Abstract
Melanoma is one of the most common skin malignancies. Both microRNAs and long non-coding RNAs (lncRNAs) have critical roles in the progression of cancers, including melanoma. However, the underlying molecular mechanism has not been fully characterized. We demonstrated that miR-34a is negatively correlated with MALAT1 in melanoma cells and tumor specimens. Interestingly, MALAT1, which contains functional sequence-specific miR-34a-binding sites, regulates miR-34a stability in melanoma cells and in vivo. Importantly, MALAT1 was significantly enriched in the Ago2 complex, but not when the MALAT1-binding site of miR-34a was mutated. Furthermore, MALAT1 could be shown to regulate c-Myc and Met expression by functioning as a miR-34a sponge. Our results reveal an unexpected mode of action for MALAT1 as an important regulator of miR-34a.
Subject terms: Medical research, Genetics research
Introduction
Melanoma is an aggressive cancer involving pigment-containing cells known as melanocytes that are found predominantly in the skin. Melanoma is the leading cause of skin cancer death in the United States, with an estimated 87,110 new cases and 9730 deaths in 20171. In China, it is estimated that there were 8000 new cases of melanoma and 3200 deaths due to this disease in 20152. The molecular mechanisms of melanoma need to be further studied, and the identification of molecular drivers may enable the development of novel melanoma therapies.
Recent studies have revealed that non-coding RNAs, including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), play important roles in various pathophysiological processes, and are frequently dysregulated in many types of cancer3–9. Several studies have demonstrated that miRNAs, which regulate target mRNAs at the post-transcriptional level, are involved in the pathogenesis of all tumor types. In addition, lncRNAs comprising non-protein-coding transcripts longer than 200 nucleotides contribute to diverse biological functions in human diseases, including cancer10–13.
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), is one of the most abundant and evolutionarily conserved lncRNAs. The deregulation and function of MALAT1 have been established in several solid tumors, including bladder cancer, lung cancer, colorectal cancer, esophageal squamous cell carcinoma, melanoma, breast cancer, and hepatocellular carcinoma14–20. MALAT1 levels were markedly higher in melanomas than in paired adjacent normal tissues21. MALAT1 was also identified as a competing endogenous RNA (i.e., miR-183 sponge), and thus regulates the molecular expression of ITGB1 in melanoma22. Furthermore, MALAT1 promotes melanoma cell growth and invasion by silencing miR-140 or miR-2217,23.
In an earlier investigation, miR-34a was characterized as a tumor suppressor, in part, via the SIRT1-p53 pathway24. Therefore, miR-34 dysregulation may be involved in the development of some cancers25,26. The c-Myc gene is often constitutively expressed in malignant tumors, and the encoded protein is believed to regulate the expression of 15% of all genes in the human genome27. Some of the regulated genes are involved in cell proliferation, metastasis, and apoptosis, thereby contributing to cancer development28–31. Meanwhile, Met is a receptor tyrosine kinase family member encoded by the proto-oncogene Met. Abnormal activation of Met in cancer is correlated with poor prognosis, likely because aberrantly active Met triggers tumor growth, angiogenesis, and metastasis32–35. In addition, Met is deregulated in many types of human malignancies, including breast cancer, lung cancer, bladder cancer, hepatocellular carcinoma, and melanoma36–40. To date, whether MALAT1 affects c-Myc and Met expression levels via a competing endogenous RNA of miR-34a remains unclear.
In this study, we investigated the MALAT1 and miR-34a levels in melanoma, and explored the correlation between MALAT1 and miR-34a production. Importantly, we identified miR-34a as a target of MALAT1, the latter of which contains functional sequence-specific miR-34a-binding sites. These findings imply that MALAT1 helps regulate miR-34a expression, thereby expanding the functions of MALAT1 to include post-transcriptional regulatory activities.
Materials and methods
Cell culture
The melanoma cell line A375 was purchased from the National Infrastructure of Cell Line Resource (Cell Resource Center of the Chinese Academy of Medical Sciences, Beijing, China) and cultured in DMEM supplemented with 2 mM l-glutamine and 10% FBS. Cells were grown at 37 °C under humid conditions with 5% CO2.
Tissue samples
Tissues were obtained from patients who were diagnosed with melanoma and treated between Mar 2015 and Feb 2017 at the Department of Dermatology, Air Force Hospital, People’s Liberation Army. Skin tissues were collected from 20 patients with melanocytic nevi (matched by sex and age) as controls. Every patient involved in the study provided written informed consent that was approved by the Ethics Committee of the Air Force Hospital. All signed consent forms were saved by the Ethics Committee. The tissue samples were frozen within 30 min of surgery and stored in liquid nitrogen until use. Tissue specimens were cut into blocks (3–4 mm thick) and then fixed in fresh 10% neutral-buffered formalin for 16–32 h at room temperature (25 °C) before being embedded in paraffin for a subsequent RNA scope analysis.
Oligonucleotides, plasmids, and transfection
The following miRNAs were synthesized by Integrated Biotech Solutions (Shanghai, China): miR-NC, miR-34a mimics, miR-34a-mut, anti-miR-34a, anti-miR-34a-mut, biotin-miR-NC, biotin-miR-34a-mut, and biotin-miR-34a. Control siRNA and MALAT1 siRNA were purchased from Bioneer (Shanghai, China). The full-length 3′ untranslated regions (3′-UTR) of c-Myc, c-Met, wt-MALAT1, and mut-MALAT1 were subcloned into the psiCHECK-2 plasmid. For mismatch constructs, seven mismatches, which are indicated in bold letters in the following MALAT1 sequence: ACCGUCAGACGGGAGUUUUCGA (Fig. 4), were introduced into the putative target site, which was modified to UGGCAGUGACGGGAGUUUUCGA. For the transfections involving DNA plasmids and oligonucleotides, Lipofectamine 2000 (Life Technologies Corporation, Grand Island, NY) was used according to the manufacturer’s instructions.
