Abstract
Background
Gene variants are dependable and sensitive biomarkers for target-specific therapies in breast cancer (BC). However, detection of mutations within tissues has many limitations. Plasma circulating free DNA (cfDNA) has been reported in many studies as an alternative tool for detection of mutations. But the diagnostic accuracy of cfDNA for most mutations in BC needs to be reviewed. This study was designed to perform comparative assessment of the diagnostic performance of cfDNA and DNA extracted from tissues for detection of single nucleotide variants (SNV) and copy number variants (CNV).
Methods
True-positive (TP), false-positive (FP), false-negative (FN), and true-negative (TN) values were extracted from each selected study. Pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio (DOR) were calculated. Subgroup analysis and single study omitted analysis were performed to quantify and explain the study heterogeneity.
Results
Twenty eligible studies that involved 1055 cases were included in this meta-analysis. SNV studies in early breast cancer (EBC) subgroup are not suitable for meta-analysis owing to high heterogeneity. However, in advanced breast cancer (ABC) subgroup, the pooled sensitivity and specificity of detection of SNVs were 0.78 (0.71–0.84) and 0.92 (0.87–0.95), respectively. The summary receiver operative curve (SROC) exhibited an area under the curve (AUC) of 0.91(0.88–0.93). The pooled results of studies involving subgroups of PIK3CA, TP53, and ESR1 indicate that the diagnostic value of different genes is different, such as AUC for PIK3CA and TP53 were reported to be 0.96 (0.94–0.98) and 0.94 (0.91–0.95), respectively, and ESR1 had the lowest diagnostic value of 0.80 (0.76–0.83). Owing to the low sensitivity and AUC in the cases of CNV, there is no value for cfDNA-based detection of CNV based on insufficient amount of CNV data.
Conclusion
This meta-analysis suggests that the detection of gene mutations in cfDNA have adequate diagnostic accuracy and can be used as an alternative to the tumor tissue for detection of SNV but not for CNV in BC yet.
Keywords: PIK3CA, TP53, ESR1, cfDNA, Mutation, Breast cancer, Single nucleotide variation, Copy number variation
Background
Breast cancer (BC) is the most common malignant tumor and the leading cause of cancer-associated death in women worldwide [1]. Studies have shown that mutations in genes related to BC can be used as biomarkers and allow personalized therapy for BC patients [2].These genes include PIK3CA, TP53, ESR1, and ERBB2 [3–12]. Sensitivity to specific drugs such as everolimus is determined by the somatic mutational status of PIK3CA [10, 13]. APR-246 (PRIMA-1 MET) can target mutant TP53 [14, 15] and ESR1 gene mutations govern the use of anti-estrogen drugs for breast cancer treatment. Single nucleotide variants (SNV) and copy number variants (CNV) are the most common types of mutation in these genes related to BC [5, 16–18].
Traditionally, the identification of somatic mutations associated with cancer relies on the sequencing of the DNA isolated from the biopsy specimens. However, there are many disadvantages in this method, since it is invasive and repeated biopsies often yield variable results owing to intra-tumor heterogeneity [19]. Recent studies have shown that the genomic mutations in solid malignant tumors can be identified using cell-free DNA (cfDNA) released from cancer cells into blood circulation. This method forms a noninvasive blood test named “liquid biopsy” [20]. The analysis of cfDNA for detection of mutations may play a major role in personalized cancer treatment owing to many advantages including: (i) a noninvasive method for the detection of clinically useful mutations to guide therapy selection [21]; (ii) early detection of mutations related to resistance to a targeted treatment [20, 22]; (iii) a sensitive method for tracking patient’s response to therapy [23]; (iv) minimization of the influences from tumor heterogeneity.
A large number of studies confirm that cfDNA can be used as an alternative tool for the identification of BC biomarkers that provides the ability to overcome the drawbacks of invasive tissue biopsies but the results of these studies are variable. A systematic review and meta-analysis has been published for the analysis of cfDNA based detection accuracy of PIK3CA mutations [24]. However, this study does not review the literature available for detection of mutations in other genes related to breast cancer. In this study, we will perform a systematic review and meta-analysis to integrate the findings of different studies involving the use of cfDNA for the identification of SNVs and CNVs in the most common genes related to BC to comprehensively evaluate the accuracy of cfDNA-based detection of gene mutations in BC.
Method
Literature research strategy
This meta-analysis was performed and reported according to the guidelines about the diagnostic studies [25, 26]. PubMed, EMBASE were searched to identify suitable studies up to the July 30, 2018 and no start data limit was applied. A systematic and comprehensive search was performed with the combination of search terms “ circulating tumor DNA ” or “ circulating tumor-specific DNA ” or “ circulating DNA ” or “ Cell-free DNA ” or “ free DNA ” or “ plasma DNA ”, and “ breast ” or “ breast carcinoma ” or “ tumor of breast ” or “ breast neoplasms ” or “ breast tumor ”. No language restriction was set for a more comprehensive analysis, but only English articles were included.
Inclusion and exclusion criteria
Eligible studies were selected based on the following inclusion criteria: i) studies that involve the evaluation of the accuracy of detecting gene mutations in BC patients using cfDNA; ii) studies that include the verification of gene mutations identified with cfDNA following the analysis of tumor tissues; iii) the studies that carry enough data to construct a diagnostic 2 × 2 table; and iv) studies that include that data for more than five patients.
The exclusion criteria included: i) Lack of verification of gene mutations by the analysis of tumor tissues; ii) insufficient data for constructing the 2 × 2 table; iii) reviews, comments, retracted studies, studies in languages other than English and those not on humans; and iv) evaluation of samples from less than five patients.
All the records were reviewed by the two authors (XY and KZ) independently and the consensus was drawn from each eligible study.
Data extraction
The data were independently extracted from the included studies by three authors (XY, KZ and RXP). The fourth author (CJZ) input the data and the fifth author (CMS) assessed the data as well as resolved any disagreements. The data extracted or calculated from the articles included the author’s name, publication year, age and pathological stage of the participants, detection methods for different kinds of samples, assay indicators and mutation type, true positive (TP), false positive (FP), false negative (FN), and true negative (TN). With various detection methods, those with best sensitivity or specificity were preferred. In some studies without the original data for TP, FP, TN, FN, the accordance, sensitivity and specificity of gene mutation detection in tissue and plasma were available. Then according to the total number of samples (n = TP + FP + TN + FN), sensitivity [= TP/ (TP + FN) × 100%], specificity [= TN/ (TN + FP) × 100%] and overall coincidence rate [= (TP + TN) / (TP + FP + TN + FN) × 100%], the original TP / FP / FN / TN data can be calculated.
Quality assessment
Quality of methodology of the included studies was evaluated based on quality assessment of diagnostic accuracy studies-2 (QUADAS-2) [27]. QUADAS-2 encompasses four key points that include patient selection, index test, reference standard, and flow and timing. According to the Standards for Reporting of Diagnostic Accuracy (STARD), the reference standard is considered to be the best available method for establishing the presence or absence of the condition of interest [28]. Various signaling questions, risk of bias and applicability concerns were judged as “low,” “high,” or “unknown”. Summary of QUADAS plot was generated by Review Manager Software (version 5.3.3, The Cochrane Collaboration).
Statistical analysis
The pooled sensitivity, specificity, positive likelihood ratio [PLR, calculated as sensitivity / (1-specificity)], negative likelihood ratio [NLR, calculated as (1-sensitivity) / specificity], diagnostic odds ratio (DOR) and corresponding 95% confidence intervals (95% CI) were calculated from the TP, FP, FN, and TN values. DOR value is calculated as PLR/NLR [29]. The higher the value of DOR, the higher the diagnostic performance [30]. SROC and AUC were also generated. The effect of threshold was determined through the Spearman correlation between the logit of sensitivity and logit of 1-specificity. Cochran’s Q test was used to assess the heterogeneity caused by the non-threshold effect. The P value ≤0.05 and an inconsistency index (I2) value ≥50% indicated significant heterogeneity.
