Summary
Induction of pluripotency in somatic cells with defined genetic factors has been successfully used to investigate the mechanisms of disease initiation and progression. Cellular reprogramming and oncogenic transformation share common features; both involve undergoing a dramatic change in cell identity, and immortalization is a key step for cancer progression that enhances reprogramming. However, there are very few examples of complete successful reprogramming of tumor cells. Here we address the effect of expressing an active oncogene, RAS, on the process of reprogramming and found that, while combined expression with reprogramming factors enhanced dedifferentiation, expression within the context of neoplastic transformation impaired reprogramming. RAS induces expression changes that promote loss of cell identity and acquisition of stemness in a paracrine manner and these changes result in reprogramming when combined with reprogramming factors. When cells carry cooperating oncogenic defects, RAS drives cells into an incompatible cellular fate of malignancy.
Key words: Ras, oncogenes, cell reprogramming, iPSC, cell plasticity
Graphical Abstract

Highlights
-
•
Oncogenic Ras enhances cell reprogramming in a wild-type context
-
•
Ras induces gene expression changes that favor reprogramming
-
•
Ras expression in immortal cells impairs cell reprogramming
-
•
Oncogenic transformation and cellular reprogramming are incompatible cell fates
Cellular reprogramming and oncogenic transformation share common features. Collado and colleagues show that oncogenic Ras enhances reprogramming to pluripotency in normal somatic cells in a paracrine manner. In contrast, oncogenically transformed cells are resistant to reprogramming. Oncogenic transformation raises reprogramming barriers that can be reverted by turning off the expression of the driving oncogene. Transformation and reprogramming are incompatible cell fates.
Introduction
The development of an in vitro cellular system allowing the reprogramming of differentiated somatic cells into induced pluripotent stem cells (iPSCs) by expression of defined genetic elements, represents an opportunity to advance in many different areas of biomedical research (Takahashi and Yamanaka, 2016). Apart from providing pluripotent cells to develop cell therapies, reverting the differentiated state of the cell offers an opportunity to create faithful disease models and to develop powerful cellular platforms in which to efficiently screen pharmacological interventions (Onder and Daley, 2012).
The application of cellular reprogramming to the study of cancer is just beginning to be explored (Papapetrou, 2016). One particularly interesting aspect of the application of cellular reprogramming to the study of cancer is the similarity between reprogramming and neoplastic transformation (Goding et al., 2014). During reprogramming, cells need to overcome barriers that oppose the drastic change in cell identity characterizing this process and gain the capacity to proliferate indefinitely. Tumor cells, on the other hand, are generally immortal and typically display the features of an undifferentiated state, especially in more advanced cancers. For example, poorly differentiated tumors present an embryonic stem-like gene signature that is considered a hallmark of aggressiveness (Ben-Porath et al., 2008), and cancer cell dedifferentiation has been proposed as a means to become more malignant (Bradner et al., 2017). Elucidating the common mechanisms and barriers shared by reprogramming and transformation could illuminate the molecular bases underlying the pathogenesis of cancer.
Illustrating that common barriers prevent cell transformation and cell reprogramming is the observation that cells deficient in tumor suppressor genes which regulate immortality, renders cells susceptible to the transforming activity of activated oncogenes and enhances reprogramming (Hong et al., 2009, Kawamura et al., 2009, Li et al., 2009, Marión et al., 2009, Utikal et al., 2009). Actually, the expression of a single oncogene on a normal differentiated cell does not lead to neoplastic transformation. Immortality is required to overcome the barriers that block the transformation into a cancer cell (Land et al., 1983, Ruley, 1983). Since immortalization is a pre-requisite for transformation, one would expect cancer cells to be more susceptible to reprogramming. However, there are strikingly few examples of successful complete reprogramming to pluripotency in cancer cells (Ramos-Mejia et al., 2012).
Using the system of cellular reprogramming has already proved extremely useful to identify previously unrecognized activities of tumor suppressors, such as the transcriptional control over pluripotency gene Sox2 exerted by cell-cycle inhibitors p27Kip1 and the retinoblastoma family of pocket proteins (Kareta et al., 2015, Li et al., 2012, Vilas et al., 2015). Similarly, it could also represent an opportunity to gain insight into the molecular mechanisms of cellular transformation driven by oncogenes.
In this work, we decided to address the effect of expressing oncogenic RAS on the process of cellular reprogramming. RAS was the first human oncogene isolated from a tumor and it is one of the most frequently mutated genes in human cancer (Malumbres and Barbacid, 2003). First, we evaluated the consequences of introducing RAS as part of the reprogramming cocktail together with Oct4, Sox2, Klf4, and c-Myc (OSKM). Introduction of activated RAS alone on normal differentiated somatic cells does not lead to neoplastic transformation and requires the presence of cooperating oncogenes to allow progression into malignancy (Serrano et al., 1997). Interestingly, in our case the combined expression of RAS and the reprogramming factors resulted in enhanced reprogramming. This effect of RAS is non-cell autonomous and seems to be a reflection of an endogenous activity played by the oncogene during early stages of a normal reprogramming process. In contrast, expression of oncogenic RAS in the context of full transformation blocks reprogramming. Using in vivo systems, we conclude that oncogene activation generates a tissue microenvironment that renders cells in the vicinity susceptible to dedifferentiation, while transformation and reprogramming seem to be alternative non-compatible cell fates.
Results
RAS Expression Enhances Cellular Reprogramming
To address the effect of oncogene expression on the process of cellular reprogramming, we overexpressed an oncogenic mutant Hras, HrasV12, in mouse embryo fibroblasts (MEFs) derived from i4F mice, a transgenic animal carrying a polycistronic tetracycline-inducible cassette expressing the reprogramming factors: Oct4, Sox2, Klf4, and c-Myc (Abad et al., 2013). After 14 days in doxycycline, iPSC colonies were stained using alkaline phosphatase (AP). The number of colonies was increased when we overexpressed oncogenic Hras compared with the overexpression of GFP as a control (Figure 1A). Similar results were obtained when we co-expressed the reprogramming factors together with HrasV12 using retroviral transduction (Figure S1A). To verify pluripotency, colonies were picked and expanded for further analysis. Embryonic stem cell markers, their capacity to differentiate in vitro, and the ability to form embryoid bodies or teratomas with the expression of markers of the three germ layers in vivo confirmed their pluripotency (Figures S1B–S1E). Thus, expression of oncogenic Hras can enhance reprogramming by OSKM.
Figure 1.
RAS Expression Enhances Cellular Reprogramming
(A) Representative picture of AP staining after reprogramming of i4F MEFs overexpressing HrasV12, or GFP as a control (left), and calculation of efficiency of reprogramming relative to GFP (right).
