Summary
The neural crest (NC) gives rise to a multitude of fetal tissues, and its misregulation is implicated in congenital malformations. Here, we investigated molecular mechanisms pertaining to NC-related symptoms in Bohring-Opitz syndrome (BOS), a developmental disorder linked to mutations in the Polycomb group factor Additional sex combs-like 1 (ASXL1). Genetically edited human pluripotent stem cell lines that were differentiated to NC progenitors and then xenotransplanted into chicken embryos demonstrated an impairment of NC delamination and emigration. Molecular analysis showed that ASXL1 mutations correlated with reduced activation of the transcription factor ZIC1 and the NC gene regulatory network. These findings were supported by differentiation experiments using BOS patient-derived induced pluripotent stem cell lines. Expression of truncated ASXL1 isoforms (amino acids 1–900) recapitulated the NC phenotypes in vitro and in ovo, raising the possibility that truncated ASXL1 variants contribute to BOS pathology. Collectively, we expand the understanding of truncated ASXL1 in BOS and in the human NC.
Keywords: ASXL1, neural crest, Bohring-Opitz syndrome, Polycomb, ZIC1
Highlights
-
•
Generation of human pluripotent stem cell models for Bohring-Opitz syndrome
-
•
ASXL1 mutations perturb neural crest (NC) emigration in vitro and in vivo
-
•
ASXL1 mutations correlate with impaired activation of ZIC1
-
•
Expression of truncated ASXL1 variants mirrors NC perturbation in vitro and in vivo
In this study, Drukker and colleagues developed human pluripotent stem cell models for the rare congenital disorder Bohring-Opitz syndrome, which is caused by mutations in the Polycomb factor ASXL1. In these lines, they found impaired neural crest emigration in vitro and in vivo and link this phenotype to impaired activation of ZIC1.
Introduction
The neural crest (NC) is an embryonic progenitor population that gives rise to multiple derivatives, including craniofacial cartilage and bones and peripheral neurons, many of which are associated with birth defects such as craniofacial and cardiac malformations (Etchevers et al., 2006, Mayor and Theveneau, 2013, Snider and Mishina, 2014). Animal studies have shown that initiation of NC development at the neural plate border, as well as subsequent delamination, migration, and terminal differentiation of the NC progenitors, are orchestrated by specific gene regulatory networks (GRNs) (Simoes-Costa and Bronner, 2015). Advancing the understanding of these GRNs in human NC development necessitates modeling by pluripotent stem cells, in particular for investigating the epigenetic mechanisms that regulate them (Bajpai et al., 2010). Nevertheless, the involvement of mutations in histone-modifying enzymes in pathological NC development has not yet been studied.
Bohring-Opitz syndrome (BOS) is a severe congenital disorder associated with de novo mutations in the Additional sex combs-like 1 (ASXL1) gene, characterized by symptoms that include developmental delay and musculoskeletal and neurological features (Hastings et al., 2011). ASXL1 is a co-factor of the Polycomb repressive complex 2 (PRC2), and loss of function (Abdel-Wahab et al., 2012) and expression of dominantly acting truncated ASXL1 variants (Balasubramani et al., 2015, Guo et al., 2018) have been associated with transcriptional misregulations in hematological disorders. Whether and how these mechanisms relate to the molecular mechanism of BOS is unknown.
Here, we reasoned that craniofacial malformations such as palate anomalies indicate a potential perturbation of NC development in BOS (Hastings et al., 2011). By analyzing a comprehensive set of genetically edited human embryonic stem cell (hESC) and patient induced pluripotent stem cell (iPSC) lines that we differentiated into NC progenitors, we identified transcriptional perturbations associated with defects in NC development, which we propose provide a putative mechanism for the craniofacial and plausibly other symptoms in BOS patients.
