Abstract
The prespliceosome, comprising U1 and U2 snRNPs bound to the pre-mRNA 5′ splice site (5′SS) and branch point sequence, associates with the U4/U6.U5 tri-snRNP to form the fully-assembled precatalytic pre-B spliceosome. Here, we report cryo-EM structures of the human pre-B complex captured before U1 snRNP dissociation at 3.3 Å core resolution, and the human tri-snRNP at 2.9 Å resolution. U1 snRNP inserts the 5′SS-U1 snRNA helix between the two RecA domains of the Prp28 DEAD-box helicase. ATP-dependent closure of the Prp28 RecA domains releases the 5′SS to pair with the nearby U6 ACAGAGA-box sequence presented as a mobile loop. The structures suggest that formation of the 5′SS-ACAGAGA helix triggers remodeling of an intricate protein-RNA network to induce Brr2 helicase relocation to its loading sequence in U4 snRNA, enabling Brr2 to unwind the U4/U6 snRNA duplex to allow U6 snRNA to form the catalytic center of the spliceosome.
Pre-mRNA splicing consists of removal of non-coding introns and ligation of coding exons by two transesterification reactions catalyzed by an immense ribonucleoprotein particle called the spliceosome. First, the pre-mRNA 5′ splice site (5′SS) is attacked by the conserved branch-point (BP) adenosine upstream of the 3′SS, yielding the cleaved 5′-exon and lariat-intron/3′-exon intermediate. Then, the freed 3′ hydroxyl of the 5′-exon attacks the 3′SS, ligating the exons and releasing the lariat intron (1–3).
The spliceosome assembles in a stepwise manner from five snRNPs, each containing an snRNA (U1, U2, U4, U5 or U6) and then undergoes an extensive remodeling process to form its active site (1–5). The accuracy of pre-mRNA splicing relies on splice site recognition by the spliceosome, which is mainly achieved through interactions with the snRNAs. The 5′SS is recognized by pairing with U1 and the BP is recognized by pairing with U2 within the prespliceosome (6), which recruits the pre-formed U4/U6.U5 tri-snRNP (7–10) to yield the fully assembled pre-B complex (11–13). In this pre-catalytic spliceosome, U6 snRNA, which will ultimately fold to form the active site, is chaperoned in an inactive conformation through pairing with U4 snRNA, and the 5′SS is still paired with U1 snRNA. Spliceosome activation is initiated by disruption of the 5′SS-U1 snRNP interaction by the DEAD-box helicase Prp28 (14) to transfer the 5′SS to the invariant ACAGAGA box of U6 snRNA and the 5′ exon to U5 snRNA loop 1. 5′SS transfer results in the formation of the B complex, and is followed by extensive reorganization of the spliceosome promoted by unwinding of the U4/U6 snRNA duplex by Brr2 helicase (15,16), which dissociates U4 snRNP and allows U6 snRNA to fold and pair with U2 snRNA, forming the catalytic center.
Over the past few years, cryo-EM studies of human and Saccharomyces cerevisiae (hereafter referred to as yeast) spliceosomes have greatly advanced our structural understanding of pre-mRNA splicing (1–3). Yet the early activation process, particularly the mechanism of Prp28-mediated 5′SS transfer from U1 snRNP to the U6 and U5 snRNAs, which ultimately ensures its correct positioning during catalysis, is poorly understood. The cryo-EM structure of yeast U4/U6.U5 tri-snRNP showed that U4 snRNA is already loaded in the active site of Brr2 helicase prior to associating with the prespliceosome to form the pre-B complex (7–9,17), whereas it was evident in the earlier cryo-EM structure of human U4/U6.U5 tri-snRNP that Brr2 is kept away from U4 snRNA by Sad1 (10) and relocates to bind U4 snRNA during the pre-B to B transition (13). In the previous cryo-EM structure of the human pre-B complex, the loosely associated U1 snRNP was located in the cryo-EM density map without a precise orientation (13).
In this study, we report high-resolution cryo-EM structures of the human tri-snRNP and pre-B spliceosome which reveal the interaction of the U1 snRNP with the tri-snRNP within pre-B as well as an intricate network of interactions between the RNA and protein components within the tri-snRNP. Our structures provide crucial insights into the mechanism of Prp28-dependent transfer of the 5′SS from U1 snRNP to the U6 snRNA ACAGAGA sequence, and how this is coupled to Brr2 relocation to allow spliceosome activation.
Structure determination of the human U4/U6.U5 tri-snRNP and pre-B complex
Human spliceosomes were assembled on an AdML pre-mRNA substrate in HeLa nuclear extract in the presence of a Prp28 helicase mutant, where the DEAD motif is replaced by AAAD and consequently ATP-dependent RecA domain closure is prevented. This mutant is dominant negative and stalls the spliceosome at the pre-B stage (Supplementary Methods; fig. S1) (11). The purified sample used for cryo-EM studies contained all five snRNAs as well as the pre-mRNA. Initial data processing revealed a mixture of pre-B complex and tri-snRNP, indicating some dissociation during sample preparation, consistent with the loose association of tri-snRNP at the pre-B stage (11) (fig. S1). Cryo-EM data analysis yielded reconstructions for tri-snRNP and the pre-B complex at average resolutions of 2.9Å and 4.0Å, respectively (Fig. 1, fig. S2 to S6, Table S1-S2, Supplementary Material PyMOL sessions 1 and 2). The resolution of the pre-B complex core was improved to 3.3Å by applying a soft mask, and focused refinement yielded reconstructions of the U1 and U2 lobes at sufficient resolution to position the two snRNPs unambiguously (fig. S6).
