Abstract
It is becoming generally accepted in recent literature that the Warburg effect in cancer depends on inhibition of M2PYK, the pyruvate kinase isozyme most commonly expressed in tumors. We remain skeptical. There continues to be a general lack of solid experimental evidence for the underlying idea that a bottle neck in aerobic glycolysis at the level of M2PYK results in an expanded pool of glycolytic intermediates (which are thought to serve as building blocks necessary for proliferation and growth of cancer cells). If a bottle neck at M2PYK exists, then the remarkable increase in lactate production by cancer cells is a paradox, particularly since a high percentage of the carbons of lactate originate from glucose. The finding that pyruvate kinase activity is invariantly increased rather than decreased in cancer undermines the logic of the M2PYK bottle neck, but is consistent with high lactate production. The “inactive” state of M2PYK in cancer is often described as a dimer (with reduced substrate affinity) that has dissociated from an active tetramer of M2PYK. Although M2PYK clearly dissociates easier than other isozymes of pyruvate kinase, it is not clear that dissociation of the tetramer occurs in vivo when ligands are present that promote tetramer formation. Furthermore, it is also not clear whether the dissociated dimer retains any activity at all. A number of non-canonical functions for M2PYK have been proposed, all of which can be challenged by the finding that not all cancer cell types are dependent on M2PYK expression. Additional in-depth studies of the Warburg effect and specifically of the possible regulatory role of M2PYK in the Warburg effect are needed.
Introduction
Many researchers have concluded that the pyruvate kinase (PYK) reaction, the last reaction in glycolysis, plays a pivotal role in controlling metabolism in cancer tissues. In fact, even in 1975 it was known that tumor cell respiration can be stimulated by the use of PYK inhibitors [46, 47]. More recent discussions on the controllers of Warburg metabolism often reference Christofk, et al. to indicate PYK’s role in tumor growth [48]. Given the appeal of targeting Warburg metabolism to treat cancer and the growing acknowledgment of the role of M2PYK in controlling that metabolism, it is not surprising that there has now been considerable effort to directly target M2PYK activity for drug design [49–68].
Unfortunately, the race to identify a M2PYK-targeting cancer drug has led to several poorly supported explanations for how M2PYK functions in cancer and many of those explanations are based on data that have not been fully scrutinized (or speculated ideas that were never supported by data!). Therefore, the goal of this review is to critically evaluate M2PYK functions in the context of Warburg metabolism. However, rather than simply dismiss M2PYK as an important enzyme in Warburg metabolism, we conclude with a speculation about the role of increased glycolysis in cancer metabolism.
A quick overview of PYK
Historically, the unique properties of the pyruvate kinase protein in cancers were identified simultaneously with isozyme expression patterns in normal tissues [75–84]. There are four isozymes expressed in mammals [75, 87, 88]. LPYK (found in liver and pancreas) and PYK from erythrocytes (R-PYK) are products of one gene as a result of alternative start sites [89–93]. M1PYK (found in heart, muscle and brain and characterized by a hyperbolic response of activity over a concentration range of PEP) and M2PYK (also referred to as KPYK due to its presence in kidney and characterized by a sigmoidal response of activity over a concentration range of PEP), originate from a second gene via alternative RNA splicing [75, 87, 88, 96, 97], and differ by only 22 amino acids. In early fetal tissue, M2-PYK is the only isozyme detected. Near the time of birth, expression of M2PYK is displaced by tissue specific isozymes in many tissues [75, 88, 100, 101]. Despite this general trend for a change in isozyme expression, several adult tissue types continue to express M2-PYK, including adult lung, kidney and many smooth muscle organs [75, 88, 100, 104–106]. Re-expression of M2PYK is an early event in transformation of normal tissue into cancer [112]; therefore, the serum level of M2PYK has been evaluated as a marker for many types of cancers [113–115]. These well-established facts about M2PYK set a background for the discussion of a role of M2PYK in Warburg metabolism.
M2PYK in cancer tissue/cells is NOT an “inhibited” isozyme of pyruvate kinase
The often-repeated assumption that the M2PYK present in cancer cells is an inhibited form of pyruvate kinase seems to have been derived from a comparison of the enzymatic properties of M2PYK to those of M1PYK [96, 97]. Indeed, in the absence of post-translational modifications, protein:protein interacting partners, or allosteric effectors, M1PYK has a lower higher apparent affinity (i.e., We use apparent affinity to refer to the concentration of substrate at ½ Vmax activity. For a sigmoidal response fit to the Hill equation, this parameter is sometimes referred to as S0.5 or K0.5. When data are hyperbolic, the parameter is a KM. Our use of apparent affinity is intended to collectively refer to both.) for the substrate, phosphoenolpyruvate (PEP), compared to M2PYK (Table 1) [88, 123]. In addition, M1PYK is often used in comparative studies of an enzyme that increases flux through the PYK reaction [48]. However, when considered in the context of all the various types of regulatory mechanisms that can alter substrate affinity, it is better to state that M2PYK has a larger range of substrate affinities that can be accessed via various regulatory mechanisms. In contrast, inhibition by hydrophobic amino acids is the only regulatory mechanism for M1PYK [124] and given the low affinities for these ligands, it is unclear if the collective concentration of all hydrophobic amino acids provides a mechanism for control of M1PYK that is physiologically meaningful. (M1PYK often is reported as not being allosterically regulated.) Unfortunately, in this comparison to M1PYK, the “tunability” of M2PYK’s affinity for PEP seems to have been completely overlooked in the assignment of M2PYK as “inhibited.”
Table 1:
Properties of pyruvate kinase isozymes in the absence of regulators
| M1PYK | LPYK | RPYK | M2PYK (normal tissue) | M2PYK (cancer tissue or cell) | ||
|---|---|---|---|---|---|---|
| In the absence of regulatorsa | ||||||
| Specific Activityb | 230–780 | 60–560 | 60–300 | 130–500 | 770 | |
| KM or S0.5 PEP (mM) | 0.04–0.09 | 0.3–1.0 | 0.5–0.6 | 0.09–0.4c | 0.4 | |
| KM ADP(mM) | 0.3–0.4 | 0.1–0.6 | 0.4–0.6 | 0.2–0.4 | ?? | |
| Full ranges when including outcomes of regulations | ||||||
| Specific Activityb | Unregulated | Unregulated | Unregulated | 34–500f | ?? | |
| KM or S0.5 PEP (mM) | Unregulatedd | 0.02–10e | ?? | 0.078–3.0f | ?? | |
| Allosteric regulations | ||||||
| Allosteric activator (lowest effective conc.) | None | Fru-1,6-BP (0.00006 mM)n | Fru-1,6-BP (0.003mM)o | Fru-1,6-BP (0.00005 mM)l | Fru-1,6-BP (0.00005 mM)h | |
| Serine (0.8mM)m | Serine?? | |||||
| SAICAR (0.1mM)p | SAICAR?? | |||||
| Allosteric inhibitor (lowest effective conc.) | Phenylalanined (5mM) | Alanine (0.5mM)n | Alanine | Phenylalanine (0.4mM)m | Phenylalanine (4.8mM)h | |
| Alanine (0.8mM)m | Alanine?? | |||||
| Cystine (0.05mM)g | ||||||
| Physiological ranges | ||||||
| Normal tissues | Cancer tissues | |||||
| Intracellular pH | 7.0–7.2i | 7.12–7.65 | ||||
| phosphoenolpyruvate | 0.0035–0.397 μmole/g fresh wtj | 1.02 × normalq | ||||
| Fru-1,6-BP | 0.009–0.120j μmole/g fresh wtj,k | 0.99 × normalq | ||||
| Serine | 0.19–1.27 μmole/g fresh wtj | 1.26 × normalq | ||||
| Alanine | 0.38–1.64 μmole/g fresh wtj | 1.01 × normalq | ||||
| Phenylalanine | 7.87 nmol/106 cellsr | 1.82 × normalq | ||||
Data in this section are from [88, 89, 252] and represents a summary of findings from many studies and from multiple mammalian species.
Specific activity is the number of μmoles of pyruvate formed per min per mg of enzyme.
The affinity of M1PYK for the allosteric inhibitor, phenylalanine, is low at pH values below 7.5 [124]. Therefore, under normal physiological conditions, this enzyme may not be regulated and values are the same as in the absence of regulators. Phenylalanine concentration that produces an effect [124, 253].
The highest value is in the presence of alanine and the lowest is in the presence of Fru-1,6-BP, however, these values are dependent on pH (7.5 used as the upper limit for considerations here), monovalent cation type, divalent cation type, and ion concentrations [254, 255].
Although inhibition is shown, estimation of PEP affinity in the presence of Cys was not included. [130]
Like control of M1PYK activity by phenylalanine, the concentration required to inhibit M2PYK is likely above the physiological range. [82]
Data are from [259] and represent a summary of findings from many studies and from many mammalian tissues.
Excludes the very low concentration of 0.0009 (μmole/g fresh wt.) found in adipose.
[116]
[262]
[156]
[263]
Given that the cell is 70% water, μmole/g fresh wt ÷ 0.7 approximates the mM concentration.
In fact, a decrease in the apparent affinity of M2PYK does not in itself imply reduced PYK activity in the cell. A few different scenarios (or combinations of these) could result in the same or increased activity in the cell, despite a reduced substrate affinity for one isozyme. For simplicity in these hypothetical considerations, we will consider a true KM instead of an S0.5. 1) The activity in a cell depends upon the cellular concentration of substrate. If PEP concentrations are sufficiently high relative to KM values for both isozymes, both the M1PYK and M2PYK enzymes could have the same level of catalytic turnover, despite differences in KM (Figure 1). 2) If an isozyme with lower substrate affinity also has a higher Vmax, then the cell could have equal or higher activity compared an enzyme with tighter substrate affinity, but lower Vmax activity (Figure 2). 3) When an enzyme with lower substrate affinity (e.g., M2PYK) is expressed at higher concentrations in the cell, the cell can have equal or higher total activity compared to when an isozyme with higher substrate affinity (e.g., M1PYK) is expressed at a lower concentration. Therefore, an isozyme switch to an enzyme with a reduced substrate affinity does not imply reduced enzyme activity and metabolic flux in the cell (Figure 2). Therefore, the reduced affinity of M2PYK for PEP, when compared to M1PYK does not imply reduced PYK activity in the cell.
Figure 1.
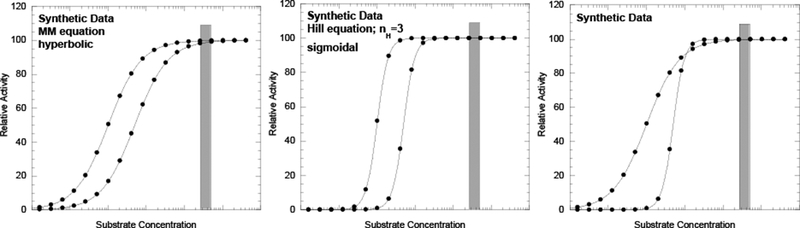
Synthetic data for two isozymes with different KM values as a demonstration that reduced substrate affinity of an isozyme alone is not sufficient to predict activity in the cell. If substrate concentrations are sufficiently high in the cell (indicated by gray column) two isozymes with different affinity for substrate can give rise to the same level of activity. The Michaelis Menten equation was used to generate synthetic data in the left panel here and in Figures 2, and 4. Data in the left panel are hyperbolic (consistent with M1PYK data) on a linear x-axis, but appear sigmoidal in this figure due to the logarithmic scale of the x-axis. Data in the middle panel were derived from the Hill equation (with a nH value equal to 3). The resulting sigmoidal response is more representative of that found for M2PYK. The panel on the right compares a hyperbolic response with a higher substrate affinity (representing M1PYK) to a sigmoidal response with a lower substrate affinity (representing M2PYK). As exemplified in this figure, the ideas discussed in the text can be represented by the simpler comparison of hyperbolic data. Therefore, all other figures in this review include only data generated from the simpler Michaelis Menten equation. Nonetheless, the reader should keep in mind that M2PYK has a sigmoidal response.
Figure 2.
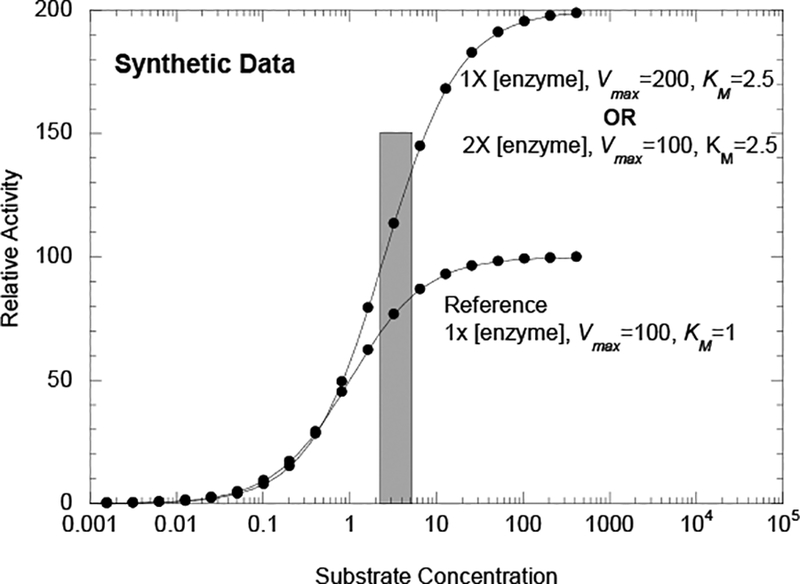
Synthetic Data for two isozymes with altered KM and enzyme increased enzyme concentration and/or increased Vmax activity to demonstrate the possibility that an isozyme with lower substrate affinity can give rise to higher activity even at sub-saturating substrate concentrations, if it also has either higher specific activity or if it is expressed at higher concentrations.
Despite the often-quoted idea that M2PYK is inhibited, the similar kinetic/enzymatic properties of M2PYK, LPYK, and RPYK (in the absence of any other regulatory mechanism) give little reason to consider M2PYK to be “inhibited” [88, 128]. LPYK and M2PYK have similar apparent affinities for PEP and similar specific activity values in the absence of regulatory effectors. Despite the M2PYK vs. LPYK similarities, the LPYK expressed in hepatocytes is displaced by M2PYK expression in heptocarcinomas, much like M2PYK expression is favored in most other cancer tissues. Therefore, pointing to the apparent affinity of M2PYK for PEP (in the absence of effectors) as the primary property that makes M2PYK important for cancer survival seems unwarranted. Furthermore, if comparisons between isozymes can be used to define one isozyme as inhibited or not inhibited and given that LPYK is not commonly referenced as an “inhibited” form, then the LPYK vs. M2PYK comparison would support that M2PYK is also not inhibited.
Many past reviews of the role of M2PYK in cancer biology have focused primarily on the isozyme expression switch that results in M2PYK being expressed in cancer tissues. However, that focus overlooks the fact that M2PYK is expressed in several normal adult tissues (e.g., lung, kidney, many smooth muscles) [75, 100, 104–106]. An isolated focus on the PYK isozyme expression switch also ignores the evidence that the “cancer” form of M2PYK has properties that differ from those of M2PYK expressed in normal adult tissues [82, 128–140]. A search of the primary literature for direct comparisons of the kinetic/enzyme properties of M2PYK isolated from normal vs. from cancer tissue/cells reveals relatively few studies. Nonetheless, those reports that are available indicate that the M2PYK isolated from cancer has higher (NOT lower) affinity for PEP when compared to M2PYK isolated from normal tissue [128, 138, 141]. In total, there is very little evidence to support the often-repeated idea that M2PYK isolated from cancer cells is inhibited.
Focusing on functional outcomes of regulatory mechanism is more important than focusing on tetrameric M2PYK dissociating into dimer/monomer
At the protein level in vitro, the M2PYK subunits exist in an equilibrium between a tetramer and dimers/monomers. The observations of a quaternary structure change in vitro, with knowledge that dissociated dimers of other PYK isozymes lack activity, have led to the acceptance that M2PYK can easily be inhibited. In fact, that idea can be extended as a mechanism of control if the percentage of the protein that exists in each quaternary state can be modulated by the various regulatory mechanisms: post translational modifications (PTMs), protein:protein interactions, or small molecule allosteric effectors [6, 34, 39, 44, 142–151].
