Significance
Nuclear receptors (NRs) are sequence-specific DNA binding proteins that act as intracellular receptors for small molecules such as hormones. Prior work has shown that NRs function as ligand-dependent switches that initiate a cascade of gene expression changes. The extent to which NRs function as direct regulators of downstream genes in these hierarchies remains incompletely understood. Here, we study the role of the NR EcR in metamorphosis of the Drosophila wing. We find that EcR directly regulates many genes at the top of the hierarchy as well as at downstream genes. Furthermore, we find that EcR binds distinct sets of target genes at different developmental times. This work helps inform how hormones elicit tissue- and temporal-specific responses in target tissues.
Keywords: hormone, transcription factor, CUT&RUN, ecdysone, temporal gene regulation
Abstract
The ecdysone pathway was among the first experimental systems employed to study the impact of steroid hormones on the genome. In Drosophila and other insects, ecdysone coordinates developmental transitions, including wholesale transformation of the larva into the adult during metamorphosis. Like other hormones, ecdysone controls gene expression through a nuclear receptor, which functions as a ligand-dependent transcription factor. Although it is clear that ecdysone elicits distinct transcriptional responses within its different target tissues, the role of its receptor, EcR, in regulating target gene expression is incompletely understood. In particular, EcR initiates a cascade of transcription factor expression in response to ecdysone, making it unclear which ecdysone-responsive genes are direct EcR targets. Here, we use the larval-to-prepupal transition of developing wings to examine the role of EcR in gene regulation. Genome-wide DNA binding profiles reveal that EcR exhibits widespread binding across the genome, including at many canonical ecdysone response genes. However, the majority of its binding sites reside at genes with wing-specific functions. We also find that EcR binding is temporally dynamic, with thousands of binding sites changing over time. RNA-seq reveals that EcR acts as both a temporal gate to block precocious entry to the next developmental stage as well as a temporal trigger to promote the subsequent program. Finally, transgenic reporter analysis indicates that EcR regulates not only temporal changes in target enhancer activity but also spatial patterns. Together, these studies define EcR as a multipurpose, direct regulator of gene expression, greatly expanding its role in coordinating developmental transitions.
Hormones function as critical regulators of a diverse set of physiological and developmental processes, including reproduction, immune system function, and metabolism. During development, hormones act as long-range signals to coordinate the timing of events between distant tissues. The effects of hormone signaling are mediated by nuclear receptors, which function as transcription factors that differentially regulate gene expression in a hormone-dependent manner. Whereas many of the coregulators that contribute to nuclear receptor function have been identified, the mechanisms used by these factors to generate distinct, yet appropriate, transcriptional responses in different target tissues are incompletely understood.
Ecdysone signaling has long served as a paradigm to understand how hormones generate spatial and temporal-specific biological responses. In Drosophila, ecdysone is produced by the ring gland and secreted into the hemolymph, where it is converted into its active form, 20-hydroxyecdysone (20E), before reaching target tissues (1, 2). Pulses of ecdysone are required for transitions between developmental stages, such as the larval molts. A high-titer pulse of ecdysone triggers the end of larval development and the beginning of metamorphosis (1, 2). Ecdysone effects transcriptional changes through binding to its receptor, a heterodimer of the proteins ecdysone receptor (EcR) (homolog of the mammalian farnesoid X receptor) and ultraspiracle (Usp) (homolog of mammalian RXR) (3). In the absence of ecdysone, EcR/Usp is nuclear localized and bound to DNA where it is thought to act as a transcriptional repressor (4, 5). Upon ecdysone binding, EcR/Usp switches to a transcriptional activator (4). Consistent with the dual regulatory capacity of EcR/Usp, a variety of coactivator and corepressor complexes have been shown to function with this heterodimer to regulate gene expression (5–8).
Understanding how ecdysone exerts its effects on the genome has been heavily influenced by the work of Ashburner and colleagues in the 1970s. By culturing larval salivary glands in vitro, Ashburner (9) described a sequence of visible puffs that appear in the giant polytene chromosomes upon addition of ecdysone. A small number of puffs appeared immediately after ecdysone addition, followed by the appearance of more than 100 additional puffs over the next several hours (9). The appearance of early puffs was found to be independent of protein synthesis, suggesting direct action by EcR/Usp, whereas the appearance of late puffs was not, suggesting they require the protein products of early genes for activation (9). These findings, and decades of subsequent work elucidating the molecular and genetic details, have led to a hierarchical model of ecdysone signaling in which EcR/Usp directly induces expression of a small number of early response genes. Many of these early response genes encode transcription factors, such as the zinc finger protein Broad, the nuclear receptor Ftz-f1, and the pipsqueak domain factor E93 (2). The early response transcription factors are required, in turn, to induce expression of the late response genes, which encode proteins that impart temporal and tissue-specific responses in target tissues.
Although the framework of the ecdysone pathway was established through work in salivary glands, additional studies affirmed an essential role for ecdysone signaling in many other tissues. Similar to other hormones, the physiological response to ecdysone is often profoundly specific to each target tissue. For example, ecdysone signaling triggers proliferation, changes in cell and tissue morphology, and eventual differentiation of larval tissues fated to become part of the adult fly, such as the imaginal discs (2, 10). By contrast, ecdysone signaling initiates the wholesale elimination of obsolete tissues, such as the larval midgut and salivary glands through programmed cell death (1, 2, 10). Ecdysone is also essential for remodeling larval neurons that persist until adulthood and specifying the temporal identity of neural stem cell progeny born during this time (11). While it is clear that ecdysone signaling triggers the gene expression cascades that underlie these events, the molecular mechanisms by which ecdysone elicits diverse transcriptional responses in target tissues remains poorly understood.
A key step in delineating the mechanisms by which ecdysone signaling regulates target gene expression involves identification of EcR/Usp DNA binding sites. Given the hierarchical structure of the ecdysone pathway, it is unclear whether EcR acts primarily at the top of the transcriptional cascade, or whether it also acts directly on downstream effector genes. Several early response genes such as br, Eip74EF, and the glue genes have been shown to be directly bound by EcR in vivo (12, 13). At the genome-wide level, polytene chromosome staining revealed ∼100 sites bound by EcR in larval salivary glands (14). DamID and ChIP-seq experiments have identified roughly 500 sites directly bound by EcR in Drosophila cell lines (15, 16). Thus, the available evidence, albeit limited, indicates that EcR binds to a limited number of target genes, consistent with hierarchical models wherein the response to ecdysone is largely driven by early response genes and other downstream factors.
We recently identified the ecdysone-induced transcription factor E93 as being essential for the proper temporal sequence of enhancer activation during pupal wing development (17). In the absence of E93, early-acting enhancers fail to turn off, and late-acting enhancers fail to turn on. Moreover, ChIP-seq identified thousands of E93 binding sites across the genome. These data support the hierarchical model of ecdysone signaling in which early response transcription factors like E93 directly regulate a significant fraction of ecdysone-responsive genes in target tissues.
Here, we sought to determine the role that EcR performs in temporal gene regulation during the larval-to-prepupal transition of the wing. Using wing-specific RNAi, we find that EcR is required for proper morphogenesis of prepupal wings, although it is largely dispensable for wing disc patterning at earlier stages of development. RNA-seq profiling reveals that EcR functions as both a temporal gate to prevent the precocious transition to prepupal development as well as a temporal trigger to promote progression to next stage. Using CUT&RUN, we map binding sites for EcR genome-wide before and after the larval-to-prepupal transition. Remarkably, we find that EcR binds extensively throughout the genome, including at many genes with wing-specific functions that are not part of the canonical ecdysone signaling cascade. Moreover, EcR binding is highly dynamic, with thousands of binding sites gained and lost over time. Finally, transgenic reporter analyses demonstrate that EcR is required not only for temporal regulation of enhancer activity but also for spatial regulation of target enhancers. Together, these findings indicate that EcR does not control gene expression solely through induction of a small number of downstream transcription factors, but instead plays a direct and widespread role in regulating tissue-specific transcriptional programs.
Results
Temporal Changes in Gene Expression During the Larval-to-Prepupal Transition.
