Abstract
PURPOSE
Genetic testing for cancer risk has expanded rapidly. We examined clinical genetic testing and results among population-based patients with breast and ovarian cancer.
METHODS
The study included all women 20 years of age or older diagnosed with breast or ovarian cancer in California and Georgia between 2013 and 2014 and reported to the SEER registries covering the entire state populations. SEER data were linked to results from four laboratories that performed nearly all germline cancer genetic testing. Testing use and results were analyzed at the gene level.
RESULTS
There were 77,085 patients with breast cancer and 6,001 with ovarian cancer. Nearly one quarter of those with breast cancer (24.1%) and one third of those with ovarian cancer (30.9%) had genetic test results. Among patients with ovarian cancer, testing was lower in blacks (21.6%; 95% CI, 18.1% to 25.4%; v whites, 33.8%; 95% CI, 32.3% to 35.3%) and uninsured patients (20.8%; 95% CI, 15.5% to 26.9%; v insured patients, 35.3%; 95% CI, 33.8% to 36.9%). Prevalent pathogenic variants in patients with breast cancer were BRCA1 (3.2%), BRCA2 (3.1%), CHEK 2 (1.6%), PALB2 (1.0%), ATM (0.7%), and NBN (0.4%); in patients with ovarian cancer, prevalent pathogenic variants were BRCA1 (8.7%), BRCA2 (5.8%), CHEK2 (1.4%), BRIP1 (0.9%), MSH2 (0.8%), and ATM (0.6%). Racial/ethnic differences in pathogenic variants included BRCA1 (ovarian cancer: whites, 7.2%; 95% CI, 5.9% to 8.8%; v Hispanics, 16.1%; 95% CI, 11.8% to 21.2%) and CHEK2 (breast cancer: whites, 2.3%; 95% CI, 1.8% to 2.8%; v blacks, 0.1%; 95% CI, 0% to 0.8%). When tested for all genes that current guidelines designate as associated with their cancer type, 7.8% of patients with breast cancer and 14.5% of patients with ovarian cancer had pathogenic variants.
CONCLUSION
Clinically-tested patients with breast and ovarian cancer in two large, diverse states had 8% to 15% prevalence of actionable pathogenic variants. Substantial testing gaps and disparities among patients with ovarian cancer are targets for improvement.
INTRODUCTION
Germline genetic testing has become an integral part of the care of patients with breast and ovarian cancer and their families since BRCA1 and BRCA2 (BRCA1/2) were identified in 1994 to 1995.1,2 Epidemiologic studies have defined the prevalence of BRCA1/2 pathogenic variants in patient subgroups such as triple-negative breast cancer (TNBC), pancreatic cancer, and prostate cancer,3-5 and testing guidelines have evolved.6 Recent advances in next-generation technology and regulatory changes have enabled sequencing of more genes at a lower cost using multiple-gene panels.7-9
Studies in clinic-based samples suggest that multiple-gene sequencing panels may double the prevalence of pathogenic variants detected by testing BRCA1/2 alone.10-16 However, such studies often used convenience samples limited to a single institution or laboratory. Almost nothing is known about the prevalence of pathogenic variants on multiple-gene panels among clinically tested, population-based patients with breast cancer and patients with ovarian cancer. Yet, such knowledge is essential to inform population-wide health policy, resource planning, and development of testing guidelines.
We established the Georgia-California Surveillance, Epidemiology and End Results (SEER) Genetic Testing Linkage Initiative with the SEER registries of Georgia and California and the four laboratories that performed nearly all cancer susceptibility testing in these regions during the study period. In this article, we examine testing use and results among all patients with breast cancer (N = 77,085) and all patients with ovarian cancer (N = 6,001) diagnosed in California and Georgia between 2013 and 2014.
METHODS
Creation of Study Cohort and Analytic Data Set
Using a third-party honest broker (Information Management Services, Rockville, MD), all female patients with breast and ovarian cancer diagnosed between 2013 and 2014 from the Georgia Cancer Registry and the California Cancer Registry were linked with germline genetic testing information from four laboratories (Ambry Genetics, Aliso Viejo, CA; GeneDx, Gaithersburg, MD; Invitae, San Francisco, CA; Myriad Genetics, Salt Lake City, UT) that performed the majority of clinical testing. Probabilistic methods were implemented to optimize linkage in the presence of uncertainty or errors in any individual linkage covariable.
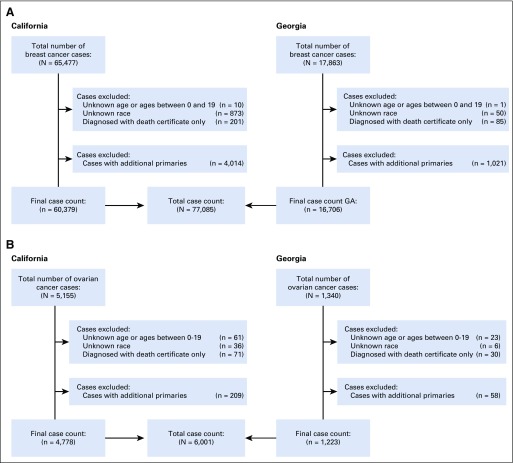
The analytic data set combined testing data from all laboratories; all variables other than genetic tests and results were from SEER registries. Second or later breast or ovarian cancers in the same patient were excluded (6.1% for breast cancer; 4.3% for ovarian cancer). Patients diagnosed before 20 years of age, at death, or with unknown race were excluded (Fig 1). Breast tumor biomarker subtype was defined by combinations of hormone receptor (HR) and human epidermal growth factor receptor 2 (HER2) expression. Genetic test use and results were captured through the first quarter of 2016. If a patient was tested more than once, results were merged, keeping the most recent result for each gene. The combined analytic file containing both registry and laboratory information was stripped of protected health information (as defined by the Health Information Portability and Accountability Act Privacy Rule),17 and some variables (age, race, marital status, poverty, insurance, histology, and test result) were collapsed to minimize the risk of reidentification. This study was approved by the institutional review boards associated with the SEER registries.
