Abstract
Neuronal populations in sensory cortex produce variable responses to sensory stimuli, and exhibit intricate spontaneous activity even without external sensory input. Cortical variability and spontaneous activity have been variously proposed to represent random noise, recall of prior experience, or encoding of ongoing behavioral and cognitive variables. Recording over 10,000 neurons in mouse visual cortex, we observed that spontaneous activity reliably encoded a high-dimensional latent state, which was partially related to the mouse’s ongoing behavior and was represented not just in visual cortex but across the forebrain. Sensory inputs did not interrupt this ongoing signal, but added onto it a representation of external stimuli in orthogonal dimensions. Thus, visual cortical population activity, despite its apparently noisy structure, reliably encodes an orthogonal fusion of sensory and multidimensional behavioral information.
In the absence of sensory inputs, the brain produces structured patterns of activity, which can be as large as or larger than sensory-driven activity (1). Ongoing activity exists even in primary sensory cortices, and has been hypothesized to reflect recapitulation or expectation of sensory experience. This hypothesis is supported by studies that found similarities between sensory-driven and spontaneous firing events (2–7). Alternatively, ongoing activity could be related to behavioral and cognitive states. The firing of sensory cortical and sensory thalamic neurons correlates with behavioral variables such as locomotion, pupil diameter, and whisking (8–23). Continued encoding of non-visual variables when visual stimuli are present could at least in part explain the trial-to-trial variability in cortical responses to repeated presentation of identical sensory stimuli (24).
The influence of trial-to-trial variability on stimulus encoding depends on its population-level structure. Variability that is independent across cells – such as the Poisson-like variability produced in balanced recurrent networks (25) – presents little impediment to information coding, as reliable signals can still be extracted as weighted sums over a large enough population. In contrast, correlated variability has consequences that depend on the form of the correlations. If correlated variability mimics differences in responses to different stimuli, it can compromise stimulus encoding (26, 27). Conversely, correlated variability in dimensions orthogonal to those encoding stimuli has no adverse impact on coding (28), and might instead reflect encoding of signals other than visual inputs.
Spontaneous cortical activity reliably encodes a high-dimensional latent signal
To distinguish between these possibilities, we characterized the structure of neural activity and sensory variability in mouse visual cortex. We simultaneously recorded from 11,262 ± 2,282 (mean ± s.d.) excitatory neurons, over nine sessions in six mice using 2-photon imaging of GCaMP6s in an 11-plane configuration (29) (Fig. 1A,B, Movie S1). These neurons’ responses to classical grating stimuli revealed robust orientation tuning as expected in visual cortex (Fig. S1).
Figure 1. Structured ongoing population activity in visual cortex.
(A,B) Two-photon calcium imaging of 10,000 neurons in visual cortex using multi-plane resonance scanning of 11 planes spaced 35 μm apart. (C) Distribution of pairwise correlations in ongoing activity, computed in 1.2 second time bins (yellow). Gray: distribution of correlations after randomly time-shifting each cell’s activity. (D) Distribution of pairwise correlations for each recording (showing 5th and 95th percentile). (E) First PC versus running speed in 1.2 s time bins. (F) Example timecourse of running speed (green), pupil area (gray), whisking (light green), first principal component of population activity (magenta dashed). (G) Neuronal activity, with neurons sorted vertically by 1st PC weighting, same time segment as F. (H) Same neurons as in G, sorted by a manifold embedding algorithm. (I) Shared Variance Component Analysis (SVCA) method for estimating reliable variance. (J) Example timecourses of SVCs from each cell set in the test epoch (1.2 s bins). (K) Same as J, plotted as scatter plot. Title is Pearson correlation between cell sets: an estimate of that dimension’s reliable variance. (L) % of reliable variance for successive dimensions. (M) Reliable variance spectrum, power law decay of exponent 1.14. (N) % of each SVC’s total variance that can be reliably predicted from arousal variables (colors as in E). (O) Percentage of total variance in first 128 dimensions explainable by arousal variables.
We began by analyzing spontaneous activity in mice free to run on an air-floating ball. Six of nine recordings were performed in darkness, but we did not observe differences between these recordings (shown in red on all plots) and recordings with gray screen (yellow on all plots). Mice spontaneously performed behaviors such as running, whisking, sniffing, and other facial movements, which we monitored with an infrared camera.
