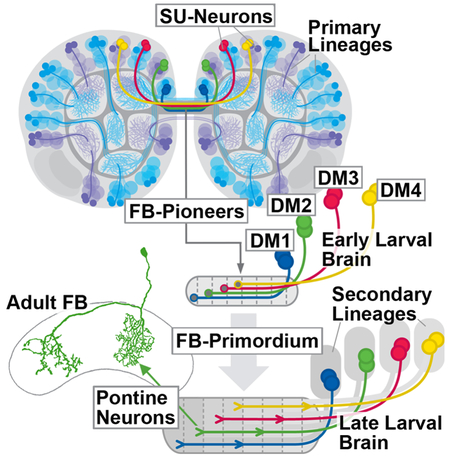
Summary
Serial electron microscopic analysis shows that the Drosophila brain at hatching possesses a large fraction of developmentally arrested neurons with a small soma, heterochromatin-rich nucleus, and unbranched axon lacking synapses. We digitally reconstructed all 802 “small undifferentiated” (SU) neurons and assigned them to the known brain lineages. By establishing the coordinates and reconstructing trajectories of the SU neuron tracts we provide a framework of landmarks for the ongoing analyses of the L1 brain circuitry. To address the later fate of SU neurons we focused on the 54 SU neurons belonging to the DM1–4 lineages, which generate all columnar neurons of the central complex. Regarding their topologically ordered projection pattern these neurons form an embryonic nucleus of the fan-shaped body (“FB pioneers”). FB pioneers survive into the adult stage where they develop into a specific class of bicolumnar elements, the pontine neurons. Later born, unicolumnar DM1–4 neurons fasciculate with the FB pioneers. Selective ablation of the FB pioneers altered the architecture of the larval FB primordium,but did not result in gross abnormalities of the trajectories of unicolumnar neurons, indicating that axonal pathfinding of the two systems may be controlled independently. Our comprehensive spatial and developmental analysis of the SU neurons adds to our understanding of the establishment of neuronal circuitry.
Keywords: Drosophila, brain, central complex, circuitry, serial electron microscopy, lineage, development
Graphical Abstract
In Brief:
Andrade et al. analyze the large set of developmentally arrested Drosophila larval brain neurons lacking a neurite tree and synapses (SU neurons) that form part of almost all larval brain lineages. SU neurons of lineages DM1-DM4 pioneer the architecture of the central complex where they differentiate into the distinct class of pontine neurons.
Introduction
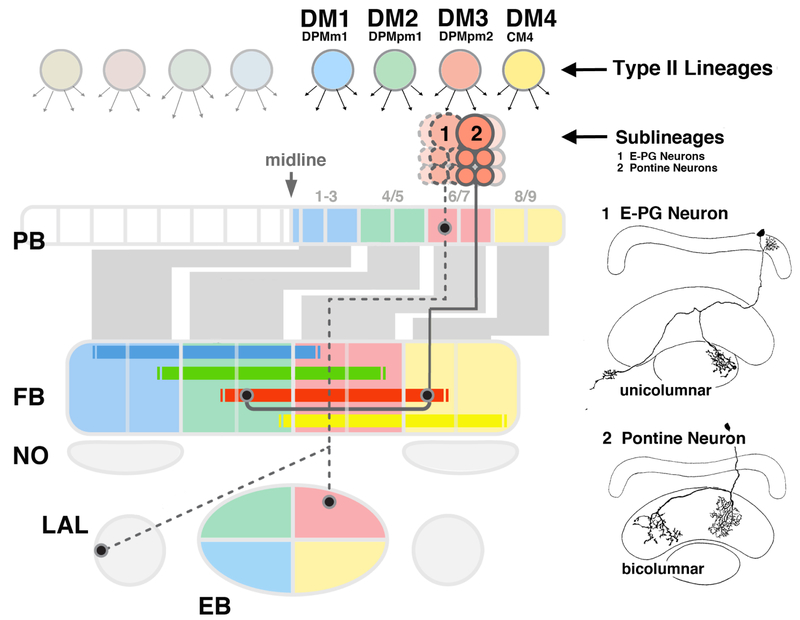
The central complex (CX) of the insect brain plays an important role in a variety of different behaviors, including the fine control of motor movement and spatial orientation [1–7]. The CX is comprised of four major compartments, the ellipsoid body (EB), fan-shaped body (FB) with noduli (NO), and protocerebral bridge (PB; [8]; Figure 1). CX circuitry is dominated by an orthogonal scaffold of transversally oriented (“tangential”) widefield neurons, and longitudinally oriented columnar small field neurons. Tangential neurons provide input to the CX from other brain areas. Best understood among these input neurons are the TL-neurons in locust [9] and their Drosophila counterparts, the R-neurons, that conduct retinotopically ordered visual information to the ellipsoid body [10, 11]. Columnar neurons, which interconnect the different CX neuropils along the antero-posterior axis, are characterized by highly localized dendritic and axonal endings in narrow volumes (“columns”) of the respective compartments. Most classes of columnar neurons, within a given CX neuropil, are confined to a single column (“unicolumnar neurons”; Figure 1). Projections are characterized by a strict homotopic order, whereby columns within the lateral half of the PB are connected to columns of the ipsilateral PB and EB, and medial PB columns project to the contralateral FB and EB (Figure 1). One class of columnar neurons, the so-called pontine neurons of the fan-shaped body [8], behave differently. Their projection is restricted to the FB, interconnecting two FB columns located on either side of the midline (Figure 1).
Figure 1.
Lineages DM1-DM4 form two types of columnar neurons. The compartments of the central and lateral accessory complex are schematically shown in a dorsal view (PB: protocerebral bridge; FB: fan-shaped body; NO: noduli; EB: ellipsoid body; LAL: lateral accessory lobe). Four bilateral pairs of lineages, DM1-DM4, located in the posterior brain cortex (top), generate the columnar neurons whose axons (grey bars and lines oriented along the vertical axis; filled circles symbolize terminal arborizations) interconnect the compartments of the central complex in a strict topographical order. The location of DM1–4 cell bodies within the cortex is reflected in the position at which their corresponding tracts enter and terminate within the central complex (20–23), as indicated by the color code. DM1 axons (blue) enter through the medial segments of the PB (1–3, after [34]), followed by DM2 (green, PB segments 4–5), DM3 (red, segments 6–7), and DM4 (yellow, segments 8–9). Each DM lineages generates multiple sublineages. These fall into two main types, unicolumnar neurons (light colors) and pontine neurons (saturated colors). Unicolumnar neurons (representing the large majority of DM neurons) interconnect compartments of the central complex along the anterior-posterior axis. They subdivide the PB, FB, and EB into four quadrants, as indicated by the coloring. For example, DM4 interconnects the lateral quarter of the PB (segments 8–9) with the lateral half of the ipsilateral FB, and the ventral half of the ipsilateral EB. This is followed by DM3, that projects from PB segments 6–7 to the medial half of the ipsilateral FB and dorsal half of the ipsilateral EB. DM2 and DM1 neurons interconnect the medial PB with the contralateral PB and FB, respectively, as indicated. The projection pattern of one class of unicolumnar neurons, E-PG (nomenclature according to [46]; “1” at top of figure), is shown representatively for lineage DM3; this neuron type is illustrated as a line drawing at the upper right of the figure (after [11], with permission). The second main type of neurons generated from DM1–4 are the pontine neurons (“2”, solid line). Axons of pontine neurons enter the central complex alongside the unicolumnar neurons, but pass through the PB without branching (see line drawing at lower right of figure, after [11], with permission). Projections of pontine neurons are confined to the FB, where they interconnect two columns along the transverse axis. The topology of this connectivity, as elaborated by Hanesch et al. [11], is indicated by horizontal bars rendered in saturated colors.
Developmental studies provide a valuable approach to unravel the circuitry of the brain, including the central complex. A hallmark of the Drosophila brain is its composition of invariant neuronal and glial lineages, originating from stem cells (neuroblasts) that appear in the early embryo. Embryonic neuroblasts express specific combinations of regulatory genes, which are thought to provide each lineage with the information needed to shape the connectivity of its neurons [12]. As a result, lineages become structural modules: Neurons of the same lineage generally project together in one or two fiber tracts, and form synapses in specific, spatially restricted brain compartments. For example, the R-neurons of the ellipsoid body are derived from one lineage, DALv2 (also called EBa1; [11, 13–16]). The columnar neurons of the CX are produced by four pairs of lineages located in the dorso-medial brain, called DM1/DPMm1, DM2/DPMpm1, DM3/DPMpm2, DM4/CM4 (called DM1-DM4 henceforward; [15–17]). The spatial pattern of these lineages is reflected in the position at which their corresponding tracts enter and terminate within the CX. In this manner, the four lineages subdivide the CX neuropils into four evenly sized quadrants, as illustrated in Figure 1.
The brain of Drosophila and other holometabolous insects arises in two distinct phases. During the first phase, neuroblasts of the embryo produce a relatively small set of primary neurons which differentiate and form the larval brain. Most neuroblasts then enter a dormant phase that lasts towards the end of the first larval instar. Subsequently they reactivate and produce secondary, adult specific neurons. These cells form axon bundles that form a “blue print” of the later connections established within the adult brain [16]. In general, primary and secondary neurons of a given lineage show fundamental structural similarities, whereby projections of secondary neurons follow those of earlier formed primary neurons [13]. Remarkably, the central complex, as defined anatomically for the adult, is the one major set of compartments of the fly brain that lack a larval counterpart. Thus, the large majority of tangential and columnar neurons with their highly ordered connections outlined above are secondary neurons born in the larva [21], even though the four DM lineages have been distinguished in the embryo [18–20]. This prompts the question of what guidance mechanisms control CX connectivity, and what part primary neurons play during this process.
Previous work [21] described for the late larval stage a set of embryonically born (i.e., primary) neurons belonging to the DM1–4 lineages. These neurons, visualized by the expression of R45F08-Gal4 [22], form a commissural tract that becomes incorporated into the fan-shaped body. Along with the emerging tracts and filopodia extended by secondary neurons of DM1–4 the R45F08-Gal4-positive neurons form a “fan-shaped body primordium” (prFB; [21, 23]). In the present paper, we undertook a detailed analysis of the structure, differentiative fate, and developmental role of the primary neurons that form the fan-shaped body primordium, using serial electron microscopy of the early larval brain in combination with confocal analysis of all stages covering early larva to adult. We show that the neurons of the fan-shaped body primordium, that we called fan-shaped body pioneers (FB pioneers), form part of a much larger population of early larval brain neurons that are arrested in development, projecting a simple, thin, unbranched process lacking synapses into the neuropil. Virtually every brain lineage possesses a complement of the small undifferentiated (SU) neurons. SU neurons differentiate in the late larva and pupa and give rise to distinct adult neuron populations; FB pioneers produce the pontine neurons of the fan-shaped body. Later born secondary neurons of the DM1–4 lineages, destined to form the various classes of unicolumnar neurons of the central complex, fasciculate with the FB pioneers on their pathway towards and across the midline. However, selective ablation of FB pioneers did not result in gross abnormalities of the trajectories of unicolumnar neurons, suggesting that the initial axonal pathfinding of the two system of columnar neurons may be controlled independently.
