Abstract
Purpose:
Fluorescence of co-enzyme reduced nicotinamide adenine dinucleotide (NADH) and oxidized flavoproteins (Fp) provides a sensitive measure of the mitochondrial redox state and cellular metabolism. By imaging NADH and Fp, we investigated the utility of optical redox imaging (ORI) to monitor cellular metabolism and detect early metabolic response to cancer drugs.
Procedures:
We performed ORI of human melanoma DB-1 cells in culture and DB-1 mouse xenografts to detect the redox response to lonidamine (LND) treatment.
Results:
For cultured cells, LND treatment for 45 min significantly lowered NADH levels with no significant change in Fp, resulting in a significant increase in the Fp redox ratio (Fp/(NADH+Fp)); 3-h prolonged treatment led to a decrease in NADH and an increase in Fp and a more oxidized redox state compared to control. Significant decrease in the mitochondrial redox capacity of LND-treated cells was observed for the first time. For xenografts, 45-min LND treatment resulted in a significant reduction of NADH content, no significant changes in Fp content, and a trend of increase in the Fp redox ratio. Intratumor redox heterogeneity was observed in both control and LND-treated groups.
Conclusion:
Our results support the utility of ORI for evaluating cellular metabolism and monitoring early metabolic response to cancer drugs.
Keywords: NADH, Flavoproteins, FAD, Optical imaging, Redox ratio, Early detection, Mitochondria, DMSO
Introduction
Altered metabolism including enhanced glycolysis and glutaminolysis is a hall mark of cancer [1]. Tumor metabolism is currently a key target of cancer therapy. Since metabolic changes at the molecular level usually precede changes in tumor volume, detection of changes in tumor metabolism can serve as early predictors and detectors of therapeutic response.
Intracellular redox state is an important and sensitive metabolic indicator [2]. Nicotinamide adenine dinucleotide (NAD+) (its reduced form NADH) is a redox co-enzyme. NAD-coupled redox potential, NAD+/NADH, plays a critical role in the mitochondrial electron transfer chain (ETC), and the NADH level as measured by its florescence intensity can be used to monitor cellular metabolism. Since NAD+ does not fluoresce upon excitation but FAD (oxidized flavin adenine dinucleotide) does, alternatively, FAD/NADH ratio or (normalized redox ratio, FAD/(NADH+FAD)) can serve as a surrogate indicator for the mitochondrial redox state [3–11] and has been shown to correlate linearly with NAD+/NADH determined by biochemical assays [12, 13]. In this report, Fp (oxidized flavoproteins containing FAD) and FAD are used interchangeably.
Optical redox imaging (ORI) techniques, pioneered by Chance et al. [2, 10, 14, 15], have increasing applications in cancer research, such as discrimination between cancer and normal tissues [16, 17], differentiation among cancer aggressiveness [18–21], distinguishing among receptor status of breast cancer [22], discerning genetic mutation [23, 24], and monitoring therapeutic effects [25–30]. For example, Walsh et al. reported that NADH/FAD ratio normalized by percentage of mitotic cells correlates with a cellular glycolytic index across a panel of human breast cell lines, and the redox ratio and the fluorescence life-times of both NADH and FAD could be sensitive to herceptin-induced early metabolic changes, whereas 2-deoxy-2-[18F]fluoro-D-glucose positron emission tomography did not resolve the early changes in xenograft models [28]. Cannon et al. reported the development of high-throughput measurements of the optical redox ratio using a commercial microplate reader to differentiate drug responder from non-responder in breast cancer cell line models [31].
Lonidamine (LND) as an anti-tumor drug inhibits both mitochondrial respiration and glycolysis, leading to a decrease in cellular ATP and selectively acidifying tumors thereby sensitizing them to other anti-tumor drugs [32, 33]. LND has differential effects on cancer and normal tissues. For example, per our NMR measurements, LND acidifies tumor only, not the muscle, brain, or liver [34, 35]. ORI has not been employed to study the effects of LND on cancer. Here we report ORI studies of the effects of LND on the mitochondrial redox state of human melanoma DB-1 both in cell culture and in tumor xenografts implanted in immunosuppressed mice.
Materials and Methods
Live Cell Preparation for Redox Imaging
Oliomycin (Olg), carbonyl cyanide-p-tri-fluoro-methoxy-phenylhydrazone (FCCP), rotenone (Rot), and antimycin A (Ant) were purchased from Sigma-Aldrich and LND from Santa Cruz Biotechnology. Ant and Olg in powder form were reconstituted with ethanol, aliquoted and stored in a − 20 °C freezer. FCCP and Rot in powder form were reconstituted with DMSO, aliquoted and stored in a − 80 °C freezer. LND for cell culture treatment with a final concentration of 150 μM was freshly prepared with DMSO.
For testing LND treatment response of cultured cells, DB-1 cells were expanded in phenol-red free RPMI 1640 complete medium. Suspended DB-1 cells (4 × 104, 200 μl) were transferred to a 35-mm glass bottom dish (MatTek, MA) and incubated for 4 h to allow cells to attach to the glass surface. Fresh medium (1 ml) was then added, and the dish was incubated for 20 h. Approximately 1 h before imaging, the medium was removed and the dish were rinsed twice with PBS (with Ca2+ and Mg2+) followed by adding 1 ml live cell imaging solution (LCIS, Life Technology) spiked with 11 mM glucose and 2 mM glutamine. For the LND-treated group, the cells were treated with 150 μM LND for 45 min (0.8 % or 0.2 % DMSO in the glucose and glutamine-spiked LCIS). The control group was treated with 0.8 % or 0.2 % DMSO for 45 min.
For the mitochondrial redox capacity experiments, we adopted the scheme commonly used for cell metabolism measurements, i.e., sequentially adding the metabolic perturbation drugs to cell culture in the following order: Olg (2 μg/ml) → FCCP (8 μM) → Rot (1 μM) + Ant (1.25 μg/ml). ORI images were taken approximately 5 min after each drug administration. Once images were acquired, the next drug was immediately administered to the dish.
A DeltaVision wide field microscope (objective × 40/ 0.95NA, image size 512 × 512 pixels, bin 2 × 2, pixel size 0.32 μm) was utilized for collecting NADH and Fp fluorescence signals in cultured cells. The excitation bandpass filters for NADH and Fp channels were 360/40 nm and 470/40 nm, respectively; the emission filters of NADH and FAD channels were 455/50 nm and 520/40 nm, respectively. Exposure time was 3 s for both channels. At least five fields of view (FOV) were imaged for each dish. Five to eight dishes of each culture were imaged. Imaging was performed on at least two different days.