Ongoing population activity in visual cortex was highly structured (Fig. 1C-H). Correlations between neuron pairs were reliable (Fig. S2), and their spread was larger than would be expected by chance (Fig. 1C,D), suggesting structured activity (30). Fluctuations in the first principal component (PC) occurred over a timescale of many seconds (Fig. S3), and were coupled to running, whisking, and pupil diameter. These arousal-related variables correlated with each other (Fig. S4A,B), and together accounted for approximately 50% of the variance of the first neural PC (Fig. 1E, Fig. S4C). Correlation with the first PC was positive or negative in approximately similar numbers of neurons (57% ± 6.7% SE positive), indicating that two large sub-populations of neurons alternate their activity (Fig. 1F,G). The slowness of these fluctuations suggests a different underlying phenomenon to previously-studied up and down phases (5, 31–34), which alternate at a much faster timescale (~100 ms instead of multiple seconds) and correlate with most neurons positively. Indeed, up/down phases could not even have been detected in our recordings, which scanned the cortex every 400 ms.
Spontaneous activity had a high-dimensional structure, more complex than would be predicted by a single factor such as arousal. We sorted the raster diagram so that nearby neurons showed strong correlations (Figs. 1H, S5). Position on this continuum bore little relation to actual distances in the imaged tissue (Fig. S6), suggesting that this activity was not organized topographically.
Despite its noisy appearance, spontaneous population activity reliably encoded a high-dimensional latent signal (Fig. 1I-K). We devised a method to identify dimensions of neural variance that are reliably determined by common underlying signals, termed Shared Variance Component Analysis (SVCA). We divided the recorded neurons into two spatially segregated sets, and divided the recorded timepoints into two equal halves (training and test; Fig. 1I). The training timepoints were used to find the dimensions in each cell set’s activity that maximally covary with each other. These dimensions are termed Shared Variance Components (SVCs). Activity in the test time-points was then projected onto each SVC (Fig. 1J), and the correlation between projections from the two cell sets (Fig. 1K) provided an estimate of the reliable variance in that SVC (see Methods and Appendix). The fraction of reliable variance in the first SVC was 97% (Fig. 1K,L), implying that only 3% of the variance along this dimension reflected independent noise. The reliable variance fraction of successive SVCs decreased slowly, with the 50th SVC at ~50% reliable variance, and the 512th at ~9% (Fig. 1L).
The magnitude of reliable spontaneous variance was distributed across dimensions according to a power law of exponent 1.14 (Fig. 1M). This value is larger than the power law exponents close to 1.0 seen for stimulus responses (35), but still indicates a high-dimensional signal. The first 128 SVCs together accounted for 86% ± 1% SE of the complete population’s reliable variance, and 67% ± 3% SE of the total variance in these 128 dimensions was reliable. Arousal variables accounted for 11% ± 1% SE of the total variance in these 128 components (16% of their reliable variance), and primarily correlated with the top SVCs (Fig. 1N,O). Thousands of neurons were required to reliably characterize activity in hundreds of dimensions, and the estimated reliability of higher SVCs increased with the number of neurons analyzed (Fig. S7A-E), suggesting that recordings of larger populations would identify yet more dimensions.
Ongoing neural activity encodes multidimensional behavioral information
Although arousal measures only accounted for a small fraction of the reliable variance of spontaneous population activity, it is possible that higher-dimensional measures of ongoing behavior could explain a larger fraction (Fig. 2A-C, Movie S2). We extracted a 1,000-dimensional summary of the motor actions visible on the mouse’s face by applying principal component analysis to the spatial distribution of facial motion at each moment in time (36). The first PC captured motion anywhere on the mouse’s face, and was strongly correlated with explicit arousal measures (Fig. S4B), while higher PCs distinguished different types of facial motion. We predicted neuronal population activity from this behavioral signal using reduced rank regression: for any N, we found the N dimensions of the video signal predicting the largest fraction of the reliable spontaneous variance (Fig. 2D).
Figure 2. Multi-dimensional behavior predicts neural activity.
(A) Frames from a video recording of a mouse’s face. (B) Motion energy, computed as the absolute value of the difference of consecutive frames. (C) Spatial masks corresponding to the top three principal components (PCs) of the motion energy movie. (D) Schematic of reduced rank regression technique used to predict neural activity from motion energy PCs. (E) Cross-validated fraction of successive neural SVCs predictable from face motion (blue), together with fraction of variance predictable from running, pupil and whisking (green), and fraction of reliable variance (the maximum explainable; gray; cf. Fig. 1L). (F) Top: raster representation of ongoing neural activity in an example experiment, with neurons arranged vertically as in Fig. 1H so correlated cells are close together. Bottom: prediction of this activity from facial videography (predicted using separate training timepoints). (G) Percentage of the first 128 SVCs’ total variance that can be predicted from facial information, as a function of number of facial dimensions used. (H) Prediction quality from multidimensional facial information, compared to the amount of reliable variance. (I) Adding explicit running, pupil and whisker information to facial features provides little improvement in neural prediction quality. (J) Prediction quality as a function of time lag used to predict neural activity from behavioral traces.
