Abstract
The purpose of the present study was to examine the effects of regular exercise on the abundance of targeted circulating microRNAs (miRNAs). The present analysis examined 20 previously sedentary adults from the HERITAGE Family Study who completed 20 weeks of endurance exercise training. The expression of 53 miRNAs related to cardiovascular disease were measured in serum collected at baseline and post-training by performing RT-qPCR on the Human Cardiovascular Disease miRNA array (Qiagen, Germany). The effect of regular exercise on circulating miRNAs was assessed using paired t-tests of baseline and post-training expression levels. A false discovery rate threshold of 5% was used to determine significance. Regular exercise resulted in significantly decreased mean serum expression of nine miRNAs (miR-486-5p, let-7b-5p, miR-29c-3p, let-7e-5p, miR-93-5p, miR-7-5p, miR-25-3p, miR-92a-3p, and miR-29b-3p; fold change range: 0.64–83, p = 0.0002–0.01) and increased mean expression of five miRNAs (miR-142-3p, miR-221-3p, miR-126-3p, miR-146a-5p, and miR-27b-3p; fold change range: 1.41–3.60, p = 0.001–0.006). Enrichment analysis found that these 14 miRNAs target genes related to over 345 different biological pathways. These results provide further evidence of the effects of regular exercise on the circulating miRNA profile.
Subject terms: Risk factors, Predictive markers, Cardiovascular genetics
Introduction
MicroRNAs (miRNAs) are small non-coding regulatory RNAs that participate in the regulation of gene expression via post-transcriptional modification of messenger RNA (mRNA)1,2. MiRNAs silence mRNAs through two distinct mechanisms: mRNA degradation or repression of mRNA translation2. Over 2,500 different miRNAs have been identified in humans and despite accounting for an estimated 1–5% of the human genome, miRNAs are estimated to regulate greater than 30% of protein coding genes2–4. MiRNAs are transported into circulation within exosomes, protein complexes, or microvesicles2,5. The transport of miRNAs in these complexes prevents their breakdown and allows miRNAs to circulate and act on target cells throughout the body1,2. Thus, circulating miRNAs represent novel biomarkers for diseases such as cancer and cardiovascular disease (CVD)2,6,7.
Altered circulating miRNA profiles have been identified in patients with various diseases, including different forms of cancer, diabetes, and CVD8,9. Additionally, Zampetaki et al10. found that baseline miRNA profiles were associated with incident myocardial infarction over ten years in an elderly cohort, suggesting that miRNAs can also be used to predict future disease. MiRNAs have also been functionally related with multiple steps in the progression towards atherosclerosis and associated with acute myocardial infarction, coronary artery disease, and unstable angina6,11. Additionally, miRNAs play an essential role in many biological processes such as angiogenesis, mitochondrial metabolism, and cardiac/skeletal muscle hypertrophy12–14.
The associations of miRNAs with disease, as well as the physiological effects of miRNAs on multiple biological pathways, demonstrate the potential clinical relevance of circulating miRNAs as biomarkers of disease. Therefore, identifying treatments and interventions to alter the circulating miRNA profile are of interest. Exercise represents one such intervention, as exercise has repeatedly been shown to lower CVD, cancer, and mortality risk15–17. However, the effects of exercise on the circulating miRNA profile are not completely understood. The current literature shows an effect of acute exercise on the circulating miRNA profile, suggesting that miRNAs play a role in physiological adaptations to acute exercise18,19. However, less is known about the effects of regular exercise on the circulating miRNA profile. The current literature examining regular exercise and circulating miRNAs is somewhat limited in that many studies have examined fewer than 10 targeted miRNAs, with little overlap of miRNAs between studies19. Furthermore, little is known about how exercise affects miRNAs related to CVD. Therefore, the purpose of the current study was to explore the potential effects of regular exercise on a relatively large panel of miRNAs known to be associated with CVD. Specifically, the Human Cardiovascular Disease miRNA array (Qiagen, Germany) was examined in 20 individuals from the HERITAGE Family Study. This array includes 84 miRNAs that are differentially regulated during CVD progression and have been associated with CVD in multiple studies involving a variety of CVD models and a variety of miRNA profiling methodologies20–28. We hypothesized that regular exercise would significantly alter the circulating miRNAs related to CVD.
