Significance
Iron regulatory proteins (IRPs) control cellular iron homeostasis. Irp2 knockout mice show symptoms of neurological disorders, which are considered to result from impaired mitochondrial activity. To explore the involvement of Irp2 in mitochondrial function, we examined the metabolic pathways of Irp2-depleted mouse embryonic fibroblasts. We found that Irp2 deficiency switches cellular metabolic pathways from oxidative phosphorylation (OXPHOS) to aerobic glycolysis. We further revealed that Irp2 deficiency induces the expression of Hif1α and Hif2α; Hif1α enhances aerobic glycolysis by upregulating its target genes related to the glycolytic pathway, and Hif2α suppresses mitochondrial Fe–S biosynthesis and OXPHOS. This identified mechanism implies that high-energy-need tissues, such as the central nervous system, could be affected when Irp2 is deficient, leading to neurological disorders.
Keywords: iron regulatory protein 2, mitochondrial function, energy metabolism, hypoxia inducible factors
Abstract
The importance of the role of iron regulatory proteins (IRPs) in mitochondrial iron homeostasis and function has been raised. To understand how an IRP affects mitochondrial function, we used globally Irp2-depleted mouse embryonic fibroblasts (MEFs) and found that Irp2 ablation significantly induced the expression of both hypoxia-inducible factor subunits, Hif1α and Hif2α. The increase of Hif1α up-regulated its targeted genes, enhancing glycolysis, and the increase of Hif2α down-regulated the expression of iron–sulfur cluster (Fe–S) biogenesis-related and electron transport chain (ETC)-related genes, weakening mitochondrial respiration. Inhibition of Hif1α by genetic knockdown or a specific inhibitor prevented Hif1α-targeted gene expression, leading to decreased aerobic glycolysis. Inhibition of Hif2α by genetic knockdown or selective disruption of the heterodimerization of Hif2α and Hif1β restored the mitochondrial ETC and coupled oxidative phosphorylation (OXPHOS) by enhancing Fe–S biogenesis and increasing ETC-related gene expression. Our results indicate that Irp2 modulates the metabolic switch from aerobic glycolysis to OXPHOS that is mediated by Hif1α and Hif2α in MEFs.
Iron is essential for growth and proliferation of mammalian cells due to its important roles in protein cofactors, hemes and iron–sulfur clusters (Fe–S), which are involved in a number of biochemical pathways, including hemoglobin synthesis and the mitochondrial respiration chain. Cellular iron homeostasis is secured by two orthologous iron regulatory proteins (IRPs), IRP1 and IRP2, both of which are iron-regulated RNA-binding proteins that posttranscriptionally control the expression of a series of iron-related genes, such as ferritin, transferrin receptor 1 (TfR1), ferroportin 1 (FPN1), DMT1, and eALAS (1, 2). When cells are iron-deficient in the labile pool, IRPs bind iron-responsive elements (IREs) located in the 5′-UTR of ferritin and FPN1 mRNA to inhibit its translation, which reduces iron storage and export, and the IRE in the 3′-UTR of TfR1 and DMT1, which stabilizes the mRNA to facilitate iron import. When cells are iron-abundant, IRP1 is converted to a [4Fe-4S]-containing aconitase, and IRP2 is removed through iron-mediated proteasomal degradation (3, 4), which increases ferritin and FPN1 translation and promotes TfR1 and DMT1 mRNA degradation, preventing additional iron absorption and avoiding excess iron-induced injury.
Studies have shown that mice lacking Irp2 have abnormal iron contents in several tissues and develop microcytic anemia and erythropoietic protoporphyria (5, 6). Irp2 knockout mice also have symptoms of neurological disorders (7–9) due to the functional iron starvation in brain and spinal cord. This functional iron starvation is therefore considered to be causative of the impaired mitochondrial activity (10). However, the exact mechanism by which Irp2 sustains normal mitochondrial function is still unclear.
In our previous study, we found that the Irp1- or Irp2-null mutation in mouse embryonic fibroblasts (MEFs) caused decreased expression of frataxin (Fxn) and iron–sulfur cluster scaffold protein IscU, two important components of the Fe–S biogenesis machinery (11). Deficiency of Fxn or IscU in human and mouse cells limits mitochondrial function due to the lack of sufficient Fe–S clusters (12, 13). Furthermore, IRP depletion-induced deficiency of Fxn and IscU specifically adversely affects the activity of the Fe–S-dependent mitochondrial respiratory chain, while the activities of other Fe–S-dependent enzymes, such as aconitase and xanthine dehydrogenase, are enhanced (11). Strangely, ATP is more highly produced in Irp2−/− MEFs than in WT (the present study). This result seeming paradoxical to the low activity of the electron transport chain (ETC) and high content of ATP, suggesting a shift of the metabolic pathway in Irp2 ablation cells.
Oxidative phosphorylation (OXPHOS) and glycolysis are two key metabolic pathways for energy production. The switch from one pathway to another is controlled by a number of factors, including two important transcription factors, HIF1 and HIF2. HIFs are heterogeneous dimers that are mainly composed of an O2-labile alpha subunit (HIF1α or HIF2α) and a stable beta subunit (HIF1β, also known as ARNT). The direct connection between Irp and Hif demonstrates that Hif2α is posttranscriptionally regulated by Irp1 through binding the IRE in the 5′-UTR of Hif2α mRNA (14, 15). Irp1 ablation mice develop polycythemia, cardiac fibrosis, and pulmonary hypertension, which are attributed to a high level of Hif2α, which mediates the up-regulation of erythropoietin (16–18). Although Hif2α is up-regulated in Irp2-depleted cells (18), the physiological roles of both Hifs in Irp2 ablation mice remain unknown.
