Abstract
A defective homologous recombination (HR) repair program increases tumor incidence as well as providing a survival advantage in patients with breast and ovarian cancers. Here, we hypothesize that the tumor-promoting side of genome maintenance programs may be contributed by a self-renewal protein, nucleostemin (NS). To address this issue, we established its functional importance in mammary tumor progression in mice and showed that mammary tumor cells become highly susceptible to replicative DNA damage following NS depletion and are protected from hydroxyurea-induced damage by NS overexpression. Breast cancer cells with basal-like characters display more reliance on NS for genome maintenance than those with luminal characters. Mechanistically, NS-deficient cells demonstrate a significantly reduced HR repair activity. TCGA analyses of human breast cancers revealed that NS is co-enriched positively with HR repair proteins and that high NS expression correlates with low HR defects and predicts poor progression-free survival and resistance to knockdown of cell cycle checkpoint genes in triple-negative/basal-like breast cancers. This work indicates that NS constitutes a tumor-promoting genome maintenance program required for mammary tumor progression.
Keywords: cancer stem cells, DNA repair, homologous recombination, mammary tumors
INTRODUCTION
Genomic damage is a constant occurrence in life, happening at a rate of about 70,000 events per day1. To minimize the effect of these continuous assaults on the genome, cells are equipped with a battery of molecular tools to sense and repair damaged DNAs2. If the DNA repair pathways, particularly those of higher fidelity, are overwhelmed by the incoming genotoxic load, some genomic “scars” will accumulate, resulting in genomic instability, cell cycle arrest, and/or cell death. Genomic instability increases the transformation rate and has long been viewed as a driver for cancers3. Indeed, next-generation sequencing has revealed DNA repair deficits in multiple tumor types, including prostate and breast cancers4, 5.
A major source of DNA damage comes intrinsically as a result of genome replication or cell metabolism6. DNA replication increases the rate of mutation during translesion synthesis, which uses error-prone DNA polymerases to help the replication machinery cross pre-existing damaged sites. DNA replication can also cause double-strand breaks (DSBs) when replication forks encounter single-strand breaks (SSBs), collide with DNA-bound protein complexes, or collapse after prolonged stalling7. Compared to other cell types, proliferative tumor cells are faced with an even greater risk of replicative damage, as oncogene activation is a potent inducer of genomic stress due to nucleotide depletion and increased DNA-bound proteins. As robust as cancer cells are, unrepaired genomic damage may still result in dire consequences, such as chromosomal loss, mitotic catastrophe, and eventually cell death. A paradox thus arises: does cellular programs that help mitigate genomic instability promote or suppress tumor development? Recent studies did indeed indicate that patients with BRCA1/2-mutated breast or ovarian cancers have better overall survival than those without8-11. The survival advantage of BRCA-mutated tumors has been largely attributed to their enhanced responsiveness to chemotherapies. It is not clear whether HR-deficient tumors undergo a more benign natural course of development compared to HR-proficient tumors12.
Nucleostemin (NS) is a nucleolar GTP-binding protein that functions in stem cell self-renewal13-15. It has also been shown to play indispensable roles in embryonic development, tissue regeneration, and pluripotency reprogramming16-20. One of its key functions is to protect mitotic stem/progenitor cells from DNA damage in the S-phase18, 20-23, which highlights the importance of genome maintenance in self-renewal as well as introducing a new component to the repair of replicative DNA damage23-25. The connection between NS and mammary tumors was raised in two studies15, 26. One showed that its expression level positively correlates with the tumorigenic activity of MMTV-wnt1 mammary tumor cells15. The other found that patients with NS-positive tumors experience shorter disease-free survival than those with NS-negative tumors26. Based on these two reports, we hypothesize that NS may offer a novel platform to elucidate the tumor-promoting aspect of genome maintenance in breast cancer development.
RESULTS
NS deletion compromises in vitro tumorigenesis and self-renewal of mammary tumor cells isolated from the MMTV-wnt1 model
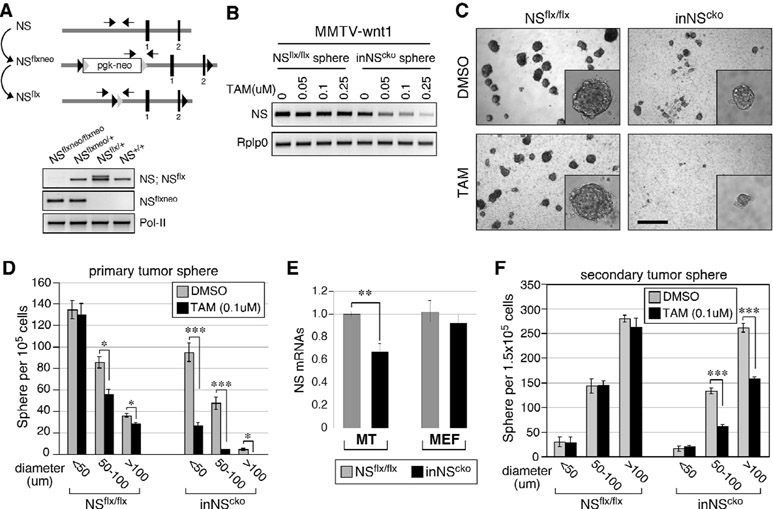
To develop a conditional strategy for NS deletion, we created an inducible CreER::NSflx/flx (inNScko) mouse model by breeding the tamoxifen (TAM)-inducible Cre (CreER) transgene into the homozygous NSflx/flx background. The design of the NSflx allele, shown in Fig.1A, has been used to study the NS deletion effect on embryogenesis18. To determine the functional importance of NS in mammary tumors, MMTV-wnt1::NSflx/flx mice were created by breeding the MMTV-wnt1 transgene into the homozygous NSflx/flx background, and then intercrossed with inNScko mice to generate trigenic MMTV-wnt1::inNScko mice. Mammary tumor cells were harvested from female bigenic (MMTV-wnt1::NSflx/flx) or trigenic (MMTV-wnt1::inNScko) mice, grown in suspension culture as tumor spheres, and treated with increasing concentrations of TAM. Semi-quantitative RT-PCR assays confirmed that TAM treatment induces a dose-dependent depletion of NS in inNScko but not in NSflx/flx mammary tumor spheres (Fig.1B). The effect of NS deletion on in vitro tumorigenesis was determined by the sphere-forming efficiency of primary mammary tumor (MT) cells in semi-solid culture. The number of spheres formed from inNScko MT cells treated with TAM at 0.1μM is significantly lower than the number of spheres formed from control DMSO-treated inNScko cells (Fig.1C and 1D). Even though TAM alone can also decrease the sphere formation of NSflx/flx MT cells, this effect is much smaller (~17% reduction) compared to its effect on inNScko cells (~79% reduction). In addition, inNScko MT cells display a lower baseline sphere-forming activity compared to NSflx/flx cells even in the absence of TAM (Fig.S1A). The decreased baseline activity of inNScko MT cells in sphere formation correlates with their lower NS expression compared to NSflx/flx MT cells (Fig.1E, left). This CreER-dependent decrease in NS baseline expression is not seen in non-mammary cells, such as mouse embryonic fibroblast (MEF) cells (Fig.1E, right). To determine the role of NS in the self-renewal of MT cells, primary tumor spheres were measured for their abilities to form secondary tumor spheres. Our data showed that TAM treatment (0.1μM) significantly decreases the 2nd sphere-forming efficiency of inNScko spheres, primarily those with diameters larger than 50μm (~46% reduction, Fig.1F). Unlike primary sphere formation, the 2nd sphere formation of NSflx/flx cells is not affected by the TAM treatment (Fig.1F, left), and there is no decrease in the baseline self-renewal activity of inNScko spheres compared to NSflx/flx spheres (Fig.S1B).