(B) Efficiency of reprogramming obtained after overexpression in MEFs of NrasV12 or BrafV600E, relative GFP as a control.
(C) Schematic representation of experimental setup to test the effect of inhibiting MEK (PD0325901) from T0-T3 compared with T10-T13 during reprogramming with overexpression of NrasV12 on i4F MEFs.
(D) Efficiency of reprogramming obtained after overexpression in i4F MEFs of NrasV12 and treatment with MEKi (PD0325901) during the first 3 days (T0-T3) or the last 3 days (T10-T13) of reprogramming, compared with vehicle (VEH), and relative to GFP control.
(E) Schematic representation of experimental setup to analyze the levels of active (GTP bound) Ras using a RalGDS-RBD pull-down from protein extracts obtained along a normal process of cellular reprogramming.
(F) Pull-down assay of active GTP-bound RAS along time points during the process of reprogramming (D0-D10) of WT MEFs transduced with OSKM. Western blot with antibodies against PAN-Ras showing the levels of active RAS in pull downs (upper panel) and inputs (lower panel). Representative of three independent experiments.
(G) Western blot showing the levels of active phosphorylated ERK (P-ERK, upper panel) and total ERK (middle panel) along the same process of reprogramming (D0-D10). Tubulin levels are shown as loading control. Representative of three independent experiments.
(H) Schematic representation of experimental setup to test the effect of inhibiting MEK (PD0325901) from T0-T3 compared with T10-T13 during normal cellular reprogramming on WT MEFs overexpressing OSKM.
(I) Efficiency of reprogramming obtained after treatment with MEKi (PD0325901) during the first 3 days (T0-T3) or the last 3 days (T10-T13) of the process in WT MEFs transduced with OSKM, compared with vehicle-treated cells (VEH).
n = 3 independent experiments. All data correspond to the average ± SD. Statistical significance was assessed by the two-tailed Student's t test: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; n.s., not significant. See also Figure S1.
Other members of the RAS family or downstream of RAS were also effective. NrasV12, and an active version of one of the RAF family members, BrafV600E, a relevant downstream effector of the RAS signaling, could also promote reprogramming (Figure 1B). In addition, treatment with MEKi (PD0325901), to block MAPK activity, during the first 3 days of reprogramming with overexpression of NrasV12 (Figure 1C) caused a dramatic drop in the reprogramming efficiency, while treatment during the previous 3 days had only a minimal impact (Figure 1D). These results show that RAS oncogene overexpression can have a positive impact on cell reprogramming efficiency, acting at least in part through MEK, during the early steps of the process.
To determine if the effect of oncogenic RAS reflected a neomorphic activity, not shared with endogenous wild-type (WT) RAS, we assessed the level of activation of endogenous RAS activity during cellular reprogramming. For this, we pulled down GTP-bound active RAS from total cell extracts obtained during consecutive days along a reprogramming process from WT MEFs overexpressing OSKM (Figure 1E). Active RAS increased during the first 2 days of reprogramming, decreasing to normal levels over the remaining process (Figure 1F). Similarly, acute increased expression of phospho-ERK and decreasing levels were noted as well, and correlated with the RAS pull-down (Figure 1G). Confirming the early involvement of this endogenous pathway on cellular reprogramming, treatment with MEKi during the first 3 days of the process (Figure 1H) caused a dramatic decrease in the number of iPSC colonies obtained, while the same treatment during the last days had a minor effect (Figure 1I).
Taken together, our results point to a positive contribution of endogenous RAS in cell reprogramming, especially early during the process, an effect that is exacerbated by the expression of oncogenic mutant RAS.
Gene Expression Changes upon RAS Expression during Reprogramming
Next, we examined how overexpression of RAS was affecting pluripotency gene expression during reprogramming. We measured the level of expression of Nanog, Ssea1, Essrb, and Dppa3, all markers of pluripotency, during cell reprogramming using OSKM expression combined with HrasV12 or GFP as a control. All these pluripotency markers consistently reached an increased expression at later time points when OSKM reprogramming was combined with HrasV12 compared with GFP expression (Figure 2A), consistent with an increased number of colonies obtained in this condition. Interestingly, the expression of Ssea1 was increased from the initial time point and in a sustained manner throughout the whole process of reprogramming when HrasV12 was expressed (Figure 2A). In addition, we checked the level of fibroblast cell marker expression because cell reprogramming implicates a change in identity from the initial differentiated cell type (in our case fibroblasts) to the final iPSCs. Both, Thy1 and Fn1 were downregulated when HrasV12 was included in the reprogramming cocktail, with levels already at initial stages clearly lower than in the control GFP-expressing cells (Figure 2B).
Figure 2.
Gene Expression Changes upon RAS Expression during Reprogramming
(A) qRT-PCR analysis of expression of pluripotency genes Nanog, Ssea1, Essrb, and Dppa3 after GFP or HrasV12 overexpression in WT MEFs transduced with OSKM along the process of cell reprogramming. Levels of expression for each gene in pluripotent ESCs (open squares) are shown as a reference.
(B) qRT-PCR analysis of expression of cell identity genes Thy1 and Fn1 after GFP or HrasV12 overexpression along the process of cell reprogramming in WT MEFs transduced with OSKM.
(C) Schematic diagram showing the experimental setup to study gene expression changes after HrasV12 expression compared with control GFP independently of cellular reprogramming.
(D) qRT-PCR analysis of expression at early time points of pluripotency gene Ssea1, and cell identity genes Thy1 and Fn1, in WT MEFs transduced with HrasV12 independently of reprogramming.
n = 3 independent experiments. All data correspond to the average ± SD. of qRT-PCR data. Statistical significance was assessed by the two-tailed Student's t test: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; n.s., not significant. See also Figure S2.
Oncogenic RAS expression in primary fibroblasts triggers a form of stable cell-cycle arrest, known as cell senescence (Serrano et al., 1997) and this is a crucial barrier opposing reprogramming (Banito and Gil, 2010). We tested if oncogenic RAS was inducing cell senescence during reprogramming by measuring the expression levels of two critical genes participating on the cell-cycle arrest that characterizes cell senescence: Cdkn1a and Ink4a (Collado and Serrano, 2006). In addition, we also checked the expression of cytokine Il6, an immune factor that has been linked to both cell senescence and reprogramming (Mosteiro et al., 2018). qRT-PCR revealed a transient peak of expression for Cdkn1a and Ink4a early during reprogramming, and a more delayed one for Il6, during reprogramming with HrasV12 (Figure S2A). To more directly assess cell senescence we measured senescence-associated beta-galactosidase (SAbGal), the most widely used marker of cell senescence (Dimri et al., 1995), employing a chemiluminescence substrate, Galacton. Cell extracts at various times of cell reprogramming were collected and assessed for SAbGal in vitro and the activity plotted relative to the number of cells. SAbGal activity was generally similar comparing oncogenic HrasV12-expressing cells and control cells, except during the first time point, when GFP-expressing cells showed even higher levels of SAbGal than HrasV12 cells (Figure S2B). To further confirm these results, we counted the cell numbers obtained at different time points during the process. HrasV12 expression caused a limited increase in the number of cells at around days 5–7, but by the end of the process of reprogramming cell numbers tended to be similar (Figure S2C).