Sub-group analyses of SNVs were performed for genes (PIK3CA, TP53, and ESR1) and stages including early breast cancer (EBC including stages I-III) and advanced breast cancer (ABC including high risk stages III and IV). According to the NCCN guidelines, BC of stage III is referred to as locally advanced breast cancer (LABC). According to the ESO-ESMO 2nd international consensus guidelines, ABC comprises both LABC and metastatic breast cancer (MBC) [31]. However, in a study by Beaver (2014), stage III BC was classified as EBC [32]. In another study [33], patients diagnosed with BC at stages I-III were grouped together. Therefore, we grouped these studies into EBC subgroup [32, 33]. All the other studies with patients classified into MBC or ABC were grouped into the ABC subgroup.
A sensitivity analysis was also performed to explore the source of heterogeneity and the stability of pooled results. Deek’s funnel plot was generated to show the publication bias and the p value < 0.05 indicated the existence of a publication bias [34]. All the statistical analyses were performed using STATA software (version 12.0; STATA Corporation, College Station, TX) with the MIDAS module.
Results
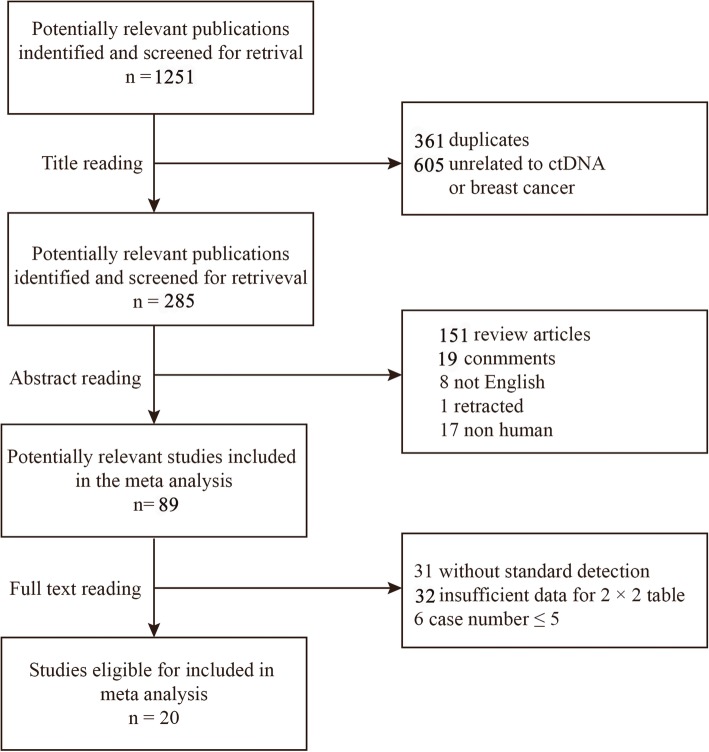
Characteristics of identified studies
Primary computerized literature search was used to identify 1251 records. However, after screening of the titles and abstracts, 1162 studies were excluded because they were either duplicate, non-English, review articles, non-human studies, retracted studies, comments, or irrelevant to the current study. Eighty-nine articles were further reviewed in detail. Out of these, 69 studies were further excluded because of insufficient data for making a 2 × 2 table or lack of standard detection. In a study by Garcia-Saenz JA [35], H1047R and E545K mutations in PIK3CA gene were detected separately. Because there is no study identifying the combination of H1047R and E545K mutations, these two data were included in this meta-analysis as independent studies. In the study by Beaver [32], although the patients came from the same population, the detection of cfDNA was conducted at “post-surgery” and “baseline”, respectively. Therefore, both studies were included. Finally, 20 studies including 1055 cases were identified as eligible (Fig. 1) for inclusion in the meta-analysis [8, 10–12, 32, 33, 35–48].
Fig. 1.
Flow diagram of study selection
All eligible studies were published between 2010 and 2018. The QUADAS-2 summary plot is presented in Fig. 2. The main features of the eligible studies are summarized in Table 1.
Fig. 2.
Methodological quality of eligible studies: Assessment of risk of bias based on the evaluation domains listed by each study (a) and presented as percentage across the included studies (b)
Table 1.
Characteristics of Eligible Studies
| Author | Year | Case | Age (range) | Stage | Detection Method | Data sources | Sampling time | Gene | Mutation Type | TP | FP | FN | TN | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gDNA (tissue) | cfDNA (plasma) | |||||||||||||
| Beaver JA | 2014 | 29 | 60 (38–77) | EBC | dPCR | dPCR | Reported in text | baseline | PIK3CA | SNV | 13 | 0 | 1 | 15 |
| Beaver JA (2) | 2014 | 29 | 60 (38–77) | EBC | dPCR | dPCR | Reported in text | post-surgery | PIK3CA | SNV | 10 | 3 | 0 | 16 |
| Dawson SJ | 2013 | 30 | 66 (43–85) | MBC | TAm-Seq/PE-WGS | dPCR,TAm-Seq | Reported in text | mid-therapy | PIK3CA | SNV | 9 | 0 | 1 | 20 |
| Dawson SJ(2) | 2013 | 30 | 66 (43–85) | MBC | TAm-Seq/PE-WGS | dPCR,TAm-Seq | Reported in text | mid-therapy | TP53 | SNV | 15 | 0 | 1 | 14 |
| Higgins MJ | 2012 | 48 | 62(39–84) | MBC | BEAMing | BEAMing | Data-extrapolated | unavailable | PIK3CA | SNV | 14 | 8 | 6 | 20 |
| Rothe F | 2014 | 17 | 48(35–62) | MBC | Ion PGM | Ion PGM | Reported in text | mid-therapy | PIK3CA | SNV | 4 | 1 | 2 | 15 |
| Rothe F (2) | 2014 | 17 | 48(35–62) | MBC | Ion PGM | Ion PGM | Reported in text | chemotherapy | TP53 | SNV | 5 | 0 | 1 | 16 |
| Spoerke J | 2016 | 156 | unavailable | MBC | dPCR | dPCR | Data-extrapolated | baseline | PIK3CA | SNV | 54 | 8 | 15 | 79 |
| Higgins MJ | 2011 | 51 | unavailable | MBC | sequencing | BEAMing | Data-extrapolated | unavailable | PIK3CA | SNV | 14 | 12 | 0 | 25 |
| Kodahl AR | 2018 | 29 | unavailable | MBC | dPCR | dPCR | Reported in text | unavailable | PIK3CA | SNV | 20 | 0 | 4 | 5 |
| Garcia-Saenz JA | 2015 | 37 | unavailable | ABC (IV 84%) | dPCR | dPCR | Data-extrapolated | unavailable | PIK3CA (p.E545K) | SNV | 4 | 2 | 0 | 31 |
| Garcia-Saenz JA (2) | 2015 | 37 | unavailable | ABC (IV 84%) | dPCR | dPCR | Data-extrapolated | unavailable | PIK3CA (p.H1047R) | SNV | 6 | 2 | 4 | 25 |
| Board RE | 2010 | 30 | 64 (39–88) | MBC | ARMS | ARMS | Data-extrapolated | unavailable | PIK3CA | SNV | 0 | 0 | 14 | 16 |
