ABSTRACT
The global outputs of annual publication in long non-coding RNAs (lncRNAs) and chemotherapeutic resistance research exponentially increased from 2 in 2008 to 176 in 2017. Using Java application CiteSpace V and VOSviewer, this study assessed the publication model of lncRNAs and chemoresistance by bibliometric analysis. Totally, 2883 authors contributed 528 publications of lncRNAs and chemoresistance in 215 academic journals in the recent decade (2008-2018). Oncotarget in the 215 academic journals published the highest number of publications (60). China had the highest number of publication outputs (358). The leading institute was Nanjing Medical University. Wang Y was the most influential author (13 counts). Gupta RA had the most cited documents (87 counts). “Gene expression” and “poor prognosis” were identified as the hotspots. “Cancer stem cell”, “HOTAIR” and “UCA1” were the frontiers of the fields in recent years. The increase of publications on lncRNAs and chemotherapeutic resistance will continue in the next years. HOTAIR and UCA1 with multiple roles in drug resistance may offer big opportunities for targeted chemoresistance in cancer therapy. These results may help us discover and explain the possible underlying laws of the subject.
Keywords: chemotherapeutic resistance, lncRNA, citespace, bibliometric analysis
INTRODUCTION
Chemotherapeutic agents, such as cisplatin, sunitinib, gemcitabine, paclitaxel and PARP inhibitors, have been widely applied in cancer treatment. While entirely eradicating the tumor and curing patients by chemotherapy alone are rare, in particular for advanced disease. The main obstacles are drug resistance that may be resulted from intrinsic and/or acquired factors. The factors of intrinsic resistance include drug breakdown, the aberrant expression and/or function of drug target, altered drug transport across membrane transporter, the ineffective interaction between drug and its molecular target [1]. Inactivation of tumor suppressor gene TP53 was also indicated to cause resistance to chemotherapeutic drugs [2]. Acquired drug resistance is usually associated with genetic or environmental factors that facilitate the development of chemoresistance cancer cell clones and/or induce gene mutations involved in relevant metabolic pathways. As the key regulatory factor of tumor stem cells, lncRNA Malat-1 can increase the ratio of pancreatic cancer stem cell, maintaining the ability of self-renewal, thus reduce the sensitivity to anticancer drugs [3]. LncRNA HOTAIR reprograms chromatin state to promote cancer metastasis [4]. The disorders or mutations of lncRNAs were indicated to be associated with cancer and chemoresistance, such as lncRNA CCAL in colorectal cancer [5], HOTTIP in osteosarcoma [6] and SNHG12 in non-small-cell lung cancer [7]. Publications focused on the research of lncRNAs and drug resistance rapidly increased in the past decade. Bibliometrics is a statistical method to assess various aspects (such as annual growth of publications, the contribution of countries, core journals and most active researchers) of scientific literature, which can help us discover and explain the possible underlying laws of the subject in lncRNAs and chemotherapeutic resistance research. There are still no reports about bibliometrics analysis of lncRNAs and chemotherapeutic resistance available in public database. Therefore, using Java application CiteSpace V [8], VOSviewer [9] and Microsoft Excel 2016 software, we carried out this study to qualitatively and quantitatively evaluate the published publications of lncRNAs and drug resistance for a better understanding on the various aspects of these publications.