This multidimensional behavior measure predicted approximately twice as much variance as the three arousal variables (Fig. 2D-J, Movie S3). To visualize how multidimensional behavior predicts ongoing population activity, we compared a raster representation of raw activity (vertically sorted as in Fig. 1H) to the prediction based on multidimensional videography (Fig. 2F, see Fig. S5 for all recordings). To quantify the quality of prediction, and the dimensionality of the behavioral signal encoded in V1, we focused on the first 128 SVCs (accounting for 86% of the population’s reliable variance). The best one-dimensional predictor extracted from the facial motion movie captured the same amount of variance as the best one-dimensional combination of whisking, running, and pupil (Fig. 2G). Prediction quality continued to increase with up to 16 dimensions of video-graphic information (and beyond, in some recordings), suggesting that visual cortex encodes at least 16 dimensions of motor information. These dimensions together accounted for 21%± 1% SE of the total population variance (31% ± 3% of the reliable variance; Fig. 2H), substantially more than the three-dimensional model of neural activity using running, pupil area and whisking (11% ± 1% SE of the total variance, 17% ± 1% SE of the reliable variance). Moreover, adding these three explicit predictors to the video signal increased the explained variance by less than 1% (Fig. 2I), even though the running signal provided information not derived from video. A neuron’s predictability from behavior was not related to its cortical location (Fig. S8). The timescale with which neural activity could be predicted from facial behavior was ~1 s (Figs. 2J, S7H), reflecting the slow nature of these behavioral fluctuations.
Behaviorally-related activity is spread across the brain
Patterns of spontaneous V1 activity were a reflection of activity patterns spread across the brain (Fig. 3A-E). To show this, we used 8 Neuropixels probes (37) to simultaneously record from frontal, sensorimotor, visual and retrosplenial cortex, hippocampus, striatum, thalamus, and midbrain (Fig. 3A,B). The mice were awake and free to rotate a wheel with their front paws. From recordings in three mice, we extracted 2,296, 2,668 and 1,462 units stable across ~1 hour of ongoing activity, and binned neural activity into 1.2 s bins, as for the imaging data.
Figure 3. Behaviorally-related activity across the forebrain in simultaneous recordings with 8 Neuropixels probes.
(A) Reconstructed probe locations of recordings in three mice. (B) Example histology slice showing orthogonal penetrations of 8 electrode tracks through a calbindin-counterstained horizontal section. (C) Comparison of mean correlation between cell pairs in a single area, with mean correlation between pairs with one cell in that area and the other elsewhere. Each dot represents the mean over all cell pairs from all recordings, color coded as in panel D. (D) Mean correlation of cells in each brain region with first principal component of facial motion. Error bars: standard deviation. (E) Top: Raster representation of ongoing population activity for an example experiment, sorted vertically so nearby neurons have correlated ongoing activity. Bottom: prediction of this activity from facial videography. Right: Anatomical location of neurons along this vertical continuum. Each point represents a cell, colored by brain area as in C,D, with x-axis showing the neuron’s depth from brain surface. (F) Percentage of population activity explainable from orofacial behaviors as a function of dimensions of reduced rank regression. Each curve shows average prediction quality for neurons in a particular brain area. (G) Explained variance as a function of time lag between neural activity and behavioral traces. Each curve shows the average for a particular brain area. (H) Same as G in 200ms bins.
Neurons correlated most strongly with others in the same area, but also correlated with neurons in other areas, suggesting non-localized patterns of neural activity (Fig. 3C). All areas contained neurons positively and negatively correlated with the arousal-related top facial motion PC, although neurons in thalamus showed predominantly positive correlations (Fig. 3D, p < 10−8 two-sided Wilcoxon sign-rank test). Sorting the neurons by correlation revealed a rich activity structure (Fig. 3E). All brain areas contained a sampling of neurons from the entire continuum (Fig. 3E, right), suggesting that a multidimensional structure of ongoing activity is distributed throughout the brain. This spontaneous activity spanned at least 128 dimensions, with 35% of the variance of individual neurons reliably predictable from population activity (Fig. S9).
Similar to visual cortical activity, the activity of brainwide populations was partially predictable from facial videography (Fig. 3F-H). Predictability of brain-wide activity again saturated around 16 behavioral dimensions, which predicted on average across areas 21.9% of the total variance (40% of the estimated maximum possible) (Fig. 3F). The amount of behavioral modulation differed between brain regions, with neurons in thalamus predicted best (35.7% of total variance, 59% of estimated maximum). The timescale of videographic prediction was again broad: neural activity was best predicted from instantaneous behavior, decaying slowly over time lags of multiple seconds (Figs. 3G-H, S10), with a full-width at half-max of 2.5±0.4 s (mean ± SE). Neural population activity showed coherent structure at timescales faster than this behavioral correlation (280 ± 43 ms, mean ± SE) (Fig. S10). The fast-timescale structure modulated nearly all neurons in the same direction, leading to rapid fluctuations in the total population rate (“up and down phases”); by contrast, the structure seen at lower time scales was dominated by alternation in the activity of different neuronal populations, and steadier total activity (Figs. S10, S11, S12 (38)).
