Significance
Cell-specific and region-specific gene expression can identify therapeutic targets in different neuroanatomic regions during neurodegenerative diseases. Multiple sclerosis (MS) is multifocal, and neuroprotective treatments are needed. Here, RNA sequencing of MS brain suggested study of the transcriptome of oligodendrocyte lineage cells (OLCs) in a location where remyelination occurs in an MS model. Cholesterol-synthesis pathways dominated as top upregulated pathways in OLCs of corpus callosum during remyelination in the cuprizone model. Estrogen receptor-β ligand treatment further increased cholesterol-synthesis pathways through direct effects on OLCs. As fetal OLCs are exposed in utero to high levels of estrogens in maternal blood, we hypothesize that remyelinating properties of estrogen treatment in adults during injury may recapitulate normal developmental myelination by targeting cholesterol homeostasis.
Keywords: multiple sclerosis, oligodendrocytes, cholesterol, remyelination, MS animal models
Abstract
Regional differences in neurons, astrocytes, oligodendrocytes, and microglia exist in the brain during health, and regional differences in the transcriptome may occur for each cell type during neurodegeneration. Multiple sclerosis (MS) is multifocal, and regional differences in the astrocyte transcriptome occur in experimental autoimmune encephalomyelitis (EAE), an MS model. MS and EAE are characterized by inflammation, demyelination, and axonal damage, with minimal remyelination. Here, RNA-sequencing analysis of MS tissues from six brain regions suggested a focus on oligodendrocyte lineage cells (OLCs) in corpus callosum. Olig1-RiboTag mice were used to determine the translatome of OLCs in vivo in corpus callosum during the remyelination phase of a chronic cuprizone model with axonal damage. Cholesterol-synthesis gene pathways dominated as the top up-regulated pathways in OLCs during remyelination. In EAE, remyelination was induced with estrogen receptor-β (ERβ) ligand treatment, and up-regulation of cholesterol-synthesis gene expression was again observed in OLCs. ERβ-ligand treatment in the cuprizone model further increased cholesterol synthesis gene expression and enhanced remyelination. Conditional KOs of ERβ in OLCs demonstrated that increased cholesterol-synthesis gene expression in OLCs was mediated by direct effects in both models. To address this direct effect, ChIP assays showed binding of ERβ to the putative estrogen-response element of a key cholesterol-synthesis gene (Fdps). As fetal OLCs are exposed in utero to high levels of estrogens in maternal blood, we discuss how remyelinating properties of estrogen treatment in adults during injury may recapitulate normal developmental myelination through targeting cholesterol homeostasis in OLCs.
Multiple sclerosis (MS) is an autoimmune disease of the CNS characterized by inflammation, demyelination, and axonal damage, with minimal remyelination (1). Current immunomodulatory treatments for MS are effective in reducing relapses, but do not repair disabilities. Neuroprotective treatments are needed that target CNS cells to improve disabilities, perhaps through increasing remyelination (2).
Insufficient remyelination in MS is related in part to the inability of oligodendrocyte precursor cells (OPCs) to differentiate into mature myelinating oligodendrocytes (3, 4). This is thought to result from a hostile CNS microenvironment that inhibits OPC differentiation, such as proinflammatory cytokines and chemokines (5, 6), leucine-rich repeat and immunoglobin-like domain-containing protein 1 (LINGO1) (7), and chondroitin sulfate proteoglycans (CSPGs) (8). Remyelination strategies will likely require modulation of extrinsic factors related to CNS inflammation, as well as targeting intrinsic factors related to oligodendrocyte maturation and myelination (2), and one without the other may not suffice (9). Further, this approach must be effective in the adult CNS in a setting of axonal damage if it is to be efficacious in patients with MS.
Molecular mechanisms intrinsic to oligodendrocytes during remyelination in vivo using a model of chronic demyelination with axonal damage may reveal therapeutic targets to facilitate remyelination in MS. The study of oligodendrocytes through single-cell and genetically engineered enrichment strategies followed by high-throughput sequencing and bioinformatics analyses has yielded valuable insights into how OPCs are regulated during developmental myelination (10, 11), as well as when OPCs from adults are activated by demyelination (10). What remains unknown is the oligodendrocyte transcriptome during remyelination in vivo in white matter of adults in the setting of axonal damage. This would provide direct insights relevant to the identification of therapeutic targets to facilitate remyelination in MS.
Here, we used RiboTag technology to determine oligodendrocyte lineage cell (OLC)-specific gene expression in vivo in corpus callosum during remyelination in a chronic cuprizone model characterized by significant axonal damage. RiboTag technology entails Cre-LoxP recombination to generate transgenic mice expressing HA-tagged ribosomes in specific cell types (12–14). Olig1-RiboTag mice permit the isolation of OLC-specific transcripts from targeted regions within brains of adult mice. When used in MS models, subsequent RNA sequencing and analyses can reveal intrinsic mechanisms in OLCs during remyelination in vivo in the adult CNS during injury. This RiboTag approach was used here to discover gene-expression pathways intrinsic to oligodendrocytes during remyelination in the chronic cuprizone and experimental autoimmune encephalomyelitis (EAE) models.
Results
RNA Sequencing in MS Brain Regions.
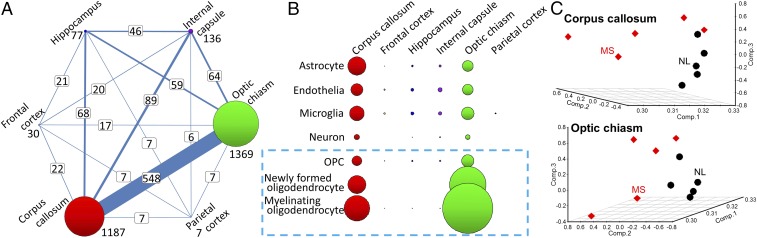
By using a high-throughput sequencing approach, we examined the impact of MS on gene expression in six different CNS regions (internal capsule, corpus callosum, optic chiasm, hippocampus, frontal cortex, and parietal cortex) of MS patients and age- and sex-matched healthy controls to identify brain regions with significant disease-related changes in the transcriptome. Corpus callosum (1,187 differentially expressed genes) and optic chiasm (1,369 differentially expressed genes) were the most significantly affected CNS regions in MS, and approximately half of these genes (548 genes) were commonly dysregulated when comparing the two regions with each other (Fig. 1A). We next classified the genes from the six CNS regions into seven different CNS cell types (astrocyte, endothelia, microglia, neuron, OPC, newly formed oligodendrocyte, and myelinating oligodendrocyte) by using the top 500 CNS cell markers from the RNA-sequencing transcriptome database (11). Although our human RNA-sequencing data contain heterogeneous CNS cell types, this approach can assess gene-expression changes in tissues by using specifically enriched markers for each cell type (15). Corpus callosum and optic chiasm both showed myelinating oligodendrocytes as the cell type most enriched with differentially expressed genes in MS (Fig. 1B). Gene-expression changes in astrocytes, endothelia, and microglia showed intermediate changes in both tissues, whereas gene-expression changes in neurons were low. The differentially expressed gene list is shown in SI Appendix, Table S1, and the cell marker gene list is shown in SI Appendix, Table S2. Principal component analysis and hierarchical clustering demonstrated regional differences in degree of separation between MS and normal, with corpus callosum and optic chiasm showing the most separation between MS and normal compared with the other four brain regions (Fig. 1C and SI Appendix, Fig. S1). Based on these results in MS, we focused hereafter on cell-specific and region-specific transcriptomics of OLCs in corpus callosum of an MS model.
Fig. 1.
Gene-expression changes in MS. High-throughput RNA sequencing was performed for six different brain regions from patients with MS (n = 5) and healthy controls (n = 5). (A) Transcriptome changes in the CNS of MS are region-specific. The numbers of differentially expressed genes in MS vs. controls are displayed as the sizes of circles, with differentially expressed gene numbers listed next to them. The numbers of shared differentially expressed genes between two different CNS tissues are shown in white boxes. The thickness of lines connecting two tissues reflects the level of sharing of differentially expressed genes. The transcriptome changes by disease were striking in corpus callosum and optic chiasm, and nearly half of differentially expressed genes were shared between these two tissues. (B) By using the top 500 CNS cell markers for seven different cell types (astrocyte, endothelia, microglia, neuron, OPC, newly formed oligodendrocyte, and myelinating oligodendrocyte) from the RNA-sequencing transcriptome database (11), the genes enriched in each cell type were selected, and the differentially expressed gene numbers (false discovery rate < 0.1) are visualized as the sizes of circles. Oligodendrocyte markers in corpus callosum and optic chiasm showed significant transcriptome changes by disease. (C) Principal component analysis of MS and control (NL) samples from corpus callosum and optic chiasm. Separation of MS and normal samples was observed in corpus callosum (Top) and optic chiasm (Bottom). MS group, five women with progressive MS, mean age, 57.6 y; control, five women, mean age, 56.2 y. Based on these results in MS, Olig1-RiboTag mice were used to define the transcriptome in OLCs (light blue dashed box in B) in corpus callosum.
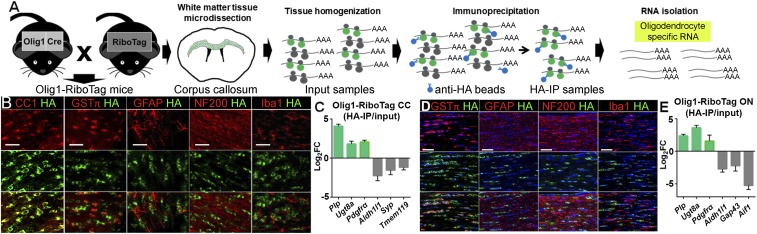
Validation of Oligodendrocyte Specificity of RNA from Olig1-RiboTag Mice.