Figure 1.
Nucleostemin (NS) deletion compromises in vitro tumorigenesis and self-renewal of MMTV-wnt1 mammary tumor cells.
(A) Schematic diagram of NS-floxneo (NSflxneo) and NS-flox (NSflx) alleles (top panel). Black bars, exons; black arrowheads, loxP; grey arrowheads, Frt; pgk-neo, neomycin-resistance cassette; arrows, PCR primers; Pol-II, RNA polymerase-II. (B) Semi-quantitative RT-PCR of NS expression in MMTV-wnt1::NSflx/flx or MMTV-wnt1:: CreER::NSflx/flx (inNScko) mammary tumor spheres grown in suspension with different concentrations of tamoxifen (TAM). Rplp0, ribosomal protein P0. (C, D) DIC images and quantifications of primary sphere formation from MMTV-wnt1::NSflx/flx or MMTV-wnt1::inNScko mammary tumors grown in semi-solid culture with TAM (0.1μM) or without (DMSO). Scale bar, 300μm. (E) qRT-PCR of baseline NS expression in mammary tumor (MT) cells (MMTV-wnt1::NSflx/flx or MMTV-wnt1::inNScko) and mouse embryonic fibroblast (MEF) cells (NSflx/flx or inNScko). (F) Secondary sphere formation from primary tumor spheres in floating culture. Bar graphs, mean±sem; p values (two-sided t-test): ≤0.01 (*), ≤0.001 (**), and ≤0.0001 (***).
Loss of NS reduces the in vivo tumorigenic activity of MMTV-wnt1 mammary tumor cells
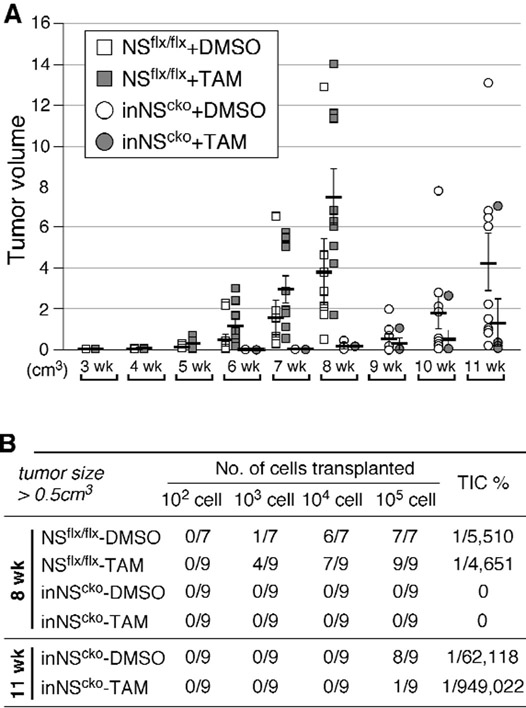
To determine how importance NS is to tumor development in vivo, primary mammary tumor spheres (NSflx/flx or inNScko) were treated with DMSO or TAM (0.1μM) for 2 days, dissociated, and grafted into the 4th inguinal mammary fat pads of nude mice at serial cell densities. We chose sphere-enriched cells as the source for xenograft because they are unaffected by TAM or CreER alone (Fig.1F). The number and size of mammary tumors formed at the transplanted sites over time are shown by the XY scatter plot in Fig.2A. The NSflx/flx groups (squares) were followed up for 8 weeks, and the inNScko groups (circles) were followed up for 11 weeks. Eight weeks after the transplantation, both DMSO-treated and TAM-treated NSflx/flx cells formed tumors ≥ 0.5cm3 in diameter at the grafted sites (Fig.2B). The estimated tumor-initiating cell (TIC) percentage is comparable between these two groups. While some tumors were formed in mice from DMSO-treated inNScko cells within 8 weeks, none of them were larger than 0.5cm3 in diameter at that time. At 11 weeks after the transplantation, 8 tumors were grown to the size of 0.5cm3 or larger in mice injected with DMSO-treated inNScko cells, but only 1 tumor did so in mice injected with TAM-treated inNScko cells. The estimated TIC percentage is 15-fold higher in DMSO-treated inNScko cells compared to TAM-treated inNScko cells (Fig.2B). These data show that NS deletion significantly reduces the in vivo tumorigenic activity of mammary tumor cells and that tumors derived from inNScko cells display a slower growth rate compared to NSflx/flx cells in vivo even without the TAM pre-treatment.
Figure 2.
Loss of NS decreases tumor formation of transplanted MMTV-wnt1 mammary tumor cells in vivo.
(A) The number and size of mammary tumors formed at the grafted sites over time by the XY scatter plot. X-axis shows the time (in weeks) after transplantation; Y-axis shows the volume (in cm3) of individual tumors. (B) Tumor incidences (numerator) from 7-9 transplanted events (denominator) tallied at 8 or 11 weeks for mice injected with NSflx/flx or inNScko mammary tumor cells, respectively. Frequencies of tumor-initiating cells (TIC%) were calculated by serial transplantation.