Results
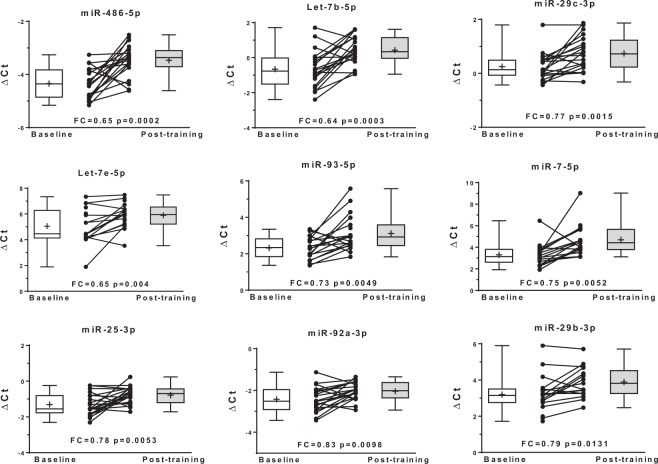
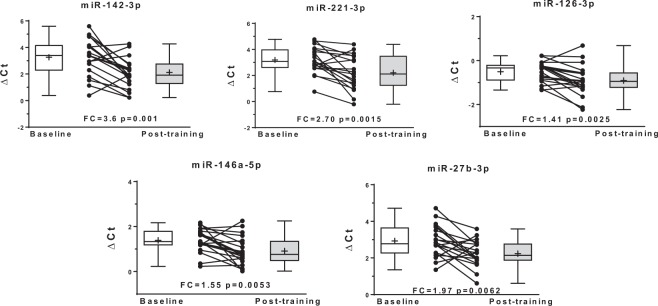
Baseline characteristics including mean values for the standard lipid panel and other cardiometabolic risk factors are shown in Table 1. No significant differences at baseline were found between sexes for these traits. The circulating expression levels of 14 miRNAs were significantly altered with regular exercise (FDR q < 0.05). Regular exercise resulted in decreased expression of nine miRNAs (miR-486-5p, let-7b-5p, miR-29c-3p, let-7e-5p, miR-93-5p, miR-7-5p, miR-25-3p, miR-92a-3p, and miR-29b-3p) (Fig. 1) and increased expression of five miRNAs (miR-142-3p, miR-221-3p, miR-126-3p, miR-146a-5p, and miR-27b-3p) (Fig. 2 and Table 2). These 14 miRNAs were entered into pathway/target analysis and combined are predicted to target over 7,500 genes, while individually each miRNA contributes to the regulation of many genes (range: 57–1406) (Supplemental Table S1). Connectivity analysis of the 14 miRNAs and their gene targets showed the minimally connected network was comprised of 47 genes and 127 edges (Supplemental Fig. S1). Enrichment analysis of the 14 miRNAs showed they were associated with over 345 different biological pathways, including PI3K-Akt, VEGFA-VEGFR2, insulin, and leptin signaling pathways (Table 3).
Table 1.
Baseline characteristics of participants (n = 20).
| Variable | Mean (SD) |
|---|---|
| Sex (Male/Female) | (10/10) |
| Age (years) | 43.7 (12.8) |
| BMI (kg/m2) | 26.2 (3.7) |
| VO2max (mL/kg/min) | 28.8 (6.9) |
| Waist circumference (cm) | 90.2 (11.4) |
| Percent fat | 28.1 (7.8) |
| SBP (mmHg) | 116.6 (12.4) |
| DBP (mmHg) | 65.9 (9.7) |
| HDL-C (mg/dL) | 43.9 (10.6) |
| LDL-C (mg/dL) | 135.4 (33.6) |
| TC (mg/dL) | 204.4 (36.7) |
| TG (mg/dL) | 156.2 (83.0) |
| CRP (mg/dL) | 0.29 (0.3) |
BMI: body mass index, HDL-C: high-density lipoprotein cholesterol, LDL-C: low-density lipoprotein cholesterol, TC: total cholesterol, TG: triglycerides, CRP: C-reactive protein, SBP: systolic blood pressure, DBP: diastolic blood pressure, VO2max: maximum rate of oxygen consumption.