It is often assumed that any reference to “inhibition” of M2PYK implies a dimeric state of the enzyme. However, it is important to note that there are many examples of enzymes/binding proteins that are inhibited (with altered substrate affinity or altered kcat) without multimeric dissociation. Therefore, the focus in the literature on a potential M2PYK dimer-tetramer equilibrium may be overemphasized. What is more relevant to metabolism are the maximal enzymatic capacity in the cell (which intrinsically includes enzyme expression level), the specifics of whether the regulatory mechanism alters substrate affinity and/or specific activity of M2PYK and how those two ideas combine to determine the metabolic flux through the PYK reaction in the cell.
In fact, the ability of enzyme expression levels to influence the metabolism flux through the PYK reaction in the cell depends on whether a given regulatory mechanism alters the enzymes KM for substrate or prevents catalytic turnover. To exemplify this concept, consider the potential of M2PYK becoming inhibited in cancer (the common consideration in much of the literature). More specifically, if the “inhibited” form of M2PYK only has reduced substrate affinity, then upon a switch from M1PYK expression to M2PYK in cancer, the reduced affinity would initially cause reduced flux through the PYK reaction and a build-up of up-stream glycolytic intermediates (Figure 3). However, once concentrations of PEP increase sufficiently (due to reduced activity at low concentrations of PEP), that increased substrate availability would drive flux through the pyruvate kinase reaction. Therefore, the new steady-state would include high concentrations of metabolites upstream of the PYK reaction, but would also include flux through the PYK reaction to result in the observed increased lactate production. In this scenario, the concentration of the M2PYK enzyme would also influence the rate of flux through the PYK reaction. In turn, it would be the build-up of glycolytic intermediates that would provide us with the most reasonable explanation for why a lack of a Pasteur effect and a high rate of glycolysis even under aerobic conditions may be beneficial to dividing cells. Due to increased substrate availability (i.e., glucose-6-phosphate), flux through the pentose phosphate pathway would be increased. The pentose phosphate pathway is largely responsible for producing the NADPH needed to regenerate the antioxidant glutathione and permit cancer cells to live with the increased oxidative stress known to exist in cancer. It follows in this logic that the rate of flux through the PYK reaction at the bottom of glycolysis influences NADPH production in the pentose phosphate pathway. This scenario emphasizes a new steady-state condition to explain a high rate of glycolysis and that new steady-state would be associated with an increase in concentrations of glycolytic intermediates, but would also allow lactate production. That new steady-state could include cancer-specific rates of glycogen turnover [152–154]. As noted, overexpression of M2PYK in this scenario could also result in increased glycolytic flux. Importantly, the scenario presented in Figure 3 is used to explain common ideas about Warburg metabolism rather than what is actually supported by data, which is that the activity of PYK is increased rather than decreased in cancer cells [128, 138, 141, 155].
Figure 3.
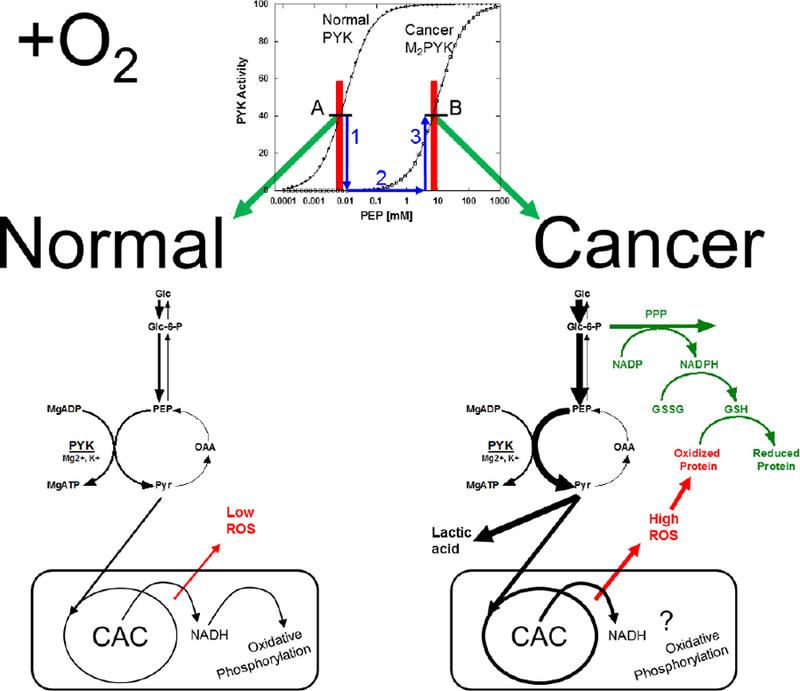
A role for altered affinity of cancer M2PYK for PEP in creating a new steady-state in Warburg metabolism. The top panel represents the activity response curves of either normal PYK or cancer M2PYK. “Normal” PYK can be M1PYK, LPYK, RPYK, or M2PYK isolated from normal tissue. Concentrations of PEP in the cell are represented by red boxes and the respective PYK activity at that PEP concentration is represented by black dashes intersecting the red box (also marked with letters A and B at the beginning and the two steady-state PYK activities) and the response curve. The blue arrows trace changes related to the progression to cancer: 1) The PYK isozyme changes to cancer M2PYK, which has little activity at the PEP concentration present in normal tissue. 2) PEP concentrations build up due to reduced PYK activity. 3) Due to increased substrate availability caused by PEP build up, the cancer form of M2PYK has activity at high PEP. This condition maintains high levels of glycolytic intermediates and allows for high flux to lactic acid production. Green arrows connect the two steady-state PYK activities (A and B) with panels representing the respective metabolism. In these two lower panels, the thickness of arrows is intended to represent the relative concentrations of intermediates in the pathway indicated by the respective arrow. Left) When normal PYK is expressed, normal metabolism is at a steady-state and in the presence of O2, that steady-state flux of carbon from sugar into the mitochondria for oxidative phosphorylation. Right) When cancer M2PYK is expressed, a new steady-state flux is established that includes higher concentrations of glycolytic intermediates. This includes high rates of flux through the cancer PYK reaction and increased production of lactic acid. We anticipate higher flux through the mitochondria as well, thus generating higher levels of reactive oxygen species (ROS) and oxidative stress. However, the increased concentration of glucose 6-phosphate in the new steady-state condition drives increased flux through the pentose phosphate pathway due to substrate availability. This change produces increased NADPH. NADPH is needed to regenerate glutathione, which counteracts the increased oxidative stress, thus allowing cancer cells to live with higher intracellular ROS concentrations.
As a further contrast, a comparison of tumor tissue with non-tumor esophageal tissue failed to identify the increase in glycolytic intermediates [156] that would be anticipated by the scenario in Figure 3. Although it is generally thought that most lactate is derived from glycolysis [109, 157, 158], Figure 3 also does not consider that lactate can be derived from the metabolism of glutamine or serine [158–164]. Therefore, although Figure 3 offers an explanation that circumvents the apparent paradox introduced by M2PYK “inhibition” vs. increased lactic acid production, the entire assumption of the M2PYK inhibition is what should be questioned. Despite the lack of support in cancer metabolism, the description in Figure 3 is not completely useless in that it is consistent with the increased levels of glycolytic intermediates identified for metabolism in other tissues that result in high levels of lactate production (e.g., ischemia in the brain and electroshock-induced seizures in the brain.) [165, 166]. Furthermore, there are indications for variable metabolism used by different types of cancer [167–169], leaving the possibility open that there may be a unique type of cancer that uses the scenario described in Figure 3.
In contrast to a K-type regulation represented in Figure 3, if “inhibition” of M2PYK means that the enzyme lacks catalytic turn over, then a regulatory mechanism that shifts M2PYK towards the inhibited form would reduce flux through the PYK reaction step, even when intermediate metabolite concentrations are increased. If indeed, the M2PYK in cancer lacks or has greatly reduced catalysis (Figure 4), then the only way to gain flux through the PYK reaction is to activate the enzyme. Even higher levels of enzyme expression would have no relevance if that enzyme has a complete lack of activity. If indeed Warburg metabolism includes both increased levels of glycolytic intermediates and increased lactate production as commonly reported, then the dynamic balance between kcat-activated and kcat-inhibited forms of M2PYK would be challenging to explain.
Figure 4.
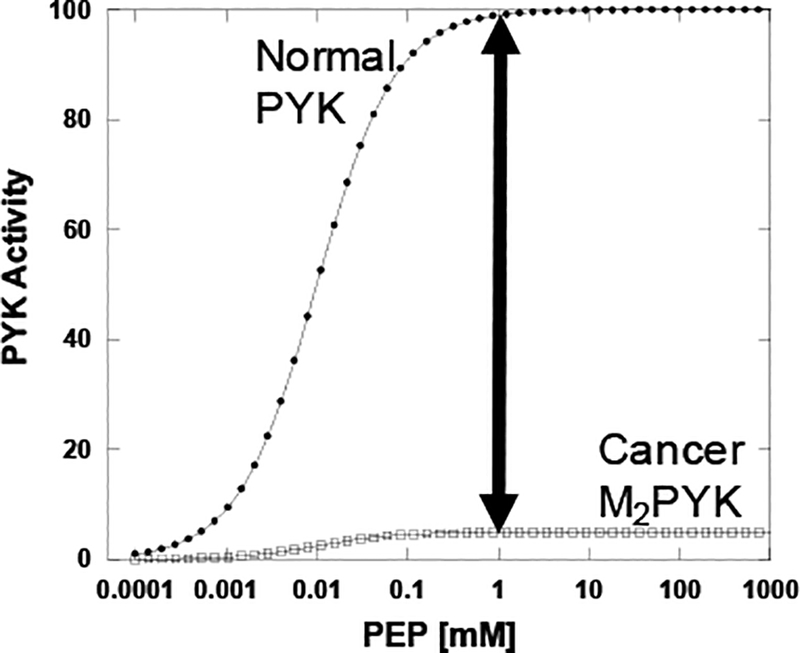
A theoretical response curve of normal PYK and cancer M2PYK, if “inhibited” cancer M2PYK involves reduced kcat. Here 5% activity compared to Normal PYK is used as an example, although the points in the text are more relevant if inhibition removes all catalytic activity. “Normal” PYK can be M1PYK, LPYK, RPYK, or M2PYK isolated from normal tissue. In the absence of changes in M2PYK expression, the only way to increase flux through the PYK reaction would be to activate the M2PYK protein, thus requiring a dynamic control of M2PYK to achieve both inhibition for build-up of glycolytic intermediates and to allow sufficient flux through the PYK reaction to generate high levels of lactic acid. The need for that dynamic regulation might predict oscillating flux of high glycolytic intermediate dispersed with periods of high lactic acid production.
Despite this thought exercise that can provide an explanation for how a reduced substrate affinity for M2PYK in cancer (a commonly repeated theme in the literature) could provide a description of why M2PYK could dictate Warburg metabolism, it is contrary to data that indicates increased PYK activity in cancer [128, 138, 141, 155]. Fructose-1,6-bisphosphate (Fru-1,6-BP) causes an approximately 8-fold increase in the apparent affinity for PEP [148, 170]. This change is also associated with a doubling of the Vmax. This change in catalytic activity does not imply a complete lack of catalysis in the non-activated or inhibited form(s), but does suggest that changes in both specific activity and substrate affinity contribute to activation/inhibition.
M2PYK tetramer-dimer equilibrium may not have physiological relevance
Unfortunately, in the studies of M2PYK, there has been considerably more focus on the tetramer dissociation upon “inhibition” than providing the clear characterizations of the functional/enzymatic outcomes associated with the form of M2PYK found in cancer. Nonetheless knowing what causes the tetramer to dissociation has the potential to indirectly contribute to our knowledge of functional outcomes if the functional properties of the dissociated dimer are well characterized. This conversation can be initiated by evaluating if there is convincing data that the M2PYK dimer retains activity, and even that conversation should be preceded with a cautionary note derived from previous studies of M1PYK.
Many more protein denaturation and refolding studies have been completed with M1PYK than with M2PYK. For the M1PYK protein, denaturation progresses from the stable tetramer to an expanded tetramer, to an inactive dimer before further unfolding steps are observed. The observation of the stable dimer is relevant to our current consideration [171–174], given the focus on the tetramer to dimer transition in studies of M2PYK in cancer. However, indications that the dimeric form of M1PYK is active are thought to be artifacts because PEP stabilizes the tetrameric form of that enzyme [173, 175, 176]. Therefore, even when isolated dimers of the M1PYK protein are added to an assay, the presence of PEP in the assay causes dimers to rapid assemble into active tetramers. Thus, the study of M1PYK exemplifies that an inactive dimeric form of a protein can easily be misinterpreted to have enzymatic activity. The conclusion of studies of M1PYK is that the dimeric form of the enzyme is not catalytically active.
Keeping in mind the lesson learned from studies of M1PYK, it is important to note that both substrates of the enzyme, ADP and PEP, stabilize the M2PYK tetramer [145, 177]. However, given that M2PYK isolated from cancer cells/tissues has properties that are different from M2PYK isolated from normal tissue [82, 128–140], a more specific consideration of M2PYK isolated from cancer may be warranted. Like the M1PYK form, there is a rapid re-association of dimers of the cancer form of M2PYK when PEP is present [149]. For all conditions studied, Hofmann et al. and co-workers found evidence for activity for the dimer, leading them to consider the dimeric form of cancer-M2PYK to be “suboptimal” (without specifying if that implied reduced kcat or reduced substrate affinity) [145, 149, 150]. Assays of the isolated dimer produced activity in the Mazurek laboratory [178], but for the reason identified in the M1PYK example, without evidence that the protein maintains a dimer form in all assay conditions, data collected from that design does not actually support that the dimer maintains activity.
Several studies probe M2PYK structure function questions with mutations, but they do not always define if the mutations cause the protein to behave like the M2PYK from normal tissues vs. from cancer tissues. For example, the G415R mutant dimer fails to tetramerize in the presence of Fru-1,6-BP but shows activity: again, the influence of PEP in the assay condition on tetramerization was not evaluated [179]. In contrast to any indication that the dimer retains activity, a K305Q mutation that prevent tetramer formation is reported to “abolish activity” [170].
More direct data indicate that detection of dimer forms of M2PYK do not correlate with cell proliferation [180]. Finally, the argument has recently been made that cellular concentrations of Fru-1,6-BP, an allosteric activator that promotes both high PEP affinity and stabilization of the tetrameric state of M2PYK, are sufficiently high to predict that unmodified M2PYK (presumably equal to that found in normal tissue) is always a tetramer in the cell [147]. Therefore, despite what is often reported, there is little to no evidence that establishes that the M2PYK dimer does or does not retain activity
Also relevant to this topic, several studies have reported the use of antibodies to detect specific oligomeric states of M2PYK (e.g., [181]). However, antibody binding to a given multimeric form of an antigen protein that is in equilibrium with some other multimeric form will of necessity influence the equilibrium of the subunit assemblies. Even in tissues that have been crosslinked (i.e., samples for histological analysis), tetramer dissociation to dimer may be possible. The idea that antibody binding causes dissociation might best be supported by considering that an antibody that specifically recognizes the monomers of M2PYK promotes dissociation into monomers [51, 144] in the same way that antibodies to dimers identifies dissociation into dimers. Furthermore, antibody staining is not quantitative: if the percentage of M2PYK in the dimer form of M2PYK changes from 0.0001% of the total M2PYK protein to 0.0003%, then stated in isolation, a 3-fold change would sound important. However, the percentage of the total protein that is under consideration is so small as to make the observation seem negligible. For these reasons, antibodies detection of dimeric M2PYK should be used with extreme caution.