RESULTS
Publication outputs and future development trend
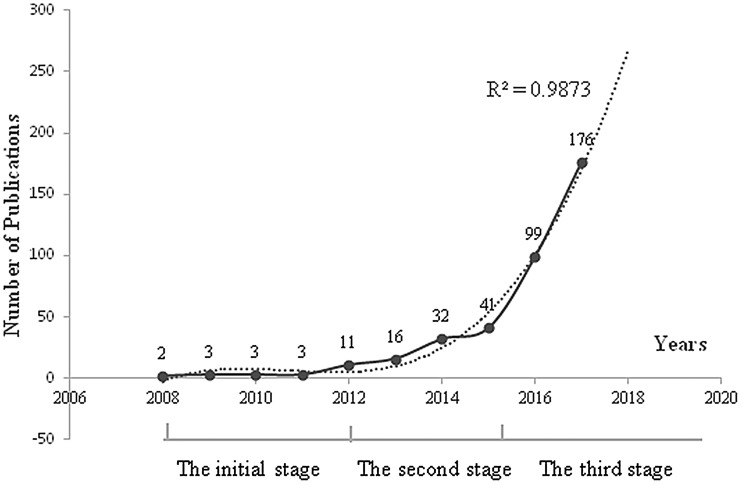
Excluding 14 non-official articles or reviews (6: Book chapter; 4: Meeting abstracts; 2: Editorial material; 2: Proceedings paper), 528 publications (398: research articles; 130: review) indexed in the WoSCC of Thomson Reuters from 2008 to 2018 were extracted for further bibliometric analysis. 526 of the 528 articles (99.621%) were published in English, one in Chinese and one in German. The annual publications in WoSCC exponentially increased from 2 in 2008 to 176 in 2017 (Figure 1). With highly reliable correlation coefficients (R2 = 0.9873), the model fitting curve revealed a significant correlation between the annual publications and publication year. Based on the equation of model fitting, it is estimated that the number of publications on lncRNAs and chemotherapeutic resistance would be about 268 in 2018. And 243 publications indexed in the WoSCC were retrievable on August 19, 2018.
Figure 1. Publication outputs and growth prediction.
The number of publications from 2008 to 2017 represented by the solid line; the dashed line represents the forecast curve, R2 = 0.9873.
Journal analysis
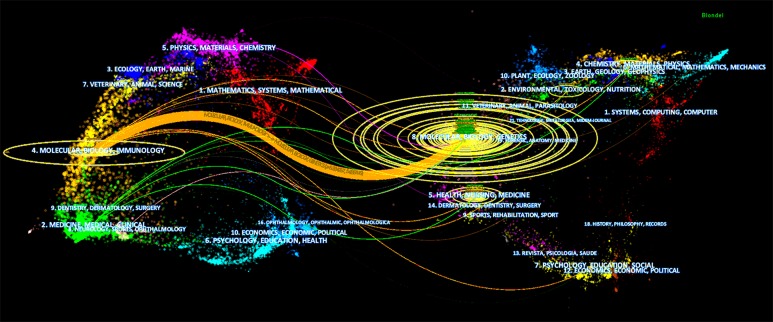
The 528 publications of lncRNAs and chemoresistance were published in 215 academic journals. And the top 15 active journals published 206 of the 528 articles, accounting for 39.016% (Table 1). With 60 counts, Oncotarget published the highest publications of lncRNAs and chemoresistance, accounting for 11.364%, followed by Oncology Reports and Tumor Biology (Table 1). Citation paths at a disciplinary level were demonstrated in the visual representation called a dual-map overlay, which effectively mapped the field and direction of the topic (Figure 2). The labels on the map represented the subject fields covered by the journals. The left of the map represented where the collected reports published and the right regions rendered where it cited from (Figure 2). Most of these publications were published in molecular journals, biology journals, and immunology journals, and cited from molecular, biology, and genetics. Citing publications and cited publications are called research frontier and knowledge base, respectively. The base map consisted of the journal/conference-level citation relationships among over 1000 venues. Major clusters were labeled by terms chosen from the titles of venues in corresponding clusters. Citation trajectories were colored according to the citing regions. The width of the paths is proportional to the z-score-scaled citation frequency (Figure 2).
Table 1. The top active 15 journals that published these publications.
| Ranking | Journal | Country | Count | Percentage (%) | IF2017 |
|---|---|---|---|---|---|
| 1 | Oncotarget | United States | 60 | 11.364 | 5.168 |
| 2 | Oncology Reports | United States | 15 | 2.841 | 2.976 |
| 3 | Tumor Biology | Switzerland | 15 | 2.841 | 3.27 |
| 4 | Cellular Physiology and Biochemistry | Switzerland | 12 | 2.273 | 5.500 |
| 5 | International Journal of Molecular Sciences | United States | 12 | 2.273 | 3.687 |
| 6 | Biomedicine and Pharmacotherapy | France | 11 | 2.083 | 3.457 |
| 7 | PLOS ONE | United States | 11 | 2.083 | 2.766 |
| 8 | Cell Death and Disease | England | 10 | 1.894 | 5.638 |
| 9 | Molecular Cancer | England | 10 | 1.894 | 7.776 |
| 10 | Oncology Letters | Greece | 9 | 1.705 | 1.664 |
| 11 | OncoTargets and Therapy | England | 9 | 1.705 | 2.656 |
| 12 | Biochemical and Biophysical Research Communications | United States | 8 | 1.515 | 2.559 |
| 13 | Cancer Chemotherapy and Pharmacology | United States | 8 | 1.515 | 2.808 |
| 14 | Cancer Letters | Netherlands | 8 | 1.515 | 6.491 |
| 15 | International Journal of Clinical and Experimental Medicine | United States | 8 | 1.515 | 0.833 |
Figure 2. Dual-map overlay of journals on lncRNA and chemoresistance.