Mammary tumor cells are protected by NS from replication-induced DNA damage
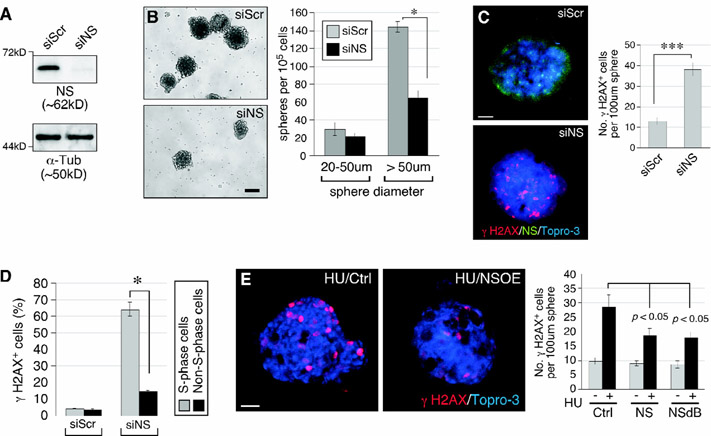
Mammary tumor cells were isolated from MMTV-wnt1::NSflx/flx tumors, grown in monolayer culture, and treated with the scrambled (siScr) or NS-specific (siNS) RNAi. Western blots confirmed that siNS treatment allows a 90% knockdown of NS protein compared to siScr treatment (Fig.3A). The in vitro tumorigenic activities of siScr and siNS-treated cells were measured by their abilities to form mammary tumor spheres in suspension culture. The results showed that NS depletion reduces the sphere-forming activity of these cells by 55% (Fig.3B). The effect of NS knockdown (NSKD) primarily affects spheres with diameters larger than 50μm, consistent with the effect of NS conditional knockout (Fig.1F). The DNA damage effect of NSKD on mammary tumors was shown by RNAi-mediated NS depletion, which significantly increases γH2AX+ cells in mammary tumor spheres (Fig.3C). To test whether NSKD-induced damage is related to genome replication, mammary tumor spheres were dissociated, grown in monolayer culture, pulse-labeled with BrdU, and double-stained with anti-γH2AX and anti-BrdU antibodies. In response to NSKD, 64.1% of the S-phase cells show γH2AX+ signals, whereas only 14.8% of the non-S-phase cells are γH2AX+ (Fig.3D), indicating that NSKD increases the susceptibility to replication-dependent DNA damage. As NSKD by itself increased spontaneous replication-dependent DNA damage, we then asked whether overexpression of NS (NSOE) could protect mammary tumor cells from drug-induced replicative DNA damage. Mammary tumor spheres were transfected with the control, NS-expressing, or NSdB-expressing plasmid, and measured for their sensitivities to hydroxyurea (HU) induced DNA damage. Our results showed that wildtype NS can protect mammary tumor spheres from HU-induced replicative damage, and so can NSdB (Fig.3E). NSdB is devoid of the N-terminal nucleolus-targeting sequence and hence distributed exclusively in the nucleoplasm27. In Fig.3C and 3E, we only counted spheres with a diameter of around 100μm to control the variable of sphere size.
Figure 3.
NS protects mammary tumor cells from replication-induced DNA damage.
(A) Western blots of RNAi-treated MMTV-wnt1::NSflx/flx mammary tumor cells in monolayer culture. α-tubulin, α-Tub. (B) DIC images and quantifications of tumor spheres from RNAi-treated cells in suspension culture. Y-axis shows the number of spheres per 105 plated cells. (C) RNAi-treated mammary tumor spheres labeled with anti-γH2AX antibody (red), anti-NS (green) antibody, and Topro-3 (blue). (D) Quantification of γH2AX+ cells in the BrdU+ (S) and BrdU− (non-S) subpopulations of RNAi-treated MMTV-wnt1 tumor cells dissociated from spheres and grown in monolayer culture. (E) Mammary tumor spheres transfected with control (Ctrl), NS-wildtype, or NSdB plasmid and examined for their DNA damage responses to hydroxyurea (HU) (2mM, 24h). Scale bars, 100μm (B) and 20μm (C, E).
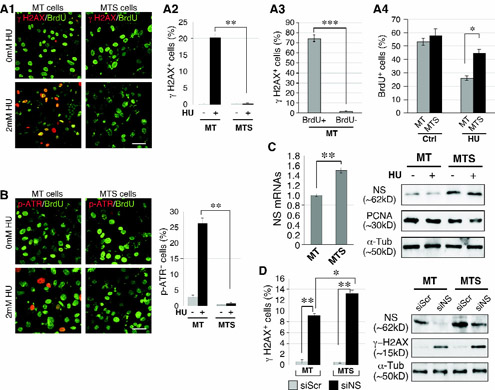
Sphere-enriched tumor cells are less sensitive to hydroxyurea (HU) treatment but more sensitive to NSKD than non-enriched mammary tumor (MT) cells.
Considering that tumor spheres are enriched in cells with basal-like properties and high NS expression15, we asked whether sphere-enriched (MTS) and non-enriched (MT) mammary tumor cells respond differently to HU and NSKD treatment. For HU sensitivity, cells were treated with 2mM HU for 24 hours, pulsed with 10μM BrdU for 1 hour before staining, and quantified for DNA damage by anti-γH2AX and S-phase cells by anti-BrdU staining (Fig.4A1). We discovered that HU treatment has a much smaller effect in inducing γH2AX+ foci in MTS cells (0.2% of total) compared to MT cells (20.3%, p<0.001) (Fig.4A2). In the MT culture, BrdU-labeled cells are much more susceptible to HU-induced DNA damage than non-BrdU-labeled cells (Fig.4A3). Before HU treatment, MT and MTS cells show comparable BrdU-labeling indices. After HU treatment, MT cells display a lower BrdU-labeling index than do MTS cells (Fig.4A4). To corroborate the γH2AX finding using a different DNA damage marker, we performed anti-phospho-ATR staining and obtained a consistent result by counting p-ATR+ cells (Fig.4B). One of the reasons that MTS cells are more resistant to replicative damage than MT cells may be related to their NS expression levels. To address this possibility, we showed that MTS cells express 50% more NS at the RNA level and 70% more at the protein level compared to MT cells (Fig.4C). Contrary to their response to HU, NS depletion significantly increases the γH2AX+ cell percentage in both MT and MTS cells, and that MTS cells are even more prone to the DNA damage effect of NSKD than are MT cells (Fig.4D, left). The different sensitivities of MT and MTS cells to the DNA damage effect of NSKD are not caused by differential KD efficiencies, as demonstrated by western blots (Fig.4D, right). Interestingly, siNS-treated MTS cells show much more γH2AX+ cells compared to siScr-treated MT cells, even though their NS protein levels are comparable, suggesting that the basal subtype of breast cancers may be more dependent on the NS function for genome maintenance than the luminal subtype.
Figure 4.
Sphere-enriched mammary tumor cells (MTS) are more sensitive to NS knockdown (NSKD) and less sensitive to HU treatment compared to non-enriched MT cells.
(A) DNA damage responses of MT and MTS cells to HU treatment by anti-γH2AX staining (red) and their mitotic indices by BrdU labeling (green). γH2AX+ cells were measured in percentages of total (A2) or BrdU+ vs. BrdU− cells (A3). BrdU+ cells were measured in percentages of total (A4). (B) DNA damage responses of MT and MTS cells to HU treatment by anti-phospho-ATR staining (red). (C) RT-PCR of NS expression in MT and MTS cells (left). Western blots of NS protein in MT and MTS cells with or without HU treatment (right). (D) Anti-γH2AX staining and western blots of NS and γH2AX proteins in RNAi-treated MT and MTS cells. Scale bars, 50μm (A, B).
