Abstract
Variants of triggering receptor expressed on myeloid cells 2 (TREM2) are associated with an increased incidence of Alzheimer’s disease, as well as other neurodegenerative disorders. Using a newly developed, highly sensitive reporter cell model, consisting of Jurkat T cells stably overexpressing a reporter gene and a gene encoding TREM2DAP12 fusion protein, we show here that TREM2-dependent signal transduction in response to apoptotic Neuro2a cells is mediated by aminophospholipid ligands, phosphatidylserine and phosphatidylethanolamine, which are not exposed on the intact cell surface, but become exposed upon apoptosis. We also show that signal-transducing TREM2 ligands different from aminophospholipids, which appear to be derived from neurons, might be present in membrane fractions of mouse cerebral cortex. These results may suggest that TREM2 regulates microglial function by transducing intracellular signals from aminophospholipids on apoptotic cells, as well as unidentified ligands in the membranes of the cerebral cortex.
Subject terms: Biological sciences, Biochemistry
Introduction
Microglia, which are resident myeloid cells in the brain, provide acute and chronic surveillance of damage- and pathogen-associated signals as a part of the innate immune system1. The role of microglia in neurodegenerative disorders associated with chronic inflammation has been investigated extensively. In the Alzheimer’s disease (AD) brain, for example, reactive microglia are associated with amyloid plaques consisting of aggregated amyloid β peptide (Aβ)2–4. Aβ stimulates microglia and induces proinflammatory phenotypes that are thought to damage neurons and exacerbate disease progression5,6. On the other hand, microglia have protective roles against AD through phagocytosis of Aβ and apoptotic neurons7,8. Thus, proper regulation of diverse microglial functions should ameliorate disease progression. Genetic studies have identified several AD-associated risk factors expressed by microglia9–12. These include triggering receptor expressed on myeloid cells-2 (TREM2)13,14, whose homozygous mutations were found in certain types of neurodegenerative disorders, such as Nasu-Hakola disease and frontotemporal dementia-like syndromes15–17. Heterozygous mutations of TREM2 show a high odds ratio for the onset of AD13,14 as well as the other neurodegenerative disorders18–20. So far, R47H and R62H variants of TREM2 have been confirmed to present a significant risk for AD21, though other variants have not reached statistical significance, probably because of their low frequency. In addition, R47H mutation is correlated with increased tau levels in cerebrospinal fluid22 and increased phosphorylated tau levels around amyloid plaques23 in AD brains. Elevated expression of TREM2 gene is reported to ameliorate pathological phenotypes in AD models24. These studies highlight that functional alterations of TREM2 due to mutation contribute to the development of neurodegenerative disorders, possibly by loss of function25,26, and indicate that a better understanding of the role of TREM2 could provide new insights for drug discovery.
TREM2 is a type I transmembrane protein associated with DNAX-activating protein of 12 kDa (DAP12) in the transmembrane domain, and DAP12 has an immunoreceptor tyrosine-based activating motif (ITAM) in the cytosolic domain. TREM2 is therefore postulated to bind ligands at its extracellular immunoglobulin V-type domain and to transduce intracellular signals that regulate microglial functions such as cytokine production, migration, proliferation, phagocytosis, cell survival, synapse elimination, and compaction of amyloid plaques23,27–34. Notably, inflammatory cytokines are downregulated in Trem2-knockout mice30,31,33, and anti-Trem2 agonistic antibody induces inflammatory cytokines in the brain35. These results suggest that TREM2 has a role in the production of inflammatory cytokines in vivo. Interestingly, a functional interaction between TREM2 and familial AD gene presenilin 1 has been reported36. Proper signal transduction might be disturbed by the risk-associated variants, leading to increased incidence of neurodegenerative disorders. Many proteins or compounds that bind to the TREM2 extracellular domain have been reported, including phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylcholine (PC), sulfoglycolipid, apolipoproteins, low-density lipoprotein, high-density lipoprotein, heat shock protein 60, DNA, E. coli, apoptotic cells, and Aβ31,37–54. Among them, apoptotic cells31,40, normal cultured cells39,40, and oligomeric Aβ53,54 have been confirmed to induce TREM2-mediated intracellular signalling. However, the ligands expressed by apoptotic or normal cultured cells have not been identified, and it is not clear whether other signal-transducing ligands of TREM2 are present in the brain. In addition, further investigations are required to understand the different pathological effects of TREM2 variants on the signal transduction50.
In this work, we have developed a highly sensitive reporter cell model to monitor signal transduction from TREM2, in order to identify the TREM2 ligands on apoptotic and normal cultured cells and to search for other ligands in the brain that may be associated with increased incidence of AD. We found that the ligands expressed on apoptotic Neuro2a cells, as well as some normal cultured cells, are aminophospholipids. Finally we suggest that other TREM2 signal-transducing ligands are present in the membranes of mouse cerebral cortex.
Results
Establishment of reporter cell models for TREM2-dependent signal transduction
To establish sensitive and specific reporter cell models to monitor TREM2 signal transduction, RAW264.7, 2B4, and Jurkat cells were stably transfected with reporter genes: nuclear factor of activated T cells (NFAT) promoter element and TREM2DAP12 fusion cDNA corresponding to TREM2 extracellular domain and DAP12 transmembrane and cytosolic domain38,55,56. RAW264.7 was chosen because it is a macrophage cell line having similar characteristics to microglia, such as phagocytosis and cytokine production. 2B4 and Jurkat cell lines were chosen because they are derived from T cells, which are the only cell type so far shown to transduce signals via TREM231,37,40. These reporter cells were incubated with apoptotic Neuro2a cells to induce signal transduction via TREM240 and luminescence or fluorescence of the cell lysates was measured. RAW264.7 reporter cells expressing TREM2DAP12 showed increased luminescence upon apoptotic cell treatment, but RAW264.7 cells not expressing TREM2DAP12 also showed increased luminescence, indicating that the signal is independent of exogenous TREM2DAP12 (not shown). On the other hand, 2B4 reporter cells expressing TREM2DAP12 showed a 2- to 3-fold increase of fluorescence upon apoptotic cell treatment, while the parent 2B4 cells (control) did not (not shown). More strikingly, Jurkat reporter cells expressing TREM2DAP12 showed a luminescence increase of more than 10-fold, while the parent cells did not (see Figs 1 to 3). These results confirmed that apoptotic cells express TREM2 ligand(s) that transduce an intracellular signal leading to NFAT activation31,40, and that T cell lines are suitable to study TREM2 signal transduction from apoptotic cells. In the following study, we used Jurkat reporter cells because of the high signal-to-background ratio.
