Abstract
HER2/ErbB2 activation turns on transcriptional processes that induce local invasion and lead to systemic metastasis. The early transcriptional changes needed for ErbB2-induced invasion are poorly understood. Here, we link ErbB2 activation to invasion via ErbB2-induced, SUMO-directed phosphorylation of a single serine residue, S27, of the transcription factor Myeloid Zinc Finger-1 (MZF1). Utilizing an antibody against MZF1-pS27, we show that the phosphorylation of S27 correlates significantly (p<0.0001) with high-level expression of ErbB2 in primary invasive breast tumors. Phosphorylation of MZF1-S27 is an early response to ErbB2 activation and results in increased transcriptional activity of MZF1. It is needed for the ErbB2-induced expression of MZF1 target genes CTSB and PRKCA, and invasion of single-cells from ErbB2-expressing breast cancer spheroids.The phosphorylation of MZF1-S27 is preceded by poly-SUMOylation of K23, which can make S27 accessible to efficient phosphorylation by PAK4. Based on our results, we suggest for an activation mechanism where phosphorylation of MZF1-S27 triggers MZF1 dissociation from its transcriptional repressors, such as the CCCTC-binding factor (CTCF). Our findings increase understanding of the regulation of invasive signaling in breast cancer by uncovering a detailed biological mechanism of how ErbB2 activation can rapidly lead to its invasion-promoting target gene expression and invasion.
Keywords: ErbB2, MZF1, PAK4, PRKCA, CTSB, MYC, CTCF, lysosomes, breast cancer, invasion, breast cancer cell spheroid, transcription factor, phosphorylation, SUMOylation
Introduction
Amplification and/or activation of the ERBB2/HER2 oncogene is detected in 20% of invasive breast cancers (BCs) and is the driving force for their aggressive phenotype [1, 2]. Although treatments directed against ErbB2-positive BC are efficient, long-term follow-up studies indicate that over 50% of advanced ErbB2-positive BCs eventually develop lethal metastases [3, 4]. This can sometimes take years, or even decades. BC is characterized as a systemic disease that undergoes early dissemination, in which the disseminated cells do not respond to the treatment and can stay dormant for long periods [5]. The metastatic capacity of ErbB2 positive BC often correlates with the expression of the NH2-terminally truncated p95 form of ErbB2, which is a more potent kinase and activator of ErbB2 downstream signaling than the full-length p185-ErbB2 [6–8]. In addition, it lacks binding sites for the clinically applied ErbB2-targeting antibodies, trastuzumab and pertuzumab [4, 9, 10].
Induction of local invasion is among the earliest changes needed for metastasis formation. The invasiveness of highly aggressive p95-ErbB2-expressing BC spheroids depends on a signaling network that activates the oncogenic transcription factor (TF) MZF1 [8]. MZF1 regulates the function and activity of lysosomes in invasive BC cells by mediating ErbB2-induced expression of lysosomal cathepsins B and L (CTSB and CTSL1). Their increased mRNA and protein levels correlate positively (p<0.0001) with high ErbB2 status in primary invasive BC [8]. Furthermore, ErbB2 activation results in the distribution of lysosomes from their normal, perinuclear position to the invadosomes at the cellular periphery [8, 11]. Peripheral lysosomes contribute to extracellular matrix (ECM) degradation both internally and externally [12, 13]. They are responsible for the degradation of internalized ECM components, and they can also secrete their hydrolytic contents, including cathepsin B, into the extracellular space [12–14]. Secreted cathepsin B degrades the ECM components of Type IV collagen, laminin and fibronectin [15], and it initiates the activation of the extracellular degradome by cleaving the pro-forms of urokinase plasminogen activator and matrix metalloproteinase (MMP)2 and MMP3, which are activators of MMP9 and MMP13 [13, 16].
Dimeric MZF1 is a member of the SCAN domain-containing zinc finger TF (SCAN-ZFP) family and a regulator of myeloid differentiation. In epithelial cancer cells MZF1 functions as an oncogene and promotes the progression of various cancers, including BC [8, 17–20]. A crucial, invasion-promoting function of MZF1 in BC cells is the activation of the transcription of CTSB and PRKCA (PKCα), which are mediators and amplifiers of ErbB2 signaling [8, 18]. In addition to CTSB and PRKCA, MZF1 also binds directly and activates the expression of cancer-promoting genes AXL, PIK3R3, TGFB1 and MYC [20–22]. The mechanisms of how MZF1 regulates invasive gene expression are unknown.
Results
MZF1 is phosphorylated and activated in response to ErbB2 signaling
MCF7 BC cells expressing an inducible 95-kDa NH2-terminally truncated, constitutively active form of ErbB2, i.e., MCF7 M8-tTAS-pTRE-Δ;NErbB2 cells (hereinafter referred to as p95-ErbB2-MCF7 cells) [23], exhibit ErbB2-dependent upregulation of CTSB, which leads to increased cysteine cathepsin activity [8]. Similarly, in BC cell lines SK-BR-3 and BT474 that express full-length p185-ErbB2, ErbB2 regulates the expression and activity of cathepsin B [8]. Since all endogenous ErbB2-overexpressing BC cell lines are either poorly invasive or not invasive at all [24], we utilized the p95-ErbB2-MCF7 cells for the analysis of ErbB2-induced invasive signaling. Moreover, the possibility to control the induction of ErbB2 expression makes them most suitable for the detection of early invasive changes.
To find out how ErbB2 signaling activates MZF1, we prepared samples for phosphor mass spectrometry. We transfected p95-ErbB2-MCF7 cells and the corresponding control cells with HA-tagged wild-type (WT) MZF1 (HA-MZF1) for 24 h to obtain low uniform, nuclear expression of HA-MZF1 (Supplementary Figure S1a). HA-MZF1 functioned as a non-tagged MZF1: it bound to the CTSB enhancer element in vivo (Supplementary Figure S1b) and positively regulated the activity of the pTAL-CTSB+126/+408-Luc reporter containing the ErbB2-inducible enhancer element of the CTSB with multiple MZF1 binding sites (hereinafter referred to as CTSB reporter) (Supplementary Figure S1c).
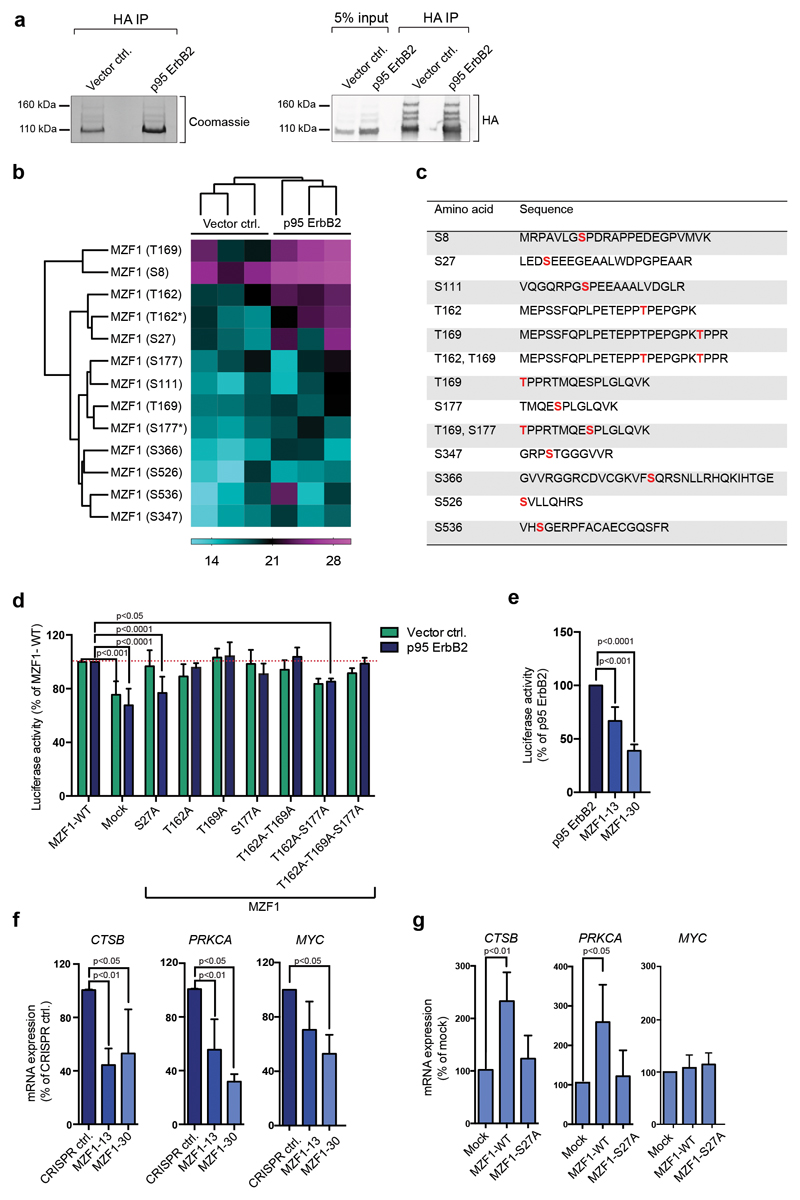
We identified the immunoprecipitated HA-MZF1 as four fractions: a prominent fraction of 100 kDa, closely corresponding to the estimated, non-modified MZF1 (80 kDa), and three less-prominent fractions of 120 kDa, 140 kDa, and 160 kDa (Fig.1a). We did the phosphopeptide analysis using the 100-kDa MZF1 for sufficient sample amount. We identified 13 peptides with 10 different phosphorylated serines and/or threonines in both cell lines (Fig.1b, Supplementary Table S1). Phosphopeptides obtained from both cell lines were clearly clustered as independent groups. Four phosphopeptides, S27, T162, S177, and T169, were identified with higher intensity in the p95 ErbB2-MCF7 cells, indicating their response to increased ErbB2 signaling (Fig.1b,c). Phosphorylation of MZF1 in the control cells was expected, since they express low levels of endogenous ErbB2 [25]. All four sites are located in the NH2-terminus of MZF1, with S27 in the acidic domain, and the other sites in the area between the leucine rich SCAN domain [26] and the transactivation domain (TAD) [27].