Thus, the expression of HrasV12 causes changes in pluripotency and cell identity genes that could facilitate cell reprogramming, with some of these occurring at very early stages of the process. We wondered whether HrasV12 expression alone, in the absence of OSKM, could account for these immediate gene expression changes that we observed. To address this question, we transduced WT MEFs with an HrasV12-expressing vector under cell reprogramming conditions of cell culture but omitting OSKM reprogramming factors (Figure 2C), and analyzed the expression of Ssea1, Thy1, and Fn1 at early time points. qRT-PCR analysis showed that HrasV12 expression alone was sufficient to cause an increase in the expression of pluripotency gene Ssea1, while at the same time causing a decrease in identity genes Thy1 and Fn1 (Figure 2D).
Collectively, our analysis of the dynamics during cell reprogramming in the presence of an active HrasV12 oncogene suggests the existence of early gene expression changes that could promote reprogramming by increasing pluripotency genes and decreasing cell identity genes.
Ras Increases Cell Reprogramming in a Paracrine Manner
Next, we wanted to address whether this effect of HrasV12 on reprogramming was cell autonomous. For this, we took advantage of a tetracycline-inducible HrasV12-expressing lentiviral vector encoding its own reverse tetracycline-controlled transactivator (rtTA). We combined HrasV12 expression from this vector with OSKM expression from a lentiviral vector that was also dependent on tetracycline but lacked rtTA. In this manner, OSKM expression (and the ensuing cell reprogramming) could only take place within cells with concomitant expression of HrasV12 (see Figure 3A for a diagram depicting the experimental setting and the vectors used). Using this setting we observed that cell reprogramming was nearly abolished when OSKM expression was forced within cells expressing HrasV12 compared with the combination of OSKM with the empty vector that provides the rtTA (Figures 3B and 3C). Introducing an extra dose of the empty vector combined with HrasV12 rescued the deficient reprogramming, since OSKM expression could now take place independently of HrasV12 (Figures 3B and 3C). We analyzed the levels of expression of Il6, a pro-inflammatory cytokine associated with oncogenic RAS expression and the secretory phenotype produced by senescent cells (known as SASP) that has been reported to favor the reprogramming process in a paracrine manner (Brady et al., 2013, Mosteiro et al., 2018). Interestingly, we observed that, in HrasV12 overexpressing cells, the levels of Il6 are higher than in the control empty vector (Figure 3D). However, this event did not correlate with reprogramming success, with similarly high levels of Il6 achieved in the condition that led to efficient reprogramming than in the one with impaired reprogramming.
Figure 3.
RAS Increases Cell Reprogramming in a Paracrine Manner
(A) Schematic diagram of the different lentiviral vectors used in this study: EV is the empty vector carrying the rtTA required for doxycycline-induced expression; OSKM stands for the inducible expression of the reprogramming factors Oct4, Sox2, Klf4, and c-Myc but lacking rtTA; HrasV12 stands for inducible expression of oncogenic Hras and carrying rtTA. The combination of EV + OSKM allows inducible expression of OSKM, while the combination of OSKM + HrasV12 forces the co-expression of the reprogramming factors and the active oncogenic Hras together, by using rtTA encoded in the HrasV12 vector.
(B) Representative picture of AP staining after reprogramming with inducible OSKM in MEFs transduced with inducible HrasV12 alone (HrasV12) compared with empty vector (EV) (left panel), and the same experiment including an extra dose of empty vector (right panel, +EV) to rescue reprogramming.
(C) Quantification of the number of AP-positive colonies obtained in (B).
(D) qRT-PCR analysis of expression of pro-inflammatory cytokine Il6 in (B).
(E) Schematic diagram showing the experimental setup to study cellular reprogramming in co-cultures of i4F MEFS grown with WT MEFs transduced with NrasV12, or GFP as control.
(F) Reprogramming efficiency (representative picture, left; and quantification relative to GFP, right panel) of i4F MEFs co-cultured with MEFs previously transduced with GFP or NrasV12.
(G) Schematic diagram showing the experimental setup to study reprogramming after addition of CM from MEFs transduced with NrasV12, or GFP as negative control, to i4F MEFs.
(H) Reprogramming efficiency (representative picture, left; and quantification relative to GFP, right panel) of i4F MEFs cultured in the presence of CM from MEFs previously transduced with NrasV12, or GFP as negative control.
(I) Schematic diagram showing the experimental setup to study the effect on the gene expression of WT MEFs exposed to soluble factors released from WT MEFs cultured on the top chamber of transwells and transduced with HrasV12 or GFP.
(J) qRT-PCR analysis of expression of pluripotency gene Ssea1 and cell identity genes Thy1 and Fn1 in MEFs cultured on the bottom chamber of transwells, with MEFs transduced with GFP or HrasV12 independently of reprogramming on the top chamber.
(K) Schematic diagram showing the experimental setup to study the effect on the gene expression of WT MEFs exposed to CM from WT MEFs transduced with HrasV12 or GFP.
(L) Gene expression analysis on MEFs cultured in the presence of CM from cells transduced with GFP or HrasV12 independently of reprogramming.
n = 3 independent experiments. All data correspond to the average ± SD. Statistical significance was assessed by the two-tailed Student's t test: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; n.s., not significant.
These results suggest that the ability of RAS to drive reprogramming might be cell non-autonomous, i.e., it might be changing cells in the neighboring environment, rather than itself. To directly test this possibility, we decided to assess the reprogramming efficiency of cells derived from the i4F mice growing on mixed cultures together with MEFs transduced with NrasV12 or GFP, as a control (Figure 3E). Using this system, we observed that the presence of cells expressing NrasV12 caused a marked increase in the efficiency of reprogramming compared with control cells expressing GFP (Figure 3F). In addition, we also performed a reprogramming experiment using i4F MEFs exposed to conditioned medium (CM) produced by MEFs transduced with NrasV12 or GFP (Figure 3G). This time, iPSC colonies were slightly more efficiently, but reproducibly, formed when i4F MEFs were reprogrammed in the presence of CM from cells expressing NrasV12 (Figure 3H).