Figure 1.
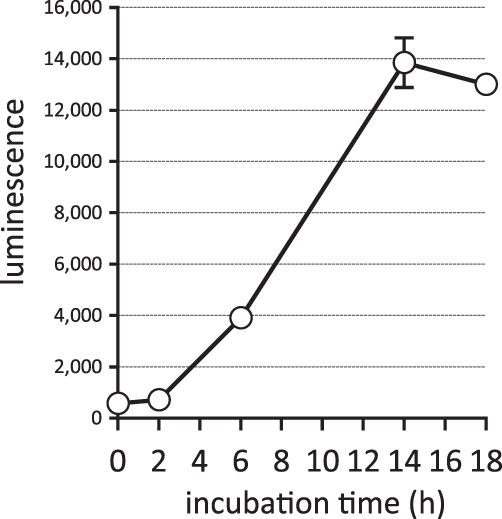
Time course of NAFT signal activation in Jurkat cell clones expressing TREM2DAP12 in the presence of apoptotic cells. Equal numbers of apoptotic cells and Jurkat reporter cell clones expressing TREM2DAP12 were incubated for the indicated times (hours) and the luminescence of the cell lysates was measured. Data are shown as means ± SD (n = 3). Where SDs are not shown, they are smaller than the symbols.
Figure 3.
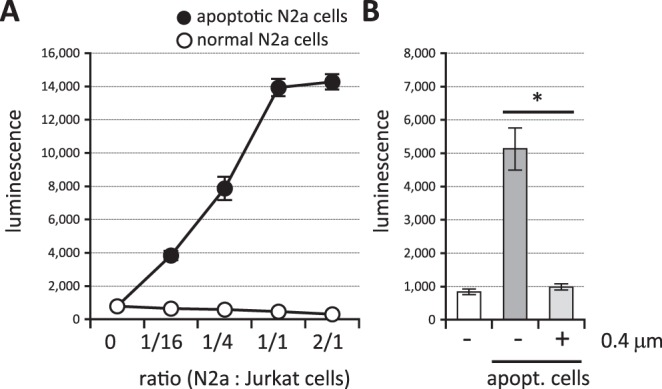
TREM2 ligands are exposed at the cell surface upon apoptosis. (A) Apoptotic (closed circles) or normal (open circles) Neuro2a cells and reporter cell clones expressing TREM2DAP12 were incubated with the indicated ratio of Neuro2a: the reporter cell numbers and the luminescence of cell lysates were measured. Data are shown as means ± SD (n = 3). Where SDs are not shown, they are smaller than the symbols. (B) Apoptotic cells and reporter cell clones expressing TREM2DAP12 were incubated at a ratio of 1/10 with or without culture inserts (0.4 μm pore size) and the luminescence of cell lysates was measured. Data are shown as means ± SD (n = 4). One-way ANOVA with the Student-Newman-Keuls test was performed. *p < 0.05.
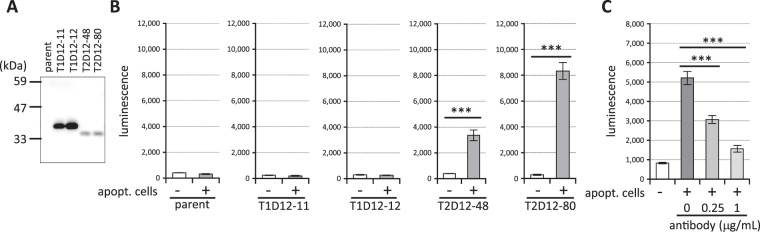
We first investigated the time course of the NFAT activation by apoptotic cells. As shown in Fig. 1, luminescence of Jurkat reporter cell clones expressing TREM2DAP12 began to increase at 6 hours and reached a plateau at 14 hours when equal numbers of apoptotic cells and reporter cells were incubated. We then examined whether or not the signal is TREM2-dependent. Reporter cell clones expressing TREM2DAP12 or TREM1DAP12 or parent reporter cells (Fig. 2A) were incubated together with apoptotic cells, and the luminescence of the cell lysates was measured. As shown in Fig. 2B, two independent reporter cell clones expressing TREM2DAP12 showed increased luminescence upon apoptotic cell treatment, while the parent reporter cells and two independent reporter cell clones expressing TREM1DAP12 did not, even though the amounts of TREM1DAP12 proteins expressed in the reporter cells were much greater than those of TREM2DAP12 proteins (Fig. 2A). All of five independent TREM2DAP12-expressing cell clones that we established showed increased luminescence in response to the apoptotic cells (not shown), although the efficacy of signal transduction was different in each case. Moreover, anti-TREM2 antibody inhibited the reporter signal in a dose-dependent manner (Fig. 2C). These results indicate that the signal transduced in the TREM2DAP12-expressing reporter cells exposed to apoptotic cells is TREM2-dependent.
Figure 2.
Establishment of TREM2-dependent reporter cells. (A) Lysates of Jurkat reporter cell clones expressing TREM1DAP12, TREM2DAP12 (11, 12, 48 and 80 indicate clone numbers) or parent reporter cells (parent) were immunoblotted by anti-DAP12 antibody. (B) Apoptotic cells (apoptot. cells) and the Jurkat reporter cell clones were incubated at a ratio of 1/2 and the luminescence of the cell lysates was measured. Data are shown as means ± SD (n = 3). Two-tailed Student’s t test was performed. (C) Apoptotic cells and Jurkat TREM2DAP12 reporter cell clones were incubated at a ratio of 1/10 together with anti-TREM2 antibodies and the luminescence of the cell lysates was measured. Data are shown as means ± SD (n = 3). One-way ANOVA with the Student-Newman-Keuls test was performed. ***p < 0.001.