| Board RE(2) | 2010 | 43 | 59(43–79) | MBC | ARMS | ARMS | Data-extrapolated | mid-therapy | PIK3CA | SNV | 8 | 1 | 2 | 32 |
| Oshiro C | 2015 | 313 |
≤50:121; > 50:192 |
EBC | real-time PCR | dPCR | Reported in text | preoperative | PIK3CA | SNV | 85 | 0 | 25 | 203 |
| Frenel,JS | 2015 | 7 | 60 (29–78) | MBC | PGM | PGM | Reported in text | mid-therapy | PIK3CA | SNV | 2 | 0 | 0 | 5 |
| Frenel,JS (2) | 2015 | 7 | 61 (29–78) | MBC | PGM | PGM | Reported in text | mid-therapy | TP53 | SNV | 4 | 0 | 2 | 1 |
| Liang,DH | 2016 | 23 | 55.5(55.5 ± 13.1) | ABC (IV/ high-risk III) | NGS | Digital Sequencing | Reported in text | mid-therapy | PIK3CA | SNV | 4 | 1 | 2 | 16 |
| Liang,DH (2) | 2016 | 23 | 55.5(55.5 ± 13.1) | ABC (IV /high-risk III) | NGS | Digital Sequencing | Reported in text | mid-therapy | TP53 | SNV | 8 | 1 | 7 | 7 |
| Schiavon G | 2015 | 31 | 58(WT);69(MT) | ABC | dPCR | dPCR | Reported in text | relapsed or progressed | ESR1 | SNV | 3 | 0 | 1 | 27 |
| Takeshita T | 2017 | 35 | 56.4 (31–84) | MBC | dPCR | dPCR | Reported in text | mid-therapy | ESR1 | SNV | 1 | 4 | 5 | 25 |
| Madic J | 2015 | 31 | unavailable | MBC | Hiseq and 454 | Hiseq and 454 | Reported in text | baseline | TP53 | SNV | 21 | 1 | 5 | 4 |
| Nakauchi C | 2016 | 17 | 57.7(32–80) | MBC | Ion-PGM | Ion-PGM | Reported in text | recurrent and primary | PIK3CA | SNV | 3 | 2 | 1 | 11 |
| Nakauchi C (2) | 2016 | 17 | 57.7(32–80) | MBC | Ion-PGM | Ion-PGM | Reported in text | recurrent or primary | TP53 | SNV | 4 | 1 | 2 | 10 |
| Sefrioui D | 2015 | 7 | 55(41–71) | MBC | dPCR | dPCR | Reported in text | mid-therapy | ESR1 | SNV | 4 | 0 | 3 | 14 |
| Janku F | 2015 | 107 | 58 (20–84) | ABC | PBDS, MPD, Ion Torrent | BEAMing | Reported in text | mid-therapy | PIK3CA | SNV | 12 | 8 | 2 | 85 |
| Chung JH | 2017 | 14 | 58 (32–85) | ABC (IV 94%) | HiSeq 2500/ 4000 | HiSeq 2500 or 4000 | Reported in text | mid-therapy | PIK3CA | SNV | 3 | 1 | 0 | 11 |
| Chung JH(2) | 2017 | 14 | 58 (32–85) | ABC (IV 94%) | HiSeq 2500/ 4000 | HiSeq 2500 or 4000 | Reported in text | mid-therapy | TP53 | SNV | 4 | 2 | 0 | 8 |
| Chung JH(3) | 2017 | 14 | 58 (32–85) | ABC (IV 94%) | HiSeq 2500/4000 | HiSeq 2500 or 4000 | Reported in text | mid-therapy | ESR1 | SNV | 4 | 3 | 1 | 9 |
| Chung JH(4) | 2017 | 14 | 58 (32–85) | ABC (IV 94%) | HiSeq 2500/ 4000 | HiSeq 2500 or 4000 | Reported in text | mid-therapy | CCND1 | CNV | 1 | 0 | 4 | 9 |
| Chung JH(5) | 2017 | 14 | 58 (32–85) | ABC (IV 94%) | HiSeq 2500/4000 | HiSeq 2500 or 4000 | Reported in text | mid-therapy | MYCN | CNV | 1 | 0 | 0 | 13 |
| Liang DH(3) | 2016 | 23 | 55.5(55.5 ± 13.1) | ABC (IV/high -risk III) | NGS | Digital Sequencing | Reported in text | mid-therapy | ERBB2 | CNV | 2 | 0 | 1 | 20 |
| Liang DH(4) | 2016 | 23 | 55.5(55.5 ± 13.2) | ABC (IV/high -risk III) | NGS | Digital Sequencing | Reported in text | mid-therapy | EGFR | CNV | 1 | 2 | 1 | 19 |
| Page K. | 2011 | 30 | unavailable | MBC | Quantitative PCR | Quantitative PCR | Reported in text | baseline | HER2 | CNV | 5 | 0 | 8 | 5 |
Abbreviation: gDNA genomic DNA, cfDNA cell free DNA, EBC early breast cancer, dPCR digital PCR, FFPE Formalin-fixed paraffin-embedded, MBC metastatic breast cancer, TAm-Seq tagged-amplicon deep sequencing, PE-WGS paired-end whole-genome sequencing, BEAMing beads, emulsion, amplification, magnetics, ARMS Amplification Refractory Mutation Testing System, WT wild type, MT mutation type, ABC advanced breast cancer, NGS Next generation sequence, PBDS PCR-based DNA sequencing
Threshold effect and heterogeneity
For detection of SNVs, as shown in the Table 2 and Fig. 4, significant heterogeneity was noticed in the data accuracy, sensitivity, and specificity when all the studies were pooled. As for the EBC subgroup, the threshold effect analysis demonstrated that the Spearman correlation coefficient and p value were 1.00 and 0.00 (< 0.05) respectively, which suggests there is significant threshold effect among the studies of the EBC subgroup and it is not suitable to pool the effect-quantity of studies. On the other hand, for the ABC subgroup, the heterogeneity was reduced significantly. The Spearman correlation coefficient and p value were 0.02 and 0.92 (> 0.05) respectively, which suggests that there is no significant threshold effect among the ABC subgroup studies and the heterogeneity was not caused by threshold. Sensitivity analysis by single-study omission analysis for ABC subgroup revealed that the pooled results were significantly affected by the studies from Higgins (2011 and 2012) (Table 3). When these two studies were excluded, the heterogeneity was decreased significantly (I2 = 28.6%, p = 0.10 and I2 = 2.81%, p = 0.42). This shows that these two studies contributed to the high level of heterogeneity observed.
Table 2.