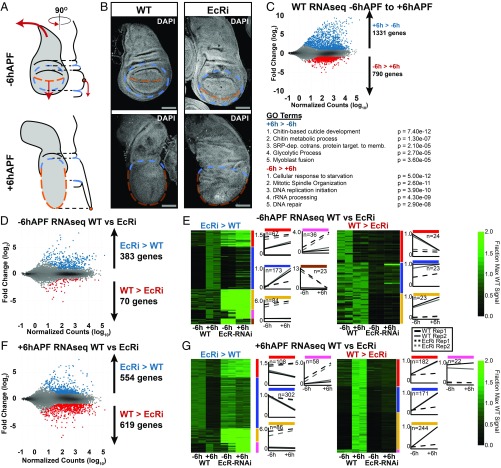
In Drosophila, the end of larval development marks the beginning of metamorphosis. Over a 5-d period, larval tissues are destroyed, and the progenitors of adult tissues, such as wing imaginal discs, undergo a series of progressive morphological and cell differentiation events to acquire their final shapes and sizes. By the end of larval development, the wing disc is comprised of a largely undifferentiated array of columnar epithelial cells (18, 19). The first 12 h after puparium formation (APF) is termed the prepupal stage. During this period, cell division is arrested, and the pouch of the wing disc everts outward, causing the dorsal and ventral surfaces of the wing to appose one another, forming the presumptive wing blade (Fig. 1 A and B) (18, 19). At the same time, the notum of the wing disc extends dorsolaterally and eventually fuses with the contralateral wing disc to form the back of the adult fly (Fig. 1 A and B). Additional events occurring during this time period include secretion of the prepupal cuticle and migration of muscle progenitor cells.
Fig. 1.
EcR is required to promote global changes in gene expression in wings between −6hAPF and +6hAPF. (A) Cartoon diagram of wild-type (WT) wing eversion between −6hAPF and +6hAPF. (B) Confocal images of WT wings and wings expressing UAS-EcR RNAi from vg-tubGAL4 (hereafter EcR-RNAi) at −6hAPF and +6hAPF. The dorsal/ventral (D/V) boundary is marked by an orange dotted line. The edge of the pouch is indicated by a blue dotted line. (C) MA plots (Top) and gene ontology terms (Bottom) of RNA-seq comparing gene WT wings at −6hAPF and +6hAPF. (D and E) MA plots and clustered heatmaps of RNA-seq data comparing EcR-RNAi wings and WT wings at −6hAPF. (F and G) MA plots and heatmaps of RNA-seq data comparing EcR-RNAi wings and WT wings at +6hAPF. (Scale bars for immunostaining: 100 µm.) For MA plots, differentially expressed genes (Padj < 0.05; absolute log2 fold change > 1) are colored red and blue. Heatmaps are represented as the fraction of max WT counts. The colored bars to the Right denote start and end of each cluster. Line plots are the mean signal for each cluster (solid, WT; dashed, EcR-RNAi; see legend between E and G).
To understand EcR’s role in promoting the larval-to-prepupal transition, we began by identifying global changes in gene expression that occur in wild-type (WT) wings before and after the onset of pupariation. We collected wing tissue from wandering, third-instar larvae, approximately 6 h before puparium formation (hereafter, −6hAPF) and from prepupae, approximately 6 h after puparium formation (hereafter, +6hAPF), and performed RNA-seq, aligning our reads to the dm3 reference sequence (20). As described previously (19), WT gene expression is highly dynamic during this time period. Using a conservative definition for differential expression (false-diskovery rate < 0.05, ≥2-fold change in expression), we identified over 1,300 genes increasing in expression and nearly 800 genes decreasing in expression (Fig. 1C). The observed gene expression changes are consistent with developmental events occurring at this time. For example, genes that increase over time are involved in cuticle deposition, cellular metabolism, and muscle development (Fig. 1C). By contrast, genes that decrease over time are involved in cell cycle regulation and DNA replication. Thus, the morphological changes that define the larval-to-prepupal transition are rooted in thousands of changes in gene expression.
EcR Is Required for the Larval-to-Prepupal Transition in Wings.
The onset of pupariation is induced by a high-titer ecdysone pulse. At the genetic level, ecdysone acts through its receptor, EcR. Null mutations in EcR are embryonic lethal. Therefore, to investigate the role that EcR plays in wing development, we used a wing-specific GAL4 driver in combination with an RNAi construct to knock down EcR expression throughout wing development (21). EcR-RNAi driven in wing discs diminished protein levels by ∼95% (SI Appendix, Fig. S1 A–C).
In agreement with previous work suggesting that EcR does not appear to be required for wing development during the first- and second-instar stages (22, 23), EcR-RNAi wings appear morphologically similar to WT wing imaginal discs at −6hAPF (Fig. 1B). However, EcR-RNAi wing discs are noticeably larger than WT wing discs, consistent with the proposed role for ecdysone signaling in cell cycle inhibition in third-instar larvae (22, 23). By contrast, EcR-RNAi wings at +6hAPF appear morphologically dissimilar to both −6hAPF EcR-RNAi wings and to WT wings at +6hAPF. The pouch fails to properly evert and larval folds remain visible. Similarly, the notum fails to extend appropriately and appears more similar to the larval notum than the notum at +6hAPF (Fig. 1B). These findings suggest that wings fail to properly progress through the larval-to-prepupal transition in the absence of EcR. Notably, this failure is likely not due to a systemic developmental arrest because legs isolated from larvae and pupae expressing EcR-RNAi in the wing exhibit no morphological defects (SI Appendix, Fig. S1D). We conclude that EcR is required tissue specifically for progression through the larval-to-prepupal transition.
To identify genes impacted by the loss of EcR, we performed RNA-seq on EcR-RNAi wings at −6hAPF and +6hAPF. Knockdown of EcR results in widespread changes in gene expression (Fig. 1D). At −6hAPF, 453 genes are differentially expressed in EcR-RNAi wings relative to WT wing imaginal discs. Remarkably, 85% of these genes (n = 383, “−6hAPF EcRi > WT”) are expressed at higher levels in EcR-RNAi wings relative to WT, suggesting that EcR is primarily required to repress gene expression at −6hAPF. To determine the expression profiles of these genes during WT development, we performed cluster analysis (Fig. 1E) and found that 72% of these −6hAPF EcRi UP genes normally increase in expression between −6hAPF and +6hAPF (Fig. 1E). Genes in this category include those involved in cuticle development as well as multiple canonical ecdysone response genes (SI Appendix, Table S1). Thus, a major role of EcR at −6hAPF is to keep genes involved in the prepupal program from being precociously activated during larval stages.
We next examined the impact of EcR knockdown in +6hAPF wings. In contrast to the effect at −6hAPF, wherein genes primarily increased in the absence of EcR, we observed approximately equal numbers of up- and down-regulated genes relative to WT wings at +6hAPF (Fig. 1F). Clustering of EcR-RNAi and WT RNA-seq data revealed distinct differences in the inferred regulatory role of EcR at +6hAPF relative to −6hAPF (Fig. 1G). Seventy-four percent of the genes expressed at higher levels in EcR-RNAi wings relative to WT normally decrease in expression between −6hAPF and +6hAPF (Fig. 1G). Genes in this category include factors that promote sensory organ development and cell cycle genes (SI Appendix, Table S2). The increased levels of these “+6hAPF EcRi > WT” genes suggest that, in addition to preventing precocious activation of the prepupal gene expression program, EcR is also required to shut down the larval gene expression program. However, we also observe a role for EcR in gene activation. For genes that are expressed at lower levels in EcR-RNAi wings (n = 619, “+6hAPF WT > EcRi”), 96% of these genes normally increase between −6hAPF and +6hAPF. Genes in this category include those involved in muscle development, metabolic genes, and regulators of cell and tissue morphology (SI Appendix, Table S2). We conclude that EcR is required not only for gene repression but also for gene activation, consistent with the demonstrated interaction of EcR with both activating and repressing gene-regulatory complexes (5–8). Collectively, these data demonstrate that the failure of EcR-RNAi wings to progress through the larval-to-prepupal transition coincides with widespread failures in temporal gene expression changes.