Figure 1. MZF1 is phosphorylated and activated in response to ErbB2 signaling.
(a) WT HA-MZF1 immunoprecipitates used for phosphopeptide analysis were immunoblotted for the detection of HA (right) and stained with Coomassie brilliant blue (left). The 100-kDa MZF1 form was used for the phosphor mass spectrometry analysis. The immunoblot is a representative of three independent experiments.
(b) Increased phosphorylation of several MZF1 peptides isolated from p95-ErbB2-MCF7 cells. Heatmap depicting the log2-transformed phosphopeptide LFQ intensities from three independent experiments. Label-free quantification was done using the MaxQuant software, followed by analysis with the Perseus software. MaxQuant was set to identify peptides with a phosphosite probability > 75%. * marks the di-phosphorylated sites.
(c) List of identified phosphopeptides. Identified phosphosites are highlighted in red.
(d) MZF1-S27A fails to activate CTSB reporter activity in response to ErbB2. Vector MCF7 and p95-ErbB2-MCF7 cells were transfected with the CTSB firefly luciferase and the Renilla luciferase construct and the empty (mock) or MZF1-WT or indicated MZF1 mutant pcDNA3.1 plasmids. Reporter activity was calculated as the firefly luciferase activity divided by the Renilla luciferase activity and presented as a percentage of the MZF1-WT control. Data shown are mean ± standard deviation of three independent experiments. Statistical significance was calculated with one-way ANOVA with Dunnett’s correction.
(e) CTSB reporter activity is decreased in p95-ErbB2-expressing CRISPR MZF1-13 and 30 cells. The cells were transfected with CTSB firefly luciferase and the Renilla control luciferase constructs, and their activities and statistical significances were calculated as in d. Results are presented as the percentages of the luciferase activity of the parental cells and the data is presented as a mean ± standard deviation of three independent experiments.
(f) MZF1 depletion in CRISPR MZF1-13 and -30 cells decreases the expression of the MZF1 target genes CTSB, PRKCA and MYC, as compared to the levels of the expression of the control cell line. Quantitative RT-PCR analysis of MZF1 target gene expression. The indicated target gene mRNA expression was normalized to the expression of PPIB and is presented as the percentage of its expression in CRISPR control cells. Values shown are mean ± standard deviation of three independent experiments. Statistical significance was calculated as in d.
(g) Ectopic expression ofMZF1-WT, but not of MZF1-S27A, rescues the expression of endogenous CTSB and PRKCA in CRISPR MZF1-30 cells; neither MZF1-WT nor MZF1-S27A rescues the expression of MYC. Quantitative RT-PCR analysis of MZF1 target gene expression in CRISPR MZF1-30 cells. Cells were transfected with empty (mock) or MZF1-WT or MZF-S27A Δt-3xHA plasmids. Indicated mRNA expression was normalized to the expression of PPIB and presented as % of its expression in mock-transfected samples. Values represent mean ± standard deviation of three independent experiments. Statistical significance was calculated with the Student’s t-test (unpaired with Welch’s correction).
As a reporter for studying the function of MZF1, we used the CTSB reporter containing the ErbB2-inducible enhancer element of CTSB with in vivo binding sites for MZF1 [8]. We prepared MZF1 expression vectors with the phosphosites mutated to alanines (Supplementary Figure S1d) for CTSB reporter activity analysis. Only the serine-to-alanine mutation at position 27 (S27A) resulted in a significant reduction of luciferase activity in the p95-ErbB2-MCF7 cells, whereas it had no effect on the luciferase activity in the control cells (Fig.1d).
To study the function of the MZF1-S27 phosphorylation, we established cell lines with no/low level of endogenous MZF1. For this, we employed the CRISPR knockout technology in p95-ErbB2-MCF7 cells. Since we did not manage to generate complete knockouts of MZF1, we derived MZF1 heterozygous cell lines with partial inactivation of the MZF1 gene (MZF1-13 and MZF1-30). These exhibited decreased expressions of MZF1 (Supplementary Figure S1e) and decreased cysteine cathepsin activity (Supplementary Figure S1f) and lower CTSB reporter activities than the control cells (Fig.1e).
In addition to CTSB, MZF1 binds directly and regulates the ErbB2-responsive gene PRKCA [28]. Consequently, both the CTSB and PRKCA mRNA levels were reduced in the MZF1 CRISPR-knockdown cell lines, as compared to their WT counterpart (Fig.1f). Introduction of MZF1-WT, but not MZF1- S27A into MZF1-30 cells rescued the expression of CTSB and PRKCA (Fig.1g). Ectopic expression of either MZF1-WT or MZF1-S27A did not rescue the decreased expression of the MZF1 target gene MYC, indicating that MZF1 depletion was not fully reversible (Fig.1f, g).
Taken together, these results indicate that ErbB2 activation induces MZF1 transcriptional activity via phosphorylation of MZF1-S27.
MZF1-S27 mediates invasive ErbB2 signaling in BC cells
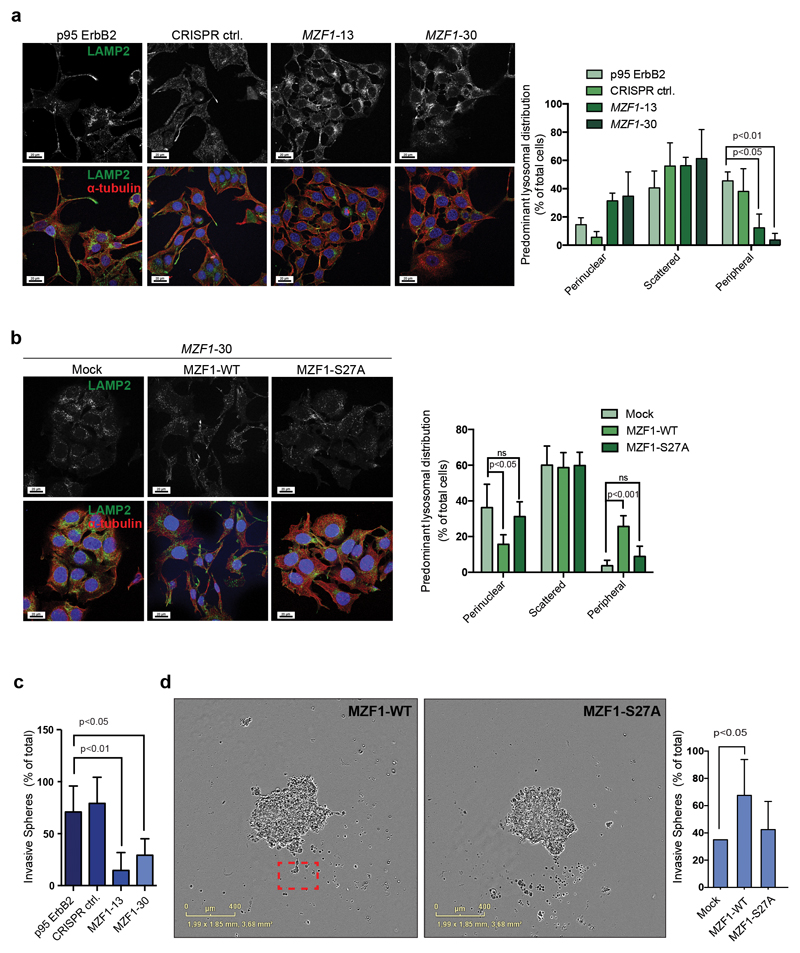
Similar to the siRNA-mediated transient depletion of ERBB2 or MZF1 (Supplementary Figure S2a; [29]), CRISPR-mediated depletion of MZF1 resulted in the redistribution of lysosomes from their ErbB2-induced, invasion-promoting peripheral location to the perinuclear area in both the MZF1-13 and MZF1-30 cell lines (Fig.2a). No redistribution of lysosomes was observed in the CRISPR control cell line (Fig.2a) expressing WT MZF1 at the level of the parental cells (Supplementary Figure S1e). Transient expression of MZF1-WT, but not of MZF1-S27A, reverted the lysosome redistribution of the MZF1-30 cells back to the cellular periphery (Fig.2b).