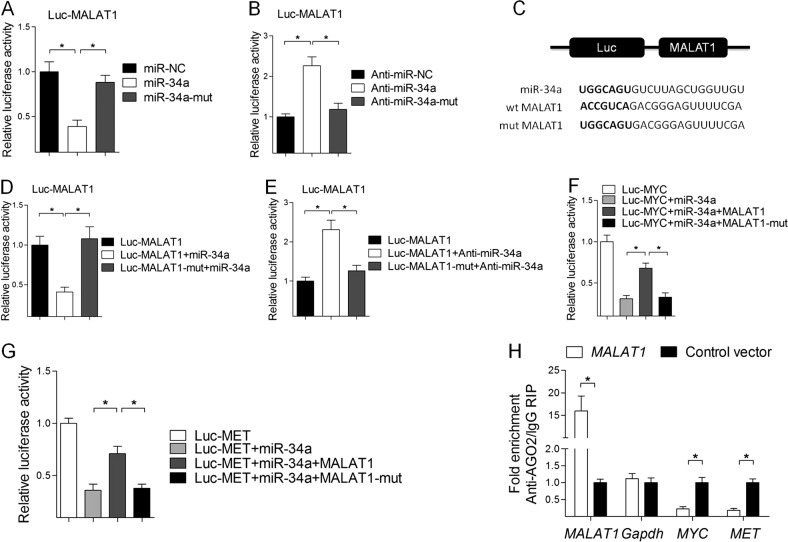
Fig. 4. MALAT1 functions as a miR-34a sponge in A375 melanoma cells.
a Luciferase reporter constructs: Wild-type MALAT1 (wt-MALAT1) and MALAT1 with mutations in the miR-34a-binding sites (Luc-MALAT1-mut) were inserted into the psiCHECK-2 vector. Letters in bold font represent mutation sites. b Relative luciferase activity of A375 cells with psiCHECK-2 containing wt-MALAT1 co-transfected with control miRNA (miR-NC), miR-34a, or mutated miR-34a. c Relative luciferase activity of A375 cells with psiCHECK-2 containing wt-MALAT1 co-transfected with control miRNA (anti-miR-NC), anti-miR-34a, or mutated miR-34a (anti-miR-34a-mut). d Relative luciferase activity of A375 cells with psiCHECK-2 containing wt-MALAT1 and mutated MALAT1 (Luc-MALAT1-mut) co-transfected with miR-34a. e Relative luciferase activity of A375 cells with psiCHECK-2 containing wt-MALAT1 and mutated MALAT1 (Luc-MALAT1-mut) co-transfected with anti-miR-34a. f Relative luciferase activity of A375 cells with psiCHECK-2 containing c-Myc co-transfected with miR-34a and wt-MALAT1 or mutated MALAT1 (MALAT1-mut) expression vectors. g Relative luciferase activity of A375 cells with psiCHECK-2 containing Met co-transfected with miR-34a and wt-MALAT1 or mutated MALAT1 (MALAT1-mut) expression vectors. h An Ago2 immunoprecipitation experiment was performed for A375 cells transfected with the control vector or MALAT1 expression vector, followed by a quantitative real-time polymerase chain reaction assay to analyze the MALAT1, GAPDH, c-Myc, and Met associated with Ago2
RNA sequencing
The A375 cells were transfected with MALAT1 siRNA, after which total RNA was isolated using the TRIzol reagent. The A375 cells transfected with control siRNA were used as the control. Each cell line was analyzed with three biological replicates. RNA-sequencing and data analysis were performed by Integrated Biotech Solutions.
RNA isolation, reverse transcription, and quantitative real-time polymerase chain reaction analysis
Total RNA was isolated from cells or tissues using a TRIzol kit (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. The extracted RNA was used as the template for a reverse transcription with the SuperScript III Reverse Transcriptase (Invitrogen). The miR-34a and MALAT1 expression levels were analyzed in a quantitative real-time polymerase chain reaction (qRT-PCR) assay, which was performed with the 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. The U6 small nuclear RNA and GAPDH gene were used as internal controls for analyzing the miRNA and mRNA levels, respectively. The following primers were designed for the qRT-PCR assay: MALAT1 forward 5′-TCCAGAAAGAGGGAGTTG-3′, reverse 5′-GAAGCCAGACCCAGTAAG-3′; GAPDH forward 5′-CCATGCCATCACTGCCACCC-3′, reverse 5′-GCCAGTGAGCTTCCCGTTCAG-3′; and miR-34a-5p forward 5′-TGGCAGTGTCTTAGCTGGTTGT-3′, reverse 5′-CTCAACTGGTGTCGTGGAGTC-3′. The resulting qRT-PCR data were analyzed using the 2−ΔΔCt method. All reactions were run in triplicate.
Biotin pull-down assay
A biotinylated-miR-34a-capture assay was carried out as previously described41. Briefly, biotin-miR-NC, biotin-miR-34a-mut, and biotin-miR-34a were separately transfected into A375 cells. At 48 h after transfection, cells were lysed and the resulting lysate was added to 30 μL beads (Dynabeads MyOne Streptavidin C1, Life Technologies). After agitating the lysate-bead mixture on a rotary shaker for 4 h at 4 °C, RNA was extracted from the beads with TRIzol Reagent (Life Technologies) and analyzed in a qRT-PCR assay.
Western blot and antibodies
Treated A375 cells were harvested and lysed in protein lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 0.1% NP-40, 5 mM EDTA, and 10% glycerol) supplemented with a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO USA). Protein concentration was determined with the BCA Protein Assay Kit (P0011, Beyotime, Shanghai, China). Proteins were separated by 12% or 9% SDS-PAGE and transferred to a PVDF membrane (IPVH00010, Millipore, Billerica, MA, USA) and immunoblotted with the following antibodies: anti-Met (ab51067, Abcam, Cambridge, MA, USA), anti-c-Myc (Abcam, ab32072), and anti-β-actin (Sigma, A5316).
Luciferase assay
Lipofectamine 2000 (Invitrogen) was used to co-transfect A375 cells with psiCHECK-2, psiCHECK-2-c-Myc, psiCHECK-2-Met, MALAT1 siRNA, and anti-miR-34a or anti-miR-34a-mut according to the manufacturer’s instructions. Three independent transfection experiments were conducted, each with three technical replicates. In all experiments, the firefly luciferase gene in psiCHECK-2 was used as a control to normalize the transfection efficiency. At 48 h after transfection, the firefly and Renilla luciferase activities were quantified with the Dual-Luciferase Reporter Assay System (Promega) and the BMG Labtech microplate reader.