To investigate cell-specific translatome changes in vivo, we used RiboTag technology (12–14). We chose Olig1-Cre mice to capture transcripts from myelinating oligodendrocytes, newly formed oligodendrocytes, and oligodendrocyte precursor cells (OPCs), as all had shown gene-expression changes in MS corpus callosum and optic chiasm (Fig. 1B, blue dotted square). We crossed Olig1-Cre mice with RiboTag mice (Olig1-RiboTag) to generate mice expressing HA-tagged ribosomal protein only in OLCs (Fig. 2A). Anti-HA antibody immunoprecipitation (IP) permitted isolation of OLC-specific ribosome-associated transcripts. When tissues from specific brain regions were used, this technology allowed isolation of actively translated mRNAs in a cell-specific and region-specific manner (12). To validate the specificity of HA labeling in Olig1-RiboTag mice, we assessed corpus callosum and optic nerve by using immunofluorescence staining and quantitative RT-PCR (qPCR). Double immunolabeling showed abundant colocalization of HA with the immature/mature oligodendrocyte marker CC1, as well as some colocalization of HA with GSTπ. Lack of colocalization was observed with other cell-specific markers, namely GFAP for astrocytes, NF200 for neurons, and Iba1 for microglia (Fig. 2 B and D). By qPCR, we confirmed enrichment of oligodendrocyte specific genes Plp, Ugt8a, and Pdgfrα in HA-IP RNA samples vs. input RNA samples, whereas astrocyte gene Aldhl1l, neuronal genes Syp and Gap43, and microglia genes Tmem119 and Aif1 were deenriched (Fig. 2 C and E). These results collectively validated RiboTag technology as a valuable tool to isolate OLC-specific RNAs from corpus callosum and optic nerve of Olig1-RiboTag mice.
Fig. 2.
Isolation and validation of oligodendrocyte-specific mRNAs from Olig1-RiboTag mice. (A) Overview of experimental strategy to isolate oligodendrocyte-specific RNAs from corpus callosum of Olig1-RiboTag mice. (B) Representative images from corpus callosum of Olig1-RiboTag mice validating the specificity of HA expression in oligodendrocytes. Immunofluorescence staining for HA showed abundant colocalization with the immature/mature oligodendrocyte marker CC1, as well as some colocalization with the mature marker GSTπ. No colocalization was shown with the astrocyte marker GFAP, neuronal marker NF200, and microglial marker Iba1. (Scale bar, 20 μm.) (C) Comparing HA-IP RNA samples vs. input RNA samples from corpus callosum (CC) of Olig1-RiboTag mice, qPCR showed enrichment of oligodendrocyte genes (Plp, Ugt8a, and Pdgfrα) and deenrichment of astrocyte (Aldh1l1), neuronal (Syp), and microglial (Tmem119) genes. (D) Representative images from optic nerve of Olig1-RiboTag mice validating the specificity of HA expression in oligodendrocytes in optic nerve. (Scale bar, 20 μm.) (E) Comparing HA-IP RNA samples vs. input RNA samples from optic nerve of Olig1-RiboTag mice, qPCR showed enrichment of oligodendrocyte genes (Plp, Ugt8a, and Pdgfrα) and deenrichment of astrocyte (Aldh1l1), neuronal (Gap43), and microglial (Aif1) genes.
Remyelination After Chronic Demyelination in the Cuprizone Diet-Induced Injury Model.
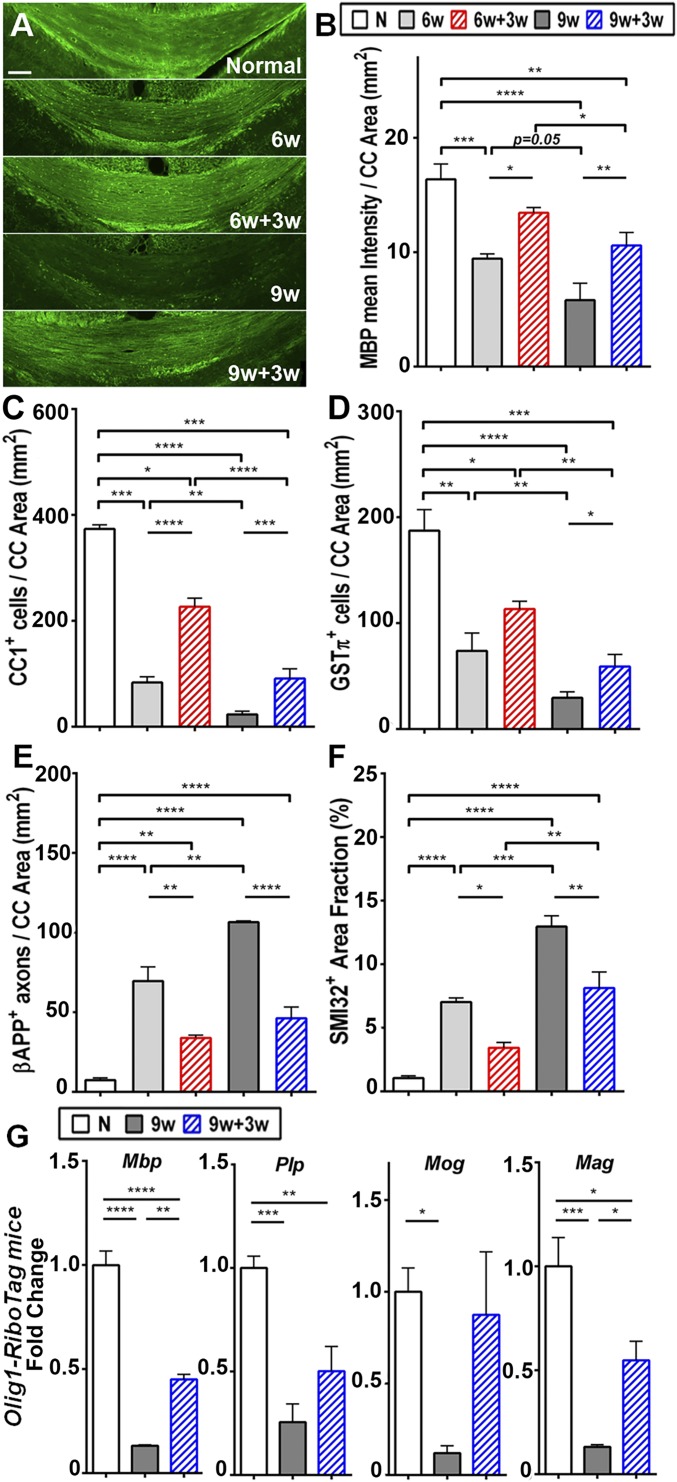
Here, we aimed to determine the transcriptome of OLCs in the corpus callosum during remyelination in the cuprizone model as a step toward identifying therapeutic targets to induce remyelination in MS. To this end, timing in the cuprizone model was first optimized to identify a time point with marked axonal damage, as axonal damage occurs in MS. We tested two different cuprizone exposure time points, 6 wk and 9 wk, each followed by a recovery period with normal diet for 3 wk (6w+3w and 9w+3w groups, respectively). Mice receiving normal diet for the duration of the entire experiment were used as normal controls. Immunofluorescence was performed on brain sections to assess myelin [myelin basic protein (MBP); Fig. 3 A and B], oligodendrocytes (CC1+ and GSTπ+ oligodendrocytes; Fig. 3 C and D), and axonal damage (βAPP and SMI32; Fig. 3 E and F) in the corpus callosum. Demyelination, oligodendrocyte loss, and axonal damage were more pronounced in the corpus callosum of mice receiving cuprizone diet for 9 wk than in those receiving it for 6 wk (Fig. 3 A–F and SI Appendix, Fig. S2). During the 3-wk normal diet recovery period, remyelination occurred to a lesser degree in 9w+3w mice compared with 6w+3w mice. Indeed, MBP intensity level was significantly lower in 9w+3w mice compared with normal controls, providing a critical window for potential treatment to enhance remyelination using this more chronic time course. In contrast, there was no window for significant further improvement with treatment in the shorter 6w+3w time course (Fig. 3B). Axonal damage after the 3-wk normal diet recovery period was also more pronounced in the 9w+3w mice compared with normal controls (Fig. 3 E and F, “N”, and SI Appendix, Fig. S2). Together, these results showed that 9 wk of cuprizone diet feeding caused demyelination and axonal damage that was not fully restored with 3 wk of normal diet. Thus, the 9w+3w time course was used hereafter to determine the transcriptome of OLCs and investigate therapeutic strategies to enhance remyelination in the setting of axonal degeneration.
Fig. 3.
Time course of myelin and axonal changes in the cuprizone model. (A) Representative images of corpus callosum (CC) sections stained for MBP in normal controls (normal), mice fed with cuprizone diet for 6 wk (6w group), mice fed with cuprizone for 6 wk and then normal diet for 3 wk (6w+3w group), mice fed with cuprizone diet for 9 wk (9w group), and mice fed with cuprizone for 9 wk then normal diet for 3 wk (9w+3w group). (Scale bar, 100 μm.) Quantitative analyses of MBP intensity (B), oligodendrocyte populations that were immature/mature CC1+ (C) and mature GSTπ+ (D), and axonal damage shown by βAPP+ axons (E) and SMI32+ area (F) in the corpus callosum. The chronic time course of demyelination (9 wk) showed worse myelin loss and axonal damage compared with the shorter time course (6 wk), and there was less robust recovery in the chronic course (9 wk + 3 wk) compared with the shorter time course (6 wk + 3 wk). (G) Oligodendrocyte-specific expression levels of myelin genes, namely Mbp, Plp, Mog, and Mag, from Olig1-RiboTag mice. During the chronic time course, myelin gene expression was significantly reduced in mice fed with cuprizone diet for 9 wk (9w group) compared with normal controls (marked as “N”). Myelin gene expression was increased in mice fed with cuprizone for 9 wk followed by normal diet for 3 wk (9w+3w group) compared with mice fed with cuprizone diet for 9 wk (9w group), but remained below levels of normal controls (“N”; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
We next assessed known OLC-specific gene expression in Olig1-RiboTag mice by using the chronic cuprizone time course. We collected OLC-specific transcripts from normal, 9w, and 9w+3w mice and performed qPCR to measure the expression level of myelin-specific genes. Expression levels of myelin genes Mbp, Mag, Plp, and Mog were significantly reduced in 9w mice compared with normal controls, whereas a significant increase in the expression levels of these genes was observed in 9w+3w mice compared with 9w mice (Fig. 3G). These results confirmed our ability to detect changes in OLC-specific gene expression during the chronic time course of cuprizone diet-induced demyelination and remyelination in Olig1-RiboTag mice.