FIG 1.
Inclusion and exclusion counts from California and Georgia SEER registries, 2013 to 2014, for (A) breast cancer and (B) ovarian cancer.
Testing Variables From Laboratories
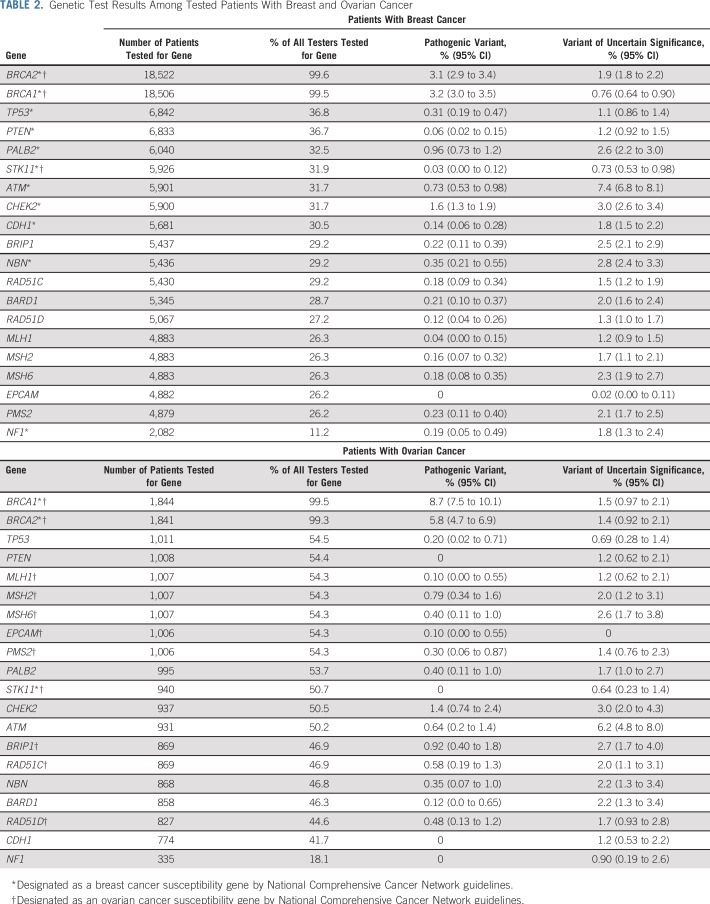
Each laboratory provided results at the gene level for 139 genes, including the interpretation, according to American College of Medical Genetics criteria,18 and sent to the ordering clinician: pathogenic or likely pathogenic (combined for analysis as pathogenic); variant of uncertain significance (VUS); and benign or likely benign (combined for analysis as normal). We categorized a test as a multiple-gene panel if it included other genes in addition to BRCA1/2, versus BRCA1/2 only. To ensure anonymity of laboratories, results from all laboratories were combined, and gene-specific results were retained in the analytic data set only for genes tested by two or more laboratories (n = 53). Results for the remaining 86 genes tested by only one laboratory were categorized collectively: pathogenic variants were rare (0% for ovarian cancer and 0.09% for breast cancer), whereas VUSs were more common (3.6% for ovarian and 2.1% for breast cancer).
Statistical Methods
We examined testing use by patient characteristics among all patients with breast cancer and all patients with ovarian cancer in Georgia and California. Gene-specific results by cancer type for a subset of genes were presented using binomial proportions with exact confidence limits. This subset included 11 genes designated by the current National Comprehensive Cancer Network guidelines as breast cancer susceptibility genes (ATM, BRCA1, BRCA2, CDH1, CHEK2, NBN, NF1, PALB2, PTEN, STK11, and TP53), one gene designated as potentially associated with breast cancer (BARD1), and 11 genes designated as ovarian cancer susceptibility genes (BRCA1, BRCA2, BRIP1, EPCAM, MLH1, MSH2, MSH6, PMS2, RAD51C, RAD51D, and STK11).6 Pathogenic variants in these genes should prompt a change in care per guidelines.6 Results were stratified by biomarker subtype (breast) and race/ethnicity (breast and ovarian). Pathogenic variant and VUS prevalence among patients tested for all 11 guideline-designated genes were examined by cancer type and race/ethnicity and tested for homogeneity by a χ2 statistic.