Citation paths at a disciplinary level were demonstrated in t a dual-map overlay. The left of the map represented the cite journals and the right of the map represented the cited journals. Citation trajectories are colored based on the citing regions. The width of the paths is proportional to the z-score-scaled citation frequency.
Country and institution analysis
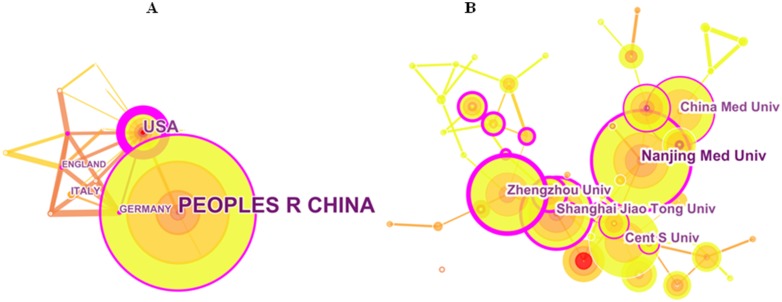
The distribution map of the country/institution can provide valuable information for researchers to better understand the community where they’re working. The distribution map indicated that the 528 publications came from 695 research institutes in 46 countries/territories (Figure 3A). The top 10 countries/territories that contributed these publications included three Asian countries, three European countries, two North America, one Middle East and one Oceania (Figure 3A and Table 2). China (358) and the United States (98) were the top two countries, accounting for 86.364% of the total publications (Figure 3A). As a developing country in this group, China had made remarkable progress in this field in the past decade. While the United States ranked the first in the citation frequency. Therefore, for the quality of research, the United States was the leading nation in this field. The cooperation between the United States and other countries was much closer than that of China (Figure 3A).
Figure 3. The distribution of countries/territories and institutions.
(A) Map of countries/territories that published these publications. (B) Map of institutions that published these publications.
Table 2. The top ten productive countries and institutions of the total publications.
| Ranking | Country | Count | Institution | Count |
|---|---|---|---|---|
| 1 | China | 358 | Nanjing Medical University | 29 |
| 2 | USA | 98 | Zhengzhou University | 22 |
| 3 | Italy | 21 | Central South University | 21 |
| 4 | Germany | 17 | Shanghai Jiao Tong University | 21 |
| 5 | England | 14 | China Medical University | 20 |
| 6 | Australia | 9 | Nanjing University | 14 |
| 7 | Japan | 9 | Tongji University | 13 |
| 8 | Taiwan | 9 | Dalian Medical University | 13 |
| 9 | Canada | 6 | Harbin Medical University | 11 |
| 10 | Iran | 6 | University Texas MD Aanderson Cancer Center | 10 |
The top 10 institutions published 174 of the 528 publications, accounting for 32.955% (Table 2). Nanjing Medical University (29) ranked first place, followed by Zhengzhou University (22) and Central South University (21) (Figure 3B and Table 2). Among the top 10 active institutions, one was from the United States (The University of Texas M.D. Anderson Cancer Center) and 9 were from China. Chinese institutions were in relatively close cooperation in the collaboration network, which may partially explain the observation that China contributed to most of the publications.
