can be transferred to infected human and mouse cells, decreasing their innate immune response (Koeppen et al.2016). Charpentier and colleagues thus propose that EVs are an important source of microbial RNAs that modulate the immune response during infection (Tsatsaronis et al.2018).
In general, it seems that mRNA is under-represented in EVs relative to the cellular RNA pool whereas RNAs originating from intergenic region are over-represented. However, Resch and colleagues reported that many mRNA species were present in Group A Streptococcus EVs and that some of them were specifically enriched in EVs. This suggests that EVs could trigger the production of new proteins in recipient cells.
Biogenesis of bacterial EVs
Although OMVs have been observed for decades (Bishop and Work 1965; Work, Knox and Vesk 1966; Chatterjee and Das 1967), the process by which diderm organisms produce them is not fully understood. The enrichment or depletion of OMV content compared to the cell suggests that it is a deliberate and regulated process (Deatherage and Cookson 2012; Schertzer and Whiteley 2012; Schwechheimer and Kuehn 2015). To date, several models for OMV biogenesis have been proposed (Mashburn-Warren and Whiteley 2006; Kulp and Kuehn 2010; Haurat, Elhenawy and Feldman 2015; Roier et al.2016a,b). However, a conserved general mechanism for biogenesis has remained elusive for a long time (Roier et al.2016a,b). This is changing as genetic and biochemical analyses over the years have begun to shed light on aspects of OMV biogenesis in diderm bacteria. Several mutants have been isolated in different species with either hypo- or hypervesiculation phenotypes. For instance, transposon mutagenesis in H. influenzae and a whole-genome knockout library of E. coli implicated 20 and 150 new genes in the process of vesiculation, respectively (Kulp et al.2015; Roier et al.2015). Most of these mutations trigger an increase in OMV formation (hypervesiculation), whereas a few others trigger a decrease (hypovesiculation). Analyses suggest that mutations in OM structures result in hypervesiculation in agreement with several proposed models for OMV production, whereas mutations in oxidative stress response pathways showed a decrease in vesiculation, in agreement with the implication of OMV production in stress response.
Earlier models suggested that OMV formation occurs due to the presence of fewer covalent linkages between the OM and the underlying peptidoglycan layer (Hoekstra et al.1976), due to the OM growing faster in certain regions (Chatterjee and Das 1967; Wensink and Witholt 1981). As a result, the OM bulges out and vesicles are formed (McBroom, Johnson and Vemulapalli 2006; Kulp and Kuehn 2010; Roier et al.2016a,b). This model is supported by the fact that deletion or truncation of genes encoding OmpA (OMP5), an abundant protein linking the OM to the peptidoglycan, triggered hypervesiculation in E. coli, V. cholera and Salmonella (Sonntag et al.1978; Song et al.2008; Deatherage et al.2009).
A subsequent model proposed that accumulation of peptidoglycan fragments (Hayashi, Hamada and Kuramitsu 2002) and misfolded proteins (McBroom and Kuehn 2007) due to stress increases turgor pressure in the periplasm and leads to the OM bulging out (Zhou et al.1998; Roier et al.2016a,b). An increase in OMV production was indeed observed when peptidoglycan fragments accumulated because of the incomplete degradation of peptidoglycan in a Porphyromonas gingivalis autolysin mutant strain (Hayashi, Hamada and Kuramitsu 2002). Additionally, deletion of the degQ gene, which encodes a periplasmic serine protease in S. oneidensis, resulted in an increased level of protein accumulation within the periplasm and subsequently a hypervesiculation phenotype (Ojima et al.2017).
A third model has been proposed based on the importance of charge–charge interactions. The LPS composition in the OM can vary in response to various environmental factors. For example, P. aeruginosa possesses two distinct LPS O-polysaccharide species, A (neutral)- and B-band (charged) LPS. In wild-type cells, the OMVs are enriched in the B-band LPS, which creates repulsion by accumulation of negative charges and therefore leads to membrane protrusion (Kadurugamuwa and Beveridge 1995). Indeed, cells expressing the neutral A band LPS produce smaller OMVs (Murphy et al.2014).
Additionally, a bilayer-couple model for OMV biogenesis was proposed in P. aeruginosa. Insertion of Pseudomonas quinolone signal (PQS) into the outer leaflet of the OM can also increase membrane curvature and lead to the formation of OMVs (Mashburn-Warren and Whiteley 2006; Schertzer and Whiteley 2012; Florez et al.2017). Mutants lacking OprF (an OmpA homolog) in P. aeruginosa have increased levels of PQS and thus increased OMV production (Schertzer and Whiteley 2012; Wessel et al.2013). However, PQS is only produced by P. aeruginosa, and therefore this model is limited to this species.
A more general model was proposed based on studies performed on two distantly related Proteobacteria, H. influenzae and V. cholerae (Roier et al.2016a,b). Among hypervesiculation mutants obtained by transposon mutagenesis in H. influenzae, Roier and colleagues focused on mutants of the VacJ/Yrb ABC transporter system, which is widely conserved in Proteobacteria and known to prevent phospholipid accumulation in the outer leaflet of the OM. A similar hypervesiculation phenotype was found in VacJ and Yrb mutants of V. cholerae. The hypervesiculation phenotype of these mutants and biochemical analysis of the OMVs they produce suggest a model in which OMV formation is induced by the accumulation of phospholipids in the outer leaflet of the OM, thereby producing an asymmetric expansion and outward bulging of this membrane. Interestingly, Roier and co-workers observed that iron limitation induced by deletion of the fur gene (an activator of iron-regulated genes), a condition frequently observed for bacterial pathogens in their host, correlates with a downregulation of vacJ/yrbB genes and an increase in OMV production. They suggest that iron-limiting conditions in hosts result in increased OMV production by pathogenic bacteria, which bind antibodies and complement attacks, consistent with previously observed immune responses (Tan et al.2007;.Perez Vidakovics et al.2010). More generally, they propose that the asymmetric distribution of phospholipids between the inner and outer leaflets of the OM represents a general mechanism that can account for OMV formation under all growth conditions.
Homologs of proteobacterial VacJ and YrbB proteins are only present in a few bacterial phyla. It is thus likely that different mechanisms controlling phospholipid asymmetry between the inner and outer leaflets of outer and inner membrane phospholipid bilayers exist in different bacterial phyla. This suggests screening for such mechanisms and their potential role in OMV or OMV-IMV formation in Bacteria may enhance our understanding of EV production. Consistent with this hypothesis, a recent study proposed that LPS remodeling exerts modifications in the curvature of the OM leading to OMV formation (Elhenawy et al.2016). Deacylation of lipid A, the hydrophobic anchor of LPS, was examined in Salmonella typhimurium. Expression of the lipid A deacylase, PagL, resulted in hypervesiculation with the deacylated lipid accumulating exclusively in OMVs (Elhenawy et al.2016) (Fig. 1a). Additionally, a ΔpagL strain showed a significant reduction in OMV secretion relative to the wild-type strain. In another study, Bonnington and Kuehn suggested that OMVs are used by the cell to remove unfavorable LPS glycoforms. Thus, OMV production may aid in remodeling of the OM—a process essential to bacterial adaptation and survival in different niches (Bonnington and Kuehn 2016).
Finally, a recent study using super-resolution microscopy revealed that explosive cell lysis in P. aeruginosa can generate membrane fragments that rapidly form EVs (Turnbull et al.2016). This phenomenon is triggered by an endolysin encoded by a prophage integrated in the genome of P. aeruginosa. Notably endolysin-deficient mutants are defective in EV production and biofilm development. Thus, cell lysis could also act as a bona fide mechanism for the production of bacterial EVs. However, the extent to which this occurs has not been established. It is not clear whether it is possible to distinguish between ‘genuine’ and reconstituted EVs produced after cell lysis, even if the latter might have an extended subset of proteins, compared to EVs produced by other mechanisms.
Cargo selection in OMVs
Selection of cargo is an important aspect of OMV biogenesis (Haurat et al.2011; Bonnington and Kuehn 2014; Tsatsaronis et al.2018). Evidence suggests that the cellular localization of a protein greatly affects its potential for inclusion into OMVs. Virulence factors such as alkaline phosphatase, phospholipase Cs, β-lactamase and Cif (CFTR inhibitory factor) are enriched in P. aeruginosa OMVs (Kadurugamuwa and Beveridge 1995; Bomberger et al.2009; Koeppen et al.2016). The loading of such virulence factors into vesicles is thought to rely on LPS subtypes. Proteins associated with charged LPS are enriched in OMVs, whereas those that co-localize with neutral LPS are retained in the OM (Haurat et al.2011; Veith et al.2014; Schwechheimer and Kuehn 2015).
Compared to OMVs, our knowledge about EV biogenesis in the Firmicutes is still in its infancy. Interestingly, Resch and colleagues discovered that production of EVs was negatively regulated by the two-component CovRS regulatory system, suggesting that CovRS could decrease EV production by triggering the expression of factors disrupting EV or preventing their release (Resch et al.2016). Resch and colleagues further observed that the phospholipid composition differs between EVs and the cytoplasmic membrane, with enrichment of phosphatidyl glycerol relative to cardiolipin, which is known to induce membrane curvature (Barák and Muchová 2013). They also reported an increase in monounsaturated fatty acid content. This indicates that modification of membrane lipid composition could play a critical role in EV production by Firmicutes, as in the case of OMVs by diderm bacteria.
Nanotubes in Bacteria
In the last six years, several laboratories have found that diverse types of bacteria produce filamentous structures called nanopods or nanotubes that are involved in cell-to-cell transfer and seem intimately connected to EVs (for a recent review, see Baidya et al.2018). Similar filamentous structures containing EVs were previously observed in the culture medium of some hyperthermophilic archaea (Soler et al.2008) (see below) and resemble eukaryotic ‘tunneling nanotubes’ (Fig. 2) (Rustom et al.2004; Lou et al.2012). The first observation of a direct connection between EVs and nanotubes in Bacteria was reported for Delftia acidovorans that produces chains of EVs that are enclosed by the S-layer forming the typical structure referred to as nanopods (Shetty et al.2011). Similar structures, usually called Nanotubes, were then reported in Firmicutes (Dubey and Ben-Yehuda 2011, Dubey et al.2016), Myxobacteria (Ducret et al.2013; Remis et al.2014; Wei et al.2014) and Proteobacteria (Francisella novicida, A. bayeli, E. coli, S. oneidensis) (McCaig, Koller and Thanassi 2013; Pirbadian et al.2014; Pande et al.2015). In Firmicutes, the membranes of nanopods correspond to an extrusion of the cytoplasmic membrane that crosses the thick peptidoglycan layer, whereas they seem to be formed by extrusion of the OM in Proteobacteria and Myxobacteria.
These similar structures, thereafter called nanotubes, can bridge neighboring cells together either between the same or different species facilitating cell-to-cell communication (see below). Remarkably, it has been shown that the so-called nanowire filaments involved in the extracellular transport of electrons produced by S. oneidensis were in fact nanotubes associated with OMVs (Pirbadian et al.2014). In Myxobacteria, it has been shown that nanotube formation increases when cultures are grown without agitation (Wei et al.2014) and is upregulated in biofilms (Remis et al.2014). It is most likely that nanotubes are not laboratory curiosities but a fundamental mechanism for bacterial communication in nature. ‘Nanotubes’ appear either as purely tubular structures or as chains of consecutive constricted segments resembling EVs but having a continuous lumen. In contrast, ‘nanopods’ contain chains of discrete EVs. The two types of structures are sometimes produced by the same species (Dubey et al.2016) and studies with Myxococcus xanthus suggest that they could be in fact different states of the same kind of structure (Wei et al.2014). These differences could be also due to the method used for nanotube preparation and visualization. Hence, Jensen and co-workers only observed nanotubes formed by chains of OMVs when they analyzed the ultrastructure of nanotubes from S. oneidensis by electron cryotomography, whereas they appeared smooth in fluorescence light microscopy (Subramanian et al.2018). In this work, the authors observed electron dense region at the junction connecting neighboring OMVs, suggesting the existence of some unknown material that facilitates nanotube formation.
Nanotubes from Firmicutes have been studied quite extensively by the group of Ben-Yehuda. These authors reported that nanotubes can bridge neighboring B. subtilis cells as well as B. subtilis and S. aureus (Dubey and Ben-Yehuda 2011). They visualized a transfer of cytoplasmic fluorescent molecules between adjacent cells and reported that plasmids can be transferred from cell to cell via these nanotubes. Additionally, their work suggests these nanotubes can deliver toxins from B. subtilis to other bacilli, and following toxin delivery, the nanotubes can even facilitate ‘looting’, importing nutrients from the competitor cell (Stempler et al.2017). These nanotubes are formed even when the cells were grown to a low density and this production of extensive elongated nanotubes greatly increases the cell surface area (Dubey et al.2016). Utilizing a combination of super-resolution, light and electron microscopy, they described nanotubes as chains of membranous segments with a continuous lumen. Importantly, Ben-Yehuda and colleagues could detect in nanotubes of B. subtilis a calcineurin-like protein, YmdB, which is required for both nanotube formation and intercellular molecular exchange (Dubey et al.2016; Stempler et al.2017). The protein YmdB, a putative sensor phosphodiesterase involved in AMPc regulation, could be involved in transmitting messages for nanotube production by an unknown mechanism (Dubey et al.2016). The YmdB protein is only present in Bacteria but highly conserved among bacteria and present in several phyla, implying that it plays a fundamental role in bacterial physiology. It will be interesting to test if this protein is also involved in nanotube formation in different species.
Observations in real time showed that nanotubes produced by B. subtilis are formed very rapidly (in the course of minutes) and display rapid movements (Dubey et al.2016). Similar observations were made with nanotubes from S. oneidensis that were described as dynamic structures capable of growth, shrinking and reversible transition between OMV chain and individual vesicle morphology (Subramanian et al.2018).
Intracellular vesicles in Bacteria
In Eukaryotes, some EV production is tightly linked to the network of intracellular vesicles that is typical of eukaryotic cells (see below). Although such intracellular vesicles are not as well known, nor as ubiquitous in Bacteria, they have been sometimes observed either within the cytoplasm or accumulating in the periplasm, see for example spectacular pictures in Dobro et al. (2017). Their function within the cell and their relationship with EVs remain unclear. Some intracellular vesicles seem to be involved in sequestration and detoxification of otherwise harmful compounds. For instance, it was recently shown that some sponge-associated bacteria mineralize both arsenic and barium on intracellular vesicles, allowing these bacteria to act as a detoxifying organ for the host (Keren et al.2017). Intracellular vesicles formed by sulfur globules surrounded or not by a proteinaceous membrane have been known to be present in Bacteria for a long time (Bazylinski et al.2004, 2013; Prange et al.2004). In some bacteria, these vesicles are transient and completely degraded after oxidation of sulfur to sulfate (Franz et al.2007). In others, they are released into the extracellular medium to avoid a toxic accumulation of sulfur (Eichinger et al.2014).
The best characterized bacterial intracellular vesicles have been observed in Planctomycetes. These bacteria form a distinct phylum and possess unusual features such as intracellular compartmentalization and proteinaceous cell walls (Fuerst and Sagulenko 2011; Devos and Ward 2014). Intriguingly, they also contain intracytoplasmic membranes which separate cells into multiple functionally distinct compartments (van Niftrik et al.2004; Gottshall et al.2014; Sagulenko et al.2017). In cells of the genus Gemmata, invagination of the cytoplasmic membrane forms a complex system of internal membranes (Lindsay et al.2001) with a network of interconnected vesicles (Acehan, Santarella-Mellwig and Devos 2014). Additionally, the ability of Gemmata obscuriglobus to internalize proteins from the extracellular environment may be reminiscent of eukaryotic endocytosis (Lonhienne et al.2010; Fuerst and Sagulenko 2011). The mechanisms by which these vesicles and membrane invaginations are formed remain unknown; however, several homologs of eukaryotic membrane coat proteins have been detected in the genomes of Planctomycetales (Santarella-Mellwig et al.2010). Despite the apparent versatility of the cell membranes of Planctomycetes, the production of OMVs by these species has not yet been reported.
EVs in Eukaryotes: microvesicles, exosomes and apoptotic bodies
Eukaryotes are composed of complex cells characterized by an extensive intracellular network of tubular membranes producing intracellular vesicles, some of these membranes being specialized in particular function (e.g. the nuclear membrane). This network is connected to a cytoplasmic membrane usually rich in glycoproteins and sometimes covered by a thick cell wall (e.g. in plants or fungi). Notably, the basic molecular mechanisms of Eukaryotes are often very divergent from those of Bacteria (sometimes even non-homologous e.g. DNA replication), showing much greater similarity to archaeal systems (e.g, translation, transcription and so on). This has triggered intense debates about the relationships between Archaea and Eukaryotes, with some authors suggesting that Eukaryotes originated from Archaea, whereas others, analyzing the same data, concluded that Eukaryotes and Archaea are two monophyletic groups that share a specific common ancestor (for recent data and discussions on this topic, see Spang et al. 2015, 2018; Da Cunha et al. 2017, 2018). Beside their archaeal-like component, all eukaryotes also share a strong bacterial heritage since they all contain mitochondria (or relics of them) that were acquired from an intracellular Alphaproteobacteria. Consequently, they exhibit a unique combination of archaeal and bacterial features associated with unique eukaryotic features, such as split genes and the spliceosomal machinery.
The release of EVs to the extracellular space is probably conserved in all types of eukaryotic cells: animals, plants, protists and fungi, be they either in unicellular or multicellular organisms; however, most studies to date have been done in animals, specifically in the two mammalian models, mouse and human. EVs produced by human cells have been studied for quite a long time now (see Yáñez-Mó et al.2015; Stahl and Raposo 2018; Tkach, Kowal and Théry 2018 for brief histories). These EVs can be found in diverse biological fluids from amniotic fluid to urine, breast milk, saliva and even cerebrospinal fluid (Mathivanan and Simpson 2009; Kalra et al.2012; Kim et al. 2013, 2015; Yáñez-Mó et al.2015; Kalra, Drummen and Mathivanan 2016; Maas, Breakefield and Weaver 2017; De la Torre Gomez et al.2018; Stahl and Raposo 2018; van Niel, D’Angelo and Raposo 2018). They are also an important component of the extracellular matrix (Rilla et al.2017). Notably, EVs produced by eukaryotic cells have the ability to deliver their cargoes not only to neighboring cells in their tissue microenvironment, but also at long distances throughout the body of multicellular organisms. In particular, they can trigger epigenetic reprogramming by delivering active RNA or DNA to recipient cells.