Results
Embryonically born neurons arrested prior to terminal differentiation prefigure adult axon tracts
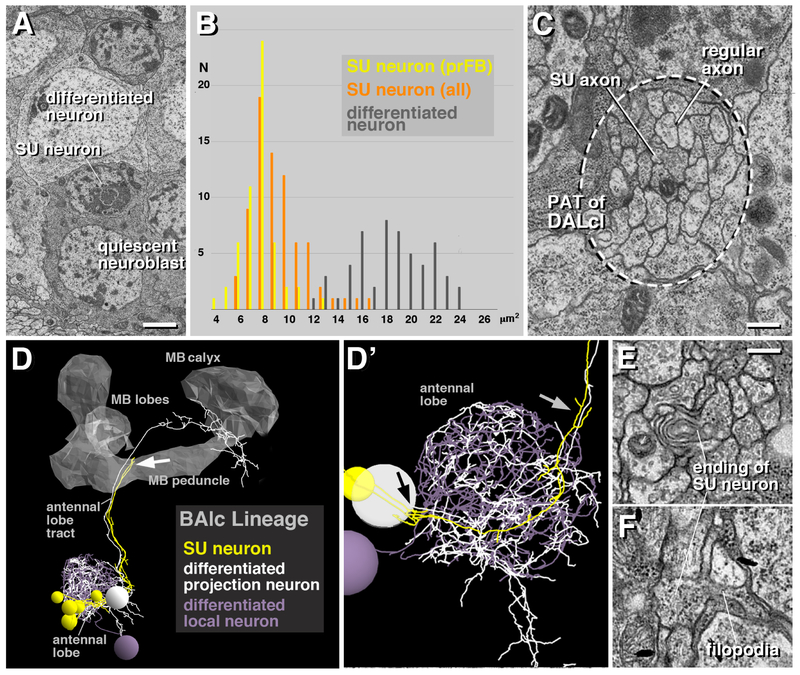
Four types of neuronal cell bodies can be distinguished ultrastructurally in the early larval brain. Most frequent are differentiated primary neurons, which measure 12–24μm2 in cross sectional area and have a relatively voluminous and electron light cytoplasm (Figure 2A, B). Processes of primary neurons of one lineage collect into cohesive bundles, the primary axon tracts (PATs), before entering the neuropil ([13, 23]; Figure 2C). Primary axon tracts continue for variable distances in the neuropil, before splitting up into individual, branched axon trees forming synapses. The second type of cell body belongs to mitotically quiescent neuroblasts. Resembling differentiated neurons in size and shape, quiescent neuroblasts can be distinguished from these cells by an electron dense cytoplasm and chromatin rich nucleus (Figure 2A). Aside from quiescent neuroblasts, the first instar brain contains five actively dividing neuroblasts, four of them associated with the mushroom body (not shown). Active neuroblasts are much larger than quiescent ones and are located at the brain surface. Finally, aside from differentiated neurons and neuroblasts, we recognized a fourth type of cell body by its small size, scanty, electron dense cytoplasm, and heterochromatin-rich nucleus (Figure 2A, B). These small neurons, measuring 4–16μm2 in cross sectional area, extend fibers which are significantly thinner and more electron-dense than the axons of differentiated neurons (Figure 2C), and which stay together as a tight bundle, without branching or forming synaptic contacts, as shown for lineage BAlc in Figure 2D, D′. Dense axons end abruptly with club-shaped endings, often producing short filopodia (Figure 2E, F). We interpret the small, heterochromatin-rich neurons as late born neurons which fail to differentiate in the embryo, and therefore call them “small undifferentiated (SU)” neurons in the following.
Figure 2.
Structural features of small undifferentiated (SU) neurons in the early larval brain. (A) Electron micrograph showing differentiated neuron (top), SU neuron (middle) and quiescent neuroblast (bottom). (B) Histogram showing size distribution of differentiated neurons (grey; number counted =50) and SU neurons (orange: all SU neurons of right brain hemisphere; n =405; yellow: SU neurons forming fan-shaped body primordium (prFB); n =54). Units on horizontal axis are in μm2 and represent area of cut surface of cell body. (C) Electron micrograph of cortex-neuropil boundary, showing bundle of axons (surrounded by hatched line) formed by the axons of the two DALcl lineages as they enter the neuropil. Within this bundle, axons of small undifferentiated neurons (“SU axon”) from a coherent contingent of thin, electron dense fibers; they are surrounded by thicker and typically more electron-lucent axons of differentiated neurons (“regular axon”). (D, D′) 3D digital model of part of one primary lineage (BAlc), illustrating structural aspects of SU neurons. BAlc includes local interneurons and projection neurons connecting the antennal lobe with higher brain centers [47]. Shown are one differentiated projection neuron (mPN iACT A1, white) and one differentiated local interneuron (Broad D1: grey) whose dendritic arborizations outline the antennal lobe; the axon of the projection neuron follows the antennal lobe tract. Shown in yellow are the SU neurons which were identified for BAlc. They each form an unbranched axon that follows the axons of the differentiated neurons as they enter the antennal lobe neuropil (black arrow in D′) and extends into the antennal lobe tract (grey arrow in D′) where they end approximately midway (white arrow in D). The model is presented in lateral view; anterior to the left, dorsal up. The mushroom body (MB) is outlined in grey for spatial reference. (E, F) Electron micrographs show club-shaped endings of SU axons, featuring membrane lamellae (E) and short filopodia (F). Bars: 1μm (A, E), 0.25μm (B, D, E), 5μm (F)
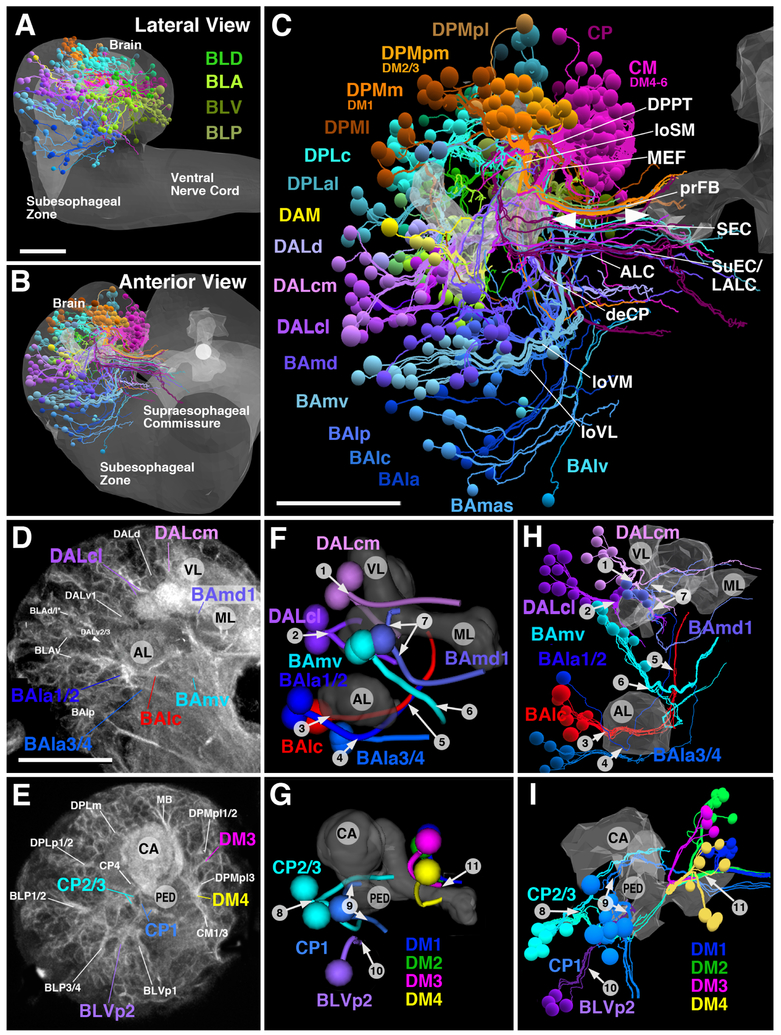
SU neurons are found scattered throughout the brain (Figure 3A–C) and amount to a number of 802, which corresponds to more than 25% of the overall number of primary neurons (DataS1A; Table S1 Related to Figure 3). Axons of SU neurons are grouped into tight bundles, each bundle surrounded by the thicker and less electron-dense axons of differentiated neurons (Figure 2C). Based on their characteristic position and trajectory, we could identify these bundles with the lineage-associated primary axon tracts (PATs) defined for the L1 brain in a previous paper [23] on the basis of confocal microscopic analysis. Once digitally reconstructed and displayed in the 3D viewer, fiber bundles of SU neurons (Figure 3H, I) can be compared with those in the light microscopic L1 brain map (Figure 3D–G), and thus identified as belonging to specific lineages, or pairs/small sets of lineages. The L1 lineage map features more than 60 (excluding the subesophageal zone and ventral nerve cord) discrete PAT fiber bundles. All of the light microscopically identified fiber bundles, with the possible exception of that of DPLpv, could be identified in the serial EM stack (Data S1A Related to Figure 3). Note, as examples shown in Figure 3F–I, the characteristic entry points and trajectories of a set of anterior lineages (BAla1/2, BAla3/4, BAlc, BAmd1, BAmv1/2, DALcl1/2, DALcm1/2) and posterior lineages (CP1, CP2/3, BLVp2, DM1–4) visible in confocal images and the EM stack. The neuropil entry points and trajectories of SU neuronal tracts provide useful landmarks for the ongoing analyses of the L1 brain circuitry using the CATMAID L1 serial TEM dataset; both sets of data are presented graphically and numerically in Table S1 (Related to Figure 3) and Figures S1–S5 (Related to Figure 3).
Figure 3.