Stimulus-evoked and ongoing activity overlap along one dimension
We next asked how ongoing activity and behavioral information relates to sensory responses (Fig. 4A-B). We thus interspersed blocks of visual stimulation (flashed natural images, presented 1 per second on average) with extended periods of spontaneous activity (gray screen), while imaging visual cortical population activity (Fig. 4A). During stimulus presentation, the mice continued to exhibit the same behaviors as in darkness, resulting in a similar distribution of facial motion components (Fig. 4B).
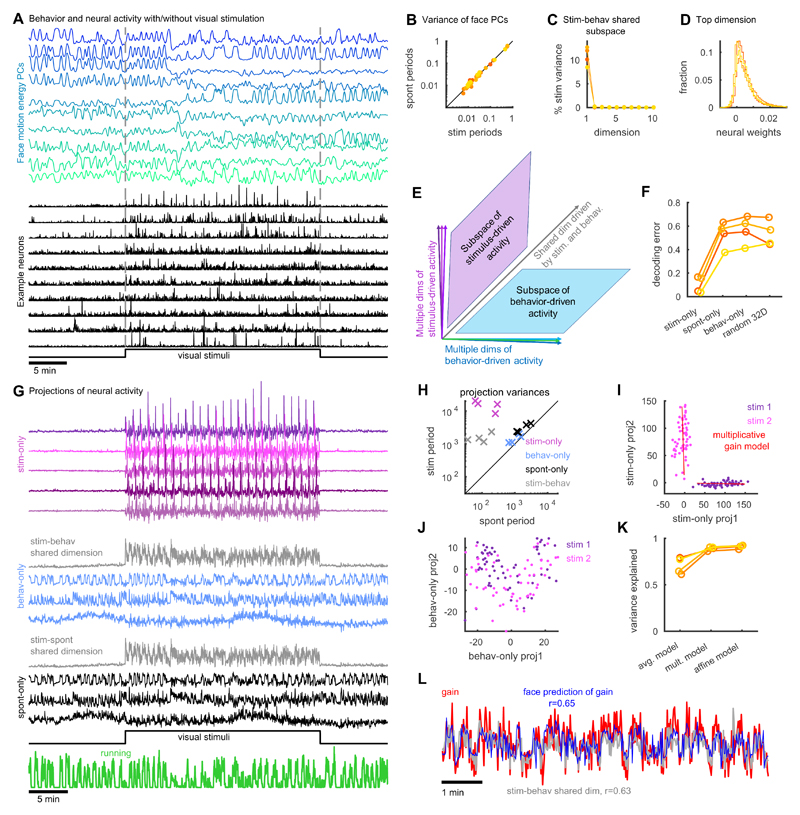
Figure 4. Neural subspaces encoding stimuli and spontaneous/behavioral variables overlap along one dimension.
(A) Principal components of facial motion energy (top) and firing of ten example V1 neurons (bottom). (B) Comparison of face motion energy for each PC during stimulus presentation and spontaneous periods. Color represents recording identity. (C) The percentage of stimulus-related variance in each dimension of the shared subspace between stimulus- and behavior-driven activity. (D) Distribution of cells’ weights on the single dimension of overlap between stimulus and behavior subspaces. (E) Schematic: stimulus- and behavior-driven subspaces are orthogonal, while a single dimension (gray; characterized in panels C,D) is shared. (F) Stimulus decoding analysis for 32 natural image stimuli from 32 dimensions of activity in the stimulus-only, behavior-only, and spontaneous-only subspaces, together with randomly-chosen 32-dimensional subspaces. Y-axis shows fraction of stimuli that were identified incorrectly. (G) Example of neural population activity projected onto these subspaces. (H) Amount of variance of each of the projections illustrated in G, during stimulus presentation and spontaneous periods. Each point represents summed variances of the dimensions in the subspace corresponding to the symbol color, for a single experiment. (I) Projection of neural responses to two example stimuli into two dimensions of the stimulus-only subspace. Each dot is a different stimulus response. Red is the fit of each stimulus response using the multiplicative gain model. (J) Same as I for the behavior-only subspace. (K) Fraction of variance in the stimulus-only subspace explained by: constant response on each trial of the same stimulus (avg. model); multiplicative gain that varies across trials (mult. model); and a model with both multiplicative and additive terms (affine model). (L) The multiplicative gain on each trial (red) and its prediction from the face motion PCs (blue).