TREM2 ligands expressed on apoptotic cells are aminophospholipids
Next, we examined the characteristics of the TREM2 ligands expressed in apoptotic cells. We incubated the reporter cells with apoptotic or normal cultured Neuro2a cells and measured the intensity of luminescence in the cell lysates. We found that apoptotic Neuro2a cells transduced the signal in a dose-dependent manner, while normal cultured Neuro2a cells transduced a signal similar to or weaker signal than that obtained with medium alone (Fig. 3A). When the reporter cells and apoptotic cells were cultured without direct contact (separated by a culture insert membrane with 0.4 μm pore size), the signal was almost completely blocked (Fig. 3B). These results suggest that TREM2 ligands are exposed on the cell surface upon apoptosis, and are not factors secreted from the cells.
These characteristics of TREM2 ligands are reminiscent of aminophospholipids, PS or PE, which are localized in the inner leaflet of the plasma membrane in intact cells, but become exposed when the cells are triggered to undergo apoptosis57. Recombinant protein MFGE8-L D89E, which retains the binding domain to aminophospholipids but cannot bind phagocytes expressing integrins58, was purified to near homogeneity (Fig. 4A) and added to the culture medium of the reporter cells and apoptotic cells. As shown in Fig. 4B, MFGE8-L D89E completely inhibited the signal induced by apoptotic cells, while control protein (BSA) did not. This strongly indicated that the TREM2 ligands on the apoptotic cells are indeed aminophospholipids. In accordance with this observation, pure aminophospholipids PS and PE induced signal transduction in the reporter cells, while PC, which is located on the exterior of intact cells, did not (Fig. 4C). The EC50 values for PS and PE could not be determined because the reporter cells died before the luminescence reached a plateau in the presence of higher concentrations of the lipids (not shown).
Figure 4.
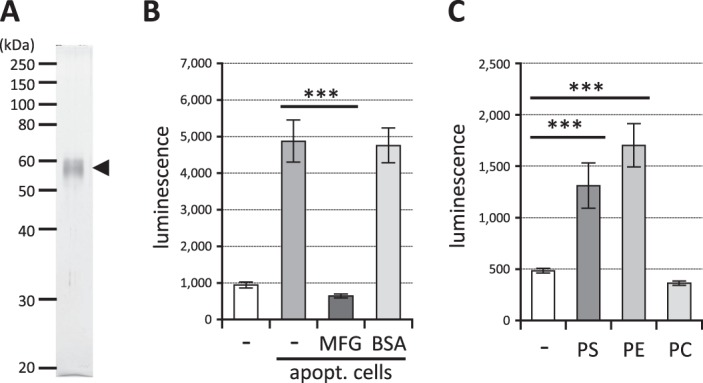
TREM2 ligands on apoptotic cells are aminophospholipids. (A) MFGE8-L D89E proteins were purified by anti-DDDDK antibody affinity column from culture media of HEK293 cells transfected with the MFGE8-L D89E cDNA. The purified proteins were electrophoresed and visualized by silver staining. (B) Apoptotic cells and reporter cell clones expressing TREM2DAP12 were incubated at a ratio of 1/10 together with 1 μg/mL MFGE8-L D89E or BSA as a control, and the luminescence of the cell lysates was measured. Data are shown as means ± SD (n = 3). One-way ANOVA with the Student-Newman-Keuls test was performed. (C) The reporter cell clones expressing TREM2DAP12 were cultured in wells coated with purified PS, PE or PC and the luminescence of the cell lysates was measured. Data are shown as means ± SD (n = 3). One-way ANOVA with the Student-Newman-Keuls test was performed. ***p < 0.001.
TREM2 ligands expressed on normal cultured cells are aminophospholipids
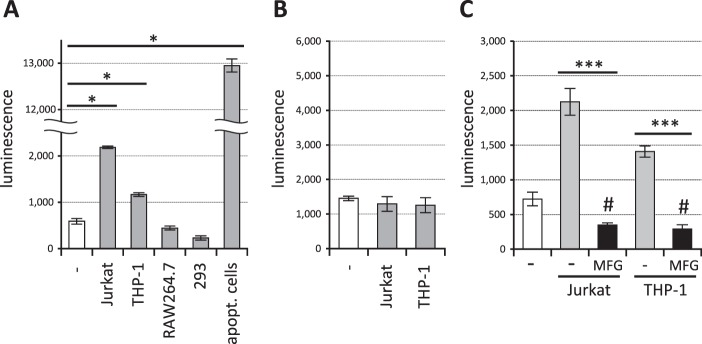
We then explored whether or not normal cultured cell lines express signal-transducing ligands of TREM2, because some cultured cell lines were shown to transduce NFAT signalling via TREM239,40. We incubated the reporter cells with various cell lines and measured the luminescence. As shown in Fig. 5A normal cultured Jurkat and THP-1 cells induced signal transduction in TREM2DAP12-expressing reporter cells although the strength of luminescence was much less than that of apoptotic cells. This signal was TREM2-dependent, because TREM1DAP12-expressing reporter cells showed no signal transduction (Fig. 5B). When MFGE8-L D89E protein was added to the cultures, the signal was inhibited (Fig. 5C), suggesting that the TREM2 ligands on these normal cultured cells are also aminophospholipids, which might have become exposed during normal culture (see discussion).
Figure 5.
TREM2 ligands on normal cultured cells are aminophospholipids. (A) Various normal cultured cell lines and reporter cell clones expressing TREM2DAP12 were incubated at a ratio of 1/1 and the luminescence of the cell lysates was measured. Data are shown as means ± SD (n = 3). One-way ANOVA with the Student-Newman-Keuls test was performed. (B) Jurkat or THP-1 cell lines and reporter cell clones expressing TREM1DAP12 were incubated at a ratio of 1/1 and the luminescence of the cell lysates was measured. Data are shown as means ± SD (n = 3). (C) Jurkat or THP-1 cells and the reporter cell clones expressing TREM2DAP12 were incubated in a ratio of 1/1 together with 1 μg/mL MFGE8-L D89E and the luminescence of cell lysates was measured. Data are shown as means ± SD (n = 3). One-way ANOVA with the Student-Newman-Keuls test was performed. *p < 0.05; ***p < 0.001; #p < 0.01 compared to the non-treated reporter cells (white column).