Taken together, these results confirm our interpretation that the increased efficiency of reprogramming associated with RAS overexpression is non-autonomous, pointing to a mechanism mediated, at least partially, through secreted factors. Since we had observed that RAS alone can cause changes in genes related with pluripotency and cell identity by itself, independently of the reprogramming process, we evaluated if these changes occur also on target cells exposed to these factors. For this, we cultured MEFs on a transwell system, seeding WT target cells on the bottom chamber and HrasV12- or GFP-transduced MEFs on the top chamber, and collected the cells after 1, 3, or 5 days of co-culture (Figure 3I). The analysis of these cells revealed higher levels of expression of pluripotency gene Ssea1 after 5 days and lower levels of Fn1 on the first day (Figure 3J). Similarly, WT MEFs cultured for 1, 3, or 5 days in the presence of CM from MEFs transduced with HrasV12 (Figure 3K) showed reduced levels of Thy1 expression compared with cells exposed to CM from GFP-expressing cells (Figure 3L). These observations are in line with the idea of these genetic changes taking place as a consequence of the exposure to factors secreted by RAS-expressing cells.
These results point to a non-autonomous effect caused by expression of oncogenic RAS on the surrounding cells, altering the levels of expression of certain genes related to the acquisition of pluripotency and the loss of cellular identity. This effect seems to be mediated in part by soluble factors induced by oncogenic RAS.
Oncogenic Activation In Vivo Promotes Dedifferentiation Events in Non-permissive Tissues
The expression of reprogramming factors in vivo using i4F mice allows the acquisition of the pluripotent state in cells of different tissues. However, although the transgenic cassette is expressed in all mouse tissues examined, not all are equally susceptible to this dedifferentiation process (Abad et al., 2013). Interestingly, previous studies showed how tissue damage and cellular senescence generate an environment that facilitates reprogramming in resistant tissues (Chiche et al., 2017, Mosteiro et al., 2016). Considering this and the results above we wanted to determine if oncogenic activation in vivo could also favor reprogramming in resistant tissues.
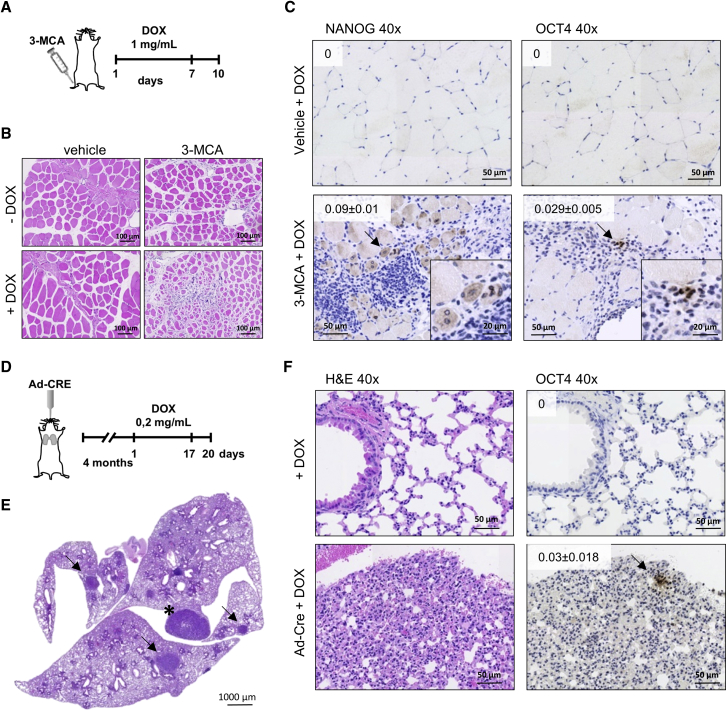
First, we injected mice intramuscularly with 3-methylcholanthrane (3-MCA), a highly carcinogenic polycyclic aromatic hydrocarbon that generates fibrosarcomas in mice when injected intramuscularly, with tumors carrying frequent oncogenic mutations in RAS oncogenes (Carbone et al., 1991, Eva and Aaronson, 1983, Watanabe et al., 1999). We combined 3-MCA injection in the tibialis anterior with the expression of OSKM in vivo in i4F mice by administration of doxycycline in the drinking water for 7 days, stopped the treatment and waited for an additional 3 days before harvesting tissue (Figure 4A). Using this protocol, we confirmed that the expression of NANOG and OCT4 by immunohistochemistry in areas affected in the pancreas, the most susceptible tissue for dedifferentiation upon OSKM expression (Figure S3A). Histologic analysis of the tibialis anterior of these mice revealed extensive damage to muscle fibers upon 3-MCA, as reflected by the smaller size of the fibers and the central localization of nuclei within fibers (Figures 4B, 4C, and S3B). This damage was exacerbated by the combination with doxycycline treatment but was not observed simply by mock injection or doxycycline alone (Figure 4B). Immunohistochemical analysis of NANOG and OCT4 expression in consecutive sections revealed cells positive for both pluripotency markers only in tissues from 3-MCA-injected i4F animals treated with doxycycline and associated with areas showing muscle fiber damage and immune cell infiltration, indicating that the combination of OSKM expression and oncogenic damage can promote dedifferentiation in the affected tissue (Figure 4C). Using a longer protocol (2.5 weeks) and lower doxycycline treatment, we obtained similar results, with positive expression of NANOG and OCT4 in damaged muscle fibers (Figures S3C and S3D). To extend our observations to a system of controlled oncogenic activation and to a different tissue refractory to dedifferentiation by OSKM expression, we decided to cross i4F mice with animals with conditional activation of KrasV12 from its endogenous allele triggered by the addition of Cre recombinase (Guerra et al., 2003). Four months after intratracheal administration of Cre, mice were treated with doxycycline in the drinking water for 2.5 weeks to induce the expression of OSKM and, 3 days later, mice were sacrificed, lungs were removed, and immunohistochemical analysis was performed (Figure 4D). Histological analysis of lung sections after induction of KrasV12 showed multiple adenomas and a few adenocarcinomas, as expected (Figures 4E and S3E), Staining with antibodies against OCT4 revealed areas with positive nuclei associated with pre-malignant, KrasV12-initiated, lesions, whereas lungs from i4F mice lacking the KrasV12 transgene were totally negative for this marker, confirming our observations obtained with 3-MCA in muscle (Figure 4F).
Figure 4.