Pattern of SU neurons and primary axon tracts (PATs) in the early larval brain. (A-C) 3D digital model of first instar larval brain in lateral view (A; anterior to the left, dorsal up) and anterior view (B, C; dorsal up). Cell bodies and axons of all SU neurons of one hemisphere are shown as colored spheres and lines. Differential coloring indicates association of SU neurons with spatially contiguous groups of lineages; labels of lineages are shown in corresponding colors (A, B; for nomenclature of lineages and fascicles see [30]). In (A, B), outline of brain and ventral nerve cord is rendered in grey; in (C), outlines of mushroom body lobes, located in center of neuropil, are shaded, with tips of left and right medial lobe pointed out by arrowheads. Note that lineage-associated bundles of SU axons follow discrete fascicles identified for the brain throughout development. Among the examples made explicit are the BAmv axons that form the longitudinal ventromedial fascicle (loVM); axons of BAlp constitute the longitudinal ventro-lateral fascicle (loVL); DALd axons form the central protocerebral descending fascicle (deCP); CM lineages the medial equatorial fascicle (MEF); DPMpm the longitudinal superior-medial fascicle (loSM) and primordium of the fan-shaped body (prFB). (D-I) Primary lineage tracts imaged light microscopically can be identified with electron microscopically reconstructed SU axon tracts. (D) and (E) show representative frontal confocal section (D: anterior section at level of MB lobes; E: posterior section at level of MB calyx); primary neurons and their axon tracts are labeled by anti-Neuroglian (white). Primary axons converge at the cortex-neuropil boundary at invariant positions to form tracts with characteristic trajectories. A subset of tracts, reconstructed from a confocal stack of an anti-Neuroglian labeled brain hemisphere, is shown in panels (F) and (G). In these models, the mushroom body lobes (ML medial lobe; VL vertical lobe) and antennal lobe (AL) are shown for reference. Representative anterior lineages (BAla1/2, BAla3/4, BAlc, BAmd1, BAmv1/2, DALcl1/2, DALcm1/2) are shown in (F)(anterior view of one hemisphere, medial to the right); representative posterior lineages (CP1, CP2/3, BLVp2, DM1–4) appear in (G)(posterior view, medial to the right). Lineage-associated tracts are shown as colored pipes; spheres indicate location of cell bodies belonging to the tracts illustrated. Panels (H, I) show digital models extracted from the serial EM stack. As in (F, G), the mushroom body and antennal lobe is shown as reference. SU neurons of the same lineages presented in (F, G) are shown as colored lines; individual cell bodies appear as spheres. Tracts formed by SU neurons can be identified with light microscopically-defined lineage-associated tracts based on specific location of entry and trajectory, as pointed out by numbered arrows that indicate the following features: (1) Entry of DALcm lineage pair dorso-laterally of anterior peduncle; upper tract passes over peduncle and turns ventrally; lower tract projects anteriorly and then medially, passing in front of MB vertical lobe; (2) Entry of DALcl lineage pair laterally of anterior peduncle; upper tract passes over peduncle and turns ventrally; lower tract turns ventrally and medially, passing underneath peduncle; (3) Entry of BAlc at lateral surface of antennal lobe (AL); (4) Entry of lineage pairs BAla1/2 and BAla3/4 at posterior-ventral surface of antennal lobe; (5) Tracts of BAlc and BAla1 converge at posterior-medial surface of antennal lobe and project dorsally, forming antennal lobe tract; (6) Entry of BAmv1/2 lineage pair medially of antennal lobe; tract turns posteriorly, forming longitudinal ventromedial fascicle (loVM); (7) Entry of BAmd1 anteriorly of MB medial lobe; tracts diverge, and upper tract passes over medial, joining upper DALcm tract on its medial turn; lower tract turns ventrally and then medially, forming antennal lobe commissure; (8) Entry of CP2/3 lineage pair laterally of MB calyx-peduncle junction; upper tract projects dorso-anteriorly, passing peduncle dorsally and forming oblique posterior fascicle; lower tract turns ventrally and then anteriorly, forming dorsal component of postero-lateral fascicle; (9) Entry of CP1 medially adjacent to CP2/3; upper tract follows CP2/3 axons in oblique posterior fascicle; lower tract projects ventrally and anteriorly, forming lateral equatorial fascicle; (10) Entry of BLVp2 ventrally of CP2/3; tract continues anteriorly, forming ventral component of postero-lateral fascicle; (11) convergence of DM1–4 forming the primordium of the fan-shaped body. See also Figures S1–S5, Table S1 for more detailed information of entry points. See also Data S1.
Bars: 20μm
Comparison between left and right brain hemisphere showed that the exact number of SU neurons accompanying lineages is relatively invariant (Data S1A Related to Figure 3). Most lineages are made up of 60–80% differentiated neuron, and 20–40% SU neurons (Data S1A Related to Figure 3). However, some lineages, notably those in the dorsal and dorso-medial brain, possess more than 50% of SU neurons (Data S1A Related to Figure 3). In terms of overall cable length, SU neurons amount to only a small fraction of the length of differentiated, which is the result of the absence of any branches in the former (Data S1B Related to Figure 3). Likewise, SU neurons possess no output synapses, and only very rarely input synapses (Data S1B Related to Figure 3).
Given our assumption that SU neurons are born late, one might surmise that they occupy a superficial position in the cortex. Thus, in the embryo, the apical (=superficial) versus basal (=deep) location of a neuron reflects its birthdate, since proliferating neuroblasts are always located at the surface. However, neuronal cell bodies reshuffle during the pronounced thinning of the cortex that occurs during the embryo-larva transition [19], and the original position of a neuron is lost. This explains why measurements of the distance between cell body and neuropil surface for SU neurons, compared to differentiated neurons, for 10 representative lineages yielded no significant difference in apical vs basal position of SU neurons vs differentiated neurons (data not shown).
SU neurons of the four lineages, DM1-DM4, form a glia ensheathed fan-shaped body primordium in the first instar larva
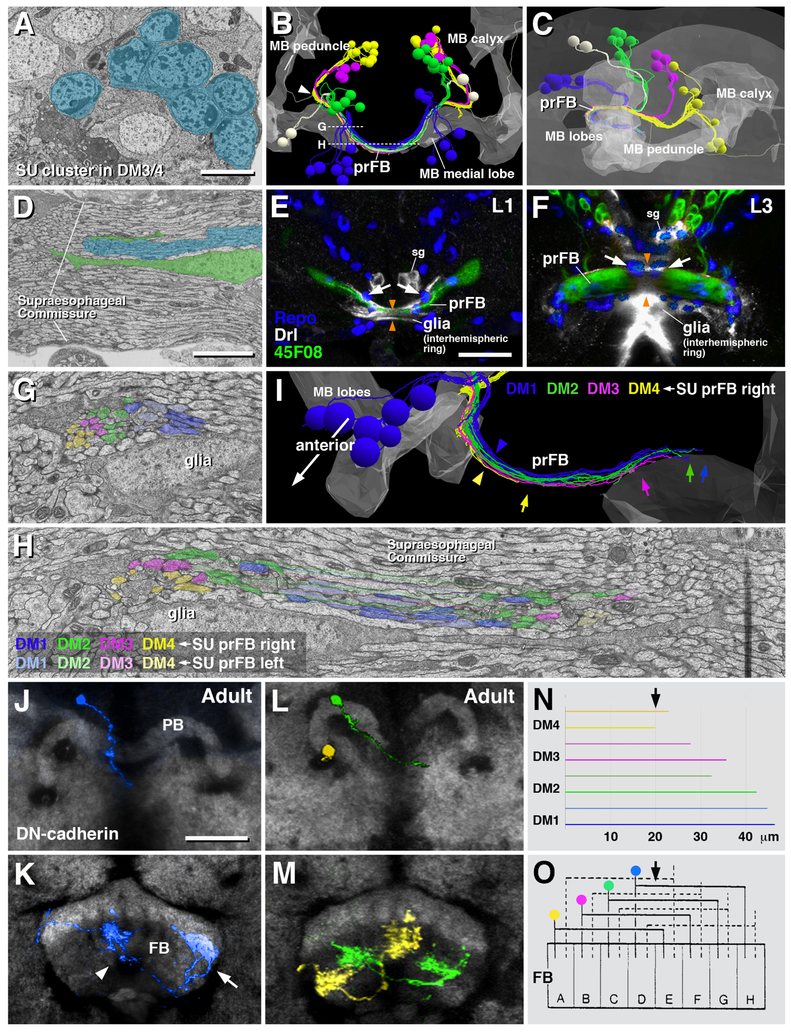
In the dorso-medial domain of the brain, which houses all lineages later giving rise to the columnar neurons of the central complex (DM1-DM4), SU neurons form large clusters of cells (Figure 4A). Based on axonal trajectory we were able to recognize the individual DM lineages and their associated SU neurons. DM1–4 are aligned along the brain midline, with DM1 occupying the most anterior, and DM4 the most posterior position (Figure 4B, C). For each of the DM lineages, SU neurons form several subpopulations with axons following different pathways (Figure 4D). One subpopulation per DM lineage projects axons that converge on a shared commissural bundle that crosses the brain midline in the center of the supraesophageal commissure, as shown in Figure 4B. This bundle stands out from surrounding tracts by its higher electron density, being composed almost entirely of thin SU axons (Figure 4D); furthermore, two paired glial cells ensheath the bundle on all sides (Figure 4D, G, H). In late larvae, the pointed insertion R45F08-Gal4 is expressed in small subpopulations of neurons of DM lineages whose axons form a glia-covered commissural structure that will enlarge into the fan-shaped body, and was therefore called fan-shaped body primordium (prFB; Figure 4F; [28]). In first instar larvae, R45F08-Gal4 highlights four paired clusters of neurons in the dorso-medial brain whose axons form a thin tract that, in regard to size, central location within the brain commissure, and glial enclosure corresponds to the structure identified electron microscopically (Figure 4E). We conclude that the fan-shaped body primordium is formed in the embryo by four paired sets of SU neurons of lineages DM1-DM4, which we will call “fan-shaped body pioneers” (FB pioneers) in the following.
Figure 4.