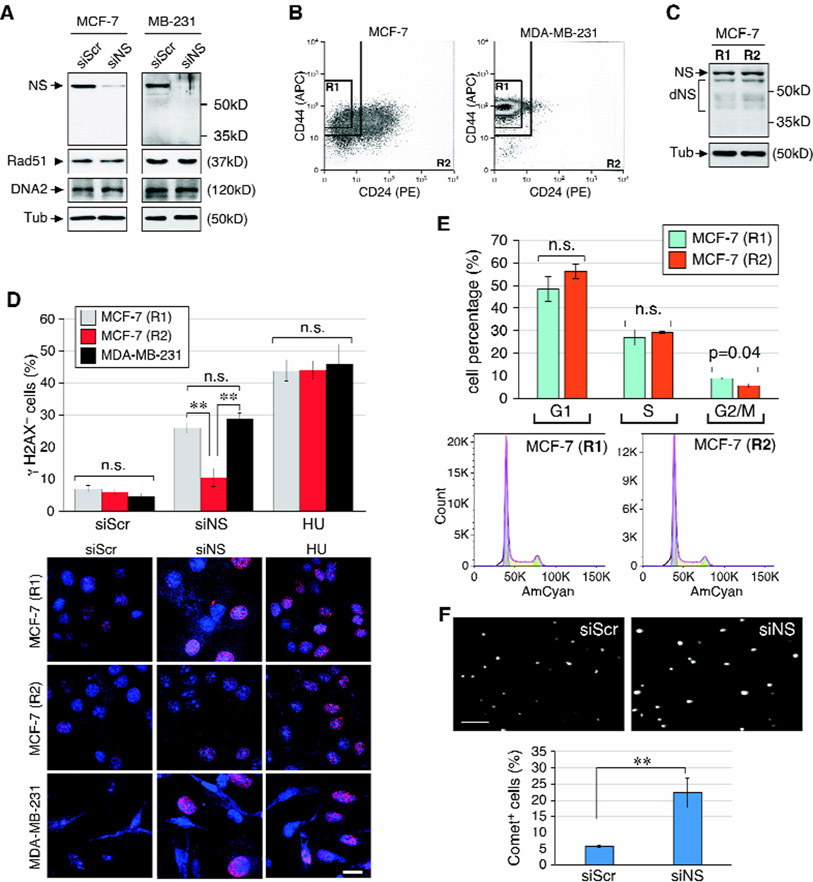
MDA-MB-231 and CD44+CD24low/- MCF-7 cells show increased reliance on NS compared to non-CD44+CD24low/- MCF-7 cells
To address the difference in NS reliance between the basal and luminal subtypes of breast cancers, we tested the NSKD response of MDA-MB-231 and MCF-7 cells, which display the basal or luminal characters, respectively. Western blots showed that growth arrest induced by serum starvation for 6 days decreases the amounts of NS and Rad51 proteins and increases the amount of Ku70 protein in both cell lines (Fig.S2A), consistent with the idea that non-proliferative cells relies mainly on NHEJ for genome maintenance. NS-specific RNAi treatment allows 90% knockdown of NS protein in both MCF-7 and MDA-MB-231 cells, but does not change the amount of Rad51 or DNA2 in either cell type (Fig.5A). A CD44/CD24-sorting paradigm has been widely used in multiple studies to purify mammary cancer stem cells (mCSCs) from human breast cancer samples28 and breast cancer cell lines29-31. Here we used this sorting scheme to subdivide MCF-7 and MDA-MB-231 cells into the R1 (CD44+CD24low/-) and R2 (non-CD44+CD24low/-) populations. FACS results showed that the R1 and R2 populations constitute 20.0% (±0.4%) and 52.7% (±2.1%) of MCF-7 cells, respectively (Fig.5B, left). In contrast, the R1 population makes up the majority of MDA-MB-231 cells (>95%) (Fig.5B, right). Western blots showed no difference in the NS protein amounts of the R1 and R2 of MCF-7 cells (Fig.5C). Yet these two populations of MCF-7 cells exhibit very distinct sensitivities to NSKD, with the R1 being much more sensitive to NSKD-induced DNA damage (26.0%) compared to the R2 (10.5%) (Fig.5D). The different sensitivities of R1 and R2 to NSKD is not a result of different proliferative activities, as both cells display equal sensitivities to HU-induced damage (44% vs. 44% γH2AX+ cells) and statistically indistinguishable cell cycle profiles (Fig.5E). Because >95% of MDA-MB-231 cells fall within the R1 domain, we used unsorted MDA-MB-231 cells to determine their susceptibility to NSKD and HU-induced DNA damage. Our results showed that MDA-MB-231 cells resemble the R1 of MCF-7 cells in their DNA damage response to NSKD (28.9%) and show no difference from either the R1 or R2 of MCF-7 cells in their baseline or HU-induced γH2AX+ cells (Fig.5D). The R1 and R2 populations of MCF-7 cells show no significant cell-cycle perturbation in response to the NSKD treatment, nor do they differ from each other (Fig. S3A), suggesting that cell cycle stoppage is not a main contributor to this finding. Finally, we demonstrated that NSKD creates physical breakage of DNAs in MDA-MB-231 cells, as measured by the increased tail moment in the comet assay (Fig.5F). These data support that the basal subtype of breast cancer cells is more reliant on NS for genome protection than the luminal subtype. Both cell types respond similarly to increasing concentrations of HU In terms of DNA damage and show no change in the protein levels of NS or RAD51 during HU treatment (Fig.S2B).
Figure 5.
CD44/CD24-sorted MCF-7 and MDA-MB-231 cells display differential sensitivities to NSKD-induced DNA damage.
(A) Western blots of NS, Rad51, and DNA2 in control (siScr) and NSKD (siNS) MCF-7 and MDA-MB-231 cells. (B) CD44/CD24 sorting of MCF-7 and MDA-MB-231 cells into a stem/basal-like population (CD44+CD24low/-, R1) and the remaining population (R2). (C) Western blots of NS protein (Ab138) in the R1 and R2 of MCF-7 cells. dNS, degraded products of NS. (D) DNA damage response of the R1 (grey) and R2 (red) of MCF-7 cells and unsorted MDA-MB-231 cells (black) under the control, NSKD, and HU treatment conditions. (E) Cell-cycle analyses of the R1 and R2 of MCF-7 cells. (F) DNA tail moments of RNAi-treated MDA-MB-231 cells by the comet assay. Scale bar, 20μm.
NS function has been previously linked to the p53 pathway32-34, and the p53 status in breast cancer varies among different subtypes, with a mutation rate of 89.5%, 70.5%, and 17.8% in the TNBC/basal-like, Her2, and luminal tumors, respectively, which is consistent with MDA-MB-231 cells being p53-mutated and MCF-7 cells being p53-wildtype. We have previously shown that NSKD triggers DNA damage independently of the p53 status in isogenic MEF cells18. To address a similar question in breast cancer cells, we measured the DNA damage effect of NSKD in BT-474 (p53-mutated luminal) and compared it with that in MCF-7 (p53-wildtype luminal) and MDA-MB-231 (p53-mutated TNBC) cells. Our results showed that the p53 status does not apparently determine the γH2AX response of breast cancer cells to NSKD either (Fig.S3B).