Results and Discussion
Generation of Human Pluripotent Stem Cell Models for BOS
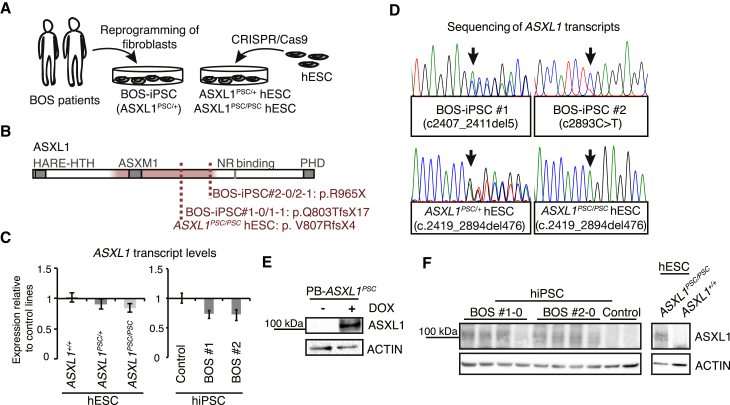
We obtained dermal fibroblasts from two BOS patients who carry monoallelic premature stop codons (PSCs) in the terminal exon of ASXL1 (Magini et al., 2012). Patient fibroblasts were reprogrammed using two techniques, by episomal plasmids and modified mRNAs, resulting in the derivation of four BOS-iPSC lines (Figure 1A, Table S1, Figure S1A). In addition, we used CRISPR/Cas9 to excise a 500 bp region from the terminal exon of ASXL1 in hESCs, generating clones that harbor heterozygous and homozygous BOS patient-like PSC mutations (ASXL1PSC/+ and ASXL1PSC/PSC; Figure 1A, Table S1, Figures S1B and S1C). BOS-iPSC and edited hESC lines expressed canonical pluripotency markers and displayed colony morphology comparable with control lines, which were the parental ASXL1+/+ hESC line and two control iPSC lines, respectively (Figure S1A and S1C–S1G, Table S1).
Figure 1.
Human Pluripotent Stem Cell Models for Bohring-Opitz Syndrome
(A) Scheme illustrating the generation of human pluripotent stem cell lines carrying premature stop codon (PSC) mutations in ASXL1.
(B) Scheme of human ASXL1 protein showing annotated domains (NR, nuclear receptor), and locations of mutations frequently reported in BOS patients (red tinted sector) and present in BOS-iPSC and ASXL1PSC/PSC/ASXL1PSC/+ hESC clones.
(C) Expression of ASXL1 in BOS-iPSC lines and ASXL1PSC/PSC/ASXL1PSC/+ hESC clones relative to the respective iPSC/hESC control lines using primers targeting exon 4 (mean ± SEM, n ≥ 3 different clones/passages).
(D) Sequences of reverse transcribed ASXL1 transcripts from ASXL1 mutant lines.
(E and F) Representative blotting (n = 3–5 independent experiments) of ASXL1, using a monoclonal antibody raised against the N terminus, in an hESC line overexpressing a truncated ASXL1 variant (PB-ASXL1PSC) (E) and in human iPSC and hESC lines (F).
See also Figure S1.
Plausible molecular mechanisms of BOS include ASXL1 haploinsufficiency and/or expression of truncated ASXL1 (Figure 1B). Testing the level of ASXL1 transcripts in undifferentiated cells did not reveal a difference between BOS-iPSC, ASXL1PSC/+, and ASXL1PSC/PSC clones and the respective control lines (Figure 1C). We confirmed the presence of mutant ASXL1 transcripts in the genetically edited hESC clones, and in both BOS-iPSC lines #1-0 and #2-0 by Sanger sequencing (Figure 1D). Furthermore, we detected comparable levels of the normal and mutant transcripts using a primer set specifically targeting the 5-bp deletion in BOS-iPSC lines #1-0/#1-1 (Figure S1H). To further investigate the expression of truncated ASXL1 variants, we raised a monoclonal antibody targeting the N terminus of ASXL1, which by western blotting detected a truncated ASXL1 variant that was overexpressed in undifferentiated hESCs (PB-ASXL1PSC; Table S1, Figure 1E). This antibody also detected putative truncated protein isoforms in BOS-iPSC lines #1 and #2, and ASXL1PSC/PSC hESC clones (Figures 1F and S1I), albeit at varying levels (Figure S1J). Collectively, this indicates that expression of truncated ASXL1 variants is a possible mode of misregulation in BOS.
ASXL1 Mutations Perturb NC Development
Craniofacial symptoms that are common in BOS patients prompted us to investigate a possible connection to the NC lineage in the hESC and iPSC models. We utilized a differentiation protocol that generated delaminating NC-like cells (Bajpai et al., 2010) from neuroepithelial spheres (neurospheres; Figure 2A), which expressed the characteristic NC markers p75NTR, SOX9, SOX10, TFAP2A, PAX3, and SNAI2 (Lee et al., 2007, Simoes-Costa and Bronner, 2015) and the proliferation marker KI67 (Figures S2A and S2B). Moreover, in accordance with the expected potency of NC progenitors, they readily gave rise to putative mesenchymal stem cells (MSCs) (Noden and Trainor, 2005) that exhibited spindle-shaped morphology, expression of consensus MSC surface markers CD73, CD90, and CD105 (Ramos et al., 2016), and terminally differentiated into osteoblasts and adipocytes (Figures S2C–S2E).
Figure 2.
ASXL1 Mutations Impair Differentiation of Neural Crest Progenitors
(A) Timeline of neural crest (NC) differentiation protocol and bright-field images during in vitro differentiation of ASXL1+/+ hESCs. Scale bar, 500 μm.