Author analysis
The 528 articles were written by 2883 authors working in the subject. The network map embodied the cooperation among authors (Supplementary Figure 1A). Among the top 10 authors with the largest number publications, WANG Y (13 publications) ranked the first, followed by LI J (11 publications) and Liu Y (11 publications) (Supplementary Figure 2 and Table 3). The author citation information was presented as a co-cited author map (Supplementary Figure 1B). Among the top 10 authors with the largest citation, Gupta RA was the top-ranked author (87), followed by Wang Y (79) and Mercer TR (58) (Table 3).
Table 3. The top 10 authors and co-cited authors working in the field.
| Ranking | Author | Count | Co-cited Author | Count |
|---|---|---|---|---|
| 1 | Wang Y | 13 | Gupta RA | 87 |
| 2 | Li J | 11 | Wang Y | 79 |
| 3 | Liu Y | 11 | Mercer TR | 58 |
| 4 | Li X | 9 | Gutschner T | 55 |
| 5 | Wang J | 8 | Fan Y | 51 |
| 6 | Wang L | 7 | Wang F | 48 |
| 7 | Li L | 7 | Tsang WP | 47 |
| 8 | Li Y | 7 | Wang KC | 46 |
| 9 | Zhang L | 7 | Li J | 43 |
| 10 | Shang C | 6 | Jemal A | 38 |
Reference co-citation analysis
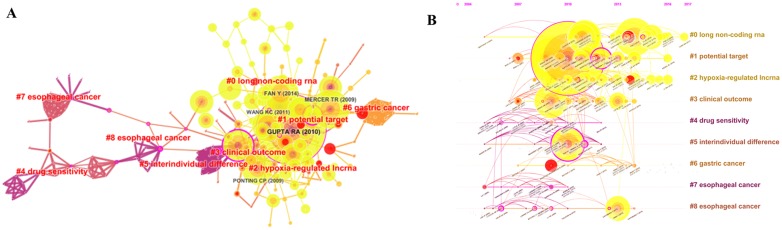
The knowledge map of citation reference of the 528 publications with 258 nodes and 539 connection lines was built (Figure 4A). The knowledge map was composed of centrality and citations, which can be used to analyze the frontier areas of this field by exploring the key clusters of publications (Figure 4A). Each cluster is a distinctive specialty or a thematic concentration. The modularity Q score was calculated to evaluate the modularity of the clustering network. The modularity Q score ranges from 0 to1, where a high value indicates that the object is well matched to the modularity. The modularity value was 0.7882, indicating that the network was reasonably divided into loosely coupled clusters. Preliminary analysis showed 51 clusters with a Mean Silhouette of 0.3891. Filtered out small clusters with a low silhouette (less than 10), the top 8 largest clusters were finally retained. Among the 8 clusters, the largest cluster #0 was “long non-coding RNA”, followed by cluster #1, labeled as “potential target”, and the third largest cluster #2 labeled as “hypoxia-regulated LncRNA “. Figure 4B was a timeline view of the top 8 largest clusters that showed the citations of each document in each cluster, and the corresponding changes of the key clusters.
Figure 4. The analysis of references.
(A) Co-citation knowledge map of references on lncRNA and chemoresistance. (B) Timeline view of references on lncRNA and chemoresistance with log-likelihood ratio (LLR) cluster labels. The color of the node represents the newness of the related reference (the red color for new studies).