Loss of NS leads to defective homologous recombination repair
The accumulation of DNA damage in NS-depleted cells may be the result of an increased flux or a decreased repair of DNA damage. To differentiate these two possibilities, we examined how NS perturbation affects the recovery of ionizing radiation (IR)-induced lesions in MDA-MB-231 cells, as they displayed a high dependence on NS for genome maintenance. Before IR treatment, siNS-treated cells show more p-ATM+ cells compared to siScr-treated cells. IR treatment equalizes the amounts of DNA damage in both control and NSKD cells to almost 100% within an hour, saturating the DNA damage flux in both groups. In the course of 24 hours, 57% of the siScr-treated cells recover, whereas only 28% of the siNS-treated cells do so (Fig.S4A). Conversely, the recruitment of RAD51 protein to IR-induced foci is reduced in siNS-treated compared to siScr-treated MDA-MB-231 cells (Fig.S4B). In gain-of-function studies, MDA-MB-231 cells expressing GFP-fused NS show a faster recovery compared to cells expressing GFP at the 12-hour and 24-hour time points after the IR treatment (Fig.S4C). These results support that NS is involved in promoting the repair rather than reducing the source of DNA damage.
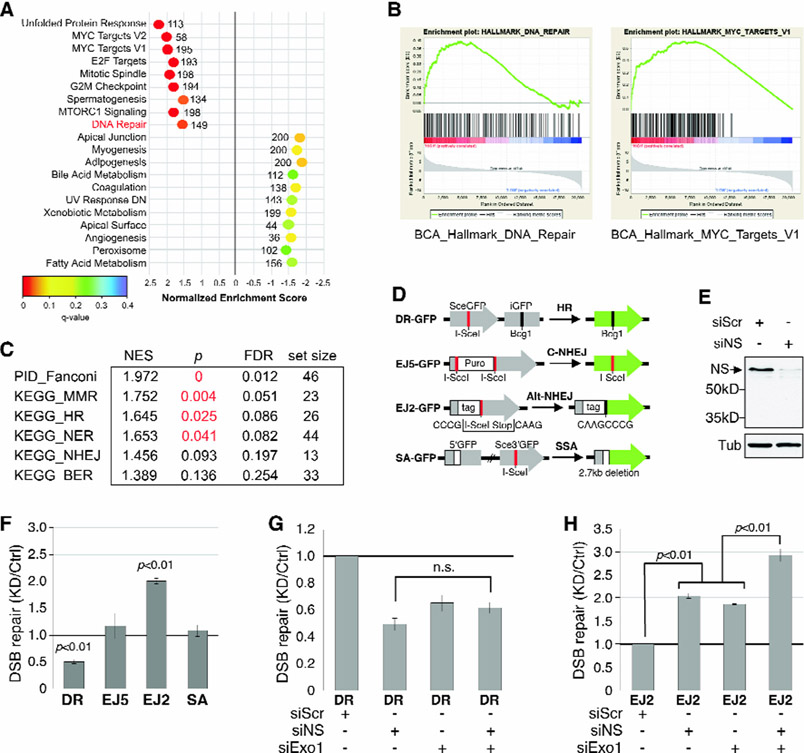
To explore the mechanism underlying the NS-promoted repair in breast cancers, we applied the Gene Set Enrichment Analysis (GSEA) on the TCGA RNAseq database to determine which of the 20,530 genes are co-enriched with NS in human BCA samples. HALLMARK pathway analyses (h.all.v5.1.symbols.gmt, 50 pathways) revealed 9 positively co-enriched gene sets and 11 negatively co-enriched gene sets above the set threshold for significance (p < 0.05) (Fig.6A). Co-enrichment of NS with the MYC targets pathway (Fig.6B, right) is a reassuring validation of this analysis, as previous studies have reported NS as a direct target of c-Myc35, 36. The pathway relevant to genome maintenance is DNA Repair (Fig.6B, left). To further dissect the connection between NS and various DNA repair pathways, we applied the canonical gene set filter (c2.cp.v5.1.symbols.gmt) that contains 1,330 pathways of smaller sizes., and showed that the Fanconi, mismatch repair (MMR), HR, and nucleotide excision repair (NER) pathways are the only ones meeting the threshold for significance in BCA samples (Fig.6C). The Notch and mTOR pathways show borderline significance (Fig.S5A). We also performed GSEA on pathways co-enriched with NS using the TCGA protein database (n=80), previously characterized by TCGA/CPTAC37. The Hallmark pathways identified as positively co-enriched with NS reiterate those identified based on the RNAseq data (Fig.S5B). In addition, the apoptosis pathway shows up with the highest negative NES score, indicating a role of NS in cell survival. Most importantly, NS is co-enriched with more members in the HR pathway at a higher level (NES>6) compared to the other three DNA repair pathways identified by the Canonical pathway analyses, indicating that NS is more likely to be co-regulated, expression-wise, with HR repair genes than with other DNA repair genes in mammary tumors in response to a persistent stress condition (Fig.S5C).
Figure 6.
NS co-enrichment with DNA repair proteins in human breast cancers and its function in promoting HR repair of double-strand breaks (DSBs).
(A) Top-ranked HALLMARK Pathways with positive (NES>1.5) or negative (NES<−1.5) co-enrichment with NS and p values <0.05 by GSEA. X-axis shows NES (−3 to 3); color scheme depicts FDR q values (0 to 0.4); numbers indicate the set size. (B) Enrichment plots for DNA repair (left) and MYC targets (right). (C) Canonical Pathway Analysis of RNAseq-based NS co-enrichment with specific DNA repair pathways in breast cancers. (D) Schematic diagrams of I-SceI-based DSB repair models. (E) Western blots of RNAi-mediated NSKD. (F) Effect of NSKD on HR, C-NHEJ, Alt-NHEJ, and SSA repair by the DR-GFP EJ5-GFP, EJ2-GFP, and SA-GFP assays, respectively. (G) Effects of NSKD and/or Exo1KD (siExo1) on HR repair by the DR-GFP assay. (H) Effects of NSKD and/or Exo1KD on Alt-NHEJ repair by the EJ2-GFP assay. ES, enrichment scores; NES, normalized ES; FDR, false discovery rate.
