Significance
EGFR cancer mutations display an astonishing tissue-specific asymmetry: in lung cancer, mutations target the intracellular kinase (KD), while in glioblastomas (GBMs), a variety of missense clusters and deletions concentrate at the ectodomain (ECD). Intriguingly, GBM-activating mutations share a paradoxical preference for inhibitors that bind the inactive kinase. By integrating simulations, small-angle X-ray scattering, and GBM models, we demonstrate that ECD mutants converge to a transition state characterized by a cryptic epitope, allosterically coupled to an intermediate kinase, and synergistically blocked by antibodies and inhibitors. Our findings indicate that apparently heterogeneous aberrations remove a similar steric restrain on KD activation. The diversity of structural tricks in ECD mutants to achieve the same conformational state constitutes a potent example of molecular mimicry and convergence.
Keywords: cancer, mutational heterogeneity, structural convergence, intermediate, cryptoepitope
Abstract
Epidermal growth factor receptor (EGFR) signaling is initiated by a large ligand-favored conformational change of the extracellular domain (ECD) from a closed, self-inhibited tethered monomer, to an open untethered state, which exposes a loop required for strong dimerization and activation. In glioblastomas (GBMs), structurally heterogeneous missense and deletion mutations concentrate at the ECD for unclear reasons. We explore the conformational impact of GBM missense mutations, combining elastic network models (ENMs) with multiple molecular dynamics (MD) trajectories. Our simulations reveal that the main missense class, located at the I-II interface away from the self-inhibitory tether, can unexpectedly favor spontaneous untethering to a compact intermediate state, here validated by small-angle X-ray scattering (SAXS). Significantly, such intermediate is characterized by the rotation of a large ECD fragment (N-TR1), deleted in the most common GBM mutation, EGFRvIII, and that makes accessible a cryptic epitope characteristic of cancer cells. This observation suggested potential structural equivalence of missense and deletion ECD changes in GBMs. Corroborating this hypothesis, our FACS, in vitro, and in vivo data demonstrate that entirely different ECD variants all converge to remove N-TR1 steric hindrance from the 806-epitope, which we show is allosterically coupled to an intermediate kinase and hallmarks increased oncogenicity. Finally, the detected extraintracellular coupling allows for synergistic cotargeting of the intermediate with mAb806 and inhibitors, which is proved herein.
Epidermal growth factor receptor (EGFR), a master switch for signaling pathways that control cell growth and differentiation, is a paradigmatic tyrosine kinase receptor and one of the first human oncogenes identified (1). Cancer mutations targeting EGFR display an astonishing tissue-specific asymmetry: in lung cancer, mutations typically target the intracellular kinase domain (KD), while in glioblastomas (GBMs), they concentrate at the extracellular ectodomain (ECD) (2, 3), which is formed by four domains (DI to DIV) arranged as two tandem repeats (TR1 and -2) (Fig. 1A) (4). Although both mutation types result in ligand-independent activation, remarkably, this mutational asymmetry translates to differential sensitivities to tyrosine kinase inhibitors (TKIs), with KD mutations responding preferentially to inhibitors that bind the active asymmetric KD (aKD) state (class I, TKI-1s) and, on the contrary, ECD mutations being more sensitive to molecules that stabilize the inactive symmetric (sKD) state (class II, TKI-2s) (5). Apart from ECD mutations’ paradoxical preference for TKI-2s—which indicates they favor an inactive-like fold, despite being constitutively active—they also show striking heterogeneity (6), with at least 4 deletion classes and about ∼130 missense variants, neatly clustered at 3 interdomain interfaces (I-II, II-IV, and II-III; Fig. 1B) in GBMs but also colon, breast, and other cancers (SI Appendix, Table S1).
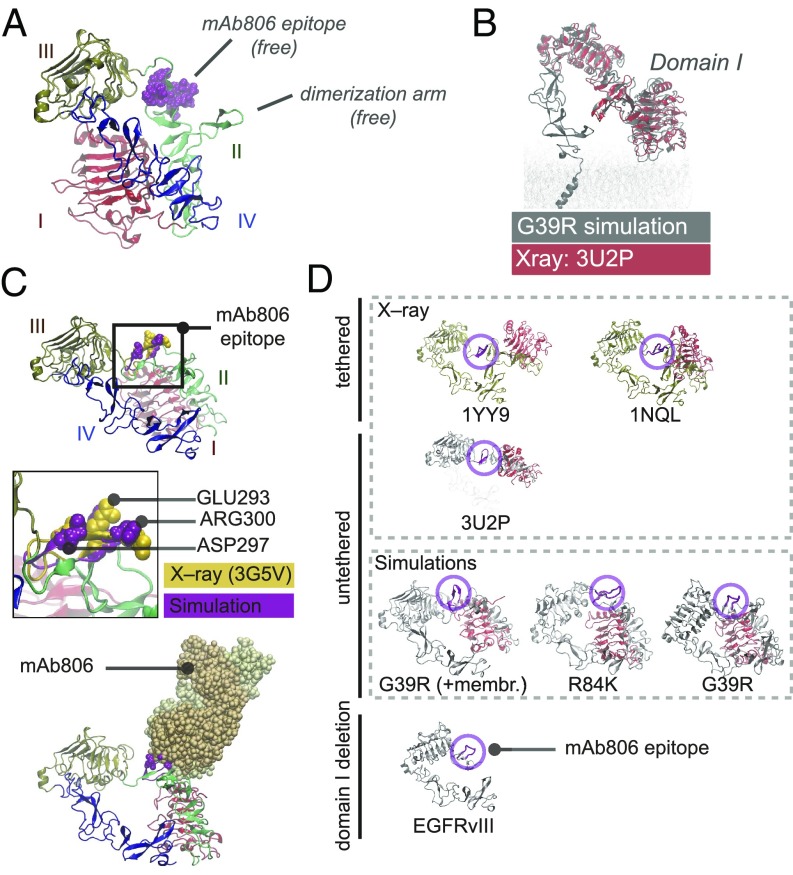
Fig. 1.
ENM and MD of tethered ECD predict that N-TR1 twisting renders the 806-epitope accessible in I-II mutants. (A) Scheme of known conformations: the closed tethered ECD (Left), which is inactive, and the active untethered ligand-bound dimer (Right). In both, the 806-epitope is buried. (B) Pattern of missense clusters (black bands) and deletions (EGFRvIII, EGFRvII) along the ECD sequence. Gray arcs indicate Cαιφ < 10Å (Left). 3D structure of the tethered ECD (1NQL) with interface clusters in dark (Right). Dashed N-TR1 fragment next to the 806-epitope is deleted in EGFRvIII. I-II interface mutations have the highest clustering (SI Appendix, Fig. S1B). (C) ENM analysis predicts large-scale twisting and bending TR motions (NMs) potentially sensitive to I-II mutations (SI Appendix, Fig. S1 D and E). Note that both NMs expose the 806-epitope, which acts as a molecular hinge, but twisting also releases the dimerization arm. (D) Free-energy landscape from 1-μs MD for clustered I-II (R84K) versus WT-EGFR, shown as projections onto NM1/NM2 together with the HER Family X-ray ensemble (black dots), reveals a highly flexible WT-ECD sampling multiple minima. The reference structure for MD frame and X-ray alignment was 1NQL (state 1a, origin), used for simulation. The WT trajectory shown (Left) departed from 1NQL to sample bent conformations (state 1b) and then visit a low-twist area with tether-truncated structures (state 2a, 3U2P) where untethering happens (>700 ns), followed by further twisting to maximally expose the 806-epitope (state 2b). For R84K (Right), untethering occurs early in the simulation (>50 ns), quickly proceeding to the twisted 806-accessible states energy minima (2a and 2b) and staying there for the remaining time. None of the ECD simulations opens in solution (states 3b and 4). (E) Diffusion accessibility of X-ray and MD conformations: the 806-epitope is maximally accessible in twisted (states 2a and 2b) and bent (1b) MD intermediates. An “open-like” conformation (state 3b) with low accessibility is observed in membranes (SI Appendix, Fig. S3B). The fully open (4) and closed (1a) X-ray structures have an occluded epitope (see SI Appendix, Figs. S2–S4).
Understanding of EGFR extracellular activation has relied on two ECD structures captured by X-ray crystallography: an inactive “tethered” monomer (“closed”) and an active “untethered” dimer (“open”), bound to ligands (Fig. 1A, Left and Right). Research on the cancer-specific antibody mAb806 indicates, however, the existence of a third ECD transitional state, characterized by exposure of a cryptic epitope (C287-C302) detected in tumors and fast dividing epithelia (7). The antibody 806 was raised against EGFRvIII, in which deletion of the N-terminal part of TR1 (N-TR1, residues 6–273) exposes this epitope, which appears buried in both X-ray conformations (8, 9). Unexpectedly, mAb806 was also found to recognize amplified WT EGFR (10). Tether mutations (9), low glycosylation (11), or quinazoline-driven dimerization (12) are also known to render the 806-epitope more accessible in the full-length receptor, while binding to EGF can bury it again reversibly (13), thus indicating that mAb806 recognizes an untethered, neither open nor closed, ECD transitional state, as EGFR activates.