Up-Regulation of Cholesterol Synthesis Pathways in OLCs During Remyelination After Chronic Demyelination.
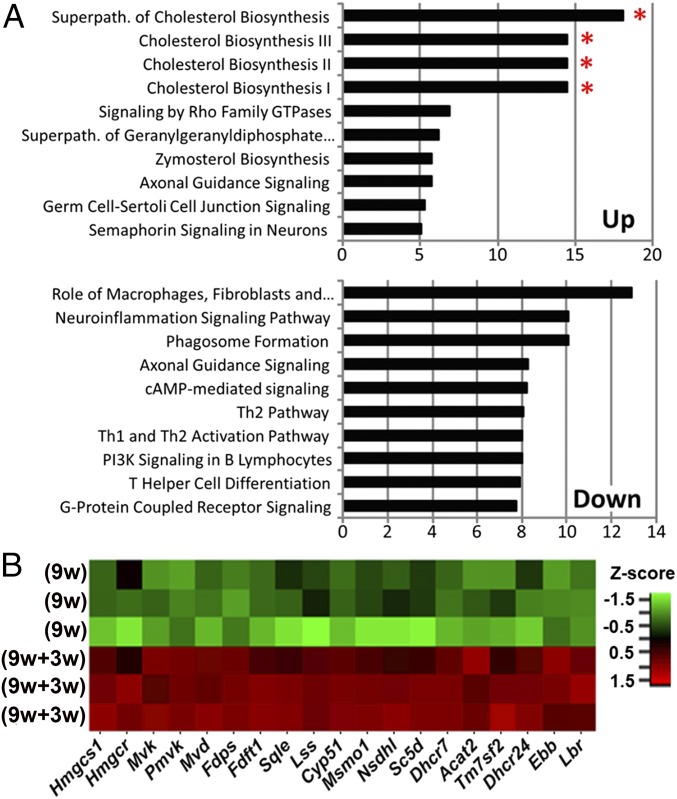
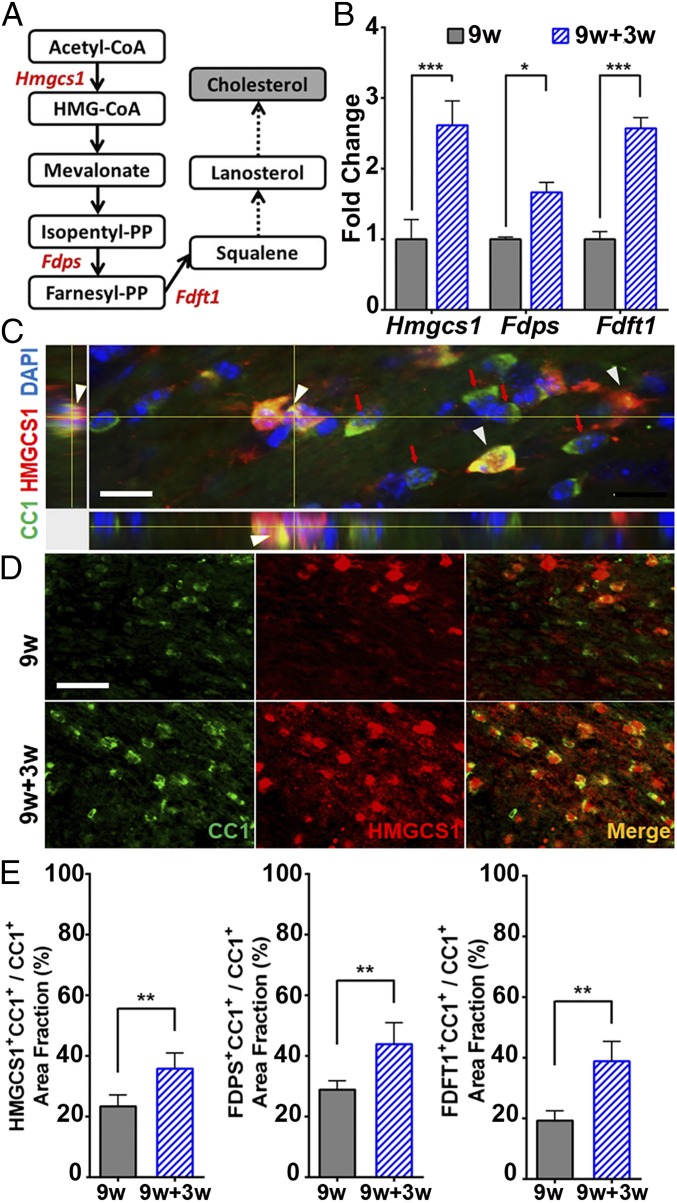
To build from a previous paper describing the OPC transcriptome activated by demyelination in adults (10), here we focused on determining the oligodendrocyte transcriptome during remyelination in adults. To this end, we used Olig1-RiboTag mice and performed high-throughput sequencing of OLC-specific ribosome-associated mRNAs. We collected corpus callosum tissues from mice receiving 9 wk of cuprizone diet followed by 3 wk of normal diet (i.e., 9w+3w group) for comparison with mice receiving 9 wk of cuprizone diet (i.e., 9w group). OLC-specific mRNAs isolated from corpus callosum tissue homogenates were used for RNA sequencing. Principal component analysis and hierarchical clustering was examined during remyelination by comparing 9w mice vs. 9w+3w mice. Clear separation of these two groups was observed (SI Appendix, Fig. S3). We next performed canonical pathway analysis of differentially expressed OLC-specific (Olig1+) genes during remyelination (Fig. 4; a full list of differentially expressed genes is provided in SI Appendix, Table S3). During remyelination, cholesterol synthesis pathways (superpathway of cholesterol biosynthesis, cholesterol biosynthesis I, II, and III) dominated as the top four up-regulated pathways (Fig. 4A, Top). The potentially deleterious neuroinflammation signaling pathway was one of the most down-regulated pathways during remyelination (Fig. 4A, Bottom). A heat map of individual genes showed that all 19 cholesterol synthesis genes were up-regulated (Fig. 4B, red) during remyelination in 9w+3w mice.
Fig. 4.
Oligodendrocyte-specific translatome changes during remyelination after chronic demyelination. Olig1-RiboTag mice were used to compare oligodendrocyte-specific gene expression in corpus callosum during the remyelination phase in mice fed with cuprizone for 9 wk followed by normal diet for 3 wk (9w+3w group) vs. the demyelination phase in mice fed with cuprizone diet for 9 wk (9w group). (A) Top 10 up-regulated and down-regulated canonical pathways from differentially expressed oligodendrocyte genes during remyelination (9w+3w group). Red asterisk indicates cholesterol synthesis pathways as the top four up-regulated pathways during remyelination. (B) Heat map shows up-regulation (red) of multiple cholesterol-synthesis pathway genes during remyelination.
To confirm the up-regulated expression of cholesterol-synthesis pathway genes during remyelination, we performed qPCR on OLC-specific RNAs isolated from corpus callosum of a different set of Olig1-RiboTag mice. As illustrated in Fig. 5A, we investigated three cholesterol-synthesis genes encoding for important enzymes in cholesterol synthesis, namely 3-hydroxy-3-methylglutaryl-CoA synthase1 (Hmgcs1), farnesyl diphosphate synthase (Fdps), and farnesyl-diphosphate farnesyltransferase 1 (Fdft1). We found that 9w+3w mice during the remyelination phase had significantly increased Hmgcs1, Fdps, and Fdft1 gene expression levels (Fig. 5B). We then showed increased expression of Hmgcs1, Fdps, and Fdft1 at the protein level by performing immunofluorescence on tissues obtained from WT mice. Double immunolabeling for HMGCS1, FDPS, or FDFT1 with the oligodendrocyte marker CC1 showed a significant increase of cholesterol synthesis protein expression in CC1+ oligodendrocytes in the corpus callosum of 9w+3w mice during the remyelination phase (Fig. 5 C–E and SI Appendix, Fig. S4). Together, these results showed that oligodendrocytes up-regulate cholesterol-synthesis gene pathways during remyelination after chronic demyelination with axonal damage.
Fig. 5.
Validation of increased cholesterol-synthesis gene expression in oligodendrocytes during remyelination. (A) Schematic illustration of cholesterol-synthesis pathway showing genes of interest (Hmgcs1, Fdps, and Fdft1; red). (B) qPCR using mRNAs from an independent set of Olig1-RiboTag mice showed that oligodendrocyte-specific Hmgcs1, Fdps, and Fdft1 expression levels were increased during remyelination in 9w+3w mice. (C) Immunofluorescence using tissues from WT mice showed that HMGCS1 (red) and CC1 (green) colocalized (yellow) in the corpus callosum during remyelination (9w+3w group). DAPI represents nuclear stain. Arrowheads indicate HMGCS1+CC1+ oligodendrocytes. Red arrows indicate HMGCS1−CC1+ oligodendrocytes. Orthogonal view. (Scale bar, 10 μm.) (D) Representative images of HMGCS1 (red) and CC1 (green) costained corpus callosum from 9w mice (Top) and 9w+3w mice (Bottom) with colocalization in merge (yellow). (Scale bar, 50 μm.) (E) Quantification of cholesterol-synthesis protein expression in CC1+ oligodendrocytes measured by area fraction as a percentage: HMGCS1 (Left), FDPS (Middle), and FDFT1 (Right). An increase in cholesterol-synthesis protein expression was shown in CC1+ oligodendrocytes during remyelination (9w+3w group; *P < 0.05, **P < 0.01, and ***P < 0.001).
Up-Regulation of Cholesterol Synthesis Pathways in OLCs Using a Second Model of Remyelination in the Setting of Axonal Damage.