Figure 2. MZF1-WT, but not MZF1-S27A, can mediate invasive ErbB2 signaling in BC cells.
(a) MZF1 depletion in CRISPR MZF1-13 and-30 cells reverse the lysosome distribution pattern from peripheral to perinuclear. Representative images of lysosomal distributions detected by immunostaining of the endogenous LAMP2 (green). Staining for α-tubulin (red) is used to mark the cytoskeleton, and Hoechst staining (blue) is used to detect the nucleus. The right-side columns show the quantification of the immunofluorescence images. Data shown are mean ± standard deviation of three independent experiments in which minimum of 4 images with 8-25 cells was analyzed. Statistical significance was calculated using one-way ANOVA with Dunnett’s correction.
(b) Ectopic expression of MZF1-WT, but not of MZF1-S27A, alters the lysosome distribution of CRISPR MZF1-30 cells from perinuclear to peripheral. Cells were transfected with empty (mock) or MZF1-WT or MZF1-S27A Δt-3xHA plasmids. Representative images of lysosomal distribution detected by immunostaining of the endogenous LAMP2 (green). Staining with α-tubulin (red) is used to indicate the cytoskeleton, and Hoechst staining (blue) the nucleus. The images were quantified as in a (right-side columns). Values shown are mean ± standard deviation of three independent experiments in which a minimum of 4 images with 8-25 cells was analyzed. Statistical significance was calculated as in a.
(c) MZF1 depletion in CRISPR MZF1-13 and -30 cells renders the cells less invasive in 3D Matrigel invasion assays than the corresponding MZF1-expressing CRISPR control cells. Invasive spheroids were counted from three biological repeats of six spheroids each. Values shown are mean ± standard deviation of the three biological repeats. Representative images are presented in Figure S3B. Statistical significance was calculated as in a.
(d) Expression of MZF1-WT, but not that of MZF1-S27A, rescues the invasion deficiency of the CRISPR MZF1-30 cells. Cells were transfected with empty (mock) or MZF1-WT or MZF1-S27A plasmids and the corresponding spheroids were analyzed for their invasiveness in the 3D MBE Matrix by imaging every 4 h. The images shown are representative of invasive (MZF1-WT) and non-invasive (MZF1-S27A) spheroids selected from corresponding movies. Invasive cells are marked with a red square. Ten spheroids from four biological repeats were quantified per treatment. Results are presented as percentage of total number of spheroids. Values shown are mean ± standard deviation of four independent experiments. Statistical significance was calculated as in a.
The altered lysosome distribution of the MZF1-13 and 30 cells correlated with decreased invasion in 3D Matrigel (Fig.2c; Supplementary Figure S2b). Ectopic expression of MZF1-WT, but not of MZF1-S27A, rescued the impaired invasion of the MZF1-30 cells (Fig.2d; Supplementary Movies 1 and 2), demonstrating that MZF1 can promote invasion of single cells from the BC spheroids.
These results show that phosphorylation of MZF1-S27 is crucial for the invasive signaling in p95-ErbB2-MCF7 BC cells.
Phosphorylation of S27 correlates significantly with high HER2 and EGFR status in primary breast tumors
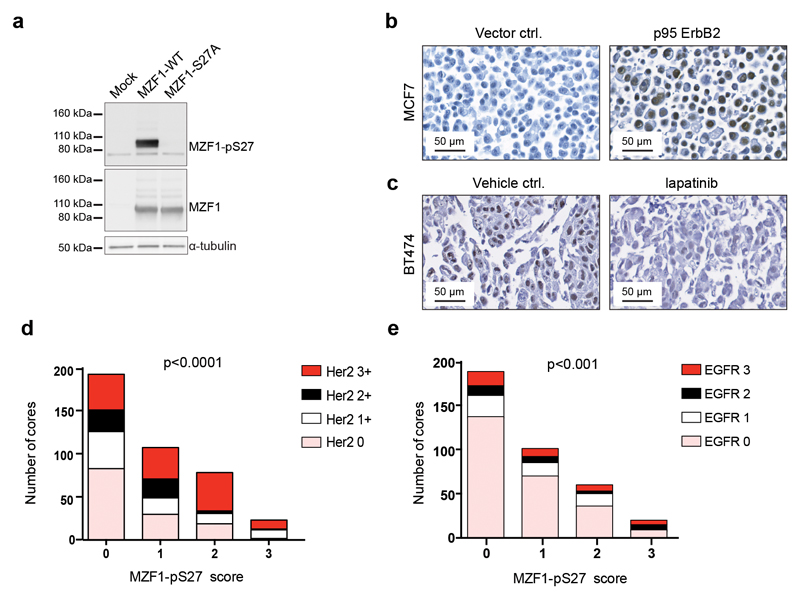
We prepared an antibody against phospho-S27 (Supplementary Figure S3a). It preferentially recognized ErbB2-activated MZF1-WT over MZF1-S27A (Fig.3a). In immunohistochemistry (IHC), it gave a strong nuclear staining in the p95-ErbB2-MCF7 cells (Fig.3b) and a weaker staining of the nuclei of the control cells (Fig.3b). Treatment of HER2-positive BT474 cells with 1 μM lapatinib abolished (Fig.3c) the nuclear phospho-MZF1 staining that was visible in the corresponding vehicle control-treated cells (Fig.3c). IHC analysis of primary BC tissue microarrays (TMAs) of 225 duplicate cores revealed that a strong MZF1-S27 phosphorylation correlated positively (p<0.0001) with a high HER2 status (Fig.3d, Supplementary Figure S3b, Supplementary Table S2). No staining was observed in the negative control, i.e., mouse lungs (Supplementary Figure S3c). Phospho-S27 staining correlated also positively (p<0.001) with high EGFR status (Fig.3e).
Figure 3. Phosphorylation of S27 correlates significantly with high HER2 and EGFR status.
(a) MZF1-S27A is not recognized by the anti-MZF1-pS27 antibody. The p95-ErbB2-MCF7 cells were transfected for 72 h with pcDNA3.1 MZF1-WT or MZF1-S27A for the detection of S27 phosphorylation. MZF1 and α-tubulin were used as loading controls.The immunoblot shown is representative of three independent experiments.
(b) The anti-MZF1-pS27 antibody stains the nuclei of ErbB2-positive cells. IHC analysis of sections from cell blocks of vector control cells (left side) and p95-ErbB2-MCF7 cells (right side) stained with the anti-MZF1-pS27 antibody (brown). The sections were counterstained with hematoxylin (blue) and the images were captured at 40x magnification.
(c) Anti-MZF1-pS27 antibody staining of the nuclei of ErbB2-positive cells is inhibited by lapatinib. IHC analysis of sections from cell blocks of ErbB2-positive BT474 cells treated for 24 h with DMSO (vehicle control; left side) or 1 mM lapatinib (right side), and then stained with anti-MZF1-pS27 antibody (brown). The sections were counterstained with hematoxylin (blue) and the images were captured at 40x magnification.
(d) MZF1-pS27 staining correlates significantly with high HER2 status in primary BC samples. Analysis of correlations between MZF1 expression and HER2 expression in BC samples. TMAs with 225 duplicate cores were stained for MZF1-pS27 and the samples were categorized into one of four groups (score of 0, 1, 2, or 3) according to the expression level of MZF1-pS27. HER2 expression levels (score of 0, 1+, 2+ and 3+) were available for these TMAs from the provider (Pantomics, USA), and were used to examine the possible associations with MZF1 expression. The Chi-square test for trend was used to assess statistical significance.
(e) MZF1-pS27 staining correlates significantly with high EGFR status in primary BC samples. Analysis of correlations between MZF1 expression and EGFR expression in BC samples. EGFR expression quantification (score of 0, 1, 2, 3) was available for these TMAs from the provider (Pantomics, USA), and was used to examine the possible association with MZF1 expression. The Chi-square test for trend was used to assess statistical significance.
These results indicate that S27 phosphorylation correlates with high levels of ErbB2/HER2, which in turn positively correlate with invasive and metastatic BC [2, 30]. Since phosphoprotein stability in tissue samples depends on their processing, and longer processing times may lead to decreases in the phosphor epitopes [31], it is possible that the positive correlations between HER2/EGFR expression and pS27 could be even better with freshly processed samples.