Lentivirus production and stable cell lines
The MALAT1 and MALAT1-mut sequences were ligated into separate pLVX-IRES-Puro vectors to construct the MALAT1 and MALAT1-mut overexpression plasmids. The HEK293T cells were co-transfected with the pLVX-IRES-Puro-MALAT1, pLVX-IRES-Puro-MALAT1-mut, psPAX2, and pMD2.G plasmids. At 48 h after transfection, the supernatant was collected and injected into nude mice. Meanwhile, a lentiviral small hairpin RNA targeting MALAT1 (sh-lncRNA-MALAT1) and sh-NC (negative control) were designed and cloned into the pLVshRNA-Puro vector according to the manufacturer’s instructions (Inovogen Tech. Co., Beijing, China). The A375 cells were grown to ~40% confluence, after which they were infected with lentiviral particles in complete medium for 48 h and then selected with puromycin.
Animal experiments
Cells carrying pLVX-IRES-Puro-MALAT1 and pLVX-IRES-Puro-MALAT1-mut, or sh-lncRNA-MALAT1 or sh-NC were injected subcutaneously into the dorsal flanks of 5-week-old male BALB/c nude mice. The xenografts were dissected and total protein and RNA were obtained to analyze MALAT1, miR-34a, c-Myc, and Met levels.
RNAscope
The MALAT1 expression level was analyzed by Advanced Cell Diagnostics with an RNAscope probe.
RNA immunoprecipitation and qRT-PCR
An immunoprecipitation experiment involving anti-Ago2 was conducted as previously described41. Briefly, A375 cells were harvested at 48 h after transfection with miR-NC, miR-34a mimics, and miR-34a-mut or the MALAT1 expression vector. The cells were lysed and centrifuged at 12,000 × g for 30 min, after which 30 μL anti-FLAG M2 magnetic beads were added to the lysate (Sigma). After agitating the lysate-bead mixture on a rotary shaker for 4 h at 4 °C. The beads were washed three times with washing buffer (50 mM Tris-HCl, 300 mM NaCl, pH 7.4, 1 mM MgCl2, and 0.1% NP-40). The pull-down complexes were analyzed by qRT-PCR.
Statistical analysis
All data are herein presented as the mean ± standard deviation. For all experiments, statistical significance of data was determined by a two-tailed Student’s t-test conducted with the SPSS 17.0 program. A P value < 0.05 was considered statistically significant.
Results
miR-34a is negatively correlated with MALAT1 in melanoma cells
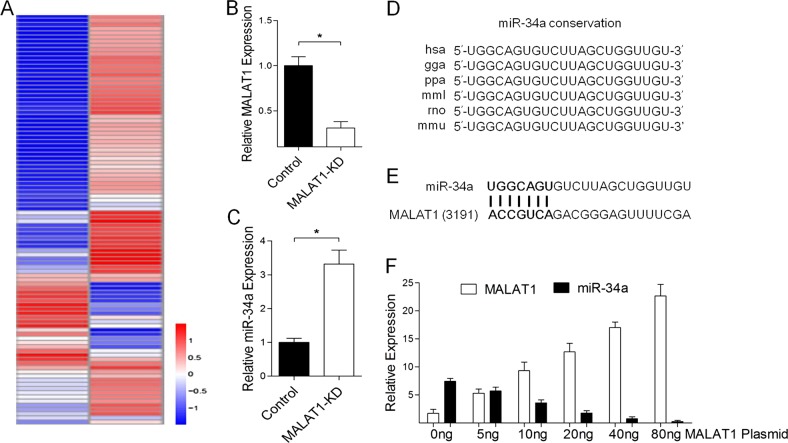
There is growing evidence of an important role for MALAT1 in tumorigenesis42–44. To investigate the potential mechanisms regulating the effects of MALAT1 on melanoma cells, we analyzed a miRNA-seq transcriptome of A375 melanoma cells transfected with MALAT1 siRNA or a scrambled control. The transcriptome data are presented in Fig. 1a, b. The eight most regulated miRNAs were hsa-miR-34a, hsa-miR-200a-3p, hsa-miR-196a-5p, hsa-miR-107, hsa-miR-196b-5p, hsa-miR-31-3p, hsa-miR-143-3p, and hsa-miR-582-3p. The largest fold change and the most-significant P value (P = 0.00027719) was observed for miR-34a (Table 1). The miR-34a expression levels in MALAT1-knockdown and control A375 cells were validated in a qRT-PCR assay, which indicated that miR-34a expression was consistent with the sequencing data (Fig. 1c). In addition, miR-34a was highly conserved among six species (Fig. 1d). Recent studies have suggested that lncRNAs may function as endogenous RNA sponges that interact with and influence the expression of miRNAs45,46. Moreover, a bioinformatics analysis revealed a predicted miR-34a response element in the MALAT1 transcript (Fig. 1e). After a 24h MALAT1 overexpression, we evaluated the miR-34a levels in A375 cells. When MALAT1 expression was enhanced, the miR-34a levels decreased in a dose-dependent manner (Fig. 1f). Thus, MALAT1 appears to negatively regulate miR-34a in A375 cells.
Fig. 1. miR-34a is negatively correlated with MALAT1 in melanoma cells.
a Heat map of differentially expressed miRNAs in A375 cells transfected with the negative control siRNA vector and MALAT1 siRNA (MALAT1-KD). A375 cells were transfected with 20 nM control siRNA or MALAT1 siRNA, and after a 48 h incubation, b MALAT1 and c miR-34a expression levels were analyzed in a quantitative real-time polymerase chain reaction (qRT-PCR) assay, with the expression data normalized against that of the control. d Aligned miR-34a sequences from nine species. e Bioinformatics analyses predicted the binding sites between MALAT1 and miR-34a. f A375 cells were transfected with different concentrations of MALAT1 expression vectors (0, 5, 10, 20, 40 , and 80 ng), after which miR-34a expression was analyzed in a qRT-PCR assay
Table 1.