Here, we extended our investigation in the cuprizone model to the EAE model. EAE is the most widely used model of MS, and is characterized by adaptive immune responses with infiltration of peripheral immune cells into the CNS, microglial and astrocyte activation, demyelination, and axonal damage. As in MS, insufficient remyelination occurs naturally in EAE (1, 9). Therefore, to study cholesterol homeostasis within oligodendrocytes during remyelination in EAE, we induced remyelination with ERβ-ligand treatment as described previously (9, 16–18).
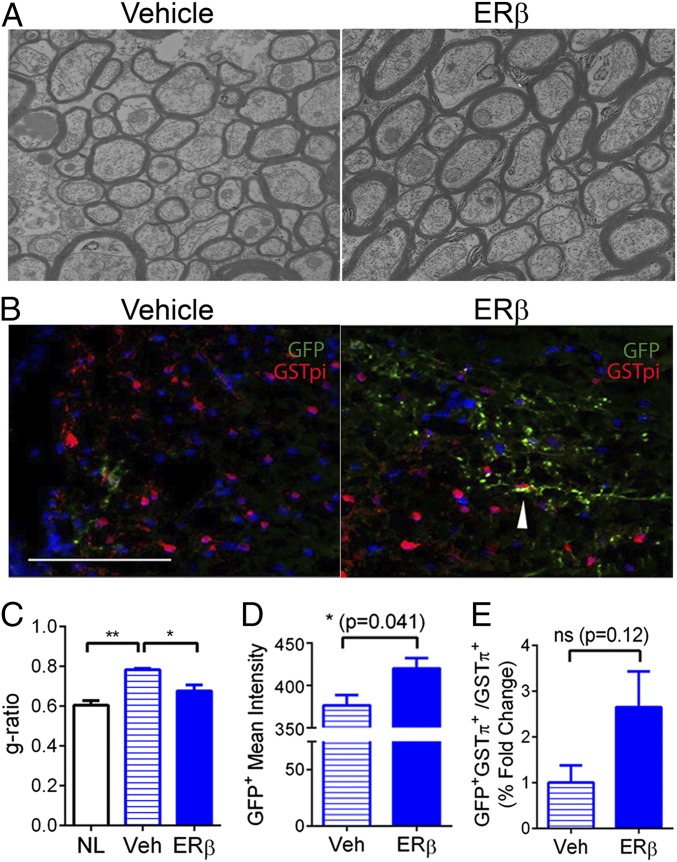
First, we assessed myelin sheath thickness in the spinal cord from EAE mice using EM to demonstrate the degree of remyelination induced by ERβ-ligand treatment (Fig. 6A). EAE mice treated with ERβ ligand showed an increase of myelin thickness and reduction of g-ratio (axon diameter divided by myelin plus axon diameter) compared with vehicle controls (Fig. 6 A and C), confirming the remyelinating effect of ERβ-ligand treatment during EAE (9, 16–18). To further distinguish between less demyelination vs. more remyelination during ERβ-ligand treatment in EAE, we used Cspg4-CreERT2/Mapt-GFP mice that enable visualization of newly formed myelin by GFP expression (2). With ERβ-ligand treatment of EAE, an increase of GFP-positive myelin was observed (Fig. 6 B, D, and E).
Fig. 6.
ERβ-ligand treatment induced remyelination during EAE. (A) Representative EM images of ultraresolution axons and myelin in the spinal cords of WT EAE mice treated with vehicle and WT EAE mice treated with ERβ ligand at EAE day 50. (B) Double labeling for GFP and GSTπ shows newly formed oligodendrocytes in EAE/vehicle and EAE/ERβ in Cspg4-CreERT2/Mapt-GFP transgenic mice at EAE day 35. (C) Quantitative analysis showed that vehicle-treated EAE mice (Veh) had increased g-ratio compared with normal controls (NL), indicative of demyelination, whereas ERβ ligand-treated EAE mice (ERβ) had decreased g-ratio compared with vehicle-treated mice (Veh), indicative of remyelination. (D) Quantification analysis in EAE Cspg4-CreERT2/Mapt-GFP transgenic mice showed a significant increase in GFP mean intensity after ERβ treatment. (E) Quantification of the percentage fold change of GFP+GSTπ+ cells showed an increase, but not a significant one, in the number of newly formed oligodendrocytes in the SC of ERβ-treated vs. vehicle-treated EAE Cspg4-CreERT2/Mapt-GFP transgenic mice (*P < 0.05 and **P < 0.01). ns, nonsignificant.
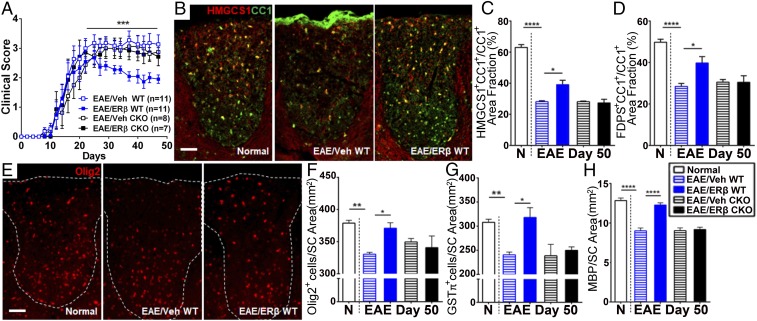
To investigate whether ERβ-ligand treatment was acting directly on OLCs to enhance remyelination in EAE, we crossed Olig1-cre mice (19) with ERβfl/fl mice (9, 17, 20) to obtain mice with conditional KO (CKO) of ERβ in Olig1+ cells (Olig1-cre+;ERβfl/fl, Olig1-CKO) (9) as well as WT littermates (Olig1-cre‒;ERβfl/fl, Olig1-WT) and treated them with ERβ ligand or vehicle. ERβ ligand-treated EAE WT mice (EAE/ERβ WT) showed protection from EAE clinical disease compared with vehicle-treated EAE WT mice (EAE/Veh WT). In contrast, ERβ ligand-treated EAE mice with a specific deletion of ERβ in Olig1+ cells (EAE/ERβ CKO) did not show clinical disease protection compared with vehicle-treated mice (EAE/Veh CKO; Fig. 7A). Further, ERβ-ligand treatment increased cholesterol-synthesis gene expression (HMGCS1 and FDPS) in CC1+ oligodendrocytes of EAE/ERβ WT mice compared with EAE/Veh WT mice (Fig. 7 B–D). In contrast, EAE/ERβ CKO mice did not show an increase in HMGCS1 and FDPS. ERβ-ligand treatment also increased the number of Olig2+ OLCs and GSTπ+ mature oligodendrocytes during EAE in EAE/ERβ WT mice compared with EAE/Veh WT mice, and these effects were also not present in EAE CKO mice (Fig. 7 E–G). MBP staining intensity in dorsal white matter of the spinal cord was higher in ERβ ligand-treated mice (EAE/ERβ WT) compared with vehicle-treated mice (EAE/Veh WT), whereas this treatment effect was not observed in mice with a specific deletion of ERβ in Olig1+ cells (EAE/ERβ CKO; Fig. 7H). Together, these results showed that ERβ-ligand treatment induced an increase in cholesterol-synthesis gene expression in OLCs during remyelination, and this was mediated by a direct effect of ERβ-ligand treatment on these cells.
Fig. 7.
ERβ-ligand treatment during EAE increases cholesterol-synthesis gene expression in oligodendrocytes. (A) EAE clinical disease scores showed that ERβ ligand-treated Olig1-WT mice (EAE/ERβ WT mice; blue solid squares) had less severe EAE scores compared with vehicle-treated (EAE/Veh WT mice; blue open squares), whereas there was no effect of treatment on clinical scores in mice with ERβ selectively deleted in OLCs (Olig1+) in the CKO mice [EAE/ERβ CKO (■) vs. EAE/Veh CKO mice (□)]. (B–D) EAE/ERβ WT mice showed increased expression of cholesterol-synthesis proteins compared with EAE/Veh WT mice, whereas this treatment effect was not present in EAE CKO mice (EAE/ERβ CKO vs. EAE/Veh CKO). (B) Representative images of HMGCS1 (red) and CC1 (green) costained (yellow) dorsal white matter of spinal cord from WT normal controls (normal), EAE/Veh WT, and EAE/ERβ WT mice. (Scale bar, 100 μm.) (C and D) Quantification of cholesterol-synthesis protein expression in CC1+ oligodendrocytes measured by area fraction (as a percentage) staining for HMGCS1 (C) and FDPS (D). (E) Representative images of Olig2-stained (red) dorsal white matter of spinal cord from normal, EAE/Veh WT, and EAE/ERβ WT mice. (Scale bar, 100 μm.) (F and G) Quantitative analysis showed that ERβ ligand-treated EAE/ERβ WT mice had significantly increased Olig2+ OLCs and GSTπ+ mature oligodendrocytes compared with vehicle-treated EAE/Veh WT mice, whereas ERβ-ligand treatment had no effect in CKO mice with EAE (EAE/ERβ CKO vs. EAE/Veh CKO). (H) Quantitative analysis of MBP intensity during EAE showed that ERβ-ligand treatment increased myelin in EAE/ERβ WT mice compared with EAE/Veh WT, whereas this treatment effect was not present in EAE CKO mice (EAE/ERβ CKO vs. EAE/Veh CKO; *P < 0.05, **P < 0.01, and ****P < 0.0001).
Insights from Cell-Specific Gene Expression Lead to Enhancement of Remyelination in the Chronic Cuprizone Model.