Figure 1.
Baseline and post-training normalized cycle threshold values (ΔCt) along with individual exercise response for nine miRNAs with decreased circulating expression (higher ΔCt) following regular exercise. FC, fold change.
Figure 2.
Baseline and post-training normalized cycle threshold values (ΔCt) along with individual exercise response for five miRNAs with increased circulating expression (lower ΔCt) following regular exercise. FC, fold change.
Table 2.
The effect of regular exercise on circulating microRNAs related to cardiovascular disease.
| miRNA | n | Fold-Change (SD) | P value a | q value b |
|---|---|---|---|---|
| miR-486-5p | 20 | 0.65 (0.4) | 0.0002 | 0.0009 |
| let-7b-5p | 20 | 0.64 (0.5) | 0.0003 | 0.0019 |
| miR-142-3p | 19 | 3.60 (3.8) | 0.001 | 0.0028 |
| miR-221-3p | 19 | 2.70 (2.6) | 0.0015 | 0.0038 |
| miR-29c-3p | 20 | 0.77 (0.3) | 0.0015 | 0.0047 |
| miR-126-3p | 20 | 1.41 (0.5) | 0.0025 | 0.0057 |
| let-7e-5p | 18 | 0.65 (0.4) | 0.004 | 0.0066 |
| miR-93-5p | 18 | 0.73 (0.5) | 0.0049 | 0.0075 |
| miR-7-5p | 16 | 0.75 (1.3) | 0.0052 | 0.0085 |
| miR-146a-5p | 20 | 1.55 (0.8) | 0.0053 | 0.0094 |
| miR-25-3p | 20 | 0.78 (0.4) | 0.0053 | 0.0104 |
| miR-27b-3p | 20 | 1.97 (1.2) | 0.0062 | 0.0113 |
| miR-92a-3p | 20 | 0.83 (0.4) | 0.0098 | 0.0123 |
| miR-29b-3p | 16 | 0.79 (0.4) | 0.0131 | 0.0132 |
| let-7d-5p | 20 | 0.84 (0.4) | 0.0175 | 0.0142 |
| miR-17-5p | 16 | 2.04 (1.7) | 0.0208 | 0.0151 |
| miR-424-5p | 19 | 1.85 (1.5) | 0.0439 | 0.016 |
| miR-23a-3p | 20 | 1.35 (0.6) | 0.0477 | 0.017 |
| miR-222-3p | 18 | 1.36 (0.6) | 0.0483 | 0.0179 |
| miR-30c-5p | 20 | 1.62 (0.9) | 0.0579 | 0.0189 |
| miR-27a-3p | 20 | 2.04 (1.6) | 0.0613 | 0.0198 |
| miR-103a-3p | 13 | 7.42 (12.3) | 0.079 | 0.0208 |
| let-7a-5p | 20 | 1.06 (1.1) | 0.0875 | 0.0217 |
| miR-29a-3p | 20 | 1.50 (1.0) | 0.0907 | 0.0226 |
| miR-24-3p | 20 | 1.47 (0.7) | 0.0917 | 0.0236 |
| miR-125a-5p | 20 | 1.46 (0.9) | 0.1006 | 0.0245 |
| miR-16-5p | 20 | 1.01 (1.0) | 0.1025 | 0.0255 |
| let-7c | 20 | 1.11 (1.2) | 0.1324 | 0.0264 |
| miR-342-3p | 20 | 1.47 (1.0) | 0.1514 | 0.0274 |
| miR-15b-5p | 20 | 0.95 (0.4) | 0.1564 | 0.0283 |
| miR-181b-5p | 14 | 1.67 (1.4) | 0.2046 | 0.0292 |
| miR-224-5p | 10 | 2.37 (2.2) | 0.2192 | 0.0302 |
| miR-125b-5p | 15 | 2.57 (2.9) | 0.2577 | 0.0311 |
| miR-130a-3p | 11 | 2.20 (1.7) | 0.3041 | 0.0321 |
| miR-195-5p | 20 | 1.48 (2.5) | 0.3096 | 0.033 |
| miR-185-5p | 18 | 1.60 (2.1) | 0.3166 | 0.034 |
| miR-10b-5p | 14 | 3.42 (4.3) | 0.3411 | 0.0349 |
| miR-150-5p | 20 | 1.40 (0.8) | 0.3693 | 0.0358 |
| miR-23b-3p | 19 | 1.34 (0.6) | 0.4056 | 0.0368 |
| miR-223-3p | 20 | 1.37 (1.3) | 0.4779 | 0.0377 |
| miR-320a | 20 | 1.13 (1.0) | 0.5106 | 0.0387 |
| miR-30e-5p | 18 | 1.72 (2.1) | 0.6095 | 0.0396 |
| miR-423-3p | 13 | 2.21 (2.3) | 0.6245 | 0.0406 |
| miR-98-5p | 18 | 1.31 (0.9) | 0.6554 | 0.0415 |
| miR-26a-5p | 20 | 1.11 (0.5) | 0.7655 | 0.0425 |
| miR-122-5p | 20 | 1.41 (1.4) | 0.7881 | 0.0434 |
| miR-26b-5p | 19 | 3.97 (13.1) | 0.82 | 0.0443 |
| miR-451a | 20 | 1.89 (2.9) | 0.8475 | 0.0453 |
| miR-100-5p | 19 | 1.59 (1.6) | 0.8936 | 0.0462 |