Oncogenic Activation In Vivo Promotes Dedifferentiation Events in Non-permissive Tissues
(A) Schematic representation of 3-MCA injection in the tibialis anterior (TA) of i4F mice followed by treatment with doxycycline for 7 days at 1 mg/mL. Mice were sacrificed 3 days after stopping doxycycline treatment.
(B) Representative pictures of H&E staining of sections of TA from mice injected with vehicle (left panels) or 3-MCA (right panels), and untreated (upper panels) or treated with doxycycline (lower panels) (20× magnification).
(C) Representative pictures of immunohistochemical analysis of NANOG (left panels) or OCT4 (right panels) expression on TA from mice injected with vehicle (upper panels) or 3-MCA (lower panels) and treated with doxycycline (40× magnification, arrows point the positive cells). Insets show amplified positive cells. Quantifications show the relative percentage of positive cells. Values correspond to average ± SD (n = 3 mice).
(D) Schematic representation of administration of Ad-Cre to i4F/KrasV12 mice followed by doxycycline treatment for 2.5 weeks at 0.2 mg/mL. Mice were sacrificed 3 days after stopping doxycycline treatment.
(E) Representative picture of a lung section from mouse showing several adenomas (arrow) and adenocarcinoma (∗), indicates the correct induction of expression of KrasV12.
(F) Representative pictures of H&E staining (left panels) or immunohistochemical analysis of OCT4 (right panels) expression on lungs from mice treated with doxycycline and treated (lower panels) or not (upper panels) with Ad-Cre to induce KrasV12 expression (40× magnification, arrow points to OCT4-positive cells in an adenoma lesion). Quantifications show the relative percentage of OCT4-positive cells. Values correspond to average ± SD (n = 3 mice). See also Figure S3.
Thus, in vivo evidence from two different mouse models of oncogene activation combined with OSKM expression show that oncogene-induced tissue damage cooperates with reprogramming factor expression to initiate a process of dedifferentiation that results in the expression of pluripotency markers.
Ras Expression in the Context of Full Oncogenic Transformation Blocks Reprogramming
Since our data show that overexpressing oncogenic RAS enhances cellular reprogramming of primary cells, we wondered whether it would have the same effect on immortal cells. For this, we transduced HrasV12 on MEFs deficient for p53 together with reprogramming factors OSK, omitting c-Myc, since this factor is a potent oncogene that efficiently cooperates with lack of p53 to transform cells and it is dispensable for reprogramming (Nakagawa et al., 2008, Wernig et al., 2008). Expression of HrasV12 in the absence of p53 did not cause increased reprogramming efficiency (Figures 5A and 5B). On the contrary, the number of iPSC colonies was reduced (Figures 5A and 5B). In parallel, HrasV12 expression enhanced reprogramming by OSKM in WT cells, as expected. Expression levels of HrasV12 were similar in both WT and p53-null MEFs (Figure 5C). Similar results were observed when MEFs derived from animals deficient in another crucial tumor suppressor, Arf, were used (Figure S4A). To confirm these results, we expressed OSKM on three different fibrosarcoma cell lines derived from tumors developed in mice after injection with 3-MCA. Despite the presence of increased levels of reprogramming factors Oct4 and Sox2, these fibrosarcoma cell lines did not produce a single iPSC colony, while control WT MEFs underwent a typical process of cell reprogramming, as expected (Figures 5D and 5E).
Figure 5.
RAS Expression in the Context of Full Oncogenic Transformation Blocks Reprogramming
(A) Representative pictures of AP staining of WT MEFs transduced with OSKM (upper panels) and p53-null MEFs transduced with OSK (lower panels) combined with expression of GFP (left panels) or HrasV12 (right panels).
(B) Normalized reprogramming efficiency of (A) relative to control condition, WT OSKM + GFP.
(C) Western blot of protein extracts from conditions assessed in (A) analyzing the expression of p53, HRAS, and actin as loading control. Representative picture of n = 3 independent experiments.
(D) Representative pictures of negative AP staining of fibrosarcoma cell lines transduced with inducible OSKM and treated with doxycycline. WT MEFs treated in parallel and showing positive staining are shown as control.
(E) qRT-PCR expression analysis of reprogramming factors Oct4 (left panel) and Sox2 (right panel) on fibrosarcoma cell lines after doxycycline-induced expression of OSKM (n = 3). WT MEFs expressing OSKM and ESC are shown as positive controls.
(F) Schematic diagram showing the experimental setup to study the effect on gene expression of p53-null MEFs transduced with HrasV12 or GFP.
(G) qRT-PCR analysis of expression at early time points of pluripotency gene Ssea1, and cell identity genes Thy1 and Fn1, in p53-null MEFs transduced with HrasV12 independently of reprogramming.
(H) Schematic diagram showing the experimental setup to study the effect of adding CM from p53-null MEFs transduced with HrasV12 or GFP, on cellular reprogramming of WT MEFs transduced with OSKM.
(I) AP staining of WT MEFs transduced with OSKM and treated with CM from p53-null MEFs expressing GFP or NrasV12 (left panel) and quantification of AP-positive colonies (right panel).
n = 3 independent experiments. All data correspond to the average ± SD. Statistical significance was assessed by the two-tailed Student's t test: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; n.s., not significant. See also Figure S4.
Since we observed that RAS was affecting cellular reprogramming by inducing changes in the expression of pluripotency and cell identity genes independently of OSKM expression in WT cells, we wondered if these same changes were occurring in p53-null cells (Figure 5F). Gene expression analysis of mRNA extracted from p53-null MEFs transduced with HrasV12 or GFP showed reduction in the expression of cell identity genes, Thy1 and Fn1 (Figure 5G), similar to what we observed when we used WT MEFs (Figure 2D). In contrast, the expression levels of pluripotency gene Ssea1 remained unchanged (Figure 5G), an observation that is in line with the decreased reprogramming efficiency observed when HrasV12 is expressed in p53-null cells. Similar gene expression changes were also observed when p53-null MEFs transduced with HrasV12 or GFP were seeded on the top chamber of transwells and normal primary MEFs were seeded on the bottom chamber (Figure S4B). Cells receiving CM from the transformed MEFs showed reduced expression of Fn1 and Ssea1, with no apparent change in Thy1 (Figure S4C). Similarly, we wanted to check the paracrine effect on reprogramming of CM from p53-null MEFs expressing NrasV12. For this, we transduced p53-null MEFs with GFP or NrasV12. CM from these cell cultures was added to WT MEFs expressing OSKM and cell reprogramming was assessed (Figure 5H). Cell reprogramming in the presence of CM from p53-null MEFs expressing NrasV12 was not increased and was even reduced (Figure 5I).
These results show that, although RAS expression in normal WT cells increases cell reprogramming, in combination with another cooperating oncogenic event it hinders reprogramming.