Overall, the M2PYK tetramer dissociates in vitro into dimers and monomers much easier than other isozymes of PYK [144, 145, 148, 149, 170]. It follows that it is challenging to prove that M2PYK dimers exist in cells, especially given the influence of various ligands and molecular crowding to promote subunit association. At least in normal tissue, and possibly in cancer tissue, dissociation may not have a physiological relevance due to ligand concentrations [147].
Level of M2PYK protein/mRNA expression and maximal enzyme capacity of the PYK reaction
Up to now, the distinction between responses to regulatory mechanisms has been emphasized. In particular, if response does not completely eliminate catalysis, then total activity levels can influence metabolic flux. It follows that a modification in the level of enzyme expression is one conceivable way to control total PYK activity. The level of M2PYK protein is upregulated in many cancer cell types, which is consistent with upregulation of the mRNA encoding the protein [182–186]. However, that upregulation is often offset by the loss of expression of some other tissue specific isozyme [75, 187]; therefore, protein expression of M2PYK alone may not correlate with an increase in total PYK activity in cancer. Furthermore, expression levels of M2PYK are not likely to correlate with cancer progression for those cancer types that are derived from normal tissues, which already express M2PYK [188, 189]. If a given range of PYK activity is what is needed for cancer metabolism, then it follows that excess PYK activity provided by some other PYK isozyme may also support rapid cell proliferation, which, in turn, may be the bases of several observations that suggest the M2PYK isozyme is not required in all types of cancers [190–195].
Despite that the level of M2PYK contribution to total PYK activity may depend on cell type, the “maximum enzyme capacity” for the PYK activity is increased in cancer cells [128, 138, 141]. Furthermore, increased pyruvate kinase activity (and other glycolytic enzyme activities) has been correlated with the growth rate of tumors/malignancy [141, 155]. This correlation again indicates more PYK activity in rapidly growing tumors, not inhibition of PYK.
Post translational modifications of M2PYK in cancer
Although we can clearly question the representation that M2PYK is “inhibited” and the “evidence” that a tetramer/reduced-activity-dimer equilibrium is central to a regulatory mechanism of M2PYK in vivo, we accept that PYK is a control site in glycolysis [10, 11]. (Although this too has been questioned in the context of cancer via flux analysis [196, 197].) It also remains that forced expression of M1PYK in place of M2PYK limits xenograph tumor growth [15] and activation of M2PYK suppresses tumor growth [26]. Therefore, M2PYK continues to be central in cancer biology and it is worth exploring what properties may be unique in cancer to cause this cancer-central role.
Again, M2PYK in cancer cells is modified compared to M2PYK in normal tissues [82, 128–140]. The early techniques used to identify these differences included isoelectric focusing, making it reasonable that the observed alterations at the protein level could be due to PTMs. Unique substrate affinities for the M2PYK protein purified from normal vs. cancer tissue could also be interpreted as consistent with PTMs that are unique in cancer [128]. As exemplified by the observation that phosphorylation promotes the dimer form of M2PYK [39, 142], modification by PTMs may work in concert with other regulatory mechanisms. Recent literature record a large number of post-translational modifications on M2PYK [198] and many more are included in the various data banks (Table 2).
Table 2:
PTMs identified on M2PYK (or a MPYK isozyme without distinguishing between M1PYK and M2PYK)
| PTM type | Amino Acid Residue Location if known | Ref |
|---|---|---|
| Glycosylation | [1, 2] | |
| O-GlcNAc (Serine/threonine) | 403 and 405 | [3–6] |
| O-methylation (glutamic acid/aspartic acid) | [7] | |
| Oxidation | 358 | [8] |
| Methylation (arginine) | 445, 447 and 455 | [9] |
| MARlation | [10] | |
| N-acetylserine | 2 | Uniprot HPRD |
| N6-acetyllysine | 3, 62, 89, 135, 162, 166, 206, 207, 230,266,305, 433 and 498 | Uniprot HPRD PhosphoSitePlus [13–18] |
| Nitrosylation (cysteine) | 49 | HPRD |
| Nitrosylation (tryptophan) | [19] | |
| Nitrosylation (tyrosine) | [20, 21] | |
| Phosphorylation (serine/threonine) | 37, 41, 45, 50, 57, 60, 77, 80, 87, 93, 95, 97, 100, 127, 129, 172, 195, 202, 222, 243, 249, 287, 328, 437 and 454 | Uniprot HPRD PhosphoSitePlus [22–38] |
| Phosphorylation (tyrosine) | 83, 105, 148, 149, 175, 370, 390 and 466 | Uniprot HPRD PhosphoSitePlus [30, 39–45] |
| Sumolation | [94] | |
| Succinylation (lysine) | 62, 66, 115, 135, 247, 311, and 498 | [95] |
| Ubiquitination (lysine) | 66, 115, 125, 136, 141, 151, 166, 186, 188, 206 224, 261, 277, 311, 322, 336, 367, 475 | PhosphoSitePlus [98, 99] |
| 4-hydroxylation (prolines | 403 and 408 | Uniprot and [70] |
| Prolyl isomerization | [102, 103] |
Unfortunately, the studies of many of these PTMs fall well short of satisfying what, if any, role they play in cancer metabolism. 1) The influence on the metabolic activity of M2PYK or their influence on cancer biology has not been detailed for most PTMs that have been identified. There are, however, some exceptions with more information available [9, 15, 23, 34, 35, 39, 44, 45, 98]. 2) Most, if not all, of these PTMs were identified from cancer cell lines. Because only a small fraction of cells from a cancer tumor are capable of being developed into a cell line, it remains unknown if regulatory mechanisms for M2PYK identified in cell lines represent those that are the most abundant in tumors. 3) Many of these PTMs were originally identified on fragments of the M2PYK protein as detected by mass spectrometry. This identification approach does not distinguish if the PTMs can be added to the whole protein or if they are added to peptide fragments that result from protein degradation. Due to the high sequence similarity, this approach also fails to distinguish if the modification was present on only M2PYK, only M1PYK, or both. If modifications can be added to both isozymes, then a stream of questions follow (e.g., Can the PTM be added to both M1PYK and M2PYK? Does the PTM regulate the two enzymes the same way? etc.). 4) Finally, the mass spectrometry techniques used to identify modifications on peptide fragments are very sensitive, leaving the possibility that modifications are only added to a negligibly small fraction of the total M2PYK present in the cell. If only a few molecules of M2PYK include a given PTM, it would be hard to imagine a regulatory role for that PTM. Even though the field seldom acknowledges that many of the identified PTMs may not have physiological roles, it remains plausible that PTMs may contribute to the unique properties of M2PYK found in tumors.
Allosteric Regulation of M2PYK by small molecule effectors
It is well established that the response of M2PYK to allosteric effectors differs from that of other mammalian PYK isozymes [88, 89, 101, 106, 136, 138, 139, 199–202]. This includes the allosteric activation by Fru-1,6-BP that results in an increased affinity of M2PYK for PEP, and alanine and phenylalanine, both allosteric inhibitors that decrease affinity of the enzyme for PEP. Although these responses are qualitatively shared with LPYK, the concentrations and the resulting substrate affinities with effector bound are unique for the two enzymes.
Yuan et al. recently extended the characterizations of amino acid regulators of M2PYK [143]. They found that in addition to phenylalanine and alanine, tryptophan, methionine, valine, and proline acted as allosteric inhibitors. The ability of hydrophobic amino acids to act as inhibitors parallels the response from M1PYK [124]. However, these inhibitory effects on M2PYK appear to be within concentration ranges that are more relevant to physiological conditions. Consistent with earlier studies [148, 170], inhibitions by amino acids were reported to include change in both Vmax and substrate affinity for PEP. Histidine and serine acted as activators in the Yuan et al. study. The activation by serine only increased Vmax without influencing substrate affinity. Importantly, these influences of allosteric amino acid effectors were measured with an M2PYK protein expressed in E. coli that would lack any cancer-specific PTMs.
Regulation by L-cysteine and L-serine can be further highlighted. An inhibition by L-cysteine [139, 140, 203] appears to be unique to the M2PYK purified out of cancer cells [128]. L-cysteine reduces both Vmax and the apparent affinity for PEP. [138]. L-serine has been known to be an activator of M2PYK for some time [135, 148, 204, 205], with some evidence for a differential response between the normal and cancer enzymes from chicken.
ATP is often listed as a common allosteric inhibitor of mammalian PYK isozymes. However, without a binding site identified via co-crystallography, it has been challenging to establish whether the inhibition is allosteric or via competitive binding with ADP in the active site. Nonetheless, the cancer form of M2PYK has been reported to lack sensitivity to ATP, in contrast to the ATP inhibition that is observed for M2PYK isolated from normal tissue [206].
Succinyl-5-amino imidazole-4-carboxamide-1-ribose-5’-phosphate (SAICAR), an intermediate of the de novo purine nucleotide biosynthesis pathway activates M2PYK, but not M1PYK [207]. SAICAR concentrations are reported higher in cancer cells than normal cells that express M2PYK (i.e., lung fibroblasts). It remains undefined if SAICAR causes a change in specific activity or modifies substrate affinity.
M2PYK metabolic functions regulated by protein:protein interactions
A number of proteins that interact directly with the M2PYK protein have been identified in recent years (Table 3). In those studies, it is not always clear if the proposed protein:protein interactions are thought to alter metabolic functions via enzyme activity in the cytosol or via nuclear functions (see below). However, some studies have directly tested the influence of protein:protein interactions on the metabolic activity of M2PYK. Interactions of M2PYK with tyrosine-phosphorylated peptides, SOCS3, PRL, JMJD8 causes reduced PYK activity [74, 110, 117, 119]. In contrast, interaction of M2PYK with DAPk results in increased PYK activity [11]. Although thought to occur in the cytoplasm, the influence of a direct interaction with PanK4 and TRIM35 on the activity of M2PYK was not determined [108, 126]. Also, whether altered activities of M2PYK are a result of changes in specific activity or substrate affinity have not typically been determined for these protein interactions.
Table 3:
Proteins reported to interact directly with M2PYK
| DAPk | [11] |
| DDB2 | [12] |
| HERC1 | [69] |
| HIF-1α | [70] |
| HPV-16 E7 | [71] |
| HSP40 | [72] |
| HSP90 | [24] |
| JMJD5 | [73] |
| JMJD8 | [74] |
| MDM2 | [85] |
| MST1 | [86] |
| Oct-4 | [107] |
| PanK4 | [108] |
| PHD3 | [70, 109] |
| Phosphotyrosine peptide | [110, 111] |
| PML | [116] |
| PRLr | [117] |
| p53 | [118] |
| SOCS3 | [119] |
| SREBP | [120–122] |
| SUMO-E3 Ligase PIAS3 | [94] |
| TGIF2 | [125] |
| TRIM35 | [126] |
| TSC22DS | [127] |
Much like PTMs, quantitation is lacking in these protein:protein interaction studies. This begs the question of whether there is a sufficient percentage of M2PYK in the protein:protein interaction to warrant consideration of that interaction as a regulatory mechanism. It should also be kept in mind that the percentage of each interacting protein that participates in a protein:protein interaction is dependent on the concentrations of both proteins. Therefore, there is a note of caution for all reports of proteins that directly interact with M2PYK when interactions are detected in systems that overexpress M2PYK or the other protein, thus artificially driving protein:protin interactions that may not occur at a meaningful level under normal expression levels. Nonetheless, given the examples of protein:protein interactions that can alter M2PYK metabolic activity, protein:protein interactions may work in concert with PTMs and/or small molecule allosteric ligands to control M2PYK activity.
The role of the affinity of M2PYK for ADP
Control of metabolic flux via M2PYK may not depend solely on changes in the enzyme’s affinity for PEP. Removal of competition between PYK and mitochondrial oxidative phosphorylation for ADP has been offered as an explanation for why tumor cell respiration can be stimulated by inhibitors of PYK [46, 47].
This idea would require M2PYK to have a higher affinity for ADP than the mitochondrial electrogenic adenine nucleotide translocator (ANT). In non-proliferating cells, the apparent affinity for ADP reported for PYK isozymes are much lower (200–400 μM) [88, 208] than values reported for ANT in non-proliferating cells (1 to 100 μM) [209]. In non-proliferating cells respiration creates a membrane potential and pH gradient across the mitochondrial inner membrane that drives release of ATP in exchange for cytosolic ADP via the electrogenic ANT located in the mitochondrial inner membrane. In turn, this leads to a high cytosolic ATP/ADP ratio and a free ADP concentration (in the range of 25 μM). That ADP range is considerably below the KM of PYK, a scenario that restricts PYK flux. Therefore, inhibition of PYK in non-proliferating cells has no effect on the respiration rate.
The conditions are much different in rapidly proliferating cancer cells. In these cells, ATP/ADP exchange is not driven by the electrogenic ANT [210], due inhibition of the mitochondrial outer membrane voltage-dependent anion channel (VDAC) [211]. This results in a low cytosolic ATP/ADP ratio and therefore a higher free cytosolic ADP concentration that promotes PYK activity, aerobic glycolysis, and the Warburg effect. Since PYK is a major user of ADP in proliferating cells, inhibition of its activity greatly impacts the free ADP concentration which overcomes inhibition of the ANT and thereby stimulates respiration. These details are consistent with tumor cell respiration being stimulated by inhibitors of PYK.
The influence of M2PYK on glycolytic flux in vivo
Throughout the discussion of M2PYK as a glycolytic enzyme, it becomes clear that many of the measurements that are easily obtained (maximal enzyme activity in a tissue, enzyme concentrations, mRNA concentrations, responses to substrates, responses to effectors and metabolite concentrations) do not easily translate into knowledge of flux through a pathway in vivo. This concept is the bases of flux analysis [212–214], which also includes increasing concentrations of other enzymes in a pathway [215]. Within flux analysis, it is recognized that a change in enzyme activity will both reduce substrate concentration and increase product for that enzymatic step. However, those changes in substrate and product concentrations will alter activity of other enzymatic steps. Thus, the ability of each enzyme to control flux is measured as a “control strength” value instead of attempting to assign a single enzyme as a “rate limiting step” in the pathway. Surprisingly, there are few flux analysis evaluations of the metabolism found in cancer cells and little evidence that M2PYK is a control point in the flux analysis studies that have been completed [196, 197].
Consistent with this lack of evidence for M2PYK as a control point and inconsistent with the highly popular speculation for inhibition of M2PYK in cancer, there is little evidence for increased concentrations of glycolytic intermediates in cancer [165, 216]. Even in a theoretical consideration, a potential increase in glycolytic intermediates, all of which are phosphorylated, must be limited to prevent depletion of inorganic phosphate: fructose intolerance is caused by phosphate depletion due to the accumulation of fructose 1-phosphate. Of special note is that most flux analysis via isotope labeling [217] can identify increased labeling of glycolytic intermediates without evaluating for increases in the concentrations of those glycolytic intermediates.
M2PYK in non-canonical role: nuclear translocation of M2PYK to act as a protein kinase
Given the many questions that have often challenged a metabolic role for M2PYK in cancer, it is not surprising that there have been many attempts to identify some function for M2PYK in cancer that is outside of the traditional metabolic function. These proposed non-canonical/non-metabolic/ moonlighting functions reported for M2PYK parallel proposals for a non-canonical functions for a large number of other enzymes [218].
Both a PEP-dependent protein kinase activity of M2-PYK [24, 134, 151, 207, 219–229] and a translocation of dimeric M2PYK into the nucleus to act as a transcription factor [34, 73, 120, 125, 230–235] have been proposed for M2PYK. The protein kinase activity appears to primarily be detected in the nucleus using nuclear proteins as substrates. However, it is less defined if all roles prescribed to the nuclear M2PYK dimer rely on that proposed protein kinase activity. The nuclear functions of a M2PYK dimer have been proposed to regulate gene transcription of both glycolytic functions and other cancer-specific properties.