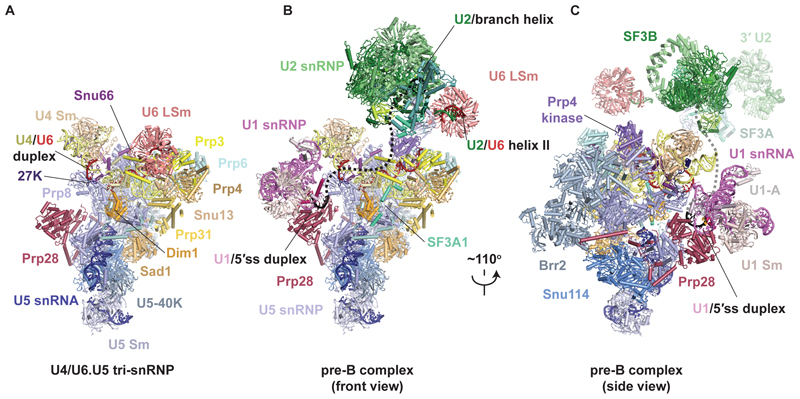
Fig. 1. Structures of human tri-snRNP and pre-B complex.
(A) Overview of human U4/U6.U5 tri-snRNP. (B and C) Overview of the human U1.U2.U4/U6.U5 pre-B complex in two orientations. The black dashed line represents the theoretical path of the pre-mRNA intron.
Organization of U4 and U6 snRNAs in human tri-snRNP
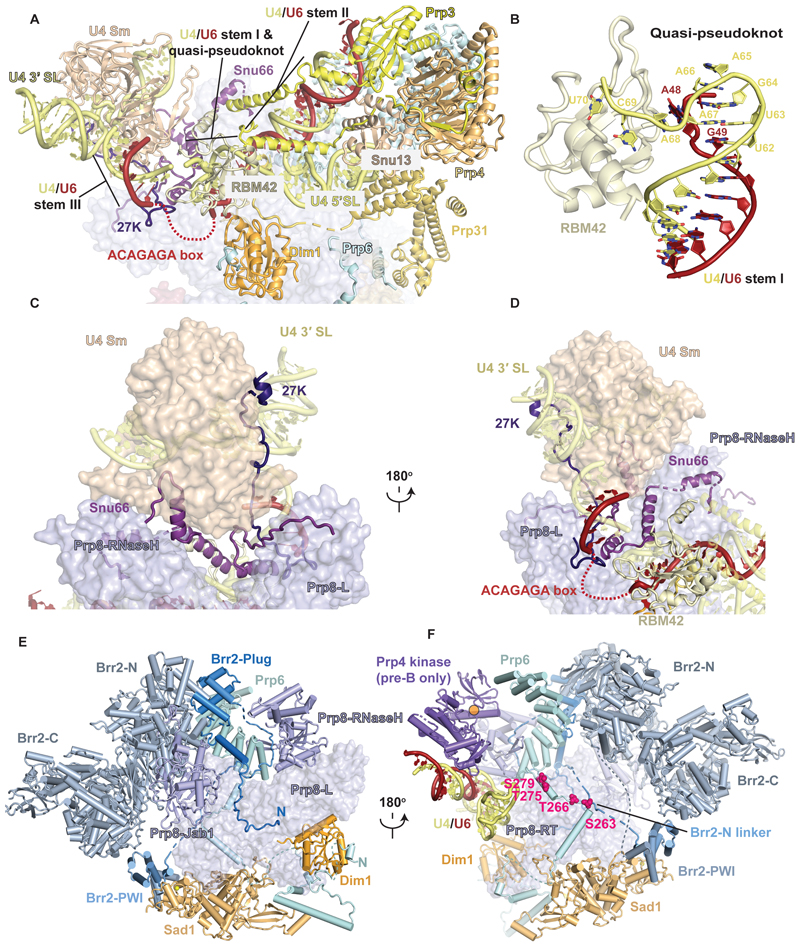
The human tri-snRNP and pre-B complex structures were previously reported at an overall resolution of 7Å (10) and 10Å (5.9Å for the pre-B core) (13). The higher resolution of our maps allowed us to build more complete models of the protein and RNA components that revealed an intricate network of interactions crucial for recognition of the 5′SS upon its transfer from U1 to U6 snRNA at the pre-B stage. The U4 snRNA nucleotides 73 – 79 base pair with nucleotides 36 – 30 of U6 snRNA (Fig. 2A, fig. S7). This helix, previously attributed incorrectly to the 5′ stem of U6 snRNA (13), is now referred to as U4/U6 stem III (18). The U4 snRNA nucleotides 63– 67 form a structure, which we refer to as a quasi-pseudoknot, stabilized by base pairing between U4 nucleotides U63 and A67 and between G64 and U6 nucleotide A47, continuous stacking of base pairs as well as cross-strand stacking of U4 A68 and U6 G49 (Fig. 2B, fig. S7, A-C). This quasi-pseudoknot is further stabilized by binding of the U4 nucleotides 68 – 70 to the β-sheet surface of the RBM42 RRM domain. The U6 A48 base is flipped out from the quasi-pseudoknot and sandwiched between Dim1 and the RBM42 RRM domain (Fig. 2 A and B). RBM42 had been identified by mass-spectrometry as an integral component of the tri-snRNP (10) and the pre-B complex (11) but had not been observed in previous cryo-EM structures (10,13).