Reversal of the Transformed Phenotype by Turning Off RAS Allows Reprogramming
To test the differential effect on cellular reprogramming of RAS oncogene expression depending on the cellular context, we decided to set up a reversible system of cellular transformation (Figure 6A). MEFs deficient for p53 were transduced with a lentiviral vector for doxycycline-inducible expression of HrasV12. In these cells, addition of doxycycline leads to the reversible expression of HrasV12 and the increased formation of colonies of transformed cells (Figures 6B, S5A, and S5B). When we expressed OSK in these cells in the presence of doxycycline, i.e., expressing HrasV12, we observed a drastic reduction in the number of iPSC colonies compared with the control reprogramming in the absence of the inducer (Figures 6C and 6D). Interestingly, withdrawal of doxycycline, i.e., shutting down HrasV12 expression, rescued this block in reprogramming and iPSC colonies were obtained (Figures 6C and 6D).
Figure 6.
Effect on Reprogramming of Reversal of the Transformed Phenotype
(A) Schematic diagram showing the experimental setup to study the effect of the transformed phenotype of cellular reprogramming. Immortal p53-null MEFs were directly transduced with OSKM for reprogramming or previously transduced with doxycycline-inducible HrasV12 to cause oncogenic transformation. HrasV12-transformed cells were then reprogrammed in the continuous presence of doxycycline (to induce HrasV12 and maintain transformation) or were transduced with OSKM for reprogramming in the absence of doxycycline (to switch off HrasV12 expression and cause reversal of transformation to an immortal state).
(B) Western blot of protein extracts from cells generated as depicted on (A) analyzing p53, HRAS, and tubulin as loading control. WT and p53-null MEFs are shown as control. Representative picture of n = 3 independent experiments.
(C) AP staining of p53-null MEFs transduced with inducible HrasV12 and OSK in the absence (–DOX), presence (+DOX) or after withdrawal (+/–DOX) of doxycycline (upper panel).
(D) Quantification of AP-positive colonies from (C).
(E) Schematic diagram showing the experimental setup to study the effect of the transformed phenotype of cellular reprogramming in vivo. Similar to that shown in (A), immortal p53-null MEFs were transduced with doxycycline-inducible HrasV12 to cause oncogenic transformation, then these cells were transduced with OSK and injected into the kidney of mice receiving doxycycline (to keep HrasV12 expression) or not (to switch off HrasV12 expression). Mice were histologically analyzed after sacrifice.
(F) Representative picture of H&E staining of a section from a tumor obtained after intrakidney injection of p53-null MEFs transduced with inducible HrasV12 and OSK in the presence of doxycycline (40× magnification).
(G) Representative pictures of H&E staining (upper panels) and immunohistochemical analysis of sections from teratomas obtained after intrakidney injection of p53-null MEFs transduced with inducible HrasV12 and OSK in the absence of doxycycline, showing the expression of CKAE1/AE3 (endoderm), Desmin (mesoderm), and GFAP (ectoderm) (40× magnification).
n = 3 independent experiments. All data correspond to the average ± SD. Statistical significance was assessed by the two-tailed Student's t test: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; n.s., not significant. See also Figure S5.
To explore this phenomenon in vivo, we recapitulated this experiment to assess the growth of these cells as tumors or teratomas when injected in animals. MEFs deficient in p53 were transduced with the inducible HrasV12 vector and OSK in the presence of doxycycline and were injected in the kidney of syngenic animals (Figure 6E). Injected mice were treated or not with doxycycline in the drinking water to allow for continuous expression of HrasV12 or its silencing, and tumor growth was evaluated. All the animals treated with doxycycline developed tumors as judged by pathological inspection (6 out of 6) (Figure 6F), whereas those without treatment developed only teratomas with a lower penetrance (2 out of 7) that showed histological structures characteristic of the three germ layers and that were positive for the expression of specific markers (Figure 6G). Tumors and teratomas were not the result of the growth of particular cell populations that did not carry the vectors for the reprogramming factors or for RAS, respectively, as observed after genotyping by PCR these tissues to detect specifically the presence of these vectors (Figure S5C).
Therefore, both in vitro and in vivo evidence shows that the reprogramming barriers raised by oncogenic transformation can be reverted by turning off the expression of the driving oncogene.
Discussion
Apart from a promising tool for regenerative medicine, cellular reprogramming is a very useful system to gain knowledge of processes involving dedifferentiation and cell identity changes. Cancer development shares intriguing similarities with reprogramming, sometimes showing similar patterns of gene expression and with common barriers opposing the drastic change in cell identity experienced during both tumorigenesis and reprogramming (Goding et al., 2014).
In this article, we addressed the effect of expressing an activated oncogene, RAS, on cellular reprogramming and found that the final outcome depends on the cellular context. In particular, RAS inclusion in the reprogramming cocktail increases the efficiency of reprogramming of normal primary cells. We obtained this same result independently of the system used to increase RAS expression and to reprogram the cells. This observation seems not to be just the result of in vitro manipulation of cells forced to overexpress RAS but a reflection of the involvement of endogenous RAS on reprogramming early during the process of reprogramming. A similar observation was reported by others through single-cell transcriptome analysis during reprogramming that revealed activation of RAS as an early molecular event (Kim et al., 2015).
Oncogenic RAS caused gene expression changes in pluripotency markers, showing earlier and higher expression than with control GFP, as well as reductions in mesenchymal identity genes, Thy1 and Fn1. The accumulation of higher numbers of reprogrammed cells at the end of the process of reprogramming with RAS could account for the increased levels of expression of some of these pluripotency genes, such as Nanog, Essrb, or Dppa3. Interestingly, however, some of these changes, such as downregulation of Thy1 and Fn1 or upregulation of Ssea1, are evident at very early stages when reprogramming has not yet occurred. This could imply that RAS alters the pattern of expression of certain genes independently of OSKM, and that these changes precede and predispose cells to the process of reprogramming. In line with this notion, we observed that these changes in gene expression are also induced in cells expressing RAS independently of the reprogramming process and could be part of the oncogene-induced plasticity that has been reported to contribute to oncogenesis (Ischenko et al., 2013). This effect seems to be exerted by RAS on a non-cell-autonomous manner, since forced expression of reprogramming factors OSKM with RAS did not result in increased efficiency, with even a negative impact on the process. Pro-inflammatory cytokine Il6 has been shown previously to be important during dedifferentiation to pluripotency (Brady et al., 2013) and was reported recently as a key factor promoting in vivo reprogramming upon OSKM expression, especially when reprogramming factors were combined with damage or injury to tissues (Chiche et al., 2017, Mosteiro et al., 2016, Mosteiro et al., 2018). In our case, RAS is capable of inducing Il6 expression independently of the co-expression of OSKM or not, but this Il6 does not contribute to reprogramming when OSKM is forced to be expressed in cells with RAS. Reinforcing the notion of non-cell-autonomous pro-reprogramming activity induced by RAS expression, we observed that mixed cultures of cells expressing either RAS or OSKM, and cellular reprogramming of cells growing in the presence of CM from RAS-expressing cells, both showed increased efficiency of reprogramming.