The transcriptional response to ecdysone has recently been examined in a set of 41 different Drosophila cell lines (24), including several wing disc-derived cell lines. To determine the extent to which these responses mirror ecdysone-triggered gene expression changes in a developing tissue, we compared them to our EcR-RNAi wings (SI Appendix, Fig. S2). In general, the overlap between differentially expressed genes for any given cell line and EcR-dependent genes in the wing was low (e.g., median of 3.97% of EcR-dependent genes at −6hAPF overlap an ecdysone-responsive gene in cell lines) (SI Appendix, Fig. S2D). A subset of wing disc-derived cell lines exhibited modestly greater similarity (e.g., median of 8.39% of ecdysone-responsive genes in wing disc-derived cell lines are categorized as EcR-dependent in −6hAPF wings); however, the overlap remained low overall. Cumulatively, only 16–21% of EcR-dependent genes in the wing were identified as ecdysone responsive in any given cell line (SI Appendix, Fig. S2 C and E). Conversely, only 6–16% of ecdysone-responsive genes in any given cell line were identified as EcR dependent in the wing (SI Appendix, Fig. S2 C and E). Thus, the transcriptional response to ecdysone is highly specific to both cell and developmental state.
EcR Directly Binds Thousands of Sites Genome-Wide.
The experiments described above reveal that ecdysone triggers thousands of gene expression changes in wings during the larval-to-prepupal transition. Because ecdysone signaling initiates a cascade of transcription factor expression, it is unclear which of these changes are mediated directly by EcR. Therefore, we sought to determine the genome-wide DNA binding profiles of EcR in developing wings. For these experiments, we utilized a fly strain in which the endogenous EcR gene product has been epitope-tagged by a transposon inserted into an intron of EcR (25). This epitope tag is predicted to be incorporated into all EcR protein isoforms (hereafter EcRGFSTF) (SI Appendix, Fig. S3A). Genetic complementation tests determined that EcRGFSTF flies are viable at the expected frequency (SI Appendix, Fig. S3B), indicating that epitope-tagged EcR proteins are fully functional. Supporting this interpretation, Western blotting demonstrated that EcRGFSTF protein levels are equivalent to untagged EcR, and immunofluorescence experiments revealed nuclear localization of EcRGFSTF as well as binding of EcRGFSTF to DNA in polytene chromosome spreads (SI Appendix, Fig. S3 C–E).
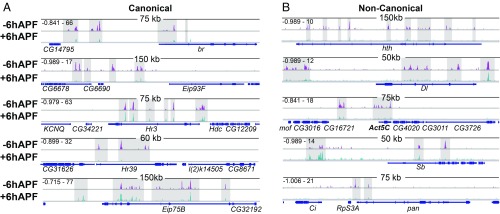
To generate genome-wide DNA binding profiles for EcR, we performed CUT&RUN on −6hAPF wings (Fig. 2A) from EcRGFSTF flies. CUT&RUN provides similar genome-wide DNA binding information for transcription factors as ChIP-seq, but requires fewer cells as input material (26), making it useful for experiments with limiting amounts of tissue. Our EcR CUT&RUN data exhibit features that are similar to those previously reported for other transcription factors, including greater DNA-binding site resolution relative to ChIP-seq (SI Appendix, Figs. S4 and S8). Wing CUT&RUN profiles at −6hAPF reveal that EcR binds extensively throughout the genome (Fig. 2). Many EcR binding sites localize to canonical ecdysone target genes, including broad, Eip93F, Hr3, Hr39, and Eip75B (Fig. 2A). Surprisingly, we also observed EcR binding to many genes that have not previously been categorized as ecdysone targets, including homothorax, Delta, Actin 5C, Stubble, and pangolin (Fig. 2B). Thus, EcR binds widely across the genome in wing imaginal discs. The widespread binding of EcR observed here contrasts with previous genome-wide DNA binding profiles obtained for EcR. For example, ChIP-seq profiles from S2 cells and DamID profiles from Kc167 cells identified 500–1,000 binding sites (15, 16). By contrast, our findings demonstrate that EcR binds both canonical and noncanonical ecdysone-target genes, raising the question as to whether EcR directly contributes to a wing-specific transcriptional program.
Fig. 2.
EcR binds extensively throughout the genome. Browser shots of EcR CUT&RUN signal (z score) at −6hAPF and +6hAPF at (A) canonical and (B) noncanonical ecdysone response genes. Signal range is indicated in Top Left corner. The shaded areas correspond to EcR peaks.
In addition to widespread DNA binding, we also observed clustering of EcR binding sites in the genome. EcR peaks are significantly closer to one another than expected by chance (SI Appendix, Fig. S5 A–C), and a large fraction of peaks are located within 5 kb of an adjacent peak (SI Appendix, Fig. S5D). In particular, canonical ecdysone target genes often exhibit clusters of EcR binding (SI Appendix, Fig. S5 E and F). These findings suggest that EcR often binds multiple cis-regulatory elements across target gene loci, consistent with the observed clustering of ecdysone-responsive enhancers in S2 cells (16).
EcR Binding Is Temporally Dynamic.
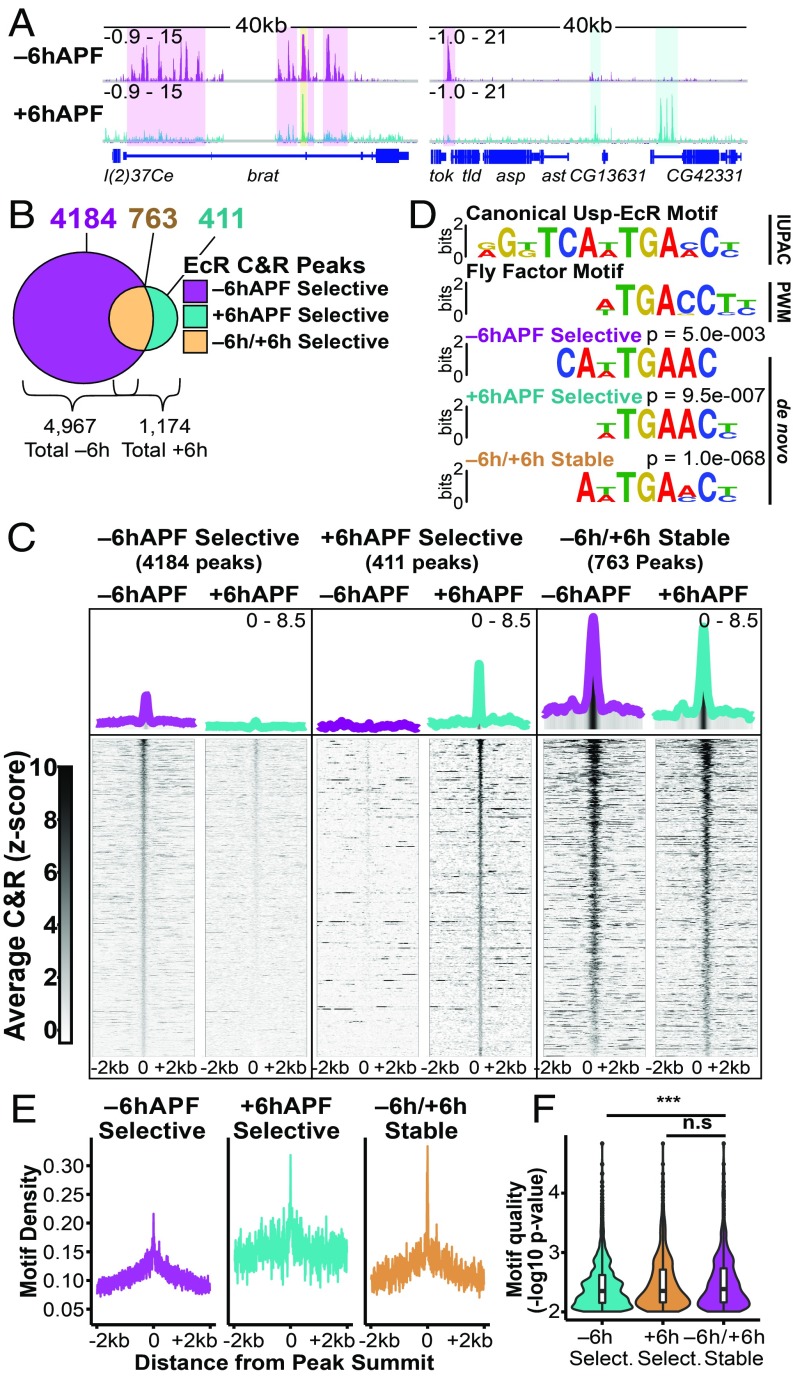
To understand the role of EcR binding in temporal progression of wing development, we performed CUT&RUN on +6hAPF wings (Figs. 2 and 3A). Similar to our findings from −6hAPF wings, we found that EcR binds widely across the genome at +6hAPF. Interestingly, there is a global decrease in the number of sites occupied by EcR over time: A total of 4,967 EcR peaks are called at −6hAPF, whereas 1,174 EcR peaks are called at +6hAPF (Fig. 3B). While many of the +6hAPF binding sites overlap with −6hAPF binding sites (763 peaks, 65%) (hereafter, −6h/+6h stable binding sites), we also identified 411 peaks that are specific to the +6hAPF time. Similar to −6hAPF peaks, +6hAPF EcR peaks are clustered genome-wide (SI Appendix, Fig. S5). Thus, the larval-to-prepupal transition in wings is marked by both a decrease in EcR occupancy at the majority of its −6hAPF binding sites, as well as an increase in EcR occupancy at hundreds of new binding sites. It is notable that many differences in EcR binding between −6hAPF and +6hAPF reflect quantitative rather than binary changes in CUT&RUN signal. Many peaks specific to −6hAPF exhibit low-level CUT&RUN signal at +6hAPF (and vice versa). Among other explanations, this suggests the propensity of EcR to occupy target DNAs is modulated over developmental time.
