Abstract
Background: Long noncoding RNAs (lncRNAs), a class of noncoding RNA nucleotides >200 bp, has been demonstrated to play vital role in the development of cancer. FEZ family zinc finger 1 antisense RNA 1 (FEZF1-AS1) has been reported as an lncRNA which acts as a tumor-promoting effect in some cancers. However, the role of it in esophageal squamous cell carcinoma (ESCC) and its potential regulatory mechanism was unclear now.
Methods: qRT-PCR was used to detect the levels of FEZF1-AS1 and mRNA CTNNB1 (β-catenin) in ESCC tissues and cells. Cell transfection experiments were used to knock down or overexpress the level of FEZF1-AS1 in EC1 and EC9706 cell lines. WST-1 assays, cell cycle assays, scratch wound assays, migration, and invasion assays were used to evaluate the function of FEZF1-AS1 in ESCC progression.
Results: FEZF1-AS1 was remarkably upregulated in ESCC tissues and cell lines. Silencing of FEZF1-AS1 significantly inhibited the migration and invasion of ESCC cells, while overexpression of FEZF1-AS1 notably accelerated ESCC migration and invasion. Meanwhile, the levels of FEZF1-AS1 had no effect on ESCC cell proliferation and cell cycle. We also found that β-catenin was upregulated in ESCC tissues, and the level of it was positively correlated with the expression of FEZF1-AS1. Silencing of FEZF1-AS1 could decrease the mRNA and protein level of β-catenin, while overexpression FEZF1-AS1 could lead to the contrary.
Conclusion: Our results suggested that the expression of lncRNA FEZF1-AS1 played an important role in ESCC progression, especially the motility of the tumor. FEZF1-AS1 may provide us with a new sight for ESCC treatment.
Keywords: esophageal squamous cell carcinoma, lncRNA, FEZF1-AS1, biological function, β-catenin
Introduction
Esophageal cancer is one of the lethal cancers worldwide with high morbidity, high fatality rate, and poor 5-year survival rates.1,2 Two subtypes of esophageal cancers are esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC).3 In 90% of the world’s countries, the proportion of ESCC is much higher than that of EAC, and men have a higher prevalence than women.4 Esophageal cancer has no obvious symptoms at earlier disease stages,5 and the gold standard techniques (endoscopy and biopsy) used in the diagnosis of esophageal cancer are invasive examinations, both of which are associated with late diagnosis and high mortality in patients with esophageal cancer.6,7 Thus, it is urgent to find better diagnosis and treatment methods of esophageal cancer.
Cancer is a disease of dysregulation of the gene networks. Genome-wide associated studies in cancer revealed that >80% of cancer-associated single nucleotide polymorphisms (SNPs) happen in noncoding regions of the genome.8 Long noncoding RNAs (lncRNAs) were defined as a kind of RNA whose transcripts >200 nucleotides in length and has no protein-coding potential.9 Accumulating research has demonstrated that lncRNAs could interact with key signaling mediators, such as DNA, RNA, and protein molecules,10–12 and their misexpression confers the cancer cell capacities for tumor initiation, growth, and metastasis.13,14 For example, Wang et al identified that lncRNA LINC00958 (BLACAT2) could induce tumor-associated lymphangiogenesis and promote lymph node (LN) metastasis via regulating VEGF-C, SNAI2, and MMP9 expression.15 lncRNA EZR-AS1 could promote the mobility and invasiveness of ESCC cells through upregulating EZR transcription via interacting with both SMYD3 and RNA polymerase II.16
lncRNA FEZ family zinc finger 1 antisense RNA 1 (FEZF1-AS1) is an lncRNA which is located in chromosome 7 and produces a 2,564 bp transcript and has been reported to be overexpressed and to facilitate proliferation and migration in some cancers such as colorectal cancer, gastric cancer, non-small cell lung cancer (NSCLC).17–19 For example, Liu et al found that FEZF1-AS1 could inhibit the expression of p21 and promote the proliferation of gastric cancer via binding with LSD1.18 Wang et al identified that FEZF1-AS1 could promote hepatocellular carcinoma cell invasion and epithelial–mesenchymal transition through JAK2/STAT3 signaling pathway.20 However, the role of FEZF1-AS1 in ESCC has not yet been reported.
In our study, we determined the expression profile of FEZF1-AS1 in ESCC and tried to reveal its function in progression and metastasis of ESCC. It may provide us with a new perspective for the in-depth study of the pathomechanism of esophageal carcinoma cancer.
Materials and methods
Patient samples and clinical data collection
45 pairs of fresh ESCC tissues with their corresponding adjacent nontumorous tissues were obtained from First Affiliated Hospital of Zhengzhou University between April 2016 and May 2017. All the patients who provided the specimens were not treated with chemotherapy or radiotherapy before surgery and were diagnosed with ESCC based on histopathological evaluation. All samples were immediately stored in liquid nitrogen until for further analysis. The study methodologies conformed to the standards set by the Declaration of Helsinki, and the study was approved by the Research Ethics Committee of the First Affiliated Hospital of Zhengzhou University. Written informed consents were obtained from all patients before the operation, and we collected clinicopathological and other clinical characteristics from the electronic medical records.