TREM2 ligands expressed in the brain are not aminophospholipids
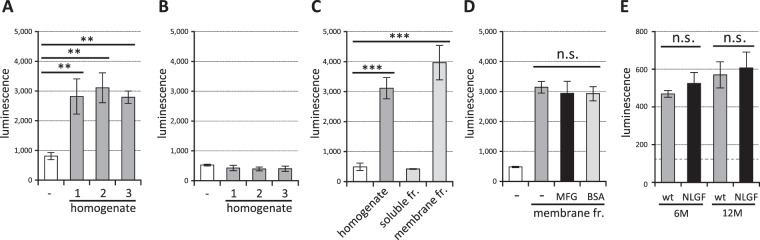
Finally, we searched for TREM2 signal-transducing ligands in the brain. We incubated homogenates of cerebral cortices from young adult mice (5 months old) with reporter cells expressing TREM2DAP12 and measured the luminescence. We found that homogenates prepared from three independent mice increased the reporter activity (Fig. 6A) suggesting that the cerebral cortex contains ligand(s) that induce NFAT signalling via TREM2. This signal was TREM2-dependent, because reporter cells expressing TREM1DAP12 (Fig. 6B) or parent control cells (not shown) showed no signal. We then found that the ligands were present in the membrane fraction, not soluble fraction of the homogenates (Fig. 6C). Unexpectedly, the signal induced by the membrane fraction was not inhibited by MFGE8-L D89E (Fig. 6D); this result suggests that the ligands in the mouse cerebral cortical membranes are different from aminophospholipids. To examine if the ligands are expressed by astrocytes, microglia or neurons, we compared the TREM2 signal-inducing activity of cerebral cortices derived from wild-type and App knock-in mice carrying NL-G-F mutations (AppNL-G-F)59, since the AppNL-G-F mice have increased numbers of astrocytes and microglia in an aging-dependent manner after 6 months60. However, we found no significant difference of TREM2 signal transduction between the two strains at 6 or 12 months (Fig. 6E) suggesting that the ligands might not be expressed by astrocytes or microglia.
Figure 6.
TREM2 ligands in membrane fractions of mouse cerebral cortex are not aminophospholipids. (A) Homogenates of cerebral cortices from three 5-month-old mice (no. 1 to 3) and reporter cell clones expressing TREM2DAP12 were incubated and the luminescence of cell lysates was measured. Data are shown as means ± SD (n = 3). One-way ANOVA with the Student-Newman-Keuls test was performed. (B) Homogenates of cerebral cortex from 5-month-old mice (no. 1 to 3) and reporter cell clones expressing TREM1DAP12 were incubated and the luminescence of cell lysates was measured. Data are shown as means ± SD (n = 3). (C) Homogenates, soluble fraction or membrane fraction of the cerebral cortex and reporter cell clones expressing TREM2DAP12 were incubated and the luminescence of the cell lysates was measured. Data are shown as means ± SD (n = 3). One-way ANOVA with the Student-Newman-Keuls test was performed. (D) Membrane fractions of the cerebral cortex and reporter cell clones expressing TREM2DAP12 were incubated together with 1 μg/mL MFGE8-L D89E or BSA and the luminescence of the cell lysates was measured. Data are shown as means ± SD (n = 3). **p < 0.01; ***p < 0.001; n.s. not significant. (E) Homogenates of cerebral cortices from 6- and 12-month wild-type and AppNL-G-F knock-in mice and reporter cell clones expressing TREM2DAP12 were incubated and the luminescence of cell lysates was measured. Luminescence of the reporter cells incubated with the buffer alone is shown as a broken line. Data are means ± SE (n = 3), and compared using Student’s t test. n.s. not significant.
Discussion
We have established a highly sensitive and specific reporter cell model for TREM2 signal transduction in Jurkat T cells. So far, two cell lines, BWZ and 2B4 T cells, have been reported as TREM2 signal-transducing cell models31,37,40. Hsieh et al. transfected both TREM2/DAP12 cDNAs in BWZ cells expressing lacZ gene under the control of NFAT promoter, and Wang et al. introduced TREM2/DAP12 cDNAs in 2B4 cells stably expressing EGFP under the control of NFAT promoter. In the presence of apoptotic cells, the BWZ and 2B4 reporter cells showed about 4-fold and 1.2-fold increases of the reporter activity, respectively31,40. The Jurkat reporter cell model established in this study is even more sensitive, as the luciferase activity was increased by more than 10-fold in the presence of apoptotic cells (Figs 1 to 3). Possible reasons for this are that Jurkat cells may have greater amounts of signalling molecules required for NFAT signal transduction via TREM2, or that TREM2DAP12 fusion proteins are better able to transduce the signal than independently expressed TREM2 and DAP12. Another possibility might be that the luc2P reporter gene used in this study has humanized codons, which may be favorable for higher expression and reduced anomalous transcription. The luc2P gene also contains hPEST, a protein destabilization sequence, which allows luc2P protein levels to respond more quickly. A further advantage is that enzymatic detection of the luc2P is faster (5 min) than that of LacZ40. In addition, the procedure is simpler than sorting EGFP-expressing cells31, and is suitable for high-throughput screening of the agonists/antagonists. At the beginning of this study we tried to establish reporter cell models in RAW264.7 macrophage cell lines, but these cells transduced the signal irrespective of overexpression of TREM2DAP12. Endogenous expression of TREM2 and DAP12 in RAW264.7 cells61,62 might inhibit the signal from the overexpressed TREM2DAP12. As we and others have shown, T cell lines are suitable models for TREM2 signal transduction, probably because T cells express little endogenous TREM2 or DAP12 and share common signalling molecules with microglia, considering that T cell receptors and DAP12 have the same ITAM at the cytoplasmic domains.
The Jurkat reporter cells expressing TREM2DAP12 transduced the signal when incubated with apoptotic cells, while those expressing TREM1DAP12 did not (Fig. 2B), indicating the specificity of these reporter cells. We speculate that ligand recognition involves specific amino acid residue(s) in TREM2. Identification of the ligand-binding site in TREM2 would facilitate an understanding the interaction between TREM2 and the ligands, and would be helpful for drug development. The TREM2 antibody clone 237920 works as an antagonist of the NFAT signal via TREM2 induced by apoptotic cells (Fig. 2C). This may suggest that the epitope of the antibody is located near the binding site to apoptotic cells. Thus, mapping of the epitope might help to identify the ligand-binding site in TREM2. It would be interesting to see whether or not the epitope is identical to the reported PS-binding site in TREM2 extracellular domain26.