Live Cell Imaging Data Analysis
A customized Matlab® program was used to extract NADH and Fp signals. Light source intensity fluctuations in both channels were monitored for each acquisition by a photosensor and were taken into account for the signal intensities of each channel by factoring in the ratio of the photosensor reading to a constant. To remove inhomogeneous illumination, each image was first flattened with a third-degree polynomial surface fit followed by thresholding. The mean value of a region of interest (ROI) where no cells were present was treated as background and subtracted from each image. The signal-to-noise (SNR) ratio was quantified by dividing the net signal by the standard deviation of the background. Pixels with SNR < 3 were excluded from further processing. The net intensities of NADH and Fp of each field of view (FOV) were then averaged. The Fp redox ratios computed pixel-by-pixel based on the net pixel values of NADH and Fp were averaged as the mean value for that FOV. Fp, NADH, and the Fp redox ratio for all FOVs were then averaged for each dish. The mean values of each group obtained by averaging across the dishes in the group were reported. In accordance with our experimental design, unpaired Student t test, or unpaired Student t test with Bonferroni adjustment or one-way ANOVA with Bonferroni adjustment was performed for statistical analysis and p < 0.05 was taken as statistically significant.
Tumor Preparation and Redox Imaging
The animal protocol utilized in the study was approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. DB-1 xenografts were grown subcutaneously in athymic nude mice as reported previously [36]. Ten mice bearing DB-1 tumors (volume <200 mm3) were included in the study. Five animals were randomly selected and treated by intraperitoneal injection with LND at a dose of 100 mg/kg (freshly prepared in Tris/glycine buffer at pH 8.3) 45 min prior to tumor harvest (LND group), and the other five animals were sham treated with Tris/glycine buffer for 45 min with tumors being harvested as controls (control group). The control and treated tumor groups were similar in average size. Tumors were excised from the anesthetized mice within 1 min and were immediately immersed and stored in liquid nitrogen. The snap-frozen tumors were then embedded adjacent to NADH and FAD frozen solution standards following the procedures described in detail elsewhere [19]. The embedded samples were then imaged using the Chance redox scanner, i.e., the 3D cryogenic NADH/Fp fluorescence imager [10, 37, 38]. First, the sample was milled flat to expose the embedded tumor tissue with a size of ~2 × 2 mm2. This section is the top section. Tissue depth is defined as the distance between the top section and a specific section down below. Three tissue sections spacing 400 μm were then raster-scanned for each tumor with a planar spatial resolution of 200 μm. Matlab® routines were used to compute the nominal concentrations of NADH and Fp by referencing their fluorescence intensities to that of the frozen solution standards of NADH and FAD, respectively. Images of the nominal concentration-based Fp, NADH, and the Fp redox ratio Fp/(NADH + Fp) were produced. Note that the true concentrations of NADH and Fp in tissues may be different from the nominal concentrations due to the difference in optical properties of frozen standard solution and frozen tissue. The nominal concentrations here only serve as a basis for the relative quantitative comparisons.
Xenograft Imaging Data Analysis
The imaging data were analyzed with two independent methods, the global averaging method, and an analysis of repeated measures with linear mixed-effects model (LMM). By global averaging method, for each tumor, the data were first averaged for each tissue section/layer, then further averaged across the sections of a tumor to obtain the average value of each of the redox indices (Fp, NADH, and Fp/(NADH+Fp)) for that tumor. The average values of each tumor were then futher averaged to obtain the mean value for each group. The mean values were compared between the treated and control groups using unpaired Student t test. In order to statistically control the possible dependence of redox indices on tissue depth and tumor volume [23, 39, 40], we performed LMM analysis of the redox indices for individual tumor sections at different depths. In LMM analysis, we set the dependent variable to be one of the redox indices, such as NADH, the repeated measure to be section, the fixed effect to be treatment, both depth and tumor volume to be covariates. P < 0.05 was taken as statistically significant.
For tissue redox heterogeneity analysis, two Gaussian functions were fit to the histograms of the redox indices (Fp, NADH, and Fp redox ratio) to extract subpopulation information. Specifically, the section histogram data were first summed across multiple sections of each tumor and the summed histograms were then smoothed with a moving average filter (n = 5) and fitted with two Gaussian functions (details can be found in Fig. S1 of Supplementary Material). The peak positions of two Gaussian curves obtained for each redox index were used for further analysis.
Results
Optical Redox Imaging Is Sensitive to Mitochondrial Metabolism in DB-1 Cells
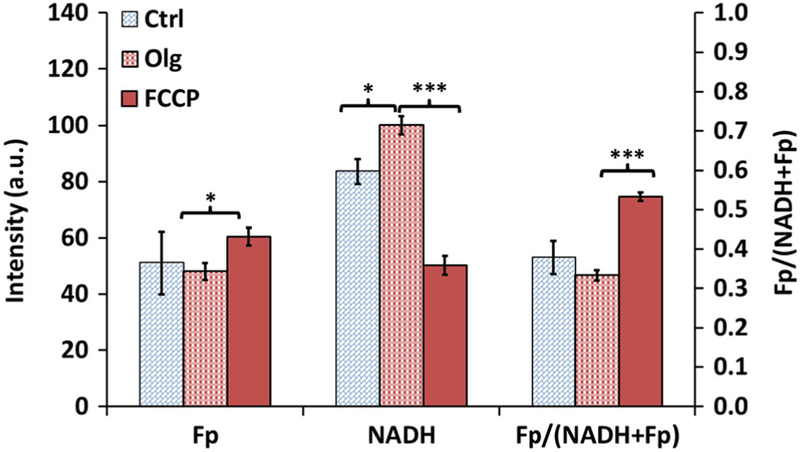
To confirm that the ORI technique is sensitive to mitochondrial metabolism in DB-1, we administered Olg followed by FCCP to observe the changes in NADH and Fp signals [41, 42]. Oligomycin is an ATP synthase inhibitor that inhibits complex V and causes buildup of NADH. Per our experimental design, we were interested in knowing the effect of Olg and FCCP. Therefore, we performed unpaired t test with Bonferroni adjustment to compare the means between control and Olg groups and the means between Olg and FCCP groups. As expected, we observed an appreciable increase in NADH caused by Olg (Fig. 1). FCCP is a mitochondrial oxidative phosphorylation uncoupler which destroys the linkage between the ETC and the phosphorylation system, resulting in unregulated oxidation of NADH without generating ATP and an accumulation of Fp as well. We observed that 8 μM FCCP led to a decrease in NADH and increase in Fp and the Fp redox ratio, i.e., a more oxidized redox state.