Keyword co-occurrence cluster analysis
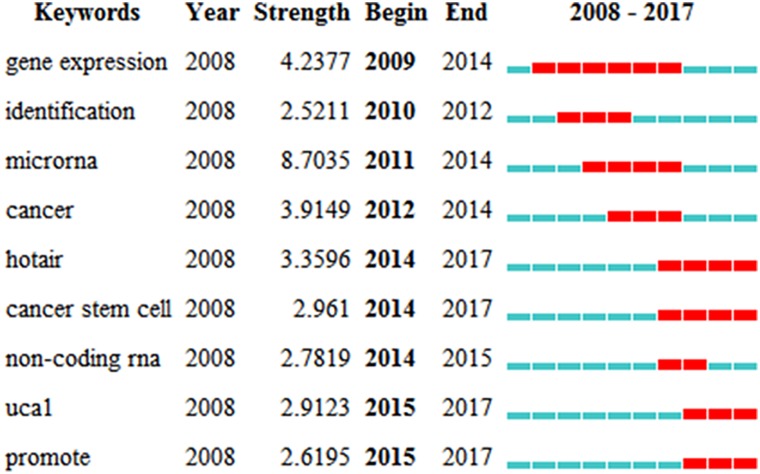
Keywords with high degree centrality and frequency represent the research hot spots over a period of time. The burst keywords represent new research frontiers. Centrality is an indicator of the relative importance of nodes in a network with respect to the total system activity. The higher frequency of co-occurrence of keywords and the stronger centrality of the points indicate the more important of the node in the field [10]. Among the 128 keywords (Indication and Class Description) extracted from the 528 publications, the top 3 high-frequency keywords were “long non-coding RNA”, “expression” and “drug resistance” (Supplementary Figure 2). The histogram showed 14 of the top 17 high-frequency keywords, excluding drug resistance, chemoresistance and chemotherapy, which were roughly divided into three parts: “gene expression”, “poor prognosis” and “cancer types” (Supplementary Figure 3). Among the 9 burst keywords, the strongest ones after 2014 included “HOTAIR” (2014–2017), “cancer stem cell” (2014–2017), “non-coding RNA” (2014–2015), “UCAL” (2015–2017) and “promote” (2015–2017) (Figure 5).
Figure 5. The top 9 keywords with the strongest citation bursts.
DISCUSSION
Co-existing analysis
The analysis of publication outputs indicated that the development of lncRNAs and chemoresistance research could be roughly divided into three stages: the initial stage (2008–2011) developing at a slow pace (2 in 2008; 3 in 2009, 2010 and 2011, respectively); the second stage with steady growth (2012–2015) (11 in 2012; 16 in 2013; 32 in 2014; 41 in 2015); the third stage with exponential growth (after 2015) (Figure 1). The prediction curve showed that the growth accompanied with the increase of our knowledge in the subject will continue in the next few years.
The impact factor (IF) of a journal is an important factor to evaluate its influence of the published articles. Among the top 15 active journals that published these publications, 6 journals had an IF > 3.0, accounting for 30.583% (6/15). Four journals (4/15, 19.417%) had an IF > 5 (Table 1). Journals with extremely high impact factor, such as Nature, Science and Cell, did not publish the topic articles. Maybe, these publications that studied the roles of lncRNA in chemotherapeutic resistance were not enough to be published in extreme high-impact journals, and thus we still need to strive for excellence in this field.
References and citation analysis
Everyone in the top 10 prolific authors had at least six publications. While the comparison of the top 10 prolific authors with the top 10 co-cited authors showed that only Wang Y and Li J in the 10 prolific authors also appeared in the list of the top 10 co-cited authors (Supplementary Figure 1 and Table 3). The observations may suggest that authors should pay more attention to the quality of their papers when trying to increase their publication number.
The analysis of references showed that Gupta RA (2010) had the highest number of references (87), followed by Mercer TR (2009) with 50 references and Fan Y (2014) with 45 references (Figure 4A). The timeline graph revealed the high-impact articles published in 2009–2016. The largest cluster #0 labeled as “long non-coding RNA” indicated the high co-citation articles for each year (Figure 4B). Analysis of high co-citation articles in cluster #0 revealed the diverse biological functions of lncRNAs. For example, as a part of the complex, lncRNA H19 associated with EZH2 enhancer and inhibited E-cad expression, which resulted in the activation of the Wnt/β-catenin pathway enhancing bladder cancer metastasis [11]. The increased level of lncRNA UCA1 activated Wnt signaling and resulted in the cisplatin resistance of bladder cancer cells in a Wnt6-dependent manner [12].
Research hot spots
Keywords are the author’s generalization and refinement of the article core. Keywords with a high degree centrality were “gene expression”, “poor prognosis” and “cancer types”, indicating that research topics represented by these keywords are at the core of lncRNA and chemotherapeutic resistance research in the past decade (Supplementary Figure 3). To date, about 20 lncRNAs were demonstrated to mediate chemotherapeutic resistance by various and often undefined mechanism in different cancer [13]. Through competitively binding mR-33a-5P, miR-33b-5P, miR-1-3P, miR-206 and miR-613, ANCR increased AXL expression and activated PI3K/Akt/NF-κB signaling pathway that caused cisplatin resistance in glioma cells [14]. Breast cancer patients with poor response to chemotherapy had fourfold higher CRALA level than responding patients [15]. Chronic oxymatrine treatment induced chemotherapy resistance and epithelial-mesenchymal transition through targeting MALAT1 in colorectal cancer cells [16]. The increased expression of HOTAIR and its DNA methylation appeared to induce carboplatin resistance [17], indicating that HOTAIR may contribute to resistance by influencing methylation.