A few considerations introduce a strong caution for the proposed PEP-dependent protein kinase activity. Presumably, the substrate peptide must bind in the nucleotide portion of the active site. However, that nucleotide binding site is similar in all pyruvate kinase isozymes, begging the question of why the other mammalian isozymes do not also have a protein kinase activity. (A protein kinase activity is also reported for yeast PYK [236].) A second caution is derived from the PYK control included in efforts to identify a protein kinase that uses PEP as a phosphate donor [237, 238]. In those studies, even in a dialyzed protein extract, there is a probability that assays include contaminating concentrations of both PYK and ADP, resulting in a transfer of a phosphate from PEP to ADP (via the PYK reaction) and, in turn, ATP being the true phosphate donor. It follows for studies of M2PYK as a protein kinase that even undetectable contamination levels of ADP and an ATP-dependent protein kinase could result in substrate protein phosphorylation and be interpreted as a PEP-dependent protein kinase activity for PYK. This becomes increasingly relevant given that cellular concentrations of ADP are largely bound to proteins [208], hence tight binding and a high possibility of proteinaceous extracts including contamination by ADP. It has also been shown that “enzyme cycling” systems, such as the one possible with PYK, ADP and a protein kinase, amplifies signal [239], an outcome that may be relevant if the protein kinase activity for PYK is an artifact due to contamination. Therefore, absolute proof of a protein kinase activity for M2PYK is challenging. Co-crystallization of a substrate peptide in the active site of M2PYK would greatly strengthen an argument for M2PYK acting as a protein kinase. Despite these considerations, there continues to be large numbers of studies reporting detection of protein kinase activity for M2PYK. However, there has also been a challenge that these protein kinase detections are not reproducible [240].
M2PYK in non-canonical roles: M2PYK translocation into the mitochondria and circulating in the blood
Several non-canonical functions have been proposed that do not have obvious dependencies on nuclear translocation. It has been proposed that M2PYK alters mitochondrial fusion via a protein:protein interaction with p53 [85, 241]. Translocation of M2PYK into the mitochondria, followed by a protein-protein interaction with and phosphorylation of Bcl2 to regulate apoptosis has also been proposed [229].
M2PYK circulating in the blood may also have a role in cancer [242, 243]. M2PYK in blood promotes angiogenesis in tumors. Much like other non-canonical functions, this activity is reported to depend on a dimer form of the enzyme. Interestingly, the caveats regarding cellular concentrations of metabolites that were used to question the dissociation of tetramer in the cytoplasm are not likely to apply to dissociation of the M2PYK tetramer in circulating blood or in other cellular compartments beyond the cytoplasm. Rather than or in addition to circulating M2PYK, M2PYK released by neutrophils at a wound site may facilitate angiogenesis [244].
Studies of M2PYK are plagued with a lack of quantitative studies
All non-canonical functions of M2PYK and many proposed PTM and protein:protein interactions can be collectively questioned by the observation that M2PYK is not required in all cancer types [190–195]. The recent report that M1PYK can promote tumor cell growth [195] may offer an even more specific challenge to these proposed non-metabolic/non-canonical roles for M2PYK in cancer (as well as many of the identified regulatory mechanisms).
A second reason that all of the various PTM, protein:protein interactions and non-canonical functions of M2PYK can collectively be questioned is the lack of quantitation and good techniques to probe only regulatory features of the M2PYK protein. Most of these non-canonical functions are heavily influenced by antibody/western blot detection techniques, which will give a result when combined with sensitive chemiluminescent detection (even if they are non-meaningful physiologically). Observations from those non-quantitative antibody-based approaches are often paired with knock-out, knock-down and overexpression strategies, which seldom neatly probe regulatory features, a third reason to question many reported observations. To fully appreciate why changing expression levels is not a good probe of function, consider the role of M2PYK in controlling glycolytic flux: Would knock-out, knock-down, or overexpression of the enzyme identify if a protein phosphorylation subtly alters glycolytic flux? The answer no because the entire glycolytic pathway would be perturbed and alterations in many downstream metabolite would be expected to act as further signals for change. It follows that knockout, knock-down and overexpression strategies may not be good tools to evaluate the potential roles of non-canonical roles of M2PYK. Cell culture and in vivo studies that include M2PYK proteins with point mutations that mimic or prevent individual types of the regulatory mechanisms starts to address the need for better probing approaches. However, even that design is challenged: mutant M2PYK introduced in vivo might mimic chronic changes in enzyme function, but they don’t recapitulate the reversible acute responses that would be possible with PTMs, allostery, and protein:protein interactions in vivo. Therefore, inducible regulatory mechanisms may have an important role in the future [245].
What can we speculate about Warburg metabolism based on primary literature
The Warburg effect is the incomplete switch from glycolytic production of lactic acid to complete glucose oxidation when O2 is present. In other words, the Warburg effect is an incomplete Pasteur Effect, or perhaps even better characterized as rapid glycolysis on top of oxidative phosphorylation [246]. Rather than increasing the concentrations of glycolytic intermediates to force flux (by mass action) into side pathways that are important for cell proliferation and growth, we visualize that increased expression of enzymes at the branch points of these side pathways, namely glucose-6-phosphate dehydrogenase [247] and 3-phosphoglycerate dehydrogenase [248], readily explains increased synthesis of NADPH, ribose 5-phosphate, serine, and glycine. Increased expression of all of the glycolytic enzymes [249], including M2PYK [216], assures that generation of intermediates of the glycolytic pathway is not rate-limiting for glycolysis as well as the side reactions. We believe the Warburg effect as it relates to glucose metabolism is due to upregulation of the enzymes of the glycolytic pathway coupled with increased expression of pyruvate dehydrogenase kinases [250] to partially inhibit the pyruvate dehydrogenase complex. In turn, this partial inhibition of the pyruvate dehydrogenase complex restricts the levels of pyruvate (a result of increased glycolytic flux) that can enter the citric acid cycle and, instead, conserves sufficient quantities of pyruvate for the recycling of NADH produced by glycolysis back to NAD+ (although citric acid cycle continues to function [217, 251]). The Warburg effect further depends upon reduced mitochondrial uptake of ADP due to suppression of the activity of the mitochondrial ANT [210] which decreases the cytosolic ATP/ADP ratio and sets a higher steady state concentration of free ADP required for M2PYK activity as discussed in greater detail above.
An explanation for outcomes in studies with M2PYK
Unfortunately, the summary of M2PYK is that there are so many functions and regulatory mechanisms reported for M2PYK (beyond catalysis in glycolysis), that it is easy to question if any of these observations have valid physiological relevance. In particular, the finding that M2PYK is not required for all cancers [190–195] seems to refocus studies on the role of M2PYK back to its metabolic function.
We propose that the many types of potential regulatory mechanisms of M2PYK allows a fine tuning of activity to a level that supports the metabolic flux levels needed for cancer survival. Therefore, drug activation of M2PYK [49–56, 61–64] or expression of M1PYK [48] that lacks sensitivity to regulatory mechanisms may introduce too much PYK activity, an outcome that would prevent some glucose-6-phosphate from entering the pentose phosphate pathway to produce the required NADPH. This scenario would be toxic to the cell. As a contrast, overexpression of M2PYK can be accommodated because of cellular mechanisms that reduce activity to the cancer-specific activity range. On the other hand, any knock-down, knock-out, or drug designs that inhibit M2PYK activity [59, 60, 63, 65–68] has the potential to reduce the total PYK activity below a threshold needed for cell survival.
In Warburg metabolism, increased flux through glycolysis results from additional PYK activity, not reduced PYK activity. Although we can conceive how that increased flux could be accompanied with increased concentrations of glycolytic intermediates when there is a switch from expression of M1PYK to M2PYK (Figure 3), we anticipate that many cancer types do not have this phenotypic change (as compared to comparable normal tissue) [156].
Acknowledgments
Research in the Fenton laboratory is supported by NIH grant GM115340 and a pilot grant from the KUMC PKD Center (DK106912) and the KU Cancer Center and a pilot award from the KUMC Alzheimer’s Disease Center (AG035982).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Gbormittah FO, Haab BB, Partyka K, Garcia-Ott C, Hancapie M, Hancock WS, Characterization of glycoproteins in pancreatic cyst fluid using a high-performance multiple lectin affinity chromatography platform, J Proteome Res, 13 (2014) 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].O’Connell K, Doran P, Gannon J, Ohlendieck K, Lectin-based proteomic profiling of aged skeletal muscle: decreased pyruvate kinase isozyme M1 exhibits drastically increased levels of N-glycosylation, Eur J Cell Biol, 87 (2008) 793–805. [DOI] [PubMed] [Google Scholar]
- [3].Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW, Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications, Mol Cell Proteomics, 1 (2002) 791–804. [DOI] [PubMed] [Google Scholar]
- [4].Champattanachai V, Netsirisawan P, Chaiyawat P, Phueaouan T, Charoenwattanasatien R, Chokchaichamnankit D, Punyarit P, Srisomsap C, Svasti J, Proteomic analysis and abrogated expression of O-GlcNAcylated proteins associated with primary breast cancer, Proteomics, 13 (2013) 2088–2099. [DOI] [PubMed] [Google Scholar]
- [5].Chaiyawat P, Chokchaichamnankit D, Lirdprapamongkol K, Srisomsap C, Svasti J, Champattanachai V, Alteration of O-GlcNAcylation affects serine phosphorylation and regulates gene expression and activity of pyruvate kinase M2 in colorectal cancer cells, Oncol Rep, 34 (2015) 1933–1942. [DOI] [PubMed] [Google Scholar]
- [6].Wang Y, Liu J, Jin X, Zhang D, Li D, Hao F, Feng Y, Gu S, Meng F, Tian M, Zheng Y, Xin L, Zhang X, Han X, Aravind L, Wei M, O-GlcNAcylation destabilizes the active tetrameric PKM2 to promote the Warburg effect, Proc Natl Acad Sci U S A, 114 (2017) 13732–13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhou W, Capello M, Fredolini C, Racanicchi L, Dugnani E, Piemonti L, Liotta LA, Novel li F, Petricoin EF, Mass spectrometric analysis reveals O-methylation of pyruvate kinase from pancreatic cancer cells, Anal Bioanal Chem, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, Thomas CJ, Vander Heiden MG, Cantley LC, Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses, Science, 334 (2011) 1278–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Liu F, Ma F, Wang Y, Hao L, Zeng H, Jia C, Wang Y, Liu P, Ong IM, Li B, Chen G, Jiang J, Gong S, Li L, Xu W, PKM2 methylation by CARM1 activates aerobic glycolysis to promote tumorigenesis, Nat Cell Biol, 19 (2017) 1358–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Carter-O’Connell I, Jin H, Morgan RK, Zaja R, David LL, Ahel I, Cohen MS, Identifying Family-Member-Specific Targets of Mono-ARTDs by Using a Chemical Genetics Approach, Cell Rep, 14 (2016) 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mor I, Carlessi R, Ast T, Feinstein E, Kimchi A, Death-associated protein kinase increases glycolytic rate through binding and activation of pyruvate kinase, Oncogene, 31 (2012) 683–693. [DOI] [PubMed] [Google Scholar]
- [12].Xie X, Wang M, Mei J, Hu F, Ding F, Lv L, Pyruvate kinase M2 interacts with DNA damage-binding protein 2 and reduces cell survival upon UV irradiation, Biochem Biophys Res Commun, 467 (2015) 427–433. [DOI] [PubMed] [Google Scholar]
- [13].Xiong Y, Lei QY, Zhao S, Guan KL, Regulation of glycolysis and gluconeogenesis by acetylation of PKM and PEPCK, Cold Spring Harb Symp Quant Biol, 76 (2011) 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Marimuthu S, Chivukula RS, Alfonso LF, Moridani M, Hagen FK, Bhat GJ, Aspirin acetylates multiple cellular proteins in HCT-116 colon cancer cells: Identification of novel targets, Int J Oncol, 39 (2011) 1273–1283. [DOI] [PubMed] [Google Scholar]
- [15].Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H, Zha Z, Liu Y, Li Z, Xu Y, Wang G, Huang Y, Xiong Y, Guan KL, Lei QY, Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth, Mol Cell, 42 (2011) 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL, Regulation of cellular metabolism by protein lysine acetylation, Science, 327 (2010) 1000–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M, Lysine acetylation targets protein complexes and co-regulates major cellular functions, Science, 325 (2009) 834–840. [DOI] [PubMed] [Google Scholar]
- [18].Lv L, Xu YP, Zhao D, Li FL, Wang W, Sasaki N, Jiang Y, Zhou X, Li TT, Guan KL, Lei QY, Xiong Y, Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization, Mol Cell, 52 (2013) 340–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kawasaki H, Ikeda K, Shigenaga A, Baba T, Takamori K, Ogawa H, Yamakura F, Mass spectrometric identification of tryptophan nitration sites on proteins in peroxynitrite-treated lysates from PC12 cells, Free Radic Biol Med, 50 (2011) 419–427. [DOI] [PubMed] [Google Scholar]
- [20].Kuo WN, Kreahling JM, Shanbhag VP, Shanbhag PP, Mewar M, Protein nitration, Mol Cell Biochem, 214 (2000) 121–129. [DOI] [PubMed] [Google Scholar]