Fig. 1.
The redox indices (Fp, NADH, and Fp redox ratio) of cultured DB-1 melanoma cells under sequential treatment of 0.2 % DMSO (Ctrl), oligomycin (Olg), and FCCP. Unpaired Student’s t test with Bonferroni correction was used to compare the differences between Ctrl and Olg and between Olg and FCCP. *p < 0.05, ***p < 0.001. N = 3 dishes.
LND Treatment Effects on the Redox State of DB-1 Melanoma Cells
Redox Indices Changed by LND Treatment
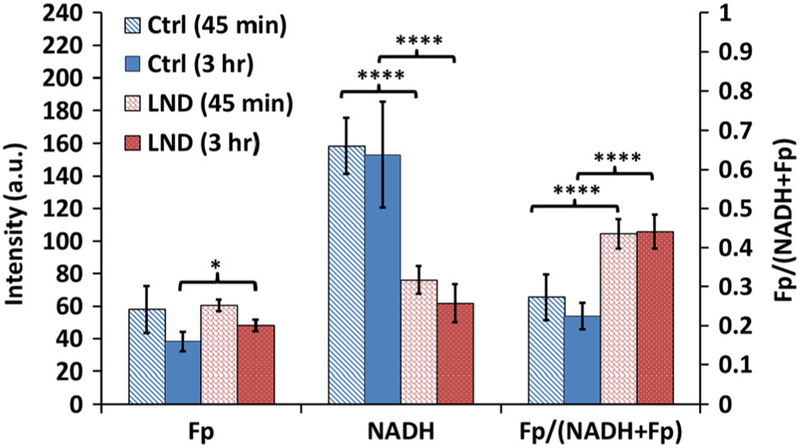
In this study, we were interested in the early metabolic/redox response to LND treatment. Thus, we imaged the treatment effect at 45-min and 3-h time points instead of the long-term effect. These time points were chosen based on our previous MRI study on tumor acidification results that showed 40-min LND treatment already caused significant decreases of intracellular bioenergetics and pH [34]. The pH stayed at the same low level until ~90 min followed by a gradual increase. At 3 h, the intracellular pH level went back to the level of 20-min treatment. However, cellular bioenergetics kept decreasing with time and stabilized by 3 h.
Fig. 2 shows typical redox images of DB-1 cells in culture. We quantified Fp, NADH, and the Fp redox ratio as summarized in Table 1 and Fig. 3. In comparison with control, LND treatment for 45 min caused a 52 % decrease in NADH (p < 0.0001) without a significant change in Fp, resulting in a more oxidized redox state of cells with a 60 % increase in the Fp redox ratio (p < 0.0001). A 3-h LND treatment resulted in 60 % decrease in NADH (p < 0.0001), 26 % increase in Fp (p < 0.05), and 97 % increase in the Fp redox ratio (p < 0.0001).
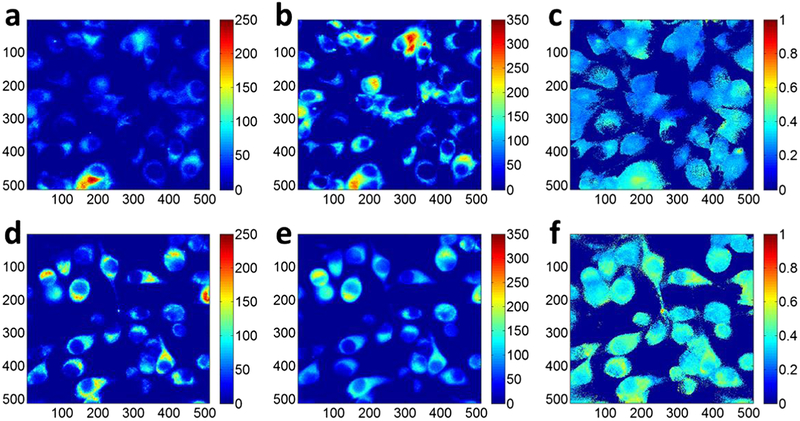
Fig. 2.
Typical redox images of the control (0.8 % DMSO): a Fp, b NADH, and c Fp/(NADH+Fp) and the treated (150 μM LND): d Fp, e NADH, and f Fp/(NADH+Fp). The image matrices were 512 × 512 (pixel size: 0.32 × 0.32 × 0.2 μm3 and binning: 2 × 2). The color bars of the Fp and NADH images indicate the signal intensity in arbitrary unit. The color bar of the Fp redox ratio image indicates the ratio range from 0 to 1.
Table 1.
LND treatment effects on DB-1 melanoma cells in culture (mean ± SD)
| Fp | NADH | Fp/(NADH+Fp) | |
|---|---|---|---|
| Control 45 min (0.8 % DMSO, N = 5) | 58 ± 15 | 158 ± 17 | 0.27 ± 0.06 |
| LND45 min (N = 5) | 60 ± 4 | 76 ± 9 | 0.44 ± 0.04 |
| Change % | 4 % | − 52 % | 60% |
| p | > 0.05 | < 0.0001 | < 0.0001 |
| Control 3 h (0.8 % DMSO, N = 6) | 38 ± 6 | 153 ± 33 | 0.22 ± 0.03 |
| LND 3 h (N = 8) | 48 ± 4 | 62 ± 12 | 0.44 ± 0.04 |
| Change % | 26 % | − 60 % | 97 % |
| p | < 0.05 | < 0.0001 | < 0.0001 |
Fig. 3.
Lonidamine treatment effects on the ORI redox indices (mean ± SD) of DB-1 melanoma cells in culture. Two treatment time windows: 45 min and 3 h for either lonidamine (LND, 150 μM) or control (Ctrl, 0.8% DMSO). One-way ANOVA with Bonferroni correction was used to compare the redox indices between LND and Ctrl for each window. *p < 0.05, ****p < 0.0001.