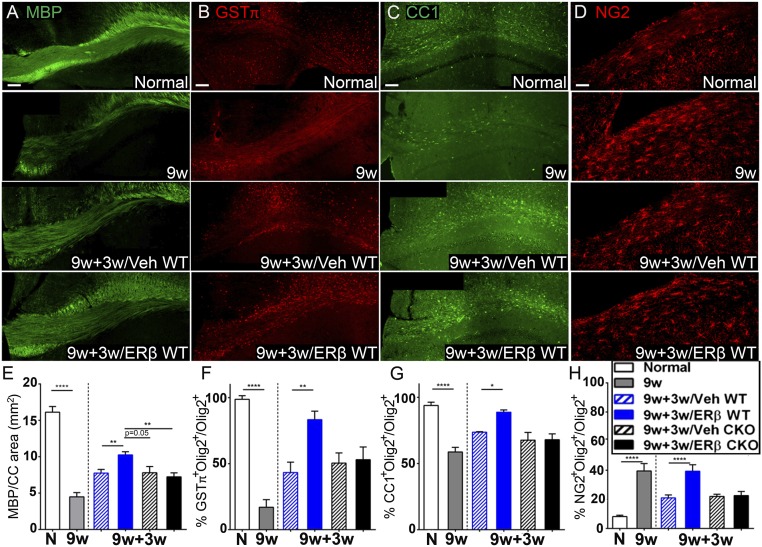
As increased cholesterol-synthesis gene expression occurred in OLCs during natural remyelination in the chronic cuprizone model, and because ERβ-ligand treatment increased cholesterol-synthesis gene expression in OLCs during remyelination in EAE, we next determined whether ERβ-ligand treatment administered during the normal-diet phase of the cuprizone model could further enhance remyelination in the chronic cuprizone model characterized by a window for potential improvement. We used C57BL/6 WT mice with demyelination (i.e., 9w mice), mice with normal diet-induced remyelination receiving vehicle treatment (9w+3w/Veh mice), mice with normal diet-induced remyelination receiving ERβ-ligand treatment (9w+3w/ERβ mice), and normal healthy control mice receiving normal diet for the entire experiment (normal mice). We assessed myelin integrity by measuring MBP intensity in the corpus callosum by immunofluorescence. ERβ-ligand treatment in 9w+3w/ERβ WT mice showed a further significant increase in MBP intensity compared with vehicle-treated 9w+3w/Veh WT mice (Fig. 8 A and E and SI Appendix, Table S4). These immunofluorescence results were confirmed by assessing myelin sheath thickness in the corpus callosum using EM (SI Appendix, Fig. S5A). Myelin thickness was quantified by using the g-ratio. The 9w+3w/Veh mice showed a reduced g-ratio compared with the 9w mice, indicative of increased myelin sheath thickness during normal diet-induced remyelination, and 9w+3w/ERβ mice exhibited a further significant decrease in g-ratio, indicative of a further increase in myelin sheath thickness, indeed close to that of normal controls (SI Appendix, Fig. S5 B and C). Together, these results showed that ERβ-ligand treatment enhanced remyelination in a chronic cuprizone model characterized by axonal damage. Double immunolabeling for GSTπ or CC1 with the pan-oligodendrocyte marker Olig2 showed that ERβ-ligand treatment compared with vehicle during the normal-diet phase increased mature GSTπ+ oligodendrocyte and immature/mature CC1+ populations in the corpus callosum (Fig. 8 B, C, F, and G and SI Appendix, Fig. S6). Next, we assessed OPC populations in the corpus callosum by double immunolabeling for the OPC marker NG2 and OLC marker Olig2. During remyelination, ERβ-ligand treatment increased the percentage of NG2+ OPCs in 9w+3w/ERβ WT mice compared with vehicle treatment in 9w+3w/Veh WT mice (Fig. 8 D and H and SI Appendix, Fig. S6).
Fig. 8.
ERβ-ligand treatment during the remyelination phase of the cuprizone model further enhances remyelination and increases mature oligodendrocytes and OPCs. (A–D) Representative images of MBP+ stained myelin (A), GSTπ+ mature oligodendrocytes (B), CC1+ immature/mature oligodendrocytes (C), and NG2+ OPCs (D) in the corpus callosum (CC) of WT mice. (Scale bars, 100 μm.) (E–H) Quantitative analysis of MBP mean intensity (E) and percentage of GSTπ+ mature oligodendrocytes (F), CC1+ immature/mature oligodendrocytes (G), and NG2+ OPCs (H) showed that ERβ ligand-treated 9w+3w/ERβ WT mice had increased myelin, mature oligodendrocytes, and OPCs compared with vehicle-treated 9w+3w/Veh WT mice, and ERβ-ligand treatment-mediated increases were not observed in Olig1 CKO mice (9w+3w/Veh CKO vs. 9w+3w/ERβ CKO; *P < 0.05, **P < 0.01, and ****P < 0.0001).
To investigate whether ERβ-ligand treatment was acting directly on oligodendrocytes to enhance remyelination in the chronic cuprizone model, we again used mice with a CKO of ERβ in Olig1+ cells (Olig1-cre+;ERβfl/fl, Olig1-CKO) as well as WT littermates (Olig1-cre−;ERβfl/fl, Olig1-WT). Whereas ERβ-ligand treatment in 9w+3w/ERβ WT mice showed a further significant increase in MBP intensity compared with vehicle-treated 9w+3w/Veh WT mice (Fig. 8 A and E and SI Appendix, Table S4), this beneficial effect of ERβ-ligand treatment was not present in mice with ERβ selectively deleted in Olig1+ cells, as 9w+3w/ERβ CKO mice were no different from 9w+3w/Veh CKO mice (Fig. 8E and SI Appendix, Table S4). When we extended the remyelination phase from 3 wk to 6 wk of normal diet, with or without ERβ-ligand treatment, each following 9 wk of cuprizone diet, myelin intensity in WT mice receiving ERβ-ligand treatment (9w+6w/ERβ WT mice) was again significantly increased compared with vehicle treatment (9w+6w/Veh WT mice), whereas this effect was not present in CKO mice (SI Appendix, Table S4).
We then investigated whether ERβ-ligand treatment has direct effects on mature oligodendrocytes. Whereas ERβ-ligand treatment significantly increased the percentages of GSTπ+ and CC1+ cells in 9w+3w/ERβ WT mice compared with 9w+3w/Veh WT mice, this protective effect was not present in 9w+3w/ERβ CKO mice compared with 9w+3w/Veh CKO mice (Fig. 8 B, C, F, and G). Next, we determined whether ERβ-ligand treatment has direct effects on OPCs. Whereas ERβ-ligand treatment increased the percentage of NG2+ OPCs in 9w+3w/ERβ WT mice compared with vehicle treatment in 9w+3w/Veh WT mice, this effect was not present in the 9w+3w CKO mice (Fig. 8 D and H). Thus, ERβ-ligand treatment had direct effects on OLCs during remyelination in the chronic cuprizone model.
Finally, we determined whether ERβ-ligand treatment modulated microglia and astrocyte reactivity in the chronic cuprizone model by assessing activation of Iba1+ microglia and GFAP+ astrocytes by double immunolabeling with MHCII. We observed a significant increase in the percentage of MHCII-expressing microglia and astrocytes in 9w mice compared with normal controls and a significant reduction during remyelination in 9w+3w mice, but there was no effect of ERβ-ligand vs. vehicle treatment (SI Appendix, Fig. S7).
Enhanced Remyelination in the Chronic Cuprizone Model Is Characterized by Further Up-Regulation of Cholesterol Synthesis Pathways in OLCs with ERβ-Ligand Treatment.
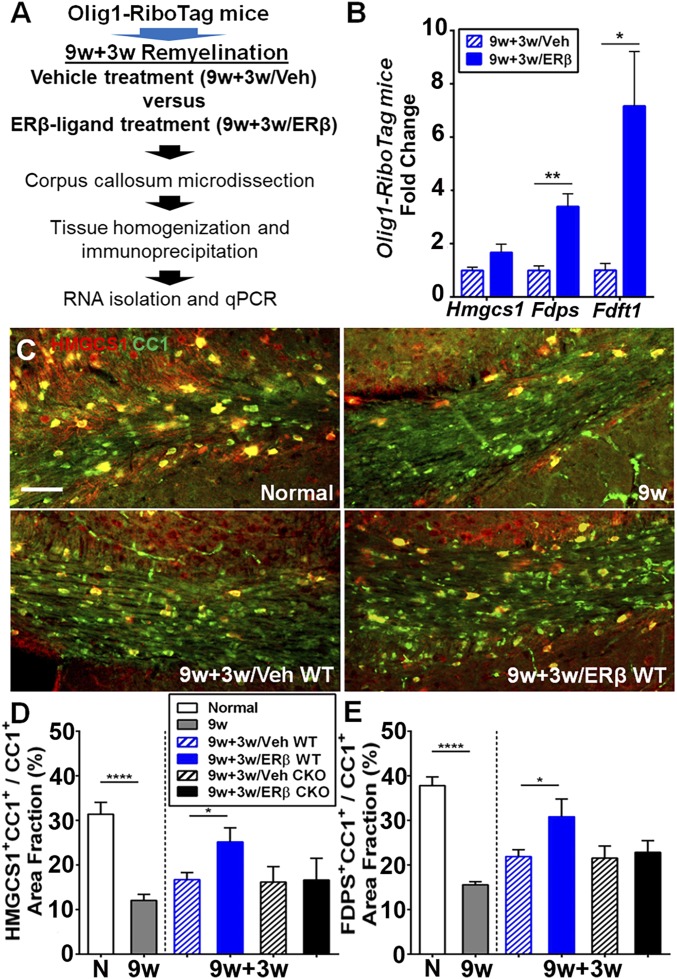
As we had found an increase in cholesterol-synthesis pathways during natural remyelination with normal diet (Figs. 4 and 5), and because ERβ-ligand treatment during normal diet further enhanced remyelination (Fig. 8), we next determined whether ERβ-ligand treatment might further increase cholesterol-synthesis gene expression in oligodendrocytes during remyelination by using Olig1-RiboTag mice. Briefly, we isolated OLC-specific RNAs from corpus callosum of ERβ ligand-treated (9w+3w/ERβ) and vehicle-treated (9w+3w/Veh) Olig1-RiboTag mice during the normal-diet remyelination phase, and then determined the expression level of cholesterol-synthesis genes by qPCR (Fig. 9A). We found that ERβ-ligand treatment increased cholesterol-synthesis gene expression in oligodendrocytes from 9w+3w/ERβ compared with 9w+3w/Veh mice (Fig. 9B). Gene expression was confirmed at the protein level by immunofluorescence. Double immunolabeling for HMGCS1 or FDPS with CC1 revealed a significant increase in cholesterol-synthesis protein expression in CC1+ oligodendrocytes in the corpus callosum of ERβ ligand-treated mice (9w+3w/ERβ WT) compared with vehicle-treated mice (9w+3w/Veh WT; Fig. 9C). In contrast, we did not observe this effect in 9w+3w/ERβ CKO vs. 9w+3w/Veh CKO mice (Fig. 9 D and E). Together, these findings demonstrated that ERβ-ligand treatment increases cholesterol-synthesis gene expression in oligodendrocytes during remyelination through direct effects on ERβ in Olig1+ oligodendrocytes.