Apoptotic cells transduce the NFAT signal via TREM231,40, while aminophospholipids such as PS and PE also transduce the signal via TREM231,48. However, it has not been reported whether the TRME2 ligands in apoptotic cells are aminophospholipids. In this study we established for the first time that the TREM2 ligands in apoptotic cells are aminophospholipids by demonstrating inhibition of the signal with MFGE8 (Fig. 4B), an aminophospholipid-binding protein that inhibits the interaction between apoptotic cells and phagocytes58. This conclusion is supported by our findings that purified aminophospholipids (PS and PE), but not PC, transduce the signal (Fig. 4C), and that direct binding between reporter cells and apoptotic cells is required for signal transduction (Fig. 3B), because aminophospholipids are exposed on the cell surface after apoptosis, while PC is already exposed in healthy cells. Actually, Bailey reported that TREM2 binds to PS but not to PC45. However, other reports suggested that PC might induce signal transduction via TREM231,48. The apparent contradiction may be due to the different reporter cell systems used (2B4 cells in their studies vs. Jurkat cells in this study). Bader Lange et al. reported that larger amounts of PS and PE are exposed on the surface of the synaptosomes in aged and AD model mouse brain63, and this may increase TREM2 signal transduction, leading to expression of microglial protective phenotypes. The increased incidence of AD associated with TREM2 R47H or R62H might be due to impaired protection of the TREM2 signal during aging or AD progression resulting from loss of function50. Although we still do not know exactly the target genes of TREM2 signal transduction responsible for the increased risk of AD, genes encoding factors involved in mTOR activation, which controls energetic and anabolic metabolism, might be candidates64. Further study will be needed to understand the link between TREM2 signal transduction and AD pathogenesis at the molecular level.
Our results indicate that the TREM2 ligands on normal cultured Jurkat and THP-1 cells are aminophospholipids (Fig. 5C). We hypothesize that small amounts of cells might die spontaneously during normal culture, resulting in exposure of previously inaccessible aminophospholipids. Since TREM2DAP12-expressing reporter cells are derived from Jurkat cells, aminophospholipids exposed on small amounts of the dying reporter cells might transduce the NFAT signal by themselves. Indeed, MFGE8 reduced the reporter activity to less than that of the normal cultured reporter cells (compare 3rd and 5th columns with 1st column in Fig. 5C), supporting this hypothesis. Unknown TREM2 ligands on normal cultured cells that transduce the NFAT signal were previously reported39,40. Our results suggest that those unidentified ligands might be aminophospholipids.
Finally, we showed that TREM2 ligands that can specifically transduce intracellular signalling are present in the membrane fractions of 5-month-old mouse cerebral cortex, and that MFGE8 protein does not inhibit the signal (Fig. 6), suggesting that the signal-transducing ligands in the mouse brain are different from aminophospholipids. However, it is not clear why aminophospholipids, which should be present in the membrane fractions, cannot transduce the signal. One possible explanation would be that signal-transducing ligands in the brain may have a higher affinity for TREM2 than aminophospholipids, thus excluding the signal from aminophospholipids. Since nuclear and soluble fractions were removed from the membrane fractions, DNA and RNA can be excluded as candidates for the endogenous signal-transducing ligands. Subtle quantitative or qualitative alterations of the ligands might occur in the brain during aging, resulting in an increase in the incidence of AD. The TREM2 signal-inducing activities in the cerebral cortex from wild-type and AppNL-G-F knock-in mice were not significantly different (Fig. 6E), suggesting that the ligands might not be derived from astrocytes or from microglia, considering that the numbers of astrocytes and microglia are robustly increased after 6 months in AppNL-G-F knock-in mice60. Thus, the ligands might be expressed by live neurons. Identification of the ligands in the cortices will be an important next step for understanding the functional interaction between microglia and neurons, as well as the relevance of TREM2 to the development of neurodegenerative disorders. It would also be interesting to investigate whether AD-related variants such as R47H affect the binding and/or signal transduction from the ligands in the mouse cerebral cortex, and we are currently comparing the signal transduction characteristics of reporter cell clones expressing wild-type TREM2DAP12 and R47H variant.
In sum, we have developed a highly sensitive reporter cell model for signal transduction via TREM2 and showed that the TREM2 ligands on apoptotic and normal cells are aminophospholipids. The model should be useful to examine signal transduction by apolipoprotein E or oligomeric Aβ and to investigate the effects of TREM2 variants on the signal transduction.
Methods
cDNAs
Human TREM265 and mouse MFGE8-L D89E58 cDNAs were as described. Human TREM1 and DAP12 cDNAs were obtained from Kazusa DNA Research Institute (Chiba, Japan) and Health Science Research Resources Bank (Osaka, Japan), respectively. cDNAs encoding the extracellular domain of TREM1 (amino acid residue 1-203) and TREM2 (amino acid residue 1-172) were amplified, fused with the transmembrane and cytosolic domain of human DAP12 (amino acid residues 30-113) with its stop codon by PCR, and inserted into the NheI and XhoI sites of pEBMulti-Neo TARGET tag-C (Fuji Film, Osaka, Japan). pGL4.30[luc2P/NFAT-RE/Hygro] vector (Promega, Madison, WI, USA) was used as a reporter gene, containing the luciferase reporter gene luc2P (Photinus pyralis) under the control of NFAT-responsive element.
Culture cells
Jurkat human T cells, 2B4 mouse T cell hybridoma cells, and THP-1 human monocytes were maintained in RPMI1640 media (Fuji Film) supplemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA) and penicillin/streptomycin (Fuji Film). RAW264.7 mouse macrophages, HEK293 human embryonic kidney cells, and Neuro2a mouse neuroblastoma cells were maintained in DMEM (Fuji Film) supplemented with 10% fetal bovine serum and penicillin/streptomycin.
