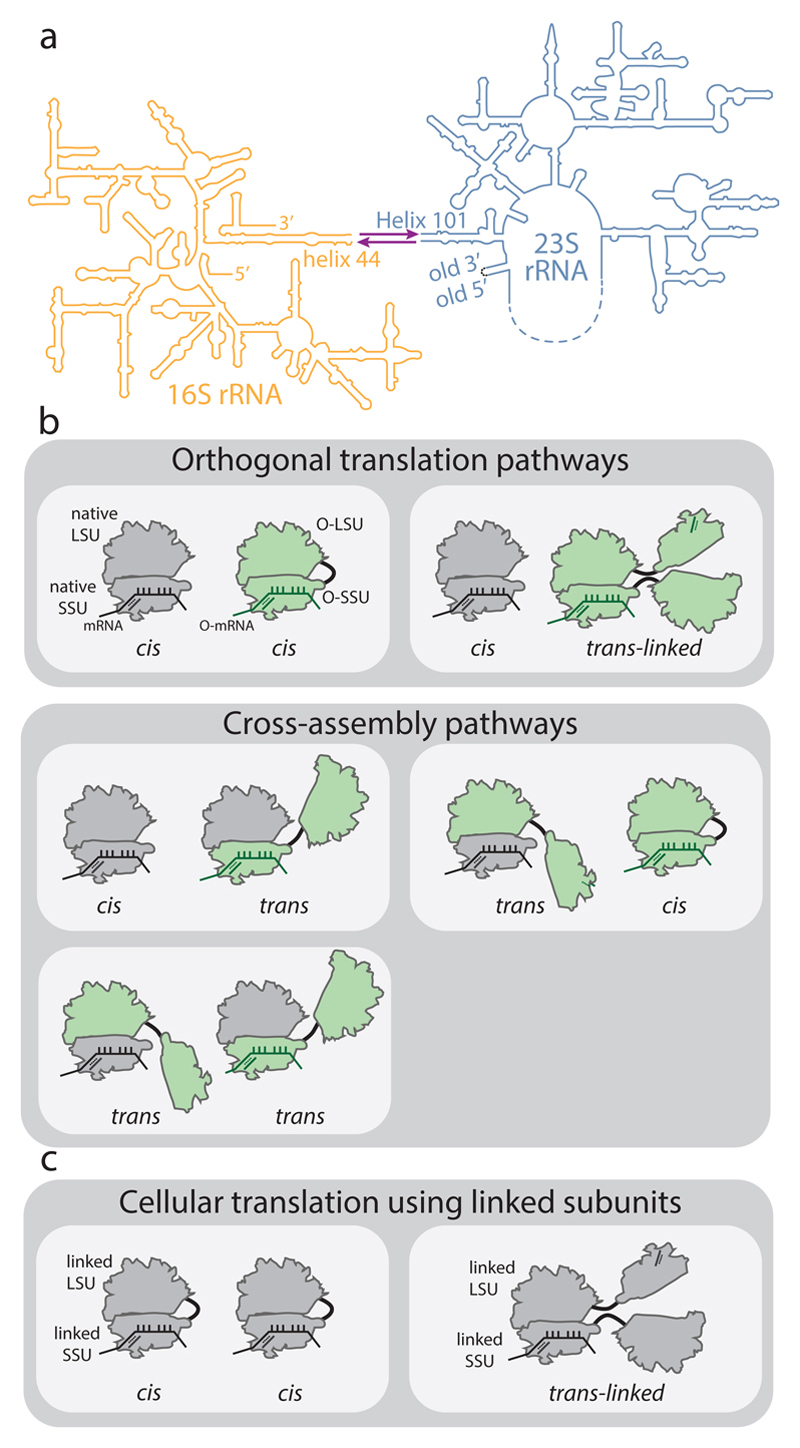
Figure 1. Ribosome stapling and potential interactions of linked subunits in vivo.
a, Secondary structure of RNA from ribosomes with linked subunits. A single rRNA transcript is generated by inserting a circularly permuted 23S rRNA (blue) into a split 16S rRNA (yellow), with the 16S and 23S rRNAs linked together by an RNA linker (purple). In stapled ribosomes the linker is a staple derived from the J5-J5a region of the Tetrahymena group I intron. The original O-stapled ribosome, referred to here as O-d0d0, directly links h44 and H101 through the staple. b, Potential interactions of linked ribosomal subunits in vivo. Cells (white boxes) containing O-ribosomes with linked subunits as well as endogenous (native) subunits may associate to create orthogonal translation pathways or cross-assembly pathways. An orthogonal translation complex is created by directing the association of an orthogonal small ribosome subunit (O-SSU) with its linked large subunit (O-LSU) in cis, or by forming trans-linked complexes. Linked ribosome subunits may interact with native ribosome subunits in trans if the linker is insufficient to direct assembly in cis. High concentrations of native ribosome subunits in the cytoplasm under physiological conditions counteract the effects of tethering and may lead to cross-assembly. c, In E. coli strains containing solely ribosomes with covalently linked subunits, cis- or trans-linked complexes may form.