Here, we address the role of up-regulated Hif1α and Hif2α in Irp2−/− MEFs in regard to energy metabolism. Using MEFs in which Irp2 is globally depleted, we demonstrated that increased Hif1α enhanced glycolysis by targeting a number of glycolytic pathway-related genes, while increased Hif2α inhibited mitochondrial OXPHOS by decreasing, likely indirectly, the expression of Fxn and IscU and affecting the mitochondrial Fe–S cluster assembly and by decreasing the expression of ETC subunits and weakening OXPHOS. Therefore, Irp2 deficiency switches energy metabolism from OXPHOS to glycolysis.
Results
Irp2 Ablation-Induced Mitochondrial Dysfunction Is Associated with the Metabolic Switch from OXPHOS to Aerobic Glycolysis in MEFs.
IRPs have been demonstrated to be important for mitochondrial iron supply and function (19). Consistent with this finding, we and other groups revealed in vivo and in vitro that a deficit of available iron and reduction of mitochondrial Fe–S biogenesis can be key factors in mitochondrial dysfunction in general (20) and in Irp2 ablated cells (10, 11). Here, we confirmed that the activities of mitochondrial complexes I, II, and III significantly decreased in Irp2−/− MEFs compared with WT (Fig. 1A), which is consistent with the levels of the Fe–S-containing subunits Ndufs1 (of complex I), SdhB (of complex II), and Uqcrfs1 (of complex III) in Irp2−/− cells (Fig. 1B and SI Appendix, Fig. S1). In addition, the amounts of other mitochondrial proteins, such as cytochrome C (CytC, an intermembrane protein) and ferrochelatase (Fech, a matrix protein), also decreased in Irp2−/− cells (Fig. 1B and SI Appendix, Fig. S1). To further detect any broad effect of Irp2 deficiency on mitochondrial function, we measured the sensitivity of Irp2−/− cells to various inhibitors of mitochondrial complexes. The results showed that the viability of Irp2−/− cells significantly decreased compared with WT (Fig. 1C), suggesting that these cells are more sensitive to perturbations of mitochondrial function after Irp2 deprivation. The mitochondrial membrane potential (MMP), as measured using a specific and sensitive dye, JC-10, was much lower in Irp2−/− cells than in WT cells (Fig. 1D), suggesting that mitochondria in Irp2−/− cells are more depolarized.
Fig. 1.
Irp2 ablation-induced mitochondrial dysfunction is associated with enhanced aerobic glycolysis in MEFs. (A) Activities of ETC complexes in Irp2-deficient MEFs. CI, CII, and CIII, complexes I, II, and III. (B) Western blot analysis of mitochondrial proteins, including Ndufs1 (a subunit of CI), SdhB (a subunit of CII), Uqcrfs1 (a subunit of CIII), Fech (a matrix enzyme ferrochelatase), CytC (an intermembrane space protein cytochrome C), and Cs (a matrix non-Fe–S citrate synthase). A representative image set is presented. Actin was used as a loading control. (C) Sensitivity of Irp2−/− cells to ETC complex inhibitors, including rotenone (10 μM, inhibitor of complex I), oxaloacetic acid (OAA) (100 μM, inhibitor of complex II), and antimycin A (10 μM, inhibitor of complex III). (D) MMP detected using JC-10. Green fluorescence represents JC-10 monomers, and red fluorescence represents JC-10 aggregates. The ratio of red fluorescence to green fluorescence represents the level of the mitochondrial membrane potential. (E) Growth curves of WT and Irp2−/− cells. (F) Intracellular ATP content of WT and Irp2−/− cells in the growth phase (day 2 after subculture). (G) A representative set for proteins Hk2, Glut1, LdhA, LdhB, Pdh(-E1α), and p-Pdh(-E1α (pSer232)) revealed by Western blot analysis. (H) Levels of medium lactic acid in WT and Irp2−/− cells cultured in medium containing 4.5 g/L glucose (H, high) or 1.0 g/L glucose (L, low). (I) A representative set of Western blot analyses of glycolytic pathway-related proteins (Hk2, Glut1, LdhA, and LdhB) and oxidative phosphorylation pathway-related proteins (Pdh, Ndufs1, and Uqcrfs1). Actin was used as a loading control. Representative blots from n = 3 experiments are shown (each with duplicates). Values represent the mean ± SEM. One-way ANOVA (H) or Student’s t test (A, C, E, and F) was performed. *P < 0.05, **P < 0.01, ***P < 0.001, mutant vs. WT. ##P < 0.01, low vs. high concentration of glucose in medium.
Interestingly, although Irp2 deletion seriously weakened mitochondrial function, the growth of Irp2−/− cells was not significantly retarded, and the difference between WT and mutant was only pronounced on the fourth day due to insufficient nutrients (Fig. 1E). Surprisingly, the level of ATP significantly increased in Irp2−/− cells compared with WT in the growth phase (day 2) (Fig. 1F). We then speculated that aerobic glycolysis was enhanced to provide enough ATP for cell growth. Therefore, we detected the levels of several proteins involved in glycolysis, such as hexokinase 2 (HK2), glucose transporter 1 (Glut1), and lactate dehydrogenase A/B (LdhA/B). The expression of these proteins was significantly increased in Irp2−/− cells (Fig. 1G and SI Appendix, Fig. S1). We also detected pyruvate dehydrogenase (Pdh), a vital regulatory enzyme that catalyzes the conversion of pyruvate into acetyl-CoA and connects glycolysis to the TCA cycle. The protein level of Pdh was significantly reduced in Irp2−/− cells (Fig. 1G and SI Appendix, Fig. S1). However, phosphorylation of the E1α subunit of Pdh (p-Pdh–E1α (pSer232)), which leads to inactivation of the Pdh complex enzymatic activity, was enhanced (Fig. 1G and SI Appendix, Fig. S1), suggesting that glycolytic metabolism was favored over mitochondria-dependent metabolism. To further verify these results, we cultured cells in medium containing a high (4.5 g/L) or low (1.0 g/L) concentration of glucose for 3 d and measured the content of lactic acid, a by-product of the postglycolysis pathway. As shown in Fig. 1H, Irp2−/− cells always produced and secreted more lactic acid than WT cells under both glucose concentration conditions and in a concentration-dependent manner. The protein levels of Hk2, Glut1, and LdhA/B all consistently increased in Irp2−/− cells under a high glucose concentration (Fig. 1I). We then evaluated cellular OXPHOS by detecting the levels of related proteins. The expression of Pdh, Ndufs1, and Uqcrfs1 was not affected by different concentrations of glucose in WT or Irp2−/− cells, although these proteins all expressed less in Irp2−/− cells (Fig. 1I). These results strongly suggest that Irp2 deficiency promotes cellular aerobic glycolysis and suppresses OXPHOS.