Fig. 3.
EcR binding is temporally dynamic. (A) Browser shots of EcR CUT&RUN data from −6hAPF and +6hAPF wings, with examples of −6hAPF-selective, +6hAPF-selective, and −6h/+6h stable peaks highlighted by colored boxes. (B) Venn diagrams showing the number of peaks in each category. (C) Heatmaps and average signal plots of EcR C&R signal (z score). (D) Sequence logos comparing the canonical EcR/Usp binding motif to the EcR half-site PWM and EcR motifs identified through de novo motif analysis. (E) Motif density plots of the number of EcR motifs around the peak summit using the EcR half-site PWM. For −6h/+6h stable peaks, the peak summit for +6h was used. (F) Violin plots showing the average motif strength (−log10 P value) of motifs within EcR peaks (***P < 0.001, Student’s t test).
To investigate the potential biological significance of temporal changes in EcR occupancy, we separated EcR peaks into three categories: −6hAPF-selective, +6hAPF-selective, and −6h/+6h stable. Gene annotation enrichment analysis identified genes involved in imaginal disc-derived wing morphogenesis as the top term for each binding site category (SI Appendix, Table S4), indicating that EcR may directly regulate genes involved in wing development at both of these developmental stages. Interestingly, we found that the amplitude of EcR CUT&RUN signal is greater at −6h/+6h stable binding sites relative to temporal-selective binding sites (Fig. 3C). To investigate the potential basis for the difference in binding intensity, we examined the DNA sequence within each class of EcR binding site. Nuclear receptors such as EcR/Usp bind palindromic motifs, with each binding partner recognizing a nearly identical 7-bp half-site (27) (SI Appendix, Fig. S6). For some nuclear receptors, the orientation and spacing of these half-sites can vary. De novo motif diskovery analysis revealed the presence of the EcR half-site in each of the three peak categories (Fig. 3D and SI Appendix, Fig. S6A). De novo searches for longer motifs identified the palindromic motif in −6h/+6h stable and +6hAPF-selective peaks (Fig. 3D and SI Appendix, Fig. S6 A and B). We did not detect variations in the orientation or spacing of half-sites, indicating that when the full palindrome is present, it preferentially exists in a 13-bp inverted repeat conformation. To determine whether differences in signal amplitude between −6h/+6h stable EcR binding sites could be caused by differences in motif content, we examined motif density around peak summits within each of the three binding site categories for the EcR and Usp half-sites, as well as for the EcR/Usp palindromic motif. On average, we observed a positive correlation between motif density and CUT&RUN signal amplitude, with −6hAPF temporal-selective binding sites having both the lowest motif density and lowest signal amplitude, and −6h/+6h stable binding sites having both the highest motif density and highest signal amplitude (Fig. 3E and SI Appendix, Fig. S6C). Furthermore, the average motif strength (i.e., the extent to which the motif matches the consensus EcR half-site) in −6h/+6h stable binding sites was also significantly higher (Fig. 3F). We observed a similar relationship in the +6hAPF-selective binding sites, which exhibit both intermediate CUT&RUN signal and intermediate motif content (Fig. 3E and SI Appendix, Fig. S6C). These data are consistent with a model in which EcR remains stably bound to target sites with high motif density and strength. Conversely, the lower motif content within temporal-selective peaks suggests EcR may rely on cooperative interactions with other transcription factors to assist binding at these sites.
In addition to motif content, we considered the possibility that temporal changes in EcR DNA-binding profiles may be a consequence of temporal changes in EcR protein isoform expression. There are three EcR protein isoforms that share the same DNA-binding domain but differ in their N-terminal domains, allowing them to differentially interact with cofactors (10, 28). The relative isoform abundance varies between tissues and developmental stages. To investigate whether changes in EcR isoform abundance could explain temporal changes in EcR DNA binding, we performed Western blots using isoform-specific antibodies. Consistent with prior studies (29), we found that EcR-A is the predominant isoform expressed in wing imaginal discs (SI Appendix, Figs. S3 and S7A). EcR-B1 is also detected, and EcR-B2 is expressed at low levels. Importantly, we observed no relative changes in EcR isoform abundance between −6hAPF and +6hAPF, nor did we observe a change in the overall levels of EcR over time (SI Appendix, Fig. S7A). Therefore, we conclude that changes in EcR isoform expression are not responsible for the observed changes in EcR binding profiles between −6hAPF and +6hAPF in the wing.
EcR Binding Is Tissue Specific.
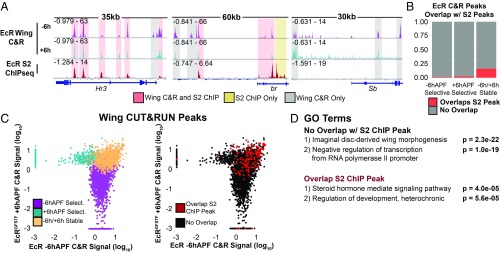
The results described above indicate that EcR binds extensively across the genome, including to many genes with wing-specific function, thus raising the question as to whether EcR binding is tissue specific. To address this question, we first examined loci that had been previously determined to contain functional EcR binding sites by in vitro DNA binding and in vivo reporter assays (13, 30, 31). Many of these sites, including the glue genes Sgs3, Sgs7, and Sgs8, the fat body protein Fbp1, and the oxidative response gene Eip71CD, show no evidence of EcR binding in wings (SI Appendix, Fig. S8), supporting the finding that EcR binds target sites in a tissue-specific manner. To examine this question more globally, we compared our wing CUT&RUN data to EcR ChIP-seq data from Drosophila S2 cells (Fig. 4A). Overall, a small fraction of wing EcR binding sites overlap an EcR binding site in S2 cells (Fig. 4 B and C). However, among the sites that are shared between wings and S2 cells, there is marked enrichment of overlap with −6h/+6h stable wing binding sites. Whereas only 0.1% of −6hAPF-selective binding sites (41 peaks) and 2% of +6hAPF-selective binding sites (9 peaks) overlap an S2 cell EcR binding site, 16% of −6h/+6h stable binding sites (122 peaks) overlap an S2 cell EcR binding site. Thus, binding sites to which EcR is stably bound over time in developing wings are more likely to be shared with EcR binding sites in other cell types, relative to temporal-selective EcR binding sites in the wing.