Fig. 9.
ERβ-ligand treatment during the remyelination phase of the cuprizone model further increases cholesterol-synthesis gene expression in oligodendrocytes. (A) Overview of experimental strategy using Olig1-RiboTag mice to study oligodendrocyte-specific gene expression in corpus callosum tissues during remyelination in ERβ ligand-treated (9w+3w/ERβ) vs. vehicle-treated (9w+3w/Veh) Olig1-RiboTag mice. (B) qPCR using mRNAs from Olig1-RiboTag mice showed that expression levels of cholesterol-synthesis pathway genes Hmgcs1, Fdps, and Fdft1 were increased in OLCs in ERβ ligand-treated (9w+3w/ERβ) compared with vehicle-treated (9w+3w/Veh) Olig1-RiboTag mice during remyelination. (C) Representative images of HMGCS1 (red) and CC1 (green) costained (yellow) corpus callosum from WT normal controls (normal), 9w, 9w+3w/Veh WT, and 9w+3w/ERβ WT mice. (Scale bar, 100 μm.) (D and E) Quantification of cholesterol-synthesis protein expression in CC1+ oligodendrocytes measured by area fraction (as a percentage) staining for HMGCS1 (D) and FDPS (E). The 9w+3w/ERβ WT mice showed increased expression of cholesterol-synthesis proteins compared with 9w+3w/Veh WT mice, whereas ERβ-ligand treatment had no effect in Olig1-CKO mice (9w+3w/Veh CKO vs. 9w+3w/ERβ CKO; *P < 0.05, **P < 0.01, and ****P < 0.0001).
ERβ Binding Profiles for Cholesterol Genes.
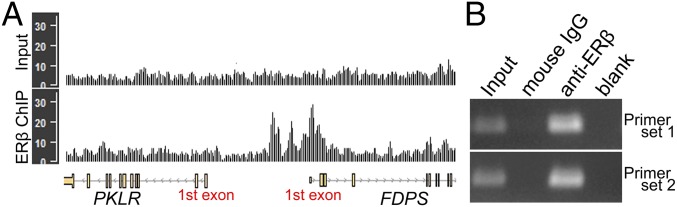
An increase in cholesterol-synthesis pathways in OLCs was shown to be mediated by a direct effect of ERβ-ligand treatment on OLCs using cell-specific KOs. How this direct effect might occur was addressed next. When estradiol or ERβ ligand binds to ERβ, it activates expression of a set of genes by interacting with the specific DNA sequences enriched around the transcription start sites. Thus, we examined the binding capacity of ERβ to the transcription start sites of target cholesterol-synthesis genes to determine whether the up-regulation of those genes with ERβ-ligand treatment could be directly mediated through ERβ. By using ChIP-sequencing (ChIP-seq) data for doxycycline-inducible ERβ-expressing human MDA-MB-231 cells treated with estradiol [Gene Expression Omnibus (GEO) database accession no. GSE108981], we examined the ERβ-binding sites across the genome. SI Appendix, Fig. S8 shows visualizations of ERβ-binding profiles for FDPS (SI Appendix, Fig. S8A), HMGCS1 (SI Appendix, Fig. S8B), FDFT1 (SI Appendix, Fig. S8C), and HMGCR (SI Appendix, Fig. S8D). Peaks of counts were located around the transcription start sites of all four cholesterol-synthesis genes in the ChIP profile, indicating that ERβ can directly bind to the transcriptional regulatory region of these cholesterol-synthesis genes. Further, ChIP assay experiments showed that, when the N20.1 oligodendrocyte cell line (21) was treated with ERβ ligand [diarylpropionitrile (DPN)], there was binding of ERβ to the putative estrogen response element (ERE) of the mouse Fdps cholesterol-synthesis gene (Fig. 10).
Fig. 10.
ERβ-binding profiles for human FDPS and ChIP assay for mouse Fdps gene. (A) ChIP-seq data for doxycycline-inducible ERβ-expressing human MDA-MB-231 cells treated with estradiol was obtained from the GEO database (accession no. GSE108981). Peaks in ERβ ChIP profiles indicate ERβ binding on the region. FDPS showed peaks around the first exons in ERβ ChIP profile, whereas the negative control input profile (DNAs from chromatin before ChIP) did not show the peak. As an additional negative control, the PKLR gene, near the FDPS gene, did not have a peak. (B) ERβ binds to the putative ERE of mouse Fdps gene. ChIP using normal mouse IgG and anti-ERβ was performed for the chromatin isolated from mouse N20.1 oligodendrocyte cell line (21) treated with ERβ ligand (DPN). Two sets of primers were designed across the region containing putative ERE of Fdps gene (51).
Discussion
Here we applied RiboTag technology to investigate molecular mechanisms within oligodendrocytes in vivo during remyelination in adult mice using two complementary models of chronic demyelination with axonal degeneration. We found that up-regulation of cholesterol-synthesis pathways dominated the transcriptome profile of OLCs during remyelination in adults after injury.
Cholesterol in the blood does not enter the healthy brain. Instead, de novo cholesterol synthesis occurs in CNS cells and is tightly regulated during development and adulthood (22–24). In the neonatal period, when synaptic plasticity and myelination are widespread, cholesterol is made by neurons to make membranes and synapses and by oligodendrocytes to make myelin. In the adult CNS, the majority of cholesterol production shifts to astrocytes. After cholesterol is made by astrocytes, it is transported via apolipoprotein E to neurons and oligodendrocytes. Here we show in a chronic cuprizone model that oligodendrocytes in adults up-regulate cholesterol-synthesis pathways during remyelination. In addition, ERβ-ligand treatment during the remyelination phase of the cuprizone model further increased cholesterol-synthesis pathways and enhanced remyelination compared with vehicle treatment. Furthermore, in the chronic EAE model, ERβ-ligand treatment increased cholesterol-synthesis pathway gene expression in oligodendrocytes and induced remyelination. As mice with a selective deletion of ERβ in OLCs did not show these effects of ERβ-ligand treatment in either model, the effects were mediated directly by ERβ signaling in OLCs. ChIP assays indicated that ERβ can directly bind to the transcriptional regulatory region of key cholesterol-synthesis genes. Interestingly, OLCs in the fetal CNS during developmental myelination are exposed in utero to the pregnant mother’s blood, which contains high levels of estriol, a naturally occurring ERβ ligand (25–27). Together, this suggests that targeting ERβ signaling to increase cholesterol-synthesis pathways in OLCs can increase remyelination in adults during injury by recapitulating a key aspect of developmental myelination.
Demyelination with insufficient remyelination occurs in MS patients and in vehicle-treated EAE mice. Cholesterol-synthesis gene expression was shown to be decreased in spinal-cord astrocytes previously (12) and in spinal-cord oligodendrocytes here (Fig. 7 B–D) in vehicle-treated EAE mice compared with normal mice. At the whole tissue level (i.e., not cell-specific) in human autopsy tissues, cholesterol-synthesis gene expression was decreased in MS patients compared with normal controls in optic chiasm (SI Appendix, Fig. S9A) (12), with more variability in corpus callosum (SI Appendix, Fig. S9B). This regional difference could result from more variability in corpus callosum subregions (anterior vs. posterior corpus callosum) compared with more uniformity in optic chiasm or from inherent differences in oligodendrocytes in optic chiasm vs. corpus callosum; each is consistent with oligodendrocyte heterogeneity (28–30). Also, MS autopsy tissues could differ in the level of demyelination and remyelination in corpus callosum vs. optic chiasm. Regardless of the level of decrease in cholesterol-synthesis gene expression in whole tissues in MS, this does not rule out an increase in cholesterol-synthesis gene expression focally in oligodendrocytes within remyelinating lesions. To show lesion-specific and cell cluster-specific differences in cholesterol-synthesis gene expression in MS tissues, single-nucleus RNA sequencing (29) would be required. A goal based on data here in MS preclinical models is to identify a treatment to increase cholesterol-synthesis gene expression in oligodendrocytes to increase the frequency of remyelinating lesions in MS.
This study provides evidence that a cell-specific gene-expression approach yields valuable insights into in vivo mechanisms relevant to the development of neuroprotective treatments in complex neurodegenerative diseases. Indeed, cell-specific translatome findings in MS models here focusing on oligodendrocytes are complementary to our previous report focusing on astrocytes (12). When the translatome of astrocytes was determined in GFAP-RiboTag mice with EAE, cholesterol-synthesis pathways dominated as the most down-regulated pathways in EAE spinal-cord and optic-nerve astrocytes compared with healthy mice. We hypothesized that, even though demyelination and synaptic loss are caused by immune attacks during EAE, the inability to repair this damage may result from decreased cholesterol synthesis in and transport by astrocytes. Treatment with an ATP-binding cassette transporter A1 (ABCA1) increased cholesterol-synthesis gene expression in astrocytes and improved EAE clinical scores compared with vehicle treatment (12) (SI Appendix, Fig. S10). At the neuropathology level, ABCA1 treatment increased healthy axons (increased NF200+ and decreased βAPP+ staining) and increased synapses (PSD95+ puncta), but ABCA1 treatment did not increase myelin (MBP+) staining. In contrast here, ERβ-ligand treatment up-regulated cholesterol-synthesis pathways in oligodendrocytes and increased remyelination. These studies reveal distinct and complementary effects of ABCA1 treatment and ERβ-ligand treatment. ABCA1 treatment increased cholesterol-synthesis pathways in astrocytes and increased synapses in EAE, whereas ERβ-ligand treatment increased cholesterol-synthesis pathways in oligodendrocytes and induced remyelination in EAE. Each treatment reduced axonal damage compared with vehicle treatment. Together, this reveals the importance of determining CNS cell type-specific mechanisms of neuroprotective treatments, as mechanism-based combinations of neuroprotective treatments may ultimately be better than monotherapy to repair neuropathology in complex neurodegenerative diseases such as MS.