There were not separate sets of neurons encoding stimuli and behavioral variables; instead, representations of sensory and behavioral information were mixed together in the same cell population. The fractions of each neuron’s variance explained by stimuli and by behavior were only slightly negatively correlated (Fig. S13; r = -0.18, p < 0.01 Spearman’s rank correlation), and neurons with similar stimulus responses did not have more similar behavioral correlates (Fig. S13; r = -0.005, p > 0.05).
The subspaces encoding sensory and behavior information overlapped in only one dimension (Fig. 4C-E). The space that encoded behavioral variables contained 11% of the total stimulus-related variance, 96% of which was contained in a single dimension (Fig. 4C) with largely positive weights onto all neurons (85% positive weights, Fig. 4D). Similarly, the space of ongoing activity, defined by the top 128 principal components of spontaneous firing, contained 23% of the total stimulus-related variance, 86% of which was contained in one dimension (85% positive weights). Thus, overlap in the spaces encoding sensory and behavioral variables arises primarily because both can change the mean firing rate of the population: the precise patterns of increases and decreases about this change in mean were essentially orthogonal (Fig. 4E). Analysis of electrophysiological recordings confirmed that the relationship between stimulus-driven and spontaneous activity was dominated by a single shared dimension: the correlation between spontaneous and signal correlations was greatly reduced after projecting out this one-dimensional activity (Fig. S14).
Stimulus decoding analysis further confirmed that information about sensory stimuli was concentrated in the stimulus-only subspace. To show this, we trained a linear classifier to identify which stimulus had been presented, from activity in different 32-dimensional neural subspaces. Decoding from the stimulus space yielded a cross-validated error rate of 10.1 ± 4.0 %; activity in the spontaneous- or behavior-only spaces yielded errors of 53.1 ± 6.4 % and 56.8 ± 6.7 %, no better than randomly-chosen dimensions (Fig. 4F).
To visualize how the V1 population integrated sensory and behavior-related activity, we examined the projection of this activity onto three orthogonal subspaces: a multidimensional subspace encoding only sensory information (stimulus-only); a multidimensional subspace encoding only behavioral information (behavior-only); and the one-dimensional subspace encoding both (stimulus-behavior shared dimension) (Fig. 4G; Fig. S15). During gray-screen periods there was no activity in the stimulus-only subspace, but when the stimuli appeared this subspace became very active. By contrast, activity in the behavioronly subspace was present prior to stimulus presentation, and continued unchanged when the stimulus appeared. The one-dimensional shared subspace showed an intermediate pattern: activity in this subspace was weak prior to stimulus onset, and increased when stimuli were presented. Similar results were seen for the spontaneous-only and stimulus-spontaneous spaces (Fig. 4G, lower panels). Across all experiments, variance in the stimulus-only subspace was 119 ± 81 SE times larger during stimulus presentation than during spontaneous epochs (Fig. 4H), while activity in the shared subspace was 19 ± 12 SE times larger; activity in the face-only and spontaneous-only subspaces was only modestly increased by sensory stimulation (1.4 ± 0.13 SE and 1.7 ± 0.2 SE times larger, respectively).
Trial-to-trial variability in population responses to repeated stimulus presentations reflected a combination of multiplicative modulation in the stimulus-space, and additive modulation in orthogonal dimensions. To visualize how stimuli affected activity in these subspaces, we plotted population responses to multiple repeats of two example stimuli (Fig. 4I-J). When projected into the stimulus-only space, the resulting clouds were tightly defined with no overlap (Fig. 4I), but in the behavior-only space, responses to the two stimuli were directly superimposed (Fig. 4J). Variability within the stimulus subspace consisted of changes in the length of the projected activity vectors between trials, resulting in narrowly elongated clouds of points (Fig. 4I), consistent with previous reports of multiplicative variability in stimulus responses (39–42). A model in which stimulus responses are multiplied by a trial-dependent factor accurately captured the data, accounting for 89% ± 0.1% SE of the variance in the stimulus subspace (Fig. 4K). Furthermore, the multiplicative gain on each trial could be predicted from facial motion energy (r = 0.61 ± 0.02 SE, cross-validated), and closely matched activity in the shared subspace (r = 0.73 ± 0.06 SE, cross-validated; Fig. 4L). Although ongoing activity in the behavior-only subspace and visual responses in the stimulus-only subspace added independently, we did not observe additive variability within the stimulus-only space itself: an “affine” model also including an additive term did not significantly increase explained variance over the multiplicative model (p > 0.05, Wilcoxon rank-sum test). Similar results were obtained when analyzing responses to grating stimuli rather than natural images (Fig. S16).