Fig. 4.
EcR binding is tissue specific. (A) Browser shots comparing EcR CUT&RUN to EcR ChIP-seq in S2 cells (16). Colored boxes highlight examples of shared (red), S2-specific (yellow), and wing-specific peaks (gray). (B) Bar plots showing the proportion of EcR C&R peaks that overlap an S2 ChIP peak in each category. (C) A comparison of the average signal within EcR C&R peaks colored by how they behave temporally (Left) and whether they overlap an S2 ChIP peak (Right). (D) GO terms of the closest gene to a wing EcR peak stratified by whether they overlap an S2 ChIP peak.
To investigate potential differences in target gene function between wing-specific binding sites and those shared with S2 cells, we performed gene annotation enrichment analysis on genes near EcR binding sites. This analysis revealed steroid hormone-mediated signaling pathway as the most significant term for genes overlapping an EcR peak in both wings and S2 cells (Fig. 4D). Genes annotated with this term include canonical ecdysone-responsive genes, such as Eip78C, Hr39, and usp. By contrast, imaginal disc-derived wing morphogenesis was identified as the top term for genes near wing-specific EcR binding sites, similar to our findings from above. These data indicate that EcR binding sites that are shared by wings and S2 cells tend to occur at canonical ecdysone target genes, whereas wing-specific EcR binding sites tend to occur at genes with wing-specific functions. Together, these data suggest EcR plays a direct role in mediating the distinct gene expression responses to ecdysone exhibited by different cell types (24).
EcR Regulates the Temporal Activity of an Enhancer for Broad, a Canonical Ecdysone Target Gene.
The results described above indicate that EcR binds to both canonical and noncanonical ecdysone target genes in the wing, and that EcR is required for temporal progression of wing transcriptional programs. We next sought to examine the relationship between EcR binding in the genome and regulation of gene expression. Because EcR both activates and represses target gene expression, we grouped all differentially expressed genes together and counted the proportion of genes that overlap an EcR binding cluster (SI Appendix, Fig. S9 A–C). We observed an enrichment of EcR binding sites near genes that are differentially expressed in EcR-RNAi wing at both −6hAPF and +6hAPF and a depletion of EcR binding sites near genes that are either temporally static or not expressed (SI Appendix, Fig. S9 A–C). These correlations support a direct role for EcR in regulating temporal changes in gene expression during the larval-to-prepupal transition.
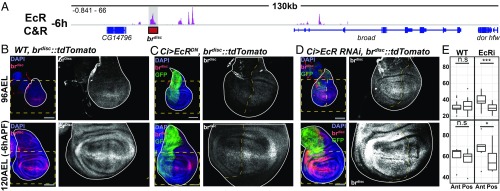
To obtain a more direct readout of EcR’s role in target gene regulation, we investigated whether EcR binding contributes to control of enhancer activity. We first examined the potential regulation of a canonical ecdysone target gene. The broad complex (br) encodes a transcription factor required for the larval-to-prepupal transition in wings and other tissues (Fig. 5A) (32, 33). Br has been characterized as a canonical ecdysone target gene that is induced early in the transcriptional response upon release of hormone (32, 34). In wing imaginal discs, Br protein levels are uniformly low in early third-instar larvae, and by late third-instar, Br levels have increased (SI Appendix, Fig. S10A). Ecdysone signaling has been proposed to contribute to this increase in Br expression in wings over time (22, 23).
Fig. 5.
EcR regulates the temporal activity of an enhancer for the gene broad. (A) Browser shots of the br locus, with the location of the brdisc highlighted by a shaded gray region. (B) brdisc activity in WT wings (red) at 96 h after egg laying (96AEL) and 120AEL (−6hAPF). (C) The effect expressing EcR-B2W650A (EcRDN) in the anterior compartment of the wing marked by GFP (green) on brdisc activity. (D and E) Comparison of brdisc activity between the anterior (Ant) and posterior (Pos) compartments of the wing in WT and EcR-RNAi wings (*P < 0.05; ***P < 0.005, paired Student’s t test). The dotted yellow boxes indicate the location of Insets. (Scale bars: 100 μm.)
Our CUT&RUN data identify multiple EcR binding sites across the br locus at both −6hAPF and +6hAPF (Fig. 5A and SI Appendix, Fig. S11). One of these binding sites corresponds to an enhancer (brdisc) we previously identified that recapitulates br activity in the wing epithelium at −6hAPF (17). Consistent with the observed increase in Br protein levels during third-instar wing development, the activity of brdisc increases with time (Fig. 5B). To investigate the potential role of EcR in controlling the activity of brdisc, we ectopically expressed an isoform of EcR with a point mutation in the ligand-binding domain that prevents it from binding ecdysone and thus functions as a constitutive repressor (EcRDN) (35). EcRDN expression in the anterior compartment of the wing results in decreased brdisc activity in both early- and late-stage wing discs (Fig. 5C), indicating that EcRDN represses brdisc. We further examined the role of EcR in regulating brdisc by knocking down EcR via RNAi, which would eliminate both activating and repressing functions of EcR. EcR knockdown resulted in a modest increase in the activity of brdisc in early wing discs compared with WT wings (Fig. 5 D and E), demonstrating that EcR is required to repress brdisc at this stage. We also observed a slight increase in brdisc activity in late wing discs (Fig. 5 D and E). Together, these findings indicate that EcR is required to keep brdisc activity low in early third-instar wing discs, but it is not required for brdisc activation in late third-instar wing discs. Additionally, the observation that brdisc is active in the absence of EcR, and continues to increase in activity over time, suggests that br requires other unknown activators that themselves may be temporally dynamic. Because the levels of Br increase with time, we conclude that release of repression by EcR functions as a temporal switch to control Br expression during the larval-to-prepupal transition.
EcR Binds to Enhancers with Spatially Restricted Activity Patterns in the Wing.
EcR’s role in controlling the timing of br transcription through the brdisc enhancer supports conventional models of ecdysone signaling in coordinating temporal gene expression. To determine whether EcR plays a similar role at noncanonical ecdysone target genes, we focused on the Delta (Dl) gene, which encodes the ligand for the Notch (N) receptor. Notch-Delta signaling is required for multiple cell fate decisions in the wing (36, 37). In late third-instar wing discs, Dl is expressed at high levels in cells adjacent to the dorsal/ventral (D/V) boundary, along each of the four presumptive wing veins, and in proneural clusters throughout the wing (37). Remarkably, despite the requirement of Notch-Delta signaling in each of these areas, no enhancers active in wing discs have been described for the Dl gene. The Dl locus contains multiple sites of EcR binding (Fig. 6A and SI Appendix, Fig. S11). Using open chromatin data from wing imaginal discs to identify potential Dl enhancers (38), we cloned two EcR-bound regions for use in transgenic reporter assays. The first of these enhancers exhibits a spatially restricted activity pattern in late third-instar wing discs that is highly reminiscent of sensory organ precursors (SOPs) (Fig. 6B). Immunostaining for the proneural factor Achaete (Ac) revealed that cells in which this Dl enhancer is active colocalize with proneural clusters (Fig. 6B). Immunostaining also confirmed these cells express Dl (Fig. 6C). We therefore refer to this enhancer as DlSOP. Notably, using DlSOP to drive expression of a destabilized GFP reporter, its activity pattern refines from a cluster of cells to a single cell (Fig. 6C), consistent with models of SOP specification in which feedback loops between N and Dl result in high levels of N signaling in the cells surrounding the SOP, and high levels of Dl expression in the SOP itself. By +6hAPF, the pattern of DlSOP activity does not change, and it remains spatially restricted to cells along the D/V boundary and proneural clusters in the notum. The second Dl enhancer bound by EcR is also active in late third-instar wing discs (Fig. 6A). This enhancer is most strongly active in Dl-expressing cells of the tegula, lateral notum, and hinge (Fig. 6 D and E) (39). In the pouch, it is active in cells that comprise the L3 and L4 proveins, which require Dl for proper development (40), although overlap with Dl in each of these regions is less precise (Fig. 6 D and E). We refer to this enhancer as Dlteg. Collectively, these data demonstrate that, in contrast to the widespread activity of brdisc, the EcR-bound enhancers in the Dl locus exhibit spatially restricted activity, raising the possibility that EcR binding may serve a different function at these binding sites.