Cell lines and culture
ESCC cell lines (EC9706, EC1) and human esophageal epithelial cell line (HET-1A) were purchased from Shanghai Institute of Life Sciences cell bank center (Shanghai, China). Cells were cultured in Roswell Park Memorial Institute-1640 medium (RPMI-1640, Hyclone, UT, USA) containing 10% FBS and incubated in a humidified incubator at 37°C with 5% CO2.
Total RNA extraction and qPCR
Total RNAs were derived from tissues and cultured cells using TriZol reagent (TakaRa, Dalian, China) following the manufacturer’s protocol. Then, cDNA was obtained through reverse transcription by using the PrimeScriptTM RT reagent Kit with gDNA Eraser (TakaRa, Dalian, China). qRT-PCR of FEZF1-AS1 and β-catenin mRNA were performed using the kit specification of SYBR® Premix Ex Taq™ II (TakaRa, Dalian, China) on LightCycler 480 II Real‐Time PCR System (Roche, Basel, Switzerland) with GAPDH as internal control as described before.21 The primers used in our study are summarized below: FEZF1-AS1 primers forward: 5ʹ-AGAGGCTATGACTCAGGGTT-3ʹ, reverse: 5ʹ-TGTTGCTCCACAGTAAAGGT-3ʹ; β-catenin primers forward: 5ʹ-GCTGACCAAACTGCTAAATGACGA-3ʹ, reverse: 5ʹ-TGTAGGGTCCCAGCGGTACAA-3ʹ; GAPDH primers forward: 5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ, reverse: 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ.
Cell transfection
Plasmid vectors (GV219-FEZF1-AS1 and GV219-NC) were purchased from Genechem Co.,Ltd. (Shanghai, China) and transfected into EC1 and EC9706 cells using lipo8000TM (Beyotime Biotechnology, Shanghai, China) according to the manufacturer’s instructions. The two si-RNA lncRNA FEZF1-AS1 oligos (si-FEZF1-AS1#1-1:5ʹ-GCCAATCGTTAACTTCCAA-3ʹ and si-FEZF1-AS1#2-2:5ʹ-GTCACTTACAAGGGTGTTA-3ʹ) were purchased from RiboBio Co., Ltd (Guangzhou, China) and transfected into cells using RFect sRNA Transfection Reagent (Changzhou Biogenerating Biotechnologies Co., Ltd, Jiangsu, China). Then, the cells were collected and used for further studies.
WST-1 assays
WST-1 assay kit (Beyotime Biotechnology) was conducted to evaluate the proliferation rate of cells in our study. Briefly, transfected log-phase cells were inoculated into 96-well plates at a density of 1,000 cells per well as described before.22 Then, cell proliferation rates were detected at 24 hrs, 48 hrs, 72 hrs, and 96 hrs using SpctraMax M5 (Molecular Devices, San Francisco, CA, USA).
Cell cycle assays
Transfected cells (1×106) were collected, washed with 1×PBS, and fixed in 70% ethanol at 4°C for 12 hrs. After this, cells were washed there times with 1×PBS and then stained with 1 mL propidium iodide (PI) for 30 mins at 37°C according to the specification. Subsequently, DNA content of the cells was detected using a BD FACSCalibur™ flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) and analyzed with ModFitLT.
In vitro migration and invasion assays
Cell motility was measured using the transwell 24-well chambers with or without Matrigel.
Transfected logarithmic growth phase cells (5×105) were resuspended in serum-free RPMI-1640 medium (200 µL). Then, the cell suspensions were seeded in the upper chamber (with or without Matrigel) and 600 µL medium containing 15% FBS was added at the bottom chamber. And then, these cells were cultured for 24 hrs (invasion assays for 48 hrs). Then, the penetrated cells on lower membrane surface were fixed with 4% paraformaldehyde for 10 mins and stained in crystal violet for 30 mins. The numbers of cells were measured from five random fields photographed using a microscope.
Scratch wound assays
Transfected cells were inoculated in the 6-well plates and cultured at 37°C until in 90%–100% fusion state. A 200 µL pipette tip was used to scratch the plate to create a scraped area. Then, the plate was washed twice with PBS and 2 mL serum-free RPMI-1640 medium was added into it. After that, the plates were placed at 37°C for another 72 hrs. The size of wounds was observed and photographed under the microscope for the comparison of cell motility.
Western blot assay
Proteins in the nucleus of cultured cells were extracted using the Nucleoprotein Extraction Kit (Sangon, Shanghai, China). Then, equal amounts of proteins for each group were separated by SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes. Immunoblotting of the membranes was performed using the following primary antibodies: anti-β-catenin (1:1,000), anti-GAPDH (1:1,000). Signals were revealed after incubation with secondary antibody by using chemiluminescence with imaging system (Bio-Rad, Hercules, CA,, USA).
Statistical analysis
GraphPad Prism 5 (GraphPad Software Inc., La Jolla, CA, USA) software and IBM SPSS statistics 21 (SPSS Inc., Chicago, IL, USA) software were used for statistics. Four statistical tests – Student’s t test, Fisher’s exact test, χ2 test, or Pearson’s correlation analysis – were used to assess the differences between variables depending on the state. P-values <0.05 were considered statistically significant.