Previous treatment with dietary cholesterol enhanced remyelination in demyelinated areas with a compromised blood–brain barrier (31). However, dietary cholesterol treatment may lead to adverse cardiovascular effects and comorbidities in patients with MS (32). Also, the cellular target and mechanism of action of dietary cholesterol remains unknown. Observational studies of cholesterol homeostasis in MS patients have included correlations of blood or CSF cholesterol levels with MS outcomes (33, 34), but interpretations are challenging given the lack of causality and cell specificity. Clinical trials of statins in MS remain unclear regarding mechanism and are controversial regarding outcomes. Statin-induced antiinflammatory effects (35) and cardiovascular benefits (32) would be desirable in MS, but inhibition of cholesterol synthesis in oligodendrocytes and astrocytes may be deleterious for remyelination and synaptic plasticity. Early clinical trials of statin treatment alone or in combination with IFN-β1a treatment in relapsing-remitting MS suggested a reduction in inflammatory lesions and relapses (36, 37), but other trials did not confirm these antiinflammatory effects (38–40). A beneficial effect of statin treatment on brain atrophy was also reported (41), but not confirmed (42). Statin treatment induced no effect on composite disability scores, but composites are insensitive measures of remyelination, so imaging biomarkers to quantify remyelination are needed. Whether statin treatment impairs remyelination in MS remains unknown. Only minimal remyelination occurs in MS patients treated with approved antiinflammatory disease-modifying treatments, so the power is insufficient to detect even less remyelination when a statin is used in combination. As statin use for cardiovascular indications is common in the general population, the possibility that statins may reduce cholesterol synthesis in oligodendrocytes and astrocytes with deleterious effects on remyelination and synaptic plasticity should be considered as a potential confounding variable in patients enrolled in future clinical trials of neuroprotective treatments for MS (43, 44).
High-throughput in vitro screens of libraries of small molecules can identify new candidates to test for induction of remyelination in vivo (2, 4, 45). Our findings indicate that their cell-specific and region-specific mechanism of action in vivo should be investigated in a comprehensive manner. For example, a recent high-throughput in vitro screen identified molecules targeting enzymes within the cholesterol synthesis pathway (46). Understanding which CNS cell types are targeted in which CNS regions in vivo during treatment with these compounds will be needed to understand the mechanism and align it with outcome measures in clinical trials if efficacy is ultimately to be demonstrated in MS (12). Ideally, high-throughput in vitro screening of compounds should be followed by in vivo cell-specific and region-specific gene expression for mechanism.
In conclusion, this work reveals the importance of using a cell-specific gene expression approach in the CNS during complex neurodegenerative diseases, here leading to insights into cholesterol homeostasis in oligodendrocytes during remyelination in the setting of axonal injury.
Materials and Methods
High-Throughput RNA-Sequencing Analysis of Tissues from MS.
Fresh-frozen autopsy brain tissue samples from five female patients with MS (mean age, 57.6 y) and five female age-matched healthy controls (mean age, 56.2 y) were obtained from Human Brain and Spinal Fluid Research Center (Los Angeles, CA). Regions included corpus callosum, optic chiasm, internal capsule, hippocampus, frontal cortex, and parietal cortex. Sequencing was performed on Illumina HiSeq3000 for a single-end 1 × 50 run. To estimate the differentially expressed gene numbers in various CNS cell types, lists of top 500 enriched genes in seven CNS cell types (neuron, microglia, astrocyte, endothelia, OPC, newly formed oligodendrocyte, and myelinating oligodendrocyte) from the RNA-sequencing transcriptome database (11) were used as a reference gene list (SI Appendix, Supplemental Materials and Methods).
Animals.
Mice were adult females (age 8–12 wk) on a C57BL/6 background. B6;129S4-Olig1tm1(cre)Rth/J (Olig1-Cre) mice and B6N.129-Rpl22tm1.1Psam/J (RiboTag) mice (14, 19) were crossed to obtain homozygote mice of Olig1-cre and RiboTag. To obtain Olig1-CKO mice, C57BL/6J-ERβfloxed/floxed mice (9) were crossed with B6;129S4-Olig1tm1(cre)Rth/J (Olig1-cre). To obtain Cspg4-CreERT2/Mapt-GFP mice, B6.Cg-Tg(Cspg4-cre/Esr1*)BAkik/J were crossed with B6;129P2-Mapt<tm2Arbr>/J mice (2). All procedures were done in accordance with the guidelines of the Chancellor’s Animal Research Committee of the University of California, Los Angeles, Office for the Protection of Research Subjects (SI Appendix, Supplemental Materials and Methods).
Cuprizone Model.
Female mice (age 8–10 wk) were on cuprizone diet for 6 wk or 9 wk to induce demyelination, followed by 3 wk of normal diet. Treatment with ERβ ligand (9) was during the normal-diet phase (SI Appendix, Supplemental Materials and Methods).
EAE Model.
Active EAE was induced with myelin oligodendrocyte glycoprotein amino acids 35–55 (12). ERβ-ligand treatment was initiated 1 wk before EAE induction and administered every other day (9). For Cspg4-CreERT2/Mapt-GFP transgenic mice, tamoxifen (Sigma-Aldrich) was dissolved in corn oil (75 mg/kg) and administered s.c. 2 wk before ERβ-ligand treatment for five consecutive days (SI Appendix, Supplemental Materials and Methods).
High-Throughput RNA Sequencing of Oligodendrocytes During Remyelination.
Corpus callosum oligodendrocyte-specific RNAs from normal mice or those receiving cuprizone diet for 9 wk (9w group) or for 9 wk followed by normal diet for 3 wk (9w+3w group) were isolated from the tissues of Olig1-RiboTag mice. Sequencing was performed on Illumina HiSeq3000 for a single-end 1 × 50 run. Canonical pathway enrichment analysis was performed for differentially expressed genes in each tissue by using Ingenuity Pathway Analysis (Qiagen) as described previously (12) (SI Appendix, Supplemental Materials and Methods).
qPCR.
Standard qPCR methods were applied as described previously (9). Lists of primers are provided in SI Appendix, Table S5 (SI Appendix, Supplemental Materials and Methods).
Histological Analysis.
Standard histological analysis methods were applied as described previously (9). Detailed methods are provided in SI Appendix, Supplemental Materials and Methods. A list of antibodies for immunofluorescence staining is provided in SI Appendix, Table S6.
ERβ ChIP-Seq Profiles for Cholesterol Synthesis Genes.
ChIP-seq data for doxycycline-inducible ERβ-expressing human MDA-MB-231 cells treated with estradiol were obtained from the GEO database (accession no. GSE108981) (47). R package “QuasR” (48) was used for the alignment to human genome (hg19), and ERβ-binding profiles were generated with input from R packages “GenomicFeatures” (49) and “Gviz” (50). The profiles for ERβ ChIP were each analyzed for cholesterol genes (SI Appendix, Supplemental Materials and Methods).
ChIP Assay to Assess ERβ Binding to ERE of Fdps.
Immortalized mouse N20.1 oligodendrocytic cell line (21) was treated with ERβ ligand. ChIP was performed by using EZ-Magna ChIP A/G (Millipore) with ERβ/NR3A2 antibody (Novus Biologicals). Immunoprecipitated DNAs around putative ERE of Fdps gene (51) were amplified by PCR (SI Appendix, Supplemental Materials and Methods).
Data Availability.
Datasets generated during this study are available in the GEO database (https://www.ncbi.nlm.nih.gov/geo/) under the following accession numbers: GSE123496, women with MS and age-matched healthy controls (hippocampus, frontal cortex, internal capsule, corpus callosum, and parietal cortex) (52); GSE100297, MS and control (optic chiasm) (53); and GSE118451, Oligo1 RiboTag data in cuprizone mice (54). Further details of methods and statistics are provided in SI Appendix, Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Roy Kim, Darian Mangu, Eunice Jung, Raul Alejandro-Ramirez, Kevin Herrera, and Alexandria S. Hoffman for technical assistance. This work was supported by NIH Grants R01NS096748 and R01NS109670 (to R.R.V.), Conrad N. Hilton Foundation Grants 20130231 and 20150232 (to R.R.V.), the Tom Sherak MS Hope Foundation, the Rhoda Goetz Foundation for MS, the Jewish Community Fund, and the Dunk MS Foundation. Human tissue specimens were obtained from the Human Brain and Spinal Fluid Resource Center, VA West Los Angeles Healthcare Center, which is sponsored by NIH, National Multiple Sclerosis Society, and the US Department of Veterans Affairs.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo [accession nos. GSE123496 (women with multiple sclerosis and age matched healthy controls: hippocampus, frontal cortex, internal capsule, corpus callosum, and parietal cortex), GSE100297 (MS and control: optic chiasm), and GSE118451 (Oligo1 RiboTag data in cuprizone mice)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821306116/-/DCSupplemental.