In humans, increasing evidence suggests that EVs play a fundamental biological role in the regulation of normal physiological and disease processes (Gatti et al.2011; Raposo and Stoorvogel 2013; Kowal, Tkach and Théry 2014; Yáñez-Mó et al.2015; Maas, Breakefield and Weaver 2017; Rilla et al.2017). In cancerous cells, the release of EVs is greatly enhanced and the composition of vesicular proteins, mRNAs and miRNAs varies significantly from healthy cells (Inal et al.2013; Ohno, Ishikawa and Kuroda 2013; Kreger et al.2016; Takahashi et al.2017; Xu et al.2018). EVs are thus believed to play an important role in tumor proliferation and evading the immune system (Al-Nedawi et al.2008; van Doormaal et al.2009; Muralidharan-Chari et al.2010; Lee et al.2011; Tricarico, Clancy and D'Souza-Schorey 2016; Whiteside 2016; Naito et al.2017; Weidle et al.2017; Xu et al.2018) and there is a great deal of interest in harnessing EVs as potential biomarkers for the diagnosis and monitoring of cancer (Muralidharan-Chari et al.2010; Verma et al.2015; Kinoshita et al.2017; Chen et al.2018; Xu et al.2018). Additionally, EVs have been implicated in spreading neuropathological diseases through the brain via the transport of amyloid proteins (Coleman and Hill 2015) or prions (Hartmann et al.2017; Liu et al.2017). Finally, EVs also play a role in aging, with even senescent cells effecting telomere regulation and gene expression of other tissues through EV-mediated mechanisms (Acosta et al.2013; Takasugi 2018).
Eukaryotic EVs are usually classified into three main categories, based on their mode of production in animal cells: microvesicles (50–1000 nm), exosomes (40–100 nm) and apoptotic bodies (800–5000 nm) (for recent reviews, see Kalra, Drummen and Mathivanan 2016; Maas, Breakefield and Weaver 2017; Stahl and Raposo 2018; van Niel, D’Angelo and Raposo 2018). Microvesicles (also sometimes referred to as microparticles or ectosomes) are formed by the outward budding of membrane vesicles from the cell surface (Fig. 4) (Muralidharan-Chari et al.2009). In some cases, they are released from tubular structures extending from the plasma membrane (Rilla et al. 2013, 2014). Microvesicles thus share some properties with EVs produced by some monoderm bacteria and some archaea (see below).
In contrast, exosomes are specific to eukaryotic cells, being formed through the endocytic pathway from the ‘outward’ budding of the late endosomal membrane (see Box 1) (Harding, Heuser and Stahl 1983; Pan and Johnstone 1983). They first accumulate in these endosomes that became known as multivesicular bodies (MVBs). The MVBs can either fuse with lysosomes, leading to the degradation and recycling of contents, or fuse with the plasma membrane and release their contents as exosomes into the extracellular space (Fig. 4). It is not clear what determines their fate for either degradation or fusion with the plasma membrane.
Box 1.
The term exosome, which can from ‘membrane exfoliated vesicles’, has a confusing history since it was used for the first time to name microvesicles released by different cultured cells and carrying a 5΄-nucleotidase activity (Trams et al.1981). However, in the early 1980s, a more complex EV secretion pathway, in which intraluminal vesicles formed within MVBs, was described (Harding, Heuser and Stahl 1983; Pan and Johnstone 1983). The existence of this secretion pathway was later also confirmed in antigen-presenting cells, epithelial and tumor cells (Raposo et al.1996; van Niel et al.2001; Wolfers et al.2001). From 1987 onwards, the term exosome was adopted to refer to EVs of endosomal origin (Johnstone et al.1987). For early publications on diverse eukaryotic EVs, see for instance Kerr, Wyllie and Currie (1972); Friend et al. (1978); Raposo et al. (1996); Heijnen et al. (1999), Théry et al. (2001); Hristov et al. (2004); Del Conde et al. (2005) and Ratajczak et al. (2006).
The third major type of eukaryotic EVs called apoptotic bodies is also specific to eukaryotic cells. They are produced during programmed cell death by outward budding from the surface of apoptotic cell (Fig. 4) (van der Pol et al.2012). They are usually larger than other vesicles, although their size range somewhat overlaps with that of microvesicles. Apoptotic bodies can contain organelles and/or nuclear remnants and are morphologically diverse (Bilyy et al.2012). They play an important biological role not only in development but also in the pathogenesis of several disease processes.
Additionally, several types of EVs are produced by specific cell types under specific circumstances (Fig. 4): for example, large oncosomes produced by cells from advanced cancers (Minciacchi et al.2015), migrasomes produced by migrating amoeboid cells (Ma et al.2015) and giant vesicles produced by breast cancer cells in the presence of estradiol (Wright et al.2014). Due to cell-specific nature of these EV subtypes, and their relatively small bodies of literature, our review will not discuss these cases.
EVs present in circulating fluids are likely to be mainly composed of both exosomes and microvesicles (Muralidharan-Chari et al.2010). Several studies have shown that EV populations are usually heterogeneous, even in pure cell culture, with each type of cell being able to produce different types of EVs. Moreover, it seems that specific types of EVs (or at least EVs with specific cargoes) may be produced exclusively by different cell types. For example, proteomic analysis of EVs purified from breast milk showed that 198 of the identified proteins are not present in the EV database, Vesiclepedia, suggesting that milk-derived EVs harbor proteins not observed in other EVs (van Herwijnen et al.2016). This combination of specific EVs being produced by specific cell types and multiple EV subtypes being produced by each cell highlights not only the heterogeneity in EVs, but also the difficulty in studying these enigmatic entities.
One of the major challenges today is to define methods that allow discrimination between these species of vesicles. Current methods of isolation and purification include ultracentrifugation, density gradient centrifugation, affinity chromatography, immuno-affinity methods (Mathivanan et al.2010; Gardiner et al.2016; Kowal et al. 2016, 2017), ligands reactive with EV surfaces (e.g. heparin) (Atai et al.2013), separation by charge (Graner et al.2009; Deregibus et al.2016) or size by field-flow fractionation techniques (Sitar et al.2015; Zhang and Lyden 2018) and polymer-based precipitation (Brown and Yin 2017). Exosomes and microvesicles are considered molecularly different in practice due to their different modes of production; however, it can be difficult to distinguish between them; thus, most purification techniques isolate mixed EV populations. Microvesicles (50–1000 nm) are typically larger and more heterogeneous than exosomes (40–100 nm) but their size range does overlap. Exosomes are enriched in the fraction of small EVs with diameter less than 200 nm, but this fraction does also contain microvesicles. As a consequence, EVs isolated by ultracentrifugation are likely to contain a mixed population of both. This has been clearly demonstrated by Théry and colleagues who isolated four different types of EVs from human primary dendritic cells by a combination of ultracentrifugation and density gradient centrifugation (Kowal et al.2016). They proposed the use of immunoisolation using exosome/microvesicle-specific antigens. To complicate matters further, the content of EVs varies depending on the source and original isolation or enrichment techniques. Thus, care must be taken before assigning functions to one EV type that could be due to other EVs present in the preparation (Théry et al.2006; Tkach and Théry 2016), and many authors now suggest calling vesicles sedimenting at 100 000 g as ‘small EVs’ rather than exosomes (Mateescu et al.2017).
In an attempt to identify the content of EV preparations, several proteins have been proposed as markers of exosomes including major histocompatibility complex, tetraspanins, ALIX proteins, flotillin, TSG101, heat-shock proteins or Rab5b (Chen et al.2015; see table 1 in Tkach and Théry 2016 and references therein). A position paper by ISEV suggested that studies should demonstrate a minimum of three of these marker proteins in EV preparations to confirm the presence of exosomes (Lötvall et al.2014). However, subsequent studies showed that even these markers are not exclusive to exosomes (Kowal et al.2016). An international consortium of EV scientists recently set up the EV-TRACK knowledge base (http://evtrack.org) to collect and normalize the various methodologies used to isolate EVs in the hope of increasing transparency and reproducibility (van Deun et al.2017; Witwer et al.2017).
Cargo of eukaryotic EVs
Both exosomes and microvesicles are formed through the packaging of cytoplasmic contents in membrane-bound vesicles, and thus have been shown to carry all types of cellular components: proteins, lipids, carbohydrates, DNA and RNAs (mRNA, microRNA and other non-coding RNAs) (El Andaloussi et al.2013; Penfornis et al.2016, Mateescu et al.2017; Record et al.2018). Several proteins are enriched in EVs, and these may provide clues as to the biogenesis and/or physiological roles of EVs. Enriched proteins include lipid raft-interacting proteins, tetraspanins and associated proteins; immunoglobulins and growth factor receptors; cytoskeletal proteins such as tubulin and actin; ESCRT-related proteins; heat-shock proteins; and proteins involved in vesicle trafficking such as Rab GTPase proteins, annexins and protein of the SPFH (stomatin, prohibitin, flotillin and HflK/C) superfamily, especially stomatin (Snyers, Umlauf and Prohaska 1999; Salzer, Mairhofer and Prohaska 2007; Lapatsina et al.2012; for a review on proteomic studies of exosomes and microvesicles, refer to Greening et al.2017). Interestingly, stomatin is known to be a major protein in vesicular lipid rafts, and despite being first detected in Eukaryotes, was later identified in both Bacteria and Archaea, suggesting some degree of conserved membrane dynamics across the three domains (Tavernarakis, Driscoll and Kyrpides 1999; Lee et al.2017). For an exhaustive review on cargo selection in eukaryotic EVs, see Villarroya-Beltri et al. (2014).
Despite being a major component of EVs, lipids have largely been sidelined until recently. Lipidomic studies of EVs from different cell types are required to elucidate the role of lipids in the biogenesis and biological functions of EVs. EVs are enriched in lipids, including sphingomyelin, cholesterol, ganglioside GM3, disaturated lipids, phosphatidylserine and ceramide (Subra et al.2007; Llorente et al.2013; Record et al.2014, 2018; Skotland, Sandvig and Llorente 2017). In contrast, the levels of phosphatidylcholine and diacyl-glycerol are decreased relative to the cell (Laulagnier et al.2004; Skotland, Sandvig and Llorente 2017, and references therein).
Additionally, some differences in lipid composition between exosomes and microvesicles have been observed, and are likely reflective of their different method of biogenesis, either originating from MVBs or the plasma membrane (Bicalho, Holovati and Acker 2013; Zaborowski et al.2015; Abels and Breakefield 2016). Indeed, these differences in lipid composition and protein-to-lipid ratios of EVs have been suggested to be a more reliable way to characterize EV subpopulations than protein content alone (Osteikoetxea et al.2015).
Not only are lipids implicated in the biogenesis of vesicles, they also play an important role in EV uptake by cells via lipid raft-mediated internalization (Mulcahy, Pink and Carter 2014). EV uptake is reduced when EV-producing cells are pre-treated with compounds which prevent the biosynthesis of glycosphingolipids. Additionally, sphingolipids of EVs have been shown to have an important role in binding and endocytosis by recipient cells, possibly through cholesterol-rich microdomains (Izquierdo-Useros et al.2009; Mulcahy, Pink and Carter 2014). Thus, it is clear that both the lipid content of vesicular membranes and the protein cargo which they contain are important at every step of EV function from biogenesis to uptake.
RNA in EVs
In 2007, Lötvall and colleagues demonstrated that eukaryotic EVs (described as exosomes) contain large amount of RNA, including mRNA and sRNA, and can transfer this RNA to recipient cells (Valadi et al.2007). Importantly, they reported that mouse mRNA transferred via EVs to recipient human cells could be translated into corresponding mouse proteins. This observation strongly stimulated interest for EVs among cell biologists, especially when it was demonstrated that the transferred RNA can be active in the recipient cell, modifying its phenotype (Skog et al.2008; Kosaka et al.2010; Pegtel et al.2010; Zhang et al.2010). Since that time, a number of studies have confirmed these preliminary observations (for reviews, see Abels and Breakfield 2016; Mateescu et al.2017).
Eukaryotic EVs from animals, plants, fungi and protists contain an abundance of different types of potentially active RNA, such as mRNAs, miRNAs and rRNAs, long and short non-coding RNA, tRNA fragments, piwi-interacting RNA, vault RNA and Y RNA as well as RNA-binding proteins that are probably involved in RNA selection and delivery (see below). Many of these RNAs are selectively enriched or depleted in EVs relative to their host cells, and even between different EV subpopulations, suggesting an active mechanism of RNA packaging (Abels and Breakefield 2016; Wei et al.2017). Although there is some intact mRNA and long non-coding RNAs present in EVs, most of the RNA is fragmented or of small size (Batagov and Kurochkin 2013; Wei et al.2017). As mentioned above, these vesicle-encapsulated RNAs can have a profound impact on recipient cells, transferring between different cell types causing transient transformation of recipient cells e.g. resulting in production of novel proteins (in the case of mRNA transfer), or regulation of gene expression (in the case of miRNAs) (Valadi et al.2007; Skog et al.2008; van der Vos et al.2016). This process has gained significant attention in the field of cancer research, where it has been observed that tumor-derived EVs can promote tumorigenesis in healthy cells and prime tissues to become future metastatic sites through the transfer of RNAs (Peinado et al.2012; Zomer et al.2015). Despite the importance of EV-derived RNAs, this is a field fraught with technical challenges—combining the issues of EV purification and identification (discussed above) with the delicate nature of minute RNA samples. As such, few standards are available to compare studies across various fields (Mateescu et al.2017).
Interesting results have already been obtained on the mechanisms of miRNA sorting into eukaryotic EVs. Understanding these mechanisms could eventually make it possible to selectively modify RNA cargoes for therapeutic purposes. Recognition of specific RNA nucleotide sequences motifs by exosomal RNA binding proteins is implicated in miRNA sorting. These motifs were detected in miRNA enriched in exosomes using bioinformatic (Batagov, Kuznetsov and Kurochkin 2011; Villarroya-Beltri et al.2013) or biochemical approaches (Santangelo, Giurato and Cicchini 2016). Bagatov and colleagues identified three 8 nucleotides motives enriched in exosomal RNA sequences extracted from gene expression databases. Later on, Kossinova and colleagues succeeded in isolating two human RNA-binding proteins present in exosomes (YB-1 and NSUN2) that bind short RNA hairpins containing these motifs (Kossinova et al.2017). Sanchez-Mardrid and colleagues identified GGAG as a motif enriched into miRNA of exosomes produced by human lymphoblasts (Villarroya-Beltri et al.2013). They found that an RNA-binding protein (hnRBP2B1) specifically binds miRNA through the recognition of this motif and controls their loading into exosomes. This mechanism appears to be itself controlled by sumoylation of this protein that triggers its binding to miRNA. More recently, using miRNA enriched in exosomes as baits, Santangelo and colleagues isolated from human hepatocyte cells another RNA-binding protein (SYNCRIP/hnRNP-Q) that specifically binds to miRNA containing the motif GGCU at their 3΄ end (Santangelo, Giurato and Cicchini 2016). SYNCRIP knockout prevents sorting of these miRNAs in exosomes. Remarkably, introducing a sequence (hEXO motif) containing the GGCU motif in an miRNA normally absent in exosome promoted its exosomal export. All these experiments confirm that RNA packaging in EVs is not a random but a highly regulated process, suggesting that the same should be true for all other types of cargoes present in EVs.
DNA in eukaryotic EVs
Relative to EV-RNA, less is known about the DNA content of eukaryotic EVs. There are reports of single-stranded DNA (Balaj et al.2011), mitochondrial DNA (Guescini et al.2010), plasmid DNA (Shader 2014) and double-stranded DNA (Thakur et al.2014); for recent reviews, see Cai et al. (2013, 2016). DNA appears present as small fragments (around 10–20 kb) and appears to be randomly selected from the entire genome, including mitochondrial DNA. Importantly, some DNA fragments associated with EVs contain entire genes with promoter and terminator regions. This DNA can be transported from cell to cell by endocytosis or fusion and this transfer can affect the transcription pattern of the recipient cells, inducing both upregulation and downregulation of many genes (Waldenström et al.2012). EV-mediated transfer of DNA coding for mRNA transcripts can thus affect cellular functions and could play an important role in the progression of various diseases (see many examples in the review by Cai et al.2016). Many aspects of this mechanism remained to be clarified. Notably, many studies have failed to conclusively demonstrate whether the nucleic acids are within or associated with the surface of EVs. Furthermore, whereas the presence of DNA in apoptotic bodies is easy to understand, the mechanism of DNA packaging in exosomes or microvesicles remains unclear.
Biogenesis of exosomes
The biogenesis of exosomes is a complex process that involves two main steps: their blebbing from the late endosome membrane leading to their accumulation in the MVB and the fusion of the MVB with the cytoplasmic membrane to release exosomes in the extracellular space (for a review, see Abels and Breakefield 2016, and references therein).
The role of the ESCRT (endosomal sorting complex required for transport) machinery in the first step is widely accepted. This machinery comprises four complexes (0, I, II and III) and many associated proteins (such as VPS4, VTA1, ALIX, and TSG101). Two ESCRT-dependent pathways for exosome biosynthesis have been described, with much research having been done to elucidate the mechanisms involved (Abels and Breakefield 2016).
In addition, ESCRT-independent pathways are likely be involved in exosome formation, though these are less well understood. Indeed, the depletion of key proteins in different ESCRT complexes does not abolish MVB formation (Trajkovic et al.2008; Stuffers et al.2009). These ESCRT-independent mechanisms are thought to involve lipids, tetraspanins or heat shock proteins.
Many enzymes involved in the modification of membrane lipids (such as phospholipase D) have been shown to regulate exosome secretion (Laulagnier et al.2004; Trajkovic et al.2008; Babst 2011; Record et al.2018). These lipids appear to become concentrated into endosomal regions of the membrane causing deformation and initiating vesicle production—a generalized mechanism similar to that found in some bacteria (see above).
As mentioned earlier, ESCRT proteins and other proteins involved in exosome biogenesis, such as tetraspanins, and ALIX, have been identified from purified exosomes in several proteomic studies and used as markers to distinguish exosomes from other types of EVs (reviewed in Choi et al.2015b). However, it is becoming apparent that the diversity of mechanisms of exosome formation parallels the diversity of exosome themselves. Furthermore, considering the difficulty to separate exosomes from microvesicles, and the heterogeneity of exosome, it is sometimes unclear if all reported mechanisms are really specific for exosomes.
The second step in the formation of exosomes, their release into the extracellular space, involves the fusion of the MVB endosomal membrane with the plasma membrane (Abels and Breakefield 2016). As with many other membrane fusion processes in Eukaryotes, the SNARE proteins (and proteins which modify their activity) appear to play an important role in exosome release (Gross et al.2012; Ruiz-Martinez et al.2016; Wei et al.2017).
Additionally, many studies have identified the small GTPases of the Rab and Ras families as playing a role in the regulation of exosome secretion (Hsu et al.2010; Ostrowski et al.2010; Koles et al.2012; for a review, see Hessvik and Llorente 2018). These proteins are membrane anchored and are thought to promote fusion of the MVB with the cell membrane. Indeed, cells with mutant forms of these proteins release fewer exosomes than their wild-type counterparts (Ostrowski et al.2010).
Biogenesis of microvesicles
Microvesicle biogenesis is less well understood than that of exosomes, though it is becoming increasingly evident that these processes share some of the same machinery. Microvesicles released from cancer cells were shown to be pinched from the membrane via actomyosin-based contraction, mediated in-part by the small Ras-related GTPase, ARF6 (Muralidharan-Chari et al.2009). ARF6 is known to activate phospholipase D in a pathway which results microvesicle budding (Muralidharan-Chari et al.2009; Tricarico, Clancy and D'Souza-Schorey 2016). More recently, Rab-family proteins have also been shown to co-localize to microvesicles and an overexpression of Rab22a led to increased levels of microvesicle budding (Wang et al.2014; Tricarico, Clancy and D'Souza-Schorey 2016).