RESULTS
Patient Characteristics
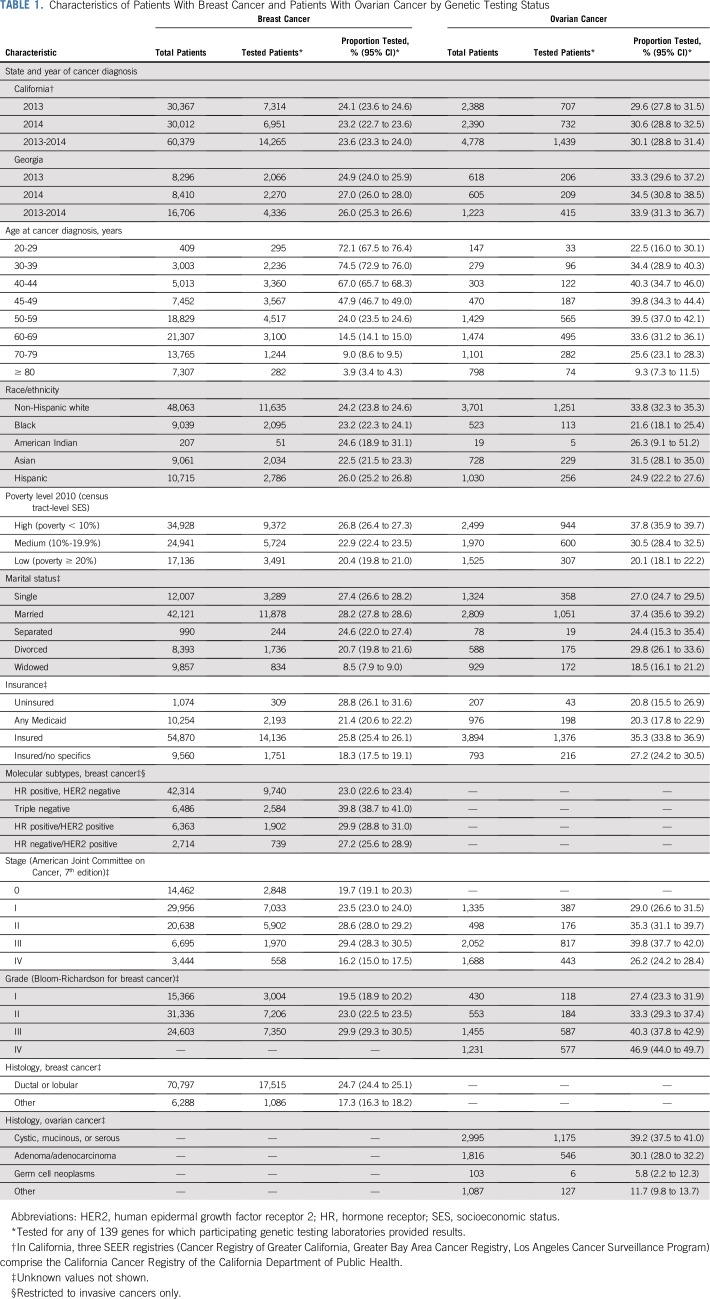
Figure 1 shows inclusion and exclusion criteria, yielding 77,085 women with breast cancer and 6,001 women with ovarian cancer. Approximately one quarter of patients with breast cancer (24.1%) and one third of patients with ovarian cancer (30.9%) had testing results linked from any laboratory. Testing rates were similar in both states and unchanged over time (Table 1).
TABLE 1.
Characteristics of Patients With Breast Cancer and Patients With Ovarian Cancer by Genetic Testing Status
Genetic Testing Rates by Demographic and Clinical Factors
Among patients with breast cancer, testing decreased with increasing age at diagnosis (72.1% among those 20 to 29 years of age; 3.9% among those ≥ 80 years of age) and increasing area-based residential poverty at the census tract level (where poverty was less than 10%: 26.8%; 95% CI, 26.4% to 27.3%; where poverty was ≥ 20%: 20.4%; 95% CI, 19.8% to 21.0%; Table 1). Testing did not vary substantially by race/ethnicity; this finding was largely consistent across age and subtypes, with some evidence for disparities in younger patients (Data Supplement). Testing rates were higher among women with the HR-negative, HER2-negative subtype (TNBC: 39.8%; 95% CI, 38.7% to 41.0%) than for the HR-positive, HER2-negative subtype (23.0%, 95% CI, 22.6% to 23.4%) and increased as both stage and grade increased, but decreased for stage IV.
Among patients with ovarian cancer, testing was highest in middle-age groups (39.7% ages 40 to 59; 95% CI, 37.6% to 41.8%; Table 1). Testing decreased with increasing area-based residential poverty (where poverty was less than 10%: 37.8%; 95% CI, 35.9% to 39.7%; where poverty was ≥ 20%: 20.1%; 95% CI, 18.1% to 22.2%). Considerable variation in testing existed by race/ethnicity (non-Hispanic white: 33.8%; 95% CI, 32.3% to 35.3%; black: 21.6%; 95% CI, 18.1% to 25.4%; Hispanic: 24.9%; 95% CI, 22.2% to 27.6%), marital status (married: 37.4%; 95% CI, 35.6% to 39.2%; single: 27.0%; 95% CI, 24.7% to 29.5%), and insurance (Medicaid: 20.3%; 95% CI, 17.8% to 22.9%; other insurance: 33.9%; 95% CI, 32.6% to 35.3%). Testing rates increased with both stage and grade, but decreased for stage IV.
Genetic Testing Results Among Patients With Breast Cancer
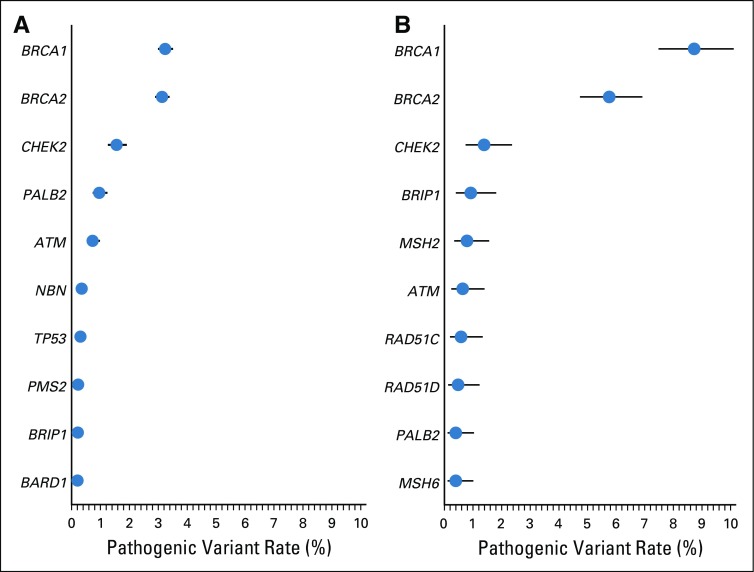
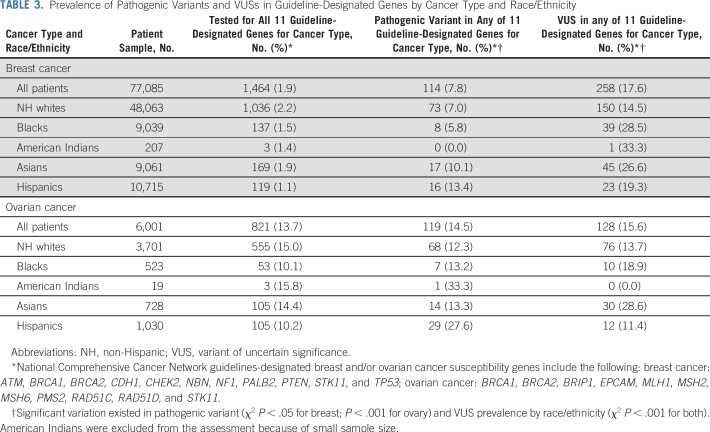
The Data Supplement lists 53 genes tested by at least two laboratories, of which 11 are currently guideline-designated as breast cancer susceptibility genes and 11 as ovarian cancer susceptibility genes.6 Among patients with breast cancer, three times more were tested for BRCA1 (18,506) and BRCA2 (18,522) than any other breast cancer gene (6,842 for TP53, fewer for others; Table 2). Pathogenic variants were most frequently detected in BRCA1 (3.2%) and BRCA2 (3.1%), followed by CHEK2 (1.6%), PALB2 (0.96%), and ATM (0.73%; Fig 2A). Pathogenic variants were detected in BARD1, CDH1, NBN, NF1, and TP53, and ranged from 0.14% to 0.35%. Few patients with breast cancer (n = 1,464; 1.9%) were tested for all 11 genes designated by current guidelines6 as breast cancer susceptibility genes (Table 3). Among patients tested for all 11 guideline-designated breast cancer genes, 7.8% had a pathogenic variant, and 17.6% had VUSs in any of these genes (Table 3). Significant variation of both proportions (pathogenic variant and VUS) existed by race/ethnicity.