Results
FEZF1-AS1 was significantly upregulated in ESCC tissue and cell lines and predicted LN metastasis
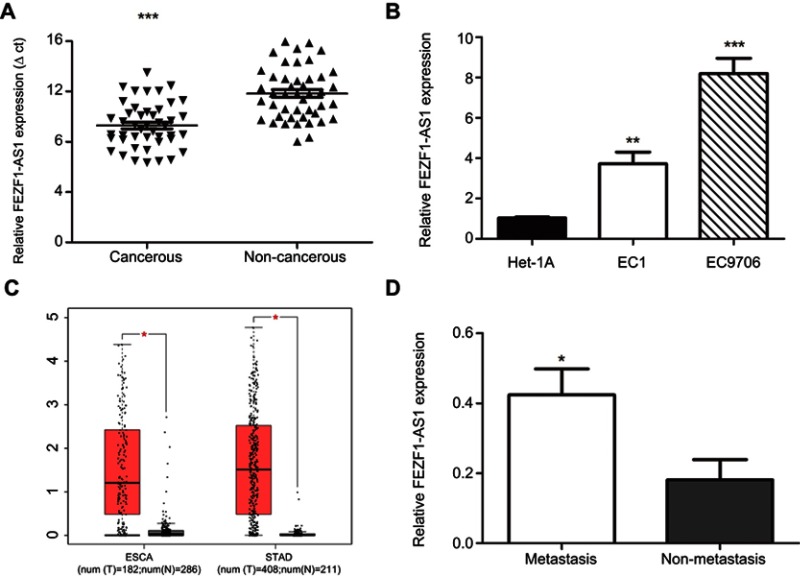
To assess the role of FEZF1-AS1 in ESCC, the expression of FEZF1-AS1 in 45 pairs of ESCC and adjacent nonneoplastic tissues was examined using qRT-PCR experiment, and we found FEZF1-AS1 was significantly upregulated in ESCC tissues compared with the adjacent non-neoplastic tissues (Figure 1A). The bioinformatic analysis data of FEZF1-AS1 on GEPIA (Gene Expression Profiling Interactive Analysis) also supported this result (Figure 1C).
Figure 1.
Expression level of lncRNA FEZF1-AS1. (A) The expression of lncRNA FEZF1-AS1 in ESCC tissues (n=45) was higher than the expression of paired adjacent noncancerous tissues; (B) The expression of FEZF1-AS1 was high ein ESCC cell lines compared to the normal esophageal epithelial cells (Het-1A); (C) FEZF1-AS1 was overexpressed in gastrointestinal tumors such as primary esophageal carcinoma (ESCA) and stomach adenocarcinoma (STAD); (D) Relative expression of FEZF1-AS1 was obviously increased in lymphatic metastasis patients compared to the patients without lymphatic metastasis. *P < 0.05, **P < 0.01, ***P < 0.001.
Abbreviations: ESCA, esophageal carcinoma; STAD, stomach adenocarcinoma.
Then, the expression of FEZF1-AS1 in Het-1A (normal esophageal epithelial cells), EC1, and EC9706 cell lines was detected. The result showed that FEZF1-AS1 was upregulated in EC1 and EC9706 cell lines compared with Het-1A (Figure 1B). To find the association between FEZF1-AS1 expression and the clinicopathological features of the ESCC patients, we defined the expression level above the median as the high-expression group and the rest as the low-expression group. The correlations between FEZF1-AS1 expression and clinicopathological characteristics of ESCC are shown in Table 1. In ESCC, there was no correlation between FEZF1-AS1 expression and patients’ age, gender, tumor size, differentiation grade, and TNM stage. FEZF1-AS1 expression was only correlated with LN metastasis status (P=0.002), and the expression of FEZF1-AS1 in metastasis group was obviously higher than that in the nonmetastasis group (Figure 1D).
Table 1.
The correlation between FEZF1-AS1 expression and clinicopathological factors of ESCC patients
| Variable | N | FEZF1-AS1 expression | P-value | ||
|---|---|---|---|---|---|
| Low (n=23) | High (n=22) | ||||
| Gender | Male | 23 | 12 | 11 | 0.884 |
| Female | 22 | 11 | 11 | ||
| Age | <65 years | 19 | 9 | 10 | 0.668 |
| ≥65 years | 26 | 14 | 12 | ||
| Tumor size | ≤3 cm | 19 | 10 | 9 | 0.862 |
| >3 cm | 26 | 13 | 13 | ||
| Differentiation grade | Poor | 12 | 7 | 5 | 0.559 |
| Well/moderate | 33 | 16 | 17 | ||
| TNM stage | I+II | 13 | 6 | 7 | 0.672 |
| III+IV | 32 | 17 | 15 | ||
| Lymphatic metastasis | Negative | 23 | 17 | 6 | 0.002** |
| Positive | 22 | 6 | 16 | ||
Note: **P < 0.01.
Abbreviation: ESCC, esophageal squamous cell carcinoma.