Meta-analysis Estimates
| SNV(Stage) | P Sen (95% CI) | Heterogeneity (I2, p value) |
P Spe (95% CI) | Heterogeneity(I2,p value) | PLR (95% CI) | NLR (95% CI) | DOR | AUC (SROC) | ADT (SCC, p) |
|---|---|---|---|---|---|---|---|---|---|
| Overall | 0.79(0.69–0.87) | 50.0%, 0.00 | 0.94(0.90–0.97) | 67.8%, 0.000 | 13.8(7.8–24.5) | 0.22(0.15–0.34) | 62(29–133) | 0.95(0.92–0.96) | 0.08, 0.5 |
| EBC | 0.79(0.04–1.00) | 92.99%, 0.00 | 1.00(0.47–1.00) | 93.30%, 0.000 | 1104.9(1.3–958,356.8) | 0.21(0.01–7.30) | 5174(22–1,242,093) | 1.00(0.99–1.00) | 1.00, 0.00 |
| ABC | 0.78(0.71–0.84) | 35.72%, 0.04 | 0.92(0.87–0.95) | 55.64%, 0.001 | 10.3(6.6–16.2) | 0.24(0.18–0.32) | 40(21–75) | 0.91(0.88–0.93) | 0.02, 092 |
| ABC* | 0.77(0.70–0.83) | 28.6%, 0.10 | 0.93(0.90–0.95) | 2.81%, 0.42 | 10.5(7.3–15.0) | 0.25(0.18–0.34) | 42(24–75) | 0.94(0.92–0.96) | −0.09, 0.69 |
| SNV(Gene) | |||||||||
| PIK3CA | 0.83(0.68–0.91) | 29.9%,0.12 | 0.95(0.90–0.98) | 78.2%,0.000 | 15.5(7.6–31.5) | 0.18(0.10–0.36) | 84(33–219) | 0.96(0.94–0.98) | 0.23, 0.37 |
| PIK3CA(EBC) | 0.79(0.04–1.00) | 92.99%, 0.00 | 1.00(0.47–1.00) | 93.30%, 0.000 | 1104.9(1.3–958,356.8) | 0.21(0.01–7.30) | 5174(22–1,242,093) | 1.00(0.99–1.00) | 1.00, 0.00 |
| PIK3CA(ABC) | 0.80(0.74–0.85) | 0.00%,0.45 | 0.91(0.86–0.96) | 65.18%,0.00 | 9.0(5.3–15.5) | 0.22(0.17–0.29) | 41(21–80) | 0.83(0.79–0.86) | 0.35, 0.25 |
| PIK3CA(ABC)a | 0.80(0.73–0.85) | 0.00%,0.78 | 0.93(0.90–0.95) | 0.00%,0.91 | 11.1(7.6–16.1) | 0.22(0.16–0.30) | 50(29–88) | 0.94(0.91–0.96) | 0.10, 0.75 |
| TP53 | 0.78(0.64–0.88) | 39.72%,0.13 | 0.92(0.81–0.97) | 3.56%,0.40 | 10.3(3.9–27.8) | 0.24(0.13–0.42) | 44(11–169) | 0.94(0.91–0.95) | 0.09, 0.87 |
| ESR1 | 0.56(0.30–0.79) | 45.67%, 0.14 | 0.95(0.69–0.99) | 72.84%, 0.01 | 10.8(1.3–89.7) | 0.47(0.25–0.88) | 23(2–282) | 0.80(0.76–0.83) | −0.05, 0.94 |
| CNV | P Sen (95% CI) |
Heterogeneity (I2,p value) |
P Spe (95% CI) | Heterogeneity(I2,p value) | PLR (95% CI) | NLR (95% CI) | DOR | AUC (SROC) | ADT (SCC, p) |
| 0.42 (0.24–0.62) | 0.0%,0.52 | 0.98(0.71–1.00) | 13.27%, 0.33 | 19.9(1.1–365.1) | 0.60(0.42–0.84) | 33(2–702) | 0.45(0.41–0.50) | −0.50, 0.39 | |
Note: a, Studies of Higgins MJ.2011 and Higgins MJ. 2012 were excluded
Abbreviation: P Sen Pooled Sensitivity, CI confidence interval, P Spe Pooled Specificity, PLR Positive Likelihood Ratio, NLR Negative Likelihood Ratio, DOR Diagnostic Odds Ratio, AUC Area Under Curve, ADT Analysis of Diagnostic Threshold, SCC Spearman correlation coefficient, EBC Early breast cancer, ABC Advanced breast cancer
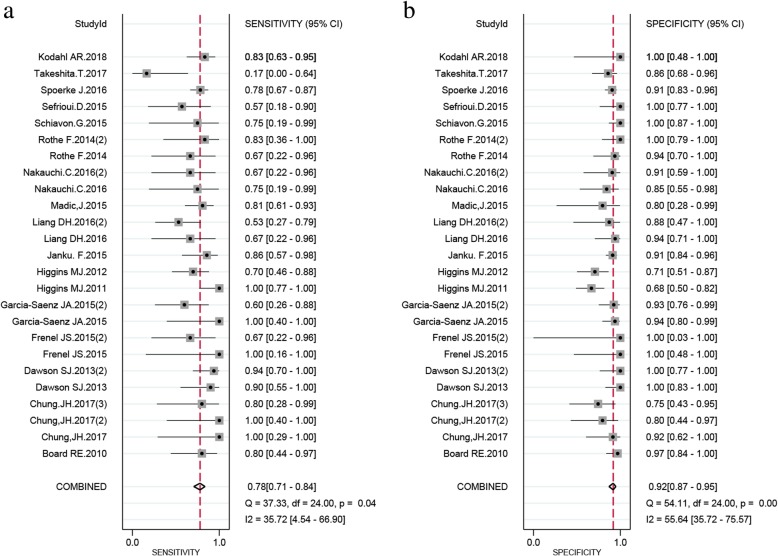
Fig. 4.
The pooled sensitivity (a) and specificity (b) of cfDNA for SNV (ABC) detection were 0.78 (0.71–0.84) and 0.92 (0.87–0.95), respectively which suggests that cfDNA has a higher specificity for SNV detection
Table 3.
Sensitivity Analysis
| SNV (ABC) |
Author(Study) | Year | Sensitivity | Heterogeneity (I2, p value) | Specificity | Heterogeneity (I2, p value) |
|---|---|---|---|---|---|---|
| Board RE.2010(2) | 2010 | 0.774(0.715–0.818) | 40.6%, 0.021 | 0.894(0.871–0.923) | 53.1%, 0.001 | |
| Chung.JH.2017 | 2017 | 0.772(0.720–0.819) | 38.3%, 0.031 | 0.898(0.876–0.922) | 55.4%, 0.001 | |
| Chung.JH.2017(2) | 2017 | 0.771(0.719–0.818) | 36.4%, 0.035 | 0.905(0.878–0.927) | 54.9%, 0.001 | |
| Chung.JH.2017(3) | 2017 | 0.774(0.716–0.818) | 40.6%, 0.024 | 0.906(0.880–0.929) | 53.4%, 0.000 | |
| Dawson SJ.2013 | 2013 | 0.770(0.717–0.817) | 38.9%, 0.028 | 0.895(0.867–0.919) | 51.3%, 0.002 | |
| Dawson SJ.2013(2) | 2013 | 0.760(0.704–0.810) | 34.0%, 0.054 | 0.896(0.88–0.920) | 52.6%, 0.001 | |
| Frenel JS.2015 | 2015 | 0.773(0.721–0.818) | 39.8%, 0.024 | 0.898(0.870–0.922) | 54.2%, 0.001 | |
| Frenel JS.2015(2) | 2015 | 0.777(0.724–0.823) | 40.1%, 0.031 | 0.899(0.871–0.922) | 55.2%, 0.000 | |
| Garcia-Saenz JA.2015 | 2015 | 0.773(0.719–0.821) | 39.4%, 0.026 | 0.896(0.876–0.926) | 59.5%, 0.000 | |
| Garcia-Saenz JA.2015(2) | 2015 | 0.780(0.728–0.827) | 36.4%, 0.040 | 0.897(0.869–0.922) | 55.2%, 0.000 | |