To establish the NS function in HR repair, we used an I-SceI-based reporter cell line to determine the NSKD effect on HR (DR-GFP). In addition to HR, I-SceI-induced double-strand breaks (DSBs) can also be repaired by classical non-homologous end-joining (C-NHEJ), alternative NHEJ (Alt-NHEJ), or single-strand annealing (SSA), which can be measured by the EJ5-GFP, EJ2-GFP, and SA-GFP models, respectively38-41 (Fig.6D). Western blots confirmed the knockdown efficiency of siNS in the parental (U2OS) cell line (Fig.6E). In support of the GSEA findings, HR repair, as measured by the GFP+ cell percentage of I-SceI-transfected DR-GFP cells, is significantly decreased by NSKD by 51% (siNS) (Fig.6F). In contrast, NSKD increases the GFP+ cell percentage of I-SceI-transfected EJ2-GFP cells by 2 folds and has no effect on EJ5-GFP or SA-GFP cells (Fig.6F). Next, we compared the HR effects of NSKD, Exo1 knockdown (Exo1KD), and combined NSKD/Exo1KD, as Exo1 is a key factor in HR initiation. Our results showed that NSKD reduces the HR-based repair as much as Exo1KD does, and that a combined depletion of NS and Exo1 does not further decrease HR (Fig.6G). For the EJ2-dependent repair, Exo1KD also increases its activity as does NSKD, and the effects of Exo1KD and NSKD are additive (Fig.6H).
NS predicts progression-free survival and synthetic lethality to gene inactivation or drug treatment in human triple-negative/basal-like breast cancers (TNBC)
To determine the clinical relevance of NS, we analyzed its expression in breast cancer samples available through TCGA. Our results showed that NS is expressed at a higher level in patients with TNBC compared to patients with luminal or HER2+ breast cancers (Fig.7A) but does not vary among different tumor stages (Fig.7B). Within the TNBC population, NS loss is associated with an increase in HR defect (HRD) score, which measures the amount of genomic scars accumulated as a result of HR repair deficiency (Fig.7C). This finding is consistent with the role of NS in promoting HR repair in breast cancers. Importantly, patients with NS-high TNBC show worse prognosis compared to patients with NS-low TNBC (Fig.7D, log-rank p=0.0018). The Cox proportional hazard model was used to control for tumor stage and showed that the level of NS expression is predictive of survival as robustly as but independently of tumor stage (Fig.7E, p=0.0017). We then took advantage of the recent ACHILLES genome-wide pooled CRISPR screening data in breast cancer cell lines to explore the relative vulnerabilities of NS-high versus NS-low tumors. We determined the Spearman correlation coefficient between NS expression levels and relative changes in cell abundance following gene knockout and used the resulting gene lists for GSEA. Negative NES values indicate synthetic lethality between NS-high cells and gene knockout, whereas positive NES values indicate the opposite. This analysis showed that blocking EGF signaling is synthetically lethal to NS-high breast cancer cells (Fig.7F). In support, knockdown of ErbB2/3 or proteins downstream of the PI3K/mTOR pathway are more deleterious to NS-high cells compared to NS-low cells (Fig.7G). Pharmacologically, the NS-high cell lines are also more sensitive to pharmacological inhibition of the ErbB and PI3K/mTOR pathways (Fig.7H).
Figure 7.
NS predicts HR defects, progression-free survival, and synthetic lethality or viability in triple-negative/basal-like breast cancers (TNBC).
TCGA analyses of RNAseq-based NS expression by breast cancer subtypes (A) or stages (B). Significance by Wilcoxon rank-sum test (A) or Kruskal-Wallis H test (B). Numbers in parentheses show sample sizes. (C) HR defect scores plotted against RNAseq-based NS expression in TNBC. Line represents the linear regression line; shaded region shows 95% confidence interval; each dot represents an individual patient (n=148). Correlation coefficient (r) and significance (P) by Spearman correlation analysis. (D) Progression-free survival of TNBC with NS-high and NS-low expression by the Kaplan Meier plot (n=149). High NS is defined as the upper tertile of NS expression and the remaining two-thirds is defined as low NS. Significance by log-rank test. (E) Multi-variate survival analysis of NS and stage in TNBC by the Cox proportional hazards model. (F) GSEA of correlation between NS expression and reduction in viability in breast cancer cells from the genome-wide ACHILLES CRISPR knockdown screen. Table lists top-ranked Reactome pathways whose knockdown shows synthetic viability (NES>0) or lethality (NES<0) with NS-high breast cancers. (G) Relative viability of NS-high vs. NS-low breast cancer cells following knockout of ERBB2/3, PIK3CA, or RICTOR. NS-high and NS-low are defined as 1 SD above and 0.5 SD below the mean, respectively. (H) Relative viability of NS-high vs. NS-low breast cancer cells following treatment with EGFR/ERBB2 or PI3K inhibitors from CTRPv2, as quantified by area under the curve (AUC) following dose-dependent treatment. Lower AUC values indicate increased sensitivity. Error bars, mean±SD; *, p<0.05; **; p<0.01; ***, p<0.001; ****, p≤0.0001; n.s., not significant; Haz, hazard ratio; PTM, post-translational modification.
DISCUSSION
The implication of NS in tumor development has been driven primarily by its increased expression in multiple cancer types and its loss-of-function effect on the proliferation and survival of cancer cell lines. In this study, we establish the functional importance of NS in mammary tumor development in vivo. Our in vitro data reveal that NS depletion decreases the number of large-sized spheres more than the number of small-sized ones. TCGA analyses of human breast cancer samples determine that NS is expressed at a higher level in the TNBC compared to the luminal or HER2+ tumors. Notably, high NS expression shows a strong correlation with low progression-free survival in TNBC, and the prognostic power of NS is close to that of the current clinical staging system. In predicting progression-free survival, NS operates independently of clinical stages, shown by the multi-variate survival analysis of NS and stage and supported by the same NS expression levels in four stages of breast cancers. Together, our data support that NS is involved in mammary tumor progression and may serve a new prognostic marker for TNBC clinically.
To create a mammary tumor model with NS conditional-null capability, we chose the TAM-CreER strategy based on the consideration of feasibility and efficiency in generating enough female mice with triple genetic modifications. Although this strategy had worked in our previous study18, two background issues were encountered when being applied to mammary tissues. First, TAM is an anti-cancer drug for ER+ mammary tumors and may inhibit tumor growth by itself. To avoid with this problem, we only used TAM transiently at a low dose (0.1μM) to avoid the reported cytostatic and cytotoxic effects of TAM, which occur at the 2-100μM and >100μM concentration, respectively42. We found that TAM by itself still has a mild inhibitory effect on the growth of primary tumor spheres (~17% reduction, Fig.1D), but has no effect on the formation of secondary tumor spheres (Fig.1F). The differential response of primary versus secondary tumor spheres to low-dose TAM can be explained by the preferential enrichment of basal-like cells over luminal cells via the sphere culture15 and that basal-like cells are ER-negative and hence TAM-resistant. Second, even without the TAM treatment, the CreER transgene itself decreases NS expression (~33% reduction, Fig.1E) and hence the baseline tumorigenic activity (Fig.S1A). The background leakage in NS deletion occurs mainly in mammary tissues and is more evident in the formation of primary spheres than secondary spheres (Fig.S1B). This leakage deletion of NS in mammary tumors may be triggered by endogenous ligands for CreER, and the lack of CreER-dependent background in sphere-enriched cells may be explained by less endogenous ligands or a higher baseline expression of NS in the basal-like subpopulation of mammary tumor cells. Interestingly, the in vivo tumor progression of sphere-enriched cells is still affected by the CreER transgene even without the TAM treatment but not by the TAM treatment alone (Fig.2), suggesting that these cells either are exposed endogenous ligands or undergo partial differentiation after being grafted back into the mammary fat pad. Despite these two issues, the NS-depleted groups invariably show significantly less tumors with smaller sizes compared to their respective controls (~79% reduction).