In Caenorhabditis elegans, ESCRT-I is required for early stages of microvesicle formation (Wehman et al.2011), whereas ESCRT-II and ESCRT-III subunits are dispensable for microvesicle biogenesis (Wehman et al.2011). Additionally, in humans ESCRT-I is recruited for microvesicle release from the plasma membrane (Nabhan et al.2012) supporting a connection between the ESCRT pathway and microvesicle formation.
EV release has been shown to take place at specific locations on the cell membrane that are enriched in assorted lipids and proteins. Lipids with similar shape cluster together forming a monolayer that adopts the spontaneous curvature of the local lipids (McMahon and Gallop 2005; Cooke and Deserno 2006). Lipid bilayers in eukaryotic cell membranes employ active mechanisms to resist spontaneous curvature, and these have been conserved throughout evolution. Thus, the likelihood of spontaneous membrane curvature being the driving force behind de novo vesiculation is assumed to be low due to energy constraints. However, membrane curvature could be triggered by active alterations in lipid and/or protein composition. An uneven distribution of lipids between the two membrane leaflets may affect membrane rigidity and curvature and result in budding. It has also been reported that Ca2+-dependent aminophospholipid translocases (flippase and floppases) contribute to the formation of membrane curvature during microvesicle formation and during formation and fission of Golgi vesicles in vitro (D’Souza-Schorey and Clancy 2012; Tricarico, Clancy and D'Souza-Schorey 2016; van Niel, D’Angelo and Raposo 2018). Additional components of lipid bilayers have been implicated in vesicle biogenesis. For example, cholesterol—a key component of membrane lipid rafts—could play a role in the ‘pinching’ events (Muralidharan-Chari et al.2010). Depletion of cholesterol has been shown to impair microvesicle shedding (Del Conde et al.2005; van Niel, D’Angelo and Raposo 2018). On the other hand, proteins may exert a localized normal force, pushing on the membrane to generate the curvature needed to begin the budding process (Boulbitch 1998; Farsad and De Camilli 2003). Proteins can bend membranes by binding to the membrane surface and force curvature as a result of increasing the surface area of one leaflet (Sheetz, Painter and Singer 1976). Protein crowding at the surface of the cell membrane can also generate pressure via protein–protein interactions and drive membrane bending (Stachowiak et al.2012). A recent study showed that even green fluorescent protein was capable of driving fission when attached to membrane surfaces under crowded conditions (Snead et al.2017). This raises the possibility that the simple enrichment of protein cargo at sites of microvesicle budding could be sufficient to drive de novo microvesicle formation (Tricarico, Clancy and D'Souza-Schorey 2016).
It is interesting to note that the release of microvesicles also resembles the events associated with viral budding (see below) (Chazal and Gerlier 2003; Morita and Sundquist 2004; Meckes and Raab-Traub 2011; Wurdinger et al.2012) and the release of apoptotic bodies, both of which form by outward protrusion of the plasma membrane. However, unlike apoptotic bodies, microvesicles do not contain fragments of cytosolic organelles (Taylor and Gercel-Taylor 2008; Crescitelli et al.2013).
Biogenesis of apoptotic bodies
Apoptotic bodies are produced when cells undergo fragmentation during apoptosis. Their production was originally believed to be a stochastic process; however, recent work has demonstrated that the formation of apoptotic bodies is a highly regulated multistep process (Chekeni et al.2010; Poon et al.2014; Atkin-Smith et al.2015). Interestingly, the group of Poon also observed vesicles resembling ‘beads-on-a-string’ being formed in apoptotic cells. They referred to these string-like structures as apoptopodia (Atkin-Smith et al.2015).
Curiously a study by the group of Lötvall (Crescitelli et al.2013) showed that RNAs isolated from apoptotic bodies, membrane vesicles and exosomes have very different profiles. Indeed, they observed that membrane vesicles had the least amount of RNA, with rRNA being primarily found in apoptotic bodies. Perhaps this reflects the more active cargo selection of microvesicles, relative to the packaging of a fragmented cytoplasm (containing many ribosomes) in apoptotic bodies.
Nanotubes in Eukaryotes
Nanotubes were first described in Eukaryotes as tunneling nanotubes (TNTs) and have been observed to connect cells over long distances and transfer vesicles (Fig. 2a), membrane proteins, cellular components (including mitochondria), prions, miRNA and viruses from cell to cell (Rustom et al.2004; Davis and Sowinski 2008; Lou et al.2012; for recent reviews, see Austefjord, Gerdes and Wang 2014; Rustom 2016; Nawaz and Fatima 2017). TNTs formed between animal cells have been likened not only to Plasmodesmata connecting plant cells but also to bacterial nanopods (Fig. 2b) (Rustom 2016). These nanotubes seem to be formed from intracellular vesicles, and their shape and movement are determined by the action of cytoskeletal proteins (Rustom 2016). Most of them contain actin filaments along their entire length, possibly explaining their rigidity. Inhibitors of actin polymerization significantly decrease the number and length of TNTs that occur between trabecular meshwork cells and reduce EV transfer, whereas compounds that stabilize actin filaments increase EV transfer (Keller et al.2017). Some eukaryotic TNTs also contain tubulin and are usually larger. Eukaryotic nanotubes are indeed variable, with diameters varying from 50 to 700 nm (Austefjord, Gerdes and Wang 2014; Bénard et al.2015). Bénard et al. (2015) have shown that the same cells can produce different types of TNTs that can be discriminated by their size and protein content, with larger TNTs (diameter 100–650 nm) containing both actin and tubulin and smaller ones (70–200 nm) containing only actin. These structures appear larger than archaeal and bacterial nanotubes.
Nanotube-like structures containing actin were also described in some cancer cells as ‘protrusions’ related to filipodia and directly associated with the production of microvesicles. Rilla et al. (2013) reported that the tip of these protrusions can detach into the culture medium as microvesicles. The formation of both protrusions and microvesicles was enhanced by overexpression of hyaluronan synthase in human mesothelial cells suggesting active regulation of these structures (Rilla et al.2013; Koistinen et al.2017). Despite their similarity, the relationship between these protrusions and TNTs is unclear.
Recently, the effect of vesicle size on nanotube formation in vitro and in vivo was investigated using lysosomes and autolysosomes (Su et al.2016). The results suggested that tubulation was dependent on the size of the vesicles in the cell, reinforcing the link between nanotubes and vesicles.
The ubiquity of EVs in Eukaryotes
The study of EVs in Eukaryotes other than humans (and a few animal models) is lagging far behind the study of human exosomes and microvesicles. However, it is already clear that all eukaryotic cells, from both unicellular and multicellular organisms, produce these two types of EVs.
Plant cells have also been shown to be producers of exosome-like vesicles, especially in response to pathogen attack (Stanly et al.2016; Rutter and Innes 2017). For instance, Arabidopsis can deliver small siRNAs into the fungal pathogen Botrytis cinecrea via EVs. These siRNAs silence genes critical for pathogenicity (Cai et al.2018). Proteomic analysis on EVs purified from apoplastic fluids of Arabidopsis leaves revealed that they are highly enriched in biotic and abiotic stress response proteins (Rutter and Innes 2017). Indeed, increased levels of EVs were observed when plants were infected with P. syringae or treated with salicylic acid. Thus, EVs may play an important role in the immune responses of plants.
Fungi were hypothesized to release EVs outside their cell membrane as early as the 1970s following observations in Saccharomyces cerevisiae and Cryptococcus neoformans (Takeo, Uehira and Nishiura 1973; Novick and Schekman 1979). Later, EV production by Candida albicans was observed by TEM and scanning electron microscopy (SEM) (Anderson, Mihalik and Soll 1990). For a long time, these observations were not followed up, largely due to the belief that the structure of the fungal cell wall was too rigid to allow budding. However, production of EVs by C. neoformans was finally confirmed (Rodrigues et al.2007) and EV production was subsequently associated with a variety of other fungi (Rodrigues et al.2007, 2014; Albuquerque et al.2008; Nosanchuk et al.2008; Oliveira et al. 2010a,b ). EVs produced by fungal cells are similar to mammalian exosomes, (Albuquerque et al.2008; Rodrigues et al.2008) including their ability to modulate the function of immune cells (Oliveira et al. 2010a,b) and export RNA (Peres da Silva et al.2015; Bielska et al.2018). Recently, it was shown that EVs produced by the human pathogen Cryptococcus gattii can be delivered to intracellular fungal cells within macrophages and trigger their division (Bielska et al.2018). The RNA and proteins associated with these EVs are essential for this long-distance pathogen-to-pathogen communication.
The EVs are probably excreted from fungal cells through the pores (from 50 to 500 nm) that are present in the thick cell wall. The size and abundance of these pores is constantly remodeled during the cell cycle, possibly controlling the release of EVs. In agreement with the hypothesis that EV release is an active process, EVs in fungi are produced exclusively from living cells. Finally, EVs from fungi contain β-glucosidase and endochitinase that can help to reduce the thickness of the wall to facilitate EV crossing (Brown et al.2015 and references therein).
A broad range of eukaryotic microbes, such as the ‘algae’ Emiliania huxleyi, the Amoeobozoa Dictyostelium discoideum and Acanthamoeba, including several extracellular parasites (e.g. Trichomonas vaginalis, Trypanosoma cruzi, Leishmania spp. and helminths) and intracellular parasites (Plasmodium falciparum, Toxoplasma gondii and Leishmania spp.), have been shown to produce EVs (Bhatnagar et al.2007; Silverman et al.2008, 2010; Martin-Jaular et al.2011; Cestari et al.2012; Marcilla et al.2012, 2014; Mantel et al.2013; Pope and Lasser 2013; Regev-Rudzki et al.2013; Twu et al.2013; Arantes et al.2016; Marti and Johnson 2016; Schatz et al.2017; Ribeiro et al.2018). These EVs can play a major role in the interactions between the parasites and their host; for example, Leishmania spp. have been shown to constitutively secrete exosomes within the lumen of the sand fly midgut and, following their ingestion into a new host during the insect's bite, these vesicles play an important role in modulation of host immunity, and thus promote the parasitic infection (Atayde et al.2015). In T. cruzi, it has been shown recently that differences in EV production between strains correlate with their infectivity and virulence (Ribeiro et al.2018). The production of EVs has been also observed in studying the infection of eukaryotic microbes by viruses. The production of EVs by Acanthamoeba was dramatically illustrated by the production of large vesicles containing virions from cells infected by the giant virus Marseillevirus (Arantes et al.2016). Additionally, the interplay between viruses and EVs has been nicely illustrated by analyzing EV production during the infection of the E. huxleyi by EhV, Phycodnaviridae (Schatz et al.2017). These are discussed further below.
The Amoebozoa D. discoideum has been proposed as a model organism for the study of eukaryotic EVs (Tatischeff 2013). Besides producing EVs implicated in the types of intercellular communication observed in other systems, it was demonstrated that Dictyostelium was able to lessen the toxicity of drugs, such as Hoechst 33342 (HO342), by secretion into EVs (Lavialle et al.2009).
EVs in Archaea
Archaea are unicellular microorganisms that look like bacteria at the phenotypic level. In particular, they lack nuclei and are often grouped with Bacteria as being prokaryotes. However, their informational systems (transcription, translation, and DNA replication) as well as some mechanisms associated with their membranes (e.g. the Sec secretion system, the ATP synthase complex, the signal recognition particles) are much more similar to those of eukaryotes. Some archaea even contain proteins closely related to eukaryotic actin and tubulins (Ettema, Lindås and Bernander 2011; Yutin and Koonin 2012; Lindås et al. 2017) as well as proteins involved in membrane remodeling and eukaryotic EV formation such as ESCRT III proteins and/or VPS4 ATPases (Makarova et al.2010). All cultured archaea lack peptidoglycan and most of them are monoderm (Ellen et al.2010; Albers and Meyer 2011), with the exception of Ignicoccus hospitalis and Methanomassilicocales which have an OM and a large periplasmic space. In most archaea, the cytoplasmic membrane is surrounded by a crystalline protein S-layer. The S-layer is usually composed of a single main protein type (40–200 kDa) arranged into a highly order structure that is capable of self-assembly, both on cell surfaces and in solution in vitro as cell-free S-layer ‘ghosts’ (Kish et al.2016). Some archaea lack this S-layer, such as Thermoplasmatales, whereas others exhibit an additional peptidoglycan-like structure formed from pseudomurein below the S-layer resulting in positive Gram staining (Methanobacteriales and Methanopyrales) (Steenbakkers et al.2006; Albers and Meyer 2011; Klingl 2014).
EV production is most likely a general phenomenon in Archaea, as in the other two domains of life (Fig. 7). Archaeal EVs were first observed in the thermoacidophilic archaeon Sulfolobus islandicus (Prangishvili et al.2000), a member of the archaeal phylum Crenarchaeota. Sulfolobus EVs (90–230 nm in diameter) are enclosed by cytoplasmic-like membrane and coated with the S-layer. Characterization of EVs produced by three other Sulfolobus species, S. acidocaldarius, S. solfataricus and S. tokodai, has shown that the lipid and protein profiles of parental cells membranes and secreted vesicles were different, suggesting a selective mechanism for protein packaging in EVs (Ellen et al.2009). Interestingly, the proteomic analyses of EVs from the three species revealed the presence of proteins homologous to eukaryotic ESCRT-III subunits and to the VPS4 ATPase. Studies have shown that Sulfolobus homologs of ESCRT-III and VPS4 (also called CdvB and CdvC, respectively) are involved in cell division (Lindås et al.2008; Samson et al.2008), and it has been suggested that these proteins could be also involved in EV formation by a mechanism similar to that of exosome production in Eukaryotes (Ellen et al.2009; Caspi and Dekker 2018). In several reviews, authors have indeed assumed that the archaeal homologs of eukaryotic components are involved in EV formation as if it was a well-established result (see for instance figure 4 in Deatherage and Cookson 2012). However, this remains a hypothesis that needs to be proved conclusively using genetic approaches.
Figure 7.
EV production in Archaea. EVs originate from membrane budding and nanotubes. Functions include HGT, intercellular competition, disposal/detoxification, biomineralization and possibly viral defense.
Notably, Sulfolobus EVs carry an antimicrobial protein, termed ‘sulfolobicin’, that inhibits the growth of related Sulfolobus species (Prangishvili et al.2000). In fact, they were first confused with viral particles because spotting EV preparations on Sulfolobus lawns resulted in halos similar to lysis plaques. Later, Driessen and colleagues showed that sulfolobicins are in fact composed of two proteins whose genes are organized in operon (Ellen et al.2011).
A recent study revealed that Sulfolobus EVs could also promote biomineralization (Kish et al.2016). Whilst S-layers have long been associated with mineral formation, the mechanisms by which this occurs remain unresolved. A study using S. acidocaldarius, a hyperthermophilic archaeon native to metal-enriched environments, demonstrated a passive process of iron phosphate nucleation and growth within the S-layer of cells and cell-free S-layer ‘ghosts’ during incubation in a Fe-rich medium. In addition, EVs of ∼ 175 nm in diameter were formed and released as a response to S-layer encrustation by minerals. These vesicles were fully encrusted by minerals, even when cells were only partially encrusted (Kish et al.2016). The authors propose that these EVs are produced in an attempt to remove sections of damaged S-layer.
Archaeal EVs have been also studied in hyperthermophilic (and neutrophilic) archaea of the genus Thermococcus (Fig. 1c and d), which are members of the phylum Euryarchaeota. Extensive screening revealed that most of the strains in this genus released EVs (50–150 nm) that are morphologically very similar (albeit a bit smaller) to those produced by Sulfolobus (Soler et al.2008; Gaudin et al.2013; Marguet et al.2013). The major protein present in both the cell membranes and EVs of Thermococcus species is the oligopeptide-binding protein OppA (Gaudin et al.2013), which is also found in Sulfolobus EVs (Ellen et al.2009). However, unlike the situation observed with Sulfolobus, the protein profiles of EVs from different species of Thermococcales showed that both EVs and cell membranes from the same species have a similar composition, suggesting that the mode of EV production could be different in Sulfolobus and Thermococcus. Indeed, homologs of CdvB are missing in Thermococcus and other Euryarchaeota (Makarova et al.2010).
Thin-section analyses by electron microscopy revealed that these EVs from Thermococcales are produced by protrusion of the cell membrane along with the S-layer (Figs 1c and d and 7). During these studies, the authors reported for the first time the presence of rows of EVs enclosed within the S-layer (Soler et al.2008) resembling the aforementioned nanopods or nanotubes later observed in Bacteria (Fig. 2) (Shetty et al.2011). EVs present within nanotubes are usually smaller than free EVs but large EVs are frequently observed at the extremities of nanopods, suggesting that these structures could be involved in the transport and/or formation of EVs. These nanopods from Thermococcales sometimes connect cells together (Marguet et al.2013) and could be involved in transfer of material between cells as in the case of their bacterial counterparts. Finally, these authors also observed ‘nanospheres’ budding from the cell surface, in which several EVs are enclosed by S-layer in a spherical structure (Marguet et al.2013).
EVs produced by Thermococcus species are often associated with genomic DNA (Soler et al.2008, Gaudin et al.2014; Choi et al.2015a) and RNA (Choi et al.2015a; Gill and Forterre unpublished observation). The association of DNA with EVs might be significant for hyperthermophilic species since this vesicle-associated DNA appears to be more resistant to thermodegradation than free DNA (Soler et al.2008). Notably, whereas HGT between hyperthermophilic species (including transfer between Archaea and Bacteria) has been clearly demonstrated in silico by comparative genomics, it is unclear how these transfers can occur at temperatures that provoke DNA melting/degradation. It thus makes sense to propose that EVs could play a major role in gene transfer between hyperthermophilic species. Forterre and colleagues demonstrated that EVs can transfer DNA at least between cells of the same species using a genetically tractable strain Thermococcus kodakaraensis (Gaudin et al.2013). After transformation with a reporter plasmid, this strain produces EVs containing plasmids and these EVs can be used to transfer the plasmid into plasmid-free cells. EVs produced by T. nautili naturally harbor DNA of the endogenous plasmids pTN1 and pTN3 suggesting a possible role for EVs in plasmid transfer in vivo. Interestingly, the plasmid pTN3 appears to be a viral genome, carrying genes encoding the major capsid protein and the packaging ATPases characteristic of the adenovirus/PRD1 lineage. This suggests that EVs may also be involved in the generation of new viral forms (Soler et al.2011; Gaudin et al.2014).