| Higgins MJ.2011 | 2011 | 0.777(0.722–0.825) | 37.4%, 0.034 | 0.902(0.875–0.925) | 59.5%, 0.000 | |
| Higgins MJ.2012 | 2012 | 0.763(0.709–0.808) | 25.2%, 0.129 | 0.914(0.892–0.939) | 43.9%, 0.012 | |
| Janku. F.2015 | 2015 | 0.780(0.726–0.825) | 39.0%, 0.027 | 0.912(0.886–0.934) | 42.7%, 0.001 | |
| Liang DH.2016 | 2016 | 0.770(0.717–0.815) | 38.8%, 0.028 | 0.896(0.865–0.926) | 55.1%, 0.000 | |
| Liang DH.2016(2) | 2016 | 0.777(0.719–0.821) | 39.4%, 0.026 | 0.897(0.875–0.925) | 55.4%, 0.000 | |
| Madic.J.2015 | 2015 | 0.787(0.729–0.831) | 32.3%, 0.066 | 0.899(0.871–0.926) | 55.4%, 0.000 | |
| Nakauchi.C.2016 | 2016 | 0.771(0.717–0.820) | 39.5%, 0.025 | 0.900(0.872–0.923) | 55.0%, 0.001 | |
| Nakauchi.C.2016(2) | 2016 | 0.775(0.717–0.819) | 39.9%, 0.024 | 0.904(0.877–0.927) | 55.1%, 0.000 | |
| Rothe F.2014 | 2014 | 0.777(0.719–0.821) | 39.4%, 0.026 | 0.899(0.871–0.926) | 55.4%, 0.001 | |
| Rothe F.2014(2) | 2014 | 0.777(0.719–0.821) | 39.4%, 0.026 | 0.898(0.875–0.925) | 55.1%, 0.001 | |
| Schiavon.G.2015 | 2015 | 0.773(0.721–0.820) | 40.5%, 0.022 | 0.896(0.867–0.920) | 52.2%, 0.000 | |
| Sefrioui.D.2015 | 2015 | 0.775(0.723–0.821) | 40.7%, 0.021 | 0.894(0.85–0.918) | 49.6%, 0.003 | |
| Spoerke J.2016 | 2016 | 0.779(0.727–0.826) | 38.6%, 0.034 | 0.896(0.874–0.924) | 52.6%, 0.001 | |
| Takeshita.T.2017 | 2017 | 0.787(0.712–0.825) | 19.3%, 0.021 | 0.901(0.867–0.927) | 55.3%, 0.001 | |
| Kodahl AR.2018 | 2018 | 0.769(0.715–0.818) | 39.8%, 0.024 | 0.898(0.878–0.928) | 54.4%, 0.001 | |
| CNV | Author(Study) | Year | Sensitivity | Heterogeneity (I2, p value) | Specificity | Heterogeneity (I2, p value) |
| Chung.JH.2017(4) | 2017 | 0.474(0.244–0.711) | 0.00%, 0.499 | 0.966(0.883–0.996) | 29.6%, 0.235 | |
| Chung.JH.2017(5) | 2017 | 0.391(0.197–0.615) | 0.00%, 0.600 | 0.964(0.875–0.996) | 24.5%, 0.264 | |
| Liang DH.2016(3) | 2016 | 0.381(0.181–0.616) | 0.00%, 0.422 | 0.958(0.857–0.995) | 12.3%, 0.331 | |
| Liang DH.2016(4) | 2016 | 0.409(0.207–0.636) | 17.1%, 0.305 | 1.000(0.925–1.000) | 0.00%, 1.000 | |
| Page.K.2011 | 2011 | 0.455(0.167–0.766) | 15.8%, 0.210 | 0.968(0.890–0.996) | 33.7%, 0.210 |
For CNV, the heterogeneity of sensitivity and specificity were 0.0% (p = 0.52) and 13.27% (p = 0.33), respectively. The Spearman correlation coefficient and p value were − 0.50 and 0.39 (> 0.05), respectively, which suggests there is no significant heterogeneity and threshold effect among the studies involving detection of CNVs.
Publication bias
For SNV, the publication bias tested using the Deek’s funnel plot was 0.70 (> 0.05) (Fig. 3b). This suggests that there is no evidence of publication bias for SNV studies. Since CNV detection studies are less than 10, it is not suitable to perform this analysis on CNV studies.
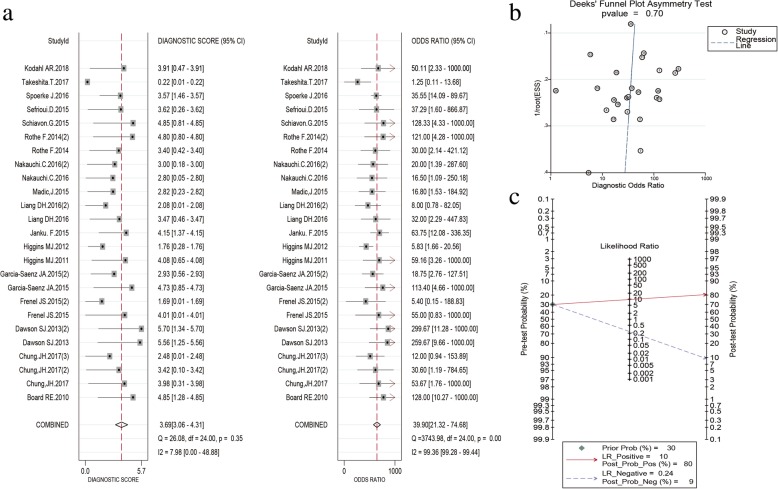
Fig. 3.
Diagnostic odds ratio (DOR), Deek’s funnel plot and Fagan’s Nomogram of cfDNA for SNV (ABC): a. The DOR of SNV was 40 (21–75) indicating high diagnostic performance; b. The p value was 0.70 (> 0.05) which suggests that there was no evidence of publication bias for SNV. c. The post-test probability of positive result was raised from 30 to 80%, which indicates a high diagnosis utility of cfDNA for detection of SNV
Diagnostic accuracy
For SNV (ABC), compared to the reference standard test, the pooled sensitivity and specificity were 0.78 (0.71–0.84) and 0.92 (0.87–0.95), respectively. The PLR, NLR and DOR were 10.3 (6.3–17.2), 0.24 (0.18–0.33), and 40 (21–75), respectively. The SROC exhibited an AUC of 0.91 (0.88–0.93) (Table 2 and Figs. 3a, 4, and 5a). After the studies by Higgins (2011 and 2012), which contributed mainly to the heterogeneity were excluded, the results of these indicators changed very slightly (Table 2). The pooled results of different genes subgroups are shown in Table 2. The diagnostic performance of different genes was different, such as AUC, PIK3CA and TP53 exhibited the values of 0.96 (0.94–0.98), 0.94 (0.91–0.96) respectively, while ESR1 showed the lowest value 0.80 (0.76–0.83).
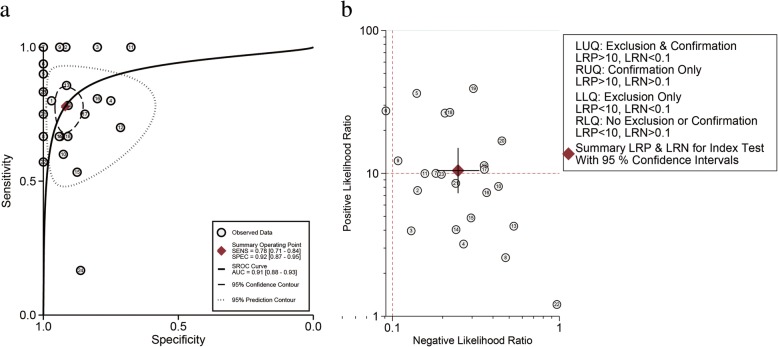
Fig. 5.