Differential Redox Response to ETC Inhibitors and Mitochondrial Uncoupler
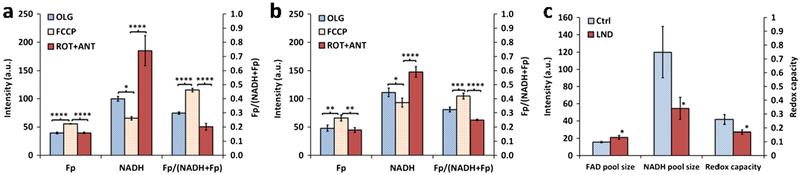
We performed mitochondrial redox capacity measurement by investigating how the redox indices of DB-1 cells respond to various perturbations of mitochondrial metabolism. As shown in Fig. 4, we quantified Fp, NADH, and the redox ratios at each stage after sequential addition of Olg, FCCP, and Rot+Ant to both LND-treated and control groups. Both groups exhibit similar patterns of change in the redox indices. Fp increased, NADH decreased, and the Fp redox ratio increased significantly due to the uncoupling effects of 8 μM FCCP. Rot and Ant inhibit complexes I and III of the ETC, respectively. As a result, NADH cannot be oxidized, leading to NADH buildup. As expected, we observed that 1 μM Rot and 1.25 μg/ml Ant markedly raised the NADH level to its maximum and decreased redox ratios significantly for both the LND-treated and control groups.
Fig. 4.
Results from redox capacity measurement on cultured DB-1 melanoma cells pretreated with either DMSO or LND. a and b Differential redox indices (mean ± SD) under sequential treatment of (i) oligomycin (Olg), (ii) mitochondrial uncoupler FCCP, (iii) rotenone plus antimycin (Rot+Ant) for a control group (0.2 % DMSO) and b LND group; one-way ANOVA with Bonferroni adjustment was used to compare the differences between Olg and FCCP and between Rot+Ant and FCCP. c Mitochondrial FAD and NADH pool sizes and the mitochondrial redox capacities (mean ± SD) for the LND and control groups; unpaired Student’s t test was used to compare the difference between LND and control groups. *p < 0.05, **p < 0.005, ***p < 0.001, ****p < 0.0001, N = 3 dishes.
However, the control and LND-treated group exhibited a differential response to the ETC inhibitors and the mitochondrial uncoupler. NADH increase caused by Rot+Ant was much larger in the control group than that in the LND-treated group. By subtracting the minimum NADH (after FCCP addition) from this maximum NADH, we obtain the mitochondrial NADH pool size (mtNADH pool) [42]. For the control group, the mtNADH pool is 120 a.u. (arbitrary unit), whereas it is 55 a.u. for the LND-treated group (~ 100 min treatment time). Thus, LND treatment decreased mitochondrial NADH pool size by 65 units, approximately 55 % decrease (p = 0.046, Fig. 4c). Note that this analysis is based on the working assumption that mitochondrial NADH contributes to the majority of signal (as demonstrated previously [11, 43, 44]), and these inhibitors are relatively specific for perturbing mitochondrial pathways. However, considering that the redox shuttles couple NADH between mitochondria and cytosol [45, 46], the latter might contribute to NADH fluorescence changes as well. This remains to be investigated in the future.
Since FCCP allows only mitochondrial FAD (mtFAD) to increase, and Rot+Ant triggers only mtFAD to decrease due to the inhibition of respiration [42], the mtFAD pool size obtained from the Fp signal difference under these two conditions is 16 and 21 a.u. for the control and treated groups, respectively, differing by ~ 30 % (p = 0.032, Fig. 4c). Thus, LND treatment significantly increased the mitochondrial FAD pool size.
By subtracting the Fp redox ratio after Rot+Ant addition from that after FCCP addition, we obtained the Fp redox ratio difference, which we herein define as the mitochondrial redox capacity (mtRedoxCap). LND-treated cells had ~ 35% lower mtRedoxCap (p = 0.033), indicating LND significantly decreased the mtRedoxCap of DB-1 cells (Fig. 4c).
In summary, LND treatment affected DB-1 cell’s mitochondrial function of maintaining redox balance by decreasing its reducing agent mitochondrial NADH pool size and increasing its oxidizing agent mitochondrial FAD pool size, accompanied with a decreased mitochondrial redox capacity.
LND Treatment Effects on DB-1 Xenografts
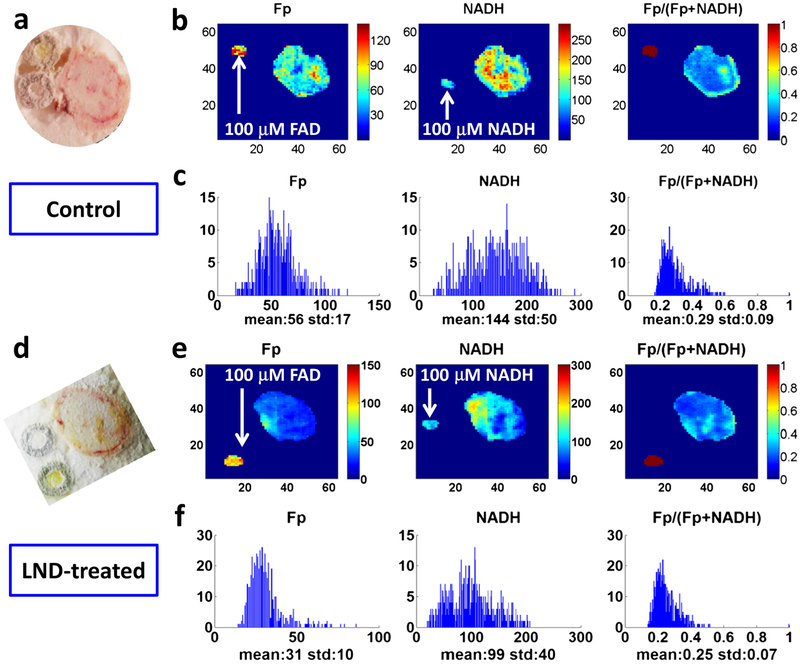
We examined how LND affects the DB-1 tumor redox state. Fig. 5 displays typical redox images (Fp and NADH nominal concentrations and redox ratio Fp/(Fp+NADH)) and the white light images of a tissue section from a control and a treated xenograft at a similar tissue depth.
Fig. 5.
Redox images and their corresponding histograms of typical sections in DB-1 melanoma xenografts. a–c A section of a control DB-1 tumor at 1100 μm depth. a White light photo and b images of Fp, NADH, and Fp/(Fp+NADH) corresponding to the section. The image matrices were 64 × 64, and the step size was 200 μm. The color bars of the Fp and NADH images indicate the nominal concentrations in micromolar referenced to the corresponding snap-frozen solution standards. The color bar of the Fp redox ratio image indicates the ratio range from 0 to 1. c The x-axes of the histograms represent the nominal concentration of Fp or NADH or the Fp redox ratio in that section. The y-axes of the histograms represent the number of pixels having a specific value of Fp or NADH or the Fp redox ratio. d–f A section of a LND-treated DB-1 tumor at 1200 μm depth. d–f as described for a–c.