FEZF1-AS1 knockdown had no effect on ESCC cell proliferation and cell cycle in vitro
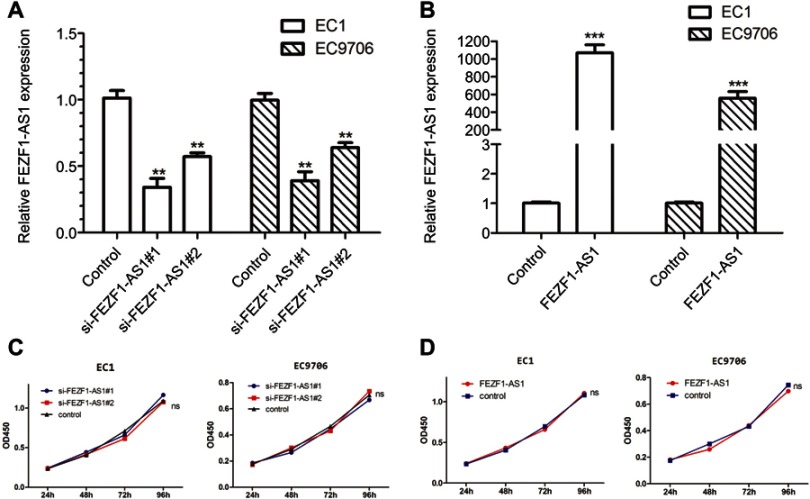
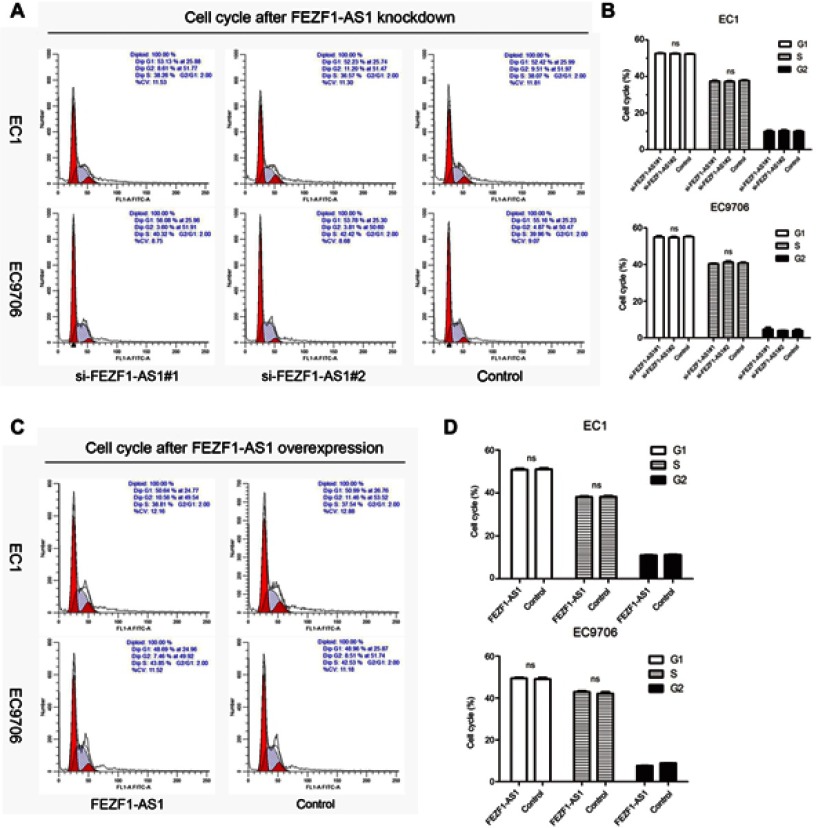
To investigate the biological function of FEZF1-AS1 in ESCC, FEZF1-AS1 was downregulated by siRNAs in EC1 and EC9706 cells. The transfection efficiency was over 80% in each group, and both si-FEZF1-AS1#1 and si-FEZF1-AS1#2 could decrease the expression of FEZF1-AS1 significantly (Figure 2A). Next, GV219-FEZF1-AS1 was transfected into EC1 and EC9706 cell lines to improve the expression of FEZF1-AS1 (Figure 2B). Then, we performed WST-1 assays to confirm the impact of FEZF1-AS1 on ESCC cell proliferation. Our result showed that FEZF1-AS1 had no effect on proliferation of EC1 and EC9706 cell lines (Figure 2C, D). For further research, we also performed the cell cycle experiment. Transfected cells were digested and stained with PI dye and detected by FCM (flow cytometry). As shown in Figure 3, there were no significant changes in the cell cycle of FEZF1-AS1 knockdown group and overexpression group.
Figure 2.
Intracellular expression of FEZF1-AS1 after transfection and the effect of FEZF1-AS1 on proliferation of esophageal cancer cells. (A) FEZF1-AS1 levels of si-FEZF1-AS1#1 and si-FEZF1-AS1#2 groups were notably decreased compared to that in control groups; (B) FEZF1-AS1 levels of FEZF1-AS1 overexpression groups were notably increased compared to that in control groups; (C) silencing of FEZF1-AS1 had no effect on ESCC cell proliferation; (D) upregulated FEZF1-AS1 levels had no effect on ESCC cell proliferation.