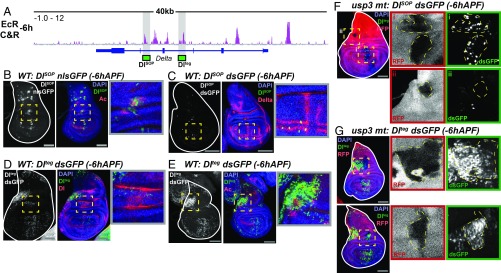
Fig. 6.
EcR regulates the spatial activity of enhancers for the gene Dl. (A) Browser shots of the Dl locus, with the location of the DlSOP and Dlteg highlighted by a gray box. (B and C) Enhancer activity of DlSOP (green) showing overlap with Ac and Dl. (D and E) Enhancer activity of Dlteg showing overlap with Dl and Ac. (F and G) Enhancer activity of DlSOP and Dlteg in usp3 mitotic clones, which are marked by the absence of RFP. The dotted yellow boxes indicate the location of Insets. (Scale bars: 100 µm.)
Ultraspiracle Clones Display Changes in the Spatial Pattern of Enhancer Activity.
We next sought to determine whether EcR regulates the activity of these enhancers. Since the Dl enhancers drive GAL4 expression, we could not use the EcRDN and EcR-RNAi lines employed above. Therefore, we generated loss-of-function clones of Usp, the DNA binding partner of EcR. Clones of usp were induced at 48–60 h and enhancer activity was assayed at −6hAPF. Surprisingly, usp loss of function results in an increased number of cells in which DlSOP is active in the pouch of wing discs (Fig. 6 F, Inset i), suggesting that EcR/Usp are required to repress DlSOP activation. Notably, clones of usp in other regions of the wing (Fig. 6 F, Inset ii) do not activate DlSOP, indicating that EcR/Usp are not necessary for repression of DlSOP in all cells of the wing. We also note that regions exhibiting ectopic DlSOP activity in usp clones tend to be near regions of existing DlSOP activity, suggesting that localized activating inputs are required to switch the DlSOP enhancer on, and that EcR/Usp binding to DlSOP acts as a countervailing force to restrict its activation to certain cells within these regions. Because the pattern of DlSOP activity does not expand between −6hAPF and +6hAPF in WT wings, the ectopic activation of this enhancer in usp clones supports the conclusion that EcR/Usp regulate the spatial pattern of DlSOP activation rather than its temporal activity pattern, as in the case of the brdisc enhancer.
We observed a similar effect of usp loss of function on activity of the Dlteg enhancer. Dlteg activity expands in usp clones adjacent to regions in which Dlteg is active in WT cells (Fig. 6G). As with DlSOP, however, loss of usp function does not appear to be sufficient to cause ectopic Dlteg activity, as clones that are not adjacent to existing Dlteg activity do not ectopically activate the enhancer. Notably, we did not observe expanded expression of Ac within usp clones, suggesting that the expanded activity pattern of the clones is not due to an expanded proneural domain (SI Appendix, Fig. S12). These results suggest that EcR primarily functions to repress these enhancers at −6hAPF to spatially restrict their activity. The observation that usp loss of function is not sufficient to cause ectopic enhancer activity may be because the activation of these enhancers requires other inputs.
Discussion
Decades of work have established the central role that ecdysone signaling, acting through its nuclear receptor, EcR/Usp, plays in promoting developmental transitions in insects. In this study, we investigate the genome-wide role of EcR during the larval-to-prepupal transition in Drosophila wings. Our findings validate existing models of ecdysone pathway function, and they extend understanding of the direct role played by EcR in coordinating dynamic gene expression programs.
The Role of EcR in Promoting Gene Expression Changes During Developmental Transitions.
Our RNA-seq data reveal that EcR controls the larval-to-prepupal transition by activating and repressing distinct sets of target genes. In larval wing imaginal discs, we find that EcR is primarily required to prevent precocious activation of the prepupal gene expression program. This finding is consistent with previous work that demonstrated precocious differentiation of sensory neurons in the absence of ecdysone receptor function (41). Since ecdysone titers remain low during most of the third larval instar, these data are also consistent with prior work that demonstrated that EcR functions as a transcriptional repressor in the absence of hormone (4, 31). Later in prepupal wings, we find that loss of EcR results in failure to activate the prepupal gene expression program. Indeed, many of the genes that become precociously activated in wing discs fail to reach their maximum level in prepupae. Since rising ecdysone titers at the end of third larval instar trigger the transition to the prepupal stage, this finding is consistent with a hormone-induced switch in EcR from a repressor to an activator (4, 31). We also find that loss of EcR results in persistent activation of the larval gene expression program in prepupal wings. This finding is not clearly explained by a hormone-induced switch in EcR’s regulatory activity. However, it is possible that EcR activates a downstream transcription factor, which represses genes involved in larval wing development. Overall, these findings indicate that EcR functions both as a temporal gate to ensure accurate timing of the larval-to-prepupal transition and as a temporal switch to simultaneously shut down the preceding developmental program and initiate the subsequent program. Finally, it is of particular note that these genome-wide results fit remarkably well with the model of ecdysone pathway function predicted by Ashburner (9) 45 y ago.
Widespread Binding of EcR Across the Genome.
Existing models describe EcR as functioning at the top of a transcriptional cascade, in which it binds a relatively small number of primary-response genes. These factors then activate downstream effectors that mediate the physiological response to ecdysone. Consistent with this model, attempts to assay EcR binding genome-wide in S2 cells and Kc167 cells identified relatively few EcR binding sites. However, this model does not adequately explain how ecdysone elicits distinct transcriptional responses from different target tissues. Our data reveal that EcR binds to thousands of sites genome-wide. While many genes bound by EcR have been previously identified as direct targets, the majority of EcR binding we observe occurs near genes with essential roles in wing development. These data support a model in which EcR directly mediates the response to ecdysone both at the top of the hierarchy and at many of the downstream effectors. Interestingly, comparison of our wing binding profiles with ChIP-seq from S2 cells revealed that shared EcR binding sites are enriched in canonical ecdysone-response genes, suggesting that the top tier of genes in the ecdysone hierarchy are direct targets of EcR across multiple tissues, while the downstream effectors are direct EcR targets only in specific tissues. These data neatly account for the observation that parts of the canonical ecdysone transcriptional response are shared between tissues, even as many other responses are tissue specific. Aside from assay-specific issues, it is possible that the greater number of EcR binding sites identified in the wing relative to cell lines is due to the presence of multiple cell types in the wing that possess distinct EcR binding profiles. Additionally, the extent of EcR binding may directly scale with the magnitude of the physiological response to ecdysone, which in wing imaginal discs is arguably greater (i.e., transformation into pupal wings) than in Kc167 cells (i.e., change in cell shape) (19). In any case, it will be important to identify the factors that contribute to EcR’s tissue-specific DNA targeting in future work. It is possible that tissue-specific transcription factors facilitate EcR binding, as suggested by recent DNA-binding motif analysis of ecdysone-responsive enhancers in S2 and OSC cell lines (16).
Temporally Dynamic Binding of EcR.
Pulses of ecdysone mediate distinct transcriptional responses at different times in development. Some of this temporal selectivity is mediated by the sequential activation of transcription factors that form the core of the ecdysone cascade (32, 42, 43). Our data suggest that changes in EcR binding over time may also be involved. The mechanisms responsible for these changes remain unclear. One potential explanation is that changes in the expression of EcR isoforms could allow recruitment to new sites in the genome. However, we do not observe changes in protein isoform abundance, indicating that this is unlikely to account for changes in EcR DNA-binding profiles. An alternative possibility is that ecdysone titers could induce ligand-dependent changes in EcR structure or affect ligand-dependent interactions with coregulator proteins that influence EcR’s DNA binding. It is also possible that overall EcR levels or the nuclear-to-cytoplasmic ratio of EcR changes with time, as has been previously proposed (44). However, we do not observe changes in EcR protein levels, and while nuclear export of EcR could explain the global reduction in the number of EcR binding sites, it cannot explain the appearance of new EcR binding sites at +6hAPF. For this reason, it is notable that temporal-selective binding sites contain lower motif content on average relative to temporally stable EcR binding sites. This suggests that temporal-selective binding may be more dependent on external factors. An intriguing possibility is that stage-specific transcription factors activated as part of the canonical ecdysone cascade may contribute to recruitment or inhibition of EcR binding at temporal-selective sites.