To verify this hypothesis, the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR), as indicators of mitochondrial respiration and the glycolytic rate, respectively, were measured using an Agilent Seahorse Analyzer. As illustrated in Fig. 2 A and B, Irp2−/− cells had lower resting OCR or OXPHOS and a lower maximal mitochondrial capacity than WT cells after treatment with carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone (FCCP), suggesting that Irp2−/− cells are less oxidative, consistent with the lower mitochondria-derived ATP content. Simultaneously, Irp2−/− cells had a much higher ECAR after glucose and oligomycin treatment (Fig. 2 C and D), suggesting that they are more glycolytic. Collectively, these results demonstrated that Irp2 deficiency induced a metabolic switch from OXPHOS to aerobic glycolysis.
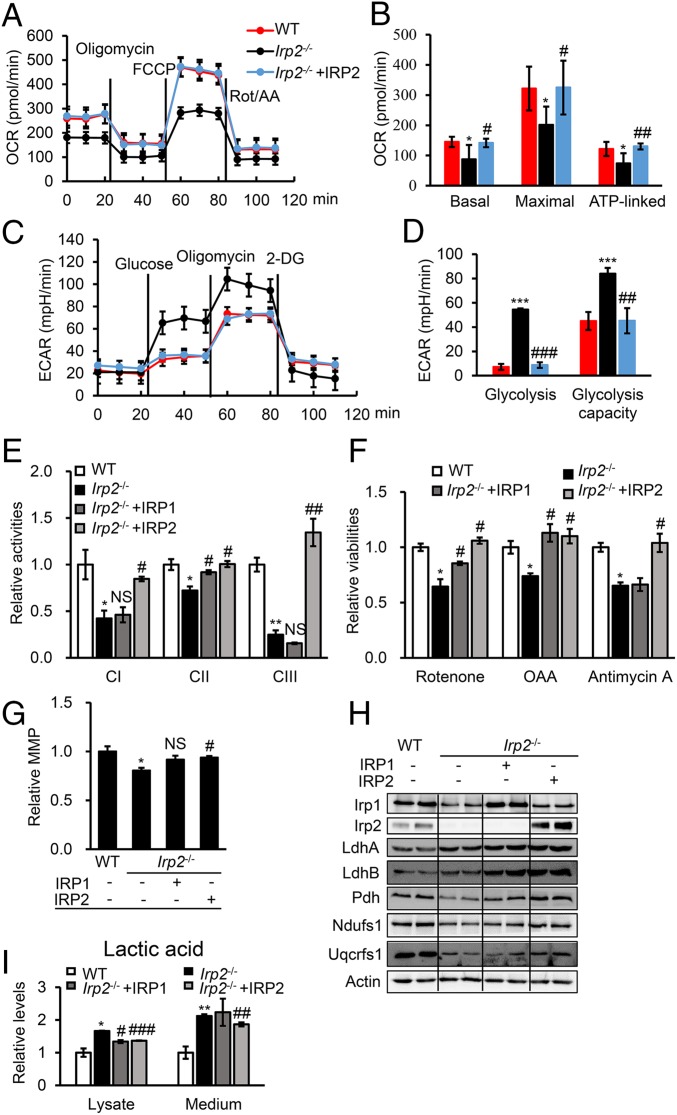
Fig. 2.
Human IRP2 rescues Irp2 ablation-induced mitochondrial dysfunction and reverses energy metabolism in MEFs. (A) Profiles of the OCR in WT and Irp2−/− cells. Oligomycin, 1 μM; FCCP, 1 μM; rotenone/antimycin (Rot/AA), 0.5 μM. (B) The calculated OCR for basal and maximal respiration and ATP production. (C) Profiles of the ECAR in WT and Irp2−/− cells.. Glucose, 10 mM; oligomycin, 1 μM; 2-deoxyglucose (2-DG), 50 mM. (D) The calculated ECAR for glycolysis and the glycolytic capacity. (E) Enzymatic activities of CI, CII, and CIII determined in Irp2-deficient MEFs after transfection with pcMV-HA-IRP1 or pDEST-his-IRP2. (F) Sensitivities to ETC complex inhibitors after IRP1 or IRP2 expression in Irp2−/− cells. The treatment with complex inhibitors was the same as in Fig. 1. (G) MMP of Irp2−/− cells after expression of IRP1 or IRP2. *P = 0.0443; #P = 0.0274; NS, P = 0.0819. (H) Protein levels of IRP1, IRP2, LdhA, LdhB, Pdh, Ndufs1, and Uqcrfs1 determined by Western blot analysis. (I) Levels of lactic acid in the medium or cell lysate of Irp2−/− cells after expression of IRP1 or IRP2. Actin was used as a loading control in Western blot analysis. Values represent the mean ± SEM (n = 3–5, each with duplicates). In B, D, E, F, G, and I, *P < 0.05, **P < 0.01, ***P < 0.001, mutant vs. WT; #P < 0.05, ##P < 0.01, ###P < 0.001, IRP rescue vs. nonrescue. NS, no significance, IRP1 rescue vs. nonrescue.