- [21].Kanski J, Hong SJ, Schoneich C, Proteomic analysis of protein nitration in aging skeletal muscle and identification of nitrotyrosine-containing sequences in vivo by nanoelectrospray ionization tandem mass spectrometry, J Biol Chem, 280 (2005) 24261–24266. [DOI] [PubMed] [Google Scholar]
- [22].Koo N, Kim KM, Distinct effects on M2-type pyruvate kinase are involved in the dimethylsulfoxide-induced modulation of cellular proliferation and degranulation of mast cells, Arch Pharm Res, 32 (2009) 1637–1642. [DOI] [PubMed] [Google Scholar]
- [23].Kumar B, Bamezai RN, Moderate DNA damage promotes metabolic flux into PPP via PKM2 Y-105 phosphorylation: a feature that favours cancer cells, Mol Biol Rep, 42 (2015) 1317–1321. [DOI] [PubMed] [Google Scholar]
- [24].Xu Q, Tu J, Dou C, Zhang J, Yang L, Liu X, Lei K, Liu Z, Wang Y, Li L, Bao H, Wang J, Tu K, HSP90 promotes cell glycolysis, proliferation and inhibits apoptosis by regulating PKM2 abundance via Thr-328 phosphorylation in hepatocellular carcinoma, Mol Cancer, 16 (2017) 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M, Global, in vivo, and site-specific phosphorylation dynamics in signaling networks, Cell, 127 (2006) 635–648. [DOI] [PubMed] [Google Scholar]
- [26].Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP, A probability-based approach for high-throughput protein phosphorylation analysis and site localization, Nat Biotechnol, 24 (2006) 1285–1292. [DOI] [PubMed] [Google Scholar]
- [27].Zahedi RP, Lewandrowski U, Wiesner J, Wortelkamp S, Moebius J, Schutz C, Walter U, Gambaryan S, Sickmann A, Phosphoproteome of resting human platelets, J Proteome Res, 7 (2008) 526–534. [DOI] [PubMed] [Google Scholar]
- [28].Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M, Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle, Mol Cell, 31 (2008) 438–448. [DOI] [PubMed] [Google Scholar]
- [29].Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP, A quantitative atlas of mitotic phosphorylation, Proc Natl Acad Sci U S A, 105 (2008) 10762–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, Rodionov V, Han DK, Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions, Sci Signal, 2 (2009) ra46. [DOI] [PubMed] [Google Scholar]
- [31].Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M, Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis, Sci Signal, 3 (2010) ra3. [DOI] [PubMed] [Google Scholar]
- [32].Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B, System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation, Sci Signal, 4 (2011) rs3. [DOI] [PubMed] [Google Scholar]
- [33].Siwko S, Mochly-Rosen D, Use of a novel method to find substrates of protein kinase C delta identifies M2 pyruvate kinase, Int J Biochem Cell Biol, 39 (2007) 978–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, Lu Z, ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect, Nat Cell Biol, 14 (2012) 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yu Z, Zhao X, Huang L, Zhang T, Yang F, Xie L, Song S, Miao P, Zhao L, Sun X, Liu J, Huang G, Proviral insertion in murine lymphomas 2 (PIM2) oncogene phosphorylates pyruvate kinase M2 (PKM2) and promotes glycolysis in cancer cells, J Biol Chem, 288 (2013) 35406–35416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liang J, Cao R, Zhang Y, Xia Y, Zheng Y, Li X, Wang L, Yang W, Lu Z, PKM2 dephosphorylation by Cdc25A promotes the Warburg effect and tumorigenesis, Nat Commun, 7 (2016) 12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lee KM, Nam K, Oh S, Lim J, Lee T, Shin I, ECM1 promotes the Warburg effect through EGF-mediated activation of PKM2, Cell Signal, 27 (2015) 228–235. [DOI] [PubMed] [Google Scholar]
- [38].Park YS, Kim DJ, Koo H, Jang SH, You YM, Cho JH, Yang SJ, Yu ES, Jung Y, Lee DC, Kim JA, Park ZY, Park KC, Yeom YI, AKT-induced PKM2 phosphorylation signals for IGF-1-stimulated cancer cell growth, Oncotarget, 7 (2016) 48155–48167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, Xie J, Gu TL, Polakiewicz RD, Roesel JL, Boggon TJ, Khuri FR, Gilliland DG, Cantley LC, Kaufman J, Chen J, Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth, Sci Signal, 2 (2009) ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, Thomas CJ, Vander Heiden MG, Cantley LC, Inhibition of Pyruvate Kinase M2 by Reactive Oxygen Species Contributes to Antioxidant Responses, Science, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Presek P, Reinacher M, Eigenbrodt E, Pyruvate kinase type M2 is phosphorylated at tyrosine residues in cells transformed by Rous sarcoma virus, FEBS Lett, 242 (1988) 194–198. [DOI] [PubMed] [Google Scholar]
- [42].Ballif BA, Carey GR, Sunyaev SR, Gygi SP, Large-scale identification and evolution indexing of tyrosine phosphorylation sites from murine brain, J Proteome Res, 7 (2008) 311–318. [DOI] [PubMed] [Google Scholar]
- [43].Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ, Immunoaffinity profiling of tyrosine phosphorylation in cancer cells, Nat Biotechnol, 23 (2005) 94–101. [DOI] [PubMed] [Google Scholar]
- [44].Kachel P, Trojanowicz B, Sekulla C, Prenzel H, Dralle H, Hoang-Vu C, Phosphorylation of pyruvate kinase M2 and lactate dehydrogenase A by fibroblast growth factor receptor 1 in benign and malignant thyroid tissue, BMC Cancer, 15 (2015) 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bettaieb A, Bakke J, Nagata N, Matsuo K, Xi Y, Liu S, AbouBechara D, Melhem R, Stanhope K, Cummings B, Graham J, Bremer A, Zhang S, Lyssiotis CA, Zhang ZY, Cantley LC, Havel PJ, Haj FG, Protein tyrosine phosphatase 1B regulates pyruvate kinase M2 tyrosine phosphorylation, J Biol Chem, 288 (2013) 17360–17371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gosalvez M, Lopez-Alarcon L, Garcia-Suarez S, Montalvo A, Weinhouse S, Stimulation of tumor-cell respiration by inhibitors of pyruvate kinase, Eur J Biochem, 55 (1975) 315–321. [DOI] [PubMed] [Google Scholar]
- [47].Gosalvez M, Perez-Garcia J, Weinhouse S, Competition for ADP between pyruvate kinase and mitochondrial oxidative phosphorylation as a control mechanism in glycolysis, Eur J Biochem, 46 (1974) 133–140. [DOI] [PubMed] [Google Scholar]
- [48].Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC, The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth, Nature, 452 (2008) 230–233. [DOI] [PubMed] [Google Scholar]
- [49].Chen C, Wang T, Wu F, Huang W, He G, Ouyang L, Xiang M, Peng C, Jiang Q, Combining structure-based pharmacophore modeling, virtual screening, and in silico ADMET analysis to discover novel tetrahydro-quinoline based pyruvate kinase isozyme M2 activators with antitumor activity, Drug Des Devel Ther, 8 (2014) 1195–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Parnell KM, Foulks JM, Nix RN, Clifford A, Bullough J, Luo B, Senina A, Vollmer D, Liu J, McCarthy V, Xu Y, Saunders M, Liu XH, Pearce S, Wright K, O’Reilly M, McCullar MV, Ho KK, Kanner SB, Pharmacologic activation of PKM2 slows lung tumor xenograft growth, Mol Cancer Ther, 12 (2013) 1453–1460. [DOI] [PubMed] [Google Scholar]
- [51].Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, Yang H, Mattaini KR, Metallo CM, Fiske BP, Courtney KD, Malstrom S, Khan TM, Kung C, Skoumbourdis AP, Veith H, Southall N, Walsh MJ, Brimacombe KR, Leister W, Lunt SY, Johnson ZR, Yen KE, Kunii K, Davidson SM, Christofk HR, Austin CP, Inglese J, Harris MH, Asara JM, Stephanopoulos G, Salituro FG, Jin S, Dang L, Auld DS, Park HW, Cantley LC, Thomas CJ, Vander Heiden MG, Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis, Nature chemical biology, 8 (2012) 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Auld D, Shen M, Skoumbourdis AP, Jiang J, Boxer M, Southall N, Inglese J, Thomas C, Identification of activators for the M2 isoform of human pyruvate kinase, Probe Reports from the NIH Molecular Libraries Program (2010). [PubMed] [Google Scholar]
- [53].Boxer MB, Jiang JK, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, Veith H, Leister W, Austin CP, Park HW, Inglese J, Cantley LC, Auld DS, Thomas CJ, Evaluation of substituted N,N’-diarylsulfonamides as activators of the tumor cell specific M2 isoform of pyruvate kinase, J Med Chem, 53 (2010) 1048–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Jiang JK, Boxer MB, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, Veith H, Leister W, Austin CP, Park HW, Inglese J, Cantley LC, Auld DS, Thomas CJ, Evaluation of thieno[3,2-b]pyrrole[3,2-d]pyridazinones as activators of the tumor cell specific M2 isoform of pyruvate kinase, Bioorg Med Chem Lett, 20 (2010) 3387–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Walsh MJ, Brimacombe KR, Anastasiou D, Yu Y, Israelsen WJ, Hong BS, Tempel W, Dimov S, Veith H, Yang H, Kung C, Yen KE, Dang L, Salituro F, Auld DS, Park HW, Vander Heiden MG, Thomas CJ, Shen M, Boxer MB, ML265: A potent PKM2 activator induces tetramerization and reduces tumor formation and size in a mouse xenograft model, (2010). [PubMed]
- [56].Boxer MB, Jiang J, Vander Heiden MG, Shen M, Veith H, Cantley LC, Thomas CJ, Identification of activators for the M2 isoform of human pyruvate kinase Version 3, (2010). [PubMed]
- [57].Li W, Liu J, Zhao Y, PKM2 inhibitor shikonin suppresses TPA-induced mitochondrial malfunction and proliferation of skin epidermal JB6 cells, Mol Carcinog, 53 (2014) 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chen J, Jiang Z, Wang B, Wang Y, Hu X, Vitamin K(3) and K(5) are inhibitors of tumor pyruvate kinase M2, Cancer Lett, (2011). [DOI] [PubMed] [Google Scholar]
- [59].Chen J, Xie J, Jiang Z, Wang B, Wang Y, Hu X, Shikonin and its analogs inhibit cancer cell glycolysis by targeting tumor pyruvate kinase-M2, Oncogene, 30 (2011) 4297–4306. [DOI] [PubMed] [Google Scholar]
- [60].Vander Heiden MG, Christofk HR, Schuman E, Subtelny AO, Sharfi H, Harlow EE, Xian J, Cantley LC, Identification of small molecule inhibitors of pyruvate kinase M2, Biochem Pharmacol, 79 (2010) 1118–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Li R, Ning X, Zhou S, Lin Z, Wu X, Chen H, Bai X, Wang X, Ge Z, Li R, Yin Y, Discovery and structure-activity relationship of novel 4-hydroxy-thiazolidine-2-thione derivatives as tumor cell specific pyruvate kinase M2 activators, Eur J Med Chem, 143 (2018) 48–65. [DOI] [PubMed] [Google Scholar]
- [62].Kim DJ, Park YS, Kim ND, Min SH, You YM, Jung Y, Koo H, Noh H, Kim JA, Park KC, Yeom YI, A novel pyruvate kinase M2 activator compound that suppresses lung cancer cell viability under hypoxia, Mol Cells, 38 (2015) 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Aslan E, Adem S, In vitro effects of some flavones on human pyruvate kinase isoenzyme M2, J Biochem Mol Toxicol, 29 (2015) 109–113. [DOI] [PubMed] [Google Scholar]
- [64].Zhang Y, Liu B, Wu X, Li R, Ning X, Liu Y, Liu Z, Ge Z, Li R, Yin Y, New pyridin-3-ylmethyl carbamodithioic esters activate pyruvate kinase M2 and potential anticancer lead compounds, Bioorg Med Chem, 23 (2015) 4815–4823. [DOI] [PubMed] [Google Scholar]
- [65].You L, Zhu H, Wang C, Wang F, Li Y, Li Y, Wang Y, He B, Scutellarin inhibits Hela cell growth and glycolysis by inhibiting the activity of pyruvate kinase M2, Bioorg Med Chem Lett, 27 (2017) 5404–5408. [DOI] [PubMed] [Google Scholar]
- [66].Ning x., Qi H, Li R, Jin Y, McNutt MA, Yin Y, Synthesis and antitumor activity of novel 2,3-didithiocarbamate substituted naphthoquinones as inhibitors of pyruvate kinase m2 isoform, Journal of Enzyme Inhibition and Medicinal Chemistry, 33 (2017) 126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Chen J, Jiang Z, Wang B, Wang Y, Hu X, Vitamin K(3) and K(5) are inhibitors of tumor pyruvate kinase M2, Cancer Lett, 316 (2012) 204–210. [DOI] [PubMed] [Google Scholar]
- [68].Spoden GA, Mazurek S, Morandell D, Bacher N, Ausserlechner MJ, Jansen-Durr P, Eigenbrodt E, Zwerschke W, Isotype-specific inhibitors of the glycolytic key regulator pyruvate kinase subtype M2 moderately decelerate tumor cell proliferation, Int J Cancer, 123 (2008) 312–321. [DOI] [PubMed] [Google Scholar]
- [69].Garcia-Gonzalo FR, Cruz C, Munoz P, Mazurek S, Eigenbrodt E, Ventura F, Bartrons R, Rosa JL, Interaction between HERC1 and M2-type pyruvate kinase, FEBS Lett, 539 (2003) 78–84. [DOI] [PubMed] [Google Scholar]
- [70].Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, Cole RN, Pandey A, Semenza GL, Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1, Cell, 145 (2011) 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Mazurek S, Zwerschke W, Jansen-Durr P, Eigenbrodt E, Metabolic cooperation between different oncogenes during cell transformation: interaction between activated ras and HPV-16 E7, Oncogene, 20 (2001) 6891–6898. [DOI] [PubMed] [Google Scholar]
- [72].Huang L, Yu Z, Zhang T, Zhao X, Huang G, HSP40 interacts with pyruvate kinase M2 and regulates glycolysis and cell proliferation in tumor cells, PLoS One, 9 (2014) e92949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wang HJ, Hsieh YJ, Cheng WC, Lin CP, Lin YS, Yang SF, Chen CC, Izumiya Y, Yu JS, Kung HJ, Wang WC, JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1alpha-mediated glucose metabolism, Proc Natl Acad Sci U S A, 111 (2014) 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Boeckel JN, Derlet A, Glaser SF, Luczak A, Lucas T, Heumuller AW, Kruger M, Zehendner CM, Kaluza D, Doddaballapur A, Ohtani K, Treguer K, Dimmeler S, JMJD8 Regulates Angiogenic Sprouting and Cellular Metabolism by Interacting With Pyruvate Kinase M2 in Endothelial Cells, Arterioscler Thromb Vasc Biol, 36 (2016) 1425–1433. [DOI] [PubMed] [Google Scholar]
- [75].Ibsen KH, Interrelationships and functions of the pyruvate kinase isozymes and their variant forms: a review, Cancer Res, 37 (1977) 341–353. [PubMed] [Google Scholar]
- [76].van Veelen CW, Verbiest H, Vlug AM, Rijksen G, Staal GE, Isozymes of pyruvate kinase from human brain, meningiomas, and malignant gliomas, Cancer Res, 38 (1978) 4681–4687. [PubMed] [Google Scholar]
- [77].Guderley H, Hochachka PW, Catalytic and regulatory properties of muscle pyruvate kinase from Cancer magister, J Exp Zool, 212 (1980) 461–469. [DOI] [PubMed] [Google Scholar]
- [78].Lopez-Alarcon L, Ruiz P, Gosalvez M, Quantitative determination of the degree of differentiation of mammary tumors by pyruvate kinase kinetic analysis, Cancer Res, 41 (1981) 2019–2020. [PubMed] [Google Scholar]
- [79].Schapira F, Resurgence of fetal isozymes in cancer: study of aldolase, pyruvate kinase, lactic dehydrogenase, and beta-hexosaminidase, Isozymes Curr Top Biol Med Res, 5 (1981) 27–75. [PubMed] [Google Scholar]