TABLE 2.
Genetic Test Results Among Tested Patients With Breast and Ovarian Cancer
FIG 2.
The 10 most prevalent pathogenic variants from genetic test results of genes related to (A) breast cancer and (B) ovarian cancer. The lines represent the 95% confidence intervals around the point estimates.
TABLE 3.
Prevalence of Pathogenic Variants and VUSs in Guideline-Designated Genes by Cancer Type and Race/Ethnicity
Genetic Testing Results Among Patients With Ovarian Cancer
Among tested patients with ovarian cancer, almost twice as many were tested for BRCA1 (1,844) and BRCA2 (1,841) as for any other National Comprehensive Cancer Network–designated ovarian cancer gene (1,007 for MSH2, fewer for others; Table 2). Pathogenic variants were most prevalent in BRCA1 (8.7%) and BRCA2 (5.8%). After BRCA1/2, pathogenic variants in ovarian cancer genes were most prevalent in BRIP1 (0.92%), MSH2 (0.79%), and RAD51C (0.58%). However, pathogenic variants were similarly prevalent among patients with ovarian cancer as among patients with breast cancer in CHEK2 (1.4%) and ATM (0.64%), genes not known to confer elevated ovarian cancer risk (Fig 2B; Table 2). Overall, 13.7% of patients with ovarian cancer were tested for all 11 genes currently guideline-designated as ovarian cancer susceptibility genes (Table 3). Among patients tested for all 11 guideline-designated ovarian cancer genes, 14.5% had a pathogenic variant, and 15.6% had VUSs in any of them.
Genetic Testing Results by Breast Cancer Subtype
Breast cancer subtype-specific differences in pathogenic variant prevalence were observed in various genes (Data Supplement). Statistically significant examples include BRCA1 (HR positive, HER2 negative: 2.1%; 95% CI, 1.8% to 2.3%; TNBC: 11.4%; 95% CI, 10.2% to 12.7%) and CHEK2 (HR positive, HER2 negative: 1.7%; 95% CI, 1.3% to 2.3%; TNBC: 0.25%; 95% CI, 0.03% to 0.90%).
Genetic Testing Results by Race/Ethnicity
There were racial/ethnic differences in pathogenic variant prevalence in various genes (Data Supplement). Statistically significant examples include BRCA1 in patients with breast cancer (whites: 2.5%; 95% CI, 2.2% to 2.8%; blacks: 4.0%; 95% CI, 3.2% to 5.0%; Asians: 3.1%; 95% CI, 2.4% to 4.0%; Hispanics: 5.8%; 95% CI, 4.9% to 6.7%) and in patients with ovarian cancer (whites: 7.2%; 95% CI, 5.9% to 8.8%; blacks: 13.4%; 95% CI, 7.7% to 21.1%; Asians: 6.2%; 95% CI, 3.4% to 10.1%; Hispanics: 16.1%; 95% CI, 11.8% to 21.2%), and CHEK2 in patients with breast cancer (whites: 2.3%; 95% CI, 1.8% to 2.8%; blacks: 0.15%; 95% CI, 0% to 0.82%; Asians: 0.45%; 95% CI, 0.09% to 1.3%; Hispanics: 0.46%; 95% CI 0.13% to 1.2%).
DISCUSSION
To our knowledge, this is the first US study of hereditary cancer genetic testing at the population level with laboratory-confirmed testing results. We took advantage of SEER registries that cover the entire populations of California and Georgia, two regions of racial/ethnic and socioeconomic diversity that together comprise approximately 50 million people. We investigated the clinical use of genetic testing from the four major laboratories that provide it in the regions under study. We found that nearly one quarter of patients with breast cancer and fewer than one third of patients with ovarian cancer diagnosed between 2013 and 2014 had germline genetic testing. Among those tested for all genes designated by current guidelines as associated with their cancer type, 7.8% of patients with breast cancer and 14.5% of patients with ovarian cancer carried a pathogenic variant that warrants a potential change in care, such as secondary breast cancer screening incorporating magnetic resonance imaging, earlier colonoscopy, or risk-reducing surgery.6
Few population-level studies have evaluated trends, patterns, and correlates of genetic testing, and most focused on BRCA1/2 testing only.19,20 We previously published articles from a survey, the iCanCare study, of more than 5,000 patients diagnosed with breast cancer in 2013 to 2015 from Georgia and Los Angeles County.9,21-24 However, that sample of testers was far too small to evaluate correlates of variants in specific genes. Furthermore, that previous study did not include patients with ovarian cancer. In this study, our expanded population-level focus on more than 83,000 patients with breast cancer or ovarian cancer offers granular detail on testing gaps, disparities, and gene-specific results at a major inflection point in the implementation of precision oncology.