To further confirm that the metabolic switch was due to Irp2 deficiency, human IRP1 or IRP2, homologs of mouse Irp1 and Irp2, respectively, was expressed in Irp2-depleted MEFs to assess the recovery of iron and energy metabolism. Cellular iron metabolism was evaluated (SI Appendix, Fig. S2), and the results supported the conserved iron regulatory function of human IRP1 and IRP2. Evaluation of energy metabolism revealed that the expression of IRP2 increased the activities of respiration complexes I, II, and III, while the expression of IRP1 in Irp2−/− cells only increased the activities of complexes I and II (Fig. 2E). This result was in agreement with the sensitivity of Irp2−/− cells to inhibitors of mitochondrial ETC complexes (Fig. 2F). We also found that only IRP2, and not IRP1, significantly improved the MMP of Irp2−/− cells (Fig. 2G). Biochemical evidence revealed that all of the tested OXPHOS-related protein levels, such as Pdh, Ndufs1, and Uqcrfs1, significantly increased in Irp2−/− cells after IRP2 expression, but only the level of Pdh increased after IRP1 expression (Fig. 2H and SI Appendix, Fig. S3). Interestingly, the levels of the postglycolysis-related proteins LdhA and LdhB remained high after either IRP1 or IRP2 expression (Fig. 2H and SI Appendix, Fig. S3). However, the contents of lactic acid in both the cell lysate and medium of Irp2−/− cells were reduced by IRP2 expression, not by IRP1 expression in medium (Fig. 2I). Thus far, we have provided evidence that Irp2 can shift cellular respiration in favor of OXPHOS over aerobic glycolysis.
Irp2 Absence-Induced Up-Regulation of Hif2α in MEFs Affects Mitochondrial Biogenesis.
We further investigated the mechanism that drives the switch from OXPHOS to glycolysis in Irp2-deprived MEFs. HIF is a key transcriptional factor in regulating a number of genes involved in glycolytic respiration, including some of the genes tested in this study. Our previous study (11) and this work revealed that Irp2 deficiency resulted in a significant reduction of the iron content in MEFs (SI Appendix, Fig. S2), which might stabilize Hif1α and Hif2α in Irp2−/− cells. Indeed, the results shown in Fig. 3A and the SI Appendix, Fig. S4 A and B, confirmed our hypothesis. We then treated Irp2−/− cells with specific inhibitors of Hif1α (PX-478) and Hif2α (PT-2385). PX-478 transcriptionally and translationally inhibits Hif1α expression, and PT-2385 selectively disrupts the heterodimerization of Hif2α with Hif1β, although their mechanisms of action have yet to be fully elucidated (see review in ref. 21). The concentrations of the drugs were optimized (SI Appendix, Fig. S4C), and the effects of the inhibition were confirmed by the down-regulation of the Hif-targeted genes LdhA, endothelin 1 (Edn1), and Glut1 (Fig. 3B). The results suggested that LdhA and Glut1 were mainly targeted by Hif1 and Edn1 was mainly targeted by Hif2 in MEFs. Surprisingly, Fxn and IscU expression was significantly up-regulated by PT-2385 but not by PX-478 (SI Appendix, Fig. S4C). We further examined a number of genes involved in iron metabolism and OXPHOS. Inhibition of Hif1α in Irp2−/− cells did not change the protein levels of iron-related genes, such as Fxn and IscU, or mitochondrion-related genes, such as Pdh, Ndufs1, SdhB, and Uqcrfs1 (Fig. 3C and SI Appendix, Fig. S5). By contrast, suppression of Hif2α in Irp2−/− cells significantly increased the protein levels of Pdh, Ndufs1, SdhB, Uqcrfs1, Fxn, and IscU (Fig. 3D and SI Appendix, Fig. S6). The effects of the specific inhibitors PX-478 and PT-2385 were also verified by an unspecific inhibitor, 2-methoxyestradiol (SI Appendix, Fig. S7), suggesting that Irp2 absence-induced up-regulation of Hif2α in MEFs inhibits mitochondria-dependent metabolism.
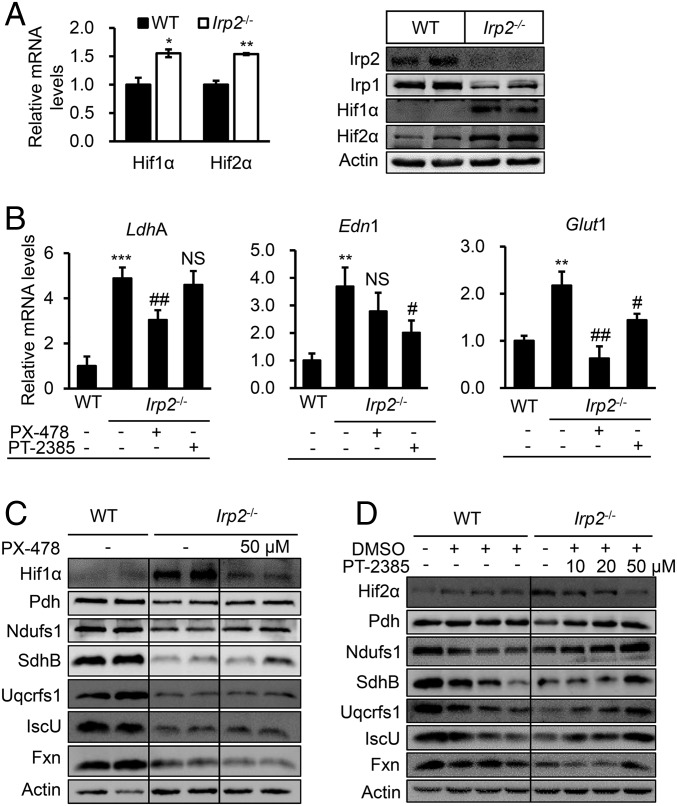
Fig. 3.