**P < 0.01, ***P < 0.001.
Figure 3.
Impacts of FEZF1-AS1 on ESCC cell cycle. (A, B) The percentage of cells arresting in G1 phase and S phase in EC1 and EC9706 cell lines had no significant (ns) changes after FEZF1-AS1 knockdown. (C, D) There were no significant changes in EC1 and EC9706 cell cycles after FEZF1-AS1 overexpression.
FEZF1-AS1 knockdown significantly inhibited the migration and invasion of ESCC cells in vitro
To investigate the role of FEZF1-AS1 in the metastasis of esophageal carcinoma cancer, we performed transwell assays to detect the migration and invasion of two cell lines after they were transfected by two si-FEZF1-AS1. As shown in Figure 4A and B, knockdown of FEZF1-AS1 could obviously suppress the migration and invasion ability of two cell lines. Moreover, we also demonstrated scratch wound assays to evaluate the compromised migration elicited by FEZF1-AS1 knockdown, the result showed that the wound closure was decelerated in si-FEZF1-AS1 groups compared to the control group (Figure 4E). In summary, our results confirmed that silencing of FEZF1-AS1 caused the negative effect of FEZF1-AS1 on ESCC cell metastatic capacity (Figure 4C, D, F).
Figure 4.
Knockdown of lncRNA FEZF1-AS1 expression inhibited ESCC cell migration and invasion in vitro. (A, B) Transwell assays demonstrated that silencing FEZF1-AS1 resulted in reduced EC1 and EC9706 cell migration and invasion; (C, D) statistical analysis showed that downregulated FEZF1-AS1 decreased the migration and invasion capacity of ESCC cells in vitro. (E, F) Scratch wound assay was performed in EC9706 cells, wound closure was reduced in si-FEZF1-AS1 groups compared to the control group. *P < 0.05, **P < 0.01, ***P < 0.001.
FEZF1-AS1 overexpression significantly increased the migration and invasion of ESCC cells in vitro
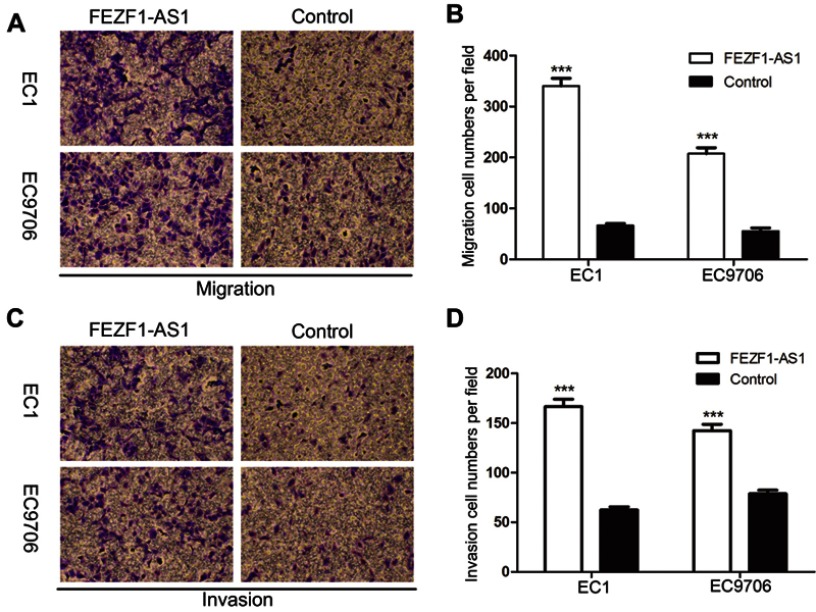
For further research, EC1 and EC9706 cells were transfected with FEZF1-AS1 overexpression plasmids (GV219-FEZF1-AS1) with GV129-NC as control, and we utilized classical transwell assays to assess the effect of FEZF1-AS1 overexpression on the invasion and migration of ESCC. The result revealed that the invasion and migration of ESCC cell lines increased significantly after FEZF1-AS1 overexpression (Figure 5A and C), which meant overexpression of FEZF1-AS1 caused the positive effect on ESCC cell metastatic capacity (Figure 5B and D). Taken together, our result confirmed that FEZF1-AS1 played an important role in the movability of ESCC.
Figure 5.
Overexpression of lncRNA FEZF1-AS1 significantly accelerated ESCC cell migration and invasion in vitro. (A, C) Transwell assays were performed in ESCC cell lines, upregulated FEZF1-AS1 resulted in boosted cell invasion and invasion; (B, D) statistical analysis indicated that upregulated FEZF1-AS1 increased the migration and invasion capacity of ESCC cells in vitro. ***P < 0.001.