The context of cancer genetic testing changed in mid-2013, after a US Supreme Court decision on gene patenting, decreasing costs, and growing public awareness.25-28 A study of medically underserved patients with breast and ovarian cancer who met criteria for Medicare coverage of BRCA1/2 testing from 2002 to 2014 reported a testing rate of 9%.29 Our study evaluated testing from 2012 to 2016 for all California and Georgia patients with breast and ovarian cancer diagnosed in 2013 to 2014. The observed testing rate of 24% among unselected patients with breast cancer demonstrates the broad penetration of testing into community practice. We found little regional variation, suggesting that these results are broadly generalizable. We also found little racial/ethnic variability in testing of patients with breast cancer, although there was an access gradient by poverty level. Higher testing rates among patients with TNBC reflect guideline recommendations to test nearly all such patients, whereas more restrictive criteria on the basis of family history are advised for other subtypes.6,30,31 Although testing rates were higher among younger women, rates lower than 100% for women diagnosed with breast cancer younger than 45 years of age are suboptimal.
A 30% genetic testing rate for patients with ovarian cancer is inadequate, because guidelines have recommended testing all patients with high-grade, serous ovarian cancer for a decade.6,32,33 We could not isolate high-grade serous cancers within this data set, but 39.2% testing among patients with tumors in the combined categories of cystic, mucinous, and serous suggests a major shortfall. Furthermore, large differences in testing rates across stage and grade subgroups suggest inappropriate targeting of testing that is indicated for all. Low testing rates among patients with earlier-stage ovarian cancer, who may survive this diagnosis, reflect a missed opportunity to provide risk-adapted screening for second cancers, such as breast, colon, and melanoma associated with pathogenic variants in specific ovarian cancer susceptibility genes. Substantial undertesting of patients with ovarian cancer may reflect their high morbidity and/or a relatively low public awareness of and advocacy for ovarian cancer. We report on a period just before US Food and Drug Administration approval of the first BRCA1/2-targeted therapy for ovarian cancer, the poly (ADP-ribose) polymerase inhibitor olaparib, in December 2014.34 With three poly (ADP-ribose) polymerase inhibitors now approved for BRCA1/2-associated ovarian cancer,34-36 two for BRCA1/2-associated metastatic breast cancer,37,38 and other targeted agents in development,39 appropriate testing of patients with metastatic breast and/or ovarian cancer is increasingly critical both for them and to inform their at-risk relatives. Finally, the large socioeconomic disparities in test receipt after ovarian cancer diagnosis highlight the challenges of ensuring universal testing access.
Among patients tested for all genes that current guidelines designate for their cancer type, 7.8% of patients with breast cancer and 14.5% of patients with ovarian cancer carried at least one pathogenic variant that warrants a potential change in care.6 These results do not constitute a true population prevalence among all patients with breast and ovarian cancer: patients were undoubtedly selected for testing according to clinician knowledge, patient preferences, and access factors.8,23,24,28,40,41 Instead, these prevalence estimates apply to the average patient with breast or ovarian cancer who undergoes testing as implemented in community practice; thus, they are highly relevant for genetic counseling. Notably, only a small minority of 2013 to 2014 patients were tested for all genes designated by current guidelines as associated with their cancer type, reflecting the fact that testing and guidelines have rapidly evolved. Moreover, guidelines do not definitively endorse multiple-gene testing for all patients for whom BRCA1/2 testing is advised. We previously reported rapid replacement of BRCA1/2-only with multiple-gene panel testing in a subsample of patients,9 concurrent with the falling costs of sequencing. Thus, our prevalence estimates for non-BRCA1/2 genes have a smaller denominator of tested patients than we would expect today. As the population of tested patients changes over time (eg, if patients with less family history of breast and/or ovarian cancers are increasingly tested for more genes), prevalence estimates for both higher-risk genes (eg, PALB2) and lower-risk genes (eg, ATM) may shift.
Testing guidelines were developed to identify carriers of BRCA1/2 pathogenic variants.6 As data emerge about the clinical presentation of other pathogenic variants, guidelines should evolve. For example, although current guidelines state that BARD1 pathogenic variants (found here in 0.2% of patients with breast cancer) have insufficient evidence to guide management,6 recent results showing a four- to five-fold associated increase in TNBC risk may inform future guidelines.3 Among patients with breast cancer, pathogenic variants in ATM, CHEK2, and PALB2 were relatively common, with prevalence approximately 1% each; these results resemble those of prior studies.11,12,15,16,30 However, we observed similar frequencies of ATM, CHEK2, and PALB2 pathogenic variants among patients with ovarian cancer, although most studies have not reported an ovarian cancer risk association with these genes.16,42,43 The prevalence of ATM, CHEK2, and PALB2 pathogenic variants among patients with ovarian cancer may merely reflect their relatively high population carrier frequency, but additional study of their ovarian cancer risk association is warranted. Pathogenic variants in BRIP1, MSH2, and RAD51C also approached 1% prevalence each among patients with ovarian cancer, consistent with their guideline designation as ovarian cancer susceptibility genes.6
We observed little racial/ethnic variation in overall pathogenic variant prevalence, but did see differences by specific gene: significantly more CHEK2 pathogenic variants among whites than blacks, and more BRCA1 pathogenic variants among Hispanics. The prevalence of CHEK2 pathogenic variants among whites likely reflects the Central European1100delC founder variant.44,45 Others have noted a high BRCA1 pathogenic variant prevalence in Hispanic and black patients with breast cancer.46 Another racial/ethnic difference is VUS rates, more common in blacks, Asians, and Hispanics than whites. We and others previously reported that this VUS difference likely reflects racial bias in the definition of the normal gene sequence along with less clinical testing of minority patients,9,47-50 because an access disparity perpetuates a genetic information disparity. Ongoing efforts in germline VUS reclassification51,52 are essential to promoting equity in cancer care.