Fig. 2. Structural features of the human tri-snRNP.
(A) Organization of U4 snRNA in the tri-snRNP prior to Brr2 relocation. SL, stem loop. The mobile ACAGAGA loop is depicted as a red dashed line. (B) Close-up view of U4/U6 stem I capped by the quasi-pseudoknot and its stabilization by RBM42 RRM. (C) and (D) U4 region in two orientations. Note how Snu66 and SNRNP-27K wrap around the U4 Sm ring, Prp8 Endonuclease and RNaseH domains, U4/U6 stem III and the quasi-pseudoknot thereby solidifying the organization of the U4 region prior to Brr2 relocation. (E) Complex interaction network of Brr2 N-terminal domain with tri-snRNP components. Brr2-C, C-terminal helicase cassette of Brr2. Brr2-N, N-terminal helicase cassette of Brr2. Prp8-L, large domain of Prp8. (F) Another view of the complex interaction network of the Brr2 N-terminal domain with tri-snRNP components. Prp8-RT, Prp8 reverse transcriptase domain. Note the extended N-terminal domain of Prp6. On this panel, the Prp4 kinase (active site is represented as an orange circle) as it interacts in pre-B and its phosphorylation targets on Prp6 (magenta spheres) are represented.
The formation of these RNA structures has important functional implications. Firstly, stem III formation occludes U4 snRNA nucleotides 68 – 81, which are loaded in the active site of Brr2 helicase after relocation in the B complex for U4/U6 duplex unwinding (13,19). Secondly, the formation of the quasi-pseudoknot brings U6 snRNA nucleotides A36 and A47 closer to enable the ACAGAGA-containing intervening sequence to loop out and project towards the U1 snRNP in the pre-B complex for 5′SS pairing (Fig. 1B and C, Fig. 2A, fig S7B).
The U4 Sm core domain is packed against the RNaseH and Endonuclease (Endo) domains of Prp8 and the U4/U6 helix stem III, and Snu66 cements the interaction between Prp8 and the U4 core domain (Fig. 2C and D). Two α-helices of Snu66 also interact extensively with the U4 snRNA quasi-pseudoknot and the U4 core domain. The C-terminus of SNRNP-27K (20) traverses from the ACAGAGA loop, interacting with Snu66 and the flat face of the U4 core domain (21), to the central hole of the Sm core domain where it interacts with U4 snRNA and projects its disordered N-terminal RS domain (Fig. 2C and D, fig. S5E). SNRNP-27K had been detected in human tri-snRNP by mass spectrometry (10) but had not been identified in previous structures. Its highly-conserved C-terminus forms a loop immediately adjacent to the flexible ACAGAGA-box, suggesting that it may contribute to orienting U6 snRNA during 5′SS transfer (Fig S7D). Indeed, mutations in this loop can activate cryptic 5′SSs that lack complementarity to U6, suggesting a role for SNRNP-27K in fidelity during 5′SS transfer (20). The extensive network of interactions described above cooperatively stabilizes the organization of the U4/U6 snRNP domain, which differs substantially from that of the yeast tri-snRNP (7–9). The U4/U6 stem III and the quasi-pseudoknot do not form in yeast (6–8) which lacks RBM42 and SNRNP-27K.
Interaction of Brr2 within human tri-snRNP
Brr2 helicase occupies two very different positions during spliceosome assembly. In yeast tri-snRNP (7–9) and in yeast (17) and human B complexes (13,19), Brr2 is loaded on U4 snRNA ready to unwind the U4/U6 duplex. In human tri-snRNP (10) and pre-B complex (13), Brr2 is kept in a pre-catalytic position away from its U4 substrate by multiple interactions (Fig. 2, E and F). Brr2 is tightly bound to the Prp8 Jab domain which interacts with the Reverse Transcriptase (RT) and Linker domains of Prp8. The Brr2 N-terminal helicase cassette is supported by Prp6 bound to the RNaseH and RT domains of Prp8, and Snu13, whereas the Brr2 C-terminal helicase cassette is packed against its PWI domain, which in turn contacts Sad1 bound to Snu114 and the Prp8 RT domain. Our high-resolution maps of the human tri-snRNP and pre-B reveal that the pre-catalytic position of Brr2 is further stabilized by numerous contacts involving its long (~400 aa) N-terminus (fig. S5D). The extreme N-terminus of Brr2 wraps around the Prp8 Large domain and forms a helix that bridges the Brr2 helicase domain to the Prp6 helical repeats, before turning back and folding into the Plug domain (22) which blocks the Brr2 N-terminal helicase cassette (Fig 2E). A long N-terminal linker connects to the PWI domain by binding first between the Prp8 RNaseH domain and the Prp6 helical repeats and then to a composite interface formed by the Prp8 RT domain and a Prp6 peptide (Fig. 2, E and F). Interestingly, this Prp6 peptide is part of an extensive N-terminal domain upstream of the Prp6 helical repeat domain that winds through the tri-snRNP and adopts a different conformation in B complex (fig. S5B, fig. S11). This complex network of interactions rationalizes the effects of various Brr2 N-terminal deletions that cause slow growth or lethality in yeast and weaken the interactions of Brr2 with the spliceosome (22).