The eight most changed miRNAs regulated by MALAT1
| No. | Reads of KD | Reads of Control | log2.fold_change |
|---|---|---|---|
| hsa-miR-34a | 149.893009 | 75.25562828 | 0.994061709 |
| hsa-miR-200a-3p | 58.4784386 | 32.17034491 | 0.862173387 |
| hsa-miR-196a-5p | 106.874388 | 60.31939671 | 0.825222254 |
| hsa-miR-107 | 70.5774259 | 42.51081292 | 0.731376966 |
| hsa-miR-196b-5p | 68.560928 | 41.36187203 | 0.729085154 |
| hsa-miR-31-3p | 131.744528 | 85.02162584 | 0.631841288 |
| hsa-miR-143-3p | 118.301209 | 76.40456917 | 0.630733995 |
| hsa-miR-582-3p | 493.369815 | 321.1289787 | 0.619516593 |
MALAT1 binds directly to miR-34a in melanoma cells
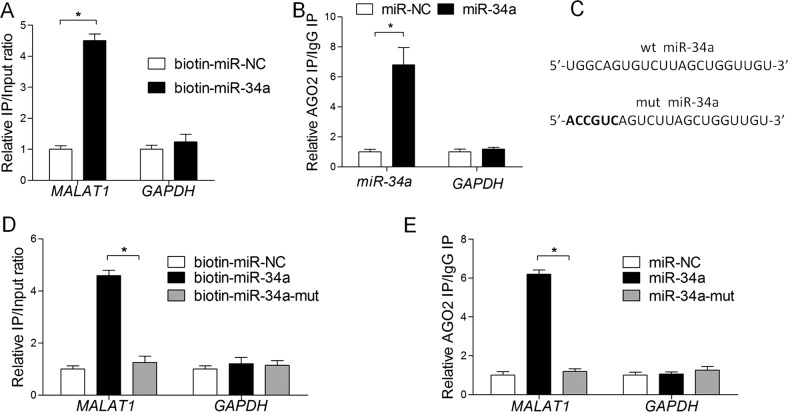
In this study, MALAT1 was detected following the affinity purification of miR-34a-interacting transcripts from A375 cells transfected with biotinylated miR-34a, but not from cells transfected with the biotinylated scrambled control. These results suggest that MALAT1 functions as a miRNA sponge that negatively regulates miR-34a levels. Moreover, neither miR-34a nor the control associated with GAPDH mRNA, which was used as a negative control (Fig. 2a). When the MALAT1-binding site of miR-34a was mutated, MALAT1 was not pulled down, indicating that MALAT1 regulated miR-34a in a sequence-specific manner (Fig. 2c, d). Because miRNAs regulate gene expression at post-transcriptional level through the RNA-induced silencing complex (RISC) containing Ago2, an RNA-binding protein immunoprecipitation assay was completed to verify whether miR-34a and MALAT1 are present in the same RISC. We observed that MALAT1 was significantly enriched in the Ago2 complex (Fig. 2b), but not when the MALAT1-binding site of miR-34a was mutated (Fig. 2e). These data indicate that miR-34a binds directly to MALAT1 in A375 cells.
Fig. 2. MALAT1 directly binds to miR-34a in melanoma cells.
a A streptavidin-capture assay was performed for A375 cells transfected with biotin-miR-NC or biotin-miR-34a, followed by a quantitative real-time polymerase chain reaction (qRT-PCR) assay to analyze MALAT1 and GAPDH mRNA levels. b An Ago2 immunoprecipitation experiment was completed for A375 cells transfected with control miRNA (miR-NC) or miR-34a, followed by a qRT-PCR assay to analyze the MALAT1 associated with Ago2. c Schematic representation of the wild-type miR-34a and mutated miR-34a. d Streptavidin -capture assay was performed for A375 cells transfected with biotin-miR-NC, biotin-miR-34a, or mutated biotin-miR-34a, followed by a qRT-PCR assay to analyze MALAT1 and GAPDH mRNA levels. e An Ago2 immunoprecipitation experiment was performed for A375 cells transfected with control miRNA (miR-NC), miR-34a, or mutated miR-34a, followed by a qRT-PCR assay to analyze the MALAT1 associated with Ago2
miR-34a target genes are regulated by MALAT1 in melanoma cells
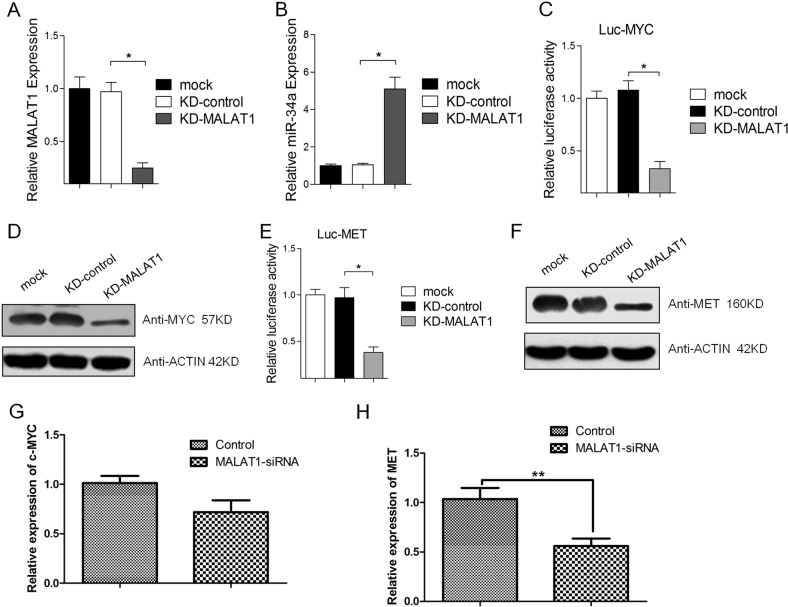
Previous studies confirmed that miR-34a can bind directly to many oncogenes, including c-Myc and Met, to regulate expression47–50. In this study, we used a luciferase reporter assay, qRT-PCR, and a western blot to verify whether c-Myc and Met are regulated by MALAT1. The knockdown of MALAT1 significantly increased the miR-34a level (Fig. 3b). Luciferase reporters containing the c-Myc and Met 3′-UTR were also constructed, and the knockdown of MALAT1 suppressed the luciferase activity of the c-Myc (Fig. 3c) and Met (Fig. 3e) reporter vectors. Additional studies showed that the knockdown of MALAT1 may decrease the c-Myc (Fig. 3d) and Met (Fig. 3f) protein levels. Thus, MALAT1 may regulate the expression of the miR-34a target genes c-Myc and Met. Interestingly, knockdown of MALAT1 suppressed the transcription of Met (Fig. 3g) but not c-Myc (Fig. 3h).