Irp2 deficiency-induced mitochondrial dysfunction is mediated by up-regulated Hif2α. (A) Irp2 deficiency-induced expression of Hif1α and Hif2α determined by qRT-PCR (Left) and Western blot analysis (Right). (B) Effects of Hif1α and Hif2α inhibition by PX-478 (a specific inhibitor of Hif1α, 50 μM) and PT-2385 (a specific inhibitor of Hif2α, 50 μM), respectively, on their target genes LdhA, endothelin 1 (Edn1), and Glut1 in Irp2−/− cells. (C and D) Expression of OXPHOS or Fe–S biogenesis-related proteins, Pdh, Ndufs1, SdhB, Uqcrfs1, IscU, and Fxn, after inhibition of Hif1α by PX-478 (50 μM) (C) and inhibition of Hif2α by PT-2385 (10–50 μM) (D) determined by Western blot analysis. DMSO as a vehicle of PT-2385 was added at an identical volume when cells were treated. A representative image set is presented, and the quantitative data of the protein levels are shown in the SI Appendix, Figs. S5 and S6. Values represent the mean ± SEM (n = 3, each with duplicates). *P < 0.05, **P < 0.01, ***P < 0.001, mutant vs. WT. #P < 0.05, ##P < 0.01, with inhibitor vs. without inhibitor. NS, no significance, with inhibitor vs. without inhibitor.
Both Hif1 and Hif2 Collaboratively Mediate the Metabolic Switch of Irp2−/− Cells.
To further demonstrate whether the increased expression of Hif1α and Hif2α induced by Irp2 deficiency was the cause of the cellular metabolic shift, we examined whether the phenotypes could be reversed after inhibition of Hif1α and Hif2α. We measured the OCR and ECAR in Irp2−/− cells after treatment with PX-478 or PT-2385. The results showed that basal respiration, maximal respiration, and OXPHOS-dependent ATP production were efficiently reversed by inhibiting Hif2α but not by inhibiting Hif1α (Fig. 4 A and B). By contrast, both glycolysis and the glycolysis capacity were significantly suppressed by inhibition of Hif1α but not by inhibition of Hif2α (Fig. 4 C and D). These results further validate that Irp2 deficiency enhances glycolysis by inducing the expression of Hif1α and suppresses OXPHOS and mitochondrial biogenesis by inducing the expression of Hif2α, thereby switching energy metabolism in MEFs.
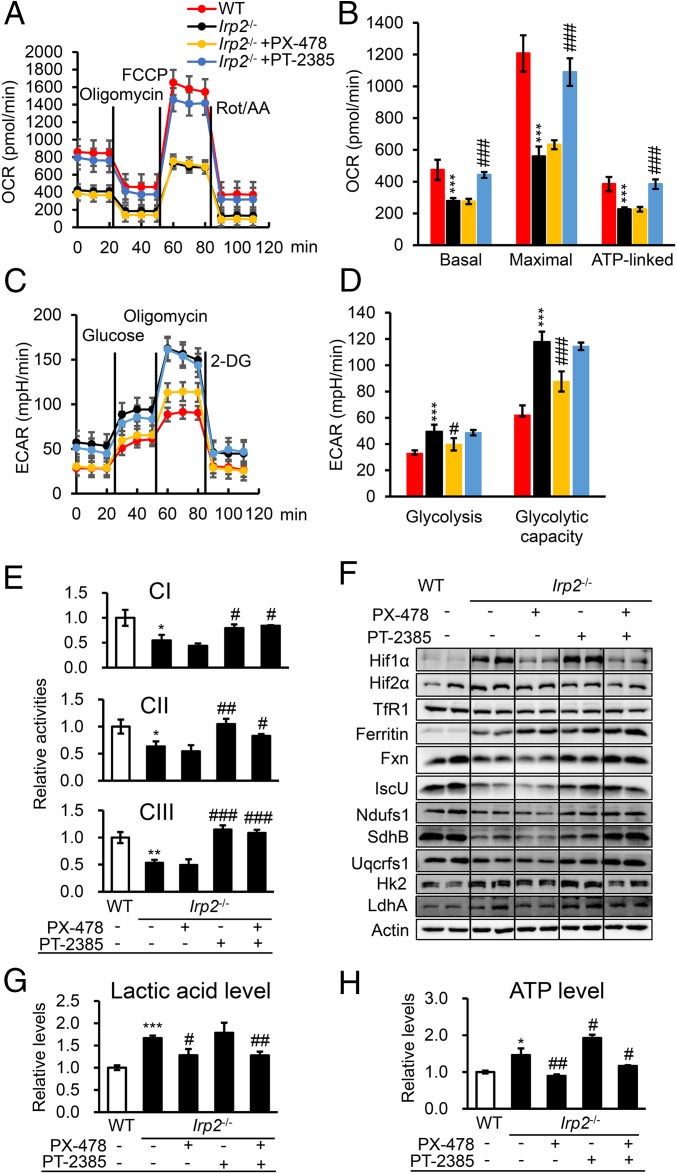
Fig. 4.