EVs have been also characterized in T. onnurineus (Choi et al.2015a). The authors separated different populations of EVs showing different buoyant densities by density gradient centrifugation. All of the recovered populations were associated with DNA fragments estimated to be around 14 kb long. Surprisingly, sequencing of the associated DNA revealed that all regions of the T. onnurineus genome were present except for a 9.4-kb region. The authors then speculated that this region could encode proteins involved in the mechanism of EV production or in EV-mediated DNA transfer. Interestingly, a T. onnurineus mutant in which this region has been deleted still produces vesicles but without associated DNA, favoring the second hypothesis (Kim YK, personal communication). This 9.4-Kb region encodes various enzymes involved in sulfur metabolism and/or hydrogen production and their possible role in DNA packaging is unclear.
Another role that these EVs play in Thermococcales is that of detoxification. Cryo-electron microscopy of T. prieurii cells revealed the presence of numerous intracellular dark vesicles that bud from the host cells (Gorlas et al.2015). These dark vesicles are exclusively found in association with intact cells and are never observed in preparations of purified membrane vesicles. Furthermore, the presence of these vesicles is exclusively observed when elemental sulfur is added into the growth medium. Energy-dispersive-X-ray analyses revealed that these dark vesicles are filled with sulfur, and hence they have been termed ‘sulfur vesicles’ (Fig. 2). These dark vesicles were also observed in T. kodakaraensis albeit in lower numbers. Curiously they are lacking in T. nautili suggesting that Thermococcales species exhibit significant differences in their sulfur metabolic pathways.
Recently, Erdmann et al. (2017) described a novel type of EV containing plasmid in Archaea. These EVs, which are produced by the psychrophilic halophilic archaea Halorubrum lacusprofundi (member of the phylum Euryarchaeota), contain a plasmid of 50 kb, pR1S1. These EVs can infect a plasmid-free strain and transform this strain into a producer of EVs containing the plasmid pR1S1. Notably, this plasmid encodes several proteins that are present in the EVs themselves. The authors suggested that these proteins could be involved in plasmid packaging inside EVs and in the formation of an apparatus responsible for the transfer of these EVs from cell to cell. Forterre, Da Cunha and Catchpole (2017) proposed that these EVs be called plasmidions (mimicking virions) to distinguish them from canonical plasmid vesicles that do not contain plasmid-encoded proteins.
The production of EVs is most likely a universal process in Archaea as in the other two domains. EVs and structures resembling nanopods have been observed in cultures of the euryarcheon Aciduliprofundum boonei (Reysenbach et al.2006; Reysenbach and Flores 2008). Additionally, large numbers of EVs were observed in the periplasm of the diderm archaeon I. hospitalis harboring several cells of the tiny archaeon Nanoarchaeum equitans on its surface (Huber et al.2002; Küper et al.2010). The volume of the Ignicoccus periplasm can be large (20–1000 nm in width) and contains numerous EVs that bud from the inner membrane and fuse with the OM (Näther and Rachel 2004). Nanoarchaeum equitans has the smallest known genome size for an archaeon (0.49 Mb) and cannot synthesize many essential components, including lipids (Waters et al.2003). Recent evidence suggests that these components are delivered from the cytoplasm of I. hospitalis to N. equitans via vesicles that reach the OM at the position of the symbiont attachment, or through nanotube-like endomembrane connections between the two cytoplasms (Heimerl et al.2017).
The mechanisms of EV production remain unknown in Archaea. The lack of simple screening methods for the detection of hypo- and hypervesiculation mutants is a major obstacle. It is likely that different types of EV production mechanisms exist in this domain as in the two others, depending on the species and/or the vesicle types. By analogy with the modes of EV production known in Bacteria and Eukarya, it is possible that EV production in Archaea is controlled by the regulation of enzymes involved in the incorporation of polar lipids in order to create asymmetry between the outer and inner membrane leaflet. However, since many archaea have monolayer cytoplasmic membranes formed by giant tetraether bipolar lipids (e.g. caldarchaeol), classical models of membrane split and fusion which involve the transient opening of the bilayer cannot be applied (Relini et al.1996).
Monolayer membranes seem to be especially prevalent in hyperthermophilic archaea in which EV production has been widely studied, such as Sulfolobales and Thermococcales. It has been shown that fusion of artificial vesicles made of Sulfolobus tetraether lipids is still possible, but is much more difficult than fusion of classical bilayer vesicles (Relini et al.1996). The authors of this study have shown that fusion is only possible if the lipid vesicles contain monosubstituted molecules in which one of the glycerol head groups is not substituted, and requires trace diether lipids. Importantly, the ratio between tetraether (caldarchaeol) and diether lipids in archaeal membranes can vary depending on the physiological conditions (Meador et al.2014; Cario et al.2015 and references therein); thus, it would be interesting to determine if there is a correlation between EV production and lipid composition. In the case of archaea with a predominant monolayer membrane, a hypothetical model could be that membrane curvature is induced by the accumulation of larger polar head groups at the outer surface of the monolayer. Another non-exclusive possibility is that patches of membranes adopt bilayer or simple monolayer structure by favoring diether lipids (archaeol) or horseshoe structures of tetraether, as recently observed in the membrane of the archaeal virus (Kasson et al.2017).
EVs and viruses
The size and morphology of EVs are strongly reminiscent of the morphology of some viral particles (virions). A virion (the hallmark of viruses, see Krupovič and Bamford 2010) contains at least one protein encoded by the viral genome. For instance, exosomes strikingly resemble virions of retroviruses and other enveloped viruses (Izquierdo-Useros et al.2011). Similarly, the virions of Plasmaviridae infecting Mycoplasma, or Pleolipoviridae infecting halophilic archaea, resemble EVs, being formed by a lipid envelope containing inserted viral-encoded proteins (Demina et al.2016; Erdmann et al.2017). Some EVs contain viral DNA (viral vesicles) or plasmids (plasmid vesicles and plasmidions) (Fig. 3). These similarities between EVs and viral particles raises a number of interesting questions relative to their respective origins, possible common production mechanisms and possibilities for physiological interactions that we will explore here. They also raise some challenging questions and problems in terms of nomenclature and discrimination between virions and various types of EVs in the environment.
Physical interactions between EVs and virions have been observed in the three domains of life. In Bacteria, Manning and Kuehn (2011) first showed that co-incubation of T4 virions and OMVs showed fast and irreversible binding of these virions to OMVs. Later, Biller and co-workers observed interactions between virions and OMVs in the case of Prochlorococcus and the cyanophage PHM-1. They further demonstrated that many vesicle-attached virions had a shortened stalk and altered capsid staining density, suggesting that they had injected their DNA into the vesicle (Biller et al.2014). More recently, Camilli and colleagues observed by cryo-electron tomography direct attachment of virions from diverse viruses infecting Vibrio cholerae to OMVs produced by this bacterium (Reyes-Robles et al.2018).
All these observations suggest that EVs could be used as antiviral systems by cells, acting as decoys to reduce infectious virus–cell interactions. The efficiency of T4 infection was indeed significantly reduced by the formation of complexes with OMVs (Manning and Kuehn 2011). The same result was obtained with V. cholerae infected by the bacteriophage ICP1 (Reyes-Robles et al.2018). If EVs are used as traps by cells to protect themselves against infection, one expects that cells increase EV production as a response to infection. This is indeed the case: as early as 1978, Loeb and colleagues demonstrated a dramatic increase in OMV production from E. coli in the presence of T4 virus (Loeb and Kilner 1978). It has now been reported in several eukaryotic systems that viral infection can increase EV production (Nolte-‘t Hoen et al.2016). However, in that case, the EVs produced did not act as decoys inhibiting viral infection, but more like Trojan horses favoring infection, facilitating viral infections by delivering viral proteins, RNA and/or DNA from infected to non-infected cells (Altan-Bonnet 2016) (see below).
Direct interactions between viral particles and EVs have been also observed in Archaea, especially between EVs and the lemon-shaped particles produced by Fuselloviridae (Geslin et al.2003) (Fig. 3). However, it is not known whether EVs can interfere with viral infection and how this may occur. Interestingly, the peptide-binding protein OppA detected in EVs of Sulfolobus and Thermococcus species has been identified as a putative receptor for the Sulfolobus virus ATV (Erdmann, Scheele and Garrett 2011). It would be interesting to test if EVs are generally enriched in viral receptors thus promoting virion–vesicle interactions.
Recently, Schatz et al. (2017) observed an intriguing interaction between EVs and virions in Eukaryotes when studying the infection of E. huxleyi by a phycodnavirus. Notably, this interaction favors the infection process. They show that mixing EVs produced by infected cells with virion preparations prolonged the half-life of the virion from 10 to 60 h. This could increase the chance of infectious encounters between virions and their cellular targets. They proposed that ‘EVs are exploited by viruses to sustain efficient virus infectivity and propagation across E. huxleyi blooms’ (Schatz et al.2017) with important environmental consequences.
Several studies have shown that EVs produced by virus-infected cells can favor viral infection by making other cells in the population more sensitive to viral infection (similar to that described for cancer metastasis above). For instance, OMVs can transfer viral receptors from sensitive to resistant cells, helping the virus to bypass bacterial resistance due to the lack of receptors in target cells (Tzipilevich, Habusha and Ben-Yehuda 2017). Exchange of phage receptor recognition proteins could even occur between different species, enabling phage adsorption to non-host species, providing an unexplored route for HGT. The fact that viral-infected cells produce EVs that favor viral infection can be understood in the framework of the virocell concept (Forterre 2013), which posits that the infected cell is no more a bacterial, an archaeal or a eukaryotic cell, but a cellular form of the virus (a virocell) whose metabolism and physiology has been remodeled by the virus to fulfill its own objectives. In that view, EVs produced by virus-infected cells can be considered to be viral EVs produced by the virocell, even if they do not contain bona fide viral components.
In many cases, EVs produced by virocells have modified biochemical composition, containing various viral elements—either proteins or nucleic acids. In particular, human cells infected by different types of viruses contain viral proteins, segments of viral RNA, microRNA or mRNA (Pegtel et al.2010; Meckes and Raab-Traub 2011; Altan-Bonnet 2016; Kouwaki et al.2017). They can also carry and transport cellular components that make recipient cells more susceptible to viral infection. These viral EVs can modulate the innate immune response system of the host and facilitate viral infection by carrying molecules implicated in virus binding and entry (György et al.2011; Kouwaki et al.2017).
In a previously mentioned study, Vardi and colleagues have shown that infection of E. huxleyi by its virus (EhV, Physodnaviridae) induces the production of specific EVs (Schatz et al.2017). These EVs have a unique lipid composition, being strongly enriched in triacylglycerols. Remarkably, the number of EVs produced by virus-infected cell was similar to the number of bona fide viral particles. These EVs can be internalized by virus-free cells, making them more sensitive to viral infection. Cells infected by these EVs are lysed faster than control cells and produce more viral particles. The authors observed modification of cellular sphingolipid metabolism and cell-cycle pathways of ‘EV-infected cells’, probably via a regulatory effect of the EVs sRNA. Notably, the production of EVs can be also triggered by heat-sensitive virus-free lysate from virus-infected cells, indicating that the virus produces one or more proteins that play a direct role in EV production. Addition of this virus-free lysate also increases the formation of triacylglycerols in control cells, again illustrating the link between cellular lipid composition and EV formation. This model system could become especially interesting to decipher the mechanism of EV production in Alga.
EVs containing viral DNA have been called viral vesicles when, unlike virions, they do not contain proteins encoded by the virus (Gaudin et al.2014). The production of such viral vesicles has been observed in the three domains of life. This definition could even be extended to eukaryotic vesicles containing viral RNA, such as exosomes containing full-length RNA from hepatitis C viruses (Bukong et al.2014; Longatti, Boyd and Chisari 2015). It has been suggested that viral vesicles could help in the dissemination of viruses in the absence of viral receptors by their capacity to transfer DNA from cell to cell (Soler et al.2015). The production of viral vesicles has been observed in the three domains of life. Viral vesicles containing genomes from Caudovirales have been described in Bacteria (Yaron et al.2000), whereas viral vesicles containing a plasmid (pTN3) corresponding to a defective viral genome and plasmid vesicles (or plasmidions) containing infectious plasmids have been described more recently in Archaea (Gaudin et al.2013; Erdmann et al.2017). Notably, in silico analysis of DNA associated with EVs isolated from diverse marine environments has suggested that viral vesicles could be abundant in nature in addition to true virions and EVs containing cellular DNA (Soler et al.2015). Further work is now required to determine if viral vesicles and plasmidions indeed play an important role in the co-evolution of viruses, plasmids and their hosts in natural environments.
In Eukaryotes, both RNA and DNA viruses can also use the EV secretion system to package a huge number of virions (Altan-Bonnet and Chen 2015; Altan-Bonnet 2016). The production of such atypical EVs (hereafter referred to as virions packaging vesicles, VPV) was observed for the first time with polioviruses and rhinoviruses that can accumulate hundreds to thousands of virions in large EVs (Chen et al.2015). These VPV are produced from autophagosomes i.e. intracellular vesicles with double membranes that originate from the endoplasmic reticulum (Chen et al.2015). Recently, VPVs carrying huge numbers of virions were also observed during infection of amoeba with Marseillevirus, a large DNA virus of the NCLDV superfamily (Fig. 3) (Arantes et al.2016), suggesting that production of VPVs carrying virions is probably a widespread strategy of eukaryotic viruses. Amoeba infected by Marseillevirus produce giant VPVs containing thousands of virions (Fig. 3) that can promote phagocytosis. These VPVs are also apparently formed by the recruitment of membranes from the endoplasmic reticulum and can be surrounded by one of two membranes, resembling the autophagosome. Their release into the environment appears to require cell lysis.
VPVs containing either Enterovirus or Marseillevirus virions are infectious and can trigger high multiplicity infections after fusion with the host cell membrane (Altan-Bonnet 2016; Arantes et al.2016). Notably, in both systems, the infection efficiency of VPVs turned out to be much higher than those of free virions (Chen et al.2015; Arantes et al.2016). The high multiplicity of infection could enhance the overall fitness of the viral population, facilitating biochemical complementation and genetic recombination, and promoting genetic cooperativity among viral quasi-species (Altan-Bonnet 2016). VPVs are presently unknown in Archaea and Bacteria.
The structural similarity between virions and EVs raises a priori practical problems for counting viral particles in environmental samples since one cannot discriminate by electron microscopy between EVs and tailless viruses (Soler et al.2015). Furthermore, the association of EVs with DNA makes EVs fluorescent after DNA staining, such that EVs and virions can be confused in epifluorescence microscopy, a technique widely used by microbial ecologists (Soler et al.2008). As a result, it has been suggested that the number of true virions has possibly been overestimated in ecological studies (Forterre et al.2013; Soler et al.2015). However, Biller et al. (2017) concluded that this might not be the case in marine environments since they only observed a small decrease of epifluorescence-visible EVs upon chloroform treatment from natural marine samples. Chloroform treatment allows discrimination of EVs from tailed viruses (Caudovirales) that do not contain lipids in their virions. Recently, a new method based on interferometry was used to discriminate between EVs and caudovirales virions in environmental samples (Roose-Amsaleg et al.2017). In this case, they found that tailed virus particles represented between 40 and 70% of all particles present in their river samples, suggesting that the ratio between EVs and viral particles could differ in various environments.
Strikingly, evolutionarily unrelated enveloped viruses infecting eukaryotes have converged in their use of the host machinery for exosome formation to promote their own budding. For example, some non-enveloped RNA viruses (picornavirus) exit from the cell without lysis by hijacking the cellular membrane to cover their capsid, thereby protecting the virion from antibody-mediated neutralization by their host (Feng et al.2013). Similarly, human immunodeficiency virus, Ebola virus, rabies virus and herpes simplex virus 1 all have well-characterized strategies to hijack members of the ESCRT pathway. These enveloped viruses carry structural Gag proteins that package an RNA genome and present specific viral glycoproteins on their surface, thus enabling them to infect target cells (Votteler and Sundquist 2013). The diversity of EV populations and components suggests that EVs enter cells through various mechanisms similar to the multiple pathways identified for viruses (Marsh and Helenius 2006).
The relationship between mechanisms involved in the production of retroviruses and exosomes and other aspects of their physiology is an active topic of investigation (Izquierdo-Useros et al.2011; Madison and Okeoma 2015). The formation of intraluminal vesicles within the MVB and the budding of enveloped virions share many features. Both processes require induction of membrane curvature, inclusion of specific cargo and membrane fission for release. Several models have been proposed to explain the similarities found between retroviruses and exosomes: either exosomes are the ancestors of retroviruses or retroviruses merely exploit the same cellular machinery designated for exosome biosynthesis (Gould, Booth and Hildreth 2003; Pelchen-Matthews, Raposo and Marsh 2004). In the latter case, one can even imagine a possible viral origin of the EV system or perhaps an evolutionary conserved system of virus-vesicle co-dependence (Izquierdo-Useros et al.2011).
A recent study describing eukaryotic-like virus budding in Archaea provides a new interesting model to study the similarities between EVs and virion production (Quemin et al.2016). The authors reported that the assembly and egress processes of the fusellovirus SV1 infecting Sulfolobus are concomitant and occur at the cellular cytoplasmic membrane via a process highly reminiscent of the budding of enveloped viruses that infect eukaryotes. This includes a step in which the lipid containing envelope of the virions fuse or split with the cytoplasmic membrane. Interestingly, this work also revealed the formation of constricted ring-like structures which resemble the budding necks observed prior to ESCRT machinery-mediated membrane scission (Quemin et al.2016). Since fuselloviruses have also been isolated from Thermococcales (Geslin et al.2003; Gorlas et al.2012), they could be useful models to study EV production in these hyperthermophilic archaea.
Considering the similarities between EVs and viral particles, it is tempting to make a link between EVs and the origin of viruses. Two different scenarios involving lipid vesicles have been proposed to explain this origin. Jalasvuori and Bamford (2008) suggested that protocells containing RNA produced vesicles that were used to infect ‘empty’ lipid vesicles, promoting the dissemination of RNA. They called these vesicles ‘protoviruses’. However, defining viruses without referring to capsid proteins is problematic because it makes it impossible to distinguish between viruses and other mobile elements, such as plasmids, transposons and so on (Krupovič and Bamford 2010). If viruses are defined as ‘capsid-encoding organisms’, i.e. producing at least one protein (Raoult and Forterre 2008), they should have only appeared after the emergence of the ribosome. In that framework, Forterre and Krupovič (2012) have suggested that the first viruses originated in the period between formation of the ribosome and LUCA. In that scenario, the emergence of a mechanism for virion formation, excretion and dissemination via transfer is the crucial step in virus origin. Forterre and Krupovic (2012) suggested different mechanisms for virion formation by exaptation of ancient cellular systems, explaining the different types of virions present in modern viruses. Among them, the production of EVs could have been at the origin of modern viruses, such as retroviruses, plasmaviruses or pleolipoviruses.