| miR-30d-5p | 20 | 1.30 (1.1) | 0.9146 | 0.0472 |
| miR-30a-5p | 20 | 1.31 (1.0) | 0.9536 | 0.0481 |
| miR-21-5p | 20 | 1.06 (0.4) | 0.9546 | 0.0491 |
| miR-365a-3p | 15 | 1.65 (1.8) | 0.9907 | 0.05 |
n represents the number of subjects in which the miRNA was detectable in both baseline and post-training samples.
aP values are the result of paired t-tests of normalized baseline and post-training miRNA expression levels.
bWhen P < q the change in expression with training is deemed significant, which is indicated in bold.
Table 3.
Select pathways associated with the 14 microRNAs significantly altered in response to regular exercise and the number of genes per pathway targeted by these microRNAs.
| Pathway | Number of miRNAs Involved | Genes Targeted |
|---|---|---|
| PI3K-Akt Signaling | ||
| PI3K-Akt-mTOR Signaling | ||
| VEGFA-VEGFR2 Signaling | ||
| Insulin Signaling | ||
| Leptin Signaling |
Pathways were identified from the exRNA atlas database (http://exrna-atlas.org).
Discussion
This exploratory study found that regular exercise significantly altered the circulating expression levels of 14 miRNAs related to CVD in 20 previously sedentary, but otherwise healthy adults from the HERITAGE Family Study. The 14 miRNAs altered with exercise potentially contribute to the regulation of thousands of genes and are associated with hundreds of different biological pathways. Thus, the potential downstream biological effects of altered miRNA levels with regular exercise are enormous.
Little is known about the potential mechanism(s) by which exercise could alter circulating miRNA expression. MiRNA expression can certainly be regulated and altered, but the regulation of miRNAs is complex and not fully understood29. One potential mechanism could be an augmented degradation of the miRNAs in circulation with exercise. However, miRNAs are transported within vesicles or within protein complexes that prevent degradation1,2, thus this seems unlikely. Another potential mechanism could be the promotion of selective uptake of these miRNAs by skeletal muscle with exercise, as several studies have demonstrated the ability of target cells, such as skeletal muscle, to take up circulating miRNAs1,2,30. Similarly, it has been proposed that exercise may cause the release of certain miRNAs from skeletal muscle, either through muscle damage or selective release in response to acute or regular exercise31,32. Thus, one plausible explanation is that the increased or decreased expression of these 14 miRNAs in circulation with regular exercise may be due in part to increased uptake of miRNAs by skeletal muscle (9 of 14 decreased with regular exercise) and the converse increased release of certain miRNAs by skeletal muscle (5 of 14 increased with regular exercise). Baggish et al33. found that regular exercise resulted in elevated levels of circulating miR-222, miR-21, and miR-221. Interestingly, expression levels of these same three miRNAs were also elevated following acute exercise, indicating a potential additive effect of repeated acute bouts, as with training, that serves to increase circulating expression of certain miRNAs33.