Despite the clear 3D pattern of ECD mutations (Fig. 1B), it is hard to understand how both large deletions like EGFRvIII (the most frequent GBM mutation) and classical missense mutations (14) result in similar KD sensitivities. Seeking to understand the mechanism of ECD mutations, we focused on the lesser understood but most common missense class, located at an interface (I-II) away from both the tether and the EGF-binding site (15). We performed elastic network model (ENM) analyses to identify potentially “hot” mutations, which upon further molecular dynamics (MD) (16) (SI Appendix, Fig. S1A), revealed that I-II mutants increase N-TR1 motions, favoring untethering to mostly compact structures. Moreover, in such conformers, the N-TR1 fragment (deleted in EGFRvIII) rotates or “twists” (Fig. 1C) exposing the 806-epitope. This observation suggested that missense mutations and the main GBM deletion could be “structurally equivalent” in the sense that they remove a similar steric hindrance that potentially prevents self-activation.
Using small-angle X-ray scattering (SAXS), we corroborate the in silico prediction that I-II mutants untether and also validate the proposed “structural convergence” detected by mAb806 using in vitro and in vivo GBM models. Our data demonstrate that radically different deletions and missense mutations, representative of all clinically relevant classes, converge to a state where the 806-epitope is accessible and allosterically coupled to an inactive-like intermediate KD. As a consequence, we show that heterogeneous ECD mutations, and not only EGFRvIII, are responsive to mAb806, which, importantly, has a synergistic tumor-inhibitory effect when combined with TKI-2s binding to the same conformational state. Our findings unexpectedly bring together two independent lines of research on GBM mutations (5) and mAb806 (10), hinting at a much broader spectrum for this antibody, far beyond our previous study (17) and, importantly, paving the way for the rational extraintracellular cotargeting of EGFR-mutated tumors.
Results
The ECD Is Intrinsically Prepared to Perform Closed⇔Open Transitions Controlled by Interfacial Molecular Joints.
To understand whether and which extracellular point mutations alter ECD stability, first we explored its global motions by computing its ENM normal modes (NMs) (16). The structural transition between closed [Protein Data Bank (PDB) ID code 1NQL] and open (PDB ID code 3NJP) ECDs is dramatic (rmsd = 25.1 Å) and involves large-scale rigid-body motions with minimal intradomain changes, with the exception of DII, which bends as a spinal backbone to allow dimerization. Significantly, this transition is predicted with high accuracy by low-frequency NMs from either the closed (75%) or open (82%) ECD, which indicates is intrinsically imprinted in the 3D shape (SI Appendix, Table S2). To identify potential regions acting as hinges or “molecular joints” for transitions among the top 3D-clustered mutations (SI Appendix, Fig. S1B), we further analyzed the ENM-derived residue rms fluctuations and cross-correlations (Materials and Methods). From 3NJP, a single mode dominates ECD closure, explained by limited DI oscillations coupled to large-scale C-terminal fluctuations (SI Appendix, Fig. S1 C–E). In contrast, from 1NQL, ENM predicts two large-scale twisting and bending TR motions around a central DII hinge (Fig. 1C and SI Appendix, Fig. S1 C and D) encompassing the dimerization arm and the nearby 806-epitope next to the II-III linker. Significantly, I-II interface mutation clusters (3×, 6×, 8×; SI Appendix, Table S1) target with precision a series of secondary hinge points for oscillations of N-TR1 (SI Appendix, Fig. S1D), which, in turn, is coupled to the tether through a sparse but long-range network of interdomain interactions (SI Appendix, Fig. S1E). Altogether, this suggested that the complex motion pattern of the closed state could be strongly disrupted by interfacial I-II mutations.
I-II Clustered Mutations Sample Untethered Conformations That Displace N-TR1 Away from the 806-Epitope.
Hence, based on ENM observations, a clustered mutant central at the I-II hinge, R84K [also a known activator (14)], was selected for preliminary MD, both from 1NQL and 3NJP. Sampling was monitored by comparison with the HER Family X-ray ensemble with NMs as collective variables (SI Appendix, Fig. S2A and Table S3), as in refs. 18 and 19. As ENM had suggested, ECD mutations had lesser impact on the open than on the closed conformation, and we focused on the latter for further studies. While the stable open state became further stabilized by mutations, the closed state proved extremely flexible for the WT-ECD, being strongly destabilized by R84K (SI Appendix, Fig. S2 B and C) as ENM had suggested. In the submicrosecond scale, the WT-ECD remained nevertheless tethered (Fig. 1 D and E; state 1a-b, SI Appendix, Fig. S2C, Top), while R84K performed larger-scale DI oscillations (SI Appendix, Fig. S2C) that triggered fast untethering (<50 ns; SI Appendix, Fig. S3A) toward a previously unseen conformation (Fig. 1 D and E, state 2a-b). Seeking to corroborate this transition, we simulated an additional high-score mutation at the I-II interface, G39R, occasionally reported (20) but never tested in vitro. As R84K, G39R displayed altered TR dynamics and quick untethering to an identical intermediate (SI Appendix, Fig. S3A). Upon time extension, G39R/R84K intermediates remained stably untethered; notably, the WT also transitioned to the intermediate after 700 ns (Fig. 1D and SI Appendix, Fig. S3A). Mechanistically, the untethering process was identical for WT, R84K, and G39R simulations (SI Appendix, Fig. S3A). In ECD constructs with the transmembrane (TM) domain and KD in 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine membranes (SI Appendix, Fig. S3 B–D), untethering to twisted conformers was again reproduced for I-II mutants, including now the deadly mutant A265V (17) and, remarkably, also evolved occasionally to open-like conformations, e.g., for R84K (Fig. 1E, state 2a-3b and SI Appendix, Fig. S3C). Overall, the observed N-TR1 oscillations and untethering were reproducible in solution and lipid bilayers for diverse interface mutations (SI Appendix, Fig. S4A and see below), rendering conformations bent, twisted, or partially open.
Remarkably, the twisted arrangement of domains I-III (Fig. 2A) is similar to that observed in structures of tether-truncated ECDs (Fig. 2B), which in the absence of a tether still retain a tethered-like configuration. In this new state, the ECD is still compact, but N-TR1 rotation bends DII and releases the dimerization arm in a configuration poised to self-interact (SI Appendix, Fig. S4B). Also relevant is the dramatic reshaping of the bonds formed by charged residues at the II-III linker as the different in silico conformers are sampled, which heavily changes the curvature and thus solvent accessibility of the nearby 806-epitope (SI Appendix, Fig. S4 C and D). In twisted conformers, N-TR1 rotation not only removes the hindrance on the 806-epitope, but it also results in a convex surface where key residues for antibody binding (21) protrude to the solvent as in epitope-Fab complexes (Fig. 2C; rmsd ∼ 2–2.5 Å). Altogether, this indicates that MD untethered conformers may correspond to the not-open/not-closed 806-intermediate state. Importantly, comparison with EGFRvIII shows that N-TR1 deletion and rotation are structurally equivalent with respect to the epitope (Fig. 2D), posing the question of whether other ECD variants may also converge to acquire a similar feature. Given these in silico observations, we sought to test if increased mAb806 binding is shared by I-II and other ECD variants and explore any relation of the 806-epitope with the KD oncogenic activity.
Fig. 2.
Twisted untethered intermediates suggest structural convergence of N-TR1 reposition and its deletion in EGFRvIII. (A) MD twisted conformers display intermediate conformations characterized by N-TR1 rotation that renders the 806-epitope and the dimerization arm free. (B) The tethered-like arrangement of domains I to III in MD intermediates overlaps with that observed for tether-truncated ECDs (e.g., 3U2P in red). (C) In twisted intermediates, the MD 806-epitope (purple) has a convex surface with key residues for mAb806 binding protruding to the solvent and aligning with X-ray structures of antigen–antibody complexes (3G5V, yellow) (Top). N-TR1 rotation removes stereochemical overlap for docking onto 3G5V, rendering stable MD complexes with mAb806 (Bottom). (D) Domain I (red) sterically occludes 806-epitope (magenta) in tethered conformations; the hindrance is greater in planar configurations (1YY9) than in low-twist tethered structures (1NQL) and minimal in the truncated ECD (3U2P). Both deletion (EGFRvIII) and rotation of N-TR1 (G39R/R84K intermediates) removes domain I from the tethered position that hinders the epitope. In I-II mutations, N-TR1 reposition allows receptor–receptor docking via dimerization arms (SI Appendix, Fig. S4B).
FACS Confirms That I-II Mutations Increase Exposure of the 806-Epitope in Relation to Interface Flexibility.