FEZF1-AS1 exerted tumor-promoting functions via Wnt/β-catenin signaling pathway in ESCC
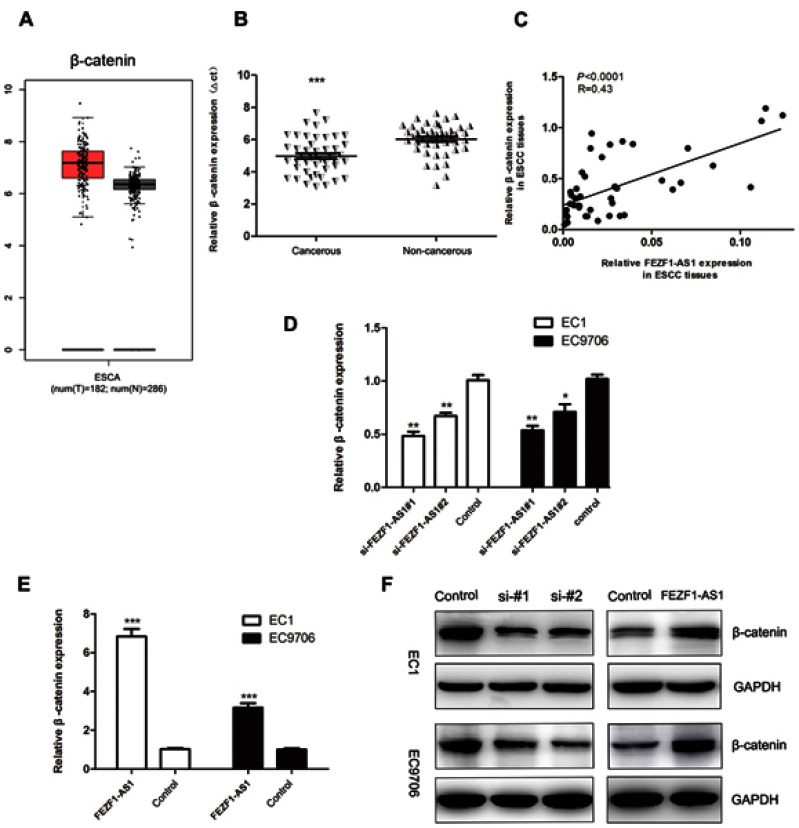
To further elucidate the mechanism underlying the role of FEZF1-AS1 in ESCC, we detected the expression levels of the mRNA which were reported closely involved in tumor progression and metastasis. We noted that β-catenin was overexpression in ESCA (Figure 6A), which was the same as the result of our qPCR assays (Figure 6B), and the expression of it displayed a significant positive correlation with the levels of FEZF1-AS1 in ESCC tissues (Figure 6C). The levels of β-catenin mRNA and protein were obviously downregulated in cells which were transfected with the si-FEZF1-AS1, while they were upregulated in cells which were transfected with the GV219-FEZF1-AS1 (Figure 6D–F).
Figure 6.
The expression levels of β-catenin in ESCC tissues, adjacent noncancerous tissues, and cell lines. (A) Expression profile of β-catenin in primary ESCC tissues and normal esophageal epithelium tissues (from GEPIA); (B) β-catenin was overexpressed in ESCC tissues (n=45); (C) The expression of β-catenin displayed an obvious positive correlated with the levels of lncRNA FEZF1-AS1 in ESCC tissues; (D) β-catenin levels in si-FEZF1-AS1#1 and si-FEZF1-AS1#1 cells were both decreased; (E) β-catenin levels in ESCC cell lines were increased after FEZF1-AS1 overexpression; (F) the levels of β-catenin protein were obviously decreased in cells which were transfected with si-FEZF1-AS1#1 and si-FEZF1-AS1#1, while they were increased in cells which were transfected with the GV219-FEZF1-AS1 *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Recently, lncRNAs have attracted much attention and demonstrated pivotal roles in the regulation of various tumor cellular processes.23–25 There is growing evidence that suggests that lncRNAs are associated with tumor invasiveness and metastatic potential.26,27 For example, Chen et al found that LNMAT1 overexpression was relevant to the lymphatic metastasis of human bladder cancer via CCL2-mediated modulation of the tumor microenvironment;28 Xiao et al found that FILNC1 deficiency in renal cancer cells alleviated energy stress-induced apoptosis through upregulation of c-Myc and promoted renal tumor development.29 Nevertheless, thus far, little is known about the clinical and biological function of lncRNA FEZF1-AS1 in ESCC.