SU neurons of lineages DM1–4 form the primordium of the fan-shaped body (prFB) and give rise to pontine neurons. (A) Electron micrograph of dorso-medial brain cortex showing clusters of contiguous SU neurons (shaded in blue) associated with lineages DM1–4. (B, C) Digital 3D models of L1 brains (B: dorsal view; C: lateral view) showing SU neurons associated with DM1–4 rendered in different colors (DM1 blue; DM2 green; DM3 magenta; DM4 yellow). Outline of mushroom body (representing center of brain neuropil) is rendered in light gray. Only subsets of SU neurons contributing to the prFB are shown. Note that in addition to DM1–4, lineages DPMm2 and CP2 contribute three fibers (white) to the prFB. (D) Electron micrograph of brain midline, showing supraesophageal commissure. The prFB appears as a bundle of electron-dense axons embedded in fascicles of lighter axons belonging to differentiated neurons (shaded blue). Glial lamella (shaded green) flanks the prFB. Note conspicuous, medially located nucleus of glial cell (white arrow) right adjacent to the prFB. (E, F) Z-projections of confocal sections of first instar brain (E) and late third instar brain (F). Glial nuclei are labeled by anti-Repo (blue); glia surrounding supraesophageal commissure (=interhemispheric ring glia) is labeled by Drl-lacZ (white); neurons of prFB are marked by driver line R45F08-Gal4>UAS-mcd8GFP (green). Interhemispheric ring glia forms several channels containing individual commissural fascicles. One channel contains the R45F08-Gal4-positive axon bundle constituting the prFB [orange arrowheads in (E, F)]. This channel widens as prFB gains in volume towards late larval stages (F; arrowheads). Note pair of large glial nuclei (arrows) located near the midline, posteriorly adjacent to the prFB. These cells correspond in shape, size and position to the prFB-associated glial cells that appear in electron micrographs (see panels D, G). (G-I) Topographical order of DM1–4 SU axons in the fan-shaped body primordium. Panels (G, H) show electron micrographs of prFB sectioned at the two planes shown by hatched lines in panel (B). Sectioned profiles of FB pioneers were assigned to their cell bodies of origin and are shaded in colors following the same scheme used in the 3D digital models in (B, C). Axons belonging to a lineage form a coherent bundle within the prFB. Note that bundles of corresponding lineages of the left and right hemisphere project together (e.g., light blue profiles of left DM1 are closest to saturated blue profiles of right DM1). Panel (I) shows digital 3D model of FB pioneers rendered of right hemisphere in different colors; antero-medial view. Note relationship between lineage identity and axonal projection (axon position within prFB, length of axons) among FB pioneers. DM1 fibers are located dorsally (blue arrowhead) and project furthest in the contralateral hemisphere (blue arrow); DM4 is located ventrally (yellow arrowhead) and barely reaches the midline (yellow arrow). DM2 and DM3 fall in between these extremes. (J-M) Frontal confocal sections of central complex of adult brain, showing single cell clones of neurons descended from FB pioneers. (J, L) are sections at the level of the protocerebral bridge (PB), (K, M) at the level of the fan-shaped body (FB). Labeled neurons are color coded following the same scheme used in panels (G-I). (J, K) show a pontine neuron of lineage DM1 (blue), with a medially located cell body and axon entering at the medial PB (J), and terminal axons branching in an ipsilateral medial column (arrowhead in K) and a contralateral lateral column (arrow in K). (L, M) show a DM4 neuron (yellow) and DM2 neuron (green). (N) Histogram depicting average length of FB pioneer axons belonging to lineages DM1–4. Upper horizontal bar of each pair belongs to left hemispheric lineage, lower bar to corresponding right hemispheric lineage. The sharp medial turn of axon as it enters the prFB (white arrowhead in panel B) was taken as origin on x-axis; arrow at top demarcates brain midline. (O) Schematic of Hanesch et al. [11; with permission], showing fan-shaped body (FB) divided into eight columns (A-H; arrow at top indicates midline). Based on Golgi-preparations, Hanesch et al. [11] distinguished between four classes of bi-columnar pontine neurons, indicated by solid lines on left side, and hatched lines on right side. Our data indicate that the four classes described by Hanesch et al. [11] correspond to the four lineages of origin of the pontine neurons. Other abbreviations: sg secondary glia labeled by Drl-lacZ. Bars: 2μm (A, D); 10μm (E, F); 2μm (G, H); 25μm (J-M)
FB pioneers are topologically ordered and give rise to the pontine neurons of the fan-shaped body
FB pioneers exhibit a strict topological order with regard to the position and average length of axons (Figure 4G–I). The DM1 bundle enters anteriorly and dorsally, and remains at the dorsal edge of the prFB, followed by DM2, DM3 and DM4, which is located most ventrally (Figure 4I). DM4 axons are the shortest, most of them ending before reaching the midline (Figure 4H, I). Axons of DM1-DM3 cross the midline but extend across for various distances, in correlation to their lineage identity: DM1 axons cross furthest, followed by DM2 and DM3 (Figure 4H, I). Using the R45F08-Gal4 driver we followed the prFB neurons through larval and pupal development into the adult, and generated single cell MCFO clones to visualize their morphology as differentiated cells. All neurons forming the early larval prFB give rise to pontine neurons, recognizable by their regular bi-columnar organization within the fan-shaped body (Figure 4J–M). Labeled neurons match the four-fold symmetric scheme elaborated by Hanesch et al. [8] for pontine neurons, whereby neurons entering the FB most laterally (DM4; Yellow in Figure 4O) branch near their point of entry in the lateral column “A”, then cross the midline, and have their second arborization in a column right next to the midline (“E” in Figure 4O; from [8]). Neurons of DM1 (blue in Figure 4O), as well as DM2 and DM3 (green and magenta, respectively) have a similar structure, innervating staggered pairs of columns, with one column always ipsilaterally relative to the location of cell bodies, and one contralaterally (Figure 4O). Note that this adult pattern corresponds the length distribution of early larval SU axons of DM1-DM4: DM4 axons are shortest, barely reaching the midline; DM1 axons are longest (Figure 4N, O).
Later born columnar neurons of DM1-DM4 follow the trajectory prefigured by the FB pioneers
R45F08-Gal4-positive axons of FB pioneers can be followed throughout larval and pupal development into the adult stage. Until 48h after hatching, the prFB formed by these axons remains as a single axon bundle located in the center of the brain commissure (Figure 5A). At this stage, the secondary phase of neuron production has commenced [24]. DM1-DM4 form secondary axon tracts, labeled by anti-Neurotactin, that grow along the R45F08-Gal4-positive (and Neurotactin-negative) FB pioneers (Figure 5B, B′, C, C′).
Figure 5.
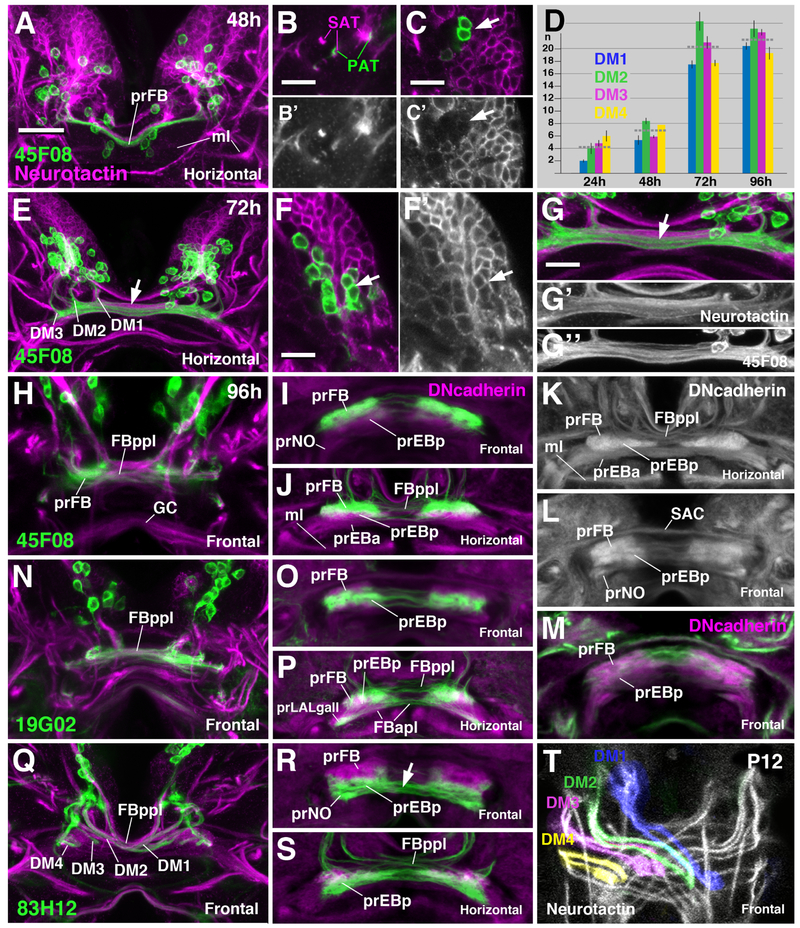
Growth and development of the prFB during the larval period. Panels show Z projections of frontal or horizontal confocal sections of larval brains at different stages (48h, 72h, 96h after hatching). Global labeling of secondary (larvally born) neurons with anti-Neurotactin (magenta in A-C, E-G, H, N, Q; white in B′, C′, F′, G′, T) or neuropil with anti-DNcadherin (magenta in I, J, M, O, P, R, S; white in K, L). FB pioneers are labeled by R45F08-Gal4>mcd8UAS-GFP (green in A-C, E-J). (A-C) At 48h (second larval instar) FB pioneers form a single bundle, as in freshly hatched first instar (see Figure 4E). The outgrowing secondary axons tracts (SATs) of DM1–4 (magenta in B) fasciculate with FB pioneers (green in B). At this stage, R45F08-Gal4 expression is still confined to Neurotactin-negative primary FB pioneers (C, C′). (D) Average numbers and standard deviations of R45F08-Gal4-positive DM1–4 neurons between 24 and 96h after hatching (n=5). Horizontal hatched lines give average for all four DM lineages. (E-G′′) At 72h after hatching (early third instar) the FB pioneer axons have split into several commissural bundles (arrow in E, G). Gaps between these bundles are filled with Neurotactin-positive secondary fibers formed by lineages DM1–4 (G-G′′). Numerous Neurotactin-positive secondary neurons have joined the set of R45F08-Gal4-positive FB pioneers (arrow in F, F′). (H-L) At 96h after hatching (late third instar) secondary DM1–4 axons have increased in number and form a system of crossing bundles, the posterior plexus of the fan-shaped body (FBppl in H, J, K). Both R45F08-Gal4-positive FB pioneers and secondary axons display tufts of filopodia which appear as DN-cadherin-positive domains (prFB, prEBp, prEBa in panels I-L). (M-P) Labeling of secondary E-PG neuron precursors (nomenclature after [46]) by R19G02-Gal4 driver. Terminal filopodia of these neurons are found in ventral prFB, prEBp and prLALgall (see also [35]). (Q-S) R83H12-Gal4 is expressed in a different subset of secondary neuronal precursors, mainly PB-FN and P-EN (nomenclature after [46]). Note in (Q) chiasmatic architecture of projection of labeled neurons towards the fan-shaped body, with all four bundles turning medially, but only DM1 and DM2 crossing towards contralaterally, and DM3/4 remaining ipsilaterally. This chiasmatic projection is typical for all unicolumnar neurons, as demonstrated in panel (T) (12h after puparium formation), where anti-Neurotactin (white) globally labels massive bundles of secondary axons of DM1–4. Bar: 25μm (for all panels except B-C′, F-G′′); 10μm (for B-C′, F-G′′). See also Figure S6.