Previous studies have examined the response to exercise of some of the same miRNAs included in this study. We found that regular exercise significantly decreased the expression of circulating miR-486-5p. Similarly, a study of 11 men found a single cycling exercise session and four weeks of regular exercise both significantly decreased circulating levels of miR-486-5p31. A 90-day period of rowing training in 10 males increased circulating miR-221-3p and miR-146a-5p expression33, the same responses found with cycle ergometer training in the current study. Circulating miR-92a-3p and miR-29b-3p expression levels were both significantly decreased in seven males following 12 weeks of regular exercise on cycle ergometers34. Both of these miRNAs also significantly decreased with training in the present analysis. The agreement with the current literature on the effects of regular exercise on these select miRNAs is encouraging and strengthens the existing evidence of a significant effect of exercise on the circulating miRNA profile. However, many of the 14 circulating miRNAs significantly altered with regular exercise in the current study have not been previously examined.
Our study benefitted from the use of a miRNA PCR array that included 84 miRNAs known to be associated with CVD. Additionally, the entire assay from RNA extraction to miRNA quantification was done using a fully robotic automated platform (QIAcube workstation) minimizing any potential pipetting errors. The study also benefitted from a fully standardized and supervised exercise program, thus all included subjects received the same relative dose of exercise and fully adhered to the program. Further research is also needed to determine the mechanisms by which exercise alters circulating miRNA expression and the association of changes in the circulating miRNA profile with changes in clinical outcomes. Importantly, the present study must be seen as a small, exploratory study. The same research questions should be investigated in greater detail with a much larger sample size and an untargeted approach to accurately and comprehensively examine the effects of regular exercise on the circulating miRNA profile. This study was also limited by a lack of technical replicates in sample analysis. Finally, further research in the field of circulating miRNAs is needed to create clear standards for data handling. Specifically, the difficulty in differentiating between missing data due to a true lack of expression in the sample and missing data due to various experimental issues is a major limitation in the field. Current conventions suggest ignoring Ct values above detection threshold limits, however these ‘missing’ data may have physiological relevance as the appearance or disappearance of a miRNA in response to an intervention is potentially even more meaningful than changes in expression. Similarly, further research is needed to determine an ideal normalization strategy for circulating miRNAs, as different normalization methodologies may lead to different conclusions and impact reproducibility and comparability of studies.
In summary, regular exercise may alter the circulating miRNA profile of select CVD related miRNAs and these alterations may have physiological relevance given the number of genes regulated by and diverse roles of these miRNAs. The current study provides further support for the effects of regular exercise on circulating miRNAs miR-486-5p, miR-146a-5p, miR-92a-3p, miR-29b-3p, and miR-221-3p and new evidence for other miRNAs. The existing evidence suggests that circulating miRNAs should be thoroughly investigated as biomarkers of regular exercise response. Further research is needed on the effects of exercise on the global circulating miRNA profile to potentially identify novel miRNAs associated with exercise and confirm the existing findings.
Materials and Methods
The HERITAGE Family Study was designed to examine the role of genetic factors on cardiometabolic responses to endurance regular exercise. The HERITAGE cohort is composed of 481 whites (232 men and 249 women) from 99 families and 250 blacks (88 men and 162 women) from 105 family units that completed a 20-week endurance exercise program at one of four clinical centers (Indiana, Minnesota, Québec, Texas). For the present analysis, 20 non-related white subjects (10 males, 10 females) from the Québec center that completed the 20-week exercise program were selected as part of an ongoing ancillary study. The study design, training protocol, and measurements have been described in detail elsewhere35. Briefly, the training program consisted of three weekly exercise sessions on cycle ergometers starting at the heart rate associated with 55% VO2max and progressing to the heart rate associated with 75% baseline VO2max for the final six weeks of the study. The study protocol was approved by the Institutional Review Boards at each of the five participating centers of the HERITAGE Family Study consortium (Indiana University, Laval University, University of Minnesota, Texas A&M University, and Washington University at St. Louis). Written informed consent was obtained from each study participant. All research was performed in accordance with the Declaration of Helsinki.