Interaction of the Prp28 N-terminus with the spliceosome
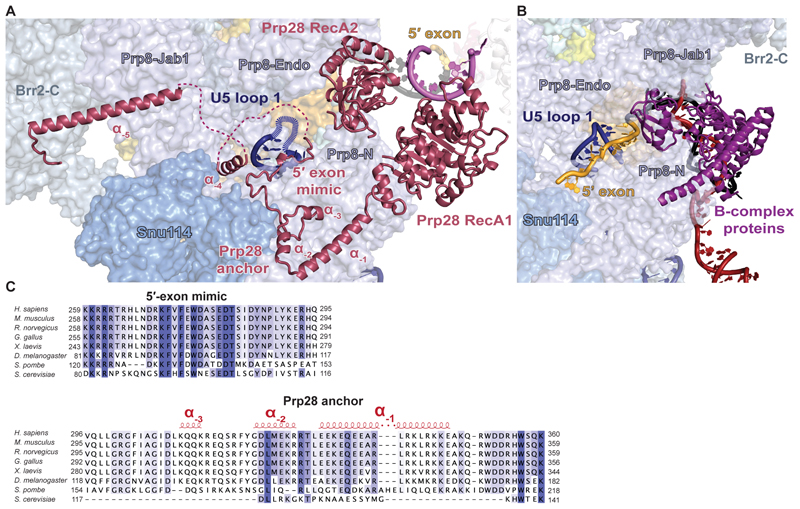
In the previous structures of human tri-snRNP (10) and pre-B complex (13) only the crystal structure of the two RecA domains was docked into the low-resolution maps. Our high-resolution map enabled us to place not only the RecA1 and RecA2 domains but also the N-terminal region of Prp28 (Prp28-N) (Fig.3, fig. S5C). Immediately upstream of the RecA1 domain, residues 286 – 356 form an “anchor” at the interface between Prp8-N and Snu114 (Fig. 3A). This anchor domain is well conserved in higher eukaryotes but largely absent in yeast (Fig. 3C), suggesting that in yeast Prp28 is recruited to the pre-B complex by a distinct mechanism. Upstream of the anchor, Prp28-N polypeptide runs along Prp8 to U5 snRNA loop 1, strikingly following the exact same path as the 5′ exon in B complex (Fig. 3, A and B). Upstream of this peptide is a disordered loop rich in basic residues that may cover the equally disordered U5 snRNA loop 1. This “5′-exon mimic” peptide is highly conserved (Fig. 3C) and essential in yeast (23). We speculate that that upon 5′SS transfer, the 5′ exon would displace Prp28-N from Prp8, weakening the interaction of Prp28 with the spliceosome and allowing its release and subsequent binding of B-complex proteins at the same position (Fig. 3B). Finally, upstream of the “5′-exon mimic” Prp28-N forms a long alpha helix that connects the Prp8 Jab1 domain and Brr2 C-terminal helicase cassette. This enhances Prp28 binding to spliceosomes where Brr2 is not relocated, and in turn means Prp28-N may stabilize Brr2 helicase in its inactive position so that dissociation of Prp28 and relocation of Brr2 are highly cooperative. Interestingly, the B-complex protein Spp381 uses the same binding surface on the Brr2 C-terminal helicase cassette (17), providing an additional example of mutually exclusive interactions of the spliceosome with Prp28 or B-complex proteins.
Fig. 3. Interaction of Prp28 with the tri-snRNP.
(A) Interaction of the N-terminus of Prp28 with the tri-snRNP. Brr2-C, C-terminal helicase cassette of Brr2. Prp8-N, N-terminal domain of Prp8. (B) Same view but of the 5′ exon and B-complex proteins in human B complex (PDB 6AHD). (C) Sequence alignment of Prp28 N-termini from different species.