At later larval stages, the number of neurons labeled by R45F08-Gal4 has increased over that counted for the early larva (Figure 5D, E), and many of these newly R45F08-expressing neurons form part of the Neurotactin-positive clusters of secondary neurons (Figure 5F–G′). In addition, a notable change in the structure of the prFB has taken place, whereby the initially coherent tract of primary FB pioneers has split into several thinner bundles (Figure 5G, G′′). The spaces in between these is filled by the secondary, Neurotactin-positive axons of DM1–4 (Figure 5G′), which give rise to the unicolumnar neurons of the central complex. Several markers for the major classes of unicolumnar neurons exist, among them 9D11-Gal4 (an insertion in the earmuff gene; [21, 22]), 19G02 [22, 25], and 83H12 ([11, 22, 25]; this study). 19G02 is expressed in several classes of neurons connecting PB, dorsal FB, and EB with the lateral accessory lobe (LAL; Figure S6B. Related to Figure 5); 83H12 is expressed in neurons connecting the PB, ventral FB, EB and the NO (Figure S6C. Related to Figure 5). In conjunction with R45F08-Gal4, expressed in pontine neurons (Figure S6A. Related to Figure 5) these markers allowed us to discern the distribution of pontine versus unicolumnar neurons in the prFB of the late larva, as presented below.
In the late larva, the fan-shaped body primordium occupies a sizeable volume as a result of the ingrowth of secondary axons, in conjunction with the formation of tufts of filopodia that prefigure the neuropil of the fan-shaped body and noduli. Primary and secondary axon tracts of DM1-DM4 form a plexus of fiber bundles at the posterior surface of the fan-shaped body primordium [posterior plexus of the fan-shaped body (FBppl) in Figure 5H, J, K]. Within this plexus, the R45F08-Gal4-positive pontine neuron precursors appear as thin axon bundles crossing the midline (Figure 5H, J). Also secondary tracts of uni-glomerular neurons, including the 83H12-positive P-FN and P-EN neurons and 19G02-positive E-PG neurons, cross in the posterior plexus, forming a system of chiasms that establish the characteristic, modular connectivity of the fan-shaped body and ellipsoid body (Figure 5N, P, Q, S, T). Tracts of DM1 and DM2 cross the midline to terminate in the lateral and medial half of the contralateral prFB, respectively; those of DM3 and DM4 remain ipsilateral, targeting the medial and lateral half of the ipsilateral prFB, respectively (Figure 5Q, T; see also [18]).
In addition to the posterior plexus of axons, the late larval fan-shaped body contains voluminous tufts of filopodia, visible by their high expression levels of DN-cadherin [21, 26, 27]. These tufts outline three bar-shaped neuropil domains (Figure 5K, L, M). The domain located dorsally and posteriorly will give rise to the neuropil of the fan-shaped body proper (prFB). Accordingly, filopodia of R45F08-Gal4-positive precursors of pontine neurons are restricted to this dorsal compartment (Figure 5I, J). Located further anteriorly and ventrally is the primordium of the posterior ellipsoid body (prEBp, labeled by 19G02 in Figure 5O, P); the primordia of the noduli (prNO) form two ventrally-directed appendages of the prFB expressing 83H12 (Figure 5L, R, S).
Ablation of the FB pioneers
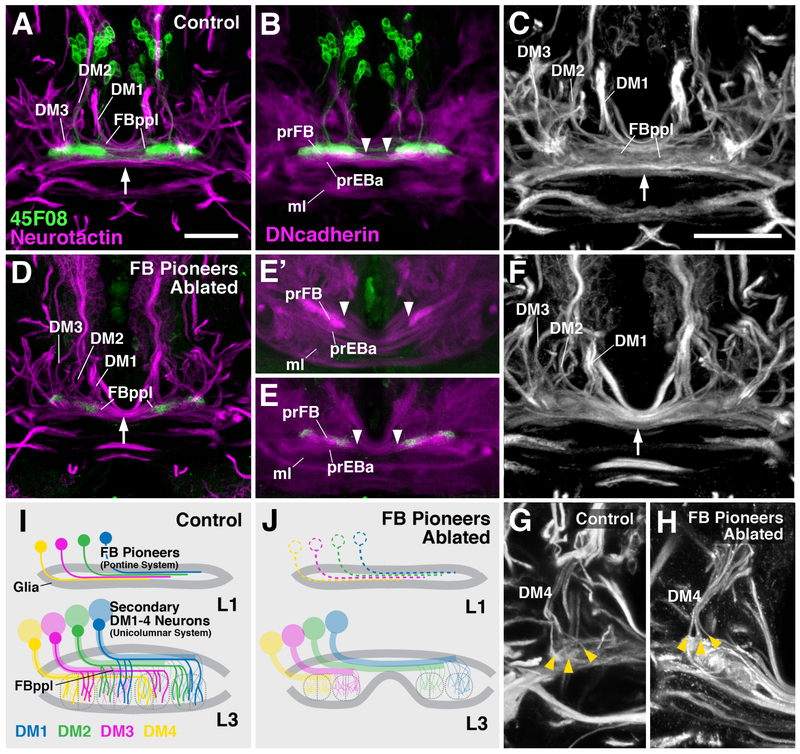
Given the early appearance of precursors of the pontine system, and its intimate spatial relationship with the subsequently generated unicolumnar neurons, we sought to establish the effect of ablating the former by using the R45F08-Gal4 construct to express UAS-hid;rpr in the FB pioneers. After activation of the proapoptotic construct between 24h and 48h after hatching (inactivation of temperature-sensitive Gal80ts by shifting to 29°C), larvae showed a high rate of lethality, which is likely due to expression of R45F08-Gal4 in other tissues (V.H., unpublished observation). No late pupa or adult flies survived under these circumstances, whereas non-temperature shifted controls were completely viable. This finding, in conjunction with previous results where the same UAS-hid;rpr construct was effective in completely ablating muscle cells [28], supported the conclusion that our experimental regimen efficiently killed the FB pioneers. However, we cannot exclude that a small number of FB pioneers might escaped UAS-hid;rpr-induced apoptosis. The late larval prFB of the surviving FB pioneer-ablated larvae showed a characteristic structural phenotype. All elements of the prFB described in the previous section could still be recognized; however, the DN-cadherin-rich primordia of fan-shaped body and posterior ellipsoid body/noduli were separated by a wider gap (arrowheads in Figure 6B, E, E′), compared to control (Figure 6A, B). In addition, the posterior plexus of the fan-shaped body, normally formed by a loose assembly of “wavy”, thin bundles (Figure 6A, C), appeared as thick, smooth cable of axons (Figure 6D, F). We conclude that FB pioneers, possibly by establishing an early connection between the left and right half of the prFB through their crossing axons, play a structural role in pulling these halves together into a united structure (Figure 6I, J). Aside from this function, there does not appear to be an effect on the formation and pathway choices of the later born secondary axons: these split into several sub bundles and projected into the prFB in a pattern that showed no gross abnormalities compared to the control (Figure 6G, H). This result suggests that axonal pathfinding of the two main systems of small field neurons of the central complex, i.e., the unicolumnar and pontine neurons, may be controlled independently.
Figure 6.
Structure of the prFB after ablation of FB pioneers. (A-C) Z-projections of horizontal confocal sections of wild-type late third instar brains, illustrating entry of lineages DM1–3, posterior plexus of fan-shaped body (FBppl), and filopodial tufts forming primordia of fan-shaped body neuropil (prFB) and ellipsoid body neuropil (prEBa). For a clear depiction of the FB pioneers, males lacking UAS-hid;rpr and tub-Gal80ts are shown. (D-F) Same views of late larval brain as in (A-C) after ablation of FB pioneers by activating UAS-hid;rpr from 24–48h with R45F08-Gal4. (E and E′) show two different representative specimens. Note that FBppl is reduced in diameter (arrow in D, F) compared to control (arrow in A, C). Also primordium of fan-shaped body neuropil (prFB in B, E, E′) is separated by wider gap in ablated specimens (arrowheads in B, E, E′). (G, H) Z-projection of horizontal confocal sections of one brain hemisphere of control (G) and ablated (H) specimen, showing normal branching pattern of secondary axon bundles of DM4 lineage (yellow arrowheads). The neurotactin and DNcadherin pattern of non-temperature shifted female controls (which contain UAS-hid;rpr and tub-Gal80ts), in which no ablation occured, were indistinguishable from A-C, G (not shown). (I, J) Schematic fan-shaped body primordium at first larval instar (L1) and late third instar (L3) in control (I) and ablated specimen (J). Bars: 25μm (A, B, D-E′); 10μm (C, F, G, H)
Discussion
Primary small undifferentiated neurons form a large neuron population of the early larval brain
Our analysis demonstrates that a large fraction of the neurons of the early Drosophila larval brain does not elaborate a branched neurite arbor and synaptic connections. This finding came as a surprise; it had been well established that the large number of neurons produced during the secondary, larval phase of neurogenesis remain undifferentiated until the onset of metamorphosis, resembling in many ways the SU neurons described here [16,29], but the same was not assumed for so many of the embryonically generated primary neurons. Previous studies had shown that in the thoracic ganglia, a subset of presumptive adult peptidergic neurons [30] and motor neurons [31] show a SU phenotype, extending a truncated axon into the peripheral nerve, but failing to form synaptic connections to the musculature. These embryonically born neurons, which transiently express the transcription factor Broad-Z3, differentiate along with the larvally born motor neurons and form dendritic and axonal branches and synapses in the pupa. Due to their delayed differentiation, typical for secondary neurons, Zhou et al. 2009 [31] term these embryonically born thoracic SU neurons “embryonically born secondary neurons”. To avoid confusion, we will stick to the convention that defines all neurons born during the embryonic period as primary neurons and call them “primary SU neurons”.
Drosophila SU neurons described in this paper exhibit structural characteristics that are similar to those described for neuronal precursors in the developing vertebrate brain. Thus, postmitotic neuronal precursors of the neocortex or hippocampus, while migrating along radial glia, are small, electron-dense cells with hetero-chromatin-rich nuclei and scant cytoplasm. Typically, they exhibit a bipolar shape, extending a leading and trailing process that is in contact with the radial glia [32]. The same phenotype is observed in neuronal precursors (“D-cells”) that are generated in the subgranular zone of the hippocampus in adult mammals [33, 34]. As neurons mature, forming dendrites and axons, nuclear and cytoplasmic size increase, and cells become transcriptionally more active, with a concurrent reduction in heterochromatin. Experimental studies have shown that a variety of signaling pathways and receptors for neurotrophic factors become activated by proteins forming part of the complex cell cycle-controlling molecular machinery [35]. However, the specific mechanism that drives the transition from small, heterochromatin-rich neural precursor to differentiated neuron is little understood. In human and mouse, mutations in the MECP2 protein, which encodes a transcriptional repressor, is associated with a reduction or delay of neuronal maturation (Rett Syndrome; [36]). The gene network (of which MECP2 may form part) that accompanies neural precursor maturation has not been established. In Drosophila, this hypothetical mechanism has to be embedded into the ecdysone hormonal cycle which has been shown to be expressed and required for different developmental changes that occur in the nervous system [37–40]. SU neurons in the Drosophila larval brain may present a favorable paradigm to study the process of neuronal maturation downstream of the ecdysone cycle. SU neurons represent a major population at the early larval stage (primary SU neurons) and late larval stage (primary and secondary SU neurons) and can be labeled by specifically expressed factors (e.g., Broad-Z3), which should make them amenable to FACS sorting and systematic gene expression screens.