Fig. 3. miR-34a target genes are regulated by MALAT1 in melanoma cell.
A375 cells were transfected with 20 nM control siRNA or MALAT1 siRNA. After a 48-h incubation, a MALAT1 and b miR-34a expression levels were analyzed in a quantitative real-time polymerase chain reaction (qRT-PCR) assay, with the expression data normalized against that of the control. After a 72-h incubation post-transfection, the luciferase activities of c Luc-c-Myc and e Luc-Met were analyzed in a luciferase reporter assay. After a 72-h incubation post-transfection, d c-Myc and f Met protein levels were analyzed in a western blot, and g) c-Myc and (h) Met mRNA levels were analyzed in a qRT-PCR assay
MALAT1 functions as a miR-34a sponge in A375 melanoma cells
A Dual-Luciferase Reporter Assay System involving the wild-type (WT) and mutant-type (Mut) MALAT1-binding sites was used to investigate whether miR-34a is targeted by MALAT1. The assay results indicated that the overexpression of miR-34a, but not miR-34a-mut, suppressed the luciferase activity of the WT reporter vector (Fig. 4a). Meanwhile, the overexpression of anti-miR-34a, but not anti-miR-34a-mut, increased the luciferase activity of the WT reporter vector (Fig. 4b). Subsequent studies revealed that only the WT MALAT1 target site is recognized by miR-34a (Fig. 4d) and anti-miR-34a (Fig. 4e). Rescue experiments were conducted to determine whether the effect of MALAT1 is dependent on miR-34a. We observed that MALAT1 rescued the luciferase activity associated with c-Myc and Met in the presence of miR-34a (Fig. 4f, g). Furthermore, the overexpression of MALAT1 resulted in the increased enrichment of Ago2 on MALAT1, but substantially decreased the enrichment on c-Myc and Met (Fig. 4h). These data demonstrate that MALAT1 contains functional miR-34a-binding sites.
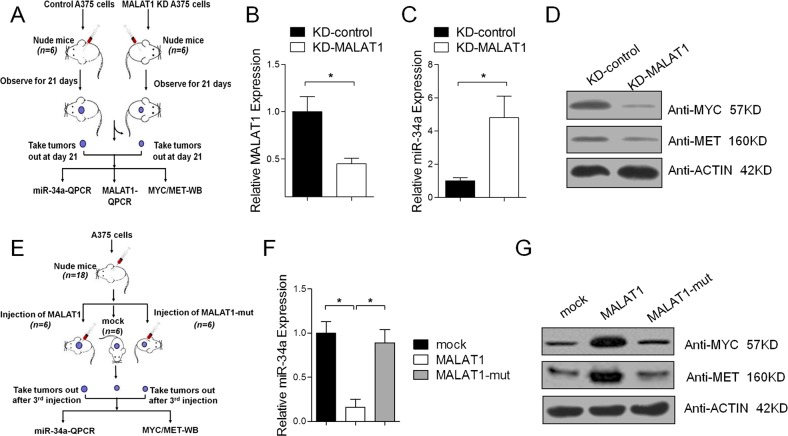
In vivo confirmation that MALAT1 functions as a miR-34a sponge
To determine whether MALAT1 functions as a molecular sponge for miR-34a in vivo, qRT-PCR and western blot experiments were completed to analyze the expression of MALAT1 and miR-34a in mice. Compared with the negative control, the relative expression of miR-34a was higher in the MALAT1 knockdown xenograft (Fig. 5c). The western blot indicated that the c-Myc and Met protein levels decreased when MALAT1 was knocked down (Fig. 5d). Conversely, MALAT1 overexpression decreased miR-34a expression, but not when the miR-34a-binding site of MALAT1 was mutated (Fig. 5f). Moreover, the overexpression of MALAT1 led to increased c-Myc and Met levels, but not if the miR-34a-binding site of MALAT1 was mutated (Fig. 5g). These results suggested that MALAT1 functions as a molecular sponge for miR-34a in vivo.
Fig. 5. In vivo confirmation that MALAT1 functions as a miR-34a sponge.
a Schematic diagram of the animal experimental design. The A375 cells stably expressing either sh-lncRNA-MALAT1 or sh-NC were injected subcutaneously into the dorsal flanks of the indicated numbers of BALB/c nude mice. (b–c) At 21 days after injection, the xenografts were dissected and examined in a quantitative real-time polymerase chain reaction (qRT-PCR) assay to analyze MALAT1 (b) and miR-34a (c) expression levels. d c-Myc and Met protein levels were analyzed in a western blot. e Schematic diagram of the animal experimental design. Stable A375 cells with pLVX-IRES-Puro-MALAT1 or pLVX-IRES-Puro-MALAT1-mut were injected subcutaneously into the dorsal flanks of 5-week-old male BALB/c nude mice. f Xenografts were dissected 21 days after injections, and MALAT1 and MALAT1-mut expression levels were analyzed in a qRT-PCR assay. g c-Myc and Met protein levels were analyzed in a western blot
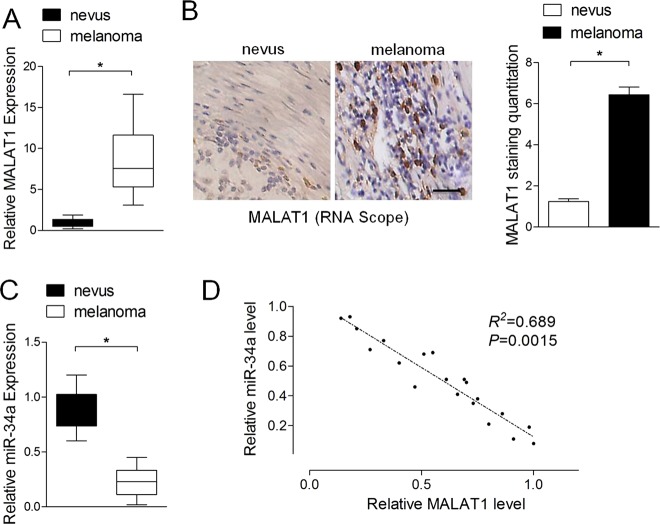
The expression of miR-34a is inversely associated with MALAT1 in melanoma tissues
To explore whether MALAT1 regulates miR-34a in clinical tissue samples, we analyzed miR-34a expression and assessed its correlation with MALAT1 in 20 melanoma tissues and 20 nevi. The qRT-PCR results showed that MALAT1 was more highly expressed in the melanoma tissues than in the benign nevi (Fig. 6a). The results of the RNAscope assay were consistent with those of the qRT-PCR, with significantly higher MALAT1 levels in melanoma tissues than in benign nevi (Fig. 6b). In contrast, the miR-34a expression level was lower in melanoma tissues than in benign nevi (Fig. 6c). Pearson’s correlation analyses revealed that miR-34a expression was inversely associated with MALAT1 in melanoma tissues (r2 = 0.689, P = 0.0015) (Fig. 6d). These data suggest that MALAT1 regulates miR-34a in melanoma tissues.