Exercise tests
Sub-maximal and maximal exercise tests on a stationary cycle ergometer were administered prior to training and additional tests were given following the completion of training35. Maximal cycle ergometer tests were performed twice on separate days at least 48 hours apart. VO2max was determined by the average of the two tests if the values were within 5% of each other, or the higher of the two values if the measures differed by more than 5%36.
Blood collection
Blood samples were obtained in the morning after a 12 hour fast, both at baseline and again 24 hours after the last training session. Serum was collected using standard methods36.
RNA extraction
Total RNA was extracted on the QIAcube workstation from baseline and post-training whole serum samples using miRNeasy Serum/Plasma Advanced Kits (Qiagen, Hilden, Germany). Samples were spiked with a known amount of synthetic C. elegans miR-39-3p (cel-miR-39-3p).
MiRNA quantification
MiRNA quantification was performed using reverse transcription real-time polymerase chain reaction (RT-qPCR). Briefly, cDNA was transcribed from the extracted RNA and then aliquoted into the Human Cardiovascular Disease miScript miRNA PCR Array (Qiagen, Hilden, Germany). This array contains primers for 84 miRNA sequences, an individual miRNA primer sequence per well, identified as exhibiting altered expression during cardiovascular disease. The selected miRNAs are listed in Fig. 3. All cDNA steps and PCR setup were performed by the QIAgility instrument (Qiagen, Hilden, Germany) using an automated pipetting protocol. RT-qPCR was performed on the miRNA PCR array in the Rotor-Disc 100 format by the Rotor-Gene Q real-time PCR cycler (Qiagen, Hilden, Germany). Rotor-Gene PCR cycling was performed according to the manufacturer’s suggested protocol and conditions. Cycle threshold (Ct) represents the cycle number at which there is an exponential increase in miRNA fluorescence. Individual miRNAs were determined to be detected when Ct values were lower than 33, while miRNAs with a Ct value ≥ 33 were considered not detected. Only miRNAs detected in at least 50% of subjects were retained for analysis, resulting in 53 total miRNAs analyzed.
Figure 3.
The 84 miRNA sequences on the Human Cardiovascular Disease miScript miRNA PCR Array grouped by their functional domains.
MiRNA validation
To validate the miRNA quantification methods, samples from six participants were randomly selected from the study inventory for analysis. Total RNA was isolated and spiked with a known amount of synthetic cel-miR-39-3p and miRNA analyzed as described above. Analysis demonstrated that cel-miR-39-3p exhibited excellent linearity between the target input and measured values with regression coefficients approaching 1 and the qPCR-based extraction efficiency (Efficiency = 10(−1/slope)−1) was approximately 80% (79.08% ± 11.26%). Additionally, as a positive control for RT-qPCR performed with the Human Cardiovascular Disease miScript miRNA PCR Array, three wells of the array contained cel-miR-39-3p. Using this internal control, the Ct coefficient of variation (CV) values were determined to be 1.14% ± 0.49% intraplate and 0.9% ± 0.62% interplate. Using these automated methods and repeating this miRNA array in triplicate for these six samples, the overall CV for each miRNA was less than 20%.
MiRNA normalization and fold change calculation
Normalized miRNA expression at baseline and post-training are represented by ΔCt. ΔCt values were calculated by subtracting the global geometric mean signal of miRNAs that were commonly expressed in the present study from individual miRNA Ct values as previously described37,38. Commonly expressed miRNAs were identified as those with Ct values < 33 in all samples, utilizing the GeneGlobe Data Analysis Center PCR software (Qiagen). ΔΔCt was then calculated by subtracting baseline ΔCt values from post-training ΔCt values. Fold change was calculated as 2−∆∆Ct.
Statistical analysis
The effects of regular exercise on circulating miRNAs were assessed using paired t-tests of baseline and post-training ΔCt levels. A false discovery rate (FDR) threshold of 5% using the Benjamini-Hochberg procedure was implemented to define statistical significance39. All analyses were performed using SAS 9.4 (Cary, NC).
MiRNA target prediction and pathway analysis
Experimentally validated miRNA targets were identified using miRNet (https://www.mirnet.ca), which utilizes data from well-annotated databases: miRTarBase v7.0, TarBase V7.0, and miRecords40,41. The connectivity of the 14 miRNAs and their target genes was visualized using miRNet with a minimum network adjustment to simplify the network visualization. Pathway analysis was performed using the extracellular RNA Atlas (http://exrna-atlas.org)42. All 14 miRNAs significantly altered with regular exercise were entered into the pathway analysis.