The direct connection between NS and DNA damage response/repair was previously implicated by its chromatin immunoprecipitation binding to DSB sites and its knockdown effect on reducing RAD51 foci formation in embryos and regenerating hepatocytes18, 20. The involvement of NS in genome maintenance in response to a wide range of DNA damage sources appears to converge on the HR repair pathway more so than on other DNA repair mechanisms. We establish that the NS function in protecting breast cancer cells from replicative DNA damage and that loss of NS perturbs HR-based repair but not that of C-NHEJ or SSA. The involvement of NS in HR repair in human breast cancers is further supported by its co-enrichment with multiple HR proteins in human breast cancers as well as by the strong negative correlation between the NS expression and HRD score in TNBC. Notably, NS-deficient EJ2-GFP cells display an increased repair. The increase in EJ2-dependent repair by both NSKD and Exo1KD suggests that it may be a secondary event to HR impairment. The additive effect of NS/Exo1 double-KD compared to single-KD suggests that NS may also suppress Alt-NHEJ independently of its HR regulation. Since NS does not increase the level of HR proteins, it may promote the HR machinery by regulating their compartmentalization, recruitment to damage sites, and/or enzymatic activities. Further investigation is required to understand the molecular detail of NS action.
Different subtypes of tumors may use different strategies to cope with genomic. Here, we address the issue of tumor heterogeneity by comparing the NS reliance between: 1) sphere-enriched vs. primary mammary tumor cells, and 2) CD44+CD24low/- vs. non-CD44+CD24low/- MCF-7 cells vs. MDA-MB-231 cells. We found that sphere-enriched cells are more resistant to HU-induced DNA damage compared to primary mammary tumor cells. Sphere culture is known to enrich for NS-high cells with basal or stem cell-like properties15. Since sphere-enriched and primary mammary tumor cells show comparable levels of BrdU incorporation and PCNA expression, their different sensitivities to HU treatment cannot be explained simply by their S-phase cell percentages, as represented by the mitotic index. We speculate that the higher HU resistance of sphere-enriched cells may be contributed by multiple factors that include higher NS expression, more multidrug resistance gene expression, as well as difference in cell cycle kinetics not revealed by the mitotic index. For human breast cancer cell lines, we found that the R1 of MCF-7 cells are more reliant on NS for genome maintenance compared to the R2, even though both of them express comparable levels of NS, display similar cell cycle profiles, and are equally sensitive to HU-induced DNA damage. Most of MDA-MB-231 cells are CD44+CD24low/- and resemble the R1 of MCF-7 cells in their NS dependence. The idea that basal-like breast cancers are more reliant on NS compared to luminal breast cancers is also supported by their higher NS expression in patients’ samples. Finally, the DNA damage effect of NSKD does not depend on the p53 status, although the pathways by which cells choose to react to NSKD-induced DNA damage are influenced by their p53 status34.
While the increased expression of NS in tumors suggests that it may act as an oncogene, its ability to promote HR repair is reminiscent of the role of a tumor suppressor, as HR deficiency is a known cancer-predisposing factor. The resolution of the seemingly dichotomous roles of genome maintenance in tumor development may lie in the differences in DNA damage types and their repair pathways. While some mutations may drive the evolution of cancers, others (such as DSBs) may be detrimental to their survival. Recent studies have indeed shown that HR deficiency can sensitize the already transformed tumors to replicative and genotoxic stress at the late stage. On one hand, DNA repair proteins whose loss of functions is relatively compatible with life may behave like tumor suppressors. On the other hand, DNA repair proteins whose loss of function is definitely incompatible with life may act like oncogenes. Since tumors evolve over time, they may also develop an essential reliance on certain genome maintenance programs to help them avoid catastrophic genomic damage. We propose that NS serves as a compensatory mechanism essential for the survival of tumors as they progress. Assuming this to be correct, one may infer that NS-enriched tumors should be less responsive to chemotherapies than NS-deficient tumors and that NS-targeting therapies might render chemo-resistant tumors to become chemo-sensitive. Our genome-wise synthetic lethal screens support this idea by showing that NS-high breast cancers are more resistant to knockdown of genes involved in cell cycle checkpoint regulation compared to NS-low tumors. Conversely, NS-high breast cancers are predicted to be more sensitive to knockdown of the EGF pathway as well as to drugs that inhibit EGFR and PI3 kinase, compared to NS-low breast cancers.
In conclusion, this work establishes the importance of NS in breast cancer development in vivo and reveals the tumor-promoting side of a HR-related genome maintenance mechanism. It also indicates that some breast cancers (e.g. TNBC) may adapt to the hyperactive mitotic state of malignancy by ramping up the NS program to compensate for pre-existing HR deficiency and hence minimizing catastrophic damage caused by replication stress and drug treatment.
MATERIALS AND METHODS
Animal care and procurement
All animals were housed by the Program for Animal Resources at the TAMHSC-Houston campus, handled in accordance with the principles described by the Guide for the Care and Use of Laboratory Animals, and approved by the institutional IACUC.
Tumor spheres in semi-solid and suspension culture
To grow tumor spheres in suspension, dissociated cells were harvested from MMTV-wnt1 tumors, plated on ultra-low attachment plates in supplemented DMEM/F12. To grow tumor spheres in semi-solid culture, 0.6% low melting-point agar in DMEM/F12 was plated at the bottom of 24-well plate. Three biological and two technical replicates were analyzed by two-sided t-test.
In vivo tumorigenic assay
Cells were isolated from MMTV-wnt1 tumors, grown in spheres, and plated on monolayer culture for TAM treatment. Cells were dissociated, resuspended at serially diluted densities, randomly injected into the 4th inguinal mammary fat pads of 6-8 week-old athymic nude female mice, and tallied at 8 and 11 weeks. Sample size for injection was estimated at 7-9 for each cell density, totaling 28-36 for each cell group.
Comet assay
DNA strand breaks were analyzed by using the alkaline CometAssay kit (Trevigen, 4250-050-K). The amounts of DNA damage were quantified by the tail moments of > 100 cells per slide using the CometScore software and analyzed from four biological replicates.
TCGA, Gene Set Enrichment Assay (GSEA), and HRD
Processed RNAseq-based gene expression data were acquired through the GDC data portal. Proteomic data were acquired from CPTAC37. Genomic homologous recombination defect (HRD) scores were obtained based on the methods described previously43, 44. All other data were acquired through cBioPortal45. Correlations between genomic HRD score and NS expression were determined based on the Spearman correlation coefficient. For survival analysis, patients were stratified into high-NS (highest 33% of samples) and low-NS (bottom 66% of samples).