MZF1 activation involves SUMOylation
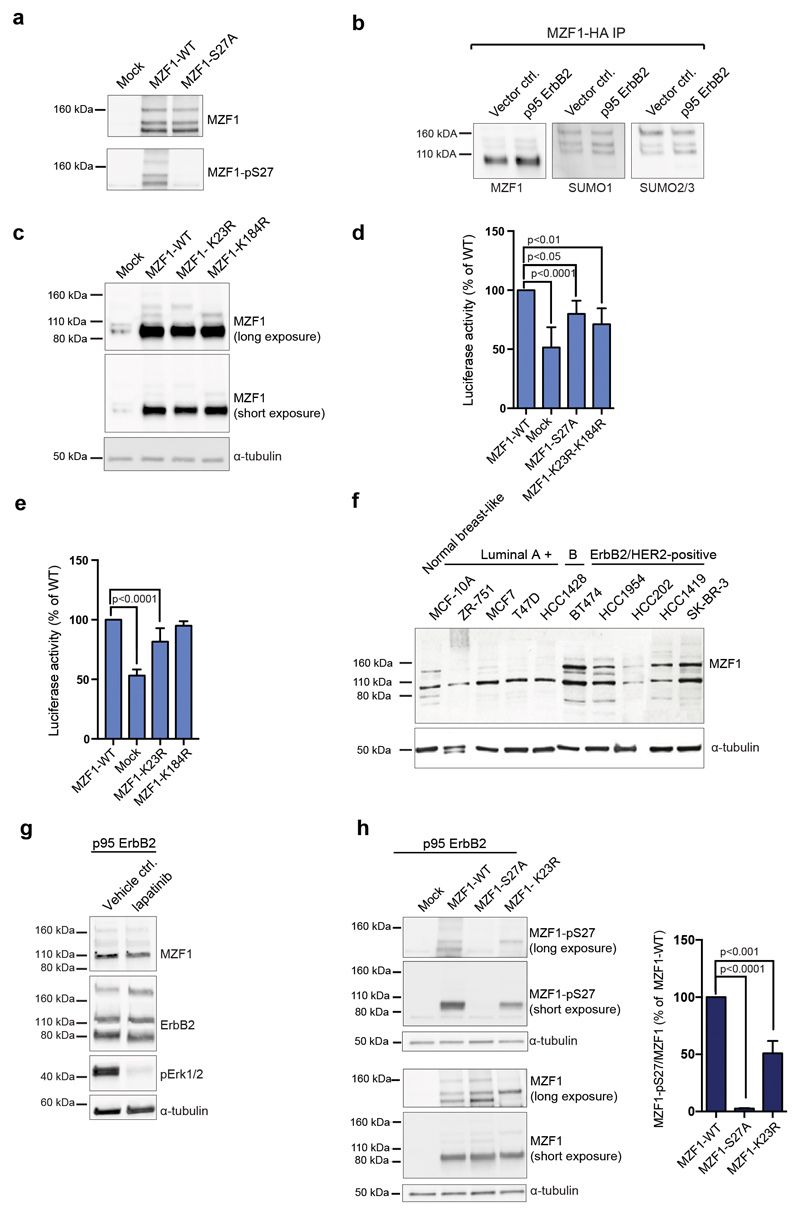
The 100-kDa form of MZF1 is often accompanied by multiple, larger forms (Fig.1a, 3a). These forms were recognized by the anti-phospho-S27-MZF1 antibody (Fig.4a). Small ubiquitin-like modifier (SUMO) is a group of small proteins that are covalently added to other proteins. Increased SUMOylation facilitates cancer development and progression, and is associated with BCs with poor prognosis [32, 33]. To identify the potential SUMO acceptor sites on MZF1, we transfected p95-ErbB2 cells with HA-MZF1 to immunoprecipitate MZF1 and its SUMOylated variants. All our SUMOylation experiments relied exclusively on the endogenous SUMO pathway, since no overexpression of SUMO pathway members was used. Besides the major form of 3xHA-tagged MZF1 (100 kDa) (Fig.4b, lanes 1 and 2), we detected three SUMO1- and SUMO2/3-positive MZF1 variants of 120, 140 and 160 kDa (Fig.4b, lanes 3-6), corresponding to its multi- or poly-SUMOylated forms.
Figure 4. SUMOylation of MZF1-K23 cooperates with the phosphorylation of S27 in MZF1 activation.
(a) High-molecular-weight forms of MZF1 are phosphorylated at S27 in ErbB2-expressing cells. The p95-ErbB2-MCF7 cells were transfected with empty vector (mock) pcDNA3.1 MZF1-WT or MZF1-S27A for 72 h, and blotted with the anti-MZF1-pS27 antibody. MZF1 and α-tubulin used as a loading controls. The immunoblot is a representative of three independent experiments.
(b) SUMO1 and SUMO2/3 are associated with the high molecular weight forms of MZF1. Membranes of HA-immunoprecipitated HA-MZF1 (pcDNA3.1) in vector and p95-ErbB2-MCF7 cells were blotted with MZF1, SUMO1 and SUMO 2/3 antibodies. The presented immunoblot is a representative of three independent experiments.
(c) K23 and K184 are MZF1 SUMO acceptor sites. Immunoblot analysis for the detection of MZF1 in lysates of p95-ErbB2-MCF7 cells transiently transfected with empty (mock), MZF1-WT or indicated mutant MZF1 pcDNA3.1 plasmids for 72h. The membrane was blotted with MZF1 and α-tubulin antibodies. The presented immunoblot is a representative of three independent experiments.
(d) MZF1-K23R-K184R expression decreases CTSB reporter activity. CRISPR MZF1-30 cells were transfected with the CTSB reporter, Renilla luciferase construct and either empty (mock), MZF1-WT or indicated mutant MZF1 Δt-3xHA plasmids. Reporter activity was calculated as firefly luciferase activity divided with Renilla luciferase activity and presented as percentage of the activity of the MZF1-WT. Data is presented as mean ± standard deviation of three independent experiments. Statistical significance was calculated with one-way ANOVA with Dunnett’s correction.
(e) MZF1-K23R expression decreases CTSB reporter activity. CRISPR MZF1-30 cells were transfected with the CTSB firefly luciferase reporter, Renilla luciferase construct and either empty (mock), MZF1-WT or indicated mutant MZF1 pcDNA3.1 plasmids. Reporter activity was calculated and presented as in d. Data shown are mean ± standard deviation of three independent experiments. Statistical significance was calculated as in d.
(f) Increased expression levels of MZF1 SUMO forms in ErbB2-expressing BC cell lines compared to normal and luminal type cells. The membrane was blotted, with MZF1 and α-tubulin was used as a loading control. The presented immunoblot is representative of three technical replicates.
(g) ErbB2 inhibition does not inhibit MZF1 SUMOylation. Immunoblot analysis of MZF1 expression in p95-ErbB2-MCF7 cells treated with DMSO (vehicle control) or 10 μg/ml lapatinib for 24 h. The membrane was blotted, with MZF1, and ErbB2 and α-tubulin were used as a loading controls, and pErk1/2 as a control for the activity/inhibition of ErbB2 signaling. The presented immunoblot is representative of three independent experiments.
(h) Phosphorylation of MZF1-S27 is decreased in MZF1-K23R-transfected cells. The p95-ErbB2-MCF7 cells were transfected with empty (mock) or MZF1-WT or MZF1-K23R pcDNA3.1 plasmids for 48 h. Lysates were subjected to SDS-PAGE, blotted, and then stained with anti-MZF1-pS27 antibody. MZF1 and α-tubulin were used as loading controls. Quantification was based on the 100-kDa MZF1 forms using the ImageJ software (right-side columns). Values shown are mean ± standard deviation of three independent immunoblots. Statistical significance was calculated as in d.
In silico analysis with SUMOplot (http://www.abgent.com/sumoplot) and SUMOsp (http://sumosp.biocuckoo.org) identified MZF1 lysine residues at positions 23 and 184 as putative SUMO acceptor sites. We expressed SUMOylation-deficient MZF1 mutants MZF1-K23R and MZF1-K184R in p95-ErbB2-MCF7 cells. Upon overexpression of MZF1-K23R, the MZF1 isoforms of 120 kDa and 160 kDa disappeared, and upon overexpression of MZF1-K184R, the MZF1 isoforms of 140 kDa and 160 kDa disappeared (Fig.4c). Therefore, K23 and K184 are the main SUMO acceptor sites in MZF1. SUMOylation at K23 resulted in increased molecular mass of around 20-30 kDa, corresponding to two SUMO moieties of 11-13 kDa each, and SUMOylation at K184 resulted in increased molecular mass of around 40-50 kDa, corresponding to four SUMO moieties. Occupation of both would result in 160-kDa form of MZF1 (Fig.4c).