Even though senescence was initially described as a barrier for cell reprogramming in vitro (Banito and Gil, 2010), OSKM expression in vivo leads to the activation of senescence, and secreted factors from these damaged cells contribute to the dedifferentiation process experienced by neighboring cells culminating in cellular reprogramming (Chiche et al., 2017, Mosteiro et al., 2016). The dazzling observation of cellular reprogramming in vivo implies that tissues present niches that are permissive to complete dedifferentiation, allowing the reversion of adult somatic cells back to an embryonic pluripotent state. However, not all tissues seem to be capable of nurturing this complete dedifferentiation, despite verifiable expression of OSKM (Abad et al., 2013). Interestingly, tissue injury such as bleomycin-induced fibrosis in the lung or cardiotoxin-induced muscle damage, renders resistant tissues susceptible to OSKM-induced reprogramming (Chiche et al., 2017, Mosteiro et al., 2016). Given our results showing increased efficiency of reprogramming after RAS oncogene expression in vitro, we decided to test the effect of oncogenic activation in vivo on the putative reprogramming of these same tissues. Interestingly, using two different mouse models, we observed that expression of OSKM after oncogene activation in muscle and lung results in the appearance of several cells positive for NANOG and OCT4, pluripotency markers that are not detected in these tissues when OSKM are expressed alone.
RAS is known to confer on cells a large degree of phenotypic plasticity that predisposes them to neoplastic transformation and acquisition of stem cell characteristics (Chaffer et al., 2011, Ischenko et al., 2013). Our data support this idea of an increased plasticity induced by RAS underlying either transformation or reprogramming, depending on the combined action of cooperating genes. In keeping with this notion, although combined expression of active RAS and OSKM leads to increased efficiency of reprogramming, expression of RAS in the context of full neoplastic transformation does not have a positive impact on reprogramming and can even have a detrimental effect on the process. Cell identity markers Thy1 and Fn1 show reduced levels after RAS expression during transformation, but in this case pluripotency markers are not upregulated. Interestingly, reversion of the transformed phenotype by switching off RAS restores the capacity of OSKM-expressing cells to dedifferentiate and generate iPSCs. This effect leads in vivo to the formation of malignant tumors when RAS is constantly expressed and to teratomas when it is silenced, clearly showing that neoplastic transformation and reprogramming to pluripotency are incompatible cellular fates.
In summary, cellular reprogramming offers an excellent opportunity to better understand processes involving cellular plasticity and cell identity change such as cancer. Applying induced dedifferentiation by OSKM expression to cells expressing oncogenic RAS allows the discrimination of two incompatible cellular fates, pluripotency and transformation, depending on the cellular context. This implies that full transformation imposes new barriers to reprogramming that might be fundamental for tumor cell identity. New findings in this direction could lead us to novel anticancer therapies, by allowing us to redirect cancer cells to a less malignant state through targeting cancer cell identity.
Experimental Procedures
Animals
All animal procedures were approved by University of Santiago de Compostela Bioethics Committee in compliance with Principles of Laboratory Animal Care of national laws. Transgenic i4F animals and KrasV12-KI were described previously (Abad et al., 2013, Guerra et al., 2003).
Reprogramming to iPSCs
For reprogramming to iPSCs, we used three different systems: i4F MEFs were cultured in the presence of doxycycline at a concentration of 1 μg/mL, and medium was replaced every 48 h during 14 days; WT MEFs were transduced with lentiviral plasmids Tet-O-FUW-OSKM and FUW-M2rtTA and cultured with doxycycline as above; or WT MEFs were transduced with retroviral plasmids: pMXs-Oct4, pMXs-Sox2, pMXs-Klf4, and pMXs-c-Myc (see Supplemental Experimental Procedures for plasmid details), and cultured in iPSC medium.
After 14 days, cell cultures were fixed with 4% paraformaldehyde and the AP activity was detected with Vector Red Substrate Kit (Vector Labs) or Alkaline Phosphatase Blue Membrane Substrate Solution (Sigma-Aldrich) according to the manufacturer's instructions. Plates were scanned with Epson Perfection V550 Photo (Epson) and quantification of positive colonies was performed using ImageJ software.
Statistical Analyses
The statistical significance of the data obtained was analyzed using the two-tailed Student's t test: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; n.s., not significant.
For protein and RNA analysis and extra details see Supplemental Experimental Procedures in the Supplemental Information.
Author Contributions
A.F. performed and designed most of the experiments, interpreted results and wrote the manuscript. P.P., S.D.S.-A., F.T.-M., J.M.V., and P.P.-R. helped with experiments. P.G. and M.G. performed immunohistochemistry analysis. H.L. helped with experiments and provided reagents. T.G.-C. helped with immunohistochemistry and histological analysis. M.G.-B. helped with experiments and provided reagents. A.V. and M.C. coordinated the work, designed the experiments, interpreted results and wrote the manuscript.
Acknowledgments
We acknowledge Manuel Serrano for i4F mice, Carmen Guerra and Mariano Barbacid for Kras-KI mice, Matthias Drosten, Frank Essmann, and Xose R Bustelo for plasmids, and Andrew Koff for critical reading of the manuscript. A.F. is an FPU predoctoral fellow from MECD; P.P. and J.M.V. are predoctoral fellows from Xunta de Galicia; F.T.-M. is a postdoctoral fellow from CONACYT (cvu 268632). M.C. is a “Miguel Servet II” investigator (CPII16/00015). Work in the laboratory of M.C. is funded by an ISCIII and EU-FEDER grant (PI14/00554). Work in the laboratory of A.V. is funded by Xunta de Galicia (ED431B 2016) and MINECO (MAT2017-89678-R; cofinanced with FEDER Funds).
Published: May 2, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.04.006.
Contributor Information
Anxo Vidal, Email: anxo.vidal@usc.es.
Manuel Collado, Email: manuel.collado.rodriguez@sergas.es.