Fig. 6. The expression of miR-34a is inversely associated with MALAT1 in melanoma tissues.
a Relative MALAT1 expression levels in melanoma tissues (n = 20) and benign nevi (n = 20) were analyzed in a quantitative real-time polymerase chain reaction (qRT-PCR) assay. b RNAscope detection of MALAT1 expression in melanoma tissues (n = 20) and benign nevi (n = 20). Left panel: representative images. Scale bars: 100 µm. Right panel: data analysis. c Relative miR-34a expression levels in melanoma tissues (n = 20) and benign nevi (n = 20) were analyzed in a qRT-PCR assay. d miR-34a expression is inversely associated with MALAT1 in melanoma tissues (r2 = 0.689, P = 0.0015)
Discussion
Melanoma is the deadliest form of skin cancer, and there are limited options for treating advanced melanoma51. Hence, identifying novel targets for melanoma therapy is critical. In this study, we first determined that miR-34a is strongly and negatively regulated by MALAT1 based on experiments in which the lncRNA was overexpressed. Previous studies revealed that MALAT1 functions as a molecular sponge to inhibit miRNA expression, and plays a significant role in cancer pathogenesis52–55. This study presents strong evidence that MALAT1 is overexpressed in melanoma tissues and functions as a molecular sponge that competitively inhibits miR-34a and modulates the abundance of the targeted c-Myc and Met. The expression of miR-34a, which is a member of the miR-34 family, is downregulated in neuroblastoma, pancreatic cancer, prostate cancer, lung cancer, malignant lymphoma, retinoblastoma, and colon cancer22,56–59. In the present study, we demonstrated that the miR-34a expression level is lower in melanoma tissues than in benign nevi.
Earlier investigations concluded that MALAT1 expression is upregulated in many tumors, including those of bladder cancer, ovarian cancer, gastric cancer, osteosarcoma, and pancreatic cancer60–64. Moreover, MALAT1 is considered to be a proto-oncogene in an increasing number of human tumor tissues65–68. In this study, we revealed that MALAT1 is more highly expressed in melanoma tissues than in benign nevi, and it contains functional sequence-specific miR-34a-binding sites. Knocking down MALAT1 significantly upregulated the expression of miR-34a.
Luciferase reporter analyses with the co-transfection of miR-34a, a MALAT1 expression plasmid, and c-Myc or a Met luciferase reporter vector clearly indicated that miR-34a suppresses c-Myc and Met luciferase activity and that the miR-34a function is mediated by MALAT1. In addition, miR-34a significantly repressed WT MALAT1, but a mutated putative MALAT1-binding site (Fig. 4) did not completely abolish the repression by miR-34a (Fig. 4), suggesting that miR-34a may also bind to other sequences in MALAT1. Knocking down MALAT1 suppressed the transcription of Met (Fig. 3g), but not c-Myc (Fig. 3h). This result may be due to the following: 1) miRNAs bind to the 3ʹ-UTR of mRNAs to inhibit protein production. Perhaps only Met expression was inhibited by miR-34a in A375 cells; 2) Gene regulation is very complex, especially in various cancer cell lines. Consequently, c-Myc mRNA may have been negatively affected by miR-34a in other cells, but not in A375 cells. In fact, our results are consistent with those from other studies. For example, Christoffersen et al. observed that miR-34a targets MYC during B-RAF-induced senescence, but the overexpression of a miR-34a precursor in TIG3 TERT/DB-RAF:ER cells has little effect on MYC expression at the mRNA level69. Disayabutr et al. also reported that miR-34 targets c-Myc mRNA in type II alveolar epithelial cells, but the overexpression of miR-34a does not markedly influence c-Myc transcription in lung epithelial cells70. We also searched in TargetScan and found there were two well-conserved miR-34-binding sites (position 51–67 bp and 2165-2171 bp) in Met 3ʹ-UTR but not found in c-Myc 3ʹ-UTR.
In summary, we demonstrated that miR-34a expression is inversely associated with MALAT1 in melanoma tissues. Moreover, MALAT1 functions as a molecular sponge for miR-34a, and helps regulate the expression of c-Myc and Met in melanoma cells. We also proved that miR-34a is targeted by MALAT1. Future investigations of the crosstalk between MALAT1 and miR-34a may provide useful insights for developing new strategies to improve the efficacy of melanoma treatments.
Acknowledgements
This study was supported by grants from the Beijing NOVA program (Z161100004916152), the National Natural Science Foundation of China grants (81802728, 81722031, and 81770873), the Major Project in Guangdong Province of Science (2014KZDXM011) and the Youth Program of the PLA (13QNP077 and 16QNP027).
Author contributions
F.L. and X.-J.L. conceived and designed the experiments; Q.L., W.L., and C.-S. Xu. performed the experiments and analyzed the data; X.-G.W. conceived the project, supervised the work, and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by G. Calin
Contributor Information
Fei Li, Phone: +086 13811177062, Email: leefei@126.com.