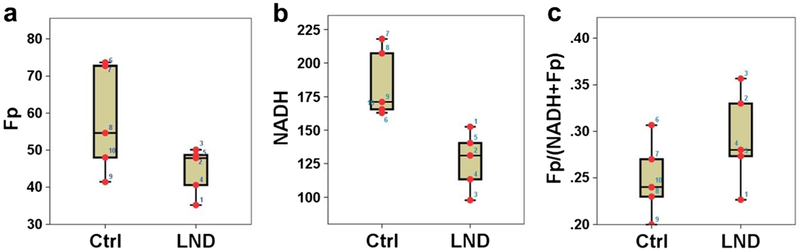
By global averaging over multiple sections of individual tumors, we obtained the mean Fp, NADH and redox ratio before and after the treatment (Table 2). We found a 31 % decrease in NADH due to LND treatment (p = 0.005). Fig. 6 shows the dotted boxplots of 45-min LND treatment effects on the redox state of DB-1 tumors.
Table 2.
45-min LND treatment effects on DB-1 melanoma tumors (mean ± SD)
| Fp (μM) | NADH (μM) | Fp/(NADH+Fp) | Vol (mm3) | |
|---|---|---|---|---|
| Control (n = 5) | 58 ± 15 | 185 ± 26 | 0.25 ± 0.04 | 174 ± 67 |
| Treated (n = 5) | 44 ± 6 | 127 ± 22 | 0.29 ±0.05 | 162 ± 84 |
| Change % due to treatment | − 24 % | − 31 % | 16 % | − 6.9% |
| p value (unpaired Student t test) | 0.11 | 0.0051 | 0.17 | 0.82 |
Fig. 6.
Effects of 45-min LND treatment on the redox indices a Fp, b NADH, and c Fp/(NADH+Fp) of DB-1 melanoma tumors shown in dotted box plots, which indicate the lower adjacent value, first quartile, median, third quartile, and upper adjacent value for the control (Ctrl) and treated (LND) groups with the numbers indicating individual tumors.
We also analyzed the section image data using LMM analysis with repeated measures (the tissue section depth and tumor volume were set as covariates). As shown in Table S1 in Supplementary Material, the LMM analysis shows that LND treatment significantly lowered NADH level (p = 0.022), consistent with the global averaging analysis. The LMM analysis also shows that LND-treated tumors had higher Fp redox ratio with a marginal significance (p = 0.096), i.e., the treated tumors were likely to be more oxidized. Additionally, it shows that LND treatment did not cause a significant change in Fp when comparing to control.
From Fig. 5, it is apparent that both control and treated tumors showed pronounced intratumor redox heterogeneity in all three indices. These heterogeneity patterns are not random and reproducible by repeatedly scanning the same tissue section or sections nearby. Previously, we have employed two-subpopulation model and Gaussian fitting to differentiate between aggressive and indolent tumors and between premalignancy and control [18, 19, 24]. Thus, although the true number of subpopulations may be more than two, here we investigate the treatment effects by using a working assumption of two subpopulations. By fitting the histograms of the globally averaged redox indices (Fp, NADH, and Fp redox ratio) with two Gaussian functions for each tumor, we identified two redox sub-populations for each redox index within the individual DB-1 tumors for both groups. Paired t tests showed that these two redox subpopulations had significant differences in their Fp, NADH, and Fp redox ratio as summarized in Table 3. The difference in the mean NADH or mean Fp between the two redox subpopulations is ~ 50 % or higher, which is larger than the measurement variability (< 20 %). These results indicate the existence of distinct redox subpopulations within individual tumors for both control and treated groups.
Table 3.
Two intratumor redox subpopulations in DB-1 melanoma xenografts (mean ± SD)
| Group | Redox indices | Sub-population 1 | Sub-population 2 | Sub-population Difference % | p |
|---|---|---|---|---|---|
| Control (N = 5) | Fp (μM) | 51 ± 9 | 82 ± 22 | 60 % | 0.008 |
| NADH (μM) | 142 ± 53 | 209 ± 47 | 47 % | 0.003 | |
| Fp redox ratio | 0.18 ± 0.05 | 0.27 ± 0.05 | 46 % | 0.0006 | |
| LND-treated (N = 5) | Fp (μM) | 30 ± 11 | 64 ± 21 | 111 % | 0.005 |
| NADH (μM) | 101 ± 15 | 187 ± 61 | 86 % | 0.021 | |
| Fp redox ratio | 0.20 ± 0.06 | 0.32 ± 0.08 | 66 % | 0.015 |
Discussion
We adopted ORI techniques to examine LND treatment effects on the redox state of DB-1 melanoma cells and tumors of matched sizes. Our results showed that 45-min LND treatment lowered NADH level at both the cellular and tissue levels. The observed responses to a single dose of drug within a short-time period support the utility of ORI as a suitable assay for early detection of therapeutic effects in cancer.
It is known that LND sensitizes tumor cells including DB-1 cells to chemo- and radio-therapies by targeting the metabolic pathways in cancer cells [47]. The mechanistic studies showed that LND inhibits the MCT1 and MCT4 to impede L-lactic acid efflux and blocks the mitochondrial pyruvate carrier (MPC) to limit pyruvate export from the cytosol into the mitochondria, thus resulting in cytosolic acidification [47, 48]. Our observation of decreased NADH after LND treatment is consistent with the results of limited pyruvate uptake by the mitochondria resulting in decreased NADH production. Furthermore, LND suppresses the pentose phosphate pathway [48, 49], reducing NADPH production. Since both NADH and NADPH have essentially the same fluorescence spectrum, the ORI techniques we employed for this investigation do not differentiate them. Therefore, NADPH decrease might also contribute to the observation of decreased blue fluorescence. Additionally, LND treatment increases glutaminolysis and reductive carboxylation flux from alpha-ketoglutarate to isocitrate in the TCA cycle [48]. Since this reversal flux from alpha-ketoglutarate to isocitrate consumes NAD(P)H, increased reductive carboxylation by LND may also contribute to the observation of a decreased NADH level. The dosage of 150 μM we used for cell culture study is an optimized dose based on our previous studies [48, 49]. Mitochondrial complex II has the lowest affinity for LND (~ 50 μM), among the key targets of LND, such as monocarboxylate transporters, mitochondrial pyruvate carrier, and mitochondrial complex II. Therefore, 150 μM (3 × 50 μM) was adopted in these studies as conventionally done. In future studies, we may perform the dose-dependent curve of the redox indices vs LND in cells.