EVs in the history of life
The production of EVs by cells from the three domains of life has suggested that the LUCA and its contemporaries already produced EVs (Gill and Forterre 2016). Comparative genome analysis indeed suggests that LUCA was already a rather sophisticated cellular organism that already harbored a cytoplasmic membrane (Pereto, Lopez-Garcia and Moreira 2004; Jekely 2006; Koonin 2006; Forterre and Gribaldo 2007). Proteins whose origin can be traced back to LUCA include several subunits of the membrane-bound ATP synthase, signal recognition particles involved in the translation of membrane proteins and Sec proteins involved in protein secretion. Phylogenomic analyses also predict that LUCA had unsaturated polyisoprenols (Lombard 2016). Since these universal molecules are used today as lipid carriers in glycosylation pathways, this suggests that LUCA possibly had some form of glycoprotein layer covering the cytoplasmic membrane. However, the fact that LUCA had a membrane does not necessarily mean that it was able to produce EVs. To clarify this point, it would be important to determine if homologous machineries for EV formation exist in Archaea and Bacteria and can be traced back to LUCA. We are thus limited in our conclusion since the mechanisms of EV production in the three domains remain poorly understood, especially in Archaea. Among proteins usually found in EVs, the only possible universal proteins could be proteins of the SPFH superfamily that are known to facilitate membrane curvature and cell fusion (Lee et al.2017) and have been involved in the production of some eukaryotic EVs (Hinderhofer et al.2009). However, these are small proteins with multiple paralogs, especially in eukaryotes, and their presence in LUCA is difficult to ascertain (Browman, Hoegg and Robbins 2007; Hinderhofer et al.2009). It would be thus important to get more insight on the possible role of these proteins in EV production and to update their phylogenomic analysis. Finally, the discovery that overexpression of hyaluronan synthase induces the formation of nanopods and the production of microvesicles in eukaryotes (Rilla et al.2013; Koistinen et al.2017) is possibly interesting because homology of hyaluronan synthase is present in Bacteria and Archaea. However, it is not known if hyaluronan is a component of EVs in these two domains. It would be interesting to know if overexpression of hyaluronan synthase also stimulates EV production in Bacteria and if homologs of this enzyme in Archaea have the same activity.
Different and possibly redundant mechanisms for EV production probably originated after LUCA, during the divergence of the three domains. The emergence of peptidoglycan in Bacteria probably led to a drastic modification in EV structure with the appearance of OMV (depleted in cytoplasmic components) and O-IMV. The emergence of the ESCRT system in the lineage common to Archaea and Eukarya suggests that this mechanism was used for EV production in the last common ancestor of both domains. However, the loss of the ESCRT system in some Archaea (e.g. Thermococcales) that produce abundant vesicles clearly indicates that several redundant systems may be involved in EV production.
Recently, two original scenarios were proposed that accord an important role for EV production in the origin of Eukaryotes. Baum and Baum (2014) suggested that the eukaryotic cytoplasm originated from the expansion of EVs and nanopods produced by an archaeon. These EVs and nanopods of increasing sizes entrapped the bacterium at the origin of mitochondria and fused to produce both a continuous cytoplasmic membrane and the endoplasmic reticulum with its extensions e.g. the nuclear membrane or the Golgi apparatus. However, it is unclear in this model how the archaeal lipids of the EVs/nanopods were transformed into the ‘bacterial-like’ lipids of the modern eukaryotic endomembrane system. Furthermore, interactions between archaeal nanopods and bacteria have not yet been observed in nature.
In a more classical model, Martin and co-workers suggested that the eukaryotic intracellular membrane system originated from OMVs produced by the bacterial ancestors of mitochondria inside the cytoplasm of an archaeal host (Gould, Garg and Martin 2016). The lipids from these OMVs progressively replaced all previous archaeal lipids of the host, including the cytoplasmic membrane. This model is based on the observation that modern mitochondria still produce EVs that can transfer materials between the mitochondrion and eukaryotic intracellular vesicles (peroxysomes, lysosomes, etc.) (Andrade-Navarro, Sanchez-Pulido and McBride 2009; Soubannier et al.2012a). Similar to OMVs produced by free-living bacteria, mitochondrial EVs are overexpressed under stress conditions and seem to be involved in the detoxification of damaged compounds such as oxidized proteins (Soubannier et al.2012b).
Although the above scenarios are somehow appealing, one should bear in mind that we have presently no examples of an archaeon engulfing a bacterium with its nanopods or of an intracellular bacteria thriving in an archaeon, as postulated by the above scenarios. The hypothesis that an archaeon was transformed into a eukaryote during eukaryogenesis was recently boosted by the discovery of the putative Asgard superphylum and phylogenetic analyses supporting their sisterhood with eukaryotes (Spang et al.2015; Zaremba-Niedzwiedzka et al.2017). However, these phylogenetic analyses have been questioned due to the inclusion of many fast-evolving species in the species dataset used for these analyses (Da Cunha et al.2017; for controversies surrounding these data, see Da Cunha et al.2018; Spang et al.2018). Universal trees of life based on the longer universal proteins, especially RNA polymerase large subunits, strongly support the classical Woese tree of life (Da Cunha et al.2017). If the bacterial ancestor of mitochondria did not invade an archaeon but a proto-eukaryote, EVs produced by ancient bacteria living as endosymbionts in proto-eukaryotes (including the mitochondrial ancestor) could still have played an important role in eukaryogenesis by interacting with the intracellular EVs network of proto-eukaryotes.
CONCLUDING REMARKS
Research on EVs from cells of all three domains of life has seen an explosion of interest in the last decade. Increasing evidence indicates that these EVs impact intra- and intercellular communication by the transfer of a myriad of biomolecules and the exchange of genetic information. Despite the increased research interest, the accumulating data on EVs are increasingly complex, leaving many fundamental questions unanswered, such as the mechanisms of cargo sorting and biogenesis. The discovery of nanotubes associated with EVs in all three domains of life is highly intriguing, suggesting that the formation of tubular structures is a universal mechanism for cell-to-cell communication and for the transport of EVs between cells.
In Bacteria, vesiculation appears to be a specific and regulated process. Given the diversity and complexity of different bacterial envelopes, it is not surprising that the mechanisms involved in biogenesis are not yet fully understood. Indeed, it may be that there is no single mechanism for all bacteria. Nonetheless, understanding how diverse bacteria achieve a common outcome will benefit both the bacterial and EV fields in general. Diverse factors such as iron and oxygen availability; biofilm versus planktonic lifestyle; SOS response; exposure to antibiotics; host factors; and growth conditions affect the composition and quantity of EVs (reviewed in Orench-Rivera and Kuehn 2016). Future studies addressing how EVs are influenced by different environmental conditions will hopefully advance our current understanding.
A major problem in the eukaryotic field is the isolation and purification of different subsets of EVs from complex mixtures. Protocols currently used for purifying EVs usually result in isolating a mixture of different EVs with various origins (microvesicles, exosomes, etc.) along with other macromolecular complexes. Moreover, as different subsets of EVs contain many common markers, this poses a problem for EV typing. In order to standardize research between different laboratories, it has been suggested that future studies should try and analyze a minimum of three or more proteins expected to be present in the EVs studied using approaches such as western blots, flow cytometry (FACS) or proteomic analysis (Lötvall et al.2014). More recently, a knowledge base termed EV-TRACK was set up to encourage increased systematic reporting of EV biology and methodology (van Deun et al.2017). Such additional measures would reinforce conclusions regarding the functions of EVs and their roles.
Though slow to start, research on archaeal EVs is showing promising results in linking mechanisms of vesicle production between domains of life. The presence of eukaryotic-like mechanisms (e.g. ESCRT) in prokaryotic organisms is a major asset in identifying a unified mechanism of EV production. However, even in Archaea, it has become apparent that many mechanisms may be required to explain the diversity of EVs observed. A major future challenge in the field of EVs will certainly be to identify specific mechanisms involved in membrane fusion and vesicle biogenesis in Archaea with monolayer membranes, since models proposed up to now are based on the ‘classical’ bacterial and eukaryotic bilayer membranes. Regardless of the mechanism, it is clear that the local structure and organization of lipid membranes plays an important role in EV biogenesis across all three domains of life.
The connection between viruses and EVs also reveals important connections in EV production and function across the three domains of life. Whether viruses have hijacked EV machinery to their own benefit, or whether EVs sit at the origin of modern viruses, it is clear that these entities are intimately linked. There appears to exist a continuum between pure vesicles and pure virions, interspersed with vesicles containing nucleic acids, plasmidions, membrane-encapsulated viruses, viral vesicles, VPV, etc. No doubt future research into both viruses and EVs will provide mutual insights to help illuminate this fascinating relationship.
Acknowledgements
We apologize to authors whose work we have not cited or only cited indirectly owing to space constraints.
FUNDING
This work was supported by an ERC grant from the European Union's Seventh Framework Program (FP7/2007-2013) [ERC grant agreement n° 340440, Project EVOMOBIL to PF].
Conflict of interest. None declared.
REFERENCES
- Abels ER, Breakefield XO. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol 2016;36:301–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acehan D, Santarella-Mellwig R, Devos DP. A bacterial tubulovesicular network. J Cell Sci 2014;127:277–80. [DOI] [PubMed] [Google Scholar]
- Acosta JC, Banito A, Wuestefeld T et al.. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol 2013;15:978–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers SV, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol 2011;9:414–26. [DOI] [PubMed] [Google Scholar]
- Albuquerque PC, Nakayasu ES, Rodrigues ML et al.. Vesicular transport in Histoplasma capsulatum: an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell Microbiol 2008;10:1695–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Nedawi K, Meehan B, Micallef J et al.. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol 2008;10:619–24. [DOI] [PubMed] [Google Scholar]
- Altan-Bonnet N. Extracellular vesicles are the Trojan horses of viral infection. Curr Opin Microbiol 2016;32:77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altan-Bonnet N, Chen YH. Intercellular transmission of viral populations with vesicles. J Virol 2015;89:12242–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altindis E, Fu Y, Mekalanos JJ. Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proc Natl Acad Sci USA 2014;111:E1548–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Mihalik R, Soll DR. Ultrastructure and antigenicity of the unique cell wall pimple of the Candida opaque phenotype. J Bacteriol 1990;172:224–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade-Navarro MA, Sanchez-Pulido L, McBride HM. Mitochondrial vesicles: an ancient process providing new links to peroxisomes. Curr Opin Cell Biol 2009;21:560–7. [DOI] [PubMed] [Google Scholar]
- Arantes TS, Rodrigues RAL, dos Santos Silva LK et al.. The large Marseillevirus explores different entry pathways by forming giant infectious vesicles. J Virol 2016;90:5246–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atai NA, Balaj L, van Veen H et al.. Heparin blocks transfer of extracellular vesicles between donor and recipient cells. J Neurooncol 2013;115:343–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atayde VD, Suau HA, Townsend S et al.. Exosome secretion by the parasitic protozoan Leishmania within the sand fly midgut. Cell Rep 2015;13:957–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin-Smith GK, Tixeira R, Paone S et al.. A novel mechanism of generating extracellular vesicles during apoptosis via a beads-on-a-string membrane structure. Nat Commun 2015;6:7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austefjord MW, Gerdes HH, Wang X. Tunneling nanotubes: diversity in morphology and structure. Commun Integr Biol 2014;7:e27934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M. MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr Opin Cell Biol 2011;23:452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baidya AK, Bhattacharya S, Dubey GP et al.. Bacterial nanotubes: a conduit for intercellular molecular trade. Curr Opin Microbiol 2018;42:1–6. [DOI] [PubMed] [Google Scholar]
- Balaj L, Lessard R, Dai L et al.. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun 2011;2:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barák I, Muchová K. The role of lipid domains in bacterial cell processes. Int J Mol Sci 2013;14:4050–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batagov AO, Kurochkin IV. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3΄-untranslated regions. Biol Direct 2013;8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batagov AO, Kuznetsov VA, Kurochkin IV. Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles. BMC Genomics 2011;12:S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum DA, Baum B. An inside-out origin for the eukaryotic cell. BMC Biol 2014;12:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazylinski DA, Dean AJ, Williams TJ et al.. Chemolithoautotrophy in the marine, magnetotactic bacterial strains MV-1 and MV-2. Arch Microbiol 2004;182:373–87. [DOI] [PubMed] [Google Scholar]
- Bazylinski DA, Williams TJ, Lefèvre CT et al.. Magnetovibrio blakemorei gen. nov., sp. nov., a magnetotactic bacterium (Alphaproteobacteria: Rhodospirillaceae) isolated from a salt marsh. Int J Syst Evol Microbiol 2013;63:1824–33. [DOI] [PubMed] [Google Scholar]
- Bénard M, Schapman D, Lebon A et al.. Structural and functional analysis of tunneling nanotubes (TnTs) using gCW STED and gconfocal approaches. Biol Cell 2015;107:419–25. [DOI] [PubMed] [Google Scholar]
- Berleman JE, Allen S, Danielewicz MA et al.. The lethal cargo of Myxococcus xanthus outer membrane vesicles. Front Microbiol 2014;5:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol 1999;181:4725–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar S, Shinagawa K, Castellino FJ et al.. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood 2007;110:3234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicalho B, Holovati JL, Acker JP. Phospholipidomics reveals differences in glycerophosphoserine profiles of hypothermically stored red blood cells and microvesicles. Biochim Biophys Acta 2013;1828:317–26. [DOI] [PubMed] [Google Scholar]
- Bielska E, Sisquella MA, Aldeieg M et al.. Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii. Nat Commun 2018;9:1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, McDaniel LD, Breitbart M et al.. Membrane vesicles in sea water: heterogeneous DNA content and implications for viral abundance estimates. ISME J 2017;11:394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, Schubotz F, Roggensack SE et al.. Bacterial vesicles in marine ecosystems. Science 2014;10:183–6. [DOI] [PubMed] [Google Scholar]
- Bitto NJ, Chapman R, Pidot S et al.. Bacterial membrane vesicles transport their DNA cargo into host cells. Sci Rep 2017;7:7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitto NJ, Kaparakis-Liaskos M. The therapeutic benefit of Bacterial membrane vesicles. Int J Mol Sci 2017;18:1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilyy RO, Shkandina T, Tomin A et al.. Macrophages discriminate glycosylation patterns of apoptotic cell-derived microparticles. J Biol Chem 2012;287:496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DG, Work E. An extracellular glycolipid produced by Escherichia coli grown under lysine-limiting conditions. Biochem J 1965;96:567–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkiron C, Simonov D, Muthukaruppan A et al.. Uropathogenic Escherichia coli releases extracellular vesicles that are associated with RNA. PLoS One 2016;11:e0160440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blesa A, Berenguer J. Contribution of vesicle-protected extracellular DNA to horizontal gene transfer in Thermus spp. Int Microbiol 2015;18:177–87. [DOI] [PubMed] [Google Scholar]
- Bomberger JM, Maceachran DP, Coutermarsh BA et al.. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog 2009;5:e1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnington KE, Kuehn MJ. Protein selection and export via outer membrane vesicles. Biochim Biophys Acta 2014;1843:1612–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnington KE, Kuehn MJ. Outer membrane vesicle production facilitates LPS remodeling and outer membrane maintenance in salmonella during environmental transitions. mBio 2016;7:e01532–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulbitch AA. Deflection of a cell membrane under application of a local force. Phys Rev E 1998;57:2123–8. [Google Scholar]
- Browman DT, Hoegg MB, Robbins SM. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol 2007;17:394–402. [DOI] [PubMed] [Google Scholar]
- Brown L, Kessler A, Cabezas-Sanchez P et al.. Extracellular vesicles produced by the Gram-positive bacterium Bacillus subtilis are disrupted by the lipopeptide surfactin. Mol Microbiol 2014;93:183–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Wolf JM, Prados-Rosales R et al.. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Micro 2015;13:620–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PN, Yin H. Polymer-based purification of extracellular vesicles. Methods Mol Biol 2017;1660:91–103. [DOI] [PubMed] [Google Scholar]
- Bukong TN, Momen-Heravi F, Kodys K et al.. Exosomes from Hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog 2014;10:e1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Han Y, Ren H et al.. Extracellular vesicle-mediated transfer of donor genomic DNA to recipient cells is a novel mechanism for genetic influence between cells. J Mol Cell Biol 2013;5:227–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Wu G, Jose PA et al.. Functional transferred DNA within extracellular vesicles. Exp Cell Res 2016;349:179–83. [DOI] [PubMed] [Google Scholar]
- Cai Q, Qiao L, Wang M et al.. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018;360:1126–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cario A, Grossi V, Schaeffer P et al.. Membrane homeoviscous adaptation in the piezo-hyperthermophilic archaeon Thermococcus barophilus. Front Microbiol 2015;6:1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi Y, Dekker C. Dividing the Archaeal Way: the ancient Cdv cell-division machinery. Front Microbiol 2018;9:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celluzzi A, Masotti A. How our other genome controls our epi-genome. Trends Microbiol 2016;24:777–87. [DOI] [PubMed] [Google Scholar]
- Cestari I, Ansa-Addo E, Deolindo P et al.. Trypanosoma cruzi immune evasion mediated by host cell-derived microvesicles. J Immunol 2012;188:1942–52. [DOI] [PubMed] [Google Scholar]
- Chatterjee SN, Das J. Electron microscopic observations on the excretion of cell-wall material by Vibrio cholerae. J Gen Microbiol 1967;49:1–11. [DOI] [PubMed] [Google Scholar]
- Chazal N, Gerlier D. Virus entry, assembly, budding, and membrane rafts. Microbiol Mol Biol R 2003;67:226–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekeni FB, Elliott MR, Sandilos JK et al.. Pannexin 1 channels mediate “find–me” signal release and membrane permeability during apoptosis. Nature 2010;467:863–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Huang AC, Zhang W et al.. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018;560:382–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Du W, Hagemeijer MC et al.. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 2015;160:619–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DH, Kwon YM, Chiura HX et al.. Extracellular vesicles of the hyperthermophilic archaeon Thermococcus onnurineus NA1T. Appl Environ Microb 2015a;81:4591–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DS, Kim DK, Choi SJ et al.. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics 2011;11:3424–9. [DOI] [PubMed] [Google Scholar]
- Choi DS, Kim DK, Kim YK et al.. Proteomics of extracellular vesicles: exosomes and ectosomes. Mass Spectrom Rev 2015b;34:474–90. [DOI] [PubMed] [Google Scholar]
- Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol 2009;19:43–51. [DOI] [PubMed] [Google Scholar]
- Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 2014;30:255–89. [DOI] [PubMed] [Google Scholar]
- Coleman BM, Hill AF. Extracellular vesicles – their role in the packaging and spread of misfolded proteins associated with neurodegenerative diseases. Semin Cell Dev Biol 2015;40:89–96. [DOI] [PubMed] [Google Scholar]
- Cooke IR, Deserno M. Coupling between lipid shape and membrane curvature. Biophys J 2006;91:487–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescitelli R, Lässer C, Szabó TG et al.. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles 2013;2 DOI: 10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cunha V, Gaia M, Gadelle D et al.. Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genet 2017;13:e1006810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cunha V, Gaia M, Nasir A et al.. Asgard archaea do not close the debate about the universal tree of life topology. PLoS Genet 2018;14:e1007215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauros-Singorenko P, Blenkiron C, Phillips A et al.. The functional RNA cargo of bacterial membrane vesicles. FEMS Microbiol Lett 2018;365:5. [DOI] [PubMed] [Google Scholar]
- Dauros Singorenko P, Chang V, Whitcombe A et al.. Isolation of membrane vesicles from prokaryotes: a technical and biological comparison reveals heterogeneity. J Extracell Vesicles 2017;6:1324731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DM, Sowinski S. Membrane nanotubes: dynamic long-distance connections between animal cells. Nat Rev Mol Cell Biol 2008;6:431–6. [DOI] [PubMed] [Google Scholar]
- Deatherage BL, Cookson BT. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet under appreciated aspect of microbial life. Infect Immun 2012;80:1948–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage BL, Lara JC, Bergsbaken T et al.. Biogenesis of bacterial membrane vesicles. Mol Microbiol. 2009;72:1395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Torre Gomez C, Goreham RV, Bech Serra JJ et al.. “Exosomics”—A review of biophysics, biology and biochemistry of exosomes with a focus on human breast milk. Front Genet 2018;9:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Conde I, Shrimpton CN, Thiagarajan P et al.. Tissue-factor-bearing microvesicles arise from lipid rafts and fuse with activated platelets to initiate coagulation. Blood 2005;106:1604–11. [DOI] [PubMed] [Google Scholar]
- Demina TA, Pietilä MK, Svirskaitė J et al.. Archaeal Haloarcula californiae Icosahedral Virus 1 highlights conserved elements in icosahedral membrane-containing DNA viruses from extreme environments. mBio 2016;7:e00699–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deregibus MC, Figliolini F, D’antico S et al.. Charge-based precipitation of extracellular vesicles. Int J Mol Med 2016;38:1359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos S, Van Oudenhove L, Stremersch S et al.. The effect of imipenem and diffusible signaling factors on the secretion of outer membrane vesicles and associated Ax21 proteins in Stenotrophomonas maltophilia. Front Microbiol 2015;6:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos S, Van Putte W, Vitse J et al.. Membrane vesicle secretion and prophage induction in multidrug-resistant Stenotrophomonas maltophilia in response to ciprofloxacin stress. Environ Microbiol 2017;19:3930–7. [DOI] [PubMed] [Google Scholar]
- Devos DP, Ward NL.. Mind the PVCs. Environ Microbiol 2014;16:1217–21. [DOI] [PubMed] [Google Scholar]
- Dobro MJ, Oikonomou CM, Piper A et al.. Uncharacterized Bacterial structures revealed by electron cryotomography. J Bacteriol 2017;199:e00100–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues S, Nielsen KM. Membrane vesicles and horizontal gene transfer in prokaryotes. Curr Opin Microbiol 2017;38:16–21. [DOI] [PubMed] [Google Scholar]
- Dorward DW, Garon CF. DNA is packaged within membrane-derived vesicles of gram-negative but not gram-positive bacteria. Appl Environ Microb 1990;56:1960–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Clancy JW. Tumor-derived microvesicles: shedding light on novel microenvironment modulators and prospective cancer biomarkers. Gene Dev 2012;26:1287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey GP, Ben-Yehuda S. Intercellular nanotubes mediate bacterial communication. Cell 2011;144:590–600. [DOI] [PubMed] [Google Scholar]
- Dubey GP, Malli Mohan GB, Dubrovsky A et al.. Architecture and characteristics of bacterial nanotubes. Dev Cell 2016;36:453–61. [DOI] [PubMed] [Google Scholar]
- Ducret A, Fleuchot B, Bergam P et al.. Direct live imaging of cell–cell protein transfer by transient outer membrane fusion in Myxococcus xanthus. Elife 2013;2:e00868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger I, Schmitz-Esser S, Schmid M et al.. Symbiont-driven sulfur crystal formation in a thiotrophic symbiosis from deep-sea hydrocarbon seeps. Environ Microbiol Rep 2014;6:364–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Andaloussi S, Mäger I, Breakefield XO et al.. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov 2013;12:347–57. [DOI] [PubMed] [Google Scholar]
- Elhenawy W, Bording-Jorgensen M, Valguarnera E et al.. LPS remodeling triggers formation of outer membrane vesicles in Salmonella. mBio 2016;7:e00940–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhenawy W, Debelyy MO, Feldman MF. Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio 2014;5:e00909–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellen AF, Albers SV, Huibers W et al.. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles 2009;13:67–79. [DOI] [PubMed] [Google Scholar]
- Ellen AF, Rohulya OV, Fusetti F et al.. The sulfolobicin genes of Sulfolobus acidocaldarius encode novel antimicrobial proteins. J Bacteriol 2011;193:4380–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellen AF, Zolghadr B, Driessen AMJ et al.. Shaping the archaeal cell envelope. Archaea 2010;2010:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol R 2010;74:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann S, Scheele U, Garrett RA. AAA ATPase p529 of Acidianus two-tailed virus ATV and host receptor recognition. Virology 2011;421:61–66. [DOI] [PubMed] [Google Scholar]
- Erdmann S, Tschitschko B, Zhong L et al.. A plasmid from an Antarctic haloarchaeon uses specialized membrane vesicles to disseminate and infect plasmid-free cells. Nat Microbiol 2017;2:1446–55. [DOI] [PubMed] [Google Scholar]
- Ettema TJ, Lindås A, Bernander R. An actin-based cytoskeleton in archaea. Mol Microbiol 2011;80:1052–61. [DOI] [PubMed] [Google Scholar]
- Fagan RP, Fairweather NF. Biogenesis and functions of bacterial S-layers. Nat Rev Microbiol 2014;12:211–22. [DOI] [PubMed] [Google Scholar]
- Farsad K, De Camilli P. Mechanisms of membrane deformation. Curr Opin Cell Biol 2003;15:372–81. [DOI] [PubMed] [Google Scholar]
- Feng Z, Hensley L, McKnight KL et al.. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 2013;7445:367–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florez C, Raab JE, Cooke AC et al.. Membrane distribution of the Pseudomonas quinolone signal modulates outer membrane vesicle production in Pseudomonas aeruginosa. mBio 2017;8:e01034–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P. The virocell concept and environmental microbiology. ISME J 2013;7:233–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, Da Cunha V, Catchpole R. Plasmid vesicles mimicking virions. Nat Microbiol 2017;2:1340–1. [DOI] [PubMed] [Google Scholar]
- Forterre P, Gribaldo S. The origin of modern terrestrial life. HFSP J 2007;1:156–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, Krupovic M. The origin of virions and virocells: the escape hypothesis revisited. In: Witzany G. (ed). Viruses: Essential Agents of Life. Netherlands: Springer Science and Business Media Dordrecht, 2012: 43–60. [Google Scholar]
- Forterre P, Soler N, Krupovic M et al.. Fake virus particles generated by fluorescence microscopy. Trends Microbiol 2013;21:1–5. [DOI] [PubMed] [Google Scholar]
- Franz B, Lichtenberg H, Hormes J et al.. Utilization of solid 'elemental' sulfur by the phototrophic purple sulfur bacterium Allochromatium vinosum: a sulfur K-edge X-ray absorption spectroscopy study. Microbiology 2007;153:1268–74. [DOI] [PubMed] [Google Scholar]
- Friend C, Marovitz W, Henie G et al.. Observations on cell lines derived from a patient with Hodgkin's disease. Cancer Res 1978;38:2581–91. [PubMed] [Google Scholar]
- Fuerst JA, Sagulenko E. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 2011;9:403–13. [DOI] [PubMed] [Google Scholar]
- Gardiner C, Vizio DD, Sahoo S et al.. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles 2016;5:32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti S, Bruno S, Deregibus MC et al.. Microvesicles derived from human adult mesenchymal stem cells protect against ischaemia-reperfusion-induced acute and chronic kidney injury. Nephrol Dial Transpl 2011;26:1474–83. [DOI] [PubMed] [Google Scholar]
- Gaudin M, Gauliard E, Schouten S et al.. Hyperthermophilic archaea produce membrane vesicles that can transfer DNA. Environ Microbiol Rep 2013;5:109–16. [DOI] [PubMed] [Google Scholar]
- Gaudin M, Krupovic M, Marguet E et al.. Extracellular membrane vesicles harbouring viral genomes. Environ Microbiol 2014;16:1167–75. [DOI] [PubMed] [Google Scholar]
- Geslin C, Le Romancer M, Erauso G et al.. PAV1, the first virus-like particle isolated from a hyperthermophilic Euryarchaeote, “Pyrococcus abyssi.” J Bacteriol 2003;185:3888–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal A, Upadhyaya BB, Fritz JV et al.. The extracellular RNA complement of Escherichia coli. Microbiol Open 2015;4:252–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S, Forterre P. Origin of life: LUCA and extracellular membrane vesicles. Int J Astrobiol 2016;15;7–15. [Google Scholar]
- Gorlas A, Koonin EV, Bienvenu N et al.. TPV1, the first virus isolated from the hyperthermophilic genus Thermococcus. Environ Microbiol 2012;14:503–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlas A, Marguet E, Gill S et al.. Sulfur vesicles from Thermococcales: a possible role in sulfur detoxifying mechanisms. Biochimie 2015;118:356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottshall EY, Seebart C, Gatlin JC et al.. Spatially segregated transcription and translation in cells of the endomembrane-containing bacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA 2014;111:11067–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Garg SG, Martin WF. Bacterial vesicle secretion and the evolutionary origin of the eukaryotic endomembrane system. Trends Microbiol 2016;24:525–34. [DOI] [PubMed] [Google Scholar]
- Gould SJ, Booth AM, Hildreth JE.. The Trojan exosome hypothesis. Proc Natl Acad Sci USA 2003;100:10592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Raposo G. As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles 2013;15:2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graner MW, Alzate O, Dechkovskaia AM et al.. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J 2009;23:1541–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granoff DM. Review of meningococcal group B vaccines. Clin Infect Dis 2010;50:S54–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening DW, Xu R, Gopal SK et al.. Proteomic insights into extracellular vesicle biology - defining exosomes and shed microvesicles. Expert Rev Proteomics 2017;14:69–95. [DOI] [PubMed] [Google Scholar]
- Gross JC, Chaudhary V, Bartscherer K et al.. Active Wnt proteins are secreted on exosomes. Nat Cell Biol 2012;14:1036–45. [DOI] [PubMed] [Google Scholar]
- Guo W, Gao Y, Li N et al.. Exosomes: new players in cancer. Oncol Rep 2017;38:665–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Mandujano A, Hernández-Cortez C, Ibarra JA et al.. The outer membrane vesicles: secretion system type zero. Traffic 2017;18:425–32. [DOI] [PubMed] [Google Scholar]
- Guescini M, Genedani S, Stocchi V et al.. Astrocytes and glioblastoma cells release exosomes carrying mtDNA. J Neural Transm 2010;117:1–4. [DOI] [PubMed] [Google Scholar]
- Gurung M, Moon DC, Choi CW et al.. Staphylococcus aureus produces membrane-derived vesicles that induce host cell death. PLoS One 2011;6:e27958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- György B, Szabó TG, Pásztói M et al.. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci 2011;68:2667–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol 1983;97:329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A, Muth C, Dabrowski O et al.. Exosomes and the prion protein: more than one truth. Front Neurosci 2017;11:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurat MF, Aduse-Opoku J, Rangarajan M et al.. Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem 2011;286:1269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurat MF, Elhenawy W, Feldman MF. Prokaryotic membrane vesicles: new insights on biogenesis and biological roles. Biol Chem 2015;396:95–109. [DOI] [PubMed] [Google Scholar]
- Hayashi JI, Hamada N, Kuramitsu HK. The autolysin of Porphyromonas gingivalis is involved in outer membrane vesicle release. FEMS Microbiol Lett 2002;216:217–22. [DOI] [PubMed] [Google Scholar]
- Heijnen HF, Schiel AE, Fijnheer R et al.. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood 1999;94:3791–9. [PubMed] [Google Scholar]
- Heimerl T, Flechsler J, Pickl C et al.. A complex endomembrane system in the archaeon Ignicoccus hospitalis tapped by Nanoarchaeum equitans. Front Microbiol 2017;8:1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci 2018;75:193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey CA, Kuhn KA, Donermeyer DL et al.. Colitogenic Bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase‐dependent manner via outer membrane vesicles. Cell Host Microbe 2015;17:672–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderhofer M, Walker CA, Friemel A et al.. Evolution of prokaryotic SPFH proteins. BMC Evol Biol 2009;9:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra D, Van der Laan J, De Leij L et al.. Release of outer membrane fragments from normally growing Escherichia coli. Biochim Biophys Acta 1976;455:889–99. [DOI] [PubMed] [Google Scholar]
- Holst J, Martin D, Arnold R et al.. Properties and clinical performance of vaccines containing outer membrane vesicles from Neisseria meningitidis. Vaccine 2009;27:B3–12. [DOI] [PubMed] [Google Scholar]
- Hristov M, Erl W, Linder S et al.. Apoptotic bodies from endothelial cells enhance the number and initiate the differentiation of human endothelial progenitor cells in vitro. Blood 2004;104:2761–6. [DOI] [PubMed] [Google Scholar]
- Hsu C, Morohashi Y, Yoshimura S et al.. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A–C. J Cell Biol 2010;189:223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H, Hohn MJ, Rachel R et al.. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 2002;417:63–67. [DOI] [PubMed] [Google Scholar]
- Inal JM, Kosgodage U, Azam S et al.. Blood/plasma secretome and microvesicles. Biochim Biophys Acta 2013;1834:2317–25. [DOI] [PubMed] [Google Scholar]
- Izquierdo-Useros N, Naranjo-Gómez M, Archer J et al.. Capture and transfer of HIV-1 particles by mature dendritic cells converges with the exosome-dissemination pathway. Blood 2009;113:2732–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo-Useros N, Puertas MC, Borràs FE et al.. Exosomes and retroviruses: the chicken or the egg? Cell Microbiol 2011;13:10–17. [DOI] [PubMed] [Google Scholar]
- Jalasvuori M, Bamford JK. Structural co-evolution of viruses and cells in the primordial world. Orig Life Evol Biosph 2008;38:165–81. [DOI] [PubMed] [Google Scholar]
- Jan AT. Outer membrane vesicles (OMVs) of Gram-negative bacteria: a perspective update. Front Microbiol 2017;8:1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang KS, Sweredoski MJ, Graham RLJ et al.. Comprehensive proteomic profiling of outer membrane vesicles from Campylobacter jejuni. J Proteomics 2014;98:90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jekely G. Did the last common ancestor have a biological membrane? Biol Direct 2006;1:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Kong Q, Roland KL et al.. Membrane vesicles of Clostridium perfringens type A strains induce innate and adaptive immunity. Int J Med Microbiol 2014;304:431–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RM, Adam M, Hammond JR et al.. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem 1987;262:9412–20. [PubMed] [Google Scholar]
- Jurkoshek KS, Wang Y, Athman JJ et al.. Interspecies communication between pathogens and immune cells via bacterial membrane vesicles. Front Cell Dev Biol 2016;4:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol 1995;177:3998–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J Bacteriol 1996;178:2767–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn ME, Barany F, Smith HO. Transformasomes: specialized membranous structures that protect DNA during Haemophilus transformation. Proc Natl Acad Sci USA 1983;80:6927–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra H, Drummen GPC, Mathivanan S. Focus on extracellular vesicles: introducing the next small big thing. Int J Mol Sci 2016;17:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra H, Simpson RJ, Ji H et al.. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol 2012;10:e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaparakis-Liaskos M, Ferrero RL. Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol 2015;15:375–87. [DOI] [PubMed] [Google Scholar]
- Kasson P, DiMaio F, Yu X et al.. Model for a novel membrane envelope in a filamentous hyperthermophilic virus. Elife 2017;6:e26268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller KE, Bradley JM, Sun YY et al.. Tunneling nanotubes are novel cellular structures that communicate signals between trabecular meshwork cells. Invest Ophthalmol Vis Sci 2017;58:5298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren R, Mayzel B, Lavy A et al.. Sponge-associated bacteria mineralize arsenic and barium on intracellular vesicles. Nat Commun 2017;8:14393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JFR, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wideranging implications in tissue kinetics. Br J Cancer 1972;26:239–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieselbach T, Zijnge V, Granström E et al.. Proteomics of Aggregatibacter actinomycetemcomitans outer membrane vesicles. PLoS One 2015;10:e0138591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DK, Kang B, Kim OY et al.. EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J Extracell Vesicles 2013;2 DOI: 10.3402/jev.v2i0.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DK, Lee J, Kim SR et al.. EVpedia: a community web portal for extracellular vesicles research. Bioinformatics 2015;31:933–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Yip KW, Spence T et al.. MicroRNAs in extracellular vesicles: potential cancer biomarkers. J Hum Genet 2017;62:67–74. [DOI] [PubMed] [Google Scholar]
- Kish A, Miot J, Lombard C et al.. Preservation of archaeal surface layer structure during mineralization. Sci Rep 2016;6:26152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klieve AV, Yokoyama MT, Forster RJ et al.. Naturally occurring DNA transfer system associated with membrane vesicles in cellulolytic Ruminococcus spp. of ruminal origin. Appl Environ Microb 2005;71:4248–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingl A. S-layer and cytoplasmic membrane - exceptions from the typical archaeal cell wall with a focus on double membranes. Front Microbiol 2014;5:624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppen K, Hampton TH, Jarek M et al.. A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog 2016;12:e1005672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koistinen V, Härkönen K, Kärnä R et al.. EMT induced by EGF and wounding activates hyaluronan synthesis machinery and EV shedding in rat primary mesothelial cells. Matrix Biol 2017;63:38–54. [DOI] [PubMed] [Google Scholar]
- Koistinen V, Kärnä R, Koistinen A et al.. Cell protrusions induced by hyaluronan synthase 3 (HAS3) resemble mesothelial microvilli and share cytoskeletal features of filopodia. Exp Cell Res 2015;337:179–91. [DOI] [PubMed] [Google Scholar]
- Koles K, Nunnari J, Korkut C et al.. Mechanism of evenness interrupted (Evi)-exosome release at synaptic boutons. J Biol Chem 2012;287:16820–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning RI, de Breij A, Oostergetel GT et al.. Cryo-electron tomography analysis of membrane vesicles from Acinetobacter baumannii ATCC19606T. Res Microbiol 2013;164:397–405. [DOI] [PubMed] [Google Scholar]
- Kosaka N, Iguchi H, Yoshioka Y et al.. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 2010;285:17442–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossinova OA, Gopanenko AV, Tamkovich SN et al.. Cytosolic YB-1 and NSUN2 are the only proteins recognizing specific motifs present in mRNAs enriched in exosomes. Biochim Biophys Acta 2017;1865:664–73. [DOI] [PubMed] [Google Scholar]
- Kouwaki T, Okamoto M, Tsukamoto H et al.. Extracellular vesicles deliver host and virus RNA and regulate innate immune response. Int J Mol Sci 2017;18:666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal EJK, Ter-Ovanesyan D, Regev A et al.. Extracellular vesicle isolation and analysis by western blotting. Methods Mol Biol 2017;1660:143–52. [DOI] [PubMed] [Google Scholar]