SUMO2/3, but not SUMO1, contains the canonical SUMOylation motif, which enables the formation of polymeric SUMO2/3 chains of varying lengths. Thus, it is likely that MZF1 is modified with polymeric SUMO2/3-SUMO1 chains at both K23 and K184, whereby SUMO1 likely acts as a SUMO2/3 polymeric chain terminator.
SUMOylation of K23 cooperates with phosphorylation of S27 for MZF1 activation
We studied the effect of SUMO modification on the CTSB reporter activity. Expression of the double-mutant MZF1-K23R-K184R decreased CTSB reporter activity (Fig.4d, Supplementary Figure S4). Most of the decrease was attributed to the K23R-mutation (Fig.4e). The differences between the MZF1-WT and lysine mutants were found significant despite a relatively high basal activity of the mock-transfected samples, which was likely to result from the partial inactivation of MZF1 gene in the CRISPR MZF1 cell lines (Supplementary Figure S1e). To clarify the extent of MZF1 SUMOylation in BC cells, we investigated the migration pattern of endogenous MZF1 in BC cells. The SUMOylated MZF1 variants were most prominent in the cells overexpressing ErbB2 (Fig.4f), suggesting that MZF1 SUMOylation correlates with ErbB2 expression. However, treatment of p95-ErbB2-MCF7 cells with 10 μM lapatinib did not affect the MZF1 SUMO forms (Fig.4g), implying that ErbB2 activation is probably not directly regulating K23 SUMOylation.
The K23 SUMOylation motif is identical to the phosphorylation-dependent SUMOylation motif (PDSM) [34]. However, SUMOylation of K23 was as efficient in MZF1-WT as in S27A-mutated MZF1 (Fig.4a). Instead, we noticed, that abrogated SUMOylation at K23 resulted in a 50% decrease in the phosphorylation of S27 when comparing the 100-kDa forms of MZF1-WT and MZF1-K23R (Fig.4h), indicating that K23 SUMOylation occurs prior to S27 phosphorylation and is likely to be a prerequisite for successful phosphorylation of MZF1-S27, and not vice versa.
These results indicate that a regulatory interplay exists between K23 SUMOylation and S27 phosphorylation in ErbB2-positive BC cells, where K23-SUMOylated MZF1 is more efficiently phosphorylated at S27 than the non-SUMOylated form.
K23 SUMOylation renders S27 accessible to PAK4 phosphorylation
To identify a kinase that can phosphorylate S27, we returned to the kinases that we earlier identified as regulators of ErbB2-induced cysteine cathepsin activity using a human kinome siRNA screen [8]. Of these, PAK4 had the highest prediction value as a kinase that phosphorylates S27, using the PhosphoNet (http://www.phosphonet.ca), NetPhorest (http://www.netphorest.info) and/or PhosphoMotif (http://www.hprd.org/PhosphoMotif). We overexpressed HA-tagged dominant-negative PAK4 (to ensure stronger kinase-substrate association) and non-tagged MZF1 in p95-ErbB2-MCF7 cells, and found that in co-immunoprecipitation assays, PAK4 associated with MZF1 (Fig.5a). Immunoprecipitated WT HA-PAK4 from p95-ErbB2-expressing cells efficiently phosphorylated a S27-containing peptide (Fig.5b). Incubation of the peptide with precipitated empty control vector resulted in a low level of phosphorylation, and phosphorylation of the peptide by PAK4 was largely abolished by the addition of the pharmacological PAK4 inhibitor PF37583909 (Fig.5b).
Figure 5. MZF1-K23 SUMOylation makes S27 accessible for PAK4 phosphorylation.
(a) PAK4 associates with MZF1. Co-immunoprecipitation of a dominant-negative HA-PAK4 (KM) with MZF1. The p95-ErbB2-MCF7 cells were transiently transfected with dominant-negative HA-PAK4 and MZF1-WT. HA-immunoprecipitates were blotted for the detection of MZF1, PAK4 and α-tubulin. The immunoblot shown is representative of three independent experiments.
(b) PAK4 phosphorylates MZF1 S27. MZF1 peptide VKLEDSEEEGEA-containing S27 was used as a substrate together with γ-ATP and subjected to PAK4 in vitro phosphorylation reaction. Precipitated HA-PAK4 was preincubated or not with 500 nM PF3758309 and an immunoprecipitated mock-transfected lysate was used as a negative control. The autophosphorylation control for HA-PAK4 (right side) was from the same blot. The blot shown is a representative of two independent experiments.
(c) Basal phosphorylation of MZF1 opens the structure of the MZF1 NH2-terminus making it accessible to SUMO groups. Left-side graph: Distribution of the radius of gyration (Rg) of unphosphorylated and phosphorylated dimeric MZF1-128 in the MD simulations. Rg gives an estimate of the compactness of the protein and it was observed that constitutive phosphorylation is sufficient to promote more open states of MZF11-128 where the K23 SUMOylation site is accessible for modification by SUMO proteins. In the plot, the Rg curves from the unphosphorylated and phosphorylated MD simulations are shown in black and red, respectively.
(d) Schematic presentation of the MZF11-128 dimer in the open state (blue), showing that adding the two SUMO moieties to K23 (yellow and orange; SUMO2 and SUMO1, respectively) causes steric constraints that are likely to open up the NH2-terminal into a conformation that makes S27 (red) is accessible to phosphorylation by kinases such as PAK4 (green).
To gain insight into the activation mechanism, we used molecular modeling and simulations of the dimeric MZF11-128. The NH2-terminal region of MZF1 is predicted to form a disordered tail. For the unphosphorylated form (Fig.5c, black line), the most favored conformation is a closed state in which the NH2-terminal tail folds back onto itself (gray dimer). This corresponds to a low radius of gyration (measure of the degree of compactness of the protein structure). When considering the size of the SUMO group at K23 with respect to the MZF1 dimer, it becomes clear that K23 is not accessible for SUMO modification in the closed state. Basal phosphorylation of the protein (Fig.1b), including local electrostatic repulsions, increases the radius of gyration (Fig.5c, red line), promoting the formation of an open conformation of the MZF1-128, where the NH2-terminal tails of the dimer are extended and located far away from the folded SCAN domain (red dimer; Fig.5c). In the open conformation, K23 is accessible to SUMO conjugation. According to our models, it is plausible that when MZF1 is SUMOylated at K23, the protein is fixed in a conformation where its NH2-terminal tail is in the extended state, making S27 accessible for phosphorylation (Fig.5d).
These results indicate that PAK4 can phosphorylate S27, especially when S27 is exposed, and that SUMOylation of K23 most likely opens up the structure of the MZF1 NH2-terminal tail to allow a kinase like PAK4 to access and phosphorylate S27.
MZF1-S27 phospho-mimetic mutant has diminished affinity for zinc finger transcription factors with potential repressor functions
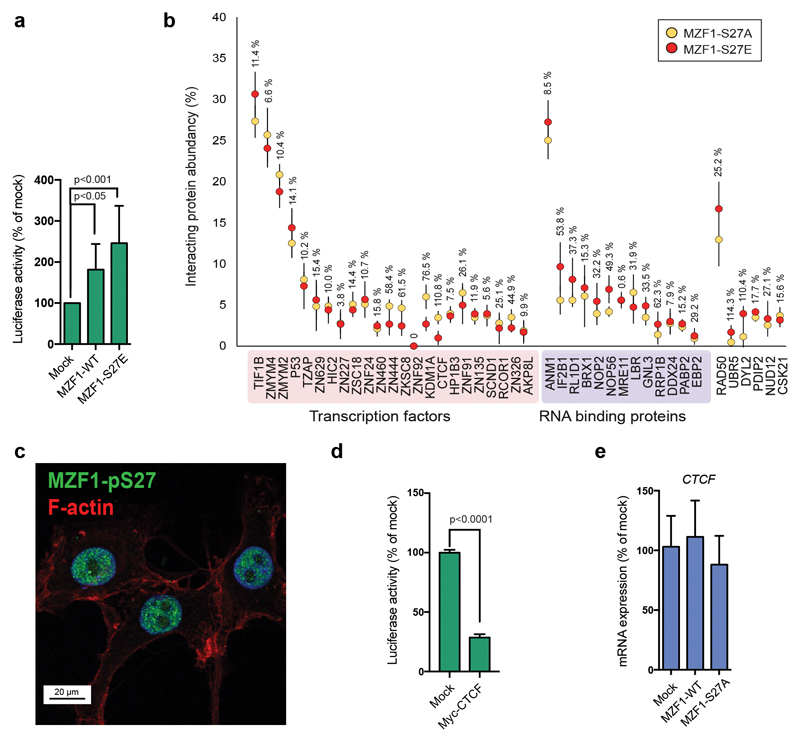
To understand the transcriptional mechanism of MZF1 activation, we mutated serine 27 to glutamic acid (MZF1-S27E), to incorporate a permanent negative charge at position 27, thereby mimicking its constitutive phosphorylation. Expression of MZF1-S27E in MCF7 cells led to 2-3-fold increase in the CTSB reporter activity in comparison to MZF1-WT (Fig.6a). TFs often co-operate by forming dynamic and spatial complexes at the active promoters and enhancers. To detect the possible MZF1 association partners involved in its activation, we utilized the Flp-In cloning of MZF1-S27A and MZF1-S27E into a tetracycline-inducible Strep-tactin vector in the 293T-REx cell line. With this, we created samples with inducible, low-level expression of MZF1 suitable for affinity-purification mass spectrometry [35]. We reasoned that this expression system could be usable for our study, since 293T cells express active EGF receptor, and EGF signaling activates S27 phosphorylation in BC tissues (Fig.3e). Thus, we tested if some of the MZF1 regulatory components could be conserved between cell lines. We compared the nuclear interactome of MZF1-S27A, identified by affinity mass spectrometry, to that of MZF1-S27E (Fig.6b; Supplementary Table S3). We focused on the nuclear interactome, since MZF1 is expressed exclusively in the nucleus (Fig.6c). We identified two major groups of with differential binding to activated MZF1: zinc finger TFs; and nuclear RNA-binding proteins. Interestingly, several zinc finger TFs exhibited significantly diminished affinity for the active form of MZF1 (Fig.6b).