- [80].Cottreau D, Rousseau-Merck MF, Nezelof C, Kahn A, Pyruvate kinase and phosphofructokinase isozymes in childhood cancers, Pediatr Res, 16 (1982) 199–202. [DOI] [PubMed] [Google Scholar]
- [81].Gali P, Hartmann L, Immunocytochemical study of pyruvate kinase isoenzymes in normal and pathologic human liver, Liver, 2 (1982) 236–243. [DOI] [PubMed] [Google Scholar]
- [82].Ibsen KH, Orlando RA, Garratt KN, Hernandez AM, Giorlando S, Nungaray G, Expression of multimolecular forms of pyruvate kinase in normal, benign, and malignant human breast tissue, Cancer Res, 42 (1982) 888–892. [PubMed] [Google Scholar]
- [83].Yanagi S, Tsuda H, Sakamoto M, Ninomiya Y, Ito N, Evaluation of a histologic classification of mouse liver tumors based on pyruvate kinase isozymes and status of host lipids, J Natl Cancer Inst, 73 (1984) 1311–1317. [PubMed] [Google Scholar]
- [84].Farina FA, Shatton JB, Morris HP, Weinhouse S, Isozymes of pyruvate kinase in liver and hepatomas of the rat, Cancer Res, 34 (1974) 1439–1446. [PubMed] [Google Scholar]
- [85].Wu H, Yang P, Hu W, Wang Y, Lu Y, Zhang L, Fan Y, Xiao H, Li Z, Overexpression of PKM2 promotes mitochondrial fusion through attenuated p53 stability, Oncotarget, 7 (2016) 78069–78082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ji F, Guo B, Wang N, Zhong C, Huang L, Huang Y, Wei L, Su M, Jiang Y, Jin Q, Liu Y, Zhang Z, Yang J, Chen T, Pyruvate kinase M2 interacts with mammalian sterile 20-like kinase 1 and inhibits tamoxifen-induced apoptosis in human breast cancer cells, Tumour Biol, 39 (2017) 1010428317692251. [DOI] [PubMed] [Google Scholar]
- [87].Ibsen KH, Krueger E, Distribution of pyruvate kinase isozymes among rat organs, Arch Biochem Biophys, 157 (1973) 509–513. [DOI] [PubMed] [Google Scholar]
- [88].Hall ER, Cottam GL, Isozymes of pyruvate kinase in vertebrates: their physical, chemical, kinetic and immunological properties, Int J Biochem, 9 (1978) 785–793. [DOI] [PubMed] [Google Scholar]
- [89].Blair JB, Regulatory Properties of Hepatic Pyruvate Kinase, in: Veneziale CM (Ed.) The Regulation of Carbohydrate Formation and Utilization in Mammals, University Park Press, Place Published, 1980, pp. 121–151. [Google Scholar]
- [90].Kahn A, Marie J, Pyruvate Kinases from Human Erythrocytes and Liver, Methods in Enzymology, 90 (1982) 131–140. [DOI] [PubMed] [Google Scholar]
- [91].Valentini G, Chiarelli LR, Fortin R, Dolzan M, Galizzi A, Abraham DJ, Wang C, Bianchi P, Zanella A, Mattevi A, Structure and function of human erythrocyte pyruvate kinase. Molecular basis of nonspherocytic hemolytic anemia, J Biol Chem, 277 (2002) 23807–23814. [DOI] [PubMed] [Google Scholar]
- [92].van den Berg GB, van Berkel TJ, Koster JF, Identification of L-type pyruvate kinase as a major phosphorylation site of endogenous cyclic AMP-dependent protein kinase in rat liver soluble fraction, FEBS Lett, 101 (1979) 289–294. [DOI] [PubMed] [Google Scholar]
- [93].Marie J, Buc H, Simon MP, Kahn A, Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases. Comparison with human liver L-type enzyme, Eur J Biochem, 108 (1980) 251–260. [DOI] [PubMed] [Google Scholar]
- [94].Spoden GA, Morandell D, Ehehalt D, Fiedler M, Jansen-Durr P, Hermann M, Zwerschke W, The SUMO-E3 ligase PIAS3 targets pyruvate kinase M2, J Cell Biochem, 107 (2009) 293–302. [DOI] [PubMed] [Google Scholar]
- [95].Xiangyun Y, Xiaomin N, Linping G, Yunhua X, Ziming L, Yongfeng Y, Zhiwei C, Shun L, Desuccinylation of pyruvate kinase M2 by SIRT5 contributes to antioxidant response and tumor growth, Oncotarget, 8 (2017) 6984–6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Noguchi T, Inoue H, Tanaka T, The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing, J Biol Chem, 261 (1986) 13807–13812. [PubMed] [Google Scholar]
- [97].Hance AJ, Lee J, Feitelson M, The M1 and M2 isozymes of pyruvate kinase are the products of the same gene, Biochem Biophys Res Commun, 106 (1982) 492–499. [DOI] [PubMed] [Google Scholar]
- [98].Viana R, Lujan P, Sanz P, The laforin/malin E3-ubiquitin ligase complex ubiquitinates pyruvate kinase M1/M2, BMC Biochem, 16 (2015) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Liu K, Li F, Han H, Chen Y, Mao Z, Luo J, Zhao Y, Zheng B, Gu W, Zhao W, Parkin Regulates the Activity of Pyruvate Kinase M2, J Biol Chem, 291 (2016) 10307–10317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Imamura K, Tanaka T, Multimolecular forms of pyruvate kinase from rat and other mammalian tissues. I. Electrophoretic studies, J Biochem (Tokyo), 71 (1972) 1043–1051. [DOI] [PubMed] [Google Scholar]
- [101].Imamura K, Tanaka T, Pyruvate kinase isozymes from rat, Methods Enzymol, 90 Pt E (1982) 150–165. [DOI] [PubMed] [Google Scholar]
- [102].Lu Z, Hunter T, Prolyl isomerase Pin1 in cancer, Cell Res, 24 (2014) 1033–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Elbers JR, van Unnik JA, Rijksen G, van Oirschot BA, Roholl PJ, Oosting J, Staal GE, Pyruvate kinase activity and isozyme composition in normal fibrous tissue and fibroblastic proliferations, Cancer, 67 (1991) 2552–2559. [DOI] [PubMed] [Google Scholar]
- [104].Guguen-Guillouzo C, Szajnert MF, Marie J, Delain D, Schapira F, Differentiation in vivo and in vitro of pyruvate kinase isozymes in rat muscle, Biochimie, 59 (1977) 65–71. [DOI] [PubMed] [Google Scholar]
- [105].Harada K, Saheki S, Wada K, Tanaka T, Purification of four pyruvate kinase isozymes of rats by affinity elution chromatography, Biochim Biophys Acta, 524 (1978) 327–339. [DOI] [PubMed] [Google Scholar]
- [106].Imamura K, Taniuchi K, Tanaka T, Multimolecular forms of pyruvate kinase. II. Purification of M 2 -type pyruvate kinase from Yoshida ascites hepatoma 130 cells and comparative studies on the enzymological and immunological properties of the three types of pyruvate kinases, L, M 1, and M 2, J Biochem (Tokyo), 72 (1972) 1001–1015. [DOI] [PubMed] [Google Scholar]
- [107].Lee J, Kim HK, Han YM, Kim J, Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription, Int J Biochem Cell Biol, 40 (2008) 1043–1054. [DOI] [PubMed] [Google Scholar]
- [108].Li Y, Chang Y, Zhang L, Feng Q, Liu Z, Zhang Y, Zuo J, Meng Y, Fang F, High glucose upregulates pantothenate kinase 4 (PanK4) and thus affects M2-type pyruvate kinase (Pkm2), Mol Cell Biochem, 277 (2005) 117–125. [DOI] [PubMed] [Google Scholar]
- [109].Chen N, Rinner O, Czernik D, Nytko KJ, Zheng D, Stiehl DP, Zamboni N, Gstaiger M, Frei C, The oxygen sensor PHD3 limits glycolysis under hypoxia via direct binding to pyruvate kinase, Cell Res, 21 (2011) 983–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC, Pyruvate kinase M2 is a phosphotyrosine-binding protein, Nature, 452 (2008) 181–186. [DOI] [PubMed] [Google Scholar]
- [111].Hacker HJ, Steinberg P, Bannasch P, Pyruvate kinase isoenzyme shift from L-type to M2-type is a late event in hepatocarcinogenesis induced in rats by a choline-deficient/DL-ethionine-supplemented diet, Carcinogenesis, 19 (1998) 99–107. [DOI] [PubMed] [Google Scholar]
- [112].Boros LG, Cascante M, Lee WN, Metabolic profiling of cell growth and death in cancer: applications in drug discovery, Drug discovery today, 7 (2002) 364–372. [DOI] [PubMed] [Google Scholar]
- [113].Ahmed AS, Dew T, Lawton FG, Papadopoulos AJ, Devaja O, Raju KS, Sherwood RA, M2-PK as a novel marker in ovarian cancer. A prospective cohort study, Eur J Gynaecol Oncol, 28 (2007) 83–88. [PubMed] [Google Scholar]
- [114].Schneider J, Tumor markers in detection of lung cancer, Adv Clin Chem, 42 (2006) 1–41. [DOI] [PubMed] [Google Scholar]
- [115].Ugurel S, Bell N, Sucker A, Zimpfer A, Rittgen W, Schadendorf D, Tumor type M2 pyruvate kinase (TuM2-PK) as a novel plasma tumor marker in melanoma, Int J Cancer, 117 (2005) 825–830. [DOI] [PubMed] [Google Scholar]
- [116].Shimada N, Shinagawa T, Ishii S, Modulation of M2-type pyruvate kinase activity by the cytoplasmic PML tumor suppressor protein, Genes Cells, 13 (2008) 245–254. [DOI] [PubMed] [Google Scholar]
- [117].Varghese B, Swaminathan G, Plotnikov A, Tzimas C, Yang N, Rui H, Fuchs SY, Prolactin inhibits activity of pyruvate kinase M2 to stimulate cell proliferation, Mol Endocrinol, 24 (2010) 2356–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Xia L, Wang XR, Wang XL, Liu SH, Ding XW, Chen GQ, Lu Y, A Novel Role for Pyruvate Kinase M2 as a Corepressor for P53 during the DNA Damage Response in Human Tumor Cells, J Biol Chem, 291 (2016) 26138–26150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Zhang Z, Liu Q, Che Y, Yuan X, Dai L, Zeng B, Jiao G, Zhang Y, Wu X, Yu Y, Zhang Y, Yang R, Antigen presentation by dendritic cells in tumors is disrupted by altered metabolism that involves pyruvate kinase M2 and its interaction with SOCS3, Cancer Res, 70 (2010) 89–98. [DOI] [PubMed] [Google Scholar]
- [120].Zhao X, Zhao L, Yang H, Li J, Min X, Yang F, Liu J, Huang G, Pyruvate kinase M2 interacts with nuclear sterol regulatory element-binding protein 1a and thereby activates lipogenesis and cell proliferation in hepatocellular carcinoma, J Biol Chem, 293 (2018) 6623–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Board M, Humm S, Newsholme EA, Maximum activities of key enzymes of glycolysis, glutaminolysis, pentose phosphate pathway and tricarboxylic acid cycle in normal, neoplastic and suppressed cells, Biochem J, 265 (1990) 503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Weinberger R, Appel B, Stein A, Metz Y, Neheman A, Barak M, The pyruvate kinase isoenzyme M2 (Tu M2-PK) as a tumour marker for renal cell carcinoma, Eur J Cancer Care (Engl), 16 (2007) 333–337. [DOI] [PubMed] [Google Scholar]
- [123].Ibsen KH, Chiu RH-C, Park HR, Sanders DA, Roy S, Garratt KN, Mueller MK, Purification and poperties of mouse pyruvate kinases K and M and of a modified K subunit, Biochmeistry, 20 (1981) 1497–1506. [DOI] [PubMed] [Google Scholar]
- [124].Williams R, Holyoak T, McDonald G, Gui C, Fenton AW, Differentiating a Ligand’s Chemical Requirements for Allosteric Interactions from Those for Protein Binding. Phenylalanine Inhibition of Pyruvate Kinase, Biochemistry, 45 (2006) 5421–5429. [DOI] [PubMed] [Google Scholar]
- [125].Hamabe A, Konno M, Tanuma N, Shima H, Tsunekuni K, Kawamoto K, Nishida N, Koseki J, Mimori K, Gotoh N, Yamamoto H, Doki Y, Mori M, Ishii H, Role of pyruvate kinase M2 in transcriptional regulation leading to epithelial-mesenchymal transition, Proc Natl Acad Sci U S A, 111 (2014) 15526–15531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Chen Z, Wang Z, Guo W, Zhang Z, Zhao F, Zhao Y, Jia D, Ding J, Wang H, Yao M, He X, TRIM35 Interacts with pyruvate kinase isoform M2 to suppress the Warburg effect and tumorigenicity in hepatocellular carcinoma, Oncogene, 34 (2015) 3946–3956. [DOI] [PubMed] [Google Scholar]
- [127].Liang F, Li Q, Li X, Li Z, Gong Z, Deng H, Xiang B, Zhou M, Li X, Li G, Zeng Z, Xiong W, TSC22D2 interacts with PKM2 and inhibits cell growth in colorectal cancer, Int J Oncol, 49 (2016) 1046–1056. [DOI] [PubMed] [Google Scholar]
- [128].Guminska M, Ignacak J, Electrophoretic pattern of cytosolic pyruvate kinase fractions A and B (type L and M2) from normal rat liver and Morris hepatoma 7777, Biochim Biophys Acta, 1292 (1996) 99–105. [DOI] [PubMed] [Google Scholar]
- [129].Guminska M, Ignacak J, Kedryna T, Stachurska MB, Tumor-specific pyruvate kinase isoenzyme M2 involved in biochemical strategy of energy generation in neoplastic cells, Acta Biochim Pol, 44 (1997) 711–724. [PubMed] [Google Scholar]
- [130].Guminska M, Stachurska MB, Ignacak J, Pyruvate kinase isoenzymes in chromatin extracts of Ehrlich ascites tumour, Morris hepatoma 7777 and normal mouse and rat livers, Biochim Biophys Acta, 966 (1988) 207–213. [DOI] [PubMed] [Google Scholar]
- [131].Ignacak J, Guminska M, Comparison of pyruvate kinase variants from rat liver and Morris hepatoma 7777, obtained by an affinity chromatography on blue sepharose CL-6B, Acta Biochim Pol, 40 (1993) 261–267. [PubMed] [Google Scholar]
- [132].Ignacak J, Guminska M, N-acetylneuraminic acid, phosphate and thiol groups of pyruvate kinase isoenzymes from Morris hepatoma 7777 and normal rat liver, Acta Biochim Pol, 44 (1997) 201–208. [PubMed] [Google Scholar]
- [133].Ignacak J, Guminska M, Steczko J, Amino-acid composition of pyruvate kinase M2 isoenzyme variants from rat liver and Morris hepatoma 7777, Acta Biochim Pol, 45 (1998) 775–780. [PubMed] [Google Scholar]
- [134].Ignacak J, Stachurska MB, The dual activity of pyruvate kinase type M2 from chromatin extracts of neoplastic cells, Comp Biochem Physiol B Biochem Mol Biol, 134 (2003) 425–433. [DOI] [PubMed] [Google Scholar]
- [135].Eigenbrodt E, Leib S, Kramer W, Friis RR, Schoner W, Structural and kinetic differences between the M2 type pyruvate kinases from lung and various tumors, Biomed Biochim Acta, 42 (1983) S278–282. [PubMed] [Google Scholar]
- [136].Ibsen KH, Basabe JR, Lopez TP, Extraction of a factor from Ehrlich ascites tumor cells that increases the activity of the fetal isozyme of pyruvate kinase in mouse liver, Cancer Res, 35 (1975) 180–188. [PubMed] [Google Scholar]
- [137].Ngo JL, Chute H, Sanders DA, Orlando RA, Ibsen KH, Regulation of pyruvate kinase expression and growth in mastocytoma cells. I. Initial observations, Exp Cell Res, 149 (1983) 565–575. [DOI] [PubMed] [Google Scholar]
- [138].Kedryna T, Guminska M, Marchut E, Pyruvate kinase from cytosolic fractions of the Ehrlich ascites tumour, normal mouse liver and skeletal muscle, Biochim Biophys Acta, 1039 (1990) 130–133. [DOI] [PubMed] [Google Scholar]
- [139].Marchut E, Guminska M, Kedryna T, Radzikowski C, Kusnierczyk H, A pyruvate kinase variant in different mouse transplanted tumors, Experientia, 44 (1988) 25–27. [DOI] [PubMed] [Google Scholar]
- [140].Guminska M, Stachurska MB, Christensen B, Tromholt V, Kieler J, Radzikowski C, Dus D, Pyruvate kinase inhibited by L-cysteine as a marker of tumorigenic human urothelial cell lines, Experientia, 45 (1989) 571–574. [DOI] [PubMed] [Google Scholar]
- [141].Weber G, Carbohydrate metabolism in cancer cells and the molecular correlation concept, Die Naturwissenschaften, 55 (1968) 418–429. [DOI] [PubMed] [Google Scholar]
- [142].Weernink PA, Rijksen G, Mascini EM, Staal GE, Phosphorylation of pyruvate kinase type K is restricted to the dimeric form, Biochim Biophys Acta, 1121 (1992) 61–68. [DOI] [PubMed] [Google Scholar]