Discussion
Ongoing population activity in visual cortex reliably encoded a latent signal of at least 100 linear dimensions, and possibly many more. The largest dimension correlated with arousal and modulated about half of the neurons positively and half negatively. At least 16 further dimensions were related to behaviors visible by facial videography, which were also encoded across the forebrain. The dimensions encoding motor variables overlapped with those encoding visual stimuli along only one dimension, which coherently increased or decreased the activity of the entire population. Activity in all other behavior-related dimensions continued unperturbed regardless of sensory stimulation. Trial-to-trial variability of sensory responses comprised additive ongoing activity in the behavior subspace, and multiplicative modulation in the stimulus subspace, resolving apparently conflicting findings concerning the additive or multiplicative nature of cortical variability (39–42).
Our data are consistent with previous reports describing low-dimensional correlates of locomotion and arousal in visual cortex (8,10–16, 33), but suggest these results were glimpses of a much larger set of non-visual variables encoded by ongoing activity patterns. 16 dimensions of facial motor activity can predict 31% of the reliable spontaneous variance. The remaining dimensions and variance might in part reflect motor activity not visible on the face or only decodable by more advanced methods (43–48), or they might reflect internal cognitive variables such as motivational drives.
Many studies have reported similarities between spontaneous activity and sensory responses (2–7). We also observed a similarity, but found it arose nearly exclusively from one dimension of neural activity. This dimension summarized the mean activity of all cells in the population, and variations along it reflected both spontaneous alternation of up and down phases and differences in mean population response between stimuli. These results therefore demonstrate that the statistical similarity of firing patterns during stimulation and ongoing activity need not imply recapitulation of previous sensory experiences, merely that cortex exhibits mean rate fluctuations with or without sensory inputs. While our results do not exclude that genuine recapitulation could occur in other behavioral circumstances, they reinforce the need for careful statistical analysis before drawing this conclusion: even a single dimension of common rate fluctuation is sufficient for some previously-applied statistical methods to report similar population activity (49).
The brain-wide representation of behavioral variables suggests that information encoded nearly any-where in the forebrain is combined with behavioral state variables into a mixed representation. We found that these multidimensional signals are present both during ongoing activity and during passive viewing of a stimulus. Recent evidence indicates that they may also be present during a decision-making task (50). What benefit could this ubiquitous mixing of sensory and motor information provide? The most appropriate behavior for an animal to perform at any moment depends on the combination of available sensory data, ongoing motor actions, and purely internal variables such as motivational drives. Integration of sensory inputs with motor actions must therefore occur some-where in the nervous system. Our data indicate that it happens as early as primary sensory cortex. This is consistent with neuroanatomy: primary sensory cortex receives innervation both from neuromodulatory systems carrying state information, and from higher-order cortices which can encode fine-grained behavioral variables (9). This and other examples of pervasive whole-brain connectivity (51–54) may coordinate the brain-wide encoding of behavioral variables we have reported here.
Supplementary Material
Acknowledgements
We thank Michael Krumin for assistance with the two-photon microscopes, and Andy Peters for comments on the manuscript.
Funding: This research was funded by Wellcome Trust Investigator grants (095668, 095669, 108726, 205093, 204915), a European Research Council award (694401) and a grant from the Simons Foundation (SCGB 325512). CS was funded by a four-year Gatsby Foundation PhD studentship. MC holds the GlaxoSmithKline/Fight for Sight Chair in Visual Neuroscience. CS and MP are now funded by HHMI Janelia. NS was supported by postdoctoral fellowships from the Human Frontier Sciences Program and the Marie Curie Action of the EU (656528).
Footnotes
Author contributions: Conceptualization, C.S., M.P., N.S., M.C. and K.D.H.; Methodology, C.S. and M.P.; Software, C.S. and M.P.; Investigation, C.S., M.P., N.S. and C.B.R.; Writing, C.S., M.P., N.S., M.C. and K.D.H; Resources, M.C. and K.D.H. Funding acquisition, M.C. and K.D.H.
Data and materials availability: All of the processed deconvolved calcium traces are available on figshare (55) (https://figshare.com/articles/Recordings_of_ten_thousand_neurons_in_visual_cortex_during_spontaneous_behaviors/6163622), together with the behavioral traces (face motion components, whisking, running and pupil). All the spike-sorting electrophysiology and the corresponding behavioral traces are also on figshare (56) (https://janelia.figshare.com/articles/Eight-probe_Neuropixels_recordings_during_spontaneous_behaviors/7739750). The code for the analysis and the figures is available on github (https://github.com/MouseLand/stringer-pachitariu-et-al-2018a)
Declaration of interests
The authors declare no competing interests.