Figure 6. S27 phosphorylation leads to dissociation of MZF1 from zinc finger transcription factors with transcriptional repressor functions.
(a) MZF1-S27E is constitutively active. MCF7 cells were transfected with the CTSB firefly luciferase reporter, the Renilla luciferase construct, and empty vector (mock) or MZF1-WT or MZF1-S27E Δt-3xHA plasmids. Reporter activity was calculated as the firefly luciferase activity divided by the Renilla luciferase activity, and is presented as the percentage of the mock-transfected control. Values shown are mean ± standard deviation of three independent experiments.Statistical significance was calculated with one-way ANOVA with Dunnett’s correction.
(b) Constitutively active MZF1-S27E binds less efficiently to some zinc-finger TFs than the MZF1-S27A (inactive form). Affinity purification mass spectrometry analysis of nuclear proteins associated with MZF1-S27E (red) and MZF1-S27A (yellow) Strep-tactin baits. The relative interacting protein abundances compared to the MZF1 bait abundance (%) were calculated from the spectral counts. The error bars represent ± the standard deviation of four experiments (SD). The interactor abundance (%) difference between the S27A and S27E mutants are shown for each interacting protein.
(c) S27-phosphorylated MZF1 resides in the nucleus. Immunofluorescence image of p95-ErbB2-MCF7 cells stained with the anti-MZF1-pS27 antibody (pS27-MZF1; green), F-actin (cytoskeleton; red), and Hoechst (nucleus; blue). Image is a representative of three independent experiments.
(d) CTCF inhibits CTSB reporter activity. MCF7 cells were transfected with the CTSB firefly luciferase reporter, the Renilla luciferase construct and empty vector (mock) or Myc-CTCF pcDNA3.1 plasmids. Reporter activity was calculated as in a. Values shown are the mean ± standard deviation of three independent experiments. Statistical significance was calculated with the Student’s t-test (unpaired with Welch’s correction).
(e) MZF1 expression levels do not affect CTCF expression. Quantitative RT-PCR of CTCF expression in CRISPR MZF1-30 cells expressing empty vector control (mock) or MZF1-WT or MZF1-S27A pcDNA 3.1 plasmids. CTCF expression is normalized to the expression of PPIB. Values shown are the mean ± standard deviation of three independent experiments. Statistical significance was calculated as in a.
To identify nuclear MZF1 interactors with the capacity to regulate its transcriptional activity in BC cells, we turned to the zinc finger TFs with potential transcriptional repressor activities. We reasoned that dissociation of a repressor could be involved in the activation of MZF1. To study whether some of these TFs could be associating with MZF1, we examined the ErbB2-inducible CTSB enhancer element for their potential binding sites. Using the Jaspar database (http://jaspar.binf.ku.dk) [36], we found two CTCF consensus binding sites (CTGCGCCCTCAGTCGG and GCCGAGGGAGGCGCCT) within the 400-bp ErbB2-inducible CTSB enhancer. In support of its function as an MZF1 repressor, the change in association between CCCTC binding TF CTCF [37] and the inactive or active form of MZF1 had the biggest difference (110.8%) in comparison to the other MZF1 associating proteins (Fig.6b). Lending further support to the putative role of CTCF as a repressor of MZF1 activity, the transient expression of CTCF in MCF7 cells repressed CTSB reporter activity (Fig.6d), while the changes in the MZF1 expression level or activation had no effect on CTCF expression (Fig.6e).
These results show that mutating MZF1-S27 to an acidic residue mimics the constitutive activation of MZF1. The active MZF1 (MZF1-S27E) has lower affinities for several zinc-finger TFs than the inactive MZF1 (MZF1-S27A). The results suggest that phosphorylation of S27 results in MZF1 activation by releasing it from its transcriptional repressor/s, such as CTCF.
Discussion
Previous large-scale proteomic studies have identified eight phosphorylated serine or threonine residues in human MZF1: S8, S27, S111, T169, S177, T210, S347 and T592 (http://www.phosphosite.org). These sites largely overlap with the 10 phosphorylated residues that we have identified. Of these sites, only four were ErbB2-responsive, suggesting that several MZF1 phosphosites exhibit constitutive phosphorylation. Molecular simulation modelling indicates that constitutive phosphorylation of MZF1 can regulate MZF1 activity, since it can affect the folding of MZF1 and thereby could open its structure to allow additional, signal-specific modifications, such as the SUMOylation of K23.
SUMOylation is significantly upregulated in the majority of cancers, including BC, due to upregulation of SUMO pathway enzymes [38], and increased SUMOylation correlates with a poor prognosis in BC [32, 33]. We show that MZF1 is endogenously modified by multiple SUMO moieties at K23 and K184. In concordance with this, an earlier study found indications of SUMOylation of unknown residues in MZF1 NH2-terminus [39]. SUMOylation is often challenging to detect without SUMO overexpression [40]. Since the detection of MZF1 SUMOylation does not require overexpression of SUMO groups or SUMO pathway enzymes, MZF1 SUMOylation in ErbB2-positive BC cells is likely to be rather stable and comprehensive. Interestingly, a mass spectrometry-based analysis reported global upregulation of SUMO2/3-modified proteins in BC cells originating from metastatic tumors [41]. We identified PAK4 as a kinase capable for phosphorylating S27 upon K23 SUMOylation. PAK4 is linked to invasion and promotion of mammary tumorigenesis [42]. PAK4 inhibitors have been developed for cancer treatments by Hoffman-La Roche, Genentech and Karyopharm Therapeutics Inc, however more studies may still needed to improve their specificity [43]. Novel PAK4 substrates, such as MZF1 could be useful for such studies and can help understanding the importance of PAK4 inhibition. A recent study reports that phosphorylation of MZF1-S27 mediates EMT by inducing the expression of N-cadherin in human esophageal cancer cells [44]. EMT is often described as a prerequisite for invasion of cancer cells. In that study, the kinase responsible for the phosphorylation was casein kinase 2, which is a constitutively active, growth factor independent kinase that is often overexpressed in cancer [44]. This suggests that the phosphorylation of MZF1-S27 can be regulating invasion in general, and that it may be mediated by different kinases depending on the tissue/cancer type.
Both K23 SUMOylation and S27 phosphorylation are important for the activation of MZF1. In a PAK4 substrate specificity study, it was shown that arginine (R) occupation in the position P4 from the PAK4 phosphorylatable serine (S) can be expected slightly more often than lysine (K), and that together their preference for occupying this position in a PAK4 substrate is approximately 25% [45]. That suggests that changing of K to R in a P4 position of a PAK4 substrate, as we did to destroy the the K23 SUMO acceptor site in MZF1, is unlikely to inhibit S27 phosphorylation merely by destroying a possible PAK4 recognition motif. Interestingly, about 9% of all SUMOylated sites in the human proteome are found to be proximal to phosphorylation sites [46], supporting a possibility for a functional SUMO-phospho interplay. This study defines a novel type of phosphorylation mechanism that can be described as “SUMO-directed phosphorylation” involving the MZF1-K23. K23 resides in a motif that is known as the “phosphorylation-dependent SUMOylation” (PDSM)-like motif [34]. Thus, occupation of this motif by both SUMO and phospho groups may play a specific biological role by itself, independent of the order of the modifications. As SUMOylation in BC is connected to increased malignancy, it is possible that SUMO-directed phosphorylation of MZF1-S27 indicates increased aggressiveness of ErbB2-positive BC.