Supplemental Information
References
- Abad M., Mosteiro L., Pantoja C., Cañamero M., Rayon T., Ors I., Graña O., Megías D., Domínguez O., Martínez D. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 2013;502:340–345. doi: 10.1038/nature12586. [DOI] [PubMed] [Google Scholar]
- Banito A., Gil J. Induced pluripotent stem cells and senescence: learning the biology to improve the technology. EMBO Rep. 2010;11:353–359. doi: 10.1038/embor.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I., Thomson M.W., Carey V.J., Ge R., Bell G.W., Regev A., Weinberg R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner J.E., Hnisz D., Young R.A. Transcriptional addiction in cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady J.J., Li M., Suthram S., Jiang H., Wong W.H., Blau H.M. Early role for IL-6 signalling during generation of induced pluripotent stem cells revealed by heterokaryon RNA-seq. Nat. Cell Biol. 2013;15:1244–1252. doi: 10.1038/ncb2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone G., Borrello M.G., Molla A., Rizzetti M.G., Pierotti M.A., Della Porta G., Parmiani G. Activation of ras oncogenes and expression of tumor-specific transplantation antigens in methylcholanthrene-induced murine fibrosarcomas. Int. J. Cancer. 1991;47:619–625. doi: 10.1002/ijc.2910470423. [DOI] [PubMed] [Google Scholar]
- Chaffer C.L., Brueckmann I., Scheel C., Kaestli A.J., Wiggins P.A., Rodrigues L.O., Brooks M., Reinhardt F., Su Y., Polyak K. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl. Acad. Sci. U S A. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiche A., Le Roux I., von Joest M., Sakai H., Aguín S.B., Cazin C., Salam R., Fiette L., Alegria O., Flamant P. Injury-induced senescence enables in vivo reprogramming in skeletal muscle. Cell Stem Cell. 2017;20:407–414.e4. doi: 10.1016/j.stem.2016.11.020. [DOI] [PubMed] [Google Scholar]
- Collado M., Serrano M. The power and the promise of oncogene-induced senescence markers. Nat. Rev. Cancer. 2006;6:472–476. doi: 10.1038/nrc1884. [DOI] [PubMed] [Google Scholar]
- Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eva A., Aaronson S.A. Frequent activation of c-kis as a transforming gene in fibrosarcomas induced by methylcholanthrene. Science. 1983;220:955–956. doi: 10.1126/science.6302839. [DOI] [PubMed] [Google Scholar]
- Goding C.R., Pei D., Lu X. Cancer: pathological nuclear reprogramming? Nat. Rev. Cancer. 2014;14:568–573. doi: 10.1038/nrc3781. [DOI] [PubMed] [Google Scholar]
- Guerra C., Mijimolle N., Dhawahir A., Dubus P., Barradas M., Serrano M., Campuzano V., Barbacid M. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischenko I., Zhi J., Moll U.M., Nemajerova A., Petrenko O. Direct reprogramming by oncogenic Ras and Myc. Proc. Natl. Acad. Sci. U S A. 2013;110:3937–3942. doi: 10.1073/pnas.1219592110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareta M.S., Gorges L.L., Hafeez S., Benayoun B.A., Marro S., Zmoos A.-F., Cecchini M.J., Spacek D., Batista L.F.Z., O’Brien M. Inhibition of pluripotency networks by the Rb tumor suppressor restricts reprogramming and tumorigenesis. Cell Stem Cell. 2015;16:39–50. doi: 10.1016/j.stem.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T., Suzuki J., Wang Y.V., Menendez S., Morera L.B., Raya A., Wahl G.M., Belmonte J.C.I. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Marinov G.K., Pepke S., Singer Z.S., He P., Williams B., Schroth G.P., Elowitz M.B., Wold B.J. Single-cell transcriptome analysis reveals dynamic changes in lncRNA expression during reprogramming. Cell Stem Cell. 2015;16:88–101. doi: 10.1016/j.stem.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H., Parada L.F., Weinberg R.A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Li H., Collado M., Villasante A., Strati K., Ortega S., Cañamero M., Blasco M.A., Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Collado M., Villasante A., Matheu A., Lynch C.J., Cañamero M., Rizzoti K., Carneiro C., Martínez G., Vidal A. p27Kip1 directly represses Sox2 during embryonic stem cell differentiation. Cell Stem Cell. 2012;11:845–852. doi: 10.1016/j.stem.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M., Barbacid M. Timeline: RAS oncogenes: the first 30 years. Nat. Rev. Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- Marión R.M., Strati K., Li H., Murga M., Blanco R., Ortega S., Fernandez-Capetillo O., Serrano M., Blasco M.A. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosteiro L., Pantoja C., Alcazar N., Marión R.M., Chondronasiou D., Rovira M., Fernandez-Marcos P.J., Muñoz-Martin M., Blanco-Aparicio C., Pastor J. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science. 2016;354:aaf4445. doi: 10.1126/science.aaf4445. [DOI] [PubMed] [Google Scholar]
- Mosteiro L., Pantoja C., de Martino A., Serrano M. Senescence promotes in vivo reprogramming through p16 INK 4a and IL-6. Aging Cell. 2018;17:e12711. doi: 10.1111/acel.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Onder T.T., Daley G.Q. New lessons learned from disease modeling with induced pluripotent stem cells. Curr. Opin. Genet. Dev. 2012;22:500–508. doi: 10.1016/j.gde.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetrou E.P. Patient-derived induced pluripotent stem cells in cancer research and precision oncology. Nat. Med. 2016;22:1392–1401. doi: 10.1038/nm.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Mejia V., Fraga M.F., Menendez P. iPSCs from cancer cells: challenges and opportunities. Trends Mol. Med. 2012;18:245–247. doi: 10.1016/j.molmed.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Ruley H.E. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature. 1983;304:602–606. doi: 10.1038/304602a0. [DOI] [PubMed] [Google Scholar]
- Serrano M., Lin A.W., McCurrach M.E., Beach D., Lowe S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016;17:183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- Utikal J., Polo J.M., Stadtfeld M., Maherali N., Kulalert W., Walsh R.M., Khalil A., Rheinwald J.G., Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilas J.M., Ferreirós A., Carneiro C., Morey L., Da Silva-Álvarez S., Fernandes T., Abad M., Di Croce L., García-Caballero T., Serrano M. Transcriptional regulation of Sox2 by the retinoblastoma family of pocket proteins. Oncotarget. 2015;6:2992–3002. doi: 10.18632/oncotarget.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Shimokado K., Asahara T., Dohi K., Niwa O. Analysis of the c-myc, K-ras and p53 genes in methylcholanthrene induced mouse sarcomas. Jpn. J. Cancer Res. 1999;90:40–47. doi: 10.1111/j.1349-7006.1999.tb00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M., Meissner A., Cassady J.P., Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.