References
- 1.Ringach DL. Current Opinion in Neurobiology. 2009;19:439. doi: 10.1016/j.conb.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffman K, McNaughton B. Science. 2002;297:2070. doi: 10.1126/science.1073538. [DOI] [PubMed] [Google Scholar]
- 3.Kenet T, Bibitchkov D, Tsodyks M, Grinvald A, Arieli A. Nature. 2003;425:954. doi: 10.1038/nature02078. [DOI] [PubMed] [Google Scholar]
- 4.Han F, Caporale N, Dan Y. Neuron. 2008;60:321. doi: 10.1016/j.neuron.2008.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luczak A, Barthó P, Harris KD. Neuron. 2009;62:413. doi: 10.1016/j.neuron.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkes P, Orbán G, Lengyel M, Fiser J. Science. 2011;331:83. doi: 10.1126/science.1195870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ONeill J, Pleydell-Bouverie B, Dupret D, Csicsvari J. Trends in neurosciences. 2010;33:220. doi: 10.1016/j.tins.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Niell CM, Stryker MP. Neuron. 2010;65:472. doi: 10.1016/j.neuron.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petreanu L, et al. Nature. 2012;489:299. doi: 10.1038/nature11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polack P-O, Friedman J, Golshani P. Nature Neuroscience. 2013;16:1331. doi: 10.1038/nn.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGinley MJ, David SV, McCormick DA. Neuron. 2015;87:179. doi: 10.1016/j.neuron.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vinck M, Batista-Brito R, Knoblich U, Cardin JA. Neuron. 2015;86:740. doi: 10.1016/j.neuron.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pakan JM, et al. eLife. 2016;5:e14985. [Google Scholar]
- 14.Dipoppa M, et al. Neuron. 2018;98:602. doi: 10.1016/j.neuron.2018.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reimer J, et al. Nature Communications. 2016;7 doi: 10.1038/ncomms12951. 13289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saleem AB, Ayaz A, Jeffery KJ, Harris KD, Carandini M. Nature Neuroscience. 2013;16:1864. doi: 10.1038/nn.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentet LJ, et al. Nature Neuroscience. 2012;15:607. doi: 10.1038/nn.3051. [DOI] [PubMed] [Google Scholar]
- 18.Peron SP, Freeman J, Iyer V, Guo C, Svoboda K. Neuron. 2015;86:783. doi: 10.1016/j.neuron.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 19.Schneider DM, Nelson A, Mooney R. Nature. 2014;513:189. doi: 10.1038/nature13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williamson RS, Hancock KE, Shinn-Cunningham BG, Polley DB. Current Biology. 2015;25:1885. doi: 10.1016/j.cub.2015.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erisken S, et al. Current Biology. 2014;24:2899. doi: 10.1016/j.cub.2014.10.045. [DOI] [PubMed] [Google Scholar]
- 22.Roth MM, et al. Nature Neuroscience. 2016;19:299. doi: 10.1038/nn.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez LM, et al. Cerebral Cortex. 2016;27:5444. doi: 10.1093/cercor/bhw311. [DOI] [PubMed] [Google Scholar]
- 24.Arieli A, Sterkin A, Grinvald A, Aertsen A. Science. 1996;273:1868. doi: 10.1126/science.273.5283.1868. [DOI] [PubMed] [Google Scholar]
- 25.van Vreeswijk C, Sompolinsky H. Science. 1996;274:1724. doi: 10.1126/science.274.5293.1724. [DOI] [PubMed] [Google Scholar]
- 26.Ruff DA, Cohen MR. Nature Neuroscience. 2014;17:1591. doi: 10.1038/nn.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno-Bote R, et al. Nature Neuroscience. 2014;17:1410. doi: 10.1038/nn.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montijn JS, Meijer GT, Lansink CS, Pennartz CM. Cell reports. 2016;16:2486. doi: 10.1016/j.celrep.2016.07.065. [DOI] [PubMed] [Google Scholar]
- 29.Pachitariu M, et al. bioRxiv. 2016 061507. [Google Scholar]
- 30.Cohen MR, Kohn A. Nature Neuroscience. 2011;14:811. doi: 10.1038/nn.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okun M, et al. Nature. 2015;521:511. doi: 10.1038/nature14273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engel TA, et al. Science. 2016;354:1140. doi: 10.1126/science.aag1420. [DOI] [PubMed] [Google Scholar]
- 33.Stringer C, et al. eLife. 2016;5:e19695. [Google Scholar]