Our MD simulations suggested that I-II interface flexibility favors N-TR1 motions and untethering, detectable by mAb806. Therefore, we analyzed I-II interdomain bond patterns (Fig. 3 A and B) to design a double mutation, R84K+A265V, in which disruption of key Arg84 H bonds and the adjacent hydrophobic patch of Ala265 (i.e., the most frequent GBM missense mutation) could further enhance I-II oscillations detected by mAb806. Simulations of the double mutation both in solution and in lipid bilayers (SI Appendix, Fig. S4A) confirmed larger-scale N-TR1 fluctuations compared with single mutants (Fig. 3C), which bend DII, exposing the 806-epitope even without untethering. Therefore, we transfected HEK293 cells to express WT, R84K, A265V, and R84K+A265V to measure mAb806 binding using FACS (Fig. 3 D and E). Normal cell surface EGFR expression was confirmed by immunostaining (Fig. 3F and SI Appendix, Fig. S5A), and receptor levels were normalized with control antibody LA22 and total protein Western blotting (WB) (SI Appendix, Fig. S5 B–D). We also examined oligomerization with cross-linking, detecting preformed WT and mutant dimers (SI Appendix, Fig. S5E). As predicted by MD, mAb806 binding was significantly increased for I-II single mutants (two- to threefold versus WT), (Fig. 3E). For R84K+A265V, the increase was additive (up to five- to sixfold; P < 0.001), reaching values close to control EGFRvIII.
Fig. 3.
Increased interfacial flexibility in I-II mutants enhances N-TR1 reposition as detected by mAb806 binding in HEK293 cells. (A) I-II interface of tethered ECD (1NQL), with protein backbone as cartoon and mutated residues as van der Waals spheres; Consurf score is in parenthesis (variable = 1, conserved = 9). (B) I-II interdomain bonds in MD from 1NQL; note that conserved mutated residues (bold) are interconnected by electrostatic and hydrophobic interactions. (C) MD trajectories of WT-ECD and I-II mutants: TR planes angle distribution for the ECD in solution (100 ns, n = 3, dark gray) and in constructs with the TM domain (350 ns, n = 3, light gray) (Left); TR2-alignment of ECD MD frames to show N-TR1 oscillations (50 ns, n = 1) versus the 806-epitope (red) (Right). The stable WT angle (∼40°) occludes the 806-epitope, while N-TR1 oscillations in the double mutant expose it at higher TR angles (∼80°). (D) FACS binding of mAb806 to HEK293 cells overexpressing WT or I-II mutant EGFR: representative overlays of the fluorescence signal for mAb806-stained cells; mAb806 binds both WT (blue) and mutants (orange) but not control HEK293 cells (red). The right shift of the mean peak in mutants indicates high 806-antigen surface expression, maximal for positive control EGFRvIII and R84K+A265V. (E) Mean fluorescence intensity normalized to total EGFR surface expression (mAbLA22) (n = 5). The double mutant, expected to further disrupt the I-II interface, raises mAb806 binding near to EGFRvIII level. *P ≤ 0.05 and ***P ≤ 0.001. (F) Immunocytochemistry of cells expressing WT or mutant EGFR, stained with mAb806 (red) and nuclear marker DAPI (blue), shows enriched localization of 806-antigen to the membrane (see SI Appendix, Fig. S5).
SAXS Confirms That I-II Mutations Untether to Twisted and Partially Open Intermediates.
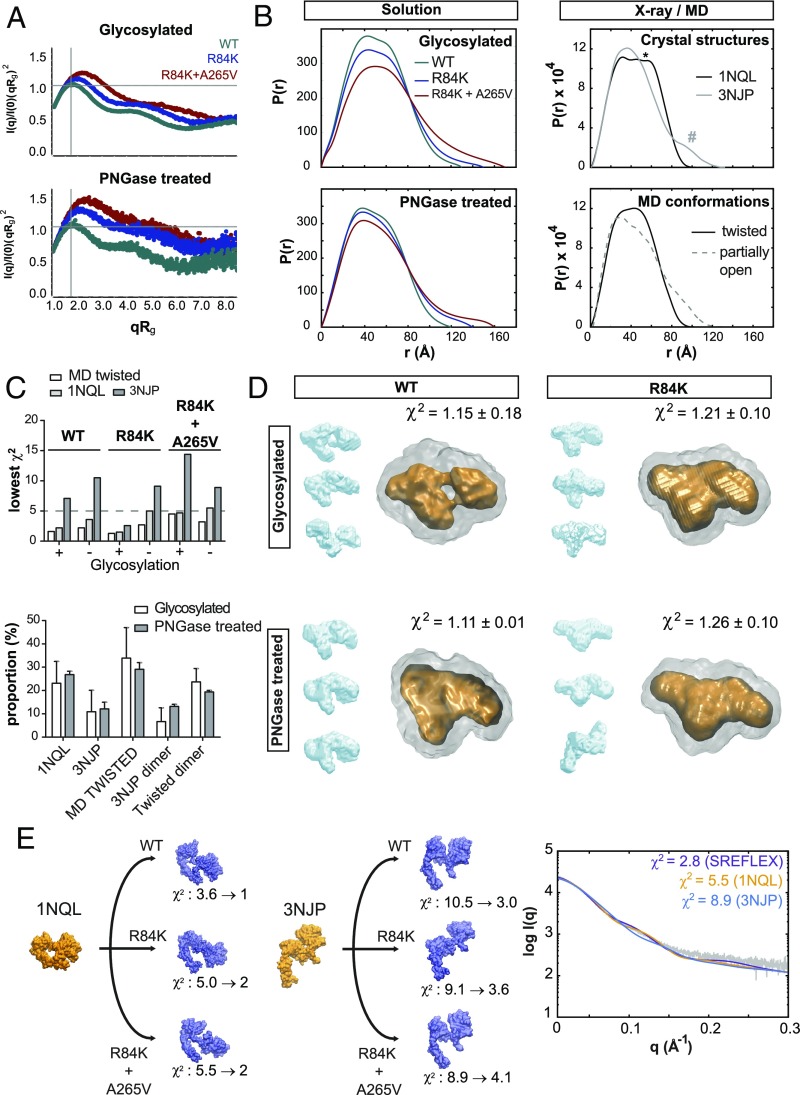
To structurally validate whether untethering occurs in I-II mutations, we collected SAXS data (SI Appendix, Fig. S6 A–C) for HEK293-expressed WT, R84K, and R84K+A265V ECDs, representative of control, intermediate, and high mAb806 binding, respectively, both with eukaryotic glycosylation and after PNGase treatment. SAXS revealed a well-folded and extremely dynamic ECD, with enhanced flexibility upon single and double mutation (Fig. 4A). Our synchrotron WT-ECD data rendered Rg and Dmax values close to theoretical ones (SI Appendix, Table S4), although higher than reported with baculovirus expression (22); upon single and double mutation, these values increased ∼10 and 20%, respectively, indicating conformational changes. For R84K+A265V, forward scattering I (0)/C (SI Appendix, Fig. S6B) was slightly increased, suggesting low dimerization. Pair distribution functions [P(r)] for mutants lacked the characteristic tether shoulder of the closed structure (*), as well as the second tail peak (#) of the open conformer, and rather resembled MD untethered profiles (Fig. 4B and SI Appendix, Fig. S7A).
Fig. 4.
SAXS of I-II mutant ECDs suggests increased flexibility and untethering to intermediate twisted and partially open conformations. Experimental synchrotron scattering data for WT and I-II mutant ECDs, collected with HEK293 glycosylation and after PNGase treatment at ESRF (Grenoble, France) and DESY (Hamburg, Germany), respectively. (A) Dimensionless Kratky plots for all ECD variants are characteristic of well-folded and flexible multidomain proteins. For mutants, and independent of glycosylation, the upright shifts versus the Guinier–Kratky point (qRg = √3, ideal globular particle) indicate transitions to less compact and more asymmetric shapes. (B) Pair distribution functions [P(r)] computed from solution scattering (Left) versus representative X-ray and MD models (Right). The P(r) curves show a conformational change from a flattened peak with a shoulder (*) in the WT (characteristic of dumbbell-shaped TRs stably held together) to a narrower peak without clear shoulder for mutants (untethered) and missing the second peak tail (#) of the fully open state. (C) Lowest-χ2 CRYSOL fittings to X-ray or MD structures (Top). Tethered 1NQL and twisted conformations render the best fittings for mutants (SI Appendix, Table S5). Presence of dimers significantly improves the fitting for the double mutant according to EOM (Bottom), reaching values as low as χ2 = 1.9 ± 0.1 (glycosylated) and 1.4 ± 0.1 (PNGase-treated) (average χ2 value of 10 runs reported). (D) SAXS-derived ab initio molecular envelopes from 50 DAMMIN runs, with representative DAMCLUST clusters (in blue), the DAMAVER average (gray), and high-density DAMFILT area (orange). Note the visible tether in the glycosylated WT-ECD DAMFILT envelope, absent upon deglycosylation. For deglycosylated R84K, as well as for the double mutant (SI Appendix, Fig. S7D), intermediate and also open-like envelopes are generated. (E) SREFLEX fittings for samples treated with PNGase (see SI Appendix, Figs. S6–S7 and Tables S4–S5).