Structure of the pre-B spliceosome
In the fully assembled pre-B complex, U2 snRNP resides on top of tri-snRNP, flexibly tethered by the newly formed U2/U6 helix II (11) and U6 LSm ring, which moves 90 Å relative to its tri-snRNP position and weakly binds SF3B1 (Fig. 1, B and C). Our high-resolution structure revealed an additional docking point of U2 snRNP to tri-snRNP, mediated by the long C-terminus of SF3A1 (hPrp21) which loosely binds along Prp4 and the U4 5′ stem-loop before forming a stable anchor between Dim1 and Prp8, consistent with cross-linking data (10) (Fig. 1, B and C, fig. S6E). This contact corresponds to an unassigned helix in human B complex (11,13), suggesting that it persists until Brr2-mediated U4/U6 unwinding. The structure of tri-snRNP remains largely unchanged upon pre-B formation except that the only pre-B–specific factor, Prp4 kinase, binds in the space between the Prp6 helical repeats, U4/U6 duplex and Prp8 RNaseH domain (Fig. 1A-C, 2F, fig. S6D, S11). In that location, Prp4 kinase is ideally positioned to phosphorylate its known substrates Prp6 and Prp31 during the pre-B to B transition ((Fig. 1A-C, fig. S11) (24).
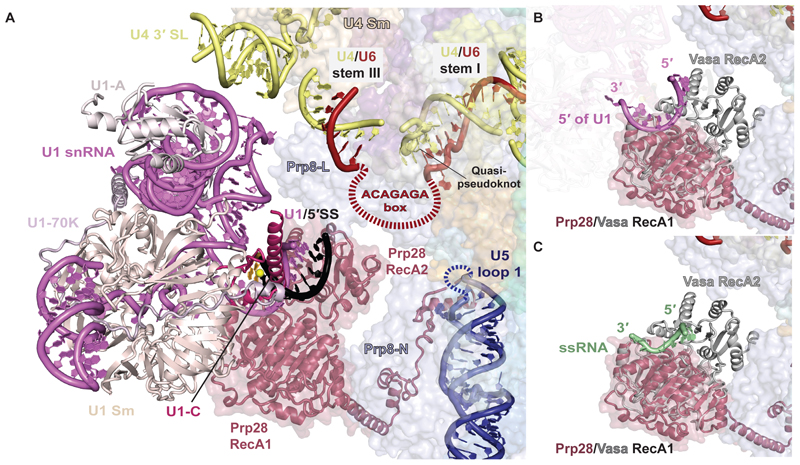
In the pre-B complex the U1 snRNP is highly mobile, which previously precluded its unambiguous assignment (13), and has poorly defined density even in subclasses of pre-B particles that nonetheless show strong signal for U2 snRNP (fig. S8). Hence, U1 snRNP is mainly anchored to tri-snRNP via the pre-mRNA substrate in pre-B and the interactions of U1 snRNP with tri-snRNP are transient (fig. S8). By extensive 3D classification, we were able to locate a subset of particles where U1 snRNP is stably bound (fig. S3 and S8), enabling us to position it unambiguously (Fig. 1, B and C, 4A, fig S6). When in contact with tri-snRNP, the U1 snRNP inserts between the U4 Sm core and Prp28 RecA1, placing U1 snRNA stem III between the U4 snRNA 3′ stem loop and the Prp8 Endo domain, while the U1 Sm proteins SmE and SmG contact Prp28 RecA1 (Fig. 1, B and C, 4A). Although the U1 and U2 snRNPs are not in direct contact in the pre-B complex, their relative orientation is similar to what is observed in the yeast prespliceosome (6), suggesting they may be bridged by auxiliary U1 components such as the PRPF39 homodimer when U1 snRNP is first presented to tri-snRNP as part of the prespliceosome (fig. S9).
Fig. 4. Binding of U1 snRNP to tri-snRNP in pre-B complex.
(A) Close-up view of the U1 / tri-snRNP interface in pre-B complex. The mobile ACAGAGA loop is depicted as a red dashed line. The disordered U5 snRNA loop 1 is depicted as a blue dashed line. The 5′ splice site (5′SS) is in striking proximity to the ACAGAGA sequence, looped out between the quasi-pseudoknot and U4/U6 stem III, and U5 snRNA loop 1. (B) A model of Prp28 with the 5′ end of U1 snRNA after ATP-dependent closure of the two RecA domains and the 5′SS release. The RecA1 domain of the closed form of the Vasa DEAD-box helicase was overlaid on the RecA1 domain of Prp28, showing the position of the 5′ end of U1 snRNA. (C) Superposition of the closed form of the Vasa DEAD-box helicase on Prp28 RecA1, showing the position of the ssRNA as observed in the original crystal structure (26). Note that the ssRNA and the 5′ end of U1 snRNA occupy a similar position suggesting that Prp28 disrupts the U1/5′SS duplex by binding the U1 snRNA, thereby freeing the pre-mRNA 5′SS to pair with the free ACAGAGA sequence.
U1/5′SS duplex disruption by Prp28 helicase in pre-B complex
Docking the U1 snRNP structure into the low-resolution map docks the U1/5′SS duplex directly into the RNA-binding pocket of the Prp28 RecA1 domain and in close proximity to the 5′SS acceptors: U5 snRNA loop 1 and the U6 snRNA ACAGAGA-loop (Fig. 4A). The presence of U1 snRNP induces a movement of RecA2, which is brought closer to RecA1 thus clamping the U1/5′SS duplex (fig. S10). In our structure, the U1/5′SS duplex is still intact, consistent with stalling at the pre-B stage. This implies that we do not observe a fully closed conformation due to our use of a DEAD-box mutant of Prp28 that is deficient in proper ATP binding (23), a canonical prerequisite for helicase domain closure (25).