Both Hif1α and Hif2α collaboratively mediate the metabolic switch of Irp2−/− cells. (A) Profiles of the OCR, reflecting OXPHOS activity in Irp2−/− cells after treatment with PX-478 (50 μM) or PT-2385 (50 μM) for 24 h. (B) The calculated OCR for basal and maximal respiration and ATP production. (C) Profiles of the ECAR, reflecting glycolytic activity in Irp2−/− cells after treatment with PX-478 or PT-2385 for 24 h. (D) The calculated ECAR for glycolysis and glycolytic capacity. (E) Activities of ETC complexes CI, CII, and CIII in Irp2−/− MEFs after treatment with PX-478, PT-2385, or both PX-478 and PT-2385 for 24 h. (F) Protein levels of a series of genes involved in either aerobic glycolysis or OXPHOS determined by Western blot analysis after inhibition of Hif1α and/or Hif2α in Irp2−/− cells. (G) Levels of lactic acid in the medium of Irp2−/− MEFs after treatment with PX-478 and/or PT-2385 for 24 h. (H) Intracellular ATP content of Irp2−/− cells after treatment with PX-478 and/or PT-2385. Values represent the mean ± SEM (n = 3–5, each with duplicates). *P < 0.05, **P < 0.01, ***P < 0.001, mutant vs. WT; #P < 0.05, ##P < 0.01, ###P < 0.001, with inhibitor vs. without inhibitor.
To reveal the biochemical basis of the above observed phenotypes, we measured the activities of mitochondrial ETC complexes I, II, and III and found that they were all significantly increased by inhibition of Hif2α alone or by inhibition of both Hif1α and Hif2α in Irp2−/− cells but not by inhibition of Hif1α alone (Fig. 4E). Although the TfR1 level remained constant for iron import, ferritin expression was significantly increased (Fig. 4F), in line with the cytoplasmic labile iron pool (LIP) level (SI Appendix, Fig. S8). In accordance with this result, the expression of Fxn, IscU, Ndufs1, SdhB, and Uqcrfs1 significantly increased in Irp2−/− cells after inhibiting Hif2α, and the effects were even more profound with simultaneous inhibition of Hif1α and Hif2α (Fig. 4F). The expression of LdhA and HK2 was reduced (Fig. 4F) after treatment with PX-478, which was in agreement with the drastically diminished production of lactic acid in the medium, while no change was observed after treatment with PT-2385 alone (Fig. 4G). The combined treatment of PX-478 and PT-2385 showed an effect on the production of lactic acid similar to that with PX-478 alone (Fig. 4G). Furthermore, the ATP content correlated very well with the levels of lactic acid and increased when Hif2 was inhibited (Fig. 4H). These results further prove that the effects of Hif1 and Hif2 are independent and that aerobic glycolysis is a major metabolic pathway in Irp2−/− MEFs.
The effects of Hif1α and Hif2α on the metabolic switch were further validated by a shRNA or siRNA knockdown approach. The knockdown efficiency was first evaluated (SI Appendix, Fig. S9 A and B), and the best shRNA and siRNA were used to knock down Hif1α and Hif2α, respectively. The Western blot results showed that the expression of the Hif1-targeted genes HK2 and LdhA was significantly reduced after Hif1α was knocked down with siRNA (SI Appendix, Fig. S9C). The consequence was the reduced production of lactic acid (SI Appendix, Fig. S9D). Similarly, the expression of Hif2-targeted genes was reduced when Hif2α was knocked down with shRNA (SI Appendix, Fig. S9E). In line with the drug inhibition of Hif2, Fe–S biogenesis-related (Fxn and IscU) and OXPHOS-related (complex I subunit Ndufs1, complex II subunit SdhB, and complex III subunit Uqcrfs1) genes were up-regulated (SI Appendix, Fig. S9F). As anticipated, the ATP content increased further (SI Appendix, Fig. S9G). Combining the results from the drug inhibition and genetic approaches, we concluded that the Irp2 ablation-induced metabolic switch is mediated by up-regulated Hif1α and Hif2α.
Discussion
In this study, we first found that Irp2-deficient MEFs had nearly normal growth despite their significantly low ETC activity. Interestingly, the ATP content was relatively higher in mutant cells. We further discovered that Irp2−/− cells favored aerobic glycolysis over OXPHOS, which was triggered by up-regulated Hif1α and Hif2α. Inhibition of both Hifs suppressed aerobic glycolysis and enhanced OXPHOS, thereby switching respiration from aerobic glycolysis to OXPHOS in Irp2−/− cells (illustrated in SI Appendix, Fig. S10). This illustration is in line with previous studies, in which Hif1 and Hif2, while sharing structural similarity and common target genes, have unique targets involved in different pathways (22, 23).
Under normal oxygen conditions, prolyl hydroxylase domain enzymes (PHDs), which are master regulators of the hypoxia response (24), hydroxylate HIFα subunits at conserved prolines, leading to HIFα degradation by the proteasome. PHD activity requires iron binding at an active site. Therefore, Hif1α is stabilized by iron starvation, which is induced by Irp2 ablation in this study. Hif1α contains a unique transactivation domain that allows preferential activation of hypoxia-responsive glycolytic genes; its downstream genes, such as HK2, Glut1, and LdhA, are up-regulated in Irp2−/− cells. This result was proven by the addition of iron, which reduced Hif1α protein levels (SI Appendix, Fig. S11). However, up-regulated Hif2α did not respond to iron treatment (SI Appendix, Figs. S11A and S12B). Hif2α stabilization seems more complex because it can be regulated both by iron, similar to Hif1α, and directly by Irps (15, 25), probably mainly by Irp1 (14). Interestingly, the Irp1 levels were reduced in Irp2−/− cells (Fig. 2H and SI Appendix, Figs. S2A and S3A). This reduction of Irp1 is presumably regulated by FBXL5, which can target both IRP1 and IRP2 for degradation (4, 26). A strong induction of FBXL5 was observed when the cytosolic Fe–S assembly system was impaired, which contributed to the degradation of the Irp1 protein (26). This degradation likely results in Hif2α up-regulation in Irp2−/− cells, in line with the unaltered protein level of Hif2α after the addition of iron (SI Appendix, Fig. S11). These results suggest that Irp1 reduction rather than iron starvation is, at least partially, causative of Hif2α stabilization in Irp2−/− cells. This result was verified by exogenous expression of IRP1 in Irp2−/− cells, which reversed the Hif2α protein levels to some extent (SI Appendix, Fig. S2), very likely through IRP1–IRE binding to the 5′-UTR of Hif2α mRNA to inhibit translation.