To further assess conformational flexibility, we applied four different SAXS analyses within the ATSAS suite (SI Appendix, Fig. S6D; see also SI Appendix, Supplementary Materials and Methods). First, scattering data were fitted to MD/X-ray structures with CRYSOL (Fig. 4C and SI Appendix, Fig. S7 B and C). We found that the closed 1NQL and twisted conformers explained the data better than open 3NJP (or partially open conformers), which usually rendered poor χ2 values (SI Appendix, Table S5). PNGase-treated samples, including the WT, were fitted better to twisted conformers than to 1NQL (Fig. 4C). Second, based on increased I (0)/C and Rg for R84K+A265V, we ran ensemble optimization (EOM) on the lowest-χ2 conformation pool, including dimers (SI Appendix, Table S5), to find that a mixture of twisted and tethered monomers with low dimerization improved fitting (Fig. 4C). Third, scattering data were independently visualized by generating averaged low-resolution envelopes from multiple (50×) DAMMIN runs. For glycosylated WT-ECD, ab initio envelopes faithfully resembled X-ray closed HER structures, including a fine-grained feature like the tether (Fig. 4D), which disappeared upon PNGase treatment or R84K mutation, although retaining a compact shape. Mutant samples however also generated open-like envelopes upon deglycosylation, especially for R84K+A265V (SI Appendix, Fig. S7D). Finally, we performed SAXS refinement through flexibility (SREFLEX) starting from X-ray structures with SREFLEX. Remarkably, this method fitted mutant scattering by deforming the 1NQL structure through N-TR1 rotation, up to reach twisted untethered states similar to MD (Fig. 4E). In contrast, when starting from 3NJP, SREFLEX partially closed the structure to fit the scattering. For R84K+A265V, SREFLEX could also fit scattering from dimers (SI Appendix, Fig. S7E). Collectively, all analyses confirm MD predictions that the WT-ECD is tethered and that I-II mutants can untether to predominantly twisted and partially, not fully open conformations; additionally, SAXS suggests slight ECD dimerization limited to the double mutant.
Heterogeneous ECD Mutants Share Exposure of the 806-Epitope Coupled to an sKD.
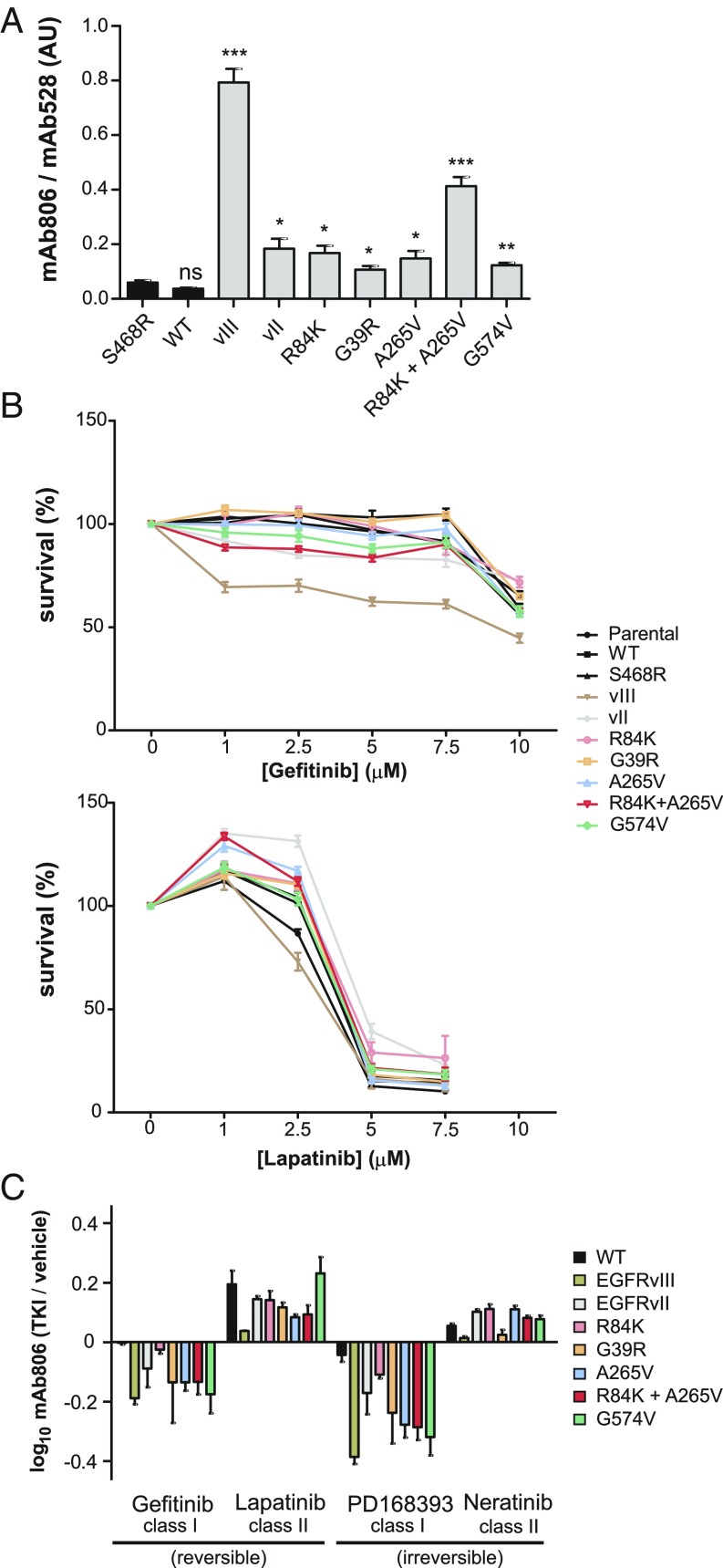
Given the accessibility of the 806-epitope in heterogeneous variants, such as EGFRvIII or I-II mutations as shown here, we investigated its potential role as a convergent hallmark beyond the I-II interface in more realistic GBM models. Thus, we generated U87 cell lines that expressed either WT EGFR, EGFRvIII, I-II point mutants, or II-IV clinically relevant tether variations (G574V and EGFRvII) at levels of amplified EGFR in GBM (SI Appendix, Supplementary Materials and Methods). We also included as negative control the Cetuximab-resistance mutation S468R (23), located out of the oncogenic interfaces and stable in simulations (SI Appendix, Fig. S4A). FACS, WB, and blue native PAGE (BN-PAGE) confirmed that all ECD variants were normally expressed at the cell membrane (SI Appendix, Fig. S8A), not aggregated (SI Appendix, Fig. S8B) and EGF-responsive (SI Appendix, Fig. S8 C and D), with the strict conformation-dependent mAb528 further validating correct folding (Fig. 5A and SI Appendix, Fig. S9A). In contrast to cross-linking (SI Appendix, Fig. S5E), BN-PAGE detected preformed dimers (SI Appendix, Fig. S8B) for all ECD variants but not for controls (WT, S468R). Confirming the results for HEK293 cells, mAb806 binding was increased for I-II single mutants versus negative controls (up to fivefold) and revealed a synergistic increase for the double mutant (13-fold, P < 0.001; Fig. 5A). Tether variants (EGFRvII and G574V) also displayed increased binding (six- and fourfold, respectively). Different binding levels could be distinguished within missense ECDs: high (R84K+A265V), intermediate (R84K, A265V, G39R, G574V), and low (WT, S468R) (SI Appendix, Table S6). This corroborated a general rise in mAb806 recognition among heterogeneous GBM mutations, supporting a free 806-epitope as a shared ECD phenotypic trait. Next, we sought to explore if mAb806 detects an ECD conformer coupled to a specific KD arrangement. As EGFR activates, the KD rearranges from an inactive sKD dimer to an aKD active dimer, which are the targets for TKI-2s and TKI-1s, respectively. Here we confirmed differential sensitivity to reversible TKIs gefitinib (TKI-1) and lapatinib (TKI-2) for all ECD variants (Fig. 5B), not limited to the classical ECD mutations (5) but including also previously untested ones (i.e., G39R, EGFRvII). This emphasizes that ECD mutations, regardless of their type (missense vs. deletions) or location (different interfaces, etc.), favor an sKD-like conformation, and raises the question of whether the EGFR-state bound by TKI-2s is the same one detected by mAb806. To answer this question, we treated the mutant panel with both TKI classes and measured mAb806 binding with FACS (Fig. 5C). Irreversible inhibitors were included to exclude the possibility that any observed changes are due to different kon/koff rates. Importantly, we found that the two TKI classes had opposite effects on mAb806 binding: the accessibility of 806-epitope was decreased by TKI-1s but increased by TKI-2s, and particularly by lapatinib, which favors an intermediate conformation with an active DFG-in [thus being specifically referred to as class I1/2 (24)]. For WT-ECD (i.e., minimal basal mAb806 binding), 806-accesibility was little affected by TKI-1s and, conversely, for EGFRvIII (i.e., maximal basal mAb806 binding), TKI-2s did not further enhance antibody recognition. However, TKI-2s increased mAb806 binding to WT, while TKI-1s decreased it for EGFRvIII [indicating the aKD rearrangement occludes the 806-epitope, as happens upon EGF binding (13)]. In contrast, the other ECD variants (i.e., mAb806 binding in between WT and EGFRvIII) responded to both TKI classes with a significant increase (TKI-2s) or decrease (TKI-1s) in 806-epitope exposure, indicative of ECD-KD allosteric coupling. For reversible inhibitors, such changes were inversely correlated (SI Appendix, Fig. S9B): the smaller the response to TKI-1s, the larger the increase for TKI-2s, with the only exception of G574V that showed similar responses to both (see Discussion). This indicates that distinct KD dimers stabilized by TKIs allosterically change inside-out the ECD conformation in antagonistic ways via N-TR1 repositioning, suggesting that mAb806 and TKI-2s target the same EGFR state in GBMs, characterized by a free 806-epitope coupled to a symmetric-like KD (ECD806+-sKD).