(B) Percentage of attached neurospheres with emigrating cells at day 7 of NC differentiation of BOS-iPSC lines and ASXL1PSC/PSC/ASXL1PSC/+ clones (mean ± SEM, n = 3–13 different passages/clones and independent experiments, all relative to respective control lines; ∗∗p<0.01).
(C) Representative bright-field images of neurospheres at day 7 of NC differentiation. Scale bar, 100 μm.
(D) Workflow of neurosphere transplantation experiments and representative bright-field and GFP images of chicken embryos directly (upper panel) and 48 h (lower panel) after transplantation of neurospheres (white arrows: exemplary transplanted GFP-ASXL1+/+ neurosphere; dorsal view). Scale bar, 500 μm (chicken embryo) and 200 μm (neurospheres).
(E) Quantification of migrating cells from experiments in (D); ASXL1+/+, n = 9; ASXL1PSC/PSC, n = 21 embryos (mean ± SEM, ∗p = 0.037). Single dots indicate outliers.
See also Figure S2.
We noted upregulation of ASXL1 and ASXL3, but not of ASXL2, in NC progenitors (Figure S2F), indicating specific roles of the two genes during NC differentiation, which was further supported by co-expression of ASXL1 and the NC regulator TFAP2A (Figure S2G). Importantly, neurospheres derived from BOS-iPSC lines with heterozygous ASXL1 mutations displayed reduced attachment and diminished emigration of NC cells in comparison with control iPSC-derived neurospheres (Figure 2B, left panel). In the genetically edited hESC lines, we observed a similar reduction in neurosphere attachment and NC emigration, an effect that was statistically significant only in homozygous ASXL1PSC/PSC clones (Figure 2B, right panel), with the presence of dead, floating cells surrounding the neurospheres (Figures 2C and S2H). ASXL1 mutations did not negatively affect the propensity to form neurospheres; conversely, an increase in the total number of neurospheres was observed in lines harboring mutant ASXL1 (Figure S2I). Thus, ASXL1 mutations perturbed the generation and/or delamination of the NC in vitro, potentially via defective induction or specification already at the neurosphere stage.
To further test the emigration of NC cells in vivo, we performed orthotopic xenotransplantation of neurospheres obtained from eGFP-labeled ASXL1PSC/PSC and ASXL+/+ hESC lines (Table S1) into Hamburger-Hamilton (HH) stage 10 chicken embryos (Figure 2D). Neurospheres derived from ASXL1PSC/PSC hESC clones exhibited a significant reduction in the number of emigrating cells compared with neurospheres derived from the parental ASXL1+/+ hESC line (Figures 2D and 2E), an effect that was not due to size differences between control and mutant neurospheres (Figure S2J). Cells that emigrated from the neurospheres of both lines transversed a similar distance (Figure S2K). Collectively, these results indicate a connection between ASXL1, NC development, and BOS.
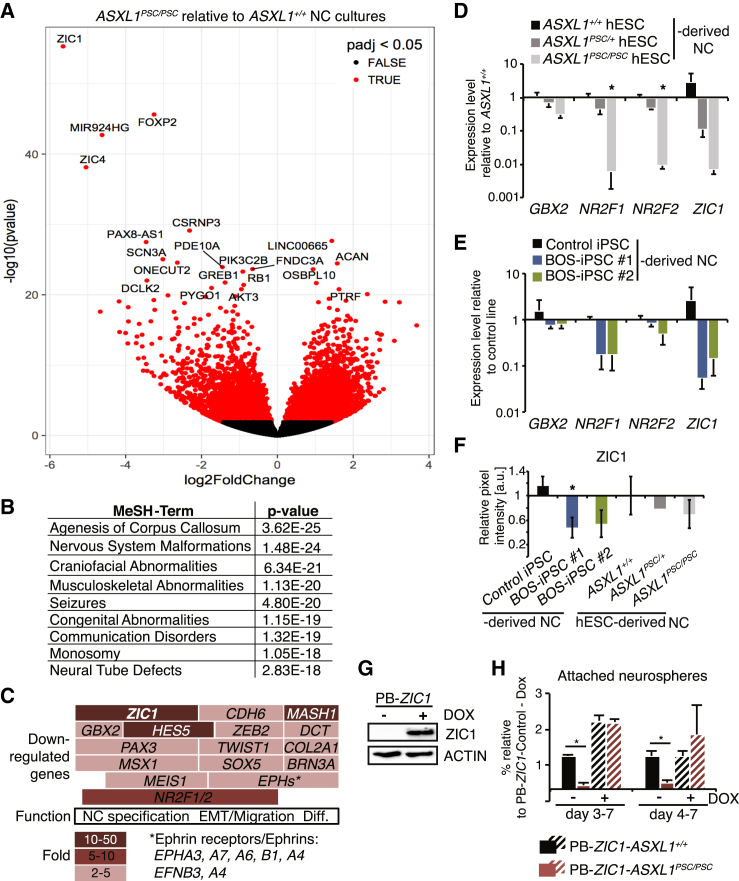
ASXL1 Mutations Impair Activation of ZIC1 and the NC Gene Regulatory Network
To delineate mechanisms underlying the paucity and migration defect of the NC progenitors, we conducted global RNA sequencing. This revealed mutually exclusive clustering of samples derived from NC progenitors harboring the ASXL1PSC/PSC genotype and the parental control cells (Figure S3A).