We observed that in the presence of FCCP, NADH level is higher in LND-treated cells compared to that of control cells (93 vs 65 units, p = 0.02). It was reported that in isolated mitochondria, LND inhibits NAD-linked electron flow and when FCCP is present, LND produces much stronger respiratory inhibition compared to that in the absence of FCCP [33]. Thus, in our case, when FCCP is present in LND-treated cells, the observed NADH level is the balance between two actions of LND. On the one hand, LND causes a NADH decrease in the TCA cycle by inhibiting MPC; on the other hand, it increases NADH level by its enhanced inhibitory effect on the ETC. The final result is an increase in NADH in LND-treated cells compared to control cells when both cells are under FCCP treatment.
We did not observe a significant change in Fp after the 45-min treatment with LND. It has been shown that Fp signals originate mainly from three components isolated in the mitochondria [7, 9, 50]: α-lipoamide dehydrogenase flavin, electron transfer flavoprotein of the fatty-acid-oxidizing system, and dithionite-reducible flavoproteins that are not involved in the flavoprotein fluorescence changes associated with the ETC. Other mitochondrial flavoproteins, such as succinate dehydrogenase (SDH), NADH dehydrogenase, α-glycerophosphate dehydrogenase, proline dehydrogenase, and choline oxidase, have a negligible effect on the overall Fp signals. It was recently demonstrated that LND affects mitochondrial complex II by inhibiting succinate-ubiquinone reductase activity without completely blocking the activity of SDH [48]. Although the FAD/FADH2 pair is involved in the SDH-catalyzed interconversion of fumarate and succinate, this FAD does not contribute significantly to the total Fp signals [9]. This may partially explain why we did not observe a change in Fp fluorescence after a 45-min LND treatment. However, with 3-h prolonged treatment, Fp increased by 26 % compared to that of 3-h control, which suggests a time-dependent effect of LND treatment on Fp.
The observed higher mtFAD pool size and lower mtNADH pool size following LND treatment indicate that mitochondria partially lost their reducing capacity and redox power. Since LND decreased the mtNADH pool size, the treated cells had a compromised reducing capacity to balance the effects of ROS. Decreased mitochondrial redox capacity (mtRedoxCap) might indicate that the treated cancer cells had a compromised capability to survive therapeutic treatment. This is in line with the reported effects of LND on sensitizing cancer to anti-tumor drugs. LND treatment shifted DB-1 cells to a more oxidized state, which might indicate more ROS, apoptosis, or suppressed proliferation [51].
Diverse effects of DMSO have been reported, and even very low doses of DMSO affect cells profoundly, ranging from change of cell morphology, attachment, and viability, as well as alteration in gene expression and protein content and functionality [52–54]. In our study, DMSO was used as a solvent for LND (and some other chemical agents such as Rot and FCCP). With use of DMSO, we observed immediate morphological changes. NADH levels were significantly raised with 0.8 % of DMSO but 0.2 % DMSO (the lowest level limited by LND solubility) did not have a significant effect on the NADH level (p = 0.8). Despite its effects on cells, DMSO should not significantly affect the main results of LND treatment in our experiments since sham treatment using the same amount of DMSO was applied to the control group. To confirm this, we also used 0.2 % DMSO and observed similar results of LND treatment compared to 0.8 % DMSO (data not shown). However, 0.2 % or lower is recommended for future redox imaging experiments.
The Chance redox scanner is capable of scanning frozen tissue immersed in liquid nitrogen and revealing treatment effects of redox changes at various tissue depths. Previously, we reported that ORI of deep tissue using the Chance redox scanner [37, 38] was able to detect CHOP (Cyclophosphamide + Hydroxydoxorubicin + Oncovin + Prednisone) treatment effects on lymphoma xenografts [25]. We observed a significant decrease in global Fp content and a more reduced redox state of lymphoma following CHOP treatment. In this study, LND treatment decreased NADH content and caused an opposite trend of change for the mitochondrial redox state of melanoma with increased Fp redox ratio, which are consistent with those obtained with cells. Whether cancer cells become more reduced or more oxidized upon drug exposure depends on how the drug targets the cells. Tumors are complex with inter- and intratumor heterogeneity and may respond differently to different drugs and even to the same drug. Also different drugs have different targets and mechanisms which may involve different signaling/metabolic pathways. Therefore, it appears to be difficult to have a unified redox response for different treatments.
In this study, there is a significant increase in Fp level in cultured cells after 3 h prolonged treatment, which was not observed in the 45-min window. Similarly, we did not observe a significant change of Fp in tumors treated for 45 min (3 h treatment not performed) although Fp mean value of the treated group was slightly higher than that of control group. It should be noted that the microenvironment is very different between 2D culture and tissue. Stromal cells also likely play significant roles. It would not be surprising even if there was a discrepancy in Fp results between these two models.
It is known that intratumor heterogeneity contributes to drug resistance [55, 56]. In this study, ORI of DB-1 xenografts revealed high levels of intratumor redox heterogeneity. Two distinct redox sub-populations have been identified for both the control and treated melanomas. In our previous publications, we have reported intratumor redox heterogeneity and that two-subpopulation modeling of the redox ratio histogram could differentiate tumor aggressiveness [18, 19], and differentiate between premalignant and control pancreases [24]. Although with the current study design, it is not possible to determine how individual redox sub-populations were affected by LND, it is clear that LND did not diminish the redox heterogeneity (no significant difference in the mean values of the standard deviations for all redox indices shown in Table 2). In the future, it would be of interest to investigate how these redox subpopulations respond to LND treatment specifically.
Conclusions
We demonstrated the utility of ORI for evaluating LND effects on the redox state of human melanoma DB-1 cells at both cellular and tissue levels. We found that, within a 45-min treatment window, LND significantly decreased NADH levels of both cultured cells and tumors without significantly affecting Fp level. LND shifted cultured cells to a more oxidized redox state; the Fp redox ratio of xenografts also had a trend of increase. Three-hour prolonged treatment of cell cultures had similar effects on NADH and the redox ratio except that a significant increase of Fp was observed. These results are consistent with the known activities of LND. We also found for the first time that LND decreased the mitochondrial reducing capacity and redox capacity in vitro, which contributes to the current knowledge on the action of LND. Our investigations support the utility of ORI as metabolic biomarkers for early therapeutic effects of anticancer drugs and for functional impairment in mitochondria.
Supplementary Material
Acknowledgements.