Supplementary information
Acknowledgements
This work was supported by multiple grants from the NIH, including P20 GM103499, P20 GM103528, U54 GM104940; HL45670, 1R01HL130972-01A1, and 1R01HL146462, as well as funding from VA 2I01-BX000168-06A1, a Merit Award from the Veterans’ Affairs Health Administration, and the USC Office of the Vice President for Research.
Author Contributions
J.B. was involved in the study design, aided in the miRNA profiling experiments, performed the data analysis, and drafted the manuscript. K.Z. performed the miRNA profiling experiments, assisted in data analysis, and reviewed the manuscript draft. K.B. was involved in study design and reviewed the manuscript draft. C.B. was involved in study design and reviewed the manuscript draft. F.S. was involved in study design and reviewed the manuscript draft. M.S. was involved in the study design, data analysis, and drafting of the manuscript. All authors read and approved the final manuscript.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-43978-x.
References
- 1.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11:537–61. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu T, et al. Circulating microRNAs in response to exercise. Scand J Med Sci Sports. 2015;25:e149–54. doi: 10.1111/sms.12421. [DOI] [PubMed] [Google Scholar]
- 6.Feinberg MW, Moore KJ. MicroRNA Regulation of Atherosclerosis. Circ Res. 2016;118:703–20. doi: 10.1161/CIRCRESAHA.115.306300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. P Natl Acad Sci USA. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogel B, et al. Multivariate miRNA signatures as biomarkers for non-ischaemic systolic heart failure. Eur Heart J. 2013;34:2812–22. doi: 10.1093/eurheartj/eht256. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 10.Zampetaki A, et al. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol. 2012;60:290–9. doi: 10.1016/j.jacc.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 11.Wang GK, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–66. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 12.Zhang C. MicroRNAs in vascular biology and vascular disease. J Cardiovasc Transl Res. 2010;3:235–40. doi: 10.1007/s12265-010-9164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan SY, et al. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10:273–84. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. 2009;21:461–9. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lear SA, et al. The effect of physical activity on mortality and cardiovascular disease in 130 000 people from 17 high-income, middle-income, and low-income countries: the PURE study. Lancet. 2017;390:2643–54. doi: 10.1016/S0140-6736(17)31634-3. [DOI] [PubMed] [Google Scholar]
- 16.Chudyk A, Petrella RJ. Effects of Exercise on Cardiovascular Risk Factors in Type 2 Diabetes A meta-analysis. Diabetes Care. 2011;34:1228–37. doi: 10.2337/dc10-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein L, Henderson BE, Hanisch R, Sullivanhalley J, Ross RK. Physical Exercise and Reduced Risk of Breast-Cancer in Young-Women. J Natl Cancer I. 1994;86:1403–8. doi: 10.1093/jnci/86.18.1403. [DOI] [PubMed] [Google Scholar]
- 18.Flowers E, Won GY, Fukuoka Y. MicroRNAs associated with exercise and diet: a systematic review. Physiol Genomics. 2015;47:1–11. doi: 10.1152/physiolgenomics.00095.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sapp RM, Shill DD, Roth SM, Hagberg JM. Circulating microRNAs in acute and chronic exercise: more than mere biomarkers. J Appl Physiol. 1985;2017(122):702–17. doi: 10.1152/japplphysiol.00982.2016. [DOI] [PubMed] [Google Scholar]
- 20.van Rooij E, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. P Natl Acad Sci USA. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong SM, et al. MicroRNA Expression Signature and the Role of MicroRNA-21 in the Early Phase of Acute Myocardial Infarction. J Biol Chem. 2009;284:29514–25. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng YH, et al. MicroRNAs are aberrantly expressed in hypertrophic heart - Do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–40. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikeda S, et al. Altered microRNA expression in human heart disease. Physiological Genomics. 2007;31:367–73. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 24.Thum T, Catalucci D, Bauersachs J. MicroRNAs: novel regulators in cardiac development and disease. Cardiovasc Res. 2008;79:562–70. doi: 10.1093/cvr/cvn137. [DOI] [PubMed] [Google Scholar]