Fig. 5.
Evidence for mAb806-binding convergence in U87 GBM cell models carrying heterogeneous ECD mutations and its allosteric coupling to symmetric KD arrangement. The conformational status of the WT-ECD, a negative control (S468R), interdomain I-II mutants (G39R, R84K, A265V, and R84K+A265V), and tether variants (G574V, EGFRvII) was tested using mAb806 binding as a reporter of N-TR1 position. (A) Mean fluorescence intensity normalized to EGFR surface expression (mAb528) in glioblastoma U87 cell lines. Note that binding is increased in ECD mutants versus WT or S468R. The double mutation at the I-II interface has a synergistic effect on mAb806 binding versus single mutations. ns, P > 0.05, *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. (B) Cell survival, as determined by crystal violet staining of ECD mutants following treatment with gefitinib (class I) or lapatinib (class II) for 48 h, demonstrates increased sensitivity to the second class. (C) FACS analysis of ECD mutants bound to mAb806 following treatment with either class I (10 μM) or class II (5 μM) TKIs, with mean FITC signal normalized to mAb528 staining. The ratio between mAb806 binding with and without TKI treatment is reported, so that a positive value indicates increased mAb806 binding versus basal levels, while a negative value indicates a decrease. ECD variants share a uniform response to TKIs, with preferred type II inhibitors further enhancing 806-binding, and type I inhibitors decreasing it (see SI Appendix, Table S6).
The ECD806+-sKD State Hallmarks Increased Oncogenic Activity and Its Extraintracellular Targeting Inhibits Tumor Growth Synergistically.
Given that GBM mutations favor an sKD-like configuration detected by mAb806, we aimed to explore if such intermediate kinase is associated with oncogenic activation in vitro and in vivo. Specifically, the designed double mutant, with 40–50% of cell surface EGFR exposing the 806-epitope (Fig. 5A), provides a unique model to investigate the biological impact of the ECD806+-sKD state, along with G39R, a naturally occurring mutation never tested to date. First, using pTyr as readout for KD activity, we checked constitutive activation after serum starvation under different conditions (Fig. 6 A and B and SI Appendix, Fig. S8 D and E). All mutations with robust constitutive activity across experiments (R84K+A265V, A265V, R84K, and EGFRvIII) shared increased mAb806 binding versus WT (>fivefold; SI Appendix, Table S6), although some ECD mutants with significant binding displayed variable (G574V) or low (G39R, EGFRvII) basal pEGFR. Importantly, R84K+A265V displayed the strongest phosphorylation in nearly every experiment. Overall, our data suggest that N-TR1 release detected by mAb806 is necessary but not always sufficient for strong activation.
Fig. 6.
Kinase activity and growth properties in U87 GBM models are influenced by the ECD806(+)-sKD state. (A) Representative WB analysis of EGFR kinase activity (pEGFR) with a pan-anti-pTyr antibody 24 h poststarvation. Phosphorylation-variable G574V scored high in this experiment (*) but, in others, shows no activity. (B) Rank classification of constitutive activity after quantification by ImageJ: pEGFR detection with three site-specific anti-Tyr antibodies 24 h poststarvation (top rows) and with pan-P-Tyr antibody after 6, 24, and 48 h poststarvation (bottom rows). WT/S468R controls score low in all readouts, while R84K+A265V typically displays the strongest activity. G574V shows variable pEGFR levels across experiments. (C) Tumor growth rate in s.c. mouse xenografts (Top); the double mutant R84K+A265V displays the fastest growth also reaching the largest volumes (SI Appendix, Fig. S9C). Zoom (gray spheres) for early tumor volumes (Bottom): R84K+A265V generates tumors before day 15 and single mutants from day 20. Slow-growing tumors appear from day 30. G574V does not generate tumors in this experiment. (D) Representative IRFP720 image at day 14 postimplantation in an orthotopic model (Right) and Kaplan–Meier median survival curves (Right) comparing mice implanted with GBMs expressing either WT EGFR or ECD mutants (n = 6 per group, P < 0.0001). Note the high activity of R84K+A265V and its negative survival impact, in a trend relating poor survival to mAb806 levels (P < 0.0001). (E) Response to mAb806 treatment reported as ratio of the average end slope of percentage change in growth curves (Left), with mAb806 versus vehicle (PBS). Tether variants (Right) are especially sensitive. (F) Tumor-growth rate upon cotreatment with either low-dose mAb806 (n = 4) (Left) or lapatinib (n = 4) (Center) alone versus cotreatment (n = 5) (Right) in mouse xenografts. Extra- and intracellular simultaneous targeting results in regression in four out of five animals (P < 0.0001) (see also SI Appendix, Figs. S8–S9 and Table S7–S8).
Next, U87 cells were s.c. grafted onto athymic nude mice to investigate how epitope detection and pEGFR level translate to in vivo oncogenicity. With the exception of G574V, which in one experiment failed to generate tumors, KD activity translated to tumor growth, with ECD mutations growing at three distinct rates: fast (R84K+A265V), intermediate (A265V, R84K, and EGFRvIII) and low (rest of the ECD variants) (Fig. 6C and SI Appendix, Fig. S9C). Strikingly, R84K+A265V generated early s.c. tumors (before day 15; Fig. 6C) that displayed accelerated growth to large volumes (SI Appendix, Fig. S9C). From day 15, tumors harboring intermediate mAb806-binding mutations with robust phosphorylation (R84K, A265V, and EGFRvIII) appeared, significantly earlier than low or variable pEGFR mutants (G39R, EGFRvII, and G574V), which nevertheless appeared earlier (from day 30; Fig. 6C) than controls (WT and S468R) (SI Appendix, Table S7). Within deletions, EGFRvIII grouped with intermediate-growing mutations, while EGFRvII led to slow tumor growth, as observed in ref. 20. These results were recapitulated in a pathologically realistic environment upon orthotopic brain implantation. The median survival followed a similar trend (Fig. 6D and SI Appendix, Table S7 and S8), with R84K+A265V standing out with a dramatic negative impact compared with lower mAb806-binding known oncogenic mutations (R84K, A265V). Notably, the brain microenvironment enhanced low/variable pEGFR tether variants (EGFRvII and G574V), so that all ECD mutants displayed reduced survival versus WT-EGFR in relation with mAb806 (SI Appendix, Fig. S9D and Table S8). Taken altogether, these results hint that ECD806+-sKD intermediate is a functionally relevant state, which above a certain threshold, associates with strong oncogenic activation. To further prove its biological impact, we tested the effect of targeting the ECD variants with low-dose mAb806, which suppresses EGFRvIII but not low mAb806-binding WT growth (25). Importantly, we found that low dose mAb806, targeting only ∼20% of the total EGFR pool in single mutants, potently inhibited tumor growth (Fig. 6E), underscoring 806-epitope detection as a relevant phenotype for oncogenic growth. Even more remarkable, as suggested from Fig. 5C, refractive WT tumors were rendered sensitive to mAb806 upon TKI allosteric induction of the ECD806+-sKD state, so that cotreatment with lapatinib and antibody resulted in synergistic inhibition (Fig. 6F and SI Appendix, Fig. S9E) and tumor regression (P < 0.0001; SI Appendix, Table S8).