SU neurons and the central complex
Most neuropil compartments of the adult Drosophila brain have a corresponding larval counterpart [41, 42]; outgrowing fibers of secondary neurons, which form much of the volume of the adult compartments, follow their primary siblings and establish dendritic and axonal branches around this primary scaffold. This principle does not apply to the secondary neurons forming the central complex, for which no anatomically defined larval counterpart exists. Small primordia of the different compartments of the central complex and associated structures (i.e., the bulb and anterior optic tubercle, which relay input to the central complex; [27]) can be first detected at the late larval stage. These larval primordia of the central complex compartments are formed by the fiber bundles and associated filopodia of the secondary lineages which will develop into the central complex of the adult brain. The minute early larval prFB, formed by the FB pioneers described in the present paper, represent an exceptional case. Thus, FB pioneers are primary neurons whose axons extend during the late embryonic phase and gather into a tight commissural bundle located in the center of the crossing fiber masses that constitute the supraesophageal commissure of the early larval brain. Several aspects of the prFB deserve special comment.
From late embryonic stages onward the FB pioneer axons are enclosed by an exclusive glial layer formed by the so-called interhemispheric ring glia [43]. Processes of these glial cells assemble into an invariant pattern of sheaths around several individual commissural bundles ([27]; this paper). The particular channel conducting the prFB stands out by a pair of glial nuclei attached to its posterior-medial wall (Figure 4); we could unequivocally identify a pair of glial nuclei at that position in the serial EM stack (Figure 4). As the larva grows and secondary tracts of DM1–4 are added to the FB pioneer bundle, the glial channel widens to eventually accommodate the entire fan-shaped body. Genetic studies indicate that interhemispheric glia does play a role in the morphogenesis of the fan-shaped body. Thus, genetic ablation of glia [44], or loss of function of molecular factors expressed specifically in the interhemispheric ring [43], result in defective shapes of the adult FB and EB. However, the pathways of DM1–4 lineages at the late larval stage did not display major defects.
FB pioneers differentiate into the pontine neurons of the adult central complex. Pontine neurons differ in their projection from all other columnar neurons, because they connect two columns on either side of the midline. For example, pontine neurons of the right hemispheric DM4 lineage connect the right lateral column of the FB with its left medial column, thereby crossing the midline. In contrast, axons of other, unicolumnar neurons of the right DM4 remain ipsilaterally, connecting only to the right lateral column of the FB (see Figure 1). The trajectories of the FB pioneers perfectly reflect this pontine-typical behavior already in the early larva (see Figure 4).
Even though primary FB pioneers and their secondary follower tracts are in close contact to each other throughout larval development, ablation of the former does not result in gross structural abnormalities of the latter. We observed a dramatic phenotype in the FB primordium in the ablated animals; however, the characteristic trajectories and branching pattern of the Neurotactin-positive secondary tracts of DM1–4 in the late larva appeared indistinguishable from the control. Filopodial tufts of secondary tracts in ablated specimens still assembled into regularly sized globular structures, representing the forerunners of fan-shaped body columns (see Figure 6B). These findings imply that separate guidance systems act on the early born pontine neurons and later born unicolumnar neurons. We should point out that following our experimental regimen it is possible that a small number of FB pioneers might have escaped UAS-hid;rpr-induced apoptosis, which might have been sufficient to play a role in the guidance of the following unicolumnar neurons.
Aside from pioneering the fan-shaped body primordium, SU neurons form part of almost all lineages of the early larval brain, but we do not yet know what fate awaits these neurons; markers similar to the one provided by R45F08-Gal4 would be required to establish what type of adult neurons the different SU neurons give rise to. Of particular interest would be SU neurons of lineages that, like DM1–4, contribute to the adult central complex. In case of DALv2, which generates all of the R-neurons of the EB, a single SU neuron exists in each hemisphere (see Table S1. Related to Figure 3). This neuron projects a short axon along the primary tract, but does not reach the EB primordium described for the late larval stage in previous works [27, 45]. Two other lineages, DALcl1 and DALcl2, form part of the anterior visual pathway, which provides input to the central complex [11]. Secondary neurons of DALcl1/2 innervate the anterior optic tubercle and the bulb of the adult brain. DALcl1/2 SU neurons extend relatively short axons that follow the differentiated DALcl1/2d neurons, cross the peduncle of the mushroom body, and terminate shortly thereafter. However, a spatially restricted territory that houses all DALcl1/2d SU terminations, and that might therefore be considered a forerunner of the bulb, does not exist in the early larva. In conclusion, the SU neurons of lineages DM1–4, described in this study, may represent a rare case where primary neurons establish a blueprint for an adult-specific brain compartment.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests for any information and requests for resources or reagents should be directed to the Lead Contact, Volker Hartenstein (volkerh@mcdb.ucla.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All fly strains were kept on standard yeast-containing fly flood at 25°C.
METHOD DETAILS
Selective ablation of fan-shaped body pioneers
FB pioneers were ablated from late first instar to second instar. The ablation experiment was carried out using the following genotype: UAS-hid UAS-rpr UAS-lacZ/+;10xUAS-mCD8::GFP/+; R45F08-Gal4/tub-Gal80ts. Freshly hatched larvae were collected and kept at 18°C degrees for two days (corresponding to 24 hours at 25°C). Subsequently, these late L1/early L2 larvae were transferred to 29°C for 24 hours. After the temperature shift, larvae were transferred back to 18°C. When reaching the third instar wandering stage, late larvae were collected, dissected and processed for analysis. The constitutive activation of UAS-hid,rpr by R45F08-Gal4 is lethal, and only survives into larval stages with Gal80-mediated suppression. Control flies were either males lacking UAS-hid UAS-rpr UAS-lacZ and tub-Gal80ts (Figure 6A–C), or females lacking a temperature shift and maintained at 18 degrees, which were completely viable (data not shown). Most experimental female larvae (heat-shifted) exhibited lethality; a small fraction were slowed in their development relative to male controls, and this was the cohort utilized for analysis.
Single cell clones
Single-cell analysis of the differentiated SU neurons in the adult was conducted using the multicolor flip-out (MCFO) method described previously [50]. Flies of the genotype R57C10-Flp-2::PEST,su(Hw)attP8::HA_V5_FLAG_1 were crossed to R45F08-Gal4 to generate the required progeny. The cross was kept at 25°C under normal light/dark cycle. 1–3 day adults were dissected, stained and imaged to screen for single-cell labeling of R45F08-Gal4-positive pontine neurons.
Immunohistochemistry
Antibodies:
The following primary antibodies were obtained from Developmental Studies Hybridoma Bank, DHSB, (Iowa City, IA): rat anti-DN-cadherin (DN-EX #8, adult, L3: 1:8, L1: 1:10), mouse anti-Brp (nc82,1:20), mouse anti-Neurotactin (BP106, 1: 6) mouse anti-Neuroglian (BP104, Adult: 1:12, L1: 1:14), mouse anti-repo (1:10), rabbit anti-βGal (1:3). We also used rabbit anti-HA (1:300, Cell Signaling Technologies), chicken anti-GFP (1:1000, Abcam) and rat anti-FLAG (1:300, Novus Biologicals). The following secondary antibodies were obtained from Jackson ImmunoResearch; Molecular Probes) and used: Alexa 568-conjugated anti-mouse (1:500), cy5-conjugated anti-mouse (1:300), cy3-conjugated anti-rat (1:300), Alexa 488- conjugated anti-rabbit (1:1000), cy5-conjugated anti-rat (1:300), cy3-conjugated anti-rabbit (1:160), and Alexa 488-conjugated anti-rabbit (1:300). We also obtained Alexa 488-conjugated anti-chicken (1:1000) from Thermo Fisher Scientific.
Adult brain preparation:
Brain samples were dissected in phosphate buffered saline (1X PBS, pH 7.4) for 30 minutes. The dissected brains were then fixed in ice-cold 4% formaldehyde fixing solution [750μl phosphate buffered saline (PBS); 250μl 16% paraformaldehyde, EM grade (Sigma)] for 2.5 hours before washing four times, 15 minutes each, with ice-cold 1X PBS. Then the brains were dehydrated in a series of ice-cold ethanol diluted with 1X PBS (5,10,20, 50, 70 and 100% ethanol; for 5 minutes at each concentration). After hydrating, the brains were stored in 100% ethanol at −20°C overnight. For rehydration, we repeated the hydration process in reverse order. These rehydrated brains then were washed with ice-cold 1X PBS 4 times, 15 minutes each. Next, they were washed 3 times, 15 minutes each, with ice-cold 0.3% PBT (PBS; 0.3% Triton X-100, pH7.4), followed by 4 washes, 15 minutes each, on the nutator at room temperature. After washing samples were blocked (10% normal goat serum in 0.3% PBT) and then stained with primary and secondary antibodies for 3 days each under gentle shaking on the nutator at 4°C. Washes were done when replacing primary with secondary antibodies with 0.3% PBT, and when removing secondary antibodies. Finally samples were transferred to Vectashield, mounted and imaged. Brains with MCFO single cell clones were processed following a shortened protocol in which the dehydration/rehydration steps were omitted.
Larval brain preparation:
Larval brains were dissected at the indicated age (24, 48, 60, 72 and 96 hours old) in phosphate buffered saline (1X PBS, pH 7.4) for 30 minutes and then fixed in 4% formaldehyde fixing solution (900μl 1X PBS, 100μl 37% paraformaldehyde) for 30 minutes with gentle shaking. The fixed brains then were washed with the indicated buffer solution 4 times, 15 minutes each. Brains were also subjected to the methanol dehydration series diluted with 1X PBS (10 minutes in 25 and 50% methanol each, and 4× 1-minute washes in 75% methanol, and one 5-minute wash in 100% methanol). Samples were then stored in 100% methanol at −20°Covernight, then rehydrated the next day by washing with 1X PBS once. Subsequently samples were washed with 0.1% PBT (PBS 0.1% Triton X-100, pH7.4) on the nutator. After washing, samples were blocked (10% normal goat serum in 0.1% PBT for 30 minutes), and incubated in primary and secondary antibodies for 2 days each at 4°C while gently shaking on the nutator. Washes with 0.1% PBT were done when replacing primary with secondary antibody and before placing the samples in VectaShield.