References
- 1.Fancy SP, et al. (2010) Overcoming remyelination failure in multiple sclerosis and other myelin disorders. Exp Neurol 225:18–23. [DOI] [PubMed] [Google Scholar]
- 2.Mei F, et al. (2016) Accelerated remyelination during inflammatory demyelination prevents axonal loss and improves functional recovery. eLife 5:e18246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang A, Tourtellotte WW, Rudick R, Trapp BD (2002) Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N Engl J Med 346:165–173. [DOI] [PubMed] [Google Scholar]
- 4.Mei F, et al. (2014) Micropillar arrays as a high-throughput screening platform for therapeutics in multiple sclerosis. Nat Med 20:954–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore CS, et al. (2015) Direct and indirect effects of immune and central nervous system-resident cells on human oligodendrocyte progenitor cell differentiation. J Immunol 194:761–772. [DOI] [PubMed] [Google Scholar]
- 6.Chew LJ, King WC, Kennedy A, Gallo V (2005) Interferon-gamma inhibits cell cycle exit in differentiating oligodendrocyte progenitor cells. Glia 52:127–143. [DOI] [PubMed] [Google Scholar]
- 7.Mi S, et al. (2005) LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci 8:745–751. [DOI] [PubMed] [Google Scholar]
- 8.Back SA, et al. (2005) Hyaluronan accumulates in demyelinated lesions and inhibits oligodendrocyte progenitor maturation. Nat Med 11:966–972. [DOI] [PubMed] [Google Scholar]
- 9.Kim RY, et al. (2018) Oestrogen receptor β ligand acts on CD11c+ cells to mediate protection in experimental autoimmune encephalomyelitis. Brain 141:132–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moyon S, et al. (2015) Demyelination causes adult CNS progenitors to revert to an immature state and express immune cues that support their migration. J Neurosci 35:4–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh N, et al. (2018) Cell-specific and region-specific transcriptomics in the multiple sclerosis model: Focus on astrocytes. Proc Natl Acad Sci USA 115:E302–E309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson MA, et al. (2016) Astrocyte scar formation aids central nervous system axon regeneration. Nature 532:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanz E, et al. (2009) Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci USA 106:13939–13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh Y, Voskuhl RR (2017) Cell specificity dictates similarities in gene expression in multiple sclerosis, Parkinson’s disease, and Alzheimer’s disease. PLoS One 12:e0181349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford DK, et al. (2010) Oestrogen receptor beta ligand: A novel treatment to enhance endogenous functional remyelination. Brain 133:2999–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khalaj AJ, et al. (2013) Estrogen receptor (ER) β expression in oligodendrocytes is required for attenuation of clinical disease by an ERβ ligand. Proc Natl Acad Sci USA 110:19125–19130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiwari-Woodruff S, Morales LB, Lee R, Voskuhl RR (2007) Differential neuroprotective and antiinflammatory effects of estrogen receptor (ER)alpha and ERbeta ligand treatment. Proc Natl Acad Sci USA 104:14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu QR, et al. (2002) Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109:75–86. [DOI] [PubMed] [Google Scholar]
- 20.Dupont S, et al. (2000) Effect of single and compound knockouts of estrogen receptors alpha (ERalpha) and beta (ERbeta) on mouse reproductive phenotypes. Development 127:4277–4291. [DOI] [PubMed] [Google Scholar]
- 21.Verity AN, Bredesen D, Vonderscher C, Handley VW, Campagnoni AT (1993) Expression of myelin protein genes and other myelin components in an oligodendrocytic cell line conditionally immortalized with a temperature-sensitive retrovirus. J Neurochem 60:577–587. [DOI] [PubMed] [Google Scholar]
- 22.Saher G, et al. (2005) High cholesterol level is essential for myelin membrane growth. Nat Neurosci 8:468–475. [DOI] [PubMed] [Google Scholar]
- 23.Mauch DH, et al. (2001) CNS synaptogenesis promoted by glia-derived cholesterol. Science 294:1354–1357. [DOI] [PubMed] [Google Scholar]
- 24.Saher G, Stumpf SK (2015) Cholesterol in myelin biogenesis and hypomyelinating disorders. Biochim Biophys Acta 1851:1083–1094. [DOI] [PubMed] [Google Scholar]
- 25.Kuiper GG, et al. (1997) Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology 138:863–870. [DOI] [PubMed] [Google Scholar]
- 26.Lindberg BS, Johansson ED, Nilsson BA (1974) Plasma levels of nonconjugated oestrone, oestradiol-17beta and oestriol during uncomplicated pregnancy. Acta Obstet Gynecol Scand Suppl 32:21–36. [DOI] [PubMed] [Google Scholar]
- 27.Voskuhl RR, et al. (2016) Estriol combined with glatiramer acetate for women with relapsing-remitting multiple sclerosis: A randomised, placebo-controlled, phase 2 trial. Lancet Neurol 15:35–46. [DOI] [PubMed] [Google Scholar]
- 28.Dutta R, Trapp BD (2012) Gene expression profiling in multiple sclerosis brain. Neurobiol Dis 45:108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jäkel S, et al. (2019) Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 566:543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viganò F, Möbius W, Götz M, Dimou L (2013) Transplantation reveals regional differences in oligodendrocyte differentiation in the adult brain. Nat Neurosci 16:1370–1372. [DOI] [PubMed] [Google Scholar]
- 31.Berghoff SA, et al. (2017) Dietary cholesterol promotes repair of demyelinated lesions in the adult brain. Nat Commun 8:14241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moccia M, et al. (2015) The Framingham cardiovascular risk score in multiple sclerosis. Eur J Neurol 22:1176–1183. [DOI] [PubMed] [Google Scholar]
- 33.Zhornitsky S, McKay KA, Metz LM, Teunissen CE, Rangachari M (2016) Cholesterol and markers of cholesterol turnover in multiple sclerosis: Relationship with disease outcomes. Mult Scler Relat Disord 5:53–65. [DOI] [PubMed] [Google Scholar]
- 34.van de Kraats C, et al. (2014) Oxysterols and cholesterol precursors correlate to magnetic resonance imaging measures of neurodegeneration in multiple sclerosis. Mult Scler 20:412–417. [DOI] [PubMed] [Google Scholar]
- 35.Waubant E, et al. ; ITN STAyCIS Study Group; ITN020AI Study Management Team (2012) Randomized controlled trial of atorvastatin in clinically isolated syndrome: The STAyCIS study. Neurology 78:1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Togha M, et al. (2010) Simvastatin treatment in patients with relapsing-remitting multiple sclerosis receiving interferon beta 1a: A double-blind randomized controlled trial. Mult Scler 16:848–854. [DOI] [PubMed] [Google Scholar]
- 37.Vollmer T, et al. (2004) Oral simvastatin treatment in relapsing-remitting multiple sclerosis. Lancet 363:1607–1608. [DOI] [PubMed] [Google Scholar]
- 38.Kamm CP, et al. (2012) Atorvastatin added to interferon β for relapsing multiple sclerosis: A randomized controlled trial. J Neurol 259:2401–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Birnbaum G, Cree B, Altafullah I, Zinser M, Reder AT (2008) Combining beta interferon and atorvastatin may increase disease activity in multiple sclerosis. Neurology 71:1390–1395. [DOI] [PubMed] [Google Scholar]
- 40.Sorensen PS, et al. ; SIMCOMBIN study investigators (2011) Simvastatin as add-on therapy to interferon β-1a for relapsing-remitting multiple sclerosis (SIMCOMBIN study): A placebo-controlled randomised phase 4 trial. Lancet Neurol 10:691–701. [DOI] [PubMed] [Google Scholar]
- 41.Chataway J, et al. (2014) Effect of high-dose simvastatin on brain atrophy and disability in secondary progressive multiple sclerosis (MS-STAT): A randomised, placebo-controlled, phase 2 trial. Lancet 383:2213–2221. [DOI] [PubMed] [Google Scholar]
- 42.Lanzillo R, et al. ; ARIANNA study group (2016) No evidence for an effect on brain atrophy rate of atorvastatin add-on to interferon β1b therapy in relapsing-remitting multiple sclerosis (the ARIANNA study). Mult Scler 22:1163–1173. [DOI] [PubMed] [Google Scholar]
- 43.Mailman T, Hariharan M, Karten B (2011) Inhibition of neuronal cholesterol biosynthesis with lovastatin leads to impaired synaptic vesicle release even in the presence of lipoproteins or geranylgeraniol. J Neurochem 119:1002–1015. [DOI] [PubMed] [Google Scholar]
- 44.Maier O, De Jonge J, Nomden A, Hoekstra D, Baron W (2009) Lovastatin induces the formation of abnormal myelin-like membrane sheets in primary oligodendrocytes. Glia 57:402–413. [DOI] [PubMed] [Google Scholar]
- 45.Lariosa-Willingham KD, et al. (2016) A high throughput drug screening assay to identify compounds that promote oligodendrocyte differentiation using acutely dissociated and purified oligodendrocyte precursor cells. BMC Res Notes 9:419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hubler Z, et al. (2018) Accumulation of 8,9-unsaturated sterols drives oligodendrocyte formation and remyelination. Nature 560:372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reese JM, et al. (2018) ERβ-mediated induction of cystatins results in suppression of TGFβ signaling and inhibition of triple-negative breast cancer metastasis. Proc Natl Acad Sci USA 115:E9580–E9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaidatzis D, Lerch A, Hahne F, Stadler MB (2015) QuasR: Quantification and annotation of short reads in R. Bioinformatics 31:1130–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrence M, et al. (2013) Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hahne F, Ivanek R (2016) Visualizing genomic data using Gviz and bioconductor. Methods Mol Biol 1418:335–351. [DOI] [PubMed] [Google Scholar]
- 51.Bourdeau V, et al. (2004) Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol 18:1411–1427. [DOI] [PubMed] [Google Scholar]
- 52.Itoh Y, Voskuhl RR(2019) Human brain tissues from healthy controls and multiple sclerosis patients. Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE123496. Deposited December 7, 2018.
- 53.Itoh N, et al. (2017) Human optic chiasm from healthy controls and multiple sclerosis patients. Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100297. Deposited June 21, 2017.
- 54.Kim RY, Tassoni A, Itoh Y, Itoh N, Voskuhl R (2019) Insights from the oligodendrocyte transcriptome during remyelination in adults. Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118451. Deposited August 13, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets generated during this study are available in the GEO database (https://www.ncbi.nlm.nih.gov/geo/) under the following accession numbers: GSE123496, women with MS and age-matched healthy controls (hippocampus, frontal cortex, internal capsule, corpus callosum, and parietal cortex) (52); GSE100297, MS and control (optic chiasm) (53); and GSE118451, Oligo1 RiboTag data in cuprizone mice (54). Further details of methods and statistics are provided in SI Appendix, Supplemental Materials and Methods.