Strikingly, the neural plate border transcription factors ZIC1 and ZIC4 (Merzdorf, 2007), which are an immediately adjacent gene pair, were among the most negatively regulated transcripts in the ASXL1PSC/PSC NC progenitors (Figure 3A). This substantiated the link to BOS and related syndromes, because heterozygous deletion of the ZIC1/ZIC4 locus is associated with Dandy-Walker malformation (Grinberg et al., 2004), a defect in cerebellar development that is observed in BOS patients (Hastings et al., 2011). Moreover, negatively regulated genes in the dataset were significantly associated with several disorders and malformations found in BOS, including agenesis of the corpus callosum, craniofacial dysmorphisms, musculoskeletal abnormalities, and seizures (Figure 3B).
Figure 3.
Misregulation of Gene Networks Associated with NC Development and BOS Symptoms in ASXL1 Mutant Lines
(A) Volcano plot exhibiting differentially expressed genes in ASXL1PSC/PSC compared with ASXL1+/+ day 7 NC progenitors (total mRNA sequencing; n = 3 ASXL1+/+ and n = 7 ASXL1PSC/PSC samples, different clones and independent differentiation experiments).
(B) Most significant Medical Subject Headings (MeSH) terms associated with downregulated gene sets (padj < 0.05) in (A).
(C) Diagram of genes with annotated functions in NC development and their degree of misregulation (padj < 0.05), based on (A). Diff., terminal differentiation.
(D and E) Relative expression levels of canonical NC genes in day 7 NC differentiation cultures of ASXL1PSC hESC lines (D) and BOS-iPSC lines (E) compared with respective control lines (mean ± SEM, n = 3–6 different clones/passages; NR2F1 in iPSC control, n = 2 different passages; ∗p < 0.05).
(F) Quantification of ZIC1 protein levels in NC cultures (day 7) derived from cell lines as in (D and E) compared with the respective control lines (based on western blotting, n = 3–4 different clones/passages, mean ± SEM, ∗p < 0.05).
(G and H) Rescue of the NC differentiation defect by ectopic expression of ZIC1. (G) Detection of ZIC1 overexpression in DOX-treated PB-ZIC1-ASXL1+/+ by western blotting. (H) Analysis of attached neurospheres with emigrating cells in PB-ZIC1-ASXL1+/+ and PB-ZIC1-ASXL1PSC/PSC lines at day 7, −/+ DOX treatment to overexpress ZIC1 starting at day 3 (n = 2 and 3 independent experiments) or day 4 (n = 3 independent experiments). All data shown as mean ± SEM, ∗p < 0.05.
See also Figure S3.
ZIC1, which was 50-fold reduced in ASXL1PSC/PSC NC cultures, is at the apex of the GRN controlling NC induction (Simoes-Costa and Bronner, 2015). Accordingly, transcription factors that promote further specification, delamination, and migration of the NC were also negatively regulated in the ASXL1PSC/PSC NC cells, including PAX3, GBX2, MSX1, NR2F1/2, ZEB2, COL2A1, TWIST1, SOX5, Cadherins, and Ephrins and their cognate receptors (Rada-Iglesias et al., 2012, Simoes-Costa and Bronner, 2015) (Figures 3C, S3B, and S3C, Table S2). qPCR, performed to confirm the transcriptional changes for a subset of NC genes, demonstrated significant downregulation in the ASXL1PSC/PSC clones and mild reduction in the patient lines (Figures 3D and 3E), and ZIC1 protein levels were significantly lower in NC cultures derived from one BOS-iPSC line (Figures 3F and S3D). To further analyze the connection of the ASXL1PSC/+ ESC line to NC defects, we performed 3′ mRNA sequencing of NC cultures derived from the heterozygous ASXLPSC/+ clones and control hESCs. This revealed mutually exclusive clustering of the two genotypes and a significant 28-fold downregulation of ZIC1 in ASXLPSC/+ NC progenitors (padj < 0.05; Figures S3E–S3G). Collectively, this supports a link between BOS-associated mutations in ASXL1 and ZIC1 misregulation.