Synthetic lethal and pharmacological inhibitor analyses
Cell line gene expression data were acquired from the Cancer Cell Line Encyclopedia46. Genome-wide pooled CRISPR screen results, calculated by the CERES method, were acquired from ACHILLES. Pharmacological screening data were acquired from CTRPv247. For validation of these findings with pharmacological inhibitors, cells were stratified using the same thresholds and quantified using area under the curve (AUC) values from drug response data in CTRPv2.
Supplementary Material
ACKNOWLEDGEMENT
We thank Margie Moczygemba at the IBT Flow Cytometry Core for her help in cell cycle analysis. This work was supported by NCI-PHS grants (R01 CA113750, R03 CA201988, R21AG052006) to RYT, National Science Council Postdoctoral Fellowship (ROC) to TCL, G. Komen PDF17483544 to DJM, and CPRIT RP140456 to GP.
Footnotes
COI: The authors declare no potential conflicts of interest.
Supplementary information is available at Oncogene’s website.
REFERENCES
- 1.Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol 2000; 65: 127–133. [DOI] [PubMed] [Google Scholar]
- 2.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell 2010; 40: 179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res 1974; 34: 2311–2321. [PubMed] [Google Scholar]
- 4.Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med 2016; 375: 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016; 534: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tubbs A, Nussenzweig A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017; 168: 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michel B, Grompone G, Flores MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proc Natl Acad Sci U S A 2004; 101: 12783–12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graeser M, McCarthy A, Lord CJ, Savage K, Hills M, Salter J et al. A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin Cancer Res 2010; 16: 6159–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009; 361: 123–134. [DOI] [PubMed] [Google Scholar]
- 10.Wiedemeyer WR, Beach JA, Karlan BY. Reversing Platinum Resistance in High-Grade Serous Ovarian Carcinoma: Targeting BRCA and the Homologous Recombination System. Front Oncol 2014; 4: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng G, Chun-Jen Lin C, Mo W, Dai H, Park YY, Kim SM et al. Genome-wide transcriptome profiling of homologous recombination DNA repair. Nat Commun 2014; 5: 3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer discovery 2015; 5: 1137–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev 2002; 16: 2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baddoo M, Hill K, Wilkinson R, Gaupp D, Hughes C, Kopen GC et al. Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J Cell Biochem 2003; 89: 1235–1249. [DOI] [PubMed] [Google Scholar]
- 15.Lin T, Meng L, Li Y, Tsai RY. Tumor-initiating function of nucleostemin-enriched mammary tumor cells. Cancer Res 2010; 70: 9444–9452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu J, Bishop JM. Nucleostemin maintains self-renewal of embryonic stem cells and promotes reprogramming of somatic cells to pluripotency. J Cell Biol 2012; 197: 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Q, Yasumoto H, Tsai RY. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol Cell Biol 2006; 26: 9279–9290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng L, Lin T, Peng G, Hsu JK, Lee S, Lin S-Y et al. Nucleostemin deletion reveals an essential mechanism that maintains the genomic stability of stem and progenitor cells. Proc Natl Acad Sci U S A 2013; 110: 11415–11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai RY. New frontiers in nucleolar research: nucleostemin and related proteins. The Nucleolus (Protein Reviews 15) 2011: 301–320. [Google Scholar]
- 20.Lin T, Ibrahim W, Peng C-Y, Finegold MJ, Tsai RY. A novel role of nucleostemin in maintaining the genome integrity of dividing hepatocytes during mouse liver development and regeneration. Hepatology 2013; 58: 2176–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu JK, Lin T, Tsai RY. Nucleostemin prevents telomere damage by promoting PML-IV recruitment to SUMOylated TRF1. J Cell Biol 2012; 197: 613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin T, Meng L, Wu LJ, Pederson T, Tsai RY. Nucleostemin and GNL3L exercise distinct functions in genome protection and ribosome synthesis, respectively. J Cell Sci 2014; 127: 2302–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai RY. Turning a new page on nucleostemin and self-renewal. J Cell Sci 2014; 127: 3885–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai RY, Meng L. Nucleostemin: A latecomer with new tricks. Int J Biochem Cell Biol 2009; 41: 2122–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai RY. Balancing self-renewal against genome preservation in stem cells: How do they manage to have the cake and eat it too? Cellular and molecular life sciences : CMLS 2016; 73: 1803–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi T, Masutomi K, Tamura K, Moriya T, Yamasaki T, Fujiwara Y et al. Nucleostemin expression in invasive breast cancer. BMC Cancer 2014; 14: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng L, Lin T, Tsai RY. Nucleoplasmic mobilization of nucleostemin stabilizes MDM2 and promotes G2-M progression and cell survival. J Cell Sci 2008; 121: 4037–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003; 100: 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH et al. CD44+/CD24− breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res 2006; 8: R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phillips TM, McBride WH, Pajonk F. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst 2006; 98: 1777–1785. [DOI] [PubMed] [Google Scholar]
- 31.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res 2008; 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol 2008; 28: 4365–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo D, Lu H. Nucleostemin: Another nucleolar “Twister” of the p53-MDM2 loop. Cell Cycle 2010; 9: 3227–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang G, Meng L, Tsai RY. p53 configures the G2/M arrest response of nucleostemin-deficient cells. Cell Death Discov 2015; 1: e15060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Connell BC, Cheung AF, Simkevich CP, Tam W, Ren X, Mateyak MK et al. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J Biol Chem 2003; 278: 12563–12573. [DOI] [PubMed] [Google Scholar]
- 36.Zwolinska AK, Heagle Whiting A, Beekman C, Sedivy JM, Marine JC. Suppression of Myc oncogenic activity by nucleostemin haploinsufficiency. Oncogene 2012; 31: 3311–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016; 534: 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 2008; 4: e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennardo N, Gunn A, Cheng A, Hasty P, Stark JM. Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet 2009; 5: e1000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gunn A, Stark JM. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol Biol 2012; 920: 379–391. [DOI] [PubMed] [Google Scholar]
- 41.Gunn A, Bennardo N, Cheng A, Stark JM. Correct end use during end joining of multiple chromosomal double strand breaks is influenced by repair protein RAD50, DNA-dependent protein kinase DNA-PKcs, and transcription context. J Biol Chem 2011; 286: 42470–42482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Etienne MC, Milano G, Fischel JL, Frenay M, Francois E, Formento JL et al. Tamoxifen metabolism: pharmacokinetic and in vitro study. Br J Cancer 1989; 60: 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watkins JA, Irshad S, Grigoriadis A, Tutt AN. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast cancer research : BCR 2014; 16: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep 2018; 23: 239–254.e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483: 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME et al. Harnessing Connectivity in a Large-Scale Small-Molecule Sensitivity Dataset. Cancer discovery 2015; 5: 1210–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.