To gain insight into U1/5′SS disruption by Prp28, we superimposed the structure of the DEAD-box helicase Vasa (26) trapped in a closed conformation bound to single-stranded RNA (ssRNA), believed to represent a post-duplex disruption state of the enzyme where the two RecA domains bind and distort one RNA strand to induce release of the other (Fig. 4, B and C). This superimposition aligns the Vasa-bound ssRNA with the U1 snRNA strand of the U1/5′SS duplex, identifying U1 snRNA as the strand gripped by the Prp28 RecA domains upon closure, which frees the 5′SS from its U1 chaperone (Fig. 4, B and C). The close proximity of the freed 5′SS to the mobile U6 snRNA ACAGAGA box and U5 snRNA loop 1, and the residues of the Prp8 Endo domain that recognize nucleotides +1 and +2 of the intron in B complex (13,19), likely ensures that once the duplex is melted, a valid 5′SS can be directly transferred before it reanneals with U1 snRNA.
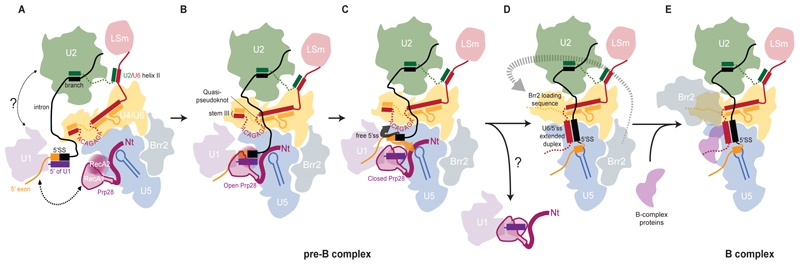
Model for 5′-splice site transfer to U6 snRNA and Brr2 relocation
Comparison of the structures of human tri-snRNP, pre-B complex, and B complex suggests a plausible mechanism for pre-B complex activation (Fig. 5 and Movie S1). At the pre-B stage, U1 snRNP is loosely tethered to the spliceosome and contacts tri-snRNP transiently (Fig. 5A). When U1 snRNP docks into the tri-snRNP, the RecA domains of Prp28 bind ATP and clamp around the 5′ end of U1 snRNA, disrupting the U1/5′SS duplex and delivering the 5′SS to U6 (Fig. 5, B and C). While the ACAGAGA box is fully exposed in pre-B, U5 snRNA loop 1 and the adjacent exon-binding channel of Prp8 are blocked by the N-terminal domain of Prp28, suggesting that initial 5′SS annealing occurs at the ACAGAGA box. This initial recognition may involve only a few nucleotides (fig. S7) but the ACAGAGA helix will extend to that observed in the B complex during activation (13,19). Converting the flexible ACAGAGA loop into a growing helix would increasingly destabilize and unfold the quasi-pseudoknot, inducing dissociation of RBM42. Pairing of the 5′SS with U6 snRNA would also destabilize stem III and displace U4 snRNA from U6 snRNA thereby freeing the Brr2 loading sequence (Fig. 5D). Moreover, the unwinding of stem III and conformational change of U4 and U6 snRNAs would destabilize the Snu66 and SNRNP-27K interaction networks and liberate the U4 core domain and the RNaseH and Endo domains of Prp8. The rotation of the RNaseH domain disrupts numerous interactions that kept Brr2 in its inactive position (fig. S11) allowing the helicase to relocate and bind U4 snRNA for unwinding. Dissociation of the Brr2 C-terminal helicase cassette would destabilize the PWI domain and facilitate Sad1 dissociation. Concomitantly the 5′ exon displaces Prp28-N from U5 snRNA and Prp8, which would trigger Prp28 and U1 snRNP dissociation and allow B-complex protein recruitment (Fig. 5, D and E).
Fig. 5. Schematic representation of pre-B complex activation.
(A) Association of the prespliceosome with the tri-snRNP. (B) Docking of U1 snRNP inserts the 5′SS-U1 snRNA duplex between the two RecA domains of Prp28. (C) Transfer of the 5′SS to the U6 snRNA ACAGAGA-loop destabilizes the binding of U1 snRNP and Prp28. (D) Formation of the 5′SS-ACAGAGA helix and dissociation of Prp28 induce movement of the U4 core domain and relocation of the Brr2 helicase. (E) Binding of Brr2 to the single stranded region of U4 snRNA made accessible by the 5′SS induced reorganization of the U4-U6 snRNA interaction.
It is intriguing that the sole binding of the 5′SS can trigger such a large reorganization of multiple protein and RNA components in agreement with (11). This transition can occur without ATP-fueled unwinding of RNA (11). Release of the 5′SS from U1 snRNP is triggered by ATP-dependent closure of the two RecA domains and ATP hydrolysis may only be necessary for separation of the two RecA domains to release the 5′ end of U1 snRNA from Prp28 (25).