Reporter cells
Jurkat cells were transfected with pGL4.30[luc2P/NFAT-RE/Hygro] in a NEPA21 super electroporator (Nepa Gene Corporation, Ltd., Chiba, Japan) and stably transfected clones were selected with 0.4 mg/mL hygromycin B (Fuji Film), followed by limiting dilution. Then, the cell clones were transfected with TREM2DAP12 (or TREM1DAP12) cDNA and stably transfected clones were selected with 1 mg/mL G418 (Fuji Film), followed by limiting dilution.
RAW264.7 cells were transfected with pGL4.30[luc2P/NFAT-RE/Hygro] using Lipofectamine2000 (Thermo Fisher Scientific, Waltham, MA, USA), and selected with 0.4 mg/mL hygromycin B. The cell clones were transfected with TREM2DAP12 cDNA and selected with 1 mg/mL G418.
2B4 cells stably transfected with enhanced green fluorescent protein (EGFP) gene under the control of NFAT-responsive element66 were transfected with TREM2DAP12 cDNA using a NEPA21 super electroporator and selected with 1 mg/mL G418.
Western blot analysis
Cell lysates were extracted with lysis buffer (1% Triton-X 100, protease inhibitor cocktail (CompleteTM, Roche Diagnostics, Indianapolis, IN, USA), 700 ng/ml pepstatin A (Peptide Institute, Osaka, Japan) in phosphate-buffered saline). Aliquots were electrophoresed on 10% polyacrylamide sodium dodecyl sulfate-gel and blotted onto polyvinylidene difluoride membrane (HybondTM-P, Amersham Biosciences, Piscataway, NJ, USA). The membranes were probed with anti-DAP12 antibodies (Santa Cruz (Dallas, TX, USA)), followed by HRP-conjugated secondary antibodies IgG (Cell Signalling technology, Danvers, MA, USA). Immunoreactive bands were visualized with ImmunoStar LD (Fuji Film) and detected on a LAS-4000 mini (Fuji Film).
Reporter assay
Neuro2a cells were treated with 100 nM staurosporin (Fuji Film) overnight to induce apoptosis, then washed and resuspended in the culture media. The apoptotic Neuro2a cells and 1 × 105 Jurkat reporter cells were incubated at 37 °C in a ratio of 1/16 to 2/1 in 48-well plates for 14 hours, unless otherwise indicated. The cell lysates were incubated with One-GloTM reagent (Promega) in a 384-well plate for 5 min, and the luminescence was measured with a SPARK 20 M (Tecan, Männedorf, Switzerland). Normal cultured Neuro2a, Jurkat, THP-1, RAW264.7, and HEK293 cells were collected by pipetting or with TrypLE™ Select CTS™ (Thermo Fisher Scientific) and resuspended in the culture media after centrifugation. Where indicated, anti-TREM2 antibody (clone 237920, R&D Systems, Minneapolis, MN, USA), purified milk fat globule-EGF factor 8 (MFGE8)-L D89E, or bovine serum albumin (BSA) (Takara Bio, Osaka, Japan) was added to the culture media. Culture inserts with 0.4 μm pore membrane (Thermo Fisher Scientific) were used to block direct interaction between the apoptotic cells and reporter cells. Phosphatidylserine (PS), phosphatidylethanolamine (PE) and phosphatidylcholine (PC) (Sigma-Aldrich) were dissolved in methanol and coated on plates by drying before the reporter cells were seeded. Homogenates of cerebral cortices from wild-type and AppNL-G-F knock-in59 mice were prepared by homogenization in buffer (50 mM Tris-HCl buffer (pH 7.4), 150 mM NaCl, protease inhibitor cocktail, 0.7 ug/ml pepstatin A). The homogenates were centrifuged at 800 g for 10 min at 4 °C to remove nuclei and cell debris, and the post nuclear supernatants were centrifuged at 70,000 rpm in a TLA110 rotor (Beckman Coulter Diagnosis, Brea, CA, USA) for 29 min at 4 °C. The supernatants (soluble proteins) and resuspended membrane pellets were incubated with the reporter cells. All experiments conducted here were approved by the Ethics Committees of Nagasaki University.
Preparation of MFGE8-L D89E protein
HEK293 cells were transfected with MFGE8-L D89E cDNA58 using Lipofectamine3000 (Thermo Fisher Scientific). After 24 hours, the medium was replaced with DMEM without fetal bovine serum and the cells were further cultured for 30 hours. MFGE8-L D89E protein in the culture media was purified with anti-DDDDK-tag mAb-Magnetic Beads (MBL, Aichi, Japan) and subjected to silver staining (Silver Stain MS kit (Fuji Film).
Statistical analysis
Statistical analysis was done by SigmaPlot software, ver.14.0 (Systat Software Inc., San Jose, CA). To compare two groups, a two-tailed Student’s t test was employed after confirming equality of variances of the groups. To compare more than two groups, we used one-way ANOVA with the Student-Newman-Keuls test. P values of less than 0.05 were considered to be significant.
Acknowledgements
This work was supported in part by funds from the Strategic Research Program for Brain Sciences (JP16dm0107068h, JP17dm0107068h), Japan Agency for Medical Research and Development (AMED), to N.I. and K.S., and from the Charitable Trust Araki Medical and Biochemical Research Memorial Fund to K.S. We thank Shigekazu Nagata at Osaka University for MFGE8-L D89E expression vector and helpful comments. We also thank Daisuke Hatta and Masashi Asai for technical assistance and discussion and Richard Steele for editing a draft of this manuscript.
Author Contributions
K.S. and N.I. designed experiments, analyzed data and wrote the manuscript; K.S., Y.H., R.Y. and E.H. performed experiments; M.C., T.S., S.H., T.S. and T.C.S. provided experimental tools and made manuscript revisions.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Keiro Shirotani, Email: keiroshiro@nagasaki-u.ac.jp.
Nobuhisa Iwata, Email: iwata-n@nagasaki-u.ac.jp.