Our study has limitations. Results are from two states only and may not be fully generalizable elsewhere, although the scant regional variation we observed is reassuring. The time period is limited (patients diagnosed in 2013 to 2014, with results complete through March 2016), with ongoing linkage under way. Some tests may have been missed because of incomplete ascertainment or uncertainty in the linkage process. However, we surveyed genetic counselors in both states who responded that the collaborating laboratories performed nearly all relevant testing in 2013 to 2014. Furthermore, testing rates from data linkage closely approximate those of patient self-report in our prior work.23,24 We did not include laboratories that offer direct-to-consumer testing with an out-of-pocket charge; had we done so, we might have observed a magnification of the reported socioeconomic testing disparities. Finally, SEER registries do not collect data on family cancer history, which influences receipt of testing.
Clinical practice and health policy on germline genetic testing of patients with cancer are evolving rapidly as studies on the utility of genetic surveillance continue to emerge.3,37-39,51,53-57 Some have called for population-based testing, either of all people or of all patients with breast cancer.57-59 Although it is possible that universal testing guidelines might reduce disparities, our results demonstrate the challenges of adhering to such inclusive guidelines in the case of ovarian cancer. Many factors conspire to limit genetic testing in those with clinical indications, including patients’ and clinicians’ attitudes about the value of genetic testing and the challenges of integrating genetic testing into the cancer treatment workflow. In particular, the surge in multiple-gene panel testing has markedly increased the demand for timely genetic expertise in the face of a limited supply of genetic counselors. More research is needed to identify optimal approaches to genetic testing delivery and results management and determine the impact of the rapidly changing landscape of genetic risk evaluation on patients with cancer and their families.
ACKNOWLEDGMENT
We thank Jill S. Dolinsky, MS, CGC, and Melissa Pronold, PhD, at Ambry Genetics; Rachel Klein, PhD, Delores Bowman, BSN, and Benjamin Solomon, MD, PhD, at GeneDx; Edward Esplin, MD, PhD, and Stephen Lincoln, PhD, at Invitae; and Johnathan Lancaster, MD, PhD, and Brian Dechairo, PhD, at Myriad Genetics for their collaboration on the genetic test data linkage to SEER data. Written permission was obtained to include the names of all acknowledged individuals.
Footnotes
Supported by the National Cancer Institute (NCI) of the National Institutes of Health under award Nos. P01 CA163233 to the University of Michigan and R01 CA225697 to Stanford University. The collection of cancer incidence data used in this study was supported by the California Department of Public Health pursuant to California Health and Safety Code Section 103885; the Centers for Disease Control and Prevention’s National Program of Cancer Registries, under cooperative agreement 5NU58DP006344; the NCI’s SEER Program under contract HHSN261201800032I awarded to the University of California, San Francisco, contract HHSN261201800015I awarded to the University of Southern California, and contract HHSN261201800009I awarded to the Public Health Institute, Cancer Registry of Greater California. The ideas and opinions expressed herein are those of the authors and do not necessarily reflect the opinions of the State of California, the Department of Public Health, the NCI, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors. Research funding to her institution for an unrelated project was provided to A.W.K. by Myriad Genetics. The authors have no conflicts of interest to declare.
Podcast by Dr Bouberhan
AUTHOR CONTRIBUTIONS
Conception and design: Allison W. Kurian, Kevin C. Ward, Dennis Deapen, Lynne S. Penberthy, Steven J. Katz
Financial support: Allison W. Kurian, Steven J. Katz
Administrative support: Dennis Deapen, Lynne S. Penberthy
Provision of study materials or patients: Kevin C. Ward, Dennis Deapen, Ann S. Hamilton, Lynne S. Penberthy
Collection and assembly of data: Allison W. Kurian, Kevin C. Ward, Dennis Deapen, Ann S. Hamilton
Data analysis and interpretation: Allison W. Kurian, Kevin C. Ward, Nadia Howlader, Dennis Deapen, Angela Mariotto, Daniel Miller, Lynne S. Penberthy, Steven J. Katz
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Genetic Testing and Results in a Population-Based Cohort of Breast Cancer Patients and Ovarian Cancer Patients
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Allison W. Kurian
Research Funding: Myriad Genetics (Inst)
Other Relationship: Ambry Genetics, Color Genomics, GeneDx/BioReference, Invitae, Genentech
No other potential conflicts of interest were reported.
REFERENCES
- 1.Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2. doi: 10.1038/378789a0. Wooster R, Bignell G, Lancaster J, et al: Identification of the breast cancer susceptibility gene BRCA2. Nature 378:789-792, 1995 [Erratum: Nature 379:749, 1996] [DOI] [PubMed] [Google Scholar]
- 3. Shimelis H, LaDuca H, Hu C, et al: Triple-negative breast cancer risk genes identified by multigene hereditary cancer panel testing. J Natl Cancer Inst 110:855-862, 2018. [DOI] [PMC free article] [PubMed]
- 4.Hu C, Hart SN, Polley EC, et al. Association between inherited germline mutations in cancer predisposition genes and risk of pancreatic cancer. JAMA. 2018;319:2401–2409. doi: 10.1001/jama.2018.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. National Comprehensive Cancer Network: Guidelines for genetic/familial risk assessment: Breast and ovarian. Version 2. 2019-July 30, 2018. https://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf.