Of the ErbB2-responsive phosphorylation sites in MZF1, only phosphorylation of S27 is responsible for its transcriptional activation and invasion of 3D BC spheroids. MZF1 target genes CTSB and PRKCA are both central mediators of ErbB2-induced invasion [8], and their expression is activated soon after induction of ErbB2 expression in p95-ErbB2-MCF7 cells. Increased expression of PRKCA is indicative of a poor prognosis in ErbB2-positive BC [47]. PKCα amplifies ErbB2 signaling by increasing the endocytic recycling of ErbB2, and it facilitates the induction of CTSB expression [8, 48]. Phosphorylation of MZF1-S27 activates the expression of both genes, indicating for its role as a regulator of a positive feedback loop between ErbB2 and PKCα. The potential importance of the phosphorylation of S27 in BC in vivo is underlined by its significant correlation with high ErbB2 status in BC. In supporting of our findings, S27 phosphorylation was identified in an ErbB2-positive breast tumor in a large-scale proteomics study comprising 105 breast tumors initially characterized for TCGA [49]. Overall these data point to a functional role for the ErbB2-responsive phosphorylation of MZF1-S27 in BC in vivo. Since high ErbB2 status correlates with metastatic BC, although not all BCs with high ErbB2 status will metastasize [2], it would be of interest to investigate whether S27 phosphorylation is connected to disease outcome.
Based on our results, we propose for a transcriptional activation mechanism in which ErbB2-induced activation of MZF1 occurs in all likelihood via S27 phosphorylation-induced dissociation of MZF1 from its transcriptional repressor/s. Of the potential repressors, the most promising is the transcriptional regulator and insulator CTCF [50], whose association with MZF1 exhibited the most dramatic decrease upon MZF1 activation and whose overexpression inhibited CTSB reporter activity. Moreover, the ErbB2-inducible enhancer sequence in the CTSB first intron contains two consequtive CTCF-binding sites close to the MZF1-binding sites, making physical association and regulation between MZF1 and CTCF possible. In addition to CTCF, three additional proteins ZN444, ZKSC8 and KDM1A exhibited over 50% less binding to consitutively active MZF1-S27E mutant than to the inactive MZF1-S27A, suggesting that they might be also involved in the regulation of MZF1 activity, which remains to be investigated.
In this study we define a detailed molecular mechanism for the regulation of ErbB2-induced gene expression and invasion, with implications for BC invasion in vivo. We show mechanistical data for a novel type of SUMO-phospho interplay and report a physical association of functional relevance between MZF1 and the transcriptional repressor and insulator CTCF. The information presented here increases understanding of cancer-induced transcriptional activation, and could potentially be used to find ways to control cancer cell invasion and metastasis that is induced by activation of ErbB2 signaling.
Materials and Methods
Most materials and methods: cell lines, clinical samples, drug treatments and plasmids are described in the Supplementary Materials and Methods
Chromatin immunoprecipitation and Real-Time PCR analysis
The chromatin immunoprecipitation (ChIP) and the real-time PCR (RT-PCR) was described previously [8]. The Pfaffl method was used to calculate the relative mRNA levels [51].
Antibody generation
The peptide, VKLEDS(p)EEEGEA-Cys (JPT Peptide Technologies GmbH, Berlin, Germany) was coupled 1:1 to mcKLH using Imject Maleimide-Activated mcKLH (Thermo Fisher, Waltham, MA, USA), following the manufacturer’s protocol. Immunization of rabbits was performed at GenScript (Piscataway, NJ, USA). Antibodies were purified using the SulfoLink Immobilization Kit for Peptides (Thermo Fisher, Waltham, MA, USA), following the manufacturer’s protocol. The antibody was affinity-purified twice: first against the phosphorylated and then against the non-phosphorylated peptide.
3D Matrigel invasion assays
3D Matrigel invasion assay was carried out as described previously [29]. The invading growth was followed for up to 3 days. Images were taken with the Olympus 1X71 light microscope using the CellaP software. Quantification of the images was done manually. Spheres were imaged at 24 and 48h. Each of the 6 spheres prepared per treatment were characterized as invasive, if invasive cells had detached from the sphere. Data is presented as the mean % of invasive spheres of total number of spheres in 4 replicates
3D BME live-imaging invasion assay and its quantification
Day after transfection, the 3D cell spheroids were made using a spinning method [52] in RPMI+GlutaMAX (GIBCO, Paisley, UK) supplemented with 6% heat-inactivated FBS (GIBCO, Paisley, UK). After 20-24 h the spheroids were embedded in Cultrex BME 2RGF Organoid Matrix (3533-010-02, Amsbio LLC, Cambridge, MA, USA) in RPMI+GlutaMAX medium with 1.5% FBS (1:1) and incubated in complete RPMI+GlutaMAX medium. The spheroids were followed by imaging using the IncuCyte ZOOM every 4 h for 48 or 72h. The images were converted to films, and each of the 10 spheres per treatment were characterized as invasive, when cells invaded completely out from the spheroids. Ten spheroids were quantified per treatment from four biological repeats. Data is presented as the mean % of invasive spheres of total number of sphere in 4 replicates.
Mass spectrometry analysis for protein phosphorylation and protein-protein association
The phosphor mass spectrometry analysis was performed by using the LTQ-Orbitrap instrument coupled to the Eksigent nano-LC 2D system and the affinity mass spectrometry was performed by using the Q Exactive ESI-quadrupole-orbitrap mass spectrometer that was coupled to an EASY-nLC 1000 nanoflow LC as described detailed in the Supplementary Materials and Methods.
Quantification of lysosomal distribution
After immunofluorescence analysis, quantification of lysosome distribution was done by assigning individual cells to one of the following groups: cells with a perinuclear-dominant lysosomal pattern, where more than 50% of the LAMP2-stained vesicles localized to the perinuclear region; cells with a peripheral-dominant pattern, where more than 50% of the LAMP2-stained vesicles localized to the peripheral region; and cells with a scattered lysosomal pattern, where more than 50% of the LAMP2-stained vesicles localized neither in the perinuclear nor the peripheral region. The quantification was based on at least three independent experiments. For each repeat a minimum of 4-5 images with 8-25 cells was analyzed.
Statistical analysis
The values for the different groups are presented as mean ± SEM. Statistical analysis was performed with the GraphPad Prism ver. 7.0 software, with one-way ANOVA with Dunnett’s corrections when comparing several treatments to one control and the Student’s t-test when comparing one treatment to one control. For the TMA results the Chi-square test for trend was used to assess statistical significance.
Supplementary Material
Acknowledgements
We acknowledge the excellent technical assistance of Anni Hadesten and Louise Vandervox and give special thanks to Professor Audrey Minden for PAK4, Professor Marin Barisic for the ΔT-3xHA-DEST CW plasmids and Dr. Vincent Collins for help with the language editing.
Funding
Novo Nordisk Foundation (NNF15OC0017324) (TK), the Danish Medical Research Council (0602-02386B) (TK), the Danish Cancer Society Scientific Committee (KBVU) (R124-A7854-15-S2 and R56-A3108-12-S2) (TK), the Danish National Research Foundation (DNRF125) (MJ), the European Research Council (AdG 340751) (MJ), EU-PRACE DECI13th grant for computational resources (EP), and the DeiC Pilot Grant 2015-2016 for the Danish Supercomputing Center Computerome (EP).
Footnotes
Data availability
The raw data from the mass spectrometry analysis, molecular simulations and TMA staining are available upon request.
Author contributions
DMB, SAT, MBH, KBC, TO, VS, ML, PP, EP and JM performed the experiments. KH, IG, PJ, MV provided materials and/or facilities, TK planned the project, and wrote the manuscript and TK, DMB and MJ edited it.
Conflicts of interest
The authors have no conflicts of interests.