Research frontiers
The burst words represent keywords that is often cited over a period of time, thereby indicating the frontier areas. The first 3 burst words after 2014 were “cancer stem cell”, “HOTAIR” and “UCA1” (Figure 5). Cancer stem cell is cancer cells that possess the characteristics of normal stem cells and can generate tumors through the stem cell processes of self-renewal and differentiation into multiple cell types. LncRNAs known to be a regulatory factor of tumor stem cells included GAS5, NEAT1, SOX2OT, Malat-1, HOTAIR, PVT1, BCAR4 and RBM5-AS1. As the key regulatory factor of tumor stem cells, Malat-1 increased the ratio of pancreatic cancer stem cell, maintained the ability of self-renewal, and thus reduced the sensitivity to anticancer drugs [3].
The burst words of “HOTAIR” and “UCA1” reflected a wide study of their roles in chemoresistance from the scientific community. HOTAIR has been shown to mediate radiosensitivity, endocrine resistance and chemotherapeutic resistance in a variety of ways [18]. The increased HOTAIR expression induced cisplatin resistance through activatingthe Wnt/b-catenin pathway and promoting cell cycle progression in ovarian cancer [19]. HOTIAR mediated chemoresistance in DNA-damage responses through the NF-κB signaling pathway [20]. Both in vitro and in vivo experiments confirmed that UCA1 induced cisplatin/gemcitabine resistance through the activation of CREB by p-AKT and the subsequent up-regulation of miR-196a-5p [21]. The increased UCA1 activated the Wnt pathway and decreased cisplatin-induced cell death [12]. UCA1 in SKOV3 cells increased SRPK1 and Bcl-2, decreased Bax, Caspase-3 and Caspase-9 expression and contributed to cisplatin resistance [22]. UCA1 induced tamoxifen resistance in breast cancer cells partly through activation of the mTOR pathway [23]. Acting as a sponge for miR-18a, UCA1 is an important modulator of tamoxifen resistance by regulating cell cycle proteins [24]. The multiple roles of HOTAIR and UCA1in drug resistance may offer big opportunities for the targets of cancer therapy.
MATERIALS AND METHODS
Data collection
The publications were retrieved on Web of Science Core Collection (WoSCC) of Thomson Reuters on August 19, 2018. The search query consisted of seven terms as follows: (TS = (chemoresistance OR chemotherapy resistance* OR chemotherapy drug resistance* OR chemotherapy-refractory drug* OR chemotherapeutic drug* OR drug resistance*)) AND (TS = (lnc RNA* OR long ncRNA* OR long non translated RNA* OR long non-coding RNA* OR long non protein coding RNA* OR long non-coding RNA* OR long untranslated RNA* OR long intergenic non protein coding RNA* OR large intergenic non coding RNA* OR large intergenic noncoding RNA* OR lincRNA* OR linc RNA*)) The wildcard character “*” captures any relevant variations of a term such as bibliometrics and bibliometric analysis. A bibliographic record was considered as relevant if any of the terms appear in its title, abstract, or keywords. The query returned 542 bibliographic records written in English between 2008 and 2018 on August 19, 2018. Excluding 14 non-official articles or reviews (6: Book chapter; 4: Meeting abstracts; 2: Editorial material; 2: Proceedings paper), 528 publications (398: research articles; 130: review) were retained for further analyzed. The pipeline of the research procedure was displayed in Supplementary Figure 4.