To investigate whether reduced ZIC1 levels could indeed underlie the observed NC differentiation phenotypes, we stably integrated an inducible ZIC1 transgene in ASXL1+/+ and ASXL1PSC/PSC hESC clones (Figure 3G, Table S1). Strikingly, overexpression of ZIC1 during NC differentiation restored the attachment and delamination phenotype of NC-like cells from ASXL1PSC/PSC neurospheres (Figure 3H). These results collectively indicate that impairment of ZIC1 activation contributes to the observed differentiation defects when ASXL1 is mutated.
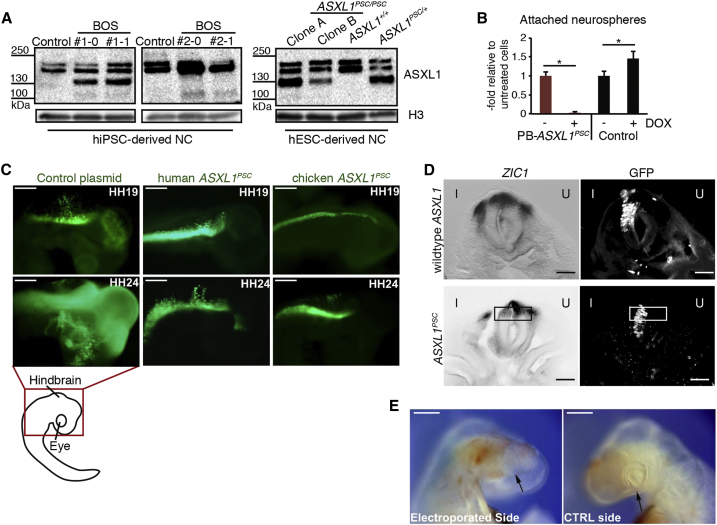
Expression of Truncated ASXL1 Recapitulates NC Developmental Effects
We next investigated how regulation of ASXL1 could be implicated in the differentiation outcomes displayed by the BOS models. We noted decreased expression of ASXL1 in NC progenitors derived from ASXL1PSC/+ and ASXL1PSC/PSC hESC, and a similar effect for the BOS-iPSC lines, which was however not deemed statistically significant (Figure S4A). Moreover, mutant and wild-type ASXL1 transcripts were found in NC cultures derived from BOS #1 and #2 lines (Figure S4B), and both alleles were expressed at comparable levels in BOS-iPSC #1 lines (Figure S4C). This argues against degradation of the mutant mRNA. We found that overexpression of wild-type ASXL1 by a stably integrated PiggyBac vector (Table S1) could not rescue the attachment defect of neurospheres derived from ASXL1PSC/PSC hESCs, nor did it rescue expression of key NC genes (Figures S4D and S4F). These findings suggest a dominant role of putative truncated ASXL1 variants over the wild-type form. Western blot analysis of early NC cells from all BOS models, using the monoclonal antibody raised against the N-terminal domain of ASXL1, revealed barely detectable levels of a putative truncated variant at 100 kDa and enhanced levels of different variants at 130 and 160 kDa (Figure 4A). Collectively, these results raise the possibility that truncated ASXL1 variants, reduction in ASXL1 levels, and potentially also misexpression of additional ASXL1 variants (Figures S1I, S1J, and 4A) contributed to the observed NC defects.
Figure 4.
Expression of Truncated ASXL1 Impairs NC Cell Emigration and ZIC1 Expression
(A) Detection of ASXL1 by western blotting in samples of day 7 NC cultures (monoclonal antibody as in Figures 1E and 1F).
(B) Percentage of attached neurospheres with emigrating cells derived at day 7 from DOX-treated ASXL1+/+ hESCs and PB-ASXL1PSC hESCs, overexpressing truncated ASXL1, relative to untreated samples (mean ± SEM, n = 3 independent experiments, ∗p < 0.05).
(C) Representative fluorescent images of chicken embryos 48 h after electroporation with plasmids expressing GFP or GFP coupled to truncated chicken or human ASXL1PSC (n = 10 embryos each); the red rectangle indicates displayed head/hindbrain region. Scale bars, 200 μm.
(D) Expression pattern of ZIC1 analyzed by in situ hybridization in neural tube sections of chicken embryos (HH15) electroporated with plasmids encoding for truncated (ASXLPSC) or full-length chicken ASXL1 coupled to GFP. I, injected side; U, control side. n = 3 sections per 2 embryos. Scale bar, 50 μm.
(E) Representative bright-field image of chicken embryo electroporated with truncated chicken ASXL1 transcript as in (C), showing craniofacial malformations (arrow, missing periocular tissue) on the injected side in comparison with the uninjected control (CTRL) side. n = 6 embryos. Scale bars, 200 μm.
See also Figure S4.