This work was supported by the NIH Grants R01CA155348 (L.Z. Li), R01CA191207 (L.Z. Li), R01-CA129544 (J. Glickson), and R01CA172820 (J. Glickson). We would also like to thank Ms. Lily Moon for technical assistance with mouse xenografts.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s11307-018-1258-z) contains supplementary material, which is available to authorized users.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.DeBerardinis RJ, Chandel NS (2016) Fundamentals of cancer metabolism. Sci Adv 2:e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chance B, Cohen P, Jobsis F, Schoener B (1962) Intracellular oxidation-reduction states in vivo. Science 137:499–508 [DOI] [PubMed] [Google Scholar]
- 3.Chance B, Ernster L, Garland PB, Lee CP, Light PA, Ohnishi T, Ragan CI, Wong D (1967) Flavoproteins of the mitochondrial respiratory chain. Proc Natl Acad Sci U S A 57:1498–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hassinen I, Chance B (1968) Oxidation-reduction properties of the mitochondrial flavoprotein chain. Biochem Biophys Res Commun 31:895–900 [DOI] [PubMed] [Google Scholar]
- 5.Garland PB, Chance B, Ernster L, Lee CP, Wong D (1967) Flavoproteins of mitochondrial fatty acid oxidation. Proc Natl Acad Sci U S A 58:1696–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ragan CI, Garland PB (1969) The intra-mitochondrial localization of flavoproteins previously assigned to the respiratory chain. Eur J Biochem 10:399–410 [DOI] [PubMed] [Google Scholar]
- 7.Kunz WS (1986) Spectral properties of fluorescent flavoproteins of isolated rat liver mitochondria. FEBS Lett 195:92–96 [DOI] [PubMed] [Google Scholar]
- 8.Kunz WS (1988) Evaluation of electron-transfer flavoprotein and alpha-lipoamide dehydrogenase redox states by two-channel fluorimetry and its application to the investigation of beta-oxidation. Biochim Biophys Acta 932:8–16 [DOI] [PubMed] [Google Scholar]
- 9.Kunz WS, Kunz W (1985) Contribution of different enzymes to flavoprotein fluorescence of isolated rat liver mitochondria. Biochim Biophys Acta 841:237–246 [DOI] [PubMed] [Google Scholar]
- 10.Chance B, Schoener B, Oshino R, Itshak F, Nakase Y (1979) Oxidation-reduction ratio studies of mitochondria in freeze-trapped samples. NADH and flavoprotein fluorescence signals. J Biol Chem 254:4764–4771 [PubMed] [Google Scholar]
- 11.Heikal AA (2010) Intracellular coenzymes as natural biomarkers for metabolic activities and mitochondrial anomalies. Biomark Med 4:241–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varone A, Xylas J, Quinn KP, Pouli D, Sridharan G, McLaughlin-Drubin ME, Alonzo C, Lee K, Munger K, Georgakoudi I (2014) Endogenous two-photon fluorescence imaging elucidates metabolic changes related to enhanced glycolysis and glutamine consumption in precancerous epithelial tissues. Cancer Res 74:3067–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozawa K, Chance B, Tanaka A, Iwata S, Kitai T, Ikai I (1992) Linear correlation between acetoacetate/beta-hydroxybutyrate in arterial blood and oxidized flavoprotein/reduced pyridine nucleotide in freeze-trapped human liver tissue. Biochim Biophys Acta 1138:350–352 [DOI] [PubMed] [Google Scholar]
- 14.Chance B, Baltscheffsky H (1958) Respiratory enzymes in oxidative phosphorylation. VII Binding of intramitochondrial reduced pyridine nucleotide. J Biol Chem 233:736–739 [PubMed] [Google Scholar]
- 15.Chance B, Schoener B (1966) Fluorometric studies of flavin component of the respiratory chain. Flavins Flavoproteins 81:510–519 [Google Scholar]
- 16.Xu HN, Tchou J, Feng M, Zhao H, Li LZ (2016) Optical redox imaging indices discriminate human breast cancer from normal tissues. J Biomed Opt 21:114003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirkpatrick ND, Brewer MA, Utzinger U (2007) Endogenous optical biomarkers of ovarian cancer evaluated with multiphoton microscopy. Cancer Epidemiol Biomark Prev 16:2048–2057 [DOI] [PubMed] [Google Scholar]
- 18.Li LZ, Zhou R, Xu HN, Moon L, Zhong T, Kim EJ, Qiao H, Reddy R, Leeper D, Chance B, Glickson JD (2009) Quantitative magnetic resonance and optical imaging biomarkers of melanoma metastatic potential. Proc Natl Acad Sci U S A 106:6608–6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu HN, Nioka S, Glickson JD, Chance B, Li LZ (2010) Quantitative mitochondrial redox imaging of breast cancer metastatic potential. J Biomed Opt 15:036010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun N, Xu HN, Luo Q, Li LZ (2016) Potential indexing of the invasiveness of breast cancer cells by mitochondrial redox ratios. Adv Exp Med Biol 923:121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alhallak K, Rebello LG, Muldoon TJ, Quinn KP, Rajaram N (2016) Optical redox ratio identifies metastatic potential-dependent changes in breast cancer cell metabolism. Biomed Opt Express 7:4364–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostrander JH, McMahon CM, Lem S et al. (2010) Optical redox ratio differentiates breast cancer cell lines based on estrogen receptor status. Cancer Res 70:4759–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu HN, Feng M, Moon L, Dolloff N, El-Deiry W, Li LZ (2013) Redox imaging of the p53-dependent mitochondrial redox state in colon cancer ex vivo. J Innov Opt Health Sci 06:1350016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu HN, Nioka S, Li LZ (2013) Imaging heterogeneity in the mitochondrial redox state of premalignant pancreas in the pancreas-specific PTEN-null transgenic mouse model. Biomarker Res 1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu HN, Zhao H, Mir TA et al. (2013) Chop therapy induced mitochondrial redox state alteration in non-Hodgkin’s lymphoma xenografts. J Innov Opt Health Sci 6:1350011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah AT, Beckler MD, Walsh AJ, Jones WP, Pohlmann PR, Skala MC (2014) Optical metabolic imaging of treatment response in human head and neck squamous cell carcinoma. PLoS One 9:e90746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh AJ, Cook RS, Sanders ME, Aurisicchio L, Ciliberto G, Arteaga CL (2014) Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer. Cancer Res 74:5184–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh AJ, Cook RS, Manning HC, Hicks DJ, Lafontant A, Arteaga CL, Skala MC (2013) Optical metabolic imaging identifies glycolytic levels, subtypes, and early-treatment response in breast cancer. Cancer Res 73:6164–6174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alam SR, Wallrabe H, Svindrych Z, Chaudhary AK, Christopher KG, Chandra D, Periasamy A (2017) Investigation of mitochondrial metabolic response to doxorubicin in prostate cancer cells: an NADH, FAD and tryptophan FLIM assay. Sci Rep 7:10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirkpatrick ND, Zou C, Brewer MA, Brands WR, Drezek RA, Utzinger U (2005) Endogenous fluorescence spectroscopy of cell suspensions for chemopreventive drug monitoring. Photochem Photobiol 81:125–134 [DOI] [PubMed] [Google Scholar]