Discussion
A key aspect of the EGFR mechanism—if and how the ECD regulates KD activation—is still under debate. While some studies support that the ECD and the KD are loosely linked (26, 27), others suggest a fine-tuned coupling (28, 29). Similarly, ECD self-inhibition by the tether has been questioned based on contradictory reports on II-IV mutation effects (13, 30). Thus, the structural basis for GBM mutations’ activity and specifically I-II mutants, along with their responses to TKIs (5), is unclear. The only study to date on I-II mutants showed that they enhance ligand affinity but not ECD dimerization (15). Here, our I-II mutant simulations unexpectedly connected two research lines on the 806-transitional state (10) and ECD mutations’ TKI sensitivity (5), inspiring experiments that altogether support that increased 806-epitope exposure is the signature of a shared activation mechanism, based on N-TR1 removal from a self-inhibitory position.
In our simulations, the WT-ECD remained tethered but proved extremely flexible (Fig. 1D), transitioning between different compact states via large-scale TR motions around the central II-III linker, which acts as a hinge. This involved dramatic linker reshaping, modifying the accessibility of the nearby 806-epitope (SI Appendix, Fig. S4 C and D). I-II mutants increased this TR flexibility, favoring untethering to a “twisted” state, which happened to fulfill the features of the 806-intermediate (i.e., neither open nor closed, untethered, with an accessible 806-epitope), inferred from multiple studies (9, 13, 31). In this untethered state, the N-TR1 region, deleted in EGFRvIII, rotates away from the 806-epitope. This suggested that deletions and missense mutations structurally converge to remove this ECD fragment from its tethered position, where it could act as steric hindrance to prevent KD activation. The antibody 806 would thus be unique, by detecting a key hinge region, whose exposure hallmarks the disruption of ECD self-inhibition. From these observations, three testable hypotheses followed: (i) increased flexibility in I-II mutants favors N-TR1 motions/untethering detected by mAb806; (ii) this reposition is a convergent trait shared by other ECD mutations; and hence (iii) N-TR1 position influences the oncogenic activity, i.e., the 806-epitope could be a KD conformational reporter.
Our SAXS data confirmed MD predictions on I-II mutant versus WT-ECD flexibility (Fig. 4A), revealing also details never seen before, e.g., the presence of the tether in solution (Fig. 4D). Previous SAXS (22) and crystallography (32) had indicated that the ECD retains its closed shape upon II-IV mutations or deletions, which contributed to question the tether inhibitory role. Here, our analyses coincide that also I-II mutants remain compact, but additionally reveal that they are untethered (Fig. 4 B–E) and sample twisted and partially open states, as seen in MD. Moreover, SAXS suggests that, along with interdomain contacts, glycosylation contributes to harness WT-ECD flexibility, as shown by the tether loss after PNGase treatment (Fig. 4D).
Apart from a compact but untethered conformation, here we corroborate that I-II mutants expose the cryptic 806-epitope, in relation with in silico N-TR1 conformational freedom (Fig. 3 C–E). We further show that clinically relevant II-IV mutants share increased mAb806 binding (Fig. 5A), which naturally raises the question of whether diverse mutants also display similar KD configurations, and how this relates to activity. Besides the fully inactive tethered state of the WT-EGFR [ECD806(-)-sKD], we show that two antagonistic ECD states for mAb806 recognition exist, allosterically coupled to distinct KDs (Fig. 5C): an ECD806(+)-sKD, twisted/partially open according to SAXS, and an ECD806(-)-aKD, with a flexible ECD (33) and distinct ligand affinities (28). Based on ECD mutants’ increased mAb806 binding, along with their preference for TKI-2s (5) and specifically lapatinib, which has intermediate active/inactive features (5) (Fig. 5B), it naturally follows that ECD806(+)-sKD must be important to drive, or is related to, oncogenic activation. This conclusion is further supported by inhibition of ECD-mutant tumors at low mAb806 doses (Fig. 6E), which will target 20% or less from the total EGFR pool. Significantly, there are evidences that only for the constitutively untethered HER2, lapatinib stabilizes ligand-responsive dimers (34), associated with tumor progression and resistance (35).
While these results indicate that the sKD-like state detected by mAb806 is functionally relevant, they also bring into question whether it just hallmarks the oncogenic transition (see ref. 13) or it can signal on its own. According to our pEGFR data, N-TR1 release is necessary but not sufficient for strong basal self-phosphorylation, although invariably, 806-epitope detection translates to higher oncogenicity in vivo (Fig. 6 C and D and SI Appendix, Fig. S9D), e.g., for untested G39R, we confirm that increased 806-binding results in higher oncogenic activity versus WT-EGFR. Significantly, for R84K+A265V, the exceptionally high mAb806-binding [>40% of receptors 806(+)] translates to superoncogenicity in vivo, while lower-binding single mutants show milder phenotypes. Tether variants display however a distinctly variable behavior, likely influenced by posttranslational factors. For G574V (with two potential glycoforms, SI Appendix, Fig. S9A), pEGFR changes heavily depending, e.g., on cell confluence (Fig. 6B and SI Appendix, Fig. S8D), and, occasionally, even fails to form tumors. However, upon brain implantation, tumor growth by G574V and EGFRvII (with surprisingly low phosphorylation; Fig. 6B) is boosted, suggesting that specific ligands enhance their activity. This is in line with the mentioned contradicting reports on II-IV mutations (13, 30) and suggests that, in comparison with the I-II interface, KD control by the tether is limited. Despite these nuances, I-II and II-IV mutants converge however on their responses to TKIs (Fig. 5B) and mAb806 (Fig. 6E), the latter being strongest for tether mutants. Altogether, these findings indicate that the population of receptors detected by mAb806 is not fully active for self-phosphorylation but is indeed biologically relevant and could be involved, e.g., in noncatalytic kinase functions (36).
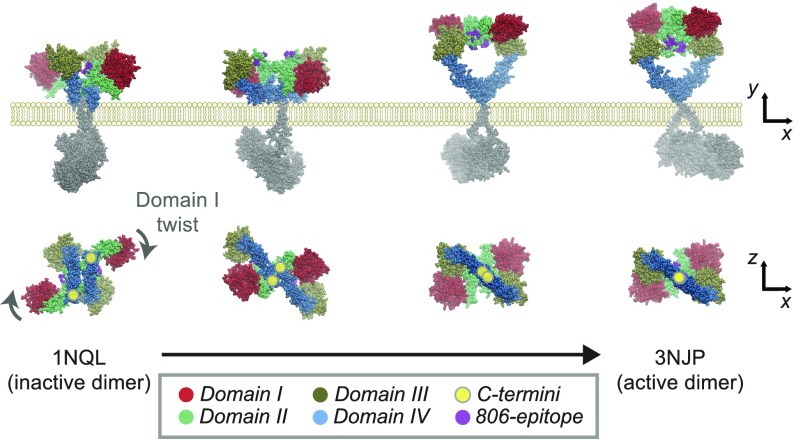
Overall, our results indicate that GBM mutations favor the release of steric constraints on the KD, revealed as a striking allosteric coupling between the ECD status, hallmarked by the 806-epitope and the KD arrangement (Fig. 5C), which is key for synergistic cotargeting (Fig. 6F). Hence, we propose that the ECD mutational pattern is consistent with a “steric” model, where N-TR1s block productive KD rearrangement, in line with that proposed for IGFR (37) or HER1 Caenorhabditis elegans homolog, LET-23 (38). Such model assumes that activation happens in preformed dimers (39) (here detected for all ECD variants; SI Appendix, Fig. S5E) can explain mutants’ dimerization-independent activity (15) and is supported by recent FRET data (40). Specifically, our data highlight N-TR1 flexibility and orientation as essential pieces to orchestrate ECD-KD coupling. Modeling of an activating transition starting from underappreciated tethered dimers (41), offers a feasible picture for this process (Fig. 7). In a self-inhibited predimer, the tether would keep the TRs in a rigid, 806-(-) conformation, acting as “spacers” to hold the ECD C-terminal apart. Increased flexibility (e.g., deglycosylation, mutation) “uncouples” the TRs (Fig. 3C), allowing N-TR1 rotation as KDs rearrange. While N-TR1–driven activation could happen without full untethering, factors restraining ECD motions (e.g., glycans) could inhibit the expression of weaker (tether) mutations. It is of note however that the greater presence of ECD mutant dimers in native conditions (SI Appendix, Fig. S8B) cannot discard a role for ECD interactions in stabilizing unbound dimers.
Fig. 7.
A plausible model for ECD activation by N-TR1 rotation in I-II mutations. Modeling of the transition from the tethered X-ray inactive dimer (1NQL) to the intermediate dimer up to reach the open dimer (3NJP; ligands have been removed) shows domain I twisting (row below) and transient epitope exposure as domain IV C termini (highlighted with yellow circles) slide and rotate to allow the rearrangement of the KD into the active conformation. Note how the 806-epitope becomes transiently accessible as the transition proceeds and is buried again when reaching the fully open state. The pathway has been generated with the coarse-grained algorithm eBDIMS (elastic-network driven Brownian dynamic important sampling; see refs. 18 and 19).