Xiaogang Wang, Phone: +086 13810228480, Email: xiaogangwang@buaa.edu.cn.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, et al. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.De Vito C, et al. A TARBP2-dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in ewing sarcoma. Cancer Cell. 2012;21:807–821. doi: 10.1016/j.ccr.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 4.Lin A, et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat. Cell. Biol. 2016;18:213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melo SA, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponting CP, Oliver PL, Reik W. Evolution and functions of long non-coding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan JH, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Zhou W, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eis PS, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl Acad. Sci. USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ginger MR, et al. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc. Natl Acad. Sci. USA. 2006;103:5781–5786. doi: 10.1073/pnas.0600745103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodrich JA, Kugel JF. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol. Cell. Biol. 2006;7:612–616. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- 14.Fan Y, et al. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 2014;20:1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 15.Gutschner T, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji Q, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br. J. Cancer. 2014;111:736–748. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luan W, et al. Long non-coding RNA MALAT1 acts as a competing endogenous RNA to promote malignant melanoma growth and metastasis by sponging miR-22. Oncotarget. 2016;7:63901–63912. doi: 10.18632/oncotarget.11564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meseure D, et al. Prognostic value of a newly identified MALAT1 alternatively spliced transcript in breast cancer. Br. J. Cancer. 2016;114:1395–1404. doi: 10.1038/bjc.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J. Biol. Chem. 2015;290:3925–3935. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan P, et al. The HIF-2α-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem. Biophys. Res. Commun. 2016;478:1067–1073. doi: 10.1016/j.bbrc.2016.08.065. [DOI] [PubMed] [Google Scholar]
- 21.Tian Y, Zhang X, Hao Y, Fang Z, He Y. Potential roles of abnormally expressed long non-coding RNA UCA1 and Malat-1 in metastasis of melanoma. Melanoma Res. 2014;24:335–341. doi: 10.1097/CMR.0000000000000080. [DOI] [PubMed] [Google Scholar]
- 22.Sun C, et al. miR-34a mediates oxaliplatin resistance of colorectal cancer cells by inhibiting macroautophagy via transforming growth factor-β/Smad4 pathway. World J. Gastroenterol. 2017;23:1816–1827. doi: 10.3748/wjg.v23.i10.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L, Sun P, Zhou QY, Gao X, Han Q. Long noncoding RNA MALAT1 promotes uveal melanoma cell growth and invasion by silencing of miR-140. Am. J. Transl. Res. 2016;8:3939–3946. [PMC free article] [PubMed] [Google Scholar]
- 24.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl Acad. Sci. USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mraz M, et al. miR-34a, miR-29c, and miR-17-5p are downregulated in CLL patients with TP53 abnormalities. Leukemia. 2009;23:1159–1163. doi: 10.1038/leu.2008.377. [DOI] [PubMed] [Google Scholar]
- 27.Gearhart J, Pashos EE, Prasad MK. Pluripotency redux--advances in stem-cell research. N. Engl. J. Med. 2007;357:1469–1472. doi: 10.1056/NEJMp078126. [DOI] [PubMed] [Google Scholar]
- 28.Campbell KJ, White RJ. MYC regulation of cell growth through control of transcription by RNA polymerases I and III. Cold Spring Harb. Perspect. Med. 2014;4:a018408. doi: 10.1101/cshperspect.a018408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong LM, et al. A regulatory loop involving miR-22, Sp1, and c-Myc modulates CD147 expression in breast cancer invasion and metastasis. Cancer Res. 2014;74:3764–3778. doi: 10.1158/0008-5472.CAN-13-3555. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, et al. The impact of C-MYC gene expression on gastric cancer cell. Mol. Cell. Biochem. 2010;344:125–135. doi: 10.1007/s11010-010-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q, et al. Domain-specific c-Myc ubiquitylation controls c-Myc transcriptional and apoptotic activity. Proc. Natl Acad. Sci. USA. 2013;110:978–983. doi: 10.1073/pnas.1208334110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon Y, Smith BD, Zhou Y, Kaufman MD, Godwin AK. Effective inhibition of c-MET-mediated signaling, growth and migration of ovarian cancer cells is influenced by the ovarian tissue microenvironment. Oncogene. 2015;34:144–153. doi: 10.1038/onc.2013.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sennino B, Ishiguro-Oonuma T, Schriver BJ, Christensen JG, Mcdonald DM. Inhibition of c-Met reduces lymphatic metastasis in RIP-Tag2 transgenic mice. Cancer Res. 2013;73:3692–3703. doi: 10.1158/0008-5472.CAN-12-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sennino B, et al. Suppression of tumor invasion and metastasis by concurrent inhibition of c-Met and VEGF signaling in pancreatic neuroendocrine tumors. Cancer Discov. 2012;2:270–287. doi: 10.1158/2159-8290.CD-11-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.You WK, et al. VEGF and c-Met blockade amplify angiogenesis inhibition in pancreatic islet cancer. Cancer Res. 2011;71:4758–4768. doi: 10.1158/0008-5472.CAN-10-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elia G, et al. Mechanisms regulating c-met overexpression in liver-metastatic B16-LS9 melanoma cells. J. Cell. Biochem. 2001;81:477–487. doi: 10.1002/1097-4644(20010601)81:3<477::AID-JCB1061>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 37.Feng Y, et al. SPSB1 promotes breast cancer recurrence by potentiating c-MET signaling. Cancer Discov. 2014;4:790–803. doi: 10.1158/2159-8290.CD-13-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozasa H, et al. Significance of c-MET overexpression in cytotoxic anticancer drug-resistant small-cell lung cancer cells. Cancer Sci. 2014;105:1032–1039. doi: 10.1111/cas.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X, et al. c-Met and CREB1 are involved in miR-433-mediated inhibition of the epithelial-mesenchymal transition in bladder cancer by regulating Akt/GSK-3β/Snail signaling. Cell Death. Dis. 2016;7:e2088. doi: 10.1038/cddis.2015.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao L, et al. c-Met identifies a population of matrix metalloproteinase 9-producing monocytes in peritumoural stroma of hepatocellular carcinoma. J. Pathol. 2015;237:319–329. doi: 10.1002/path.4578. [DOI] [PubMed] [Google Scholar]