- 31.Cannon TM, Shah AT, Walsh AJ, Skala MC (2015) High-throughput measurements of the optical redox ratio using a commercial microplate reader. J Biomed Opt 20:010503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cervantes-Madrid D, Romero Y, Due, et al. (2015) Reviving lonidamine and 6-Diazo-5-oxo-L-norleucine to be used in combination for metabolic cancer therapy. BioMed Research International 2015:690492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Floridi A, Lehninger AL (1983) Action of the antitumor and antispermatogenic agent lonidamine on electron transport in ehrlich ascites tumor mitochondria. Arch Biochem Biophys 226:73–83 [DOI] [PubMed] [Google Scholar]
- 34.Nath K, Nelson DS, Ho A et al. (2013) 31P and 1H MRS of DB-1 melanoma xenografts: lonidamine selectively decreases tumor intracellular pH and energy status and sensitizes tumors to melphalan. NMR Biomed 26:98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nath K, Nelson DS, Heitjan DF, Zhou R, Leeper DB, Glickson JD (2015) Effects of hyperglycemia on lonidamine-induced acidification and de-energization of human melanoma xenografts and sensitization to melphalan. NMR Biomed 28:395–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nath K, Nelson DS, Putt ME, Leeper DB, Garman B, Nathanson KL, Glickson JD (2016) Comparison of the lonidamine potentiated effect of nitrogen mustard alkylating agents on the systemic treatment of DB-1 human melanoma xenografts in mice. PLoS One 11:e0157125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quistorff B, Haselgrove JC, Chance B (1985) High spatial resolution readout of 3-D metabolic organ structure: an automated, low-temperature redox ratio-scanning instrument. Anal Biochem 148:389–400 [DOI] [PubMed] [Google Scholar]
- 38.Li LZ, Xu HN, Ranji M et al. (2009) Mitochondrial redox imaging for cancer diagnostic and therapeutic studies. J Innov Optical Health Sci 2:325–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu HN, Zheng G, Tchou J, Nioka S, Li LZ (2013) Characterizing the metabolic heterogeneity in human breast cancer xenografts by 3D high resolution fluorescence imaging. SpringerPlus 2:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu HN, Zhou R, Moon L, Feng M, Li LZ (2014) 3D imaging of the mitochondrial redox state of rat hearts under normal and fasting conditions. J Innov Opt Health Sci 7:1350045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang S, Heikal AA, Webb WW (2002) Two-photon fluorescence spectroscopy and microscopy of NAD(P)H and flavoprotein. Biophys J 82:2811–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartolome F, Abramov AY (2015) Measurement of mitochondrial NADH and FAD autofluorescence in live cells. Methods Mol Biol 1264:263–270 [DOI] [PubMed] [Google Scholar]
- 43.Yu Q, Heikal AA (2009) Two-photon autofluorescence dynamics imaging reveals sensitivity of intracellular NADH concentration and conformation to cell physiology at the single-cell level. J Photochem Photobiol B Biol 95:46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blinova K, Levine RL, Boja ES, Griffiths GL, Shi ZD, Ruddy B, Balaban RS (2008) Mitochondrial NADH fluorescence is enhanced by complex I binding. Biochemistry 47:9636–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo X, Li R, Yan LJ (2015) Roles of pyruvate, NADH, and mitochondrial complex I in redox balance and imbalance in beta cell function and dysfunction. J Diabetes Res 2015:512618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berg JM, Tymoczko JL, Stryer L (2002) Many shuttles allow movement across the mitochondrial membranes In: Biochemistry. 5th edition. New York: W H freeman; section 18.5, Available from: https://www.ncbi.nlm.nih.gov/books/NBK22470/ [Google Scholar]
- 47.Nath K, Guo L, Nancolas B et al. (2016) Mechanism of antineoplastic activity of lonidamine. Biochim Biophys Acta Rev Cancer 1866:151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo L, Shestov AA, Worth AJ, Nath K, Nelson DS, Leeper DB, Glickson JD, Blair IA (2016) Inhibition of mitochondrial complex II by the anticancer agent lonidamine. J Biol Chem 291:42–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nancolas B, Guo L, Zhou R, Nath K, Nelson DS, Leeper DB, Blair IA, Glickson JD, Halestrap AP (2016) The anti-tumour agent lonidamine is a potent inhibitor of the mitochondrial pyruvate carrier and plasma membrane monocarboxylate transporters. Biochem J 473:929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunz WS, Gellerich FN (1993) Quantification of the content of fluorescent flavoproteins in mitochondria from liver, kidney cortex, skeletal muscle, and brain. Biochem Med Metab Biol 50:103–110 [DOI] [PubMed] [Google Scholar]
- 51.Heaster TM, Walsh AJ, Zhao Y, et al. (2018) Autofluorescence imaging identifies tumor cell-cycle status on a single-cell level. J Biophotonics 11:e201600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pal R, Mamidi MK, Das AK, Bhonde R (2012) Diverse effects of dimethyl sulfoxide (DMSO) on the differentiation potential of human embryonic stem cells. Arch Toxicol 86:651–661 [DOI] [PubMed] [Google Scholar]
- 53.Yuan C, Gao J, Guo J, Bai L, Marshall C, Cai Z, Wang L, Xiao M (2014) Dimethyl sulfoxide damages mitochondrial integrity and membrane potential in cultured astrocytes. PLoS One 9:e107447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang C-C, Lin S-Y, Lai Y-H, Liu YJ, Hsu YL, Chen JJW (2012) Dimethyl sulfoxide promotes the multiple functions of the tumor suppressor HLJ1 through activator Protein-1 activation in NSCLC cells. PLoS One 7:e33772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saunders NA, Simpson F, Thompson EW, Hill MM, Endo-Munoz L, Leggatt G, Minchin RF, Guminski A (2012) Role of intratumoural heterogeneity in cancer drug resistance: molecular and clinical perspectives. EMBO Mol Med 4:675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lopes-Rodrigues V, Di Luca A, Mleczko J et al. (2017) Identification of the metabolic alterations associated with the multidrug resistant phenotype in cancer and their intercellular transfer mediated by extracellular vesicles. Sci Rep 7:44541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.