EcR Controls both Temporal and Spatial Patterns of Gene Expression.
EcR has been shown to act as both a transcriptional activator and repressor. This dual functionality confounded our attempts to draw genome-wide correlations between EcR binding and changes in gene expression. Therefore, we sought to examine the effect of EcR binding on individual target enhancers. We find that EcR regulates the temporal activity of an enhancer for the early-response gene, br. In WT wings, the activity of this enhancer increases between early and late third-instar stages, as do Br protein levels. Ectopic expression of a dominant-repressor isoform of EcR decreased activity of brdisc. Surprisingly, RNAi knockdown of EcR increased brdisc activity, indicating that EcR is not required for brdisc activation. Instead, these findings indicate that EcR represses brdisc in early third-instar wings, consistent with our RNA-seq data which demonstrated that EcR prevents precocious activation of the prepupal gene expression program before the developmental transition. It is not known what factors activate br or other prepupal genes.
Temporal control of gene expression by EcR is expected given its role in governing developmental transitions. However, our examination of EcR-bound enhancers from the Dl locus demonstrates that it also directly controls spatial patterns of gene expression. Loss-of-function clones for EcR’s DNA binding partner Usp exhibited ectopic activation of two Dl enhancers. However, we did not detect ectopic enhancer activity in all usp mutant clones, indicating that EcR is required to restrict activity of target enhancers only at certain locations within the wing. Examination of +6hAPF wings revealed no changes in the spatial pattern of Dl enhancer activity relative to −6hAPF, indicating that ectopic enhancer activation in usp clones does not reflect incipient changes in enhancer activity. Recently, EcR binding sites were shown to overlap with those for the Notch regulator, Hairless, supporting a potential role of EcR in regulating spatial patterns of gene expression (45). We conclude that EcR regulates both temporal and spatial patterns of gene expression. Given the widespread binding of EcR across the genome, our findings suggest that EcR plays a direct role in temporal and spatial patterning of many genes.
Hormones and other small molecules act through nuclear receptors to initiate transcriptional cascades that continue for extended periods of time. For example, thyroid hormone triggers metamorphosis in frogs and other chordates, a process that can take weeks for completion (46). Our work raises the possibility that nuclear receptors play a direct role in regulating the activity of many response genes. In particular, the widespread and temporally dynamic binding of EcR that we observed over a short interval of wing development suggests that the complete repertoire of EcR targets is vastly larger than previously appreciated.
Methods
Detailed experimental materials and methods can be found in SI Appendix, Supplementary Materials and Methods.
RNA-Seq.
RNA from a minimum of 60 wings was extracted as described previously (38). Total RNA-seq was performed with the Ovation RNA-seq system. Reads were aligned to the dm3 reference genome. DESeq2 was used to generate normalized count matrices and identify differentially expressed genes (Padj < 0.05, absolute log2 fold change > 1). Gene clustering was performed using k-medoids. Gene ontology terms used expressed genes as a background.
CUT&RUN.
A minimum of 100 wings was dissected from w; EcRGFSTF/Df(2R)BSC889. Intact wings were permeabalized using digitonin as previously described (26). MNase was activated and digestion was performed for 45 s. Soluble DNA fragments were used as input for the Rubicon Thruplex 12s DNaseq kit. Fragments between 20 and 120 bp were identified and used throughout. Peaks called in a merged file that overlap a peak from at least one replicate were used for analysis. Coverage files were z-normalized per chromosome arm. Gene ontology terms used all genes as a background. Peak clusters were created by resizing each peak to 5,000 bp, and then taking the furthest start and end coordinates of peaks that fell within each overlapping region.
Motif Analysis.
De novo motifs were identified using DREME using FAIRE peaks as a background. FIMO was used to identify EcR and Usp motifs genome-wide using position weight matrices (PWMs) from bacterial one-hybrids.
Supplementary Material
Acknowledgments
We thank Peter J. Skene and Steven Henikoff for reagents and advice on the CUT&RUN protocol. Stocks obtained from the Bloomington Drosophila Stock Center (NIH Grant P40OD018537) were used in this study. C.M.U. was supported in part by NIH Grant T32GM007092. This work was supported in part by Research Scholar Grant RSG-17-164-01-DDC (to D.J.M.) from the American Cancer Society, and in part by Grant R35-GM128851 (to D.J.M.) from the National Institute of General Medical Sciences of the NIH (https://www.nigms.nih.gov/).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE124254).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900343116/-/DCSupplemental.
References
- 1.Thummel CS. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev Cell. 2001;1:453–465. doi: 10.1016/s1534-5807(01)00060-0. [DOI] [PubMed] [Google Scholar]
- 2.Yamanaka N, Rewitz KF, O’Connor MB. Ecdysone control of developmental transitions: Lessons from Drosophila research. Annu Rev Entomol. 2013;58:497–516. doi: 10.1146/annurev-ento-120811-153608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao TP, et al. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature. 1993;366:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- 4.Dobens L, Rudolph K, Berger EM. Ecdysterone regulatory elements function as both transcriptional activators and repressors. Mol Cell Biol. 1991;11:1846–1853. doi: 10.1128/mcb.11.4.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai C-C, Kao H-Y, Yao T-P, McKeown M, Evans RM. SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol Cell. 1999;4:175–186. doi: 10.1016/s1097-2765(00)80365-2. [DOI] [PubMed] [Google Scholar]
- 6.Badenhorst P, et al. The Drosophila nucleosome remodeling factor NURF is required for ecdysteroid signaling and metamorphosis. Genes Dev. 2005;19:2540–2545. doi: 10.1101/gad.1342605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carbonell A, Mazo A, Serras F, Corominas M. Ash2 acts as an ecdysone receptor coactivator by stabilizing the histone methyltransferase Trr. Mol Biol Cell. 2013;24:361–372. doi: 10.1091/mbc.E12-04-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreher J, et al. EcR recruits dMi-2 and increases efficiency of dMi-2-mediated remodelling to constrain transcription of hormone-regulated genes. Nat Commun. 2017;8:14806. doi: 10.1038/ncomms14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashburner M. Puffs, genes, and hormones revisited. Cell. 1990;61:1–3. doi: 10.1016/0092-8674(90)90205-s. [DOI] [PubMed] [Google Scholar]
- 10.Talbot WS, Swyryd EA, Hogness DS. Drosophila tissues with different metamorphic responses to ecdysone express different ecdysone receptor isoforms. Cell. 1993;73:1323–1337. doi: 10.1016/0092-8674(93)90359-x. [DOI] [PubMed] [Google Scholar]
- 11.Syed MH, Mark B, Doe CQ. Steroid hormone induction of temporal gene expression in Drosophila brain neuroblasts generates neuronal and glial diversity. eLife. 2017;6:e26287. doi: 10.7554/eLife.26287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hitrik A, et al. Combgap promotes ovarian niche development and chromatin association of EcR-binding regions in BR-C. PLoS Genet. 2016;12:e1006330. doi: 10.1371/journal.pgen.1006330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehmann M, Wattler F, Korge G. Two new regulatory elements controlling the Drosophila Sgs-3 gene are potential ecdysone receptor and fork head binding sites. Mech Dev. 1997;62:15–27. doi: 10.1016/s0925-4773(96)00644-2. [DOI] [PubMed] [Google Scholar]
- 14.Fisk GJ, Thummel CS. The DHR78 nuclear receptor is required for ecdysteroid signaling during the onset of Drosophila metamorphosis. Cell. 1998;93:543–555. doi: 10.1016/s0092-8674(00)81184-8. [DOI] [PubMed] [Google Scholar]
- 15.Gauhar Z, et al. Genomic mapping of binding regions for the ecdysone receptor protein complex. Genome Res. 2009;19:1006–1013. doi: 10.1101/gr.081349.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shlyueva D, et al. Hormone-responsive enhancer-activity maps reveal predictive motifs, indirect repression, and targeting of closed chromatin. Mol Cell. 2014;54:180–192. doi: 10.1016/j.molcel.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 17.Uyehara CM, et al. Hormone-dependent control of developmental timing through regulation of chromatin accessibility. Genes Dev. 2017;31:862–875. doi: 10.1101/gad.298182.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fristrom D, Wilcox M, Fristrom J. The distribution of PS integrins, laminin A and F-actin during key stages in Drosophila wing development. Development. 1993;117:509–523. doi: 10.1242/dev.117.2.509. [DOI] [PubMed] [Google Scholar]
- 19.Guo Y, Flegel K, Kumar J, McKay DJ, Buttitta LA. Ecdysone signaling induces two phases of cell cycle exit in Drosophila cells. Biol Open. 2016;5:1648–1661. doi: 10.1242/bio.017525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uyehara CM, McKay DJ. 2019 Direct and widespread role for the nuclear receptor EcR in mediating the response to ecdysone in Drosophila. Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124254. Deposited December 21, 2018.