Inhibition of Hif1α in Irp2−/− cells suppressed the Hif1α target genes HK2, Glut1, and LdhA. As a result, aerobic glycolysis was repressed, and the lactic acid levels decreased, whereas OXPHOS-related genes and enzyme activities did not respond to Hif1α inhibition. Thus, the decreased ATP content caused by Hif1α inhibition is glycolysis-dependent. By contrast, Hif2α inhibition in Irp2−/− cells drastically up-regulated the OXPHOS-related genes Ndufs1, SdhB, and Uqcrfs1, leading to the restoration of the activities of complexes I–III. Meanwhile, the ATP content increased further, proving that Irp2 depletion-induced Hif2α represses OXPHOS and reduces mitochondrion-dependent ATP production. This effect is likely attributed to the suppression of IscU and Fxn (this study and refs. 11 and 27). IscU has been revealed to be a member of the miR-210 regulon during hypoxia and adversely controls mitochondrial metabolism (28). The promoter of miR-210 contains a hypoxic responsive element (HRE) for Hif binding (29). Therefore, down-regulation of IscU in Irp2−/− MEFs is presumably through the miR-210–Hif axis. Hif1 and Hif2, both in combination (30) and individually (31), have been verified to target miR-210 in various tumor cells. Remarkably, in Irp2−/− MEFs, only the inhibition of Hif2α, not that of Hif1α, increased IscU expression, suggesting that miR-210 is regulated by Hif2 in MEFs. Strikingly, Fxn exhibits very similar responses to Irp2 depletion (ref. 11 and this study), Hif inhibition (this study), and iron regulation (12, 32) as IscU. Human FXN has been reported to be directly regulated by Hif1, not by Hif2 (33). Mouse Fxn, in contrast, is controlled by Hif2, not by Hif1 (34). The up-regulation of FXN by both Hifs is thought to occur through the binding of HIF-HRE to the promoter region of FXN. However, we found that Hif2, but not Hif1, down-regulated the expression of Fxn in MEFs. The mechanism needs to be investigated further.
Consistently, we found here and previously (11) that Irp1 and Irp2 had distinct impacts on mitochondrial metabolism, although they are interchangeable in terms of iron metabolism. The rescue experiments (Fig. 2E) showed that either IRP1 or IRP2 could reverse the Irp2 deficiency-induced enzymatic defects of complexes I and II, whereas the activity of complex III could only be reversed by IRP2 expression. Comparable results have been reported in which tempol treatment restored complex I activity of Irp2−/− mice by converting Irp1 from the cytosolic aconitase to the IRE binding form for iron uptake to improve the neurodegenerative symptoms of Irp2−/− mice (35). Tempol-induced iron bioavailability, presumably, also reduces Hif1α stability. The important role of Hif1α in neurodegenerative diseases has been proposed, where Hif1α can be considered to be a therapeutic target (36). Moreover, inhibition of Hif1α blocked glycolysis, which is the main metabolic pathway to generate ATP and lactic acid in Irp2−/− cells. The high level of lactic acid or methylglyoxal, a highly reactive dicarbonyl compound inevitably formed as a by-product of glycolysis, might be toxic to neuronal function (37, 38). Despite the astrocyte-neuron lactate shuttle hypothesis (39) and the high expression of lactate dehydrogenase (this study), a burgeoning neuronal energy demand is hard to fulfill by the remarkably weakened OXPHOS due to Irp2 ablation. These results suggest that the involvement of Irp2 in energy metabolism is beyond direct iron regulation.
In conclusion, we demonstrated that Irp2 depletion increases the protein levels of Hif1α and Hif2α. Hif1α enhances aerobic glycolysis in Irp2−/− MEFs by up-regulating target genes related to the glycolytic pathway. Hif2α suppresses mitochondrial OXPHOS, at least partially, by downregulating the expression of Fxn and IscU to further reduce the biogenesis of mitochondrial Fe–S and ETC subunits. These results indicate that Irp2 may switch energy metabolism between OXPHOS and glycolysis, implying that high-energy-need tissues could be affected when Irp2 is deficient, as in Irp2−/− mice, leading to neurological disorders (8–10).
Materials and Methods
MEFs derived from WT and global Irp2-deficient mice were generously given by Dr. Tracey Rouault (Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH). Detailed information on cell lines and cell culture, antibodies and reagents, constructs and cell transfection, Western blot, determination of mitochondrial membrane potential, enzymatic activities, ATP and lactic acid contents, qRT-PCR, mitochondrial respiration and glycolytic assays, and statistical analysis is available in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Dong Wang for language assistance and Dr. Xianwei Cui for technical assistance using the Seahorse Bioscience XF24 Machine. This study was supported by grants from the National Basic Research Program of China (Grant 2015CB856300) and by the National Natural Science Foundation of China (Grant 31571218).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820051116/-/DCSupplemental.