FEZF1-AS1 is an lncRNA which is located on the opposite strand of gene FEZF1 in chromosome 7.17 It has been found to be overexpressed in cancers such as breast cancer and pancreatic ductal adenocarcinoma, and can promote the proliferation, migration and invasion of tumor cells.30,31 There are some papers reported that antisense RNA could affect the expression of its host gene, for example, FOXF1-AS1 regulates cell proliferation, migration, and invasion by affecting its host gene FOXF1 expression in osteosarcoma.32 However, in our study, no effect of FEZF1-AS1 on the expression level of FEZF1 was found, and we speculated that FEZF1-AS1 promoted the invasion and migration of ESCC cell through other channels. The activation of embryonic epithelial-to-mesenchymal (EMT) program is generally accepted as a vital component of carcinoma progression.33,34 EMT is a complicated process, and molecular reprogramming occurring during EMT is triggered by various factors. There are many signaling pathways that induce EMT in cancer, and one of the most important is the Wnt/β-catenin pathway.35 β-catenin (CTNNB1) is an intracellular scaffold protein that interacts with adhesion molecules,36 and previous studies have reported that its abnormal expression was related to lncRNA. For example, Wang et al found that lncRNA LINC00968 could promote NSCLC growth, migration, and invasion via activating β-catenin expression;37 Xiang et al found that the overexpression of lncRNA DGCR5 could repress hepatocellular carcinoma progression through inhibiting Wnt/β-catenin signaling pathway.38
In this study, we found that FEZF1-AS1 was obviously overexpressed in esophageal carcinoma tissues and cell lines and predicted LN metastasis in patients of ESCC. Additionally, in order to investigate the role of FEZF1-AS1 in ESCC progression, we further performed the loss-of-function and gain-of-function experiments. Our results demonstrated that knockdown FEZF1-AS1 contributed to significantly inhibit cell migration and invasion in vitro and upregulated FEZF1-AS1 contributed to significantly accelerate cell migration and invasion in vitro. Silencing of FEZF1-AS1 remarkably decreased the expression level of β-catenin in EC1 and EC9706 cells and overexpression of FEZF1-AS1 resulted in the contrary. Furthermore, the β-catenin expression was significantly positively correlated with the levels of FEZF1-AS1 in ESCC tissues.
In conclusion, our study indicated that FEZF1-AS1 could exert a tumor-promoting role in ESCC by targeting Wnt/β-catenin signaling pathway. FEZF1-AS1 may be a biomarker for the prediction of LN metastasis and a new therapeutic target for the cascade of invasion and metastasis in patients with ESCC. Further studies are required to determine how FEZF1-AS1 is involved in the in-depth mechanisms of the invasion-metastasis cascade in ESCC.
Acknowledgments
The authors express their thanks for the support from Academy of Medical Sciences of Zhengzhou University Translational Medicine platform and all the researchers who helped during the study. This study was supported by the Medical Scientific Research Foundation of Health Department of Henan Province of China (Grant No. 201403077), the Science and Technology Project of Henan province of China (Grant No. 192102310133) and the National Natural Science Foundation of China grants (Grant No. 81702780).
Disclosure
The authors declare that there are no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Smyth EC, Lagergren J, Fitzgerald RC, et al. Oesophageal cancer. Nat Rev Dis Primers. 2017;3:17048. doi: 10.1038/nrdp.2017.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang L, Sun K, Chu J, et al. Long non-coding RNA FTH1P3 regulated metastasis and invasion of esophageal squamous cell carcinoma through SP1/NF-kB pathway. Biomed Pharmacother. 2018;106:1570–1577. doi: 10.1016/j.biopha.2018.07.129 [DOI] [PubMed] [Google Scholar]
- 4.Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64(3):381–387. doi: 10.1136/gutjnl-2014-308124 [DOI] [PubMed] [Google Scholar]
- 5.Li DJ, Shi M, Wang Z. RUNX3 reverses cisplatin resistance in esophageal squamous cell carcinoma via suppression of the protein kinase B pathway. Thorac Cancer. 2016;7(5):570–580. doi: 10.1111/1759-7714.12370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yazbeck R, Jaenisch SE, Watson DI. From blood to breath: new horizons for esophageal cancer biomarkers. World J Gastroenterol. 2016;22(46):10077–10083. doi: 10.3748/wjg.v22.i46.10077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun K, Zhao X, Wan J, et al. The diagnostic value of long non-coding RNA MIR31HG and its role in esophageal squamous cell carcinoma. Life Sci. 2018;202:124–130. doi: 10.1016/j.lfs.2018.03.050 [DOI] [PubMed] [Google Scholar]