Digital reconstruction and analysis of SU neurons from serial EM
In order to reconstruct the SU neurons, we used a serial transmission electron microscopy (ssTEM) dataset that included the whole nervous system of a first instar larva [51]. Brains of 6-h-old female larvae were dissected and fixed in a solution containing 2% glutaraldehyde and 2% OsO4 in 0.1 M sodium cacodylate buffer (pH 7.4). Samples in solution were microwaved at 350-W, 375-W and 400-W pulses for 30 sec each, separated by 60-sec pauses. Samples were transferred into a new fixing solution that resembled the one above, except that the concentration of OsO4 was lowered to 1%. Samples were then subjected to another round of microwaving as described above. Subsequently samples were stained en bloc with 1% uranyl acetate in water while microwaving (350 W for 3×3 30 sec with 60-sec pauses). Samples were dehydrated in an ethanol series, transferred to propylene oxide and infiltrated and embedded with EPON resin. Blocks were sectioned using a Leica UC6 ultramicrotome. Sections were imaged semi-automatically with the Leginon software [52] at 3.8 × 3.8 × 50nm on a FEI Spirit BioTWIN TEM (Hillsboro) and assembled using the TrakEM2 package [53].
Digital reconstructions of neurons used a modified version of CATMAID [54] - [55], a web-based application. To reconstruct neurons in CATMAID, we first identified neurite bundles corresponding to discrete lineages or sets of lineages, using the recently published atlas of lineage-associated tracts of the first instar larval brain (23). Lineage by lineage, we then traced axons from the point of entry into the neuropil towards the cell body, which allowed us to unambiguously identify SU neurons. Subsequently we traced SU neuronal axons forward into the neuropil to obtained their full length, including rare short branches and input synapses. For obtaining the area of the nucleus and cytoplasm of a cell, a section at or near the center of the soma was chosen. Subsequently, the length of the longest and shortest radius was obtained by using a tool of CATMAID that gives the radius in nm. The average radius was taken to calculate the area.
QUANTIFICATION AND STATISTICAL ANALYSIS
For the comparison of depth of SU neurons versus differentiated neurons we calculated means for ten representative lineages and used the two-sample t-test.
DATA AND SOFTWARE AVAILABILITY
For analysis of confocal image stacks we used the ImageJ/Fiji package that can be downloaded without cost (https://imagej.net/Fiji/Downloads). Reconstruction of neurons from the serial EM dataset used the software CATMAID as described in [55]
Supplementary Material
Data S1 Quantitative aspects of SU neurons among larval brain lineages. (Related to Figure 3)(Sheet A) Listed in the first column (A, F, K) are all lineages and their associated fiber tracts; the second and third column present numbers of SU neurons and differentiated neurons associated with individual lineages; SU neurons for the left and right brain hemisphere are listed separately (columns B, G, L; SU R/L), differentiated neurons are shown for the right hemisphere only (columns C, H, M; Diff R). The fourth column (D, I, N) provides rates of SU neurons per lineage (SU %), with values from 0–10% shaded yellow, 11–29.9% light orange, 30–49.9% dark orange, 50% and above red.
(Sheet B) Average cable length (columns 2 and 3), ratio (column 4) and synapse number (input synapses: column 5 and 6; output synapses: column 7 and 8) for differentiated neurons and SU neurons, respectively, of ten lineages on the right hemisphere; reconstruction of differentiated neurons for the lineage chosen were complete or within >90% completion. For more detail on lineages and their nomenclature see Table S1.
KEY RESOURCES TABLE
| Source | Identifier | |
|---|---|---|
| Antibodies | ||
| Rat anti-DN-cadherin | Developmental Studies Hybridoma Bank | DN-EX #8, RRID: AB_528121 |
| Mouse anti-Bruchpilot | Developmental Studies Hybridoma Bank | nc82, RRID: AB_2314866 |
| Mouse anti-Neurotactin | Developmental Studies Hybridoma Bank | BP106, RRID: AB_528404 |
| Mouse anti-Neuroglian | Developmental Studies Hybridoma Bank | BP104, RRID: AB_528402 |
| Mouse 8D12 anti-Repo | Developmental Studies Hybridoma Bank | RRID: AB_528448 |
| Rabbit anti-β-galactosidase | Cappel/ICN (now MP Biomedicals) | Cat #: 55976 |
| Rabbit anti-HA | Cell Signaling Technologies | Cat#: mAb #3724 |
| Chicken anti-GFP | Invitrogen, Thermo Fisher Scientific | Cat#: A10262 |
| Rat anti-FLAG | Novus Biologicals | Cat#: NBP1–06712SS |
| Goat anti-Mouse Alexa Flour-568 | Life Technologies, Thermo Fisher Scientific | Cat#: A-11031 |
| Donkey anti-Mouse Cy5 | Jackson Immunoresearch, Molecular Probes | Code: 712-175-153 |
| Donkey anti-Rat Cy3 | Jackson Immunoresearch, Molecular Probes | Code: 712-165-153 |
| Goat anti-Rat Cy5 | Jackson Immunoresearch, Molecular Probes | Code: 112-175-143 |
| Goat anti-Rabbit Cy3 | Jackson Immunoresearch, Molecular Probes | Code: 711-165-152 |
| Goat anti-Rabbit Alexa Flour-488 | Invitrogen, Thermo Fisher Scientific | Cat#: A-11008 |
| Goat anti-Chicken Alexa Flour-488 | Thermo Fisher Scientific | Cat#: A-11039 |
| Experimental Models: Organisms/ Strains | ||
| w[1118];P{y[+t7.7]w[+mC]=GMR45F08-GAL4} attP2 | Bloomington Stock Centre (USA) | BL#49565 |
| w[1118];P{y[+t7.7]w[+mC]=10XUAS-mCD8::GFP} attP2 | Bloomington Stock Centre (USA) | BL#32184 |
| w[*]:P{y[+t7.7]w[+mC]=10XUAS-IVS-mCD8::GFP} attP40 | Bloomington Stock Centre (USA) | BL#32186 |
| w[1118];P{y[+t7.7]w[+mC]=GMR57C10-FLP2.PEST}su(Hw)attP8;PBac{[+mDint2]w[+mC]=10xUAS(FRT.stop)myr::smGdP-HA}VK00005P{y[+t7.7]w[+mC]=10xUAS(FRT.stop)myr::smGdP-V5-THS-10xUAS(FRT.stop)myr::smGdp-FLAG}su(Hw)attP1 | Gift from Mark Frye Lab | [48] |
| UAS-his UAS-rpr-UAS-lacZ | [48] | |
| w[*];sna[Sco/CyO; P{w[+mC]=tubP-GAL80[ts]}ncd[GAL80ts-7] | Bloomington Stock Centre (USA) | BL#7018 |
| Drl-tau::lacZ | Prof. H. Hing (The College at Brockport, State University of New York) | [49] |
| Chemicals | ||
| Paraformaldehyde 16% Solution, EM Grade | Electron Microscopy Sciences | Cat#: 15710 |
| Triton X-100 | MP Biomedicals, LLC | Cat#: 194854 |
| Agar | Sigma-Aldrich | A1296 |
| Software | ||
| Image-J/Fiji | Fiji | N/A |
| Adobe Illustrator | Adobe System | N/A |
| Adobe Photoshop | Adobe System | N/A |
| CATMAID | CATMAID.org | [52] |
Highlights.