- 7.Kurian AW, Ford JM. Multigene panel testing in oncology practice: How should we respond? JAMA Oncol. 2015;1:277–278. doi: 10.1001/jamaoncol.2015.28. [DOI] [PubMed] [Google Scholar]
- 8.Katz SJ, Kurian AW, Morrow M. Treatment decision making and genetic testing for breast cancer: Mainstreaming mutations. JAMA. 2015;314:997–998. doi: 10.1001/jama.2015.8088. [DOI] [PubMed] [Google Scholar]
- 9.Kurian AW, Ward KC, Hamilton AS, et al. Uptake, results, and outcomes of germline multiple-gene sequencing after diagnosis of breast cancer. JAMA Oncol. 2018;4:1066–1072. doi: 10.1001/jamaoncol.2018.0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurian AW, Hare EE, Mills MA, et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol. 2014;32:2001–2009. doi: 10.1200/JCO.2013.53.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tung N, Battelli C, Allen B, et al. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer. 2015;121:25–33. doi: 10.1002/cncr.29010. [DOI] [PubMed] [Google Scholar]
- 12.Tung N, Lin NU, Kidd J, et al. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J Clin Oncol. 2016;34:1460–1468. doi: 10.1200/JCO.2015.65.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desmond A, Kurian AW, Gabree M, et al. Clinical actionability of multigene panel testing for hereditary breast and ovarian cancer risk assessment. JAMA Oncol. 2015;1:943–951. doi: 10.1001/jamaoncol.2015.2690. [DOI] [PubMed] [Google Scholar]
- 14.Maxwell KN, Wubbenhorst B, D’Andrea K, et al. Prevalence of mutations in a panel of breast cancer susceptibility genes in BRCA1/2-negative patients with early-onset breast cancer. Genet Med. 2015;17:630–638. doi: 10.1038/gim.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaDuca H, Stuenkel AJ, Dolinsky JS, et al. Utilization of multigene panels in hereditary cancer predisposition testing: Analysis of more than 2,000 patients. Genet Med. 2014;16:830–837. doi: 10.1038/gim.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. doi: 10.1200/PO.16.00066. Kurian AW, Hughes E, Handorf E, et al: Breast and ovarian cancer penetrance estimates derived from germline multiple-gene sequencing results in women. Precision Oncol 10.1200/PO.16.00066 [epub ahead of print on June 27, 2017] [DOI] [PubMed] [Google Scholar]
- 17. US Department of Health and Human Services: Guidance regarding methods for de-identification of protected health information in accordance with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule. https://www.hhs.gov/hipaa/for-professionals/privacy/special-topics/de-identification/index.html.
- 18.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armstrong J, Toscano M, Kotchko N, et al. Utilization and outcomes of BRCA genetic testing and counseling in a national commercially insured population: The ABOUT study. JAMA Oncol. 2015;1:1251–1260. doi: 10.1001/jamaoncol.2015.3048. [DOI] [PubMed] [Google Scholar]
- 20.McCarthy AM, Bristol M, Domchek SM, et al. Health care segregation, physician recommendation, and racial disparities in BRCA1/2 testing among women with breast cancer. J Clin Oncol. 2016;34:2610–2618. doi: 10.1200/JCO.2015.66.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katz SJ, Ward KC, Hamilton AS, et al. Gaps in receipt of clinically indicated genetic counseling after diagnosis of breast cancer. J Clin Oncol. 2018;36:1218–1224. doi: 10.1200/JCO.2017.76.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz SJ, Bondarenko I, Ward KC, et al. Association of attending surgeon with variation in the receipt of genetic testing after diagnosis of breast cancer. JAMA Surg. 2018;153:909–916. doi: 10.1001/jamasurg.2018.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurian AW, Li Y, Hamilton AS, et al. Gaps in incorporating germline genetic testing into treatment decision-making for early-stage breast cancer. J Clin Oncol. 2017;35:2232–2239. doi: 10.1200/JCO.2016.71.6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurian AW, Griffith KA, Hamilton AS, et al. Genetic testing and counseling among patients with newly diagnosed breast cancer. JAMA. 2017;317:531–534. doi: 10.1001/jama.2016.16918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Offit K, Bradbury A, Storm C, et al. Gene patents and personalized cancer care: Impact of the Myriad case on clinical oncology. J Clin Oncol. 2013;31:2743–2748. doi: 10.1200/JCO.2013.49.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borzekowski DL, Guan Y, Smith KC, et al. The Angelina effect: Immediate reach, grasp, and impact of going public. Genet Med. 2014;16:516–521. doi: 10.1038/gim.2013.181. [DOI] [PubMed] [Google Scholar]
- 27.Evans DG, Barwell J, Eccles DM, et al. The Angelina Jolie effect: How high celebrity profile can have a major impact on provision of cancer related services. Breast Cancer Res. 2014;16:442. doi: 10.1186/s13058-014-0442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noar SM, Althouse BM, Ayers JW, et al. Cancer information seeking in the digital age: Effects of Angelina Jolie’s prophylactic mastectomy announcement. Med Decis Making. 2015;35:16–21. doi: 10.1177/0272989X14556130. [DOI] [PubMed] [Google Scholar]
- 29.Gross AL, Blot WJ, Visvanathan K. BRCA1 and BRCA2 testing in medically underserved Medicare beneficiaries with breast or ovarian cancer. JAMA. 2018;320:597–598. doi: 10.1001/jama.2018.8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Couch FJ, Shimelis H, Hu C, et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. 2017;3:1190–1196. doi: 10.1001/jamaoncol.2017.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee E, McKean-Cowdin R, Ma H, et al. Characteristics of triple-negative breast cancer in patients with a BRCA1 mutation: Results from a population-based study of young women. J Clin Oncol. 2011;29:4373–4380. doi: 10.1200/JCO.2010.33.6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.American College of Obstetricians and Gynecologists. et al. ACOG practice bulletin no. 103: Hereditary breast and ovarian cancer syndrome. Obstet Gynecol. 2009;113:957–966. doi: 10.1097/AOG.0b013e3181a106d4. [DOI] [PubMed] [Google Scholar]