- Kowal J, Arras G, Colombo M et al.. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA 2016;113:E968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol 2014;29:116–25. [DOI] [PubMed] [Google Scholar]
- Kreger BT, Dougherty AL, Greene KS et al.. Microvesicle cargo and function changes upon induction of cellular transformation. J Biol Chem 2016;291:19774–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovič M, Bamford DH. Order to the viral universe. J Virol 2010;84:12476–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Temporal order of evolution of DNA replication systems inferred by comparison of cellular and viral DNA polymerases. Biol Direct 2006;1:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 2010;64:163–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp AJ, Sun B, Ai T et al.. Genome-wide assessment of outer membrane vesicle production in Escherichia coli. PLoS One 2015;10:e0139200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küper U, Meyer C, Müller V et al.. Energized outer membrane and spatial separation of metabolic processes in the hyperthermophilic Archaeon Ignicoccus hospitalis. Proc Natl Acad Sci USA 2010;107:3152–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapatsina L, Jira JA, Smith E et al.. Regulation of ASIC channels by a stomatin/STOML3 complex located in a mobile vesicle pool in sensory neurons. Open Biology 2012;2:120096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulagnier K, Grand D, Dujardin A et al.. PLD2 is enriched on exosomes and its activity is correlated to the release of exosomes. FEBS Lett 2004;572:11–14. [DOI] [PubMed] [Google Scholar]
- Lavialle F, Deshayes S, Gonnet F et al.. Nanovesicles released by Dictyostelium cells: a potential carrier for drug delivery. Int J Pharm 2009;380:206–15. [DOI] [PubMed] [Google Scholar]
- Lee EY, Choi DS, Kim KP et al.. Proteomics in gram-negative bacterial outer membrane vesicles. Mass Spectrom Rev 2008;27:535–55. [DOI] [PubMed] [Google Scholar]
- Lee EY, Choi DY, Kim DK et al.. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 2009;9:5425–36. [DOI] [PubMed] [Google Scholar]
- Lee J, Kim OY, Gho Y. Proteomic profiling of Gram-negative bacterial outer membrane vesicles: current perspectives. Prot Clin Appl 2016;10:897–909. [DOI] [PubMed] [Google Scholar]
- Lee J, Lee EY, Kim SH et al.. Staphylococcus aureus extracellular vesicles carry biologically active β-Lactamase. Antimicrob Agents Ch 2013;57:2589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Lee EJ, Lee JH et al.. Klebsiella pneumoniae secretes outer membrane vesicles that induce the innate immune response. FEMS Microbiol Lett 2012;331:17–24. [DOI] [PubMed] [Google Scholar]
- Lee JH, Hsieh CF, Liu HW et al.. Lipid raft–associated stomatin enhances cell fusion. FASEB J 2017;31:47–59. [DOI] [PubMed] [Google Scholar]
- Lee TH, D’Asti E, Magnus N et al.. Microvesicles as mediators of intercellular communication in cancer-the emerging science of cellular ‘debris’. Semin Immunopathol 2011;33:455–67. [DOI] [PubMed] [Google Scholar]
- Liao S, Klein MI, Heim KP et al.. Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J Bacteriol 2014;196:2355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindås AC, Karlsson EA, Lindgren MT et al.. A unique cell division machinery in the Archaea. Proc Natl Acad Sci USA 2008;105:18942–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindås AC, Valegård K, Ettema TJG. Archaeal actin-family filament systems. In: Löwe J, Amos L. (eds) Prokaryotic Cytoskeletons. Subcellular Biochemistry, vol 84. Springer, Cham, 2017, 379–92. [DOI] [PubMed] [Google Scholar]
- Lindsay MR, Webb RI, Strous M et al.. Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell. Arch Microbiol 2001;175:413–29. [DOI] [PubMed] [Google Scholar]
- Liu S, Hossinger A, Göbbels S et al.. Prions on the run: how extracellular vesicles serve as delivery vehicles for self-templating protein aggregates. Prion 2017;11:98–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente A, Skotland T, Sylvanne T et al.. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim Biophys Acta 2013;1831:1302–9. [DOI] [PubMed] [Google Scholar]
- Loeb MR, Kilner J. Release of a special fraction of the outer membrane from both growing and phage T4-infected Escherichia coli B. Biochim Biophys Acta 1978;514:117–27. [DOI] [PubMed] [Google Scholar]
- Lombard J. The multiple evolutionary origins of the eukaryotic N-glycosylation pathway. Biol Direct 2016;11:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longatti A, Boyd B, Chisari FV. Virion-independent transfer of replication-competent Hepatitis C virus RNA between permissive cells. J Virol 2015;89:2956–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonhienne TG, Sagulenko E, Webb RI et al.. Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA 2010;107:12883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lötvall J, Hill AF, Hochberg F et al.. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 2014;3 DOI: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou E, Fujisawa S, Barlas A et al.. Tunneling nanotubes: a new paradigm for studying intercellular communication and therapeutics in cancer. Commun Integr Biol 2012;5:399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li Y, Peng J et al.. Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res 2015;25:24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas SLN, Breakefield XO, Weaver AM. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol 2017;27:172–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBroom AJ, Johnson AP, Vemulapalli S. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J Bacteriol 2006;188:5385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol 2007;63:545–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaig WD, Koller A, Thanassi DG. Production of outer membrane vesicles and outer membrane tubes by Francisella novicida. J Bacteriol 2013;195:1120–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald IA, Kuehn MJ. Offense and defense: microbial membrane vesicles play both ways. Res Microbiol 2012;163:607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald IA, Kuehn MJ. Stress-induced outer membrane vesicle production by Pseudomonas aeruginosa. J Bacteriol 2013;195:2971–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 2005;438:590–6. [DOI] [PubMed] [Google Scholar]
- Madison MN, Okeoma CM. Exosomes: implications in HIV-1 pathogenesis. Viruses 2015;7:4093–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Yutin N, Bell SD et al.. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat Rev Microbiol 2010;8:731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AJ, Kuehn MJ. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol 2011;1:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel PY, Hoang AN, Goldowitz I et al.. Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host Microbe 2013;13:521–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcilla A, Martin-Jaular L, Trelis M et al.. Extracellular vesicles in parasitic diseases. J Extracell Vesicles 2014;3:25040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcilla A, Trelis M, Cortés A et al.. Extracellular vesicles from parasitic helminths contain specific excretory/secretory proteins and are internalized in intestinal host cells. PLoS One 2012;7:e45974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maredia R, Devineni N, Lentz P et al.. Vesiculation from Pseudomonas aeruginosa under SOS. Scientific World J 2012;2012:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguet E, Gaudin M, Gauliard E et al.. Membrane vesicles, nanopods and/or nanotubes produced by hyperthermophilic archaea of the genus Thermococcus. Biochm Soc Trans 2013;41:436–42. [DOI] [PubMed] [Google Scholar]
- Marsh M, Helenius A. Virus entry: open sesame. Cell 2006;124:729–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsollier L, Brodin P, Jackson M et al.. Impact of Mycobacterium ulcerans biofilm on transmissibility to ecological niches and Buruli ulcer pathogenesis. PLoS Pathog 2007;4:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti M, Johnson PJ. Emerging roles for extracellular vesicles in parasitic infections. Curr Opin Microbiol 2016;32:66–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Jaular L, Nakayasu ES, Ferrer M et al.. Exosomes from Plasmodium yoelii-infected reticulocytes protect mice from lethal infections. PLoS One 2011;6:e26588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren LM, Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol Microbiol 2006;61:839–46. [DOI] [PubMed] [Google Scholar]
- Mateescu B, Kowal EJ, van Balkom BW et al.. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J Extracell Vesicles 2017;6:1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S, Lim JW, Tauro BJ et al.. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics 2010;9:197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S, Simpson RJ. ExoCarta: a compendium of exosomal proteins and RNA. Proteomics 2009;9:4997–5000. [DOI] [PubMed] [Google Scholar]
- Meador TB, Gagen EJ, Loscar ME et al.. Thermococcus kodakarensis modulates its polar membrane lipids and elemental composition according to growth stage and phosphate availability. Front Microbiol 2014;5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes DG, Raab-Traub N. Microvesicles and viral infection. J Virol 2011;85:12844–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milasan A, Tessandier N, Tan S et al.. Extracellular vesicles are present in mouse lymph and their level differs in atherosclerosis. J Extracell Vesicles 2016;5 DOI: 10.3402/jev.v5.31427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol 2015;40:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol 2004;20:395–425. [DOI] [PubMed] [Google Scholar]
- Mulcahy LA, Pink RC, Carter DRF. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles 2014;3 DOI: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraca M, Putignani L, Fierabracci A et al.. Gut microbiota-derived outer membrane vesicles: under-recognized major players in health and disease? Discov Med 2015;19:343–8. [PubMed] [Google Scholar]
- Muralidharan-Chari V, Clancy J, Plou C et al.. ARF6-Regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr Biol 2009;19:1875–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan-Chari V, Clancy JW, Sedgwick A et al.. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci 2010;123:1603–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Park AJ, Hao Y et al.. Influence of O polysaccharides on biofilm development and outer membrane vesicle biogenesis in Pseudomonas aeruginosa PAO1. J Bacteriol 2014;196:1306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Hu R, Oh RS et al.. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci USA 2012;109:4146–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y, Yoshioka Y, Yamamoto Y et al.. How cancer cells dictate their microenvironment: present roles of extracellular vesicles. Cell Mol Life Sci 2017;74:697–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao R, Hasegawa H, Ochiai K et al.. Outer membrane vesicles of Porphyromonas gingivalis elicit a mucosal immune response. PLoS One 2011;6:e26163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näther DJ, Rachel R. The outer membrane of the hyperthermophilic archaeon Ignicoccus: dynamics, ultrastructure and composition. Biochem Soc Trans 2004;32:199–203. [DOI] [PubMed] [Google Scholar]
- Nawaz M, Fatima F. Extracellular vesicles, tunneling nanotubes, and cellular interplay: synergies and missing links. Front Mol Biosci 2017;4:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte-'t Hoen ENM, Cremer T, Gallo RC et al.. Extracellular vesicles and viruses: Are they close relatives? Proc Natl Acad Sci USA 2016;113:9155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosanchuk JD, Nimrichter L, Casadevall A et al.. A role for vesicular transport of macromolecules across cell walls in fungal pathogenesis. Commun Integr Biol 2008;1:37–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 1979;76:1858–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue EJ, Krachler AM. Mechanisms of outer membrane vesicle entry into host cells. Cell Microbiol 2016;18:1508–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue EJ, Sirisaengtaksin N, Browning DF et al.. Lipopolysaccharide structure impacts the entry kinetics of bacterial outer membrane vesicles into host cells. PLoS Pathog 2017;13:e1006760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Ishikawa A, Kuroda M. Roles of exosomes and microvesicles in disease pathogenesis. Adv Drug Deliv Rev 2013;65:398–401. [DOI] [PubMed] [Google Scholar]
- Ojima Y, Mohanadas T, Kitamura K et al.. Deletion of degQ gene enhances outer membrane vesicle production of Shewanella oneidensis cells. Arch Microbiol 2017;199:415–23. [DOI] [PubMed] [Google Scholar]
- Olaya-Abril A, Prados-Rosales R, McConnell MJ et al.. Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumoniae. J Proteomics 2014;106:46–60. [DOI] [PubMed] [Google Scholar]
- Oliveira DL, Nakayasu ES, Joffe LS et al.. Biogenesis of extracellular vesicles in yeast: many questions with few answers. Commun Integr Biol 2010a;3:533–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DL, Nakayasu ES, Joffe LS et al.. Characterization of yeast extracellular vesicles: evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS One 2010b;5:e11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orench-Rivera N, Kuehn M. Environmentally controlled bacterial vesicle-mediated export. Cell Microbiol 2016;18:1525–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteikoetxea X, Balogh A, Szabó-Taylor K et al.. Improved characterization of EV preparations based on protein to lipid ratio and lipid properties. PLoS One. 2015;10:e0121184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski M, Carmo NB, Krumeich S et al.. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 2010;12:19–30. [DOI] [PubMed] [Google Scholar]
- Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell 1983;33:967–78. [DOI] [PubMed] [Google Scholar]
- Pande S, Shitut S, Freund L et al.. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat Commun 2015;6:6238. [DOI] [PubMed] [Google Scholar]
- Pathirana RD, Kaparakis‐Liaskos M. Bacterial membrane vesicles: biogenesis, immune regulation and pathogenesis. Cell Microbiol 2016;18:1518–24. [DOI] [PubMed] [Google Scholar]
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA et al.. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA 2010;107:6328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Alečković M, Lavotshkin S et al.. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012;18:883–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelchen-Matthews A, Raposo G, Marsh M. Endosomes, exosomes and Trojan viruses. Trends Microbiol 2004;12:310–6. [DOI] [PubMed] [Google Scholar]
- Penfornis P, Vallabhaneni KC, Whitt J et al.. Extracellular vesicles as carriers of microRNA, proteins and lipids in tumor microenvironment. Int J Cancer 2016;138:14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres da Silva R, Puccia R, Rodrigues ML et al.. Extracellular vesicle-mediated export of fungal RNA. Sci Rep 2015;5:7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereto J, Lopez-Garcia P, Moreira D. Ancestral lipid biosynthesis and early membrane evolution. Trends Biochem Sci 2004;29:469–77. [DOI] [PubMed] [Google Scholar]
- Pérez-Cruz C, Carrión O, Delgado L et al.. New type of outer membrane vesicle produced by the Gram-negative bacterium Shewanella vesiculosa M7T: implications for DNA content. Appl Environ Microb 2013;79:1874–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cruz C, Delgado L, López-Iglesias C et al.. Outer-inner membrane vesicles naturally secreted by gram-negative pathogenic bacteria. PLoS One 2015;12:e0116896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Vidakovics MLA, Jendholm J, Mörgelin M et al.. B Cell activation by outer membrane vesicles-a novel virulence mechanism. PLoS Pathog 2010;6:e1000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirbadian S, Barchinger SE, Leung KM et al.. Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc Natl Acad Sci USA 2014;111:12883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon IKH, Lucas CD, Rossi AG et al.. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol 2014;14:166–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope SM, Lasser C. Toxoplasma gondii infection of fibroblasts causes the production of exosome-like vesicles containing a unique array of mRNA and miRNA transcripts compared to serum starvation. J Extracell Vesicles 2013;2:22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prados-Rosales R, Baena A, Martinez LR et al.. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J Clin Invest 2011;121:1471–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prados-Rosales R, Brown L, Casadevall A et al.. Isolation and identification of membrane vesicle-associated proteins in Gram-positive bacteria and mycobacteria. Methods X 2014;1:124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prange A, Engelhardt H, Truper HG et al.. The role of the sulfur globule proteins of Allochromatium vinosum: mutagenesis of the sulfur globule protein genes and expression studies by real-time RT-PCR. Arch Microbiol 2004;182:165–74. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Holz I, Stieger E et al.. Sulfolobicins, specific proteinaceous toxins produced by strains of the extremely thermophilic archaeal genus Sulfolobus. J Bacteriol 2000;182:2985–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NL, Goyette-Desjardins G, Nothaft H et al.. Glycoengineered outer membrane vesicles: a novel platform for bacterial vaccines. Sci Rep 2016;6:24931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quemin ERJ, Chlanda P, Sachse M et al.. Eukaryotic-like virus budding in Archaea. mBio 2016;7:e01439–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Coyne MJ, Comstock LE. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol 2014;24:40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D, Forterre P. Redefining viruses: lessons from Mimivirus. Nat Rev Microbiol 2008;6:315–9. [DOI] [PubMed] [Google Scholar]
- Raposo G, Nijman HW, Stoorvogel W et al.. B lymphocytes secrete antigen-presenting vesicles. J Exp Med 1996;183:1161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013;200:373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashed MH, Bayraktar E, Helal GK et al.. Exosomes: from garbage bins to promising therapeutic targets. Int J Mol Sci 2017;18:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak J, Miekus K, Kucia M et al.. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006;20:847–56. [DOI] [PubMed] [Google Scholar]
- Record M, Carayon K, Poirot M et al.. Exosomes as new vesicular lipid transporters involved in cell–cell communication and various pathophysiologies. Biochim Biophys Acta 2014;1841:108–20. [DOI] [PubMed] [Google Scholar]
- Record M, Silvente-Poirot S, Poirot M et al.. Extracellular vesicles: lipids as key components of their biogenesis and functions. J Lipid Res 2018;59:1316–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev-Rudzki N, Wilson DW, Carvalho TG et al.. Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell 2013;153:1120–33. [DOI] [PubMed] [Google Scholar]
- Relini A, Cassinadri D, Fan Q et al.. Effect of physical constraints on the mechanisms of membrane fusion: bolaform lipid vesicles as model systems. Biophys J 1996;71:1789–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remis JP, Wei D, Gorur A et al.. Bacterial social networks: structure and composition of Myxococcus xanthus outer membrane vesicle chains. Environ Microbiol 2014;16:598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renelli M, Matias V, Lo RY et al.. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 2004;150:2161–9. [DOI] [PubMed] [Google Scholar]
- Resch U, Tsatsaronis JA, Le Rhun A et al.. Two-component regulatory system impacts extracellular membrane-derived vesicle production in group A Streptococcus. mBio 2016;7:e00207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewatkar PV, Parton RG, Parekh HS et al.. Are caveolae a cellular entry route for non-viral therapeutic delivery systems? Adv Drug Deliv Rev 2015;91:92–108. [DOI] [PubMed] [Google Scholar]
- Reyes-Robles T, Dillard RS, Cairns LS et al.. Vibrio cholerae outer membrane vesicles inhibit bacteriophage infection. J Bacteriol 2018. DOI: 10.1128/JB.00792-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach AL, Flores GE. Electron microscopy encounters with unusual thermophiles helps direct genomic analysis of Aciduliprofundum boonei. Geobiology 2008;6:331–6. [DOI] [PubMed] [Google Scholar]
- Reysenbach AL, Liu Y, Banta AB et al.. A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature 2006;442:444–7. [DOI] [PubMed] [Google Scholar]
- Ribeiro KS, Vasconcellos CI, Soares RP et al.. Proteomic analysis reveals different composition of extracellular vesicles released by two Trypanosoma cruzi strains associated with their distinct interaction with host cells. J Extracell Vesicles 2018;7:1463779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rilla K, Mustonen AM, Arasu UT et al.. Extracellular vesicles are integral and functional components of the extracellular matrix. Matrix Biol 2017. DOI: 10.1016/j.matbio.2017.10.003 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Rilla K, Pasonen-Seppänen S, Deen AJ et al.. Hyaluronan production enhances shedding of plasma membrane-derived microvesicles. Exp Cell Res 2013;319:2006–18. [DOI] [PubMed] [Google Scholar]