Overall, our pre-B and tri-snRNP structures reveal the mechanism of 5′SS delivery, identify the binding site for U1 snRNP in the fully-assembled spliceosome and show how a DEAD-box helicase can act within a large ribonucleoprotein complex. The structures show how dramatic compositional and conformational changes in the spliceosome are coupled to crucial checkpoints in splice site recognition, and provide a structural framework for further biochemical analysis of this process.
Supplementary Material
One Sentence Summary.
Structure of a fully assembled pre-catalytic human spliceosome explains catalytic center formation.
Acknowledgments
We thank G. Cannone, S. Chen, G. McMullan, J. Brown, J. Grimmett, and T. Darling for smooth running of the EM and computing facilities; R. Thompson and D. Maskell for assistance with data collection at Leeds and D. Chirgadze at University of Cambridge; the mass spectrometry facility for help; and the members of the spliceosome group for critical reading of the manuscript. We thank in particular Pei-Chun Lin for help and advice. We thank J. Löwe, D. Barford, S. Scheres and R. Henderson for their continuing support.
Funding: The project was supported by the Medical Research Council (MC_U105184330) and European Research Council Advanced Grant (AdG-693087-SPLICE3D). C.C. was supported by EMBO and Marie Skłodowska-Curie fellowships. M.E.W. was supported by a Cambridge-Rutherford Memorial PhD Scholarship.
Footnotes
Author contributions: C.C. and M.E.W. established complex preparation. C.C. purified the sample. C.C. and M.E.W. made EM grids, collected and processed EM data, and built the model. M.E.W. refined the structure. C.C., M.E.W. and K.N. analyzed the structure and wrote the manuscript. K.N. coordinated the spliceosome project.
Competing interests: Authors declare no competing interests.
Data and materials availability: Cryo-EM maps are deposited in the Electron Microscopy Data Bank under accession numbers EMD-4658 (tri-snRNP overall map), EMD-4672, EMD-4673, EMD-4674, EMD-4675, EMD-4676 (the five tri-snRNP multibody maps), EMD-4686 (tri-snRNP after classification on Prp28), EMD-4665 (pre-B core), EMD-4687 (pre-B with stable U1 snRNP), EMD-4688 (pre-B with stable Prp4 kinase), EMD-4689 (pre-B focused refinement on U1 snRNP), and EMD-4690 (pre-B focused refinement on SF3B). The atomic models are deposited in the Protein Data Bank under accession 6QW6 (tri-snRNP) and 6QX9 (pre-B complex).
References
- 1.Plaschka C, Newman AJ, Nagai K. Structural basis of Nuclear pre-mRNA splicing: lessons from yeast. Cold Spring Harb Perspect Biol. doi: 10.1101/cshperspect.a032391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan C, Wan R, Shi Y. Molecular mechanisms of pre-mRNA splicing through structural biology. Cold Spring Harb Perspect Biol. doi: 10.1101/cshperspect.a032409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kastner B, Will CL, Stark H, Lührmann R. Structural Insights into Nuclear pre-mRNA Splicing in Higher Eukaryotes. Cold Spring Harb Perspect Biol. doi: 10.1101/cshperspect.a032417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steitz TA, Steitz JA. A, general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013;503:229–234. doi: 10.1038/nature12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plaschka C, Lin PC, Charenton C, Nagai K. Prespliceosome structure provides insights into spliceosome assembly and regulation. Nature. 2018;559:419–422. doi: 10.1038/s41586-018-0323-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen TH, Galej WP, Bai XC, Savva CG, Newman AJ, Scheres SH, Nagai K. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature. 2015;523:47–52. doi: 10.1038/nature14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen HDT, Galej WP, Bai XC, Oubridge C, Newman AJ, Scheres SHW, Nagai K. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature. 2016;530:298–302. doi: 10.1038/nature16940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan R, Yan C, Bai R, Wang L, Huang M, Wong CC, Shi Y. The 3.8 Å structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science. 2016;351:466–475. doi: 10.1126/science.aad6466. [DOI] [PubMed] [Google Scholar]
- 10.Agafonov DE, Kastner B, Dybkov O, Hofele RV, Liu WT, Urlaub H, Lührmann R, Stark H. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science. 2016;351:1416–1420. doi: 10.1126/science.aad2085. [DOI] [PubMed] [Google Scholar]
- 11.Boesler C, Rigo N, Anokhina MM, Tauchert MJ, Agafonov DE, Kastner B, Urlaub H, Ficner R, Will CL, Lührmann R. A spliceosome intermediate with loosely associated tri-snRNP accumulates in the absence of Prp28 ATPase activity. Nat Commun. 2016;7 doi: 10.1038/ncomms11997. 11997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai R, Wan R, Yan C, Lei J, Shi Y. Structures of the fully assembled Saccharomyces cerevisiae spliceosome before activation. Science. 2018;360:1423–1429. doi: 10.1126/science.aau0325. [DOI] [PubMed] [Google Scholar]