- 41.Zhu M, et al. Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat. Commun. 2017;8:14718. doi: 10.1038/ncomms14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y, et al. Targeting the MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in multiple myeloma. Leukemia. 2018;32:2250–2262. doi: 10.1038/s41375-018-0104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji DG, et al. Inhibition of MALAT1 sensitizes liver cancer cells to 5-flurouracil by regulating apoptosis through IKKα/NF-kappaB pathway. Biochem. Biophys. Res. Commun. 2018;501:33–40. doi: 10.1016/j.bbrc.2018.04.116. [DOI] [PubMed] [Google Scholar]
- 44.Toraih EA, et al. Oncogenic long noncoding RNA MALAT1 and HCV-related hepatocellular carcinoma. Biomed. Pharmacother. 2018;102:653–669. doi: 10.1016/j.biopha.2018.03.105. [DOI] [PubMed] [Google Scholar]
- 45.Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cesana M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He X, et al. MiR-34a modulates ionizing radiation-induced senescence in lung cancer cells. Oncotarget. 2017;8:69797–69807. doi: 10.18632/oncotarget.19267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li QC, Xu H, Wang X, Wang T, Wu J. miR-34a increases cisplatin sensitivity of osteosarcoma cells in vitro through up-regulation of c-Myc and Bim signal. Cancer Biomark. 2017;21:135–144. doi: 10.3233/CBM-170452. [DOI] [PubMed] [Google Scholar]
- 49.Liu F, et al. The long non-coding RNA NEAT1 enhances epithelial-to-mesenchymal transition and chemoresistance via the miR-34a/c-Met axis in renal cell carcinoma. Oncotarget. 2017;8:62927–62938. doi: 10.18632/oncotarget.17757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maroni P, et al. In bone metastasis miR-34a-5p absence inversely correlates with Met expression, while Met oncogene is unaffected by miR-34a-5p in non-metastatic and metastatic breast carcinomas. Carcinogenesis. 2017;38:492–503. doi: 10.1093/carcin/bgx027. [DOI] [PubMed] [Google Scholar]
- 51.Balch CM, et al. Final version of 2009 AJCC melanoma staging and classification. J. Clin. Oncol. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang SM, Hu WW. Long non-coding RNA MALAT1 promotes oral squamous cell carcinoma development via microRNA-125b/STAT3 axis. J. Cell. Physiol. 2018;233:3384–3396. doi: 10.1002/jcp.26185. [DOI] [PubMed] [Google Scholar]
- 53.Chen L, Yao H, Wang K, Liu X, Long Non-Coding RNA. MALAT1 regulates ZEB1 expression by sponging miR-143-3p and promotes hepatocellular carcinoma progression. J. Cell. Biochem. 2017;118:4836–4843. doi: 10.1002/jcb.26158. [DOI] [PubMed] [Google Scholar]
- 54.Jiang LT, et al. Long noncoding RNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) promotes renal cell carcinoma progression via sponging miRNA-429. Med. Sci. Monit. 2018;24:1794–1801. doi: 10.12659/MSM.909450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia C, et al. Metformin, a first-line drug for type 2 diabetes mellitus, disrupts the MALAT1/miR-142-3p sponge to decrease invasion and migration in cervical cancer cells. Eur. J. Pharmacol. 2018;830:59–67. doi: 10.1016/j.ejphar.2018.04.027. [DOI] [PubMed] [Google Scholar]
- 56.Kong D, et al. Epigenetic silencing of miR-34a in human prostate cancer cells and tumor tissue specimens can be reversed by BR-DIM treatment. Am. J. Transl. Res. 2012;4:14–23. [PMC free article] [PubMed] [Google Scholar]
- 57.Meng Y, Quan L, Liu A. Identification of key microRNAs associated with diffuse large B-cell lymphoma by analyzing serum microRNA expressions. Gene. 2018;642:205–211. doi: 10.1016/j.gene.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 58.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 59.Zhao K, et al. Circulating microRNA-34 family low expression correlates with poor prognosis in patients with non-small cell lung cancer. J. Thorac. Dis. 2017;9:3735–3746. doi: 10.21037/jtd.2017.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiao D, et al. LncRNA MALAT1 promotes tumor growth and metastasis by targeting miR-124/foxq1 in bladder transitional cell carcinoma (BTCC) Am. J. Cancer Res. 2018;8:748–760. [PMC free article] [PubMed] [Google Scholar]
- 61.Li J, Gao J, Tian W, Li Y, Zhang J. Long non-coding RNA MALAT1 drives gastric cancer progression by regulating HMGB2 modulating the miR-1297. Cancer Cell. Int. 2017;17:44. doi: 10.1186/s12935-017-0408-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Q, et al. Long noncoding RNA MALAT1 promotes cell proliferation through suppressing miR-205 and promoting SMAD4 expression in osteosarcoma. Oncotarget. 2017;8:106648–106660. doi: 10.18632/oncotarget.20678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin Q, et al. MALAT1 affects ovarian cancer cell behavior and patient survival. Oncol. Rep. 2018;39:2644–2652. doi: 10.3892/or.2018.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou Y, et al. Study on mechanism about long noncoding RNA MALAT1 affecting pancreatic cancer by regulating Hippo-YAP signaling. J. Cell. Physiol. 2018;233:5805–5814. doi: 10.1002/jcp.26357. [DOI] [PubMed] [Google Scholar]
- 65.Gu Y, Xiao X, Yang S. LncRNA MALAT1 acts as an oncogene in multiple myeloma through sponging miR-509-5p to modulate FOXP1 expression. Oncotarget. 2017;8:101984–101993. doi: 10.18632/oncotarget.21957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang J, Yang Y, Fang F, Liu K. MALAT1 modulates the autophagy of retinoblastoma cell through miR-124-mediated stx17 regulation. J. Cell. Biochem. 2018;119:3853–3863. doi: 10.1002/jcb.26464. [DOI] [PubMed] [Google Scholar]
- 67.Malakar P, et al. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 upregulation and mTOR activation. Cancer Res. 2017;77:1155–1167. doi: 10.1158/0008-5472.CAN-16-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pa, M., Naizaer, G., Seyiti, A., Kuerbang, G. Long noncoding rna malat1 functions as a sponge of mir-200c in ovarian cancer. Oncol. Res.10.3727/096504017X15049198963076 (2017). [DOI] [PubMed]
- 69.Christoffersen NR, et al. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17:236–245. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 70.Disayabutr S, et al. miR-34 miRNAs regulate cellular senescence in type II alveolar epithelial cells of patients with idiopathic pulmonary fibrosis. PloS ONE. 2016;11:e0158367. doi: 10.1371/journal.pone.0158367. [DOI] [PMC free article] [PubMed] [Google Scholar]