To substantiate one of these possibilities, we expressed in hESCs a truncated ASXL1 isoform, which was similar to the variant detected in BOS patient 2 (Table S1). Such ectopic expression led to a nearly complete failure of neurospheres to attach and produce migratory NC-like cells (Figure 4B). Moreover, truncated human as well as chicken ASXL1 that were injected into the anterior neural region of chicken embryos inhibited the delamination of the electroporated GFP-labeled cells, an effect that was not seen in embryos injected with control plasmid encoding only eGFP (Figure 4C). Strikingly, ZIC1 expression was reduced in neural tube cells expressing truncated chicken ASXL1 but not in embryos electroporated with full-length chicken ASXL1 (Figures 4D and S4G). Under the same conditions, expression of TFAP2A was unaltered (Figure S4G), indicating a specific perturbation of ZIC1 expression that was similar to the case of ASXL1PSC/PSC NC progenitors in vitro (Figure S3B).
Finally, we analyzed electroporated embryos at later developmental stages, around HH22–25, to assess craniofacial phenotypes caused by expression of truncated ASXL1. This revealed decreased eye size and malformation of periocular tissue on the injected side compared with the uninjected side (Figure 4E). We thus concluded that expression of truncated ASXL1 in NC progenitors could lead to BOS-like phenotypes in vivo.
Collectively, in this study we created a human pluripotent stem cell toolset for investigating pathological ASXL1 mutations present in, or resembling genotypes of, BOS patients. These models enabled us to discover the impaired generation of NC cells, which might be partially caused by a defect in early neuroectoderm specification, and our results support an association with reduced induction of ZIC1. However, the question of whether the truncated ASXL1 isoforms by themselves, haploinsufficiency, other isoforms, or combinations of these mechanisms underlie BOS pathogenesis warrants additional studies. A similar question exists with respect to truncated endogenous ASXL1 variants that are associated with leukemia (Abdel-Wahab et al., 2012, Balasubramani et al., 2015, Inoue et al., 2016). Thereby, our findings and models are likely pertinent to hematological malignancies. We moreover linked the truncated isoform to craniofacial dysmorphisms, which have strong association with BOS, and therefore propose ASXL1-mediated perturbation of the NC as a potential cause of characteristic symptoms in BOS. A further medical relevance is apparent from the symptomatic similarity of BOS to Bainbridge-Roper syndrome (BRS), a congenital disorder that is associated with ASXL3 mutations (Bainbridge et al., 2013), which raises the possibility that BRS is caused by similar mechanisms. The results may also explain the occurrence of Dandy-Walker malformation in BOS patients that could take place due to ZIC1/ZIC4 repression (Grinberg et al., 2004). Finally, the speech and language learning disabilities in BOS (Hastings et al., 2011) may be linked to the reduced FOXP2 activation (Figure 3A), as this transcription factor has a primary role in speech and language development (Enard et al., 2002). Overall, the established models could help elucidate ASXL1-related pathological mechanisms in additional tissues, including the nervous system and the hematopoietic lineage.
Experimental Procedures
Generation of Monoclonal Antibodies
Expression and purification of an N-terminal fragment of ASXL1 (amino acids 1–618), followed by immunization of Lou/c rats was performed as previously described (Feederle et al., 2016, Studier, 2005).
Generation of Pluripotent Stem Cell Lines and Neural Crest Differentiation
Fibroblasts derived from skin biopsies of healthy donors and BOS patients under oversight by local institutional review boards (Magini et al., 2012) were reprogrammed by modified mRNA and episomal plasmids (Diecke et al., 2015, Kunze et al., 2018), and 500 bp deletions in the ASXL1 gene were induced in iCas9 HUES9 hESC (ASXL1+/+ control line) (Gonzalez et al., 2014). ASXL1PSC/PSC and ASXL1+/+ hESC lines were stably integrated with PiggyBac vectors harboring either the truncated ASXL1 cDNA sequence (N-terminal 2,892 bp; PB-ASXL1PSC), the full-length human ASXL1 cDNA (4,656 bp; PB-ASXL1), the ZIC1 cDNA sequence (amplified from ZIC1 human cDNA clone, Biocat; PB-ZIC1), or an expression cassette for continuous eGFP expression.
Neural crest differentiation was performed as described (Bajpai et al., 2010), and the number of attached neurospheres was quantified at day 7 of the protocol.
Manipulation of Chicken Embryos
According to German animal care guidelines, no IACUC (Institutional Animal Care and Use Committee) approval was necessary to perform chicken embryo experiments.