- Rilla K, Siiskonen H, Tammi M et al.. Hyaluronan-coated extracellular vesicles—a novel link between hyaluronan and cancer. Adv Cancer Res 2014;123:121–48. [DOI] [PubMed] [Google Scholar]
- Rivera J, Cordero RJ, Nakouzi AS et al.. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins . Proc Natl Acad Sci USA 2010;107:19002–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins PD, Dorronsoro A, Booker CN. Regulation of chronic inflammatory and immune processes by extracellular vesicles. J Clin Invest 2016;126:1173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Nakayasu ES, Almeida IC et al.. The impact of proteomics on the understanding of functions and biogenesis of fungal extracellular vesicles. J Proteomics 2014;97:177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Nakayasu ES, Oliveira DL et al.. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell 2008;7:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Nimrichter L, Oliveira DL et al.. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell 2007;6:48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roier S, Blume T, Klug L et al.. A basis for vaccine development: comparative characterization of Haemophilus influenzae outer membrane vesicles. Int J Med Microbiol 2015;305:298–309. [DOI] [PubMed] [Google Scholar]
- Roier S, Zingl FG, Cakar F et al.. Bacterial outer membrane vesicle biogenesis: a new mechanism and its implications. Microbial Cell 2016a;3:257–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roier S, Zingl FG, Cakar F et al.. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat Commun 2016b;7:10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompikuntal PK, Thay B, Khan MK et al.. Perinuclear localization of internalized outer membrane vesicles carrying active cytolethal distending toxin from Aggregatibacter actinomycetemcomitans. Infect Immun 2012;80:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose-Amsaleg C, Fedala Y, Vénien-Bryan C et al.. Utilization of interferometric light microscopy for the rapid analysis of virus abundance in a river. Res Microbiol 2017;168:413–8. [DOI] [PubMed] [Google Scholar]
- Ruiz-Martinez M, Navarro A, Marrades RM et al.. YKT6 expression, exosome release, and survival in non-small cell lung cancer. Oncotarget 2016;7:51515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbo C, Fernández-Moreira E, Merino M et al.. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Ch 2011;55:3084–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustom A. The missing link: does tunnelling nanotube-based supercellularity provide a new understanding of chronic and lifestyle diseases? Open Biol 2016;6:160057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustom A, Saffrich R, Markovic I et al.. Nanotubular highways for intercellular organelle transport. Science 2004;303:1007–10. [DOI] [PubMed] [Google Scholar]
- Rutter BD, Innes RW. Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol 2017;173:728–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko E, Nouwens A, Webb RI et al.. Nuclear pore-like structures in a compartmentalized bacterium. PLoS One 2017;12:e0169432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer U, Mairhofer M, Prohaska R. Stomatin: a new paradigm of membrane organization emerges. Dyn Cell Biol 2007;1:20–33. [Google Scholar]
- Samson RY, Obita T, Freund SM et al.. A role for the ESCRT system in cell division in Archaea. Science 2008;322:1710–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo L, Giurato G, Cicchini C. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep 2016;17:799–808. [DOI] [PubMed] [Google Scholar]
- Santarella-Mellwig R, Franke J, Jaedicke A et al.. The compartmentalized bacteria of the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum have membrane coat-like proteins. PLoS Biol 2010;8:e1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D, Rosenwasser S, Malitsky S et al.. Communication via extracellular vesicles enhances viral infection of a cosmopolitan alga. Nat Microbiol 2017;2:1485–92. [DOI] [PubMed] [Google Scholar]
- Schatz D, Vardi A. Extracellular vesicles - new players in cell-cell communication in aquatic environments. Curr Opin Microbiol 2018;43:148–54. [DOI] [PubMed] [Google Scholar]
- Schertzer JW, Whiteley M. A bilayer-couple model of bacterial outer membrane vesicle biogenesis. mBio 2012;3:e00297–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooling SR, Beveridge TJ. Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol 2006;188:5945–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooling SR, Hubley A, Beveridge TJ. Interactions of DNA with biofilm-derived membrane vesicles. J Bacteriol 2009;191:4097–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 2015;13:605–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shader RI. Cell-to-cell talk: plasmids and exosomes. Clin Ther 2014;6:817–8. [DOI] [PubMed] [Google Scholar]
- Sheetz MP, Painter RG, Singer SJ. Biological membranes as bilayer couples. III. Compensatory shape changes induced in membranes. J Cell Biol 1976;70:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty A, Chen S, Tocheva EI et al.. Nanopods: a new bacterial structure and mechanism for deployment of outer membrane vesicles. PLoS One 2011;6:e20725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Chan SK, Robinson DP et al.. Proteomic analysis of the secretome of Leishmania donovani. Genome Biol 2008;9:R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Clos J, de’Oliveira CC et al.. An exosome-based secretion pathway is responsible for protein export from Leishmania and communication with macrophages. J Cell Sci 2010;123:842–52. [DOI] [PubMed] [Google Scholar]
- Sitar S, Kejžar A, Pahovnik D et al.. Size characterization and quantification of exosomes by asymmetrical-flow field-flow fractionation. Anal Chem 2015;87:9225–33. [DOI] [PubMed] [Google Scholar]
- Sjöström AE, Sandblad L, Uhlin BE et al.. Membrane vesicle-mediated release of bacterial RNA. Sci Rep 2015;5:15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J, Wurdinger T, van Rijn S et al.. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008;10:1470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotland T, Sandvig K, Llorente A. Lipids in exosomes: current knowledge and the way forward. Prog Lipid Res 2017;66:30–41. [DOI] [PubMed] [Google Scholar]
- Snead WT, Hayden CC, Gadok AK et al.. Membrane fission by protein crowding. Proc Natl Acad Sci USA 2017;114:E3258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyers L, Umlauf E, Prohaska R. Association of stomatin with lipid-protein complexes in the plasma membrane and the endocytic compartment. Eur J Cell Biol 1999;78:802–12. [DOI] [PubMed] [Google Scholar]
- Soler N, Gaudin M, Marguet E et al.. Plasmids, viruses and virus-like membrane vesicles from Thermococcales. Biochm Soc Trans 2011;39:36–44. [DOI] [PubMed] [Google Scholar]
- Soler N, Krupovic M, Marguet E et al.. Membrane vesicles in natural environments: a major challenge in viral ecology. ISME J 2015;9:793–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler N, Marguet E, Verbavatz J-M et al.. Virus-like vesicles and extracellular DNA produced by hyperthermophilic archaea of the order Thermococcales. Res Microbiol 2008;159:390–9. [DOI] [PubMed] [Google Scholar]
- Song T, Mika F, Lindmark B et al.. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol Microbiol 2008;70:100–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag I, Schwarz H, Hirota Y et al.. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J Bacteriol 1978;136:280–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubannier V, McLelland GL, Zunino R et al.. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol 2012a;22:135–41. [DOI] [PubMed] [Google Scholar]
- Soubannier V, Rippstein P, Kaufman BA et al.. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS One 2012b;7:e52830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Eme L, Saw JH et al.. Asgard archaea are the closest prokaryotic relatives of eukaryotes. PLoS Genet 2018;14:e1007080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Saw JH, Jørgensen SL et al.. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 2015;521:173–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak JC, Schmid EM, Ryan CJ et al.. Membrane bending by protein–protein crowding. Nat Cell Biol 2012;14:944–9. [DOI] [PubMed] [Google Scholar]
- Stahl PD, Raposo G. Exosomes and extracellular vesicles: the path forward. Essays Biochem 2018;62:119–24. [DOI] [PubMed] [Google Scholar]
- Stanly C, Fiume I, Capasso G et al.. Isolation of exosome-like vesicles from plants by ultracentrifugation on sucrose/deuterium oxide (D2O) density cushions. Methods Mol Biol 2016;1459:259–69. [DOI] [PubMed] [Google Scholar]
- Steenbakkers PJM, Geerts WJ, Ayman-Oz NA et al.. Identification of pseudomurein cell wall binding domains. Mol Microbiol 2006;62:1618–30. [DOI] [PubMed] [Google Scholar]
- Stempler O, Baidya AK, Bhattacharya S et al.. Interspecies nutrient extraction and toxin delivery between bacteria. Nat Commun 2017;8:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuffers S, Sem Wegner C, Stenmark H et al.. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009;10:925–37. [DOI] [PubMed] [Google Scholar]
- Su QP, Du W, Ji Q et al.. Vesicle size regulates nanotube formation in the cell. Sci Rep 2016;6:24002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subra C, Laulagnier K, Perret B et al.. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 2007;89:205–12. [DOI] [PubMed] [Google Scholar]
- Subramanian P, Pirbadian S, El-Naggar MY et al.. Ultrastructure of Shewanella oneidensis MR-1 nanowires revealed by electron cryotomography. Proc Natl Acad Sci USA 2018;115:E3246–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi RU, Prieto-Vila M, Hironaka A et al.. The role of extracellular vesicle microRNAs in cancer biology. Clin Chem Lab Med 2017;55:648–56. [DOI] [PubMed] [Google Scholar]
- Takasugi M. Emerging roles of extracellular vesicles in cellular senescence and aging. Aging Cell 2018;17 DOI: 10.1111/acel.12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo KUI, Uehira K, Nishiura M. Fine structure of Cryptococcus neoformans grown in vivo as observed by freeze-etching. J Bacteriol 1973;113:1449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TT, Mörgelin M, Forsgren A et al.. Haemophilus influenzae survival during complement-mediated attacks is promoted by Moraxella catarrhalis outer membrane vesicles. J Infect Dis 2007;195:1661–70. [DOI] [PubMed] [Google Scholar]
- Tatischeff I. Assets of the non-pathogenic microorganism Dictyostelium discoideum as a model for the study of eukaryotic extracellular vesicles. F1000Res 2013;2:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008;110:13–21. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Driscoll M, Kyrpides NC. The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem Sci 1999;24:425–7. [DOI] [PubMed] [Google Scholar]
- Thakur BK, Zhang H, Becker A et al.. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014;24:766–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thay B, Wai SN, Oscarsson J. Staphylococcus aureus α-toxin-dependent induction of host cell death by membrane-derived vesicles. PLoS One 2013,8:e54661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C, Amigorena S, Raposo G et al.. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 2006;30:3.22.1–3.22.29. [DOI] [PubMed] [Google Scholar]
- Théry C, Boussac M, Véron P et al.. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol 2001;166:7309–18. [DOI] [PubMed] [Google Scholar]
- Thompson AG, Gray E, Heman-Ackah SM et al.. Extracellular vesicles in neurodegenerative disease — pathogenesis to biomarkers. Nat Rev Neurol 2016;12:346–57. [DOI] [PubMed] [Google Scholar]
- Tkach M, Kowal J, Théry C. Why the need and how to approach the functional diversity of extracellular vesicles. Phil Trans R Soc B 2018;373 pii: 20160479 DOI: 10.1098/rstb.2016.0479 Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell 2016;164:1226–32. [DOI] [PubMed] [Google Scholar]
- Toyofuku M, Cárcamo-Oyarce G, Yamamoto T et al.. Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat Commun 2017b;8:481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku M, Morinaga K, Hashimoto Y et al.. Membrane vesicle mediated bacterial communication. ISME J 2017a;11:1504–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S et al.. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008,319:1244–7. [DOI] [PubMed] [Google Scholar]
- Trams EG, Lauter CJ, Salem N et al.. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta 1981;645:63–70. [DOI] [PubMed] [Google Scholar]
- Tricarico C, Clancy J, D'Souza-Schorey C. Biology and biogenesis of shed microvesicles. Small GTPases 2016;5:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsatsaronis JA, Franch-Arroyo S, Resch U et al.. Extracellular vesicle RNA: a universal mediator of microbial communication? Trends Microbiol 2018;26:401–10. [DOI] [PubMed] [Google Scholar]
- Turnbull L, Toyofuku M, Hynen AL et al.. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun 2016;7:11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twu O, de Miguel N, Lustig G et al.. Trichomonas vaginalis exosomes deliver cargo to host cells and mediate host:parasite interactions. PLoS Pathog 2013;9:e1003482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzipilevich E, Habusha M, Ben-Yehuda S. Acquisition of phage sensitivity by bacteria through exchange of phage receptors. Cell 2017;168:186–199.e12. e12 [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekström K, Bossios A et al.. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654–9. [DOI] [PubMed] [Google Scholar]
- van der Pol E, Böing AN, Harrison P et al.. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol 2012;64:676–705. [DOI] [PubMed] [Google Scholar]
- van der Pol L, Stork M, van der Ley P. Outer membrane vesicles as platform vaccine technology. Biotechnol J 2015;10:1689–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vos KE, Abels ER, Zhang X et al.. Directly visualized glioblastoma-derived extracellular vesicles transfer RNA to microglia/macrophages in the brain. Neuro Oncol 2016;18:58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deun J, Mestadagh P, Agostinis P et al.. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods 2017;14:228–32. [DOI] [PubMed] [Google Scholar]
- van Doormaal FF, Kleinjan A, Di Nisio M et al.. Cell-derived microvesicles and cancer. Neth J Med 2009;67:266–73. [PubMed] [Google Scholar]
- van Herwijnen MJC, Zonneveld MI, Goerdayal S et al.. Comprehensive proteomic analysis of human milk-derived extracellular vesicles unveils a novel functional proteome distinct from other milk components. Mol Cell Proteomics 2016;15:3412–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018;19:213–28. [DOI] [PubMed] [Google Scholar]
- van Niel G, Raposo G, Candalh C et al.. Intestinal epithelial cells secrete exosome–like vesicles. Gastroenterology 2001;121:337–49. [DOI] [PubMed] [Google Scholar]
- van Niftrik LA, Fuerst JA, Sinninghe Damsté JS et al.. The anammoxosome: an intracytoplasmic compartment in anammox bacteria. FEMS Microbiol Lett 2004;233:7–13. [DOI] [PubMed] [Google Scholar]
- Vdovikova S, Luhr M, Szalai P et al.. A novel role of Listeria monocytogenes membrane vesicles in inhibition of autophagy and cell death. Front Cell Infect Microbiol 2017;7:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith PD, Chen YY, Gorasia DG et al.. Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J Proteome Res 2014;13:2420–32. [DOI] [PubMed] [Google Scholar]
- Verma M, Lam TK, Hebert E et al.. Extracellular vesicles: potential applications in cancer diagnosis, prognosis, and epidemiology. BMC Clin Pathol 2015;15:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya-Beltri C, Baixauli F, Gutiérrez-Vázquez C et al.. Sorting it out: regulation of exosome loading. Semin Cancer Biol 2014;28:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F et al.. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 2013;4:2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votteler J, Sundquist WI. Virus budding and the ESCRT Pathway. Cell Host Microbe 2013;14:232–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldenström A, Gennebäck N, Hellman U et al.. Cardiomyocyte microvesicles contain DNA/RNA and convey biological messages to target cells. PLoS One 2012;7:e34653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Thompson CD, Weidenmaier C et al.. Release of Staphylococcus aureus extracellular vesicles and their application as a vaccine platform. Nat Commun 2018;9:1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Gilkes DM, Takano N et al.. Hypoxia-inducible factors and RAB22A mediate formation of microvesicles that stimulate breast cancer invasion and metastasis. Proc Natl Acad Sci USA 2014;111:E3234–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters E, Hohn MJ, Ahel I et al.. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci 2003;100:12984–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehman AM, Poggioli C, Schweinsberg P et al.. The P4-ATPase TAT-5 Inhibits the budding of extracellular vesicles in C. elegans embryos. Curr Biol 2011;21:1951–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Batagov AO, Schinelli S et al.. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat Commun 2017;8:1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Vassallo CN, Pathak DT et al.. Myxobacteria produce outer membrane-enclosed tubes in unstructured environments. J Bacteriol 2014;196:1807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidle HU, Birzele F, Kollmorgen G et al.. The multiple roles of exosomes in metastasis. Cancer Genomics Proteomics 2017;14:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensink J, Witholt B. Outer-membrane vesicles released by normally growing Escherichia coli contain very little lipoprotein. Eur J Biochem 1981;116:331–5. [DOI] [PubMed] [Google Scholar]
- Wessel AK, Liew J, Kwon T et al.. Role of Pseudomonas aeruginosa peptidoglycan-associated outer membrane proteins in vesicle formation. J Bacteriol 2013;195:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside TL. Tumor-derived exosomes and their role in cancer progression. Adv Clin Chem 2016;74:103–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer KW, Soekmadji C, Hill AF et al.. Updating the MISEV minimal requirements for extracellular vesicle studies: building bridges to reproducibility. J Extracell Vesicles 2017;6:1396823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990;87:4576–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf P. The nature and significance of platelet products in human plasma. Br J Haematol 1967;13:269–88. [DOI] [PubMed] [Google Scholar]
- Wolfers J, Lozier A, Raposo G et al.. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med 2001;7:297–303. [DOI] [PubMed] [Google Scholar]
- Work E, Knox KW, Vesk M. The chemistry and electron microscopy of an extracellular lipopolysaccharide from Escherichia coli. Ann NY Acad Sci 1966;133:438–49. [DOI] [PubMed] [Google Scholar]
- Wright PK, Jones SB, Ardern N et al.. 17β-estradiol regulates giant vesicle formation via estrogen receptor-alpha in human breast cancer cells. Oncotarget 2014;5:3055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdinger T, Gatson N, Balaj L et al.. Extracellular vesicles and their convergence with viral pathways. Ad Virol 2012;2012:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Rai A, Chen M et al.. Extracellular vesicles in cancer — implications for future improvements in cancer care. Nat Rev Clin Oncol 2018; DOI: 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- Yáñez-Mó M, Siljander PR-M, Andreu Z et al.. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 2015;4:617–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron S, Kolling GL, Simon L et al.. Vesicle-mediated transfer of virulence genes from Escherichia coli O157: H7 to other enteric bacteria. Appl Environ Microb 2000;66:4414–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun SH, Lee SY, Choi CW et al.. Proteomic characterization of the outer membrane vesicle of the halophilic marine bacterium Novosphingobium pentaromativorans US6-1. J Microbiol 2017;55:56–62. [DOI] [PubMed] [Google Scholar]
- Yutin N, Koonin EV. Archaeal origin of tubulin. Biol Direct 2012;7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborowski MP, Balaj L, Breakefield XO et al.. Extracellular vesicles: composition, biological relevance, and methods of study. Bioscience 2015;65:783–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba-Niedzwiedzka K, Caceres EF, Saw JH et al.. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 2017;541:353–8. [DOI] [PubMed] [Google Scholar]
- Zhang H, Lyden D. A protocol for asymmetric-flow field-flow fractionation (AF4) of small extracellular vesicles. Protocol Exchange 2018. DOI: 10.1038/protex.2018.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kim MS, Jia B et al.. Hypothalamic stem cells control ageing speed partly through exosomal miRNAs. Nature 2017;548:52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu D, Chen X et al.. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell 2010;39:133–44. [DOI] [PubMed] [Google Scholar]
- Zhou L, Srisatjaluk R, Justus DE et al.. On the origin of membrane vesicles in Gram-negative bacteria. FEMS Microbiol Lett 1998;163:223–8. [DOI] [PubMed] [Google Scholar]
- Zomer A, Maynard C, Verweij FJ et al.. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 2015;161:1046–57. [DOI] [PMC free article] [PubMed] [Google Scholar]