References
- 1.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1319. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama H, et al. Inflammation and Alzheimer’s disease. Neurobiol. Aging. 2000;21:383–421. doi: 10.1016/S0197-4580(00)00124-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wyss-Coray T. Inflammation in Alzheimer disease: Driving force, bystander or beneficial response? Nat. Med. 2006;12:1005–1015. doi: 10.1038/nm1484. [DOI] [PubMed] [Google Scholar]
- 4.Heneka MT, et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015;14:388–405. doi: 10.1016/S1474-4422(15)70016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhawan G, Floden AM, Combs CK. Amyloid-β oligomers stimulate microglia through a tyrosine kinase dependent mechanism. Neurobiol. Aging. 2012;33:2247–2261. doi: 10.1016/j.neurobiolaging.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ransohoff RM. How neuroinflammation contributes to neurodegeneration. Science. 2016;353:777–783. doi: 10.1126/science.aag2590. [DOI] [PubMed] [Google Scholar]
- 7.Lee CY, Landreth GE. The role of microglia in amyloid clearance from the AD brain. J. Neural. Transm (Vienna). 2010;117:949–960. doi: 10.1007/s00702-010-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gold M, El Khoury J. β-amyloid, microglia, and the inflammasome in Alzheimer’s disease. Semin Immunopathol. 2015;37:607–611. doi: 10.1007/s00281-015-0518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertram L, et al. Genome-wide association analysis reveals putative Alzheimer’s disease susceptibility loci in addition to APOE. Am. J. Hum. Genet. 2008;83:623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambert JC, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 11.Naj, A. C. et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet. 43, 436–441 (2011). [DOI] [PMC free article] [PubMed]
- 12.Hollingworth P, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’ s disease. Nat. Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerreiro R, et al. TREM2 Variants in Alzheimer’s disease. N. Engl. J. Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonsson T, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paloneva J, et al. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am. J. Hum. Genet. 2002;71:656–662. doi: 10.1086/342259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chouery E, et al. Mutations in TREM2 lead to pure early-onset dementia without bone cysts. Hum. Mutat. 2008;29:194–204. doi: 10.1002/humu.20836. [DOI] [PubMed] [Google Scholar]
- 17.Guerreiro RJ, et al. Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia-like syndrome without bone involvement. Arch. Neurol. 2013;70:78–84. doi: 10.1001/jamaneurol.2013.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rayaprolu S, et al. TREM2 in neurodegeneration: Evidence for association of the p.R47H variant with frontotemporal dementia and Parkinson’s disease. Mol. Neurodegener. 2013;8:19. doi: 10.1186/1750-1326-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borroni B, et al. Heterozygous TREM2 mutations in frontotemporal dementia. Neurobiol. Aging. 2014;35:934.e7–934.e10. doi: 10.1016/j.neurobiolaging.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 20.Cady J, et al. TREM2 variant p.R47H as a risk factor for sporadic amyotrophic lateral sclerosis. JAMA Neurol. 2014;71:449–453. doi: 10.1001/jamaneurol.2013.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin SC, et al. Coding variants in TREM2 increase risk for Alzheimer’s disease. Hum. Mol. Genet. 2014;23:5838–5846. doi: 10.1093/hmg/ddu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cruchaga C, et al. GWAS of cerebrospinal fluid tau levels identifies risk variants for alzheimer’s disease. Neuron. 2013;78:256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan P, et al. TREM2 haplodeficiency in mice and humans impairs the microglia barrier function leading to decreased amyloid compaction and severe axonal dystrophy. Neuron. 2016;90:724–739. doi: 10.1016/j.neuron.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee CYD, et al. Elevated TREM2 gene dosage reprograms microglia responsivity and ameliorates pathological phenotypes in Alzheimer’s disease models. Neuron. 2018;97:1032–1048.e5. doi: 10.1016/j.neuron.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng-Hathaway PJ, et al. The Trem2 R47H variant confers loss-of-function-like phenotypes in Alzheimer’s disease. Mol. Neurodegener. 2018;13:1–12. doi: 10.1186/s13024-018-0262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudom A, et al. Molecular basis for the loss-of-function effects of the Alzheimer’s disease-associated R47H variant of the immune receptor TREM2. J. Biol. Chem. 2018;293:12634–12646. doi: 10.1074/jbc.RA118.002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi K, Rochford CDP, Neumann H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med. 2005;201:647–657. doi: 10.1084/jem.20041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ulrich JD, et al. Altered microglial response to Aβ plaques in APPPS1-21 mice heterozygous for TREM2. Mol. Neurodegener. 2014;9:20. doi: 10.1186/1750-1326-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, et al. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 2016;213:667–675. doi: 10.1084/jem.20151948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jay TR, et al. TREM2 deficiency eliminates TREM2 + inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J. Exp. Med. 2015;212:287–295. doi: 10.1084/jem.20142322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 2015;160:1061–1071. doi: 10.1016/j.cell.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazaheri F, et al. TREM2 deficiency impairs chemotaxis and microglial responses to neuronal injury. EMBO Rep. 2017;18:1186–1198. doi: 10.15252/embr.201743922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jay TR, et al. Disease progression-dependent effects of TREM2 deficiency in a mouse model of Alzheimer’s disease. J. Neurosci. 2017;37:637–647. doi: 10.1523/JNEUROSCI.2110-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filipello F, et al. The microglial innate immune receptor TREM2 is required for synapse elimination and normal brain connectivity. Immunity. 2018;48:979–991.e8. doi: 10.1016/j.immuni.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi M, Konishi H, Sayo A, Takai T, Kiyama H. TREM2/DAP12 signal elicits proinflammatory response in microglia and exacerbates neuropathic pain. J. Neurosci. 2016;36:11138–11150. doi: 10.1523/JNEUROSCI.1238-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao, Y. et al. Intracellular trafficking of TREM2 is regulated by presenilin 1. Exp. Mol. Med. 49 (2017). [DOI] [PMC free article] [PubMed]