- 13.Zhan X, Yan C, Zhang X, Lei J, Shi Y. Structures of the human pre-catalytic spliceosome and its precursor spliceosome. Cell Res. 2018;28:1129–1140. doi: 10.1038/s41422-018-0094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staley JP, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 15.Laggerbauer B, Achsel T, Lührmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci USA. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 17.Plaschka C, Lin PC, Nagai K. Structure of a pre-catalytic spliceosome. Nature. 2017;546:617–621. doi: 10.1038/nature22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakab G, Mougin A, Kis M, Pollák T, Antal M, Branlant C, Solymosy F. Chlamydomonas U2, U4 and U6 snRNAs. An evolutionary conserved putative third interaction between U4 and U6 snRNAs which has a counterpart in the U4atac-U6atac snRNA duplex. Biochimie. 1997;79:387–95. doi: 10.1016/s0300-9084(97)86148-2. [DOI] [PubMed] [Google Scholar]
- 19.Bertram K, Agafonov DE, Dybkov O, Haselbach D, Leelaram MN, Will CL, Urlaub H, Kastner B, Lührmann R, Stark H. Cryo-EM Structure of a Pre-catalytic Human Spliceosome Primed for Activation. Cell. 2017;170:701–713. doi: 10.1016/j.cell.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Zahler AM, Rogel LE, Glover ML, Yitiz S, Ragle JM, Katzman S. SNRP-27, the C. elegans homolog of the tri-snRNP 27K protein, has a role in 5′ splice site positioning in the spliceosome. RNA. 2018;24:1314–1325. doi: 10.1261/rna.066878.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leung AK, Nagai K, Li J. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature. 2011;473:536–539. doi: 10.1038/nature09956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Absmeier E, Wollenhaupt J, Mozaffari-Jovin S, Becke C, Lee CT, Preussner M, Heyd F, Urlaub H, Lührmann R, Santos KF, Wahl MC. The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation. Genes Dev. 2015;29:2576–87. doi: 10.1101/gad.271528.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacewicz A, Schwer B, Smith P, Shuman S. Crystal structure, mutational analysis and RNA-dependent ATPase activity of the yeast DEAD-box pre-mRNA splicing factor Prp28. Nucleic Acids Res. 2014;42:12885–12898. doi: 10.1093/nar/gku930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider M, Hsiao HH, Will CL, Giet R, Urlaub H, Lührmann R. Human PRP4 kinase is required for stable tri-snRNP association during spliceosomal B complex formation. Nat Struct Mol Biol. 2010;17:216–221. doi: 10.1038/nsmb.1718. [DOI] [PubMed] [Google Scholar]
- 25.Mallam AL, Del Campo M, Gilman B, Sidote DJ, Lambowitz AM. Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p. Nature. 2012;490:121–125. doi: 10.1038/nature11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 27.Galej WP, Oubridge C, Newman AJ, Nagai K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature. 2013;493:638–643. doi: 10.1038/nature11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu TP, Ruan KC, Liu WY. A fluorescence-labeling method for sequencing small RNA on polyacrylamide gel. Nucleic Acids Res. 1996;24:3472–3473. doi: 10.1093/nar/24.17.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayeda A, Krainer AR. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol Biol. 1999;118:309–314. doi: 10.1385/1-59259-676-2:309. [DOI] [PubMed] [Google Scholar]
- 30.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen S, McMullan G, Faruqi AR, Murshudov GN, Short JM, Scheres SH, Henderson R. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner T, Merino F, Stabrin M, Moriya T, Antoni C, Apelbaum A, Hagel P, Sitsel O, Raisch T, Prumbaum D, Quentin D, et al. SPHIRE-crYOLO: A fast and well-centering automated particle picker for cryo-EM. bioRxiv. doi: 10.1101/356584. 356584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zivanov J, Nakane T, Forsberg B, Kimanius D, Hagen WJH, Lindahl E, Scheres SHW. RELION-3: new tools for automated high-resolution cryo-EM structure determination. eLife. 2018;7 doi: 10.7554/eLife.42166. e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakane T, Kimanius D, Lindahl E, Scheres SHW. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife. 2018;7 doi: 10.7554/eLife.36861. e36861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 Å resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kondo Y, Oubridge C, van Roon AM, Nagai K. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. elife. 2015;4 doi: 10.7554/eLife.04986. e04986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fica SM, Oubridge C, Wilkinson ME, Newman AJ, Nagai K. A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation. Science. 2019;363:710–714. doi: 10.1126/science.aaw5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372(6505):432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 40.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korneta I, Magnus M, Bujnicki JM. Structural bioinformatics of the human spliceosomal proteome. Nucleic Acids Res. 2012;40:7046–7065. doi: 10.1093/nar/gks347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lebedev AA, Young P, Isupov MN, Moroz OV, Vagin AA, Murshudov GN. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Cryst. 2012;D68:431–440. doi: 10.1107/S090744491200251X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma S, Wongpalee SP, Vashisht A, Wohlschlegel JA, Black DL. Stem-loop 4 of U1 snRNA is essential for splicing and interacts with the U2 snRNP-specific SF3A1 protein during spliceosome assembly. Genes Dev. 2014;28:2518–2531. doi: 10.1101/gad.248625.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DMG, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.