- 21.Colombani J, et al. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. 2005;310:667–670. doi: 10.1126/science.1119432. [DOI] [PubMed] [Google Scholar]
- 22.Herboso L, et al. Ecdysone promotes growth of imaginal s through the regulation of Thor in D. melanogaster. Sci Rep. 2015;5:12383. doi: 10.1038/srep12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirth CK, Truman JW, Riddiford LM. The ecdysone receptor controls the post-critical weight switch to nutrition-independent differentiation in Drosophila wing imaginal s. Development. 2009;136:2345–2353. doi: 10.1242/dev.032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoiber M, Celniker S, Cherbas L, Brown B, Cherbas P. Diverse hormone response networks in 41 independent Drosophila cell lines. G3 (Bethesda) 2016;6:683–694. doi: 10.1534/g3.115.023366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagarkar-Jaiswal S, et al. A genetic toolkit for tagging intronic MiMIC containing genes. eLife. 2015;4:e08469. doi: 10.7554/eLife.08469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skene PJ, Henikoff JG, Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc. 2018;13:1006–1019. doi: 10.1038/nprot.2018.015. [DOI] [PubMed] [Google Scholar]
- 27.D’Avino PP, Crispi S, Cherbas L, Cherbas P, Furia M. The moulting hormone ecdysone is able to recognize target elements composed of direct repeats. Mol Cell Endocrinol. 1995;113:1–9. doi: 10.1016/0303-7207(95)03584-t. [DOI] [PubMed] [Google Scholar]
- 28.Cherbas L, Hu X, Zhimulev I, Belyaeva E, Cherbas P. EcR isoforms in Drosophila: Testing tissue-specific requirements by targeted blockade and rescue. Development. 2003;130:271–284. doi: 10.1242/dev.00205. [DOI] [PubMed] [Google Scholar]
- 29.Schubiger M, Tomita S, Sung C, Robinow S, Truman JW. Isoform specific control of gene activity in vivo by the Drosophila ecdysone receptor. Mech Dev. 2003;120:909–918. doi: 10.1016/s0925-4773(03)00134-5. [DOI] [PubMed] [Google Scholar]
- 30.Antoniewski C, Laval M, Dahan A, Lepesant JA. The ecdysone response enhancer of the Fbp1 gene of Drosophila melanogaster is a direct target for the EcR/USP nuclear receptor. Mol Cell Biol. 1994;14:4465–4474. doi: 10.1128/mcb.14.7.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cherbas L, Lee K, Cherbas P. Identification of ecdysone response elements by analysis of the Drosophila Eip28/29 gene. Genes Dev. 1991;5:120–131. doi: 10.1101/gad.5.1.120. [DOI] [PubMed] [Google Scholar]
- 32.Karim FD, Guild GM, Thummel CS. The Drosophila Broad-Complex plays a key role in controlling ecdysone-regulated gene expression at the onset of metamorphosis. Development. 1993;118:977–988. doi: 10.1242/dev.118.3.977. [DOI] [PubMed] [Google Scholar]
- 33.Kiss I, Beaton AH, Tardiff J, Fristrom D, Fristrom JW. Interactions and developmental effects of mutations in the Broad-Complex of Drosophila melanogaster. Genetics. 1988;118:247–259. doi: 10.1093/genetics/118.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Kalm L, Crossgrove K, Von Seggern D, Guild GM, Beckendorf SK. The Broad-Complex directly controls a tissue-specific response to the steroid hormone ecdysone at the onset of Drosophila metamorphosis. EMBO J. 1994;13:3505–3516. doi: 10.1002/j.1460-2075.1994.tb06657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown HLD, Cherbas L, Cherbas P, Truman JW. Use of time-lapse imaging and dominant negative receptors to dissect the steroid receptor control of neuronal remodeling in Drosophila. Development. 2006;133:275–285. doi: 10.1242/dev.02191. [DOI] [PubMed] [Google Scholar]
- 36.de Celis JF, Garcia-Bellido A, Bray SJ. Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal. Development. 1996;122:359–369. doi: 10.1242/dev.122.1.359. [DOI] [PubMed] [Google Scholar]
- 37.Kooh PJ, Fehon RG, Muskavitch MA. Implications of dynamic patterns of Delta and Notch expression for cellular interactions during Drosophila development. Development. 1993;117:493–507. doi: 10.1242/dev.117.2.493. [DOI] [PubMed] [Google Scholar]
- 38.McKay DJ, Lieb JD. A common set of DNA regulatory elements shapes Drosophila appendages. Dev Cell. 2013;27:306–318. doi: 10.1016/j.devcel.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang F, Dambly-Chaudière C, Ghysen A. The emergence of sense organs in the wing of Drosophila. Development. 1991;111:1087–1095. doi: 10.1242/dev.111.4.1087. [DOI] [PubMed] [Google Scholar]
- 40.Huppert SS, Jacobsen TL, Muskavitch MA. Feedback regulation is central to Delta-Notch signalling required for Drosophila wing vein morphogenesis. Development. 1997;124:3283–3291. doi: 10.1242/dev.124.17.3283. [DOI] [PubMed] [Google Scholar]
- 41.Schubiger M, Carré C, Antoniewski C, Truman JW. Ligand-dependent de-repression via EcR/USP acts as a gate to coordinate the differentiation of sensory neurons in the Drosophila wing. Development. 2005;132:5239–5248. doi: 10.1242/dev.02093. [DOI] [PubMed] [Google Scholar]
- 42.Woodard CT, Baehrecke EH, Thummel CS. A molecular mechanism for the stage specificity of the Drosophila prepupal genetic response to ecdysone. Cell. 1994;79:607–615. doi: 10.1016/0092-8674(94)90546-0. [DOI] [PubMed] [Google Scholar]
- 43.Agawa Y, et al. Drosophila Blimp-1 is a transient transcriptional repressor that controls timing of the ecdysone-induced developmental pathway. Mol Cell Biol. 2007;27:8739–8747. doi: 10.1128/MCB.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S, Wang J, Sun Y, Song Q, Li S. PKC-mediated USP phosphorylation at Ser35 modulates 20-hydroxyecdysone signaling in Drosophila. J Proteome Res. 2012;11:6187–6196. doi: 10.1021/pr3008804. [DOI] [PubMed] [Google Scholar]
- 45.Chan SKK, et al. Role of co-repressor genomic landscapes in shaping the Notch response. PLoS Genet. 2017;13:e1007096. doi: 10.1371/journal.pgen.1007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen L, Shi Y-B. Regulation of growth rate and developmental timing by Xenopus thyroid hormone receptor α. Dev Growth Differ. 2016;58:106–115. doi: 10.1111/dgd.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.