Convergent evolution drives random variations under a selection pressure to independently achieve equivalent solutions, creating analogous structures with similar form or function, and emerging evidences support its importance in cancer (42). Before our study, mAb806 was known to bind EGFRvIII, along with designed tether mutations and natural II-III mutants (43), but was not tested against I-II variants due to their unclear structural impact. Our study, together with these former evidences, supports an extracellular convergence, detected by mAb806, for GBM variants regardless of their class—deletions or missense changes, at any interface (Table 1). Moreover, we show that for the major mutations, this ECD806(+) phenotype is allosterically coupled to the KD, relevant for function and for drug sensitivity. Contrary to KD mutations, which favor the aKD state (44), we propose that ECD mutants remove a steric hindrance to favor an sKD-like state. From an evolutionary perspective, the tissue-specific accumulation of mutations at either the ECD or KD, together with their differential TKI sensitivities (5) or signaling profiles (2), are themselves markers of convergence. Since distinct KD-JM arrangements are linked to different signaling pathways (29), ECD806(+)-sKD convergence could thus reflect a selective pressure to favor specific routes. Under this view, the often puzzling diversity of ECD mutants within the same tumor (6), which can harbor multiple deletions, point variants (e.g., EGFRvIII, R84K, etc.; SI Appendix, Table S9), amplification, or even independent clonal copies of a mutation (EGFRvII) (20), is simplified to an essential EGFR biochemical equivalence in terms of their responses to state-specific drugs like mAb806 (Fig. 6E) and TKIs (Fig. 5B). Currently, EGFR amplification and EGFRvIII are the primary biomarkers for mAb806 trials (10). Growing evidences indicate that the sole presence of EGFR-KD mutations in a tumor predicts positive responses to anti-EGFR therapy (45). If the same holds true for ECD mutations, 806-convergence will open a wider application spectrum of mAb806 than ever suggested (17), and together with TKI-driven ECD conversion (Figs. 5C and 6F), will pave a rational basis for extraintracellular EGFR targeting.
Table 1.
Structural convergence of clinical ECD mutations detected by mAb806 epitope exposure
| Mutation type | ECD variant | Structural effect | mAb806 | Oncogenic activity |
| Deletion | EGFRvII | Tether deletion | 24-fold | Confirmed |
| EGFRvIII | TR1 deletion | 6-fold | Confirmed | |
| Missense | I-II interface G39R R84K A265V | TR1 flexibility/untethering | 3- to 5-fold | Confirmed |
| R84K+A265V | Enhanced TR1 flexibility | 13-fold | Enhanced oncogenicity | |
| II-III interface R300L E306K | TR1 flexibility/untethering | Increase | Confirmed (43) | |
| II-IV interface G574V | TR1 flexibility/untethering | Increase | Confirmed |
Materials and Methods
Full method details are in SI Appendix, Supplementary Materials and Methods.
Molecular Modeling.
Studies of the EGFR ECD were based on the PDB ID codes 1NQL and 3NJP, representing the closed and open conformations (Glu3-Thr614). Residues are numbered as on the mature receptor (as per common use in structural studies) instead of the initiating Met; note the 24-residue shift between schemes. See the summary of ENM data and MD trajectories in SI Appendix, Table S2.
SAXS Analysis of WT and Mutant ECDs.
All variants of the human EGFR ECD (residues 1–618) were expressed in HEK293 cells and purified at the Institute for Research in Biomedicine Protein Expression Facility. SAXS synchrotron data were collected at European Synchrotron Radiation Facility (ESRF) for glyco-ECDs and at PETRA III storage ring (DESY) for PNGase-treated ECDs. Data were processed with the ATSAS suite using four independent methods (SI Appendix, Fig. S6D). For detailed information, see SI Appendix, Supplementary Materials and Methods.
HEK293 Experiments.
All point mutations and deletions were introduced into expression vector pcDNA6A-EGFR-WT (no. 42665; Addgene). HEK293 cells were obtained from the American Type Culture Collection, maintained in DMEM and transfected, collected, and labeled for FACS using mAb806 and fluorescein-conjugated goat anti-mouse IgG (AlexaFluor 568). LA22Ab, which targets DIII (351–364), was used to measure total receptor. For immunocytochemistry, cells were transfected, fixed, and incubated with mAb806, and DNA was stained with DAPI.
U87 Glioblastoma Model Experiments.
U87 parental cells were retrovirally transfected and selected, and stable clones were verified by WB and FACS. To measure binding affinity, cells were stained separately with mAb528 and mAb806, and mean FITC intensity was normalized to the corresponding mean mAb528 FITC intensity. To address whether TKI treatment will enhance 806-epitope exposure, TKIs were tested with a cell survival assay (Fig. 5B), and then cells were pretreated with the minimal TKI concentration causing a 90% reduction of cell viability before staining with mAb806 and FACS.
WB Kinase Phosphorylation Assays.
U87 mutant cells were serum-starved for 24 h (occasionally 6 or 48 h) and treated with or without EGF. Protein concentration was determined using a standard detergent-compatible assay (Bio-Rad), and equal amounts were loaded for WB detection with anti-pTyr antibodies. ImageJ software was used to quantify band signal intensity normalized to total protein (actin bands). To compare pEGFR across gels (Fig. 6B), the ECD-variant benchmark values were ranked from highest to lowest (1–9) for each experiment.
In Vivo Tumorigenicity Assays with U87 GBM Models.
Animal experiments were performed in accordance with the Institutional Animal Care and Use Committee of the University of California, San Diego; 4- to 6-wk-old female athymic nu/nu mice were used for the s.c. tumor studies, and 6- to 8-wk-old female athymic nu/nu mice for intracranial tumor studies (Charles River Laboratories; The Jackson Laboratory) (n = 4). For intracranial orthotopic models (Fig. 6E), EGFR variants were tagged with fluorescent protein IRFP720. Fluorescence molecular tomography was used for brain tumor visualization. Log-rank tests for survival were done with Stat14.2 and GraphPad (SI Appendix, Table S8).
Intervention Models with mAb806.
Female athymic nu/nu mice (6–8 wk of age) were injected with U87 cells/100 μL PBS. When tumors reached 100 mm3, animals were treated with either PBS control or low-dose mAb806 (0.1 mg/mouse) 3×/wk, for 2–3 wk (n = 4). For combination studies, animals were treated for 2 wk with either low-dose mAb806, oral lapatinib (n = 4), or both (n = 5). For combination groups, both drugs were administered at the same time. The effect of low-dose mAb806 treatment (Fig. 6E) was evaluated by computing the average slope, S, of the curves at the end of the treatments (SI Appendix, Supplementary Material and Methods). Student t testing with GraphPad was used to compare tumor-growth curves for control versus treatment (SI Appendix, Table S8).
Supplementary Material
Acknowledgments
L.O. thanks Dr. K. M. Ferguson for kindly providing 1NQL dimer coordinates. We thank the financial support of the Ministry of Economy and Competitiveness, the Catalan Institution for Research and Advanced Studies, and Generalitat de Catalunya (M.O.); the European Research Council (M.O. and E.L.); the Vetenskapsrådet and Swedish e-Science Research Center (E.L.); and the National Brain Tumor Society, NIH Grant R01-NS080939, and James S. McDonnell Foundation (F.B.F.). Calculations were run at the Barcelona Supercomputing Center and Swedish National Infrastructure for Supercomputing, with support by H2020 BioExcel and Elixir-Accelerate. P.B. acknowledges funds from Labex, EpiGenMed, and the “Investissements d’avenir” program (Grant ANR-10-LABX-12-01). L.O. thanks support from the Sven and Lilly Lawskis Foundation, A.H.T. from the National Cancer Institute (Grant 2T32CA009523-29A1), A.D.P. from the NIH (Grant T32GM008666), T.N.C. from MostMicro (Grant LISBOA-01-0145-FEDER-007660), and A.M.S. from the National Health and Medical Research Council (Grant APP1084178). This content is solely the responsibility of the authors and does not necessarily represent the views of the NIH. We acknowledge the use of the European Molecular Biology P12 and the European Synchrotron Radiation Facility Bm29 BioSAXS beamlines.
Footnotes
Conflict of interest statement: A.M.S. is an inventor on patents related to mAb806 and has received research support from Abbvie, the National Health and Medical Research Council, and the Cancer Council Victoria. A.M.S. is also a consultant with Life Science Pharmaceuticals. A.M.S., F.B.F., and W.K.C. are inventors on US Patent 9,023,356 on a treatment method using EGFR antibodies and src inhibitors in related formulations.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821442116/-/DCSupplemental.