Neurospheres obtained at day 5 of NC differentiation were inserted into the developing anterior neural region of chicken embryos (HH10; Hamburger and Hamilton, 1992) and operated embryos were isolated at HH22 and analyzed under a fluorescence stereo microscope. To overexpress transcripts in chicken embryos, the truncated chicken (Gallus gallus) ASXL1 cDNA sequence (Gg-ASXL1PSC; N-terminal 2,445 bp), the truncated human ASXL1 cDNA sequence (hASXL1PSC; N-terminal 2,892 bp), or the full-length chicken ASXL1 cDNA sequence (Gg-ASXL1; 4,617 bp) were cloned into expression plasmids and electroporated into the developing brain and neural tube at HH9–10 as described (Rehimi et al., 2016). Manipulated chicken embryos were imaged at HH19 and HH24–25 under a fluorescence stereo microscope. In situ hybridization experiments in electroporated chicken embryos were performed as described (Rehimi et al., 2016).
Transcriptome Analysis (RNA Sequencing)
Total RNA was isolated from day 7 NC progenitor cultures and libraries were prepared using the TruSeq Stranded Total RNA LT Library Prep Kit (Illumina) followed by single-end sequencing. Differential gene expression analysis was performed with the DESeq2 package (Love et al., 2014).
Statistical Analysis
Values are expressed as means ± standard error of the mean; pairwise comparison was performed using Welch's t test or Wilcoxon’s rank-sum test.
Author Contributions
F.M. and M.D. designed the study. F.M., E.R., and L.M. performed functional experiments. E.R. and A.P. generated iPSC lines. M.M. performed library preparation and analyzed 3′ RNA sequencing experiments. R.R. and A.R.-I. contributed to the chicken embryo experiments. E.K., R.F., A.F., and A.G. produced monoclonal antibodies. V.R. performed MSC analysis. F.M. and M.D. wrote the manuscript with input from the other authors.
Acknowledgments
We thank the patients' families and following scientists for generously providing fibroblasts: Pamela Magini (S. Orsola-Malpighi Hospital, University of Bologna, Bologna, Italy), Matteo Della Monica (Meyer Children's Hospital, University of Florence, Florence, Italy), Gioacchino Scarano (Medical Genetics Unit, A.O.R.N “G. Rummo”, Benevento, Italy), and Prof. Maria Luisa Giovannucci Uzielli (Genetics and Molecular Medicine, University of Florence, Florence, Italy). We would also like to thank Meino Rohlfs and Jens Hinke (Klinikum LMU, Munich) and Christine Wurmser (TU Munich) for providing help with RNA sequencing, Lukas Simon (Helmholtz Zentrum München, Neuherberg) for support with bioinformatics data analysis, and Dmitry Shaposhnikov (Helmholtz Zentrum München) for providing plasmids.
Published: April 18, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.03.006.
Accession Numbers
Gene expression data from the RNA sequencing experiments have been deposited in NCBI's Gene Expression Omnibus (GEO) under accession numbers GEO: GSE120200 and GSE127269.
Supplemental Information
References
- Abdel-Wahab O., Adli M., LaFave L.M., Gao J., Hricik T., Shih A.H., Pandey S., Patel J.P., Chung Y.R., Koche R. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22:180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge M.N., Hu H., Muzny D.M., Musante L., Lupski J.R., Graham B.H., Chen W., Gripp K.W., Jenny K., Wienker T.F. De novo truncating mutations in ASXL3 are associated with a novel clinical phenotype with similarities to Bohring-Opitz syndrome. Genome Med. 2013;5:11. doi: 10.1186/gm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai R., Chen D.A., Rada-Iglesias A., Zhang J., Xiong Y., Helms J., Chang C.P., Zhao Y., Swigut T., Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramani A., Larjo A., Bassein J.A., Chang X., Hastie R.B., Togher S.M., Lahdesmaki H., Rao A. Cancer-associated ASXL1 mutations may act as gain-of-function mutations of the ASXL1-BAP1 complex. Nat. Commun. 2015;6:7307. doi: 10.1038/ncomms8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diecke S., Lu J., Lee J., Termglinchan V., Kooreman N.G., Burridge P.W., Ebert A.D., Churko J.M., Sharma A., Kay M.A. Novel codon-optimized mini-intronic plasmid for efficient, inexpensive, and xeno-free induction of pluripotency. Sci. Rep. 2015;5:8081. doi: 10.1038/srep08081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W., Przeworski M., Fisher S.E., Lai C.S., Wiebe V., Kitano T., Monaco A.P., Paabo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- Etchevers H.C., Amiel J., Lyonnet S. Molecular bases of human neurocristopathies. Adv. Exp. Med. Biol. 2006;589:213–234. doi: 10.1007/978-0-387-46954-6_14. [DOI] [PubMed] [Google Scholar]