Investigating the intellectual structure in bibliometrics
The publication records, including author, title, abstract, organization, country, journal, keyword, and reference, were extracted and input into CiteSpace [8] in txt format to build the knowledge network, map historical footprint and investigate citation paths at a disciplinary level. Given the records, the toolkits detect and render thematic patterns and emerging trends in science as networked in a variety of bibliographic units. Using a top-down approach, we analyzed data going from macro-level to micro-level, which allowed us to gradually move on to lower-level units of analysis such as journal-level citation paths, subject categories, keywords, titles and abstracts to cited references. In doing so, we effectively captured emerging trends, recent developments and current problems in the research of ncRNA and chemoresistance. At the same time, Microsoft Excel 2016 was used to predict the future trend of LncRNA and chemotherapeutic resistance publications, The model: f(x) = ax3 + bx2 + cx + d, x represents the publication year, f(x) represents the cumulative number of publications.
The strategies in the analysis of these publications included: (1) Clustering: Clustering is unsupervised learning that can uncover latent groups of entities sharing homogeneous characteristics. We captured thematically similar clusters on a document co-citation network by using a network clustering technique called smart local moving [25] to (2) Burst detection: burst detection models the burstiness of features which rise sharply in frequency [26]. An entity has bursting activities when it intensively appears during a specific span of time. We can overcome the limitation coming from considering cumulative, snapshot metrics as impact measures; (3) Cluster labeling: CiteSpace labels clusters with extracted terms from titles and abstracts of citing articles. There are three algorithms to serve cluster labeling: latent semantic analysis (LSA), log-likelihood ratio (LLR) and mutual information (MI). LSA captures unknown semantic relationships over all the documents while LLR and MI reflect a unique aspect of a cluster [27].
SUPPLEMENTARY MATERIALS
ACKNOWLEDGMENTS AND FUNDING
This work was supported by grants from the National Nature Science Foundation of China (31160233), the Science and Technology Foundation of Jiangxi Province (20142BAB204013) and Graduate Student Innovation Special Foundation of Jiangxi Province (YC2018-S087).
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interests.
REFERENCES
- 1.Nikolaou M, Pavlopoulou A, Georgakilas AG, Kyrodimos E. The challenge of drug resistance in cancer treatment: a current overview. Clin Exp Metastasis. 2018; 35:309–18. 10.1007/s10585-018-9903-0. [DOI] [PubMed] [Google Scholar]
- 2.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993; 74:957–67. 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 3.Jiao F, Hu H, Han T, Yuan C, Wang L, Jin Z, Guo Z, Wang L. Long Noncoding RNA MALAT-1 Enhances Stem Cell-Like Phenotypes in Pancreatic Cancer Cells. Int J Mol Sci. 2015; 16:6677–93. 10.3390/ijms16046677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010; 464:1071–6. 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, Yang Z, Liu W, Zhang H, Chen N, Wang H, Wang H, Qin H. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut. 2016; 65:1494–504. 10.1136/gutjnl-2014-308392. [DOI] [PubMed] [Google Scholar]
- 6.Li Z, Dou P, Liu T, He S. Application of Long Noncoding RNAs in Osteosarcoma: Biomarkers and Therapeutic Targets. Cell Physiol Biochem. 2017; 42:1407–19. 10.1159/000479205. [DOI] [PubMed] [Google Scholar]
- 7.Wang P, Chen D, Ma H, Li Y. LncRNA SNHG12 contributes to multidrug resistance through activating the MAPK/Slug pathway by sponging miR-181a in non-small cell lung cancer. Oncotarget. 2017; 8:84086–101. 10.18632/oncotarget.20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C, Hu Z, Liu S, Tseng H. Emerging trends in regenerative medicine: a scientometric analysis in CiteSpace. Expert Opin Biol Ther. 2012; 12:593–608. 10.1517/14712598.2012.674507. [DOI] [PubMed] [Google Scholar]