Summary roc curve (SROC) and Summary PLR and NLR for cfDNA test: a. SROC of cfDNA based detection of SNV (ABC): The area under the curve (AUC) was 0.91 (0.88–0.93) indicating an impressive overall accuracy; b. Summary PLR and NLR for cfDNA based detection of SNV (ABC): The PLR and NLR were 10.3 (6.6–16.2) and 0.24 (0.18–0.32), respectively, indicating that the detection of SNV through cfDNA has a highly significant detection rate but exhibits a poor exclusion ability
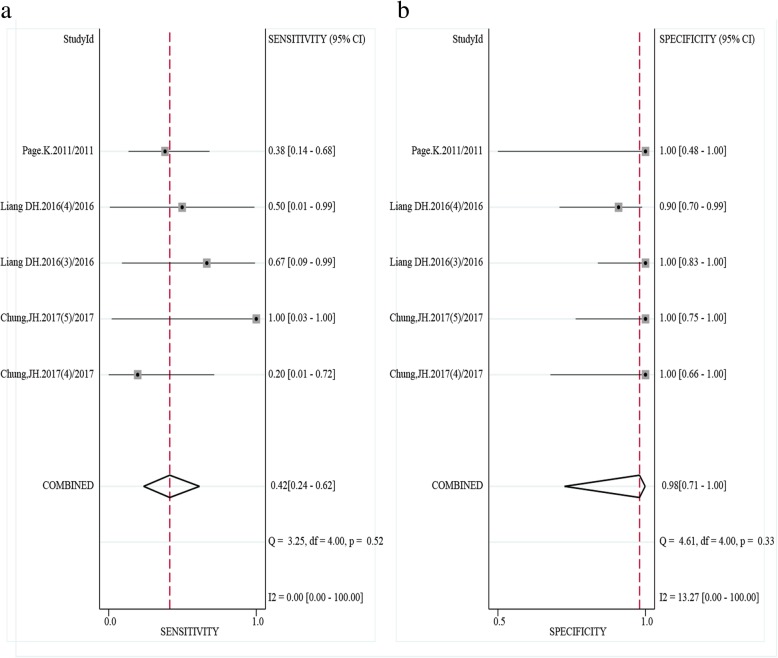
For CNV, the pooled sensitivity, specificity, PLR, NLR, DOR and AUC were 0.42 (0.24–0.62), 0.98 (0.71–1.00), 19.9 (1.1–365.1), 0.60 (0.42–0.84), 33 (2–702) and 0.45 (0.41–0.50) respectively (Table 2 and Fig. 6).
Fig. 6.
The pooled sensitivity (a) and specificity (b) of cfDNA for CNV detection were 0.42 (0.24–0.62) and 0.98 (0.71–1.00), respectively
Discussion
This study is the first study involving the evaluation of the diagnostic accuracy of cfDNA for detection of different mutation types (SNV and CNV) and for different genes. Currently, there are other meta-analysis studies on the diagnostic values of cfDNA in BC, such as studies from Wang H et al. and Lin Z et al. [49, 50]. But these studies focus on the quantitative or qualitative evaluation of cfDNA for the diagnosis of BC and the identification of benign breast disease. The results of these studies suggest that plasma cfDNA is of great importance in the screening and diagnosis of breast cancer. However, the current study was mainly designed to evaluate the consistency of non-invasive cfDNA detection of gene mutations using tissue DNA detection as a standard reference.
For SNV (ABC), analysis results of ABC subgroup show that mutation detection has a high degree of consistency between cfDNA and biopsy tissue DNA. Although the pooled results including sensitivity, specificity, PLR, DOR and AUC (0.78, 0.92, 10.3, 40 and 0.91) were all lower than the previous study (0.91, 0.98, 39.0, 428 and 0.99) [24], because the present study included more reports and more genes (PIK3CA, TP53, and ESR1), the conclusions drawn are theoretically more reliable.
Fagan’s plot was generated for the visual presentation of the clinical utility of cfDNA. The results revealed that the post-test probability of positive result was raised from 30 to 80% (Fig. 3c). PLR > 10.0 and NLR < 0.1 was defined generally as clinically useful test. In this study, the pooled PLR and NLR of SNV (ABC) reached 10.3 and 0.24, respectively, indicating that the detection of SNV through cfDNA has significantly high detection rate but exhibits a very low ability for exclusion (Fig. 5b, Table 2). In other words, SNV detection using cfDNA qualified as a confirmative assay although it may not be suitable to be used as a test for exclusion. There are also differences among the several common genes, and according to AUC, the diagnostic value of cfDNA for PIK3CA and TP53 is higher than ESR1. This study suggests that for the patients with ABC, the detection of genetic mutations by cfDNA has a high utility of being used as a surrogate of tissue DNA, yet reliable results cannot be obtained in EBC patients because of the obvious heterogeneity.
In the case of CNV, the meta-analysis results showed a good homogeneity among the studies evaluating the use of cfDNA for the detection of CNV. Owing to low sensitivity and AUC compared with the tissue DNA based detection (Table 2), cfDNA is not very suitable for the detection of CNVs. The reliable conclusions depend on more published research results which can be included in our study. However, as the primitive attempt to Meta-analyze the diagnostic value of cfDNA for detection of CNVs, it still has important significance which can attract more interested researchers to conduct further study.
False negatives observed for cfDNA mainly because of the cfDNA detection limits such as the recovery of cfDNA or non-biological errors deriving from library preparation and sequencing, represent a main barrier for employing super-sensitive cfDNA for identification of markers [51]. But this barrier can be overcome by plasma DNA extraction and new high efficiency methods for enrichment and capture in sequencing. Thus, the analytical sensitivity and specificity can be further improved.
In addition, there are some factors that may cause differences in tissue DNA and cfDNA test results, resulting in the heterogeneity between studies and a bias in the final results. For example, the time of tissue collection or surgery and/or administration of systemic therapy relative to the blood collection, differences owing to the use of stored and fresh biological specimens, differences in the detection methods used for tissue and blood in some studies, and variability of the cfDNA detection methods used. Therefore, in order to get more reliable results, more rigorous inclusion criteria should be set, and tissue and blood samples should be obtained at the same time point. More detailed subgroup design may be required, such as the before treatment, after treatment, different treatment method, different specimen storage time, and different detection method subgroups.
However, there are some limitations of this meta-analysis. Firstly, several studies were small scale, which might lead to a bias. The Deek’s funnel plot showed that there is no evidence of publication bias for SNV. But there are very few studies on CNV to test for the publication bias. Thus, more reliable results require more research reports for CNV detection using cfDNA. Secondly, significant heterogeneity was observed in the SNV detection studies. We explored the source of heterogeneity by subgroup analysis, threshold effect analysis and single-study omission analysis. Because of significant heterogeneity in EBC subgroup, these studies were not pooled into meta-analysis. For the studies of ABC subgroup, after studies of Higgins (2011 and 2012) were omitted, high level of detection accuracy was observed as shown in the Table 2, indicating that these two studies may be the primary source of heterogeneity. Thirdly, only studies in English were included in this meta-analysis, but there are still several studies written in non-English language that must be taken into consideration. Fourthly, only the studies on the gene mutation analysis using cfDNA in BC were included. There is a more sensitive method for detection of mutation in cfDNA such as integrated digital error suppression (iDES) [51]. But this study was about other cancers instead of BC so it was excluded for this meta-analysis. This may lead to the under-representation of the performance of cfDNA based mutation detection. Fifthly, owing to the significant heterogeneity, the results from EBC subgroup could not be included in the pooled analysis. More homogeneous studies are needed to evaluate the combined diagnostic value of detection of gene mutations by cfDNA. Sixthly, molecular classification of tumors is of great significance for predicting the risk of recurrence and metastasis of breast cancer and its response to treatment. BC is currently classified into four intrinsic subtypes: Luminal A, Luminal B, ‘basal-like,’ and Erb-B2 overexpression subtype [52]. But in the studies included in this meta-analysis, there is no sufficient data presented for describing or calculating sensitivity and specificity values based on the molecular classification. Therefore, in this meta-analysis study, we did not perform subgroup analyses by molecular classification.
Conclusions
In conclusion, this meta-analysis shows that SNV detection through cfDNA has a high sensitivity, specificity, and accuracy, when the detection with DNA isolated from tissue samples was used as the standard reference. Therefore, it is a promising alternative tool to the tumor tissue for detection of SNV in BC. But for CNV, there is a need for further exploration.