References
- 1.Anderson CP, Shen M, Eisenstein RS, Leibold EA (2012) Mammalian iron metabolism and its control by iron regulatory proteins. Biochim Biophys Acta 1823:1468–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hentze MW, Muckenthaler MU, Galy B, Camaschella C (2010) Two to tango: Regulation of mammalian iron metabolism. Cell 142:24–38. [DOI] [PubMed] [Google Scholar]
- 3.Salahudeen AA, et al. (2009) An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326:722–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vashisht AA, et al. (2009) Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326:718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooperman SS, et al. (2005) Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood 106:1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galy B, et al. (2005) Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2). Blood 106:2580–2589. [DOI] [PubMed] [Google Scholar]
- 7.Galy B, et al. (2006) Iron homeostasis in the brain: Complete iron regulatory protein 2 deficiency without symptomatic neurodegeneration in the mouse. Nat Genet 38:967–969. [DOI] [PubMed] [Google Scholar]
- 8.Zumbrennen-Bullough KB, et al. (2014) Abnormal brain iron metabolism in Irp2 deficient mice is associated with mild neurological and behavioral impairments. PLoS One 9:e98072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaVaute T, et al. (2001) Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat Genet 27:209–214. [DOI] [PubMed] [Google Scholar]
- 10.Jeong SY, et al. (2011) Iron insufficiency compromises motor neurons and their mitochondrial function in Irp2-null mice. PLoS One 6:e25404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, et al. (2018) Iron regulatory protein deficiency compromises mitochondrial function in murine embryonic fibroblasts. Sci Rep 8:5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong WH, Rouault TA (2006) Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab 3:199–210. [DOI] [PubMed] [Google Scholar]
- 13.Jasoliya MJ, McMackin MZ, Henderson CK, Perlman SL, Cortopassi GA (2017) Frataxin deficiency impairs mitochondrial biogenesis in cells, mice and humans. Hum Mol Genet 26:2627–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmer M, et al. (2008) Small-molecule inhibitors of HIF-2a translation link its 5'UTR iron-responsive element to oxygen sensing. Mol Cell 32:838–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez M, Galy B, Muckenthaler MU, Hentze MW (2007) Iron-regulatory proteins limit hypoxia-inducible factor-2alpha expression in iron deficiency. Nat Struct Mol Biol 14:420–426. [DOI] [PubMed] [Google Scholar]
- 16.Wilkinson N, Pantopoulos K (2013) IRP1 regulates erythropoiesis and systemic iron homeostasis by controlling HIF2α mRNA translation. Blood 122:1658–1668. [DOI] [PubMed] [Google Scholar]
- 17.Anderson SA, et al. (2013) The IRP1-HIF-2α axis coordinates iron and oxygen sensing with erythropoiesis and iron absorption. Cell Metab 17:282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh MC, et al. (2013) Deletion of iron regulatory protein 1 causes polycythemia and pulmonary hypertension in mice through translational derepression of HIF2α. Cell Metab 17:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galy B, et al. (2010) Iron regulatory proteins secure mitochondrial iron sufficiency and function. Cell Metab 12:194–201. [DOI] [PubMed] [Google Scholar]
- 20.Rensvold JW, et al. (2013) Complementary RNA and protein profiling identifies iron as a key regulator of mitochondrial biogenesis. Cell Rep 3:237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu T, Tang B, Sun X (2017) Development of inhibitors targeting hypoxia-inducible factor 1 and 2 for cancer therapy. Yonsei Med J 58:489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC (2003) Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol 23:9361–9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keith B, Johnson RS, Simon MC (2011) HIF1α and HIF2α: Sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer 12:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myllyharju J. (2013) Prolyl 4-hydroxylases, master regulators of the hypoxia response. Acta Physiol (Oxf) 208:148–165. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh MC, Zhang DL, Rouault TA (2015) Iron misregulation and neurodegenerative disease in mouse models that lack iron regulatory proteins. Neurobiol Dis 81:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson NB, Deck KM, Nizzi CP, Eisenstein RS (2017) A synergistic role of IRP1 and FBXL5 proteins in coordinating iron metabolism during cell proliferation. J Biol Chem 292:15976–15989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong WH, et al. (2018) TLR-activated repression of Fe-S cluster biogenesis drives a metabolic shift and alters histone and tubulin acetylation. Blood Adv 2:1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan SY, et al. (2009) MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab 10:273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulshreshtha R, et al. (2007) A microRNA signature of hypoxia. Mol Cell Biol 27:1859–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCormick RI, et al. (2013) miR-210 is a target of hypoxia-inducible factors 1 and 2 in renal cancer, regulates ISCU and correlates with good prognosis. Br J Cancer 108:1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dang K, Myers KA (2015) The role of hypoxia-induced miR-210 in cancer progression. Int J Mol Sci 16:6353–6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li K, Besse EK, Ha D, Kovtunovych G, Rouault TA (2008) Iron-dependent regulation of frataxin expression: Implications for treatment of Friedreich ataxia. Hum Mol Genet 17:2265–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guccini I, et al. (2011) Frataxin participates to the hypoxia-induced response in tumors. Cell Death Dis 2:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oktay Y, et al. (2007) Hypoxia-inducible factor 2alpha regulates expression of the mitochondrial aconitase chaperone protein frataxin. J Biol Chem 282:11750–11756. [DOI] [PubMed] [Google Scholar]
- 35.Ghosh MC, et al. (2008) Tempol-mediated activation of latent iron regulatory protein activity prevents symptoms of neurodegenerative disease in IRP2 knockout mice. Proc Natl Acad Sci USA 105:12028–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, Yan J, Chang Y, ShiDu Yan S, Shi H (2011) Hypoxia inducible factor-1 as a target for neurodegenerative diseases. Curr Med Chem 18:4335–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allaman I, Bélanger M, Magistretti PJ (2015) Methylglyoxal, the dark side of glycolysis. Front Neurosci 9:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walton ZE, et al. (2018) Acid suspends the circadian clock in hypoxia through inhibition of mTOR. Cell 174:72–87.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pellerin L, Magistretti PJ (1994) Glutamate uptake into astrocytes stimulates aerobic glycolysis: A mechanism coupling neuronal activity to glucose utilization. Proc Natl Acad Sci USA 91:10625–10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.