References
- 1.Hubbard SR, Miller WT. Receptor tyrosine kinases: Mechanisms of activation and signaling. Curr Opin Cell Biol. 2007;19:117–123. doi: 10.1016/j.ceb.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wykosky J, Fenton T, Furnari F, Cavenee WK. Therapeutic targeting of epidermal growth factor receptor in human cancer: Successes and limitations. Chin J Cancer. 2011;30:5–12. doi: 10.5732/cjc.010.10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2:re6, and erratum (2009) 2:er7. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 4.Leahy DJ. Structure and function of the epidermal growth factor (EGF/ErbB) family of receptors. Adv Protein Chem. 2004;68:1–27. doi: 10.1016/S0065-3233(04)68001-6. [DOI] [PubMed] [Google Scholar]
- 5.Vivanco I, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2:458–471. doi: 10.1158/2159-8290.CD-11-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer. 2015;15:302–310. doi: 10.1038/nrc3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott AM, et al. A phase I clinical trial with monoclonal antibody ch806 targeting transitional state and mutant epidermal growth factor receptors. Proc Natl Acad Sci USA. 2007;104:4071–4076, and erratum (2007) 104:15965. doi: 10.1073/pnas.0611693104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jungbluth AA, et al. A monoclonal antibody recognizing human cancers with amplification/overexpression of the human epidermal growth factor receptor. Proc Natl Acad Sci USA. 2003;100:639–644. doi: 10.1073/pnas.232686499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johns TG, et al. Identification of the epitope for the epidermal growth factor receptor-specific monoclonal antibody 806 reveals that it preferentially recognizes an untethered form of the receptor. J Biol Chem. 2004;279:30375–30384. doi: 10.1074/jbc.M401218200. [DOI] [PubMed] [Google Scholar]
- 10.Gan HK, Burgess AW, Clayton AH, Scott AM. Targeting of a conformationally exposed, tumor-specific epitope of EGFR as a strategy for cancer therapy. Cancer Res. 2012;72:2924–2930. doi: 10.1158/0008-5472.CAN-11-3898. [DOI] [PubMed] [Google Scholar]
- 11.Johns TG, et al. The antitumor monoclonal antibody 806 recognizes a high-mannose form of the EGF receptor that reaches the cell surface when cells over-express the receptor. FASEB J. 2005;19:780–782. doi: 10.1096/fj.04-1766fje. [DOI] [PubMed] [Google Scholar]
- 12.Johns TG, et al. Antitumor efficacy of cytotoxic drugs and the monoclonal antibody 806 is enhanced by the EGF receptor inhibitor AG1478. Proc Natl Acad Sci USA. 2003;100:15871–15876. doi: 10.1073/pnas.2036503100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker F, et al. CR1/CR2 interactions modulate the functions of the cell surface epidermal growth factor receptor. J Biol Chem. 2004;279:22387–22398. doi: 10.1074/jbc.M401244200. [DOI] [PubMed] [Google Scholar]
- 14.Lee JC, et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006;3:e485. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bessman NJ, Bagchi A, Ferguson KM, Lemmon MA. Complex relationship between ligand binding and dimerization in the epidermal growth factor receptor. Cell Rep. 2014;9:1306–1317. doi: 10.1016/j.celrep.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orozco M, et al. Coarse-grained representation of protein flexibility. Foundations, successes, and shortcomings. Adv Protein Chem Struct Biol. 2011;85:183–215. doi: 10.1016/B978-0-12-386485-7.00005-3. [DOI] [PubMed] [Google Scholar]
- 17.Binder ZA, et al. Epidermal growth factor receptor extracellular domain mutations in glioblastoma present opportunities for clinical imaging and therapeutic development. Cancer Cell. 2018;34:163–177.e7. doi: 10.1016/j.ccell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orellana L, Yoluk O, Carrillo O, Orozco M, Lindahl E. Prediction and validation of protein intermediate states from structurally rich ensembles and coarse-grained simulations. Nat Commun. 2016;7:12575. doi: 10.1038/ncomms12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orellana L, Gustavsson J, Bergh C, Yoluk O, Lindahl E. eBDIMS server: Protein transition pathways with ensemble analysis in 2D-motion spaces. Bioinformatics. February 19, 2019 doi: 10.1093/bioinformatics/btz104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francis JM, et al. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014;4:956–971. doi: 10.1158/2159-8290.CD-13-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chao G, Cochran JR, Wittrup KD. Fine epitope mapping of anti-epidermal growth factor receptor antibodies through random mutagenesis and yeast surface display. J Mol Biol. 2004;342:539–550. doi: 10.1016/j.jmb.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 22.Dawson JP, Bu Z, Lemmon MA. Ligand-induced structural transitions in ErbB receptor extracellular domains. Structure. 2007;15:942–954. doi: 10.1016/j.str.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Montagut C, et al. Identification of a mutation in the extracellular domain of the epidermal growth factor receptor conferring cetuximab resistance in colorectal cancer. Nat Med. 2012;18:221–223, and erratum (2012) 18:1445. doi: 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- 24.Roskoski R., Jr Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacol Res. 2016;103:26–48. doi: 10.1016/j.phrs.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 25.Luwor RB, et al. Monoclonal antibody 806 inhibits the growth of tumor xenografts expressing either the de2-7 or amplified epidermal growth factor receptor (EGFR) but not wild-type EGFR. Cancer Res. 2001;61:5355–5361. [PubMed] [Google Scholar]
- 26.Mi L-Z, et al. Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat Struct Mol Biol. 2011;18:984–989. doi: 10.1038/nsmb.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu C, et al. Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor. Mol Cell Biol. 2010;30:5432–5443. doi: 10.1128/MCB.00742-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macdonald-Obermann JL, Pike LJ. Allosteric regulation of epidermal growth factor (EGF) receptor ligand binding by tyrosine kinase inhibitors. J Biol Chem. 2018;293:13401–13414. doi: 10.1074/jbc.RA118.004139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doerner A, Scheck R, Schepartz A. Growth factor identity is encoded by discrete coiled-coil rotamers in the EGFR juxtamembrane region. Chem Biol. 2015;22:776–784. doi: 10.1016/j.chembiol.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattoon D, Klein P, Lemmon MA, Lax I, Schlessinger J. The tethered configuration of the EGF receptor extracellular domain exerts only a limited control of receptor function. Proc Natl Acad Sci USA. 2004;101:923–928. doi: 10.1073/pnas.0307286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garrett TPJ, et al. Antibodies specifically targeting a locally misfolded region of tumor associated EGFR. Proc Natl Acad Sci USA. 2009;106:5082–5087. doi: 10.1073/pnas.0811559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu P, Bouyain S, Eigenbrot C, Leahy DJ. The ErbB4 extracellular region retains a tethered-like conformation in the absence of the tether. Protein Sci. 2012;21:152–155. doi: 10.1002/pro.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu C, Mi L-Z, Schürpf T, Walz T, Springer TA. Mechanisms for kinase-mediated dimerization of the epidermal growth factor receptor. J Biol Chem. 2012;287:38244–38253. doi: 10.1074/jbc.M112.414391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claus J, et al. Inhibitor-induced HER2-HER3 heterodimerisation promotes proliferation through a novel dimer interface. eLife. 2018;7:e32271. doi: 10.7554/eLife.32271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, Chen X, Mao Y, Qu Q, Shen K. Association of epithelial-mesenchymal transition with lapatinib resistance through multiple pathways activation in HER2-positive breast cancer. J Clin Oncol. 2014;32:e11579–e11579. [Google Scholar]
- 36.Kung JE, Jura N. Structural basis for the non-catalytic functions of protein kinases. Structure. 2016;24:7–24. doi: 10.1016/j.str.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kavran JM, et al. How IGF-1 activates its receptor. Elife. 2014;3:e03772. doi: 10.7554/eLife.03772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freed DM, Alvarado D, Lemmon MA. Ligand regulation of a constitutively dimeric EGF receptor. Nat Commun. 2015;6:7380. doi: 10.1038/ncomms8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maruyama IN. Activation of transmembrane cell-surface receptors via a common mechanism? The “rotation model”. BioEssays. 2015;37:959–967. doi: 10.1002/bies.201500041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zanetti-Domingues LC, et al. The architecture of EGFR’s basal complexes reveals autoinhibition mechanisms in dimers and oligomers. Nat Commun. 2018;9:4325. doi: 10.1038/s41467-018-06632-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferguson KM, et al. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 42.Chen H, He X. The convergent cancer evolution toward a single cellular destination. Mol Biol Evol. 2016;33:4–12. doi: 10.1093/molbev/msv212. [DOI] [PubMed] [Google Scholar]
- 43.Ymer SI, et al. Glioma specific extracellular missense mutations in the first cysteine rich region of epidermal growth factor receptor (EGFR) initiate ligand independent activation. Cancers (Basel) 2011;3:2032–2049. doi: 10.3390/cancers3022032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, et al. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat Struct Mol Biol. 2011;18:1388–1393. doi: 10.1038/nsmb.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao D, et al. The prognostic role of EGFR-TKIs for patients with advanced non-small cell lung cancer. Sci Rep. 2017;7:40374. doi: 10.1038/srep40374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.