- [143].Yuan M, McNae IW, Chen Y, Blackburn EA, Wear MA, Michels PAM, Fothergill-Gilmore LA, Hupp T, Walkinshaw MD, An allostatic mechanism for M2 pyruvate kinase as an amino-acid sensor, Biochem J, 475 (2018) 1821–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Ashizawa K, Willingham MC, Liang CM, Cheng SY, In vivo regulation of monomer-tetramer conversion of pyruvate kinase subtype M2 by glucose is mediated via fructose 1,6-bisphosphate, J Biol Chem, 266 (1991) 16842–16846. [PubMed] [Google Scholar]
- [145].Hofmann E, Kurganov BI, Schellenberger W, Schulz J, Sparmann G, Wenzel KW, Zimmermann G, Association-dissociation behavior of erythrocyte phosphofructokinase and tumor pyruvate kinase, Adv Enzyme Regul, 13 (1975) 247–277. [DOI] [PubMed] [Google Scholar]
- [146].Oude Weernink PA, Rijksen G, Staal GE, Phosphorylation of pyruvate kinase and glycolytic metabolism in three human glioma cell lines, Tumour Biol, 12 (1991) 339–352. [DOI] [PubMed] [Google Scholar]
- [147].Gavriilidou AFM, Holding FP, Mayer D, Coyle JE, Veprintsev DB, Zenobi R, Native Mass Spectrometry Gives Insight into the Allosteric Binding Mechanism of M2 Pyruvate Kinase to Fructose-1,6-Bisphosphate, Biochemistry, 57 (2018) 1685–1689. [DOI] [PubMed] [Google Scholar]
- [148].Morgan HP, O’Reilly FJ, Wear MA, O’Neill JR, Fothergill-Gilmore LA, Hupp T, Walkinshaw MD, M2 pyruvate kinase provides a mechanism for nutrient sensing and regulation of cell proliferation, Proc Natl Acad Sci U S A, 110 (2013) 5881–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Schulz J, Sparmann G, Hofmann E, Alanine-mediated reversible inactivation of tumour pyruvate kinase caused by a tetramer-dimer transition, FEBS Lett, 50 (1975) 346–350. [DOI] [PubMed] [Google Scholar]
- [150].Sparmann G, Schulz J, Hofmann E, Effects of L-alanine and fructose (1,6-diphosphate) on pyruvate kinase from ehrlich ascites tumour cells, FEBS Lett, 36 (1973) 305–308. [DOI] [PubMed] [Google Scholar]
- [151].Gao X, Wang H, Yang JJ, Liu X, Liu ZR, Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase, Mol Cell, 45 (2012) 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Shulman RG, Rothman DL, The Glycogen Shunt Maintains Glycolytic Homeostasis and the Warburg Effect in Cancer, Trends Cancer, 3 (2017) 761–767. [DOI] [PubMed] [Google Scholar]
- [153].Lee WN, Guo P, Lim S, Bassilian S, Lee ST, Boren J, Cascante M, Go VL, Boros LG, Metabolic sensitivity of pancreatic tumour cell apoptosis to glycogen phosphorylase inhibitor treatment, Br J Cancer, 91 (2004) 2094–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Ma D, Wang J, Zhao Y, Lee WN, Xiao J, Go VL, Wang Q, Recker RR, Xiao GG, Inhibition of glycogen phosphorylation induces changes in cellular proteome and signaling pathways in MIA pancreatic cancer cells, Pancreas, 41 (2012) 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Weinhouse S, Glycolysis, respiration, and anomalous gene expression in experimental hepatomas: G.H.A. Clowes memorial lecture, Cancer Res, 32 (1972) 2007–2016. [PubMed] [Google Scholar]
- [156].Tokunaga M, Kami K, Ozawa S, Oguma J, Kazuno A, Miyachi H, Ohashi Y, Kusuhara M, Terashima M, Metabolome analysis of esophageal cancer tissues using capillary electrophoresis-time-of-flight mass spectrometry, Int J Oncol, 52 (2018) 1947–1958. [DOI] [PubMed] [Google Scholar]
- [157].Lazo PA, Amino acids and glucose utilization by different metabolic pathways in ascites-tumour cells, Eur J Biochem, 117 (1981) 19–25. [DOI] [PubMed] [Google Scholar]
- [158].Grabon W, Otto-Slusarczyk D, Chrzanowska A, Mielczarek-Puta M, Joniec-Maciejak I, Slabik K, Baranczyk-Kuzma A, Lactate Formation in Primary and Metastatic Colon Cancer Cells at Hypoxia and Normoxia, Cell Biochem Funct, 34 (2016) 483–490. [DOI] [PubMed] [Google Scholar]
- [159].Eigenbrodt E, Reinacher M, Scheefers-Borchel U, Scheefers H, Friis R, Double role for pyruvate kinase type M2 in the expansion of phosphometabolite pools found in tumor cells, Crit Rev Oncog, 3 (1992) 91–115. [PubMed] [Google Scholar]
- [160].McKeehan WL, Glycolysis, glutaminolysis and cell proliferation, Cell Biol Int Rep, 6 (1982) 635–650. [DOI] [PubMed] [Google Scholar]
- [161].Kovacevic Z, McGivan JD, Mitochondrial metabolism of glutamine and glutamate and its physiological significance, Physiological reviews, 63 (1983) 547–605. [DOI] [PubMed] [Google Scholar]
- [162].Perez-Escuredo J, Dadhich RK, Dhup S, Cacace A, Van Hee VF, De Saedeleer CJ, Sboarina M, Rodriguez F, Fontenille MJ, Brisson L, Porporato PE, Sonveaux P, Lactate promotes glutamine uptake and metabolism in oxidative cancer cells, Cell Cycle, 15 (2016) 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Maresperlman JA, Shrago E, Energy Substrate Utilization in Freshly Isolated Morris Hepatoma 7777 Cells, Cancer Research, 48 (1988) 602–608. [PubMed] [Google Scholar]
- [164].Reitzer LJ, Wice BM, Kennell D, Evidence That Glutamine, Not Sugar, Is the Major Energy-Source for Cultured Hela-Cells, Journal of Biological Chemistry, 254 (1979) 2669–2676. [PubMed] [Google Scholar]
- [165].Lowry OH, Passonneau JV, Hasselberger FX, Schulz D, Effect of Ischemia on Known Substrates and Cofactors of the Glycolytic Pathway in Brain, The Journal of Biological Chemistry, 239 (1964) 18–30. [PubMed] [Google Scholar]
- [166].howse DC, duffy TH, Control of the redox state of the pyridine nucleotides in the rat cerebral cortex. Effect of electroshock-induced seizures, Journal of Neurochemistry, 24 (1975) 935–940. [DOI] [PubMed] [Google Scholar]
- [167].Sahu D, Lotan Y, Wittmann B, Neri B, Hansel DE, Metabolomics analysis reveals distinct profiles of nonmuscle-invasive and muscle-invasive bladder cancer, Cancer Med, 6 (2017) 2106–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Willmann L, Schlimpert M, Halbach S, Erbes T, Stickeler E, Kammerer B, Metabolic profiling of breast cancer: Differences in central metabolism between subtypes of breast cancer cell lines, J Chromatogr B Analyt Technol Biomed Life Sci, 1000 (2015) 95–104. [DOI] [PubMed] [Google Scholar]
- [169].Flores A, Sandoval-Gonzalez S, Takahashi R, Krall A, Sathe L, Wei L, Radu C, Jolly J, Graham N, Christofk HR, Lowry WE, Increased lactate dehydrogenase activity is dispensable in squamous carcinoma cells of origin, Nat Commun, 10 (2019) 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Wang P, Sun C, Zhu T, Xu Y, Structural insight into mechanisms for dynamic regulation of PKM2, Protein Cell, 6 (2015) 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Steinmetz MA, Deal WC Jr., Metabolic control and structure of glycolytic enzymes. 3. Dissociation and subunit structure of rabbit muscle pyruvate kinase, Biochemistry, 5 (1966) 1399–1405. [DOI] [PubMed] [Google Scholar]
- [172].Cottam GL, Hollenberg PF, Coon MJ, Subunit structure of rabbit muscle pyruvate kinase, J Biol Chem, 244 (1969) 1481–1486. [PubMed] [Google Scholar]
- [173].Doster W, Hess B, Reversible solvent denaturation of rabbit muscle pyruvate kinase, Biochemistry, 20 (1981) 772–780. [DOI] [PubMed] [Google Scholar]
- [174].Edwin F, Jagannadham MV, Salt-induced folding of a rabbit muscle pyruvate kinase intermediate at alkaline pH, J Protein Chem, 19 (2000) 361–371. [DOI] [PubMed] [Google Scholar]
- [175].De Felice FG, Soares VC, Ferreira ST, Subunit dissociation and inactivation of pyruvate kinase by hydrostatic pressure oxidation of sulfhydryl groups and ligand effects on enzyme stability, Eur J Biochem, 266 (1999) 163–169. [DOI] [PubMed] [Google Scholar]
- [176].Consler TG, Lee JC, Domain interaction in rabbit muscle pyruvate kinase. I. Effects of ligands on protein denaturation induced by guanidine hydrochloride, J Biol Chem, 263 (1988) 2787–2793. [PubMed] [Google Scholar]
- [177].Friesen RH, Chin AJ, Ledman DW, Lee JC, Interfacial communications in recombinant rabbit kidney pyruvate kinase, Biochemistry, 37 (1998) 2949–2960. [DOI] [PubMed] [Google Scholar]
- [178].Mazurek S, Pyruvate kinase type M2: a key regulator within the tumour metabolome and a tool for metabolic profiling of tumours, Ernst Schering Foundation symposium proceedings, (2008) 99–124. [DOI] [PubMed] [Google Scholar]
- [179].Yan M, Chakravarthy S, Tokuda JM, Pollack L, Bowman GD, Lee YS, Succinyl-5-aminoimidazole-4-carboxamide-1-ribose 5’-Phosphate (SAICAR) Activates Pyruvate Kinase Isoform M2 (PKM2) in Its Dimeric Form, Biochemistry, 55 (2016) 4731–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Kumar Y, Mazurek S, Yang S, Failing K, Winslet M, Fuller B, Davidson BR, In vivo factors influencing tumour M2-pyruvate kinase level in human pancreatic cancer cell lines, Tumour Biol, 31 (2010) 69–77. [DOI] [PubMed] [Google Scholar]
- [181].Hardt PD, Mazurek S, Toepler M, Schlierbach P, Bretzel RG, Eigenbrodt E, Kloer HU, Faecal tumour M2 pyruvate kinase: a new, sensitive screening tool for colorectal cancer, Br J Cancer, 91 (2004) 980–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Zhou W, Capello M, Fredolini C, Piemonti L, Liotta LA, Novelli F, Petricoin EF, Proteomic analysis of pancreatic ductal adenocarcinoma cells reveals metabolic alterations, J Proteome Res, 10 (2011) 1944–1952. [DOI] [PubMed] [Google Scholar]
- [183].Zhou W, Capello M, Fredolini C, Racanicchi L, Piemonti L, Liotta LA, Novelli F, Petricoin EF, Proteomic analysis reveals Warburg effect and anomalous metabolism of glutamine in pancreatic cancer cells, J Proteome Res, 11 (2012) 554–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Huang C, Huang Z, Bai P, Luo G, Zhao X, Wang X, Expression of pyruvate kinase M2 in human bladder cancer and its correlation with clinical parameters and prognosis, Onco Targets Ther, 11 (2018) 2075–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Lu W, Cao Y, Zhang Y, Li S, Gao J, Wang XA, Mu J, Hu YP, Jiang L, Dong P, Gong W, Liu Y, Up-regulation of PKM2 promote malignancy and related to adverse prognostic risk factor in human gallbladder cancer, Sci Rep, 6 (2016) 26351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Tian S, Li P, Sheng S, Jin X, Upregulation of pyruvate kinase M2 expression by fatty acid synthase contributes to gemcitabine resistance in pancreatic cancer, Oncol Lett, 15 (2018) 2211–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Wong CC, Au SL, Tse AP, Xu IM, Lai RK, Chiu DK, Wei LL, Fan DN, Tsang FH, Lo RC, Wong CM, Ng IO, Switching of pyruvate kinase isoform L to M2 promotes metabolic reprogramming in hepatocarcinogenesis, PLoS One, 9 (2014) e115036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Bluemlein K, Gruning NM, Feichtinger RG, Lehrach H, Kofler B, Ralser M, No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis, Oncotarget, 2 (2011) 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Kobierzycki C, Pula B, Werynska B, Piotrowska A, Muszczynska-Bernhard B, Dziegiel P, Rakus D, The lack of evidence for correlation of pyruvate kinase M2 expression with tumor grade in non-small cell lung cancer, Anticancer Res, 34 (2014) 3811–3817. [PubMed] [Google Scholar]
- [190].Lau AN, Israelsen WJ, Roper J, Sinnamon MJ, Georgeon L, Dayton TL, Hillis AL, Yilmaz OH, Di Vizio D, Hung KE, Vander Heiden MG, PKM2 is not required for colon cancer initiated by APC loss, Cancer Metab, 5 (2017) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Mukherjee J, Phillips JJ, Zheng S, Wiencke J, Ronen SM, Pieper RO, Pyruvate kinase M2 expression, but not pyruvate kinase activity, is up-regulated in a grade-specific manner in human glioma, PLoS One, 8 (2013) e57610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Lunt SY, Muralidhar V, Hosios AM, Israelsen WJ, Gui DY, Newhouse L, Ogrodzinski M, Hecht V, Xu K, Acevedo PN, Hollern DP, Bellinger G, Dayton TL, Christen S, Elia I, Dinh AT, Stephanopoulos G, Manalis SR, Yaffe MB, Andrechek ER, Fendt SM, Vander Heiden MG, Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation, Mol Cell, 57 (2015) 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW, Burga LN, Xie J, Jurczak MJ, DePinho RA, Clish CB, Jacks T, Kibbey RG, Wulf GM, Di Vizio D, Mills GB, Cantley LC, Vander Heiden MG, PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells, Cell, 155 (2013) 397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Cortes-Cros M, Hemmerlin C, Ferretti S, Zhang J, Gounarides JS, Yin H, Muller A, Haberkorn A, Chene P, Sellers WR, Hofmann F, M2 isoform of pyruvate kinase is dispensable for tumor maintenance and growth, Proc Natl Acad Sci U S A, 110 (2013) 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Morita M, Sato T, Nomura M, Sakamoto Y, Inoue Y, Tanaka R, Ito S, Kurosawa K, Yamaguchi K, Sugiura Y, Takizaki H, Yamashita Y, Katakura R, Sato I, Kawai M, Okada Y, Watanabe H, Kondoh G, Matsumoto S, Kishimoto A, Obata M, Matsumoto M, Fukuhara T, Motohashi H, Suematsu M, Komatsu M, Nakayama KI, Watanabe T, Soga T, Shima H, Maemondo M, Tanuma N, PKM1 Confers Metabolic Advantages and Promotes Cell-Autonomous Tumor Cell Growth, Cancer Cell, 33 (2018) 355–367 e357. [DOI] [PubMed] [Google Scholar]
- [196].Xie J, Dai C, Hu X, Evidence That Does Not Support Pyruvate Kinase M2 (PKM2)-catalyzed Reaction as a Rate-limiting Step in Cancer Cell Glycolysis, J Biol Chem, 291 (2016) 8987–8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Marin-Hernandez A, Rodriguez-Enriquez S, Vital-Gonzalez PA, Flores-Rodriguez FL, Macias-Silva M, Sosa-Garrocho M, Moreno-Sanchez R, Determining and understanding the control of glycolysis in fast-growth tumor cells. Flux control by an over-expressed but strongly product-inhibited hexokinase, FEBS J, 273 (2006) 1975–1988. [DOI] [PubMed] [Google Scholar]
- [198].Prakasam G, Iqbal MA, Bamezai RNK, Mazurek S, Posttranslational Modifications of Pyruvate Kinase M2: Tweaks that Benefit Cancer, Front Oncol, 8 (2018) 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Consler TG, Woodard SH, Lee JC, Effects of primary sequence differences on the global structure and function of an enzyme: a study of pyruvate kinase isozymes, Biochemistry, 28 (1989) 8756–8764. [DOI] [PubMed] [Google Scholar]
- [200].Ikeda Y, Taniguchi N, Noguchi T, Dominant negative role of the glutamic acid residue conserved in the pyruvate kinase M(1) isozyme in the heterotropic allosteric effect involving fructose-1,6-bisphosphate, J Biol Chem, 275 (2000) 9150–9156. [DOI] [PubMed] [Google Scholar]
- [201].Ikeda Y, Noguchi T, Allosteric regulation of pyruvate kinase M2 isozyme involves a cysteine residue in the intersubunit contact, J Biol Chem, 273 (1998) 12227–12233. [DOI] [PubMed] [Google Scholar]