References
- 1.Arteaga CL, Engelman JA. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25(3):282–303. doi: 10.1016/j.ccr.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slamon DJ, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 3.Venur VA, Leone JP. Targeted Therapies for Brain Metastases from Breast Cancer. Int J Mol Sci. 2016;17(9) doi: 10.3390/ijms17091543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duchnowska R, et al. Quantitative HER2 and p95HER2 levels in primary breast cancers and matched brain metastases. Neuro Oncol. 2015;17(9):1241–9. doi: 10.1093/neuonc/nov012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dittmer J. Mechanisms governing metastatic dormancy in breast cancer. Semin Cancer Biol. 2017;44:72–82. doi: 10.1016/j.semcancer.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Segatto O, et al. Different structural alterations upregulate in vitro tyrosine kinase activity and transforming potency of the erbB-2 gene. Mol Cell Biol. 1988;8(12):5570–4. doi: 10.1128/mcb.8.12.5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saez R, et al. p95HER-2 predicts worse outcome in patients with HER-2-positive breast cancer. Clin Cancer Res. 2006;12(2):424–31. doi: 10.1158/1078-0432.CCR-05-1807. [DOI] [PubMed] [Google Scholar]
- 8.Rafn B, et al. ErbB2-driven breast cancer cell invasion depends on a complex signaling network activating myeloid zinc finger-1-dependent cathepsin B expression. Mol Cell. 2012;45(6):764–76. doi: 10.1016/j.molcel.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 9.Scaltriti M, et al. Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. J Natl Cancer Inst. 2007;99(8):628–38. doi: 10.1093/jnci/djk134. [DOI] [PubMed] [Google Scholar]
- 10.Tural D, et al. P95 HER2 fragments and breast cancer outcome. Expert Rev Anticancer Ther. 2014;14(9):1089–96. doi: 10.1586/14737140.2014.929946. [DOI] [PubMed] [Google Scholar]
- 11.Brix DM, et al. Screening and identification of small molecule inhibitors of ErbB2-induced invasion. Mol Oncol. 2014;8(8):1703–18. doi: 10.1016/j.molonc.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamalisto S, Jaattela M. Lysosomes in cancer-living on the edge (of the cell) Curr Opin Cell Biol. 2016;39:69–76. doi: 10.1016/j.ceb.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sevenich L, Joyce JA. Pericellular proteolysis in cancer. Genes Dev. 2014;28(21):2331–47. doi: 10.1101/gad.250647.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kallunki T, Olsen OD, Jaattela M. Cancer-associated lysosomal changes: friends or foes? Oncogene. 2013;32(16):1995–2004. doi: 10.1038/onc.2012.292. [DOI] [PubMed] [Google Scholar]
- 15.Fonovic M, Turk B. Cysteine cathepsins and extracellular matrix degradation. Biochim Biophys Acta. 2014;1840(8):2560–70. doi: 10.1016/j.bbagen.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 16.Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol. 2011;21(4):228–37. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mudduluru G, Vajkoczy P, Allgayer H. Myeloid zinc finger 1 induces migration, invasion, and in vivo metastasis through Axl gene expression in solid cancer. Mol Cancer Res. 2010;8(2):159–69. doi: 10.1158/1541-7786.MCR-09-0326. [DOI] [PubMed] [Google Scholar]
- 18.Yue CH, et al. Expression of protein kinase C alpha and the MZF-1 and Elk-1 transcription factors in human breast cancer cells. Chin J Physiol. 2012;55(1):31–6. doi: 10.4077/CJP.2012.AMM109. [DOI] [PubMed] [Google Scholar]
- 19.Tsai LH, et al. The MZF1/c-MYC axis mediates lung adenocarcinoma progression caused by wild-type lkb1 loss. Oncogene. 2015;34(13):1641–9. doi: 10.1038/onc.2014.118. [DOI] [PubMed] [Google Scholar]
- 20.Deng Y, et al. p55PIK transcriptionally activated by MZF1 promotes colorectal cancer cell proliferation. Biomed Res Int. 2013;2013 doi: 10.1155/2013/868131. 868131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eguchi T, et al. Role and Regulation of Myeloid Zinc Finger Protein 1 in Cancer. J Cell Biochem. 2015 doi: 10.1002/jcb.25203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber CE, et al. Osteopontin mediates an MZF1-TGF-beta1-dependent transformation of mesenchymal stem cells into cancer-associated fibroblasts in breast cancer. Oncogene. 2014 doi: 10.1038/onc.2014.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egeblad M, Mortensen OH, Jaattela M. Truncated ErbB2 receptor enhances ErbB1 signaling and induces reversible, ERK-independent loss of epithelial morphology. Int J Cancer. 2001;94(2):185–91. doi: 10.1002/ijc.1459. [DOI] [PubMed] [Google Scholar]
- 24.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagashima T, et al. Quantitative transcriptional control of ErbB receptor signaling undergoes graded to biphasic response for cell differentiation. J Biol Chem. 2007;282(6):4045–56. doi: 10.1074/jbc.M608653200. [DOI] [PubMed] [Google Scholar]
- 26.Sander TL, et al. Identification of a novel SCAN box-related protein that interacts with MZF1B. The leucine-rich SCAN box mediates hetero- and homoprotein associations. J Biol Chem. 2000;275(17):12857–67. doi: 10.1074/jbc.275.17.12857. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa H, et al. Regulation of myeloid zinc finger protein 2A transactivation activity through phosphorylation by mitogen-activated protein kinases. J Biol Chem. 2003;278(5):2921–7. doi: 10.1074/jbc.M207615200. [DOI] [PubMed] [Google Scholar]
- 28.Yue CH, et al. MZF-1/Elk-1 Complex Binds to Protein Kinase Calpha Promoter and Is Involved in Hepatocellular Carcinoma. PLoS One. 2015;10(5):e0127420. doi: 10.1371/journal.pone.0127420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tvingsholm SA, et al. Let-7 microRNA controls invasion-promoting lysosomal changes via the oncogenic transcription factor myeloid zinc finger-1. Oncogenesis. 2018;7(2):14. doi: 10.1038/s41389-017-0014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slamon DJ, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–12. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 31.Baker AF, et al. Stability of phosphoprotein as a biological marker of tumor signaling. Clin Cancer Res. 2005;11(12):4338–40. doi: 10.1158/1078-0432.CCR-05-0422. [DOI] [PubMed] [Google Scholar]
- 32.Bawa-Khalfe T, Yeh ET. SUMO Losing Balance: SUMO Proteases Disrupt SUMO Homeostasis to Facilitate Cancer Development and Progression. Genes Cancer. 2010;1(7):748–752. doi: 10.1177/1947601910382555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim KI, Baek SH. SUMOylation code in cancer development and metastasis. Mol Cells. 2006;22(3):247–53. [PubMed] [Google Scholar]
- 34.Hietakangas V, et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci U S A. 2006;103(1):45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varjosalo M, et al. Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nat Methods. 2013;10(4):307–14. doi: 10.1038/nmeth.2400. [DOI] [PubMed] [Google Scholar]
- 36.Mathelier A, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42(Database issue):D142–7. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marshall AD, Bailey CG, Rasko JE. CTCF and BORIS in genome regulation and cancer. Curr Opin Genet Dev. 2014;24:8–15. doi: 10.1016/j.gde.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 38.Seeler JS, Dejean A. SUMO and the robustness of cancer. Nat Rev Cancer. 2017;17(3):184–197. doi: 10.1038/nrc.2016.143. [DOI] [PubMed] [Google Scholar]
- 39.Noll L, et al. Heterodimer formation of the myeloid zinc finger 1 SCAN domain and association with promyelocytic leukemia nuclear bodies. Leuk Res. 2008;32(10):1582–92. doi: 10.1016/j.leukres.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 40.Xiao Y, et al. Can your protein be sumoylated? A quick summary and important tips to study SUMO-modified proteins. Anal Biochem. 2015;477:95–7. doi: 10.1016/j.ab.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramonian D, et al. Analysis of changes in SUMO-2/3 modification during breast cancer progression and metastasis. J Proteome Res. 2014;13(9):3905–18. doi: 10.1021/pr500119a. [DOI] [PubMed] [Google Scholar]
- 42.Minden A. The pak4 protein kinase in breast cancer. ISRN Oncol. 2012;2012 doi: 10.5402/2012/694201. 694201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abdel-Magid AF. P21-Activated Kinase 4 (PAK4) Inhibitors as Potential Cancer Therapy. ACS Med Chem Lett. 2015;6(1):17–8. doi: 10.1021/ml500445c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ko H, et al. Phosphorylation-dependent stabilization of MZF1 upregulates N-cadherin expression during protein kinase CK2-mediated epithelial-mesenchymal transition. Oncogenesis. 2018;7(3):27. doi: 10.1038/s41389-018-0035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ha BH, et al. Signaling, Regulation, and Specificity of the Type II p21-activated Kinases. J Biol Chem. 2015;290(21):12975–83. doi: 10.1074/jbc.R115.650416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hendriks IA, et al. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol. 2017;24(3):325–336. doi: 10.1038/nsmb.3366. [DOI] [PubMed] [Google Scholar]
- 47.Tan M, et al. Upregulation and activation of PKC alpha by ErbB2 through Src promotes breast cancer cell invasion that can be blocked by combined treatment with PKC alpha and Src inhibitors. Oncogene. 2006;25(23):3286–95. doi: 10.1038/sj.onc.1209361. [DOI] [PubMed] [Google Scholar]
- 48.Bailey TA, et al. A kinase inhibitor screen reveals protein kinase C-dependent endocytic recycling of ErbB2 in breast cancer cells. J Biol Chem. 2014;289(44):30443–58. doi: 10.1074/jbc.M114.608992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mertins P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534(7605):55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim S, Yu NK, Kaang BK. CTCF as a multifunctional protein in genome regulation and gene expression. Exp Mol Med. 2015;47:e166. doi: 10.1038/emm.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ivascu A, Kubbies M. Rapid generation of single-tumor spheroids for high-throughput cell function and toxicity analysis. J Biomol Screen. 2006;11(8):922–32. doi: 10.1177/1087057106292763. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.