- 8.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108(12):2419–2425. doi: 10.1038/bjc.2013.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin C, Yang L. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 2018;28(4):287–301. doi: 10.1016/j.tcb.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17(1):106–116. doi: 10.1093/bib/bbv031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ballantyne MD, McDonald RA, Baker AH. lncRNA/MicroRNA interactions in the vasculature. Clin Pharmacol Ther. 2016;99(5):494–501. doi: 10.1002/cpt.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosseini ES, Meryet-Figuiere M, Sabzalipoor H, Kashani HH, Nikzad H, Asemi Z. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol Cancer. 2017;16(1):107. doi: 10.1186/s12943-017-0671-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He W, Zhong G, Jiang N, et al. Long noncoding RNA BLACAT2 promotes bladder cancer-associated lymphangiogenesis and lymphatic metastasis. J Clin Invest. 2018;128(2):861–875. doi: 10.1172/JCI96218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang XD, Huang GW, Xie YH, et al. The interaction of lncRNA EZR-AS1 with SMYD3 maintains overexpression of EZR in ESCC cells. Nucleic Acids Res. 2018;46(4):1793–1809. doi: 10.1093/nar/gkx1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen N, Guo D, Xu Q, et al. Long non-coding RNA FEZF1-AS1 facilitates cell proliferation and migration in colorectal carcinoma. Oncotarget. 2016;7(10):11271–11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu YW, Xia R, Lu K, et al. LincRNAFEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-Mediated H3K4me2 demethylation. Mol Cancer. 2017;16(1):39. doi: 10.1186/s12943-017-0588-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He R, Zhang FH, Shen N. LncRNA FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through suppressing E-cadherin and regulating WNT pathway in non-small cell lung cancer (NSCLC). Biomed Pharmacother. 2017;95:331–338. doi: 10.1016/j.biopha.2017.08.057 [DOI] [PubMed] [Google Scholar]
- 20.Wang YD, Sun XJ, Yin JJ, et al. Long non-coding RNA FEZF1-AS1 promotes cell invasion and epithelial-mesenchymal transition through JAK2/STAT3 signaling pathway in human hepatocellular carcinoma. Biomed Pharmacother. 2018;106:134–141. doi: 10.1016/j.biopha.2018.05.116 [DOI] [PubMed] [Google Scholar]
- 21.Wan J, Liu H, Feng Q, Liu J, Ming L. HOXB9 promotes endometrial cancer progression by targeting E2F3. Cell Death Dis. 2018;9(5):509. doi: 10.1038/s41419-018-0556-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan J, Liu H, Yang L, Ma L, Liu J, Ming L. JMJD6 promotes hepatocellular carcinoma carcinogenesis by targeting CDK4. Int J Cancer. 2019;144(19):2489–2500. [DOI] [PubMed] [Google Scholar]
- 23.Wang K, Liu CY, Zhou LY, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun. 2015;6:6779. doi: 10.1038/ncomms7779 [DOI] [PubMed] [Google Scholar]
- 24.Wu X, Yan T, Wang Z, Wu X, Cao G, Zhang C. LncRNA ZEB2-AS1 promotes bladder cancer cell proliferation and inhibits apoptosis by regulating miR-27b. Biomed Pharmacother. 2017;96:299–304. doi: 10.1016/j.biopha.2017.08.060 [DOI] [PubMed] [Google Scholar]
- 25.Damas ND, Marcatti M, Come C, et al. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat Commun. 2016;7:13875. doi: 10.1038/ncomms13875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20(3):300–307. doi: 10.1038/nsmb.2480 [DOI] [PubMed] [Google Scholar]
- 28.Chen C, He W, Huang J, et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat Commun. 2018;9(1):3826. doi: 10.1038/s41467-018-06152-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao ZD, Han L, Lee H, et al. Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat Commun. 2017;8(1):783. doi: 10.1038/s41467-017-00902-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye H, Zhou Q, Zheng S, et al. FEZF1-AS1/miR-107/ZNF312B axis facilitates progression and Warburg effect in pancreatic ductal adenocarcinoma. Cell Death Dis. 2018;9(2):34. doi: 10.1038/s41419-017-0052-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, Sun L, Zhang Y, Lu G, Li Y, Wei Z. Long non-coding RNA FEZF1-AS1 promotes breast cancer stemness and tumorigenesis via targeting miR-30a/Nanog axis. J Cell Physiol. 2018;233(11):8630–8638. doi: 10.1002/jcp.26611 [DOI] [PubMed] [Google Scholar]
- 32.Kun-Peng Z, Chun-Lin Z, Xiao-Long M. Antisense lncRNA FOXF1-AS1 promotes migration and invasion of osteosarcoma cells through the FOXF1/MMP-2/-9 pathway. Int J Biol Sci. 2017;13(9):1180–1191. doi: 10.7150/ijbs.21722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goossens S, Vandamme N, Van Vlierberghe P, Berx G. EMT transcription factors in cancer development re-evaluated: beyond EMT and MET. Biochim Biophys Acta Rev Cancer. 2017;1868(2):584–591. doi: 10.1016/j.bbcan.2017.06.006 [DOI] [PubMed] [Google Scholar]
- 34.Wan J, Liu H, Ming L. Lysine crotonylation is involved in hepatocellular carcinoma progression. Biomed Pharmacother. 2019;111:976–982. doi: 10.1016/j.biopha.2018.12.148 [DOI] [PubMed] [Google Scholar]
- 35.Garg M. Epithelial-mesenchymal transition – activating transcription factors – multifunctional regulators in cancer. World J Stem Cells. 2013;5(4):188–195. doi: 10.4252/wjsc.v5.i4.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh M. Multilayered prevention and treatment of chronic inflammation, organ fibrosis and cancer associated with canonical WNT/betacatenin signaling activation (Review). Int J Mol Med. 2018;42(2):713–725. doi: 10.3892/ijmm.2018.3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Zhou J, Xu YJ, Hu HB. Long non-coding RNA LINC00968 acts as oncogene in NSCLC by activating the Wnt signaling pathway. J Cell Physiol. 2018;233(4):3397–3406. doi: 10.1002/jcp.26186 [DOI] [PubMed] [Google Scholar]
- 38.Wang XL, Shi M, Xiang T, Bu YZ. Long noncoding RNA DGCR5 represses hepatocellular carcinoma progression by inactivating Wnt signaling pathway. J Cell Biochem. 2019;120(1):275–282. [DOI] [PubMed] [Google Scholar]