- 37.Daws MR, et al. Pattern recognition by TREM-2: binding of anionic ligands. J. Immunol. 2003;171:594–599. doi: 10.4049/jimmunol.171.2.594. [DOI] [PubMed] [Google Scholar]
- 38.Hamerman JA, et al. Cutting Edge: Inhibition of TLR and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. J. Immunol. 2006;177:2051–2055. doi: 10.4049/jimmunol.177.4.2051. [DOI] [PubMed] [Google Scholar]
- 39.Piccio L, et al. Blockade of TREM-2 exacerbates experimental autoimmune encephalomyelitis. Eur. J. Immunol. 2007;37:1290–1301. doi: 10.1002/eji.200636837. [DOI] [PubMed] [Google Scholar]
- 40.Hsieh CL, et al. A role for TREM2 ligands in the phagocytosis of apoptotic neuronal cells by microglia. J. Neurochem. 2009;109:1144–1156. doi: 10.1111/j.1471-4159.2009.06042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stefano L, et al. The surface-exposed chaperone, Hsp60, is an agonist of the microglial TREM2 receptor. J. Neurochem. 2009;110:284–294. doi: 10.1111/j.1471-4159.2009.06130.x. [DOI] [PubMed] [Google Scholar]
- 42.Cannon JP, O’Driscoll M, Litman GW. Specific lipid recognition is a general feature of CD300 and TREM molecules. Immunogenetics. 2012;64:39–47. doi: 10.1007/s00251-011-0562-4. [DOI] [PubMed] [Google Scholar]
- 43.Ito H, Hamerman JA. TREM-2, triggering receptor expressed on myeloid cell-2, negatively regulates TLR responses in dendritic cells. Eur. J. Immunol. 2012;42:176–185. doi: 10.1002/eji.201141679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phongsisay V, Iizasa E, Hara H, Yamasaki S. 3-O-sulfo-β-d-galactose moiety of endogenous sulfoglycolipids is a potential ligand for immunoglobulin-like receptor LMIR5. Mol. Immunol. 2015;63:595–599. doi: 10.1016/j.molimm.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 45.Bailey CC, DeVaux LB, Farzan M. The Triggering receptor expressed on myeloid cells 2 binds Apolipoprotein E. J. Biol. Chem. 2015;290:26033–42. doi: 10.1074/jbc.M115.677286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atagi Y, et al. Apolipoprotein E is a ligand for triggering receptor expressed on myeloid cells 2 (TREM2) J. Biol. Chem. 2015;290:26043–26050. doi: 10.1074/jbc.M115.679043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawabori M, et al. Triggering receptor expressed on myeloid cells 2 (TREM2) deficiency attenuates phagocytic activities of microglia and exacerbates ischemic damage in experimental stroke. J. Neurosci. 2015;35:3384–3396. doi: 10.1523/JNEUROSCI.2620-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poliani, P. L. et al. JCI - TREM2 sustains microglial expansion during aging and response to demyelination. 125, 2161–2170 (2015). [DOI] [PMC free article] [PubMed]
- 49.Kober DL, et al. Neurodegenerative disease mutations in TREM2 reveal a functional surface and distinct loss-of-function mechanisms. Elife. 2016;5:e20391. doi: 10.7554/eLife.20391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song W, et al. Alzheimer’s disease-associated TREM2 variants exhibit either decreased or increased ligand-dependent activation. Alzheimer’s Dement. 2017;13:381–387. doi: 10.1016/j.jalz.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeh FL, Wang Y, Tom I, Gonzalez LC, Sheng M. TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron. 2016;91:328–340. doi: 10.1016/j.neuron.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 52.Zhao Y, et al. TREM2 Is a receptor for β-amyloid that mediates microglial function. Neuron. 2018;97:1023–1031.e7. doi: 10.1016/j.neuron.2018.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong L, et al. Amyloid-beta modulates microglial responses by binding to the triggering receptor expressed on myeloid cells 2 (TREM2) Mol. Neurodegener. 2018;13:15. doi: 10.1186/s13024-018-0247-71-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lessard CB, et al. High affinity interactions and signal transduction between Aβ oligomers and TREM2. EMBO Mol. Med. 2018;10:e9027. doi: 10.15252/emmm.201809027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.N’Diaye EN, et al. TREM-2 (triggering receptor expressed on myeloid cells 2) is a phagocytic receptor for bacteria. J. Cell Biol. 2009;184:215–223. doi: 10.1083/jcb.200808080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleinberger G, et al. TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci. Transl. Med. 2014;6:243ra86. doi: 10.1126/scitranslmed.3009093. [DOI] [PubMed] [Google Scholar]
- 57.Nagata S, Suzuki J, Segawa K, Fujii T. Exposure of phosphatidylserine on the cell surface. Cell Death Differ. 2016;23:952–961. doi: 10.1038/cdd.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanayama R, et al. Identification of a factor that links apoptotic cells to phagocytes. Nature. 2002;417:182–187. doi: 10.1038/417182a. [DOI] [PubMed] [Google Scholar]
- 59.Saito T, et al. Single App knock-in mouse models of Alzheimer’s disease. Nat. Neurosci. 2014;17:661–663. doi: 10.1038/nn.3697. [DOI] [PubMed] [Google Scholar]
- 60.Masuda A, et al. Cognitive deficits in single App knock-in mouse models. Neurobiol. Learn. Mem. 2016;135:73–82. doi: 10.1016/j.nlm.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 61.Humphrey MB, et al. The signaling adapter protein DAP12 regulates multinucleation during osteoclast development. J. Bone Miner. Res. 2004;19:224–234. doi: 10.1359/JBMR.0301234. [DOI] [PubMed] [Google Scholar]
- 62.Humphrey MB, et al. TREM2, a DAP12-associated receptor, regulates osteoclast differentiation and function. J. Bone Miner. Res. 2006;21:237–245. doi: 10.1359/JBMR.051016. [DOI] [PubMed] [Google Scholar]
- 63.Bader Lange ML, et al. Age-related loss of phospholipid asymmetry in APPNLh/APPNLhx PS-1P264L/PS-1P264Lhuman double mutant knock-in mice: Relevance to Alzheimer disease. Neurobiol. Dis. 2010;38:104–115. doi: 10.1016/j.nbd.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ulland TK, et al. TREM2 maintains microglial metabolic fitness in Alzheimer’s disease. Cell. 2017;170:649–663.e13. doi: 10.1016/j.cell.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bouchon A, Dietrich J, Colonna M. Cutting Edge: Inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J. Immunol. 2000;164:4991–4995. doi: 10.4049/jimmunol.164.10.4991. [DOI] [PubMed] [Google Scholar]
- 66.Ohtsuka M, et al. NFAM1, an immunoreceptor tyrosine-based activation motif-bearing molecule that regulates B cell development and signaling. Proc. Natl. Acad. Sci. USA. 2004;101:8126–8131. doi: 10.1073/pnas.0401119101. [DOI] [PMC free article] [PubMed] [Google Scholar]