Acknowledgements
Not applicable.
Funding
The collection of literatures, analysis of the data and even the language editing of this work were supported by Youth Foundation of Yantai Yuhuangding Hospital (201603) and Yantai science and technology development plan (2019YD004).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- ABC
Advanced breast cancer
- AUC
Area under the curve
- BC
Breast cancer
- cfDNA
Cell-free tumor DNA
- CNV
Copy number variant
- DOR
Diagnostic odds ratio
- EBC
Early breast cancer
- FN
False negative
- FP
False positive
- NLR
Negative likelihood ratio
- PLR
Positive likelihood ratio
- SNV
Single nucleotide variant
- SROC
Summary receiver operative curve
- TN
True negative
- TP
True positive
Authors’ contributions
XY and KZ reviewed all records independently and reached consensus at each eligible study and were major contributors in writing the manuscript. KZ and RXP independently extracted data from the articles and integrated the final results. CJZ and CMS input and assessed the data, and resolved any disagreements. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xin Yang, Phone: +86 05356691999, Email: yx9828@foxmail.com.
Chengming Sun, Phone: +86 05356691999, Email: 18953569897@163.com.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Matsumoto A, Jinno H, Ando T, Fujii T, Nakamura T, Saito J, Takahashi M, Hayashida T, Kitagawa Y. Biological markers of invasive breast cancer. Jpn J Clin Oncol. 2016;46(2):99–105. doi: 10.1093/jjco/hyv153. [DOI] [PubMed] [Google Scholar]
- 3.De Mattos-Arruda L, Weigelt B, Cortes J, Won HH, Ng CK, Nuciforo P, Bidard FC, Aura C, Saura C, Peg V, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25(9):1729–1735. doi: 10.1093/annonc/mdu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang P, Bahreini A, Gyanchandani R, Lucas PC, Hartmaier RJ, Watters RJ, Jonnalagadda AR, Trejo Bittar HE, Berg A, Hamilton RL, et al. Sensitive detection of mono- and polyclonal ESR1 mutations in primary tumors, metastatic lesions and cell free DNA of breast cancer patients. Clin Cancer Res. 2015. [DOI] [PMC free article] [PubMed]
- 5.Vasan N, Yelensky R, Wang K, Moulder S, Dzimitrowicz H, Avritscher R, Wang B, Wu Y, Cronin MT, Palmer G, et al. A targeted next-generation sequencing assay detects a high frequency of therapeutically targetable alterations in primary and metastatic breast cancers: implications for clinical practice. Oncologist. 2014;19(5):453–458. doi: 10.1634/theoncologist.2013-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, Enoksson J, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65(7):2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 7.Cizkova M, Susini A, Vacher S, Cizeron-Clairac G, Andrieu C, Driouch K, Fourme E, Lidereau R, Bieche I. PIK3CA mutation impact on survival in breast cancer patients and in ERalpha, PR and ERBB2-based subgroups. Breast Cancer Res. 2012;14(1):R28. doi: 10.1186/bcr3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Board RE, Wardley AM, Dixon JM, Armstrong AC, Howell S, Renshaw L, Donald E, Greystoke A, Ranson M, Hughes A, et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat. 2010;120(2):461–467. doi: 10.1007/s10549-010-0747-9. [DOI] [PubMed] [Google Scholar]
- 9.Robles AI, Jen J, Harris CC: Clinical outcomes of TP53 mutations in cancers. LID - 10.1101/cshperspect.a026294 [doi] LID - a026294 [pii]. (2157-1422 (Electronic)).
- 10.Higgins MJ, Jelovac D, Barnathan E, Blair B, Slater S, Powers P, Zorzi J, Jeter SC, Oliver GR, Fetting J, et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res. 2012;18(12):3462–3469. doi: 10.1158/1078-0432.CCR-11-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sefrioui D, Perdrix A, Sarafan-Vasseur N, Dolfus C, Dujon A, Picquenot JM, Delacour J, Cornic M, Bohers E, Leheurteur M, et al. Short report: monitoring ESR1 mutations by circulating tumor DNA in aromatase inhibitor resistant metastatic breast cancer. Int J Cancer. 2015;137(10):2513–2519. doi: 10.1002/ijc.29612. [DOI] [PubMed] [Google Scholar]
- 12.Schiavon G, Hrebien S, Garcia-Murillas I, Cutts RJ, Pearson A, Tarazona N, Fenwick K, Kozarewa I, Lopez-Knowles E, Ribas R, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015;7(313). [DOI] [PMC free article] [PubMed]
- 13.Takano T. Individualized treatment for HER2-positive breast cancer. Ann Oncol. 2014;25:v12. doi: 10.1093/annonc/mdu405.4. [DOI] [Google Scholar]
- 14.Fransson A, Glaessgen D, Alfredsson J, Wiman KG, Bajalica-Lagercrantz S, Mohell N: Strong synergy with APR-246 and DNA-damaging drugs in primary cancer cells from patients with TP53 mutant high-grade serous ovarian cancer. (1757–2215 (Electronic)). [DOI] [PMC free article] [PubMed]
- 15.Bykov VJ, Zhang Q, Zhang M, Ceder S, Abrahmsen L, Wiman KG: Targeting of mutant p53 and the cellular redox balance by APR-246 as a strategy for efficient Cancer therapy. (2234-943X (Print)). [DOI] [PMC free article] [PubMed]
- 16.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross JS, Ali SM, Wang K, Khaira D, Palma NA, Chmielecki J, Palmer GA, Morosini D, Elvin JA, Fernandez SV, et al. Comprehensive genomic profiling of inflammatory breast cancer cases reveals a high frequency of clinically relevant genomic alterations. Breast Cancer Res Treat. 2015;154(1):155–162. doi: 10.1007/s10549-015-3592-z. [DOI] [PubMed] [Google Scholar]
- 18.Page K, Guttery DS, Fernandez-Garcia D, Hills A, Hastings RK, Luo J, Goddard K, Shahin V, Woodley-Barker L, Rosales BM, et al. Next generation sequencing of circulating cell-free DNA for evaluating mutations and gene amplification in metastatic breast Cancer. Clin Chem. 2017;63(2):532–541. doi: 10.1373/clinchem.2016.261834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, Collisson EA, Divers SG, Hoon DSB, Scott Kopetz E, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One. 2015(10):10. [DOI] [PMC free article] [PubMed]
- 20.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 21.Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong V, Sandoval-Tan J, Ladrera G, Thongprasert S, Srimuninnimit V, Liao M, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated Erlotinib and chemotherapy. Clin Cancer Res. 2015;21(14):3196–3203. doi: 10.1158/1078-0432.CCR-14-2594. [DOI] [PubMed] [Google Scholar]
- 22.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchetti A, Palma JF, Felicioni L, De Pas TM, Chiari R, Del Grammastro M, Filice G, Ludovini V, Brandes AA, Chella A, et al. Early prediction of response to tyrosine kinase inhibitors by quantification of EGFR mutations in plasma of NSCLC patients. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2015;10(10):1437–1443. doi: 10.1097/JTO.0000000000000643. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Y, Wang C, Zhu H, Lin Y, Pan B, Zhang X, Huang X, Xu Q, Xu Y, Sun Q. Diagnostic accuracy of PIK3CA mutation detection by circulating free DNA in breast Cancer: a meta-analysis of diagnostic test accuracy. PLoS One. 2016;11(6):e0158143. doi: 10.1371/journal.pone.0158143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leeflang MM. Systematic reviews and meta-analyses of diagnostic test accuracy. Clinical microbiology and infection : the official publication of the European Society of Clinical. Microbiology and Infectious Diseases. 2014;20(2):105–113. doi: 10.1111/1469-0691.12474. [DOI] [PubMed] [Google Scholar]