- Feederle R., Gerber J.K., Middleton A., Northrup E., Kist R., Kremmer E., Peters H. Generation of Pax1/PAX1-specific monoclonal antibodies. Monoclonal antibodies in immunodiagnosis and immunotherapy. Monoclon. Antib. Immunodiagn. Immunother. 2016;35:259–262. doi: 10.1089/mab.2016.0029. [DOI] [PubMed] [Google Scholar]
- Gonzalez F., Zhu Z., Shi Z.D., Lelli K., Verma N., Li Q.V., Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg I., Northrup H., Ardinger H., Prasad C., Dobyns W.B., Millen K.J. Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy-Walker malformation. Nat. Genet. 2004;36:1053–1055. doi: 10.1038/ng1420. [DOI] [PubMed] [Google Scholar]
- Guo Y., Yang H., Chen S., Zhang P., Li R., Nimer S.D., Harbour J.W., Xu M.J., Yang F.C. Reduced BAP1 activity prevents ASXL1 truncation-driven myeloid malignancy in vivo. Leukemia. 2018;32:1834–1837. doi: 10.1038/s41375-018-0126-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V., Hamilton H.L. A series of normal stages in the development of the chick embryo. 1951. Dev. Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Hastings R., Cobben J.M., Gillessen-Kaesbach G., Goodship J., Hove H., Kjaergaard S., Kemp H., Kingston H., Lunt P., Mansour S. Bohring-Opitz (Oberklaid-Danks) syndrome: clinical study, review of the literature, and discussion of possible pathogenesis. Eur. J. Hum. Genet. 2011;19:513–519. doi: 10.1038/ejhg.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D., Matsumoto M., Nagase R., Saika M., Fujino T., Nakayama K.I., Kitamura T. Truncation mutants of ASXL1 observed in myeloid malignancies are expressed at detectable protein levels. Exp. Hematol. 2016;44:172–176.e1. doi: 10.1016/j.exphem.2015.11.011. [DOI] [PubMed] [Google Scholar]
- Kunze C., Borner K., Kienle E., Orschmann T., Rusha E., Schneider M., Radivojkov-Blagojevic M., Drukker M., Desbordes S., Grimm D. Synthetic AAV/CRISPR vectors for blocking HIV-1 expression in persistently infected astrocytes. Glia. 2018;66:413–427. doi: 10.1002/glia.23254. [DOI] [PubMed] [Google Scholar]
- Lee G., Kim H., Elkabetz Y., Al Shamy G., Panagiotakos G., Barberi T., Tabar V., Studer L. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat. Biotechnol. 2007;25:1468–1475. doi: 10.1038/nbt1365. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magini P., Della Monica M., Uzielli M.L., Mongelli P., Scarselli G., Gambineri E., Scarano G., Seri M. Two novel patients with Bohring-Opitz syndrome caused by de novo ASXL1 mutations. Am. J. Med. Genet. A. 2012;158A:917–921. doi: 10.1002/ajmg.a.35265. [DOI] [PubMed] [Google Scholar]
- Mayor R., Theveneau E. The neural crest. Development. 2013;140:2247–2251. doi: 10.1242/dev.091751. [DOI] [PubMed] [Google Scholar]
- Merzdorf C.S. Emerging roles for zic genes in early development. Dev. Dyn. 2007;236:922–940. doi: 10.1002/dvdy.21098. [DOI] [PubMed] [Google Scholar]
- Noden D.M., Trainor P.A. Relations and interactions between cranial mesoderm and neural crest populations. J. Anat. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Prescott S., Brugmann S.A., Swigut T., Wysocka J. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell. 2012;11:633–648. doi: 10.1016/j.stem.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos T.L., Sanchez-Abarca L.I., Muntion S., Preciado S., Puig N., Lopez-Ruano G., Hernandez-Hernandez A., Redondo A., Ortega R., Rodriguez C. MSC surface markers (CD44, CD73, and CD90) can identify human MSC-derived extracellular vesicles by conventional flow cytometry. Cell Commun. Signal. 2016;14:2. doi: 10.1186/s12964-015-0124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehimi R., Nikolic M., Cruz-Molina S., Tebartz C., Frommolt P., Mahabir E., Clement-Ziza M., Rada-Iglesias A. Epigenomics-based identification of major cell identity regulators within heterogeneous cell populations. Cell Rep. 2016;17:3062–3076. doi: 10.1016/j.celrep.2016.11.046. [DOI] [PubMed] [Google Scholar]
- Simoes-Costa M., Bronner M.E. Establishing neural crest identity: a gene regulatory recipe. Development. 2015;142:242–257. doi: 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider T.N., Mishina Y. Cranial neural crest cell contribution to craniofacial formation, pathology, and future directions in tissue engineering. Birth Defects Res. C Embryo Today. 2014;102:324–332. doi: 10.1002/bdrc.21075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.