- 25.El-Armouche Ali, Schwoerer Alexander Peter, Neuber Christiane, Emmons Julius, Biermann Daniel, Christalla Thomas, Grundhoff Adam, Eschenhagen Thomas, Zimmermann Wolfram Hubertus, Ehmke Heimo. Common MicroRNA Signatures in Cardiac Hypertrophic and Atrophic Remodeling Induced by Changes in Hemodynamic Load. PLoS ONE. 2010;5(12):e14263. doi: 10.1371/journal.pone.0014263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahlin F, et al. MicroRNAs as circulating biomarkers in acute coronary syndromes: A review. Vasc Pharmacol. 2016;81:15–21. doi: 10.1016/j.vph.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Vegter EL, van der Meer P, de Windt LJ, Pinto YM, Voors AA. MicroRNAs in heart failure: from biomarker to target for therapy. Eur J Heart Fail. 2016;18:457–68. doi: 10.1002/ejhf.495. [DOI] [PubMed] [Google Scholar]
- 28.Orlicka-Plocka M, Gurda D, Fedoruk-Wyszomirska A, Smolarek I, Wyszko E. Circulating microRNAs in Cardiovascular Diseases. Acta Biochim Pol. 2016;63:725–9. doi: 10.18388/abp.2016_1334. [DOI] [PubMed] [Google Scholar]
- 29.Gulyaeva LF, Kushlinskiy NE. Regulatory mechanisms of microRNA expression. J Transl Med. 2016;14:143. doi: 10.1186/s12967-016-0893-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mittelbrunn M, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aoi W, et al. Muscle-enriched microRNA miR-486 decreases in circulation in response to exercise in young men. Front Physiol. 2013;4:80. doi: 10.3389/fphys.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayr M, Zampetaki A, Kiechl S. MicroRNA biomarkers for failing hearts? European Heart Journal. 2013;34:2782–3. doi: 10.1093/eurheartj/eht261. [DOI] [PubMed] [Google Scholar]
- 33.Baggish AL, et al. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J Physiol. 2011;589:3983–94. doi: 10.1113/jphysiol.2011.213363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen S, et al. The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS One. 2014;9:e87308. doi: 10.1371/journal.pone.0087308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouchard C, et al. The HERITAGE family study. Aims, design, and measurement protocol. Med Sci Sports Exerc. 1995;27:721–9. doi: 10.1249/00005768-199505000-00015. [DOI] [PubMed] [Google Scholar]
- 36.Leon AS, et al. Blood lipid response to 20 weeks of supervised exercise in a large biracial population: The HERITAGE Family Study. Metabolism. 2000;49:513–20. doi: 10.1016/S0026-0495(00)80018-9. [DOI] [PubMed] [Google Scholar]
- 37.Vigneron N, et al. Towards a new standardized method for circulating miRNAs profiling in clinical studies: Interest of the exogenous normalization to improve miRNA signature accuracy. Mol Oncol. 2016;10:981–92. doi: 10.1016/j.molonc.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mestdagh Pieter, Van Vlierberghe Pieter, De Weer An, Muth Daniel, Westermann Frank, Speleman Frank, Vandesompele Jo. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biology. 2009;10(6):R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 40.Fan YN, et al. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;44:W135–W41. doi: 10.1093/nar/gkw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan, Y. N., Habib, M. & Xia, J. G. Xeno-miRNet: a comprehensive database and analytics platform to explore xeno-miRNAs and their potential targets. Peerj; 6 (2018) [DOI] [PMC free article] [PubMed]
- 42.Subramanian Sai Lakshmi, Kitchen Robert R., Alexander Roger, Carter Bob S., Cheung Kei-Hoi, Laurent Louise C., Pico Alexander, Roberts Lewis R., Roth Matthew E., Rozowsky Joel S., Su Andrew I., Gerstein Mark B., Milosavljevic Aleksandar. Integration of extracellular RNA profiling data using metadata, biomedical ontologies and Linked Data technologies. Journal of Extracellular Vesicles. 2015;4(1):27497. doi: 10.3402/jev.v4.27497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.