- 33.Committee on Practice Bulletins–Gynecology, Committee on Genetics, Society of Gynecologic Oncology Practice bulletin no 182: Hereditary breast and ovarian cancer syndrome. Obstet Gynecol. 2017;130:e110–e126. doi: 10.1097/AOG.0000000000002296. [DOI] [PubMed] [Google Scholar]
- 34.Kim G, Ison G, McKee AE, et al. FDA approval summary: Olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin Cancer Res. 2015;21:4257–4261. doi: 10.1158/1078-0432.CCR-15-0887. [DOI] [PubMed] [Google Scholar]
- 35.Syed YY. Rucaparib: First global approval. Drugs. 2017;77:585–592. doi: 10.1007/s40265-017-0716-2. [DOI] [PubMed] [Google Scholar]
- 36.Scott LJ. Niraparib: First global approval. Drugs. 2017;77:1029–1034. doi: 10.1007/s40265-017-0752-y. [DOI] [PubMed] [Google Scholar]
- 37.Robson M, Im SA, Senkus E, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 38.Litton JK, Rugo HS, Ettl J, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018;379:753–763. doi: 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cruz C, Llop-Guevara A, Garber JE, et al: Multicenter phase II study of lurbinectedin in BRCA-mutated and unselected metastatic advanced breast cancer and biomarker assessment substudy. J Clin Oncol 36:3134-3143, 2018. [DOI] [PMC free article] [PubMed]
- 40.Bradbury AR, Patrick-Miller LJ, Egleston BL, et al. Patient feedback and early outcome data with a novel tiered-binned model for multiplex breast cancer susceptibility testing. Genet Med. 2016;18:25–33. doi: 10.1038/gim.2015.19. [DOI] [PubMed] [Google Scholar]
- 41.Robson ME, Bradbury AR, Arun B, et al. American Society of Clinical Oncology policy statement update: Genetic and genomic testing for cancer susceptibility. J Clin Oncol. 2015;33:3660–3667. doi: 10.1200/JCO.2015.63.0996. [DOI] [PubMed] [Google Scholar]
- 42.Antoniou AC, Casadei S, Heikkinen T, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norquist BM, Harrell MI, Brady MF, et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016;2:482–490. doi: 10.1001/jamaoncol.2015.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meijers-Heijboer H, van den Ouweland A, Klijn J, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 45.Vahteristo P, Bartkova J, Eerola H, et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet. 2002;71:432–438. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.John EM, Miron A, Gong G, et al. Prevalence of pathogenic BRCA1 mutation carriers in 5 US racial/ethnic groups. JAMA. 2007;298:2869–2876. doi: 10.1001/jama.298.24.2869. [DOI] [PubMed] [Google Scholar]
- 47.Manrai AK, Funke BH, Rehm HL, et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med. 2016;375:655–665. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hall MJ, Reid JE, Burbidge LA, et al. BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer. 2009;115:2222–2233. doi: 10.1002/cncr.24200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caswell-Jin JL, Gupta T, Hall E, et al. Racial/ethnic differences in multiple-gene sequencing results for hereditary cancer risk. Genet Med. 2018;20:234–239. doi: 10.1038/gim.2017.96. [DOI] [PubMed] [Google Scholar]
- 50.Mersch J, Brown N, Pirzadeh-Miller S, et al. Prevalence of variant reclassification following hereditary cancer genetic testing. JAMA. 2018;320:1266–1274. doi: 10.1001/jama.2018.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slavin TP, Van Tongeren LR, Behrendt CE, et al. Prospective Study of Cancer Genetic Variants: Variation in Rate of Reclassification by Ancestry. J Natl Cancer Inst. 2018;110:1059–1066. doi: 10.1093/jnci/djy027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Findlay GM, Daza RM, Martin B, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217–222. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor A, Brady AF, Frayling IM, et al. Consensus for genes to be included on cancer panel tests offered by UK genetics services: Guidelines of the UK Cancer Genetics Group. J Med Genet. 2018;55:372–377. doi: 10.1136/jmedgenet-2017-105188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manchanda R, Patel S, Gordeev VS, et al. Cost-effectiveness of population-based BRCA1, BRCA2, RAD51C, RAD51D, BRIP1, PALB2 mutation testing in unselected general population women. J Natl Cancer Inst. 2018;110:714–725. doi: 10.1093/jnci/djx265. [DOI] [PubMed] [Google Scholar]
- 55. Lu HM, Li S, Black MH, et al: Association of breast and ovarian cancers with predisposition genes identified by large-scale sequencing. JAMA Oncol . [epub ahead of print on August 16, 2018] [DOI] [PMC free article] [PubMed]
- 56. doi: 10.1093/jnci/djy147. Caswell-Jin JL, Zimmer AD, Stedden W, et al: Cascade genetic testing of relatives for hereditary cancer risk: Results of an online initiative. J Natl Cancer Inst 111:95-98, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Beitsch PD, Whitworth PW, Hughes K, et al: Underdiagnosis of hereditary breast cancer: Are genetic testing guidelines a tool or an obstacle? J Clin Oncol 10.1200/JCO.18.01631 [epub ahead of print on December 7, 2018] [DOI] [PMC free article] [PubMed]
- 58.Narod S, Akbari MR. Population-based genetic testing for BRCA1 and BRCA2. J Clin Oncol. 2018;36:517. doi: 10.1200/JCO.2017.75.8490. [DOI] [PubMed] [Google Scholar]
- 59.King MC, Levy-Lahad E, Lahad A. Population-based screening for BRCA1 and BRCA2: 2014 Lasker Award. JAMA. 2014;312:1091–1092. doi: 10.1001/jama.2014.12483. [DOI] [PubMed] [Google Scholar]