Many early larval neurons lack a branched neurite tree and synapses (SU neurons)
Axons of SU neurons form bundles associated with the fiber tracts of most lineages
SU neurons of lineages DM1-DM4 pioneer the neural architecture of the central complex
SU neurons differentiate into the pontine neurons of the adult central complex
Acknowledgements
This work was supported by the NIH (grant R01 NS096290 to V.H.), an NIH Ruth L. Kirschstein National Research Service Award (to J.J.O.; GM007185), the University of California, Los Angeles Dissertation Year Fellowship (to J.J.O.), and the Swiss NSF (to N.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Martin JR, Raabe T, and HEISENBERG M (1999). Central complex substructures are required for the maintenance of locomotor activity in Drosophila melanogaster. J Comp Physiol A 185, 277–288. [DOI] [PubMed] [Google Scholar]
- 2.Neuser K, Triphan T, Mronz M, Poeck B, and Strauss R (2008). Analysis of a spatial orientation memory in Drosophila. Nature 453, 1244–1247. [DOI] [PubMed] [Google Scholar]
- 3.Ofstad TA, Zuker CS, and Reiser MB (2011). Visual place learning in Drosophila melanogaster. Nature 474, 204–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfeiffer K, and Homberg U (2014). Organization and functional roles of the central complex in the insect brain. Annu. Rev. Entomol 59, 165–184. [DOI] [PubMed] [Google Scholar]
- 5.Seelig JD, and Jayaraman V (2015). Neural dynamics for landmark orientation and angular path integration. Nature 521, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green J, Adachi A, Shah KK, Hirokawa JD, Magani PS, and Maimon G (2017). A neural circuit architecture for angular integration in Drosophila. Nature 546, 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner-Evans D, Wegener S, Rouault H, Franconville R, Wolff T, Seelig JD, Druckmann S, and Jayaraman V (2017). Angular velocity integration in a fly heading circuit. Elife 6, e04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.HANESCH U, FISCHBACH KF, and HEISENBERG M (1989). Neuronal Architecture of the Central Complex in Drosophila-Melanogaster. Cell Tissue Res. 257, 343–366. [Google Scholar]
- 9.Jundi el, B., Pfeiffer K, Heinze S, and Homberg U (2014). Integration of polarization and chromatic cues in the insect sky compass. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol 200, 575–589. [DOI] [PubMed] [Google Scholar]
- 10.Seelig JD, and Jayaraman V (2013). Feature detection and orientation tuning in the Drosophila central complex. Nature 503, 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Omoto JJ, Keleş MF, Nguyen B-CM, Bolanos C, Lovick JK, Frye MA, and Hartenstein V (2017). Visual Input to the Drosophila Central Complex by Developmentally and Functionally Distinct Neuronal Populations. Curr. Biol 27, 1098–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohwi M, and Doe CQ (2013). Temporal fate specification and neural progenitor competence during development. Nat. Rev. Neurosci 14, 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen C, Shy D, Spindler SR, Fung S, Pereanu W, Younossi-Hartenstein A, and Hartenstein V (2009). Patterns of growth, axonal extension and axonal arborization of neuronal lineages in the developing Drosophila brain. Dev. Biol 335, 289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong DC, Lovick JK, Ngo KT, Borisuthirattana W, Omoto JJ, and Hartenstein V (2013). Postembryonic lineages of the Drosophila brain: II. Identification of lineage projection patterns based on MARCM clones. Dev. Biol 384, 258–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang JS, Awasaki T, Yu H-H, He Y, Ding P, Kao J-C, and Lee T (2013). Diverse neuronal lineages make stereotyped contributions to the Drosophila locomotor control center, the central complex. J. Comp. Neurol 521, 2645–Spc1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereanu W, and Hartenstein V (2006). Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J. Neurosci 26, 5534–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito K, and Awasaki T (2008). Clonal Unit Architecture of the Adult Fly Brain In Brain Development in Drosophila melanogaster Advances in Experimental Medicine and Biology. (New York, NY: Springer, New York, NY), pp. 137–158. [DOI] [PubMed] [Google Scholar]
- 18.Boyan G, Liu Y, Khalsa SK, and Hartenstein V (2017). A conserved plan for wiring up the fan-shaped body in the grasshopper and Drosophila. Dev. Genes Evol. 227, 253–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walsh KT, and Doe CQ (2017). Drosophila embryonic type II neuroblasts: origin, temporal patterning, and contribution to the adult central complex. Development 144, 4552–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Álvarez J-A, and Díaz-Benjumea FJ (2018). Origin and specification of type II neuroblasts in the Drosophila embryo. Development 145, dev158394. [DOI] [PubMed] [Google Scholar]
- 21.Riebli N, Viktorin G, and Reichert H (2013). Early-born neurons in type II neuroblast lineages establish a larval primordium and integrate into adult circuitry during central complex development in Drosophila. Neural Dev 8, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenett A, Rubin GM, Ngo T-TB, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Rep 2, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartenstein V, Younossi-Hartenstein A, Lovick JK, Kong A, Omoto JJ, Ngo KT, and Viktorin G (2015). Lineage-associated tracts defining the anatomy of the Drosophila first instar larval brain. Dev. Biol 406, 14–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovick JK, and Hartenstein V (2015). Hydroxyurea-mediated neuroblast ablation establishes birth dates of secondary lineages and addresses neuronal interactions in the developing Drosophila brain. Dev. Biol 402, 32–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolff T, Iyer NA, and Rubin GM (2015). Neuroarchitecture and neuroanatomy of the Drosophila central complex: A GAL4-based dissection of protocerebral bridge neurons and circuits. J. Comp. Neurol 523, 997–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Omoto JJ, Yogi P, and Hartenstein V (2015). Origin and development of neuropil glia of the Drosophila larval and adult brain: Two distinct glial populations derived from separate progenitors. Dev. Biol 404, 2–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovick JK, Omoto JJ, Ngo KT, and Hartenstein V (2017). Development of the anterior visual input pathway to the Drosophila central complex. J. Comp. Neurol 525, 3458–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aghajanian P, Takashima S, Paul M, Younossi-Hartenstein A, and Hartenstein V (2016). Metamorphosis of the Drosophila visceral musculature and its role in intestinal morphogenesis and stem cell formation. Dev. Biol 420, 43–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Truman JW (1990). Metamorphosis of the central nervous system of Drosophila. J. Neurobiol 21, 1072–1084. [DOI] [PubMed] [Google Scholar]
- 30.Veverytsa L, and Allan DW (2012). Temporally tuned neuronal differentiation supports the functional remodeling of a neuronal network in Drosophila. Proc. Natl. Acad. Sci. U.S.A 109, E748–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou B, Williams DW, Altman J, Riddiford LM, and Truman JW (2009). Temporal patterns of broad isoform expression during the development of neuronal lineages in Drosophila. Neural Dev 4, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckenhoff MF, and Rakic P (1984). Radial organization of the hippocampal dentate gyrus: a Golgi, ultrastructural, and immunocytochemical analysis in the developing rhesus monkey. J. Comp. Neurol 223, 1–21. [DOI] [PubMed] [Google Scholar]
- 33.Seri B, García-Verdugo JM, Collado-Morente L, McEwen BS, and Alvarez-Buylla A (2004). Cell types, lineage, and architecture of the germinal zone in the adult dentate gyrus. J. Comp. Neurol 478, 359–378. [DOI] [PubMed] [Google Scholar]
- 34.Ngwenya LB, Rosene DL, and Peters A (2008). An ultrastructural characterization of the newly generated cells in the adult monkey dentate gyrus. Hippocampus 18, 210–220. [DOI] [PubMed] [Google Scholar]
- 35.Kawauchi T, Shikanai M, and Kosodo Y (2013). Extra-cell cycle regulatory functions of cyclin-dependent kinases (CDK) and CDK inhibitor proteins contribute to brain development and neurological disorders. Genes Cells 18, 176–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martínez de Paz A, and Ausió J (2017). MeCP2, A Modulator of Neuronal Chromatin Organization Involved in Rett Syndrome In Neuroepigenomics in Aging and Disease Advances in Experimental Medicine and Biology. (Cham: Springer International Publishing; ), pp. 3–21. [DOI] [PubMed] [Google Scholar]
- 37.Brown HLD, and Truman JW (2009). Fine-tuning of secondary arbor development: the effects of the ecdysone receptor on the adult neuronal lineages of the Drosophila thoracic CNS. Development 136, 3247–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schubiger M, Wade AA, Carney GE, Truman JW, and Bender M (1998). Drosophila EcR-B ecdysone receptor isoforms are required for larval molting and for neuron remodeling during metamorphosis. Development 125, 2053–2062. [DOI] [PubMed] [Google Scholar]
- 39.Zheng X, Wang J, Haerry TE, Wu AY-H, Martin J, O’Connor MB, Lee C-HJ, and Lee T (2003). TGF-beta signaling activates steroid hormone receptor expression during neuronal remodeling in the Drosophila brain. Cell 112, 303–315. [DOI] [PubMed] [Google Scholar]
- 40.Syed MH, Mark B, and Doe CQ (2017). Steroid hormone induction of temporal gene expression in Drosophila brain neuroblasts generates neuronal and glial diversity. Elife 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Younossi-Hartenstein A, Salvaterra PM, and Hartenstein V (2003). Early development of the Drosophila brain: IV. Larval neuropile compartments defined by glial septa. J. Comp. Neurol 455, 435–450. [DOI] [PubMed] [Google Scholar]
- 42.Pereanu W, Kumar A, Jennett A, Reichert H, and Hartenstein V (2010). Development-based compartmentalization of the Drosophila central brain. J. Comp. Neurol 518, 2996–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon AF, Boquet I, Synguélakis M, and Préat T (1998). The Drosophila putative kinase linotte (derailed) prevents central brain axons from converging on a newly described interhemispheric ring. Mech. Dev 76, 45–55. [DOI] [PubMed] [Google Scholar]
- 44.Spindler SR, Ortiz I, Fung S, Takashima S, and Hartenstein V (2009). Drosophila cortex and neuropile glia influence secondary axon tract growth, pathfinding, and fasciculation in the developing larval brain. Dev. Biol 334, 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lovick JK, Kong A, Omoto JJ, Ngo KT, Younossi-Hartenstein A, and Hartenstein V (2016). Patterns of growth and tract formation during the early development of secondary lineages in the Drosophila larval brain. Dev Neurobiol 76, 434–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franconville R, Beron C, and Jayaraman V (2018). Building a functional connectome of the Drosophila central complex. Elife 7, e04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berck ME, Khandelwal A, Claus L, Hernandez-Nunez L, Si G, Tabone CJ, Li F, Truman JW, Fetter RD, Louis M, et al. (2016). The wiring diagram of a glomerular olfactory system. Elife 5, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou L, Schnitzler A, Agapite J, Schwartz LM, Steller H, and Nambu JR (1997). Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system midline cells. Proc. Natl. Acad. Sci. U.S.A 94, 5131–5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen W, and Hing H (2008). The L1-CAM, Neuroglian, functions in glial cells for Drosophila antennal lobe development. Dev Neurobiol 68, 1029–1045. [DOI] [PubMed] [Google Scholar]
- 50.Nern A, Pfeiffer BD, and Rubin GM (2015). Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl. Acad. Sci. U.S.A 112, E2967–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohyama T, Schneider-Mizell CM, Fetter RD, Aleman JV, Franconville R, Rivera-Alba M, Mensh BD, Branson KM, Simpson JH, Truman JW, et al. (2015). A multilevel multimodal circuit enhances action selection in Drosophila. Nature 520, 633–639. [DOI] [PubMed] [Google Scholar]
- 52.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, and Carragher B (2005). Automated molecular microscopy: the new Leginon system. J. Struct. Biol 151, 41–60. [DOI] [PubMed] [Google Scholar]
- 53.Cardona A, Saalfeld S, Schindelin J, Arganda-Carreras I, Preibisch S, Longair M, Tomancak P, Hartenstein V, and Douglas RJ (2012). TrakEM2 software for neural circuit reconstruction. PLoS ONE 7, e38011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saalfeld S, Cardona A, Hartenstein V, and Tomancak P (2009). CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics 25, 1984–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider-Mizell CM, Gerhard S, Longair M, Kazimiers T, Li F, Zwart MF, Champion A, Midgley FM, Fetter RD, Saalfeld S, et al. (2016). Quantitative neuroanatomy for connectomics in Drosophila. Elife 5, 1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 Quantitative aspects of SU neurons among larval brain lineages. (Related to Figure 3)(Sheet A) Listed in the first column (A, F, K) are all lineages and their associated fiber tracts; the second and third column present numbers of SU neurons and differentiated neurons associated with individual lineages; SU neurons for the left and right brain hemisphere are listed separately (columns B, G, L; SU R/L), differentiated neurons are shown for the right hemisphere only (columns C, H, M; Diff R). The fourth column (D, I, N) provides rates of SU neurons per lineage (SU %), with values from 0–10% shaded yellow, 11–29.9% light orange, 30–49.9% dark orange, 50% and above red.
(Sheet B) Average cable length (columns 2 and 3), ratio (column 4) and synapse number (input synapses: column 5 and 6; output synapses: column 7 and 8) for differentiated neurons and SU neurons, respectively, of ten lineages on the right hemisphere; reconstruction of differentiated neurons for the lineage chosen were complete or within >90% completion. For more detail on lineages and their nomenclature see Table S1.
Data Availability Statement
For analysis of confocal image stacks we used the ImageJ/Fiji package that can be downloaded without cost (https://imagej.net/Fiji/Downloads). Reconstruction of neurons from the serial EM dataset used the software CATMAID as described in [55]