- 26.Leeflang MM, Deeks JJ, Gatsonis C, Bossuyt PM. Systematic reviews of diagnostic test accuracy. Ann Intern Med. 2008;149(12):889–897. doi: 10.7326/0003-4819-149-12-200812160-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM. Group Q-: QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 28.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig LM, Lijmer JG, Moher D, Rennie D, de Vet HC, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. Ann Intern Med. 2003;138(1):40–44. doi: 10.7326/0003-4819-138-1-200301070-00010. [DOI] [PubMed] [Google Scholar]
- 29.Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol. 2003;56(11):1129–1135. doi: 10.1016/S0895-4356(03)00177-X. [DOI] [PubMed] [Google Scholar]
- 30.Liao W, Mao Y, Ge P, Yang H, Xu H, Lu X, Sang X, Zhong S. Value of quantitative and qualitative analyses of circulating cell-free DNA as diagnostic tools for hepatocellular carcinoma: a meta-analysis. Medicine (Baltimore) 2015;94(14):e722. doi: 10.1097/MD.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardoso F, Costa A, Norton L, Senkus E, Aapro M, Andre F, Barrios CH, Bergh J, Biganzoli L, Blackwell KL, et al. ESO-ESMO 2nd international consensus guidelines for advanced breast cancer (ABC2) Breast. 2014;23(5):489–502. doi: 10.1016/j.breast.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Beaver JA, Jelovac D, Balukrishna S, Cochran RL, Croessmann S, Zabransky DJ, Wong HY, Valda Toro P, Cidado J, Blair BG, et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res. 2014;20(10):2643–2650. doi: 10.1158/1078-0432.CCR-13-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oshiro C, Kagara N, Naoi Y, Shimoda M, Shimomura A, Maruyama N, Shimazu K, Kim SJ, Noguchi S. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res Treat. 2015;150(2):299–307. doi: 10.1007/s10549-015-3322-6. [DOI] [PubMed] [Google Scholar]
- 34.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58(9):882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Saenz JA, Acosta D, Moreno F, Ayllon P, Sotelo M, Caldes T, Diaz-Rubio E, Romero A. Detection of H1047R and E545K PIK3CA mutations from peripheral blood in ER positive breast cancer patients. Cancer Res. 2015;(9):75.
- 36.Higgins MJ, Jelovac D, Barnathan E, Blair B, Slater S, Powers P, Zorzi J, Jeter SC, Oliver GR, Diehl F, et al. Improving detection methods for PIK3CA mutations in breast cancer using peripheral blood from patients with metastastic breast cancer (MBC). J Clin Oncol. 2011;(15):29.
- 37.Page K, Hava N, Ward B, Brown J, Guttery DS, Ruangpratheep C, Blighe K, Sharma A, Walker RA, Coombes RC, et al. Detection of HER2 amplification in circulating free DNA in patients with breast cancer. Br J Cancer. 2011;104(8):1342–1348. doi: 10.1038/bjc.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 39.Rothé F, Laes JF, Lambrechts D, Smeets D, Vincent D, Maetens M, Fumagalli D, Michiels S, Drisis S, Moerman C, et al. Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann Oncol. 2014;25(10):1959–1965. doi: 10.1093/annonc/mdu288. [DOI] [PubMed] [Google Scholar]
- 40.Frenel JS, Carreira S, Goodall J, Roda D, Perez-Lopez R, Tunariu N, Riisnaes R, Miranda S, Figueiredo I, Nava-Rodrigues D, et al. Serial next-generation sequencing of circulating cell-free DNA evaluating tumor clone response to molecularly targeted drug administration. Clin Cancer Res. 2015;21(20):4586–4596. doi: 10.1158/1078-0432.CCR-15-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janku F, Angenendt P, Tsimberidou AM, Fu S, Naing A, Falchook GS, Hong DS, Holley VR, Cabrilo G, Wheler JJ, et al. Actionable mutations in plasma cell-free DNA in patients with advanced cancers referred for experimental targeted therapies. Oncotarget. 2015;6(14):12809–12821. doi: 10.18632/oncotarget.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madic J, Kiialainen A, Bidard FC, Birzele F, Ramey G, Leroy Q, Frio TR, Vaucher I, Raynal V, Bernard V, et al. Circulating tumor DNA and circulating tumor cells in metastatic triple negative breast cancer patients. Int J Cancer. 2015;136(9):2158–2165. doi: 10.1002/ijc.29265. [DOI] [PubMed] [Google Scholar]
- 43.Liang DH, Ensor JE, Liu ZB, Patel A, Patel TA, Chang JC, Rodriguez AA. Cell-free DNA as a molecular tool for monitoring disease progression and response to therapy in breast cancer patients. Breast Cancer Res Treat. 2016;155(1):139–149. doi: 10.1007/s10549-015-3635-5. [DOI] [PubMed] [Google Scholar]
- 44.Nakauchi C, Kagara N, Shimazu K, Shimomura A, Naoi Y, Shimoda M, Kim SJ, Noguchi S. Detection of TP53/PIK3CA mutations in cell-free plasma DNA from metastatic breast Cancer patients using next generation sequencing. Clin Breast Cancer. 2016;16(5):418–423. doi: 10.1016/j.clbc.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Spoerke J, Gendreau S, Johnston S, Schmid P, Krop I, Qui J, Derynck M, Chan I, Walter K, Amler L, et al. High prevalence and clonal heterogeneity of ESR1 mutations (mt) in circulating tumor DNA (ctDNA) from patients (pts) enrolled in FERGI, a randomized phase II study testing pictilisib (GDC-0941) in combination with fulvestrant (F) in pts that failed a prior aromatase inhibitor (AI). Cancer Res. 2016;(4):76.
- 46.Chung JH, Pavlick D, Hartmaier R, Schrock AB, Young L, Forcier B, Ye P, Levin MK, Burris H, Gay LM, et al. Hybrid capture-based genomic profiling of circulating tumor DNA from patients with estrogen receptor-positive metastatic breast cancer. Ann Oncol. 2017. [DOI] [PMC free article] [PubMed]
- 47.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Tomiguchi M, Sueta A, Murakami K, Omoto Y, Iwase H. Comparison of ESR1 mutations in tumor tissue and matched plasma samples from metastatic breast Cancer patients. Transl Oncol. 2017;10(5):766–771. doi: 10.1016/j.tranon.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kodahl AR, Ehmsen S, Pallisgaard N, Jylling AMB, Jensen JD, Lænkholm AV, Knoop AS, Ditzel HJ. Correlation between circulating cell-free PIK3CA tumor DNA levels and treatment response in patients with PIK3CA-mutated metastatic breast cancer. Mol Oncol. 2018;12(6):925–935. doi: 10.1002/1878-0261.12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H, Liu Z, Xie J, Wang Z, Zhou X, Fang Y, Pan H, Han W. Quantitation of cell-free DNA in blood is a potential screening and diagnostic maker of breast cancer: a meta-analysis. Oncotarget. 2017;8(60):102336–102345. doi: 10.18632/oncotarget.21827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin Z, Neiswender J, Fang B, Ma X, Zhang J, Hu X. Value of circulating cell-free DNA analysis as a diagnostic tool for breast cancer: a meta-analysis. Oncotarget. 2017;8(16):26625–26636. doi: 10.18632/oncotarget.15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, Stehr H, Liu CL, Bratman SV, Say C, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34(5):547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Jeffrey Ss, Rees CA, Rees Ca, Pollack JR, Ross DT, Johnsen H, Johnsen H, Akslen LA, Fluge O et al: Molecular portraits of human breast tumours. 2000(0028–0836 (Print)). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.