- [202].Ikeda Y, Tanaka T, Noguchi T, Conversion of non-allosteric pyruvate kinase isozyme into an allosteric enzyme by a single amino acid substitution, J Biol Chem, 272 (1997) 20495–20501. [DOI] [PubMed] [Google Scholar]
- [203].Nakatsu D, Horiuchi Y, Kano F, Noguchi Y, Sugawara T, Takamoto I, Kubota N, Kadowaki T, Murata M, L-cysteine reversibly inhibits glucose-induced biphasic insulin secretion and ATP production by inactivating PKM2, Proc Natl Acad Sci U S A, 112 (2015) E1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Chaneton B, Hillmann P, Zheng L, Martin ACL, Maddocks ODK, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, Frezza C, O’Reilly M, Gottlieb E, Serine is a natural ligand and allosteric activator of pyruvate kinase M2, Nature, 491 (2012) 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Ye J, Mancuso A, Tong X, Ward PS, Fan J, Rabinowitz JD, Thompson CB, Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation, Proc Natl Acad Sci U S A, 109 (2012) 6904–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Gosalvez M, Garcia-Suarez S, Lopez-Alarcon L, Metabolic control of glycolysis in normal and tumor permeabilized cells, Cancer Res, 38 (1978) 142–148. [PubMed] [Google Scholar]
- [207].Keller KE, Doctor ZM, Dwyer ZW, Lee YS, SAICAR induces protein kinase activity of PKM2 that is necessary for sustained proliferative signaling of cancer cells, Mol Cell, 53 (2014) 700–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Veech RL, Lawson JW, Cornell NW, Krebs HA, Cytosolic phosphorylation potential, J Biol Chem, 254 (1979) 6538–6547. [PubMed] [Google Scholar]
- [209].Halestrap AP, Brenner C, The adenine nucleotide translocase: a central component of the mitochondrial permeability transition pore and key player in cell death, Current medicinal chemistry, 10 (2003) 1507–1525. [DOI] [PubMed] [Google Scholar]
- [210].Maldonado EN, DeHart DN, Patnaik J, Klatt SC, Gooz MB, Lemasters JJ, ATP/ADP Turnover and Import of Glycolytic ATP into Mitochondria in Cancer Cells Is Independent of the Adenine Nucleotide Translocator, J Biol Chem, 291 (2016) 19642–19650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].DeHart DN, Lemasters JJ, Maldonado EN, Erastin-Like Anti-Warburg Agents Prevent Mitochondrial Depolarization Induced by Free Tubulin and Decrease Lactate Formation in Cancer Cells, SLAS Discov, 23 (2018) 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Newsholme EA, Board M, Application of metabolic-control logic to fuel utilization and its significance in tumor cells, Adv Enzyme Regul, 31 (1991) 225–246. [DOI] [PubMed] [Google Scholar]
- [213].Groen AK, van Roermund CW, Vervoorn RC, Tager JM, Control of gluconeogenesis in rat liver cells. Flux control coefficients of the enzymes in the gluconeogenic pathway in the absence and presence of glucagon, Biochem J, 237 (1986) 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Hornberg JJ, Bruggeman FJ, Bakker BM, Westerhoff HV, Metabolic control analysis to identify optimal drug targets, Prog Drug Res, 64 (2007) 171, 173–189. [DOI] [PubMed] [Google Scholar]
- [215].Hennipman A, van Oirschot BA, Smits J, Rijksen G, Staal GE, Heterogeneity of glycolytic enzyme activity and isozyme composition of pyruvate kinase in breast cancer, Tumour Biol, 9 (1988) 178–189. [DOI] [PubMed] [Google Scholar]
- [216].Marin-Hernandez A, Gallardo-Perez JC, Ralph SJ, Rodriguez-Enriquez S, Moreno-Sanchez R, HIF-1alpha modulates energy metabolism in cancer cells by inducing over-expression of specific glycolytic isoforms, Mini Rev Med Chem, 9 (2009) 1084–1101. [DOI] [PubMed] [Google Scholar]
- [217].Courtney KD, Bezwada D, Mashimo T, Pichumani K, Vemireddy V, Funk AM, Wimberly J, McNeil SS, Kapur P, Lotan Y, Margulis V, Cadeddu JA, Pedrosa I, DeBerardinis RJ, Malloy CR, Bachoo RM, Maher EA, Isotope Tracing of Human Clear Cell Renal Cell Carcinomas Demonstrates Suppressed Glucose Oxidation In Vivo, Cell metabolism, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Snaebjornsson MT, Schulze A, Non-canonical functions of enzymes facilitate cross-talk between cell metabolic and regulatory pathways, Exp Mol Med, 50 (2018) 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G, Rabinowitz JD, Asara JM, Cantley LC, Evidence for an alternative glycolytic pathway in rapidly proliferating cells, Science, 329 (2010) 1492–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Diaz-Jullien C, Moreira D, Sarandeses CS, Covelo G, Barbeito P, Freire M, The M2-type isoenzyme of pyruvate kinase phosphorylates prothymosin alpha in proliferating lymphocytes, Biochim Biophys Acta, 1814 (2011) 355–365. [DOI] [PubMed] [Google Scholar]
- [221].Jiang Y, Wang Y, Wang T, Hawke DH, Zheng Y, Li X, Zhou Q, Majumder S, Bi E, Liu DX, Huang S, Lu Z, PKM2 phosphorylates MLC2 and regulates cytokinesis of tumour cells, Nat Commun, 5 (2014) 5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z, PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis, Cell, 150 (2012) 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Cheng TY, Yang YC, Wang HP, Tien YW, Shun CT, Huang HY, Hsiao M, Hua KT, Pyruvate kinase M2 promotes pancreatic ductal adenocarcinoma invasion and metastasis through phosphorylation and stabilization of PAK2 protein, Oncogene, 37 (2018) 1730–1742. [DOI] [PubMed] [Google Scholar]
- [224].Wei Y, Wang D, Jin F, Bian Z, Li L, Liang H, Li M, Shi L, Pan C, Zhu D, Chen X, Hu G, Liu Y, Zhang CY, Zen K, Pyruvate kinase type M2 promotes tumour cell exosome release via phosphorylating synaptosome-associated protein 23, Nat Commun, 8 (2017) 14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Gao X, Wang H, Yang JJ, Chen J, Jie J, Li L, Zhang Y, Liu ZR, Reciprocal regulation of protein kinase and pyruvate kinase activities of pyruvate kinase M2 by growth signals, J Biol Chem, 288 (2013) 15971–15979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].He CL, Bian YY, Xue Y, Liu ZX, Zhou KQ, Yao CF, Lin Y, Zou HF, Luo FX, Qu YY, Zhao JY, Ye ML, Zhao SM, Xu W, Pyruvate Kinase M2 Activates mTORC1 by Phosphorylating AKT1S1, Sci Rep, 6 (2016) 21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Xia L, Qin K, Wang XR, Wang XL, Zhou AW, Chen GQ, Lu Y, Pyruvate kinase M2 phosphorylates H2AX and promotes genomic instability in human tumor cells, Oncotarget, 8 (2017) 109120–109134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Jiang Y, Li X, Yang W, Hawke DH, Zheng Y, Xia Y, Aldape K, Wei C, Guo F, Chen Y, Lu Z, PKM2 regulates chromosome segregation and mitosis progression of tumor cells, Mol Cell, 53 (2014) 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Liang J, Cao R, Wang X, Zhang Y, Wang P, Gao H, Li C, Yang F, Zeng R, Wei P, Li D, Li W, Yang W, Mitochondrial PKM2 regulates oxidative stress-induced apoptosis by stabilizing Bcl2, Cell Res, 27 (2017) 329–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Stetak A, Veress R, Ovadi J, Csermely P, Keri G, Ullrich A, Nuclear translocation of the tumor marker pyruvate kinase M2 induces programmed cell death, Cancer Res, 67 (2007) 1602–1608. [DOI] [PubMed] [Google Scholar]
- [231].Hoshino A, Hirst JA, Fujii H, Regulation of cell proliferation by interleukin-3-induced nuclear translocation of pyruvate kinase, J Biol Chem, 282 (2007) 17706–17711. [DOI] [PubMed] [Google Scholar]
- [232].Giannoni E, Taddei ML, Morandi A, Comito G, Calvani M, Bianchini F, Richichi B, Raugei G, Wong N, Tang D, Chiarugi P, Targeting stromal-induced pyruvate kinase M2 nuclear translocation impairs oxphos and prostate cancer metastatic spread, Oncotarget, 6 (2015) 24061–24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Li Q, Zhang D, Chen X, He L, Li T, Xu X, Li M, Nuclear PKM2 contributes to gefitinib resistance via upregulation of STAT3 activation in colorectal cancer, Sci Rep, 5 (2015) 16082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Zhang J, Feng G, Bao G, Xu G, Sun Y, Li W, Wang L, Chen J, Jin H, Cui Z, Nuclear translocation of PKM2 modulates astrocyte proliferation via p27 and -catenin pathway after spinal cord injury, Cell Cycle, 14 (2015) 2609–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Wang F, Wang K, Xu W, Zhao S, Ye D, Wang Y, Xu Y, Zhou L, Chu Y, Zhang C, Qin X, Yang P, Yu H, SIRT5 Desuccinylates and Activates Pyruvate Kinase M2 to Block Macrophage IL-1beta Production and to Prevent DSS-Induced Colitis in Mice, Cell Rep, 19 (2017) 2331–2344. [DOI] [PubMed] [Google Scholar]
- [236].Li S, Swanson SK, Gogol M, Florens L, Washburn MP, Workman JL, Suganuma T, Serine and SAM Responsive Complex SESAME Regulates Histone Modification Crosstalk by Sensing Cellular Metabolism, Mol Cell, 60 (2015) 408–421. [DOI] [PubMed] [Google Scholar]
- [237].Jeyasingham MD, Carlson GM, Evaluation of phosphoenolpyruvate as a phosphoryl group donor for phosphoproteins in skeletal muscle, Arch Biochem Biophys, 357 (1998) 285–292. [DOI] [PubMed] [Google Scholar]
- [238].Mattoo RL, Waygood EB, Khandelwal RL, Activation of phosphoenolpyruvate-dependent protein kinase by cytidine 5’-triphosphate in rat skeletal muscle, FEBS Lett, 165 (1984) 117–120. [DOI] [PubMed] [Google Scholar]
- [239].Ueda S, Sakasegawa S, Atomi H, Development of an Enzymatic Cycling Method Using Pyruvate Kinase for Assaying Pyruvate or Phosphoenolpyruvate, Current Biotechnology, 7 (2018) 125–131. [Google Scholar]
- [240].Hosios AM, Fiske BP, Gui DY, Vander Heiden MG, Lack of Evidence for PKM2 Protein Kinase Activity, Mol Cell, 59 (2015) 850–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Wu H, Li Z, Wang Y, Yang P, Li Z, Li H, Wu C, MiR-106b-mediated Mfn2 suppression is critical for PKM2 induced mitochondrial fusion, Am J Cancer Res, 6 (2016) 2221–2234. [PMC free article] [PubMed] [Google Scholar]
- [242].Li L, Zhang Y, Qiao J, Yang JJ, Liu ZR, Pyruvate kinase M2 in blood circulation facilitates tumor growth by promoting angiogenesis, J Biol Chem, 289 (2014) 25812–25821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Hsu MC, Hung WC, Yamaguchi H, Lim SO, Liao HW, Tsai CH, Hung MC, Extracellular PKM2 induces cancer proliferation by activating the EGFR signaling pathway, Am J Cancer Res, 6 (2016) 628–638. [PMC free article] [PubMed] [Google Scholar]
- [244].Zhang Y, Li L, Liu Y, Liu ZR, PKM2 released by neutrophils at wound site facilitates early wound healing by promoting angiogenesis, Wound Repair Regen, 24 (2016) 328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Gehrig S, Macpherson JA, Driscoll PC, Symon A, Martin SR, MacRae JI, Kleinjung J, Fraternali F, Anastasiou D, An engineered photoswitchable mammalian pyruvate kinase, FEBS J, 284 (2017) 2955–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Chen X, Qian Y, Wu S, The Warburg effect: evolving interpretations of an established concept, Free Radic Biol Med, 79 (2015) 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Gao L, Mejias R, Echevarria M, Lopez-Barneo J, Induction of the glucose-6-phosphate dehydrogenase gene expression by chronic hypoxia in PC12 cells, FEBS Lett, 569 (2004) 256–260. [DOI] [PubMed] [Google Scholar]
- [248].Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, Sasaki AT, Anastasiou D, Mullarky E, Vokes NI, Sasaki M, Beroukhim R, Stephanopoulos G, Ligon AH, Meyerson M, Richardson AL, Chin L, Wagner G, Asara JM, Brugge JS, Cantley LC, Vander Heiden MG, Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis, Nat Genet, 43 (2011) 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Altenberg B, Greulich KO, Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes, Genomics, 84 (2004) 1014–1020. [DOI] [PubMed] [Google Scholar]
- [250].Kim JW, Tchernyshyov I, Semenza GL, Dang CV, HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia, Cell metabolism, 3 (2006) 177–185. [DOI] [PubMed] [Google Scholar]
- [251].Weinhouse S, The Warburg hypothesis fifty years later, Z Krebsforsch Klin Onkol Cancer Res Clin Oncol, 87 (1976) 115–126. [DOI] [PubMed] [Google Scholar]
- [252].Dombrauckas JD, Santarsiero BD, Mesecar AD, Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis, Biochemistry, 44 (2005) 9417–9429. [DOI] [PubMed] [Google Scholar]
- [253].Ibsen KH, Trippet P, Effects of amino acids on the kinetic properties of three noninterconvertible rat pyruvate kinases, Arch Biochem Biophys, 163 (1974) 570–580. [DOI] [PubMed] [Google Scholar]
- [254].Fenton AW, Hutchinson M, The pH dependence of the allosteric response of human liver pyruvate kinase to fructose-1,6-bisphosphate, ATP, and alanine, Arch Biochem Biophys, 484 (2009) 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Fenton AW, Alontaga AY, The impact of ions on allosteric functions in human liver pyruvate kinase, Methods Enzymol, 466 (2009) 83–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Shirmanova MV, Druzhkova IN, Lukina MM, Matlashov ME, Belousov VV, Snopova LB, Prodanetz NN, Dudenkova VV, Lukyanov SA, Zagaynova EV, Intracellular pH imaging in cancer cells in vitro and tumors in vivo using the new genetically encoded sensor SypHer2, Biochim Biophys Acta, 1850 (2015) 1905–1911. [DOI] [PubMed] [Google Scholar]
- [257].Damaghi M, Wojtkowiak JW, Gillies RJ, pH sensing and regulation in cancer, Front Physiol, 4 (2013) 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Webb BA, Chimenti M, Jacobson MP, Barber DL, Dysregulated pH: a perfect storm for cancer progression, Nat Rev Cancer, 11 (2011) 671–677. [DOI] [PubMed] [Google Scholar]
- [259].Bergmeyer HU, Methods of Enzymatic Analysis, Verlag Chemie Academic Press, 1974. [Google Scholar]
- [260].Rodriguez-Horche P, Luque J, Perez-Artes E, Pineda M, Pinilla M, Comparative kinetic behaviour and regulation by fructose-1,6-bisphosphate and ATP of pyruvate kinase from erythrocytes, reticulocytes and bone marrow cells, Comp Biochem Physiol B, 87 (1987) 553–557. [DOI] [PubMed] [Google Scholar]
- [261].Garreau H, Buc-Temkine H, Allosteric activation of human erythrocyte pyruvate kinase by fructose-1,6-diphosphate. Kinetic and equilibrium binding studies, Biochimie, 54 (1972) 1103–1107. [DOI] [PubMed] [Google Scholar]
- [262].Keller KE, Tan IS, Lee YS, SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions, Science, 338 (2012) 1069–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Thalhammer O, Pollak A, Lubec G, Konigshofer H, Intracellular concentrations of phenylalanine, tyrosine and alpha-aminobutyric acid in 13 homozygotes and 19 heterozygotes for phenylketonuria (PKU) compared with 26 normals, Hum Genet, 54 (1980) 213–216. [DOI] [PubMed] [Google Scholar]