- 34.Constantinople CM, Bruno RM. Neuron. 2011;69:1061. doi: 10.1016/j.neuron.2011.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stringer C, Pachitariu M, Steinmetz N, Carandini M, Harris KD. bioRxiv. 2018 374090. [Google Scholar]
- 36.Powell K, Mathy A, Duguid I, Häusser M. eLife. 2015;4 doi: 10.7554/eLife.07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jun JJ, et al. Nature. 2017;551:232. doi: 10.1038/nature24636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okun M, Steinmetz NA, Lak A, Dervinis M, Harris KD. bioRxiv. 2018 395251. [Google Scholar]
- 39.Goris RLT, Movshon JA, Simoncelli EP. Nature Neuroscience. 2014;17:858. doi: 10.1038/nn.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.I-C Lin, Okun M, Carandini M, Harris KD. Neuron. 2015;87:644. doi: 10.1016/j.neuron.2015.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dadarlat MC, Stryker MP. Journal of Neuroscience. 2017;37:3764. doi: 10.1523/JNEUROSCI.2728-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christensen AJ, Pillow JW. bioRxiv. 2017 214007. [Google Scholar]
- 43.Brown AEX, Yemini EI, Grundy LJ, Jucikas T, Schafer WR. Proceedings of the National Academy of Sciences. 2013;110:791. doi: 10.1073/pnas.1211447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Machado AS, Darmohray DM, Fayad J, Marques HG, Carey MR. eLife. 2015;4 doi: 10.7554/eLife.07892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robie AA, et al. Cell. 2017;170:393. doi: 10.1016/j.cell.2017.06.032. [DOI] [PubMed] [Google Scholar]
- 46.Wiltschko AB, et al. Neuron. 2015;88:1121. doi: 10.1016/j.neuron.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurnikova A, Moore JD, Liao S-M, Deschłnes M, Kleinfeld D. Current Biology. 2017;27:688. doi: 10.1016/j.cub.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathis A, et al. Nature Neuroscience. 2018;21:1281. doi: 10.1038/s41593-018-0209-y. [DOI] [PubMed] [Google Scholar]
- 49.Okun M, et al. Journal of Neuroscience. 2012;32:17108. doi: 10.1523/JNEUROSCI.1831-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Musall S, Kaufman MT, Gluf S, Churchland AK. bioRxiv. 2018 308288. [Google Scholar]
- 51.Gămănuţ R, et al. Neuron. 2018;97:698. doi: 10.1016/j.neuron.2017.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris JA, et al. bioRxiv. 2018 292961. [Google Scholar]
- 53.Economo MN, et al. eLife. 2016;5 doi: 10.7554/eLife.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han Y, et al. Nature. 2018;556:51. doi: 10.1038/nature26159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stringer C, Pachitariu M, Reddy C, Carandini M, Harris KD. Recordings of ten thousand neurons in visual cortex during spontaneous behaviors. 2018 doi: 10.25378/janelia.6163622.v4. [DOI] [Google Scholar]
- 56.Steinmetz N, Pachitariu M, Stringer C, Carandini M, Harris KD. Eight-probe neuropixels recordings during spontaneous behaviors. 2019 doi: 10.25378/janelia.7739750. [DOI] [Google Scholar]
- 57.Steinmetz NA, et al. eNeuro. 2017;4 ENEURO. [Google Scholar]
- 58.Deng J, et al. Computer Vision and Pattern Recognition, 2009. CVPR 2009. IEEE Conference; IEEE; 2009. pp. 248–255. [Google Scholar]
- 59.Chen T-W. Nature. 2013;499:295. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pachitariu M, Stringer C, Harris KD. Journal of Neuroscience. 2018 doi: 10.1523/JNEUROSCI.3339-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Friedrich J, Zhou P, Paninski L. PLOS Computational Biology. 2017;13:e1005423. doi: 10.1371/journal.pcbi.1005423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ludwig KA, et al. Journal of Neurophysiology. 2009;101:1679. doi: 10.1152/jn.90989.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pachitariu M, Steinmetz NA, Kadir SN, Carandini M, Harris KD. Advances in Neural Information Processing Systems. 2016:4448–4456. [Google Scholar]
- 64.Izenman AJ. Journal of Multivariate Analysis. 1975;5:248. [Google Scholar]
- 65.Harris KD, Csicsvari J, Hirase H, Dragoi G, Buzsáki G. Nature. 2003;424:552. doi: 10.1038/nature01834. [DOI] [PubMed] [Google Scholar]
- 66.Pillow JW, et al. Nature. 2008;454:995. doi: 10.1038/nature07140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.London M, Roth A, Beeren L, Häusser M, Latham PE. Nature. 2010;466:123. doi: 10.1038/nature09086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.