- 9.van Eck NJ, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics. 2010; 84:523–38. 10.1007/s11192-009-0146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong Y, Yao Q, Yang Y, Feng JJ, Wu SD, Ji WX, Yao L, Liu ZY. Knowledge structure and theme trends analysis on general practitioner research: A Co-word perspective. BMC Fam Pract. 2016; 17:10. 10.1186/s12875-016-0403-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013; 333:213–21. 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 12.Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 2014; 281:1750–8. 10.1111/febs.12737. [DOI] [PubMed] [Google Scholar]
- 13.Ayers D, Vandesompele J. Influence of microRNAs and Long Non-Coding RNAs in Cancer Chemoresistance. Genes (Basel). 2017; 8:E95. 10.3390/genes8030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma Y, Zhou G, Li M, Hu D, Zhang L, Liu P, Lin K. Long noncoding RNA DANCR mediates cisplatin resistance in glioma cells via activating AXL/PI3K/Akt/NF-kappaB signaling pathway. Neurochem Int. 2018; 118:233–41. 10.1016/j.neuint.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Wang B, Lai H, Li S, You Q, Fang Y, Li Q, Liu Y. Long non-coding RNA CRALA is associated with poor response to chemotherapy in primary breast cancer. Thorac Cancer. 2017; 8:582–91. 10.1111/1759-7714.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong Y, Wang J, Zhu H, Liu L, Jiang Y. Chronic oxymatrine treatment induces resistance and epithelial-mesenchymal transition through targeting the long non-coding RNA MALAT1 in colorectal cancer cells. Oncol Rep. 2018; 39:967–76. 10.3892/or.2018.6204. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Teschendorff AE, Lee SH, Jones A, Fiegl H, Kalwa M, Wagner W, Chindera K, Evans I, Dubeau L, Orjalo A, Horlings HM, Niederreiter L, Kaser A, et al. HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med. 2015; 7:108. 10.1186/s13073-015-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu R, Zhang J, Zhang W, Huang Y, Wang N, Zhang Q, Qu S. Circulating HOTAIR expression predicts the clinical response to neoadjuvant chemotherapy in patients with breast cancer. Cancer Biomark. 2018; 22:249–56. 10.3233/CBM-170874. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Yang S, Su N, Wang Y, Yu J, Qiu H, He X. Overexpression of long non-coding RNA HOTAIR leads to chemoresistance by activating the Wnt/β-catenin pathway in human ovarian cancer. Tumour Biol. 2016; 37:2057–65. 10.1007/s13277-015-3998-6. [DOI] [PubMed] [Google Scholar]
- 20.Özeş AR, Miller DF, Özeş ON, Fang F, Liu Y, Matei D, Huang T, Nephew KP. NF-kappaB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene. 2016; 35:5350–61. 10.1038/onc.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan J, Li X, Wu W, Xue M, Hou H, Zhai W, Chen W. Long non-coding RNA UCA1 promotes cisplatin/gemcitabine resistance through CREB modulating miR-196a-5p in bladder cancer cells. Cancer Lett. 2016; 382:64–7. 10.1016/j.canlet.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 22.Wang F, Zhou J, Xie X, Hu J, Chen L, Hu Q, Guo H, Yu C. Involvement of SRPK1 in cisplatin resistance related to long non-coding RNA UCA1 in human ovarian cancer cells. Neoplasma. 2015; 62:432–8. 10.4149/neo_2015_051. [DOI] [PubMed] [Google Scholar]
- 23.Wu C, Luo J. Long non-coding rna (lncRNA) urothelial carcinoma-associated 1 (UCA1) enhances tamoxifen resistance in breast cancer cells via inhibiting mTOR signaling pathway. Med Sci Monit. 2016; 22:3860–7. 10.12659/MSM.900689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Wu Y, Liu A, Tang X. Long non-coding RNA UCA1 enhances tamoxifen resistance in breast cancer cells through a miR-18a-HIF1α feedback regulatory loop. Tumour Biol. 2016; 37:14733–43. 10.1007/s13277-016-5348-8. [DOI] [PubMed] [Google Scholar]
- 25.Emmons S, Kobourov S, Gallant M, Börner K. Analysis of Network Clustering Algorithms and Cluster Quality Metrics at Scale. PLoS One. 2016; 11:e0159161. 10.1371/journal.pone.0159161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noto K, Brodley C, Slonim D. FRaC: a feature-modeling approach for semi-supervised and unsupervised anomaly detection. Data Min Knowl Discov. 2012; 25:109–33. 10.1007/s10618-011-0234-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Ibekwe-Sanjuan F, Hou J. The structure and dynamics of cocitation clusters: A multiple-perspective cocitation analysis. Journal of the American Society for Information Science and Technology. 2010; 61:1386–409. 10.1002/asi.21309. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.