Significance
Retinitis pigmentosa (RP) is a genetically heterogenous disease that lacks effective treatment. In RP, there is degeneration of cone photoreceptors in the eye, often leading to complete blindness. The reasons for cone degeneration remain largely unknown. Development of therapies that preserve cones and vision in patients with RP, particularly in a mutation-independent manner, would benefit from a greater understanding of the underlying pathological mechanisms. We examined immune responses in RP mice during the period of cone degeneration and identified soluble CX3CL1 (sCX3CL1) as a promising therapy for cones. Viral-mediated expression of sCX3CL1 prolonged cone survival in different RP models and improved visual function. These results establish viral delivery of sCX3CL1 as a potential treatment for RP and other retinal disorders.
Keywords: retinitis pigmentosa, cone degeneration, gene therapy, CX3CL1, microglia
Abstract
Retinitis pigmentosa (RP) is a disease that initially presents as night blindness due to genetic deficits in the rod photoreceptors of the retina. Rods then die, causing dysfunction and death of cone photoreceptors, the cell type that mediates high acuity and color vision, ultimately leading to blindness. We investigated immune responses in mouse models of RP and found evidence of microglia activation throughout the period of cone degeneration. Using adeno-associated vectors (AAVs), delivery of genes encoding microglial regulatory signals led to the identification of AAV serotype 8 (AAV8) soluble CX3CL1 (sCX3CL1) as a promising therapy for degenerating cones. Subretinal injection of AAV8-sCX3CL1 significantly prolonged cone survival in three strains of RP mice. Rescue of cones was accompanied by improvements in visual function. AAV8-sCX3CL1 did not affect rod survival, microglia localization, or inflammatory cytokine levels in the retina. Furthermore, although RNA sequencing of microglia demonstrated marked transcriptional changes with AAV8-sCX3CL1, pharmacological depletion of up to ∼99% of microglia failed to abrogate the effect of AAV8-sCX3CL1 on cone survival. These findings indicate that AAV8-sCX3CL1 can rescue cones in multiple mouse models of RP via a pathway that does not require normal numbers of microglia. Gene therapy with sCX3CL1 is a promising mutation-independent approach to preserve vision in RP and potentially other forms of retinal degeneration.
Retinitis pigmentosa (RP) is a disease of the eye that presents with progressive degeneration of rod and cone photoreceptors, the light-sensing cells of the retina (1). The disease can result from mutations in any of over 80 different genes and is the most common inherited form of blindness in the world, affecting an estimated 1 in 4,000 individuals (2–4). One approach proposed to treat RP is gene therapy, for example, using adeno-associated vectors (AAVs) to deliver a wild-type (WT) allele to complement a mutated gene (5, 6). While this approach has proven successful in other conditions, even leading to the approval of a gene therapy for RPE65-associated Leber’s congenital amaurosis (7), it is difficult to implement for the majority of patients with RP, given the extensive heterogeneity of the genetic lesions and the fact that a causal mutation is not always identified (2). A broadly applicable gene therapy that is agnostic to the genetic lesion would provide a treatment option for a greater number of patients with RP. Presently, there is no effective therapy of any kind for RP, and despite more than a dozen clinical trials to date, none have been able to demonstrate an improvement in visual function (8).
In patients with RP, there is an initial loss of rods, the photoreceptors that mediate vision in dim light. Clinically, this results in the first manifestation of RP, poor or no night vision, which usually occurs between birth and adolescence (1). Daylight vision in RP is largely normal for decades but eventually deteriorates, beginning when most of the rods have died. This is due to dysfunction and then death of the cone photoreceptors, which are essential for high acuity and color vision, and are the major source of morbidity in the disease (1). Importantly, while the vast majority of genes implicated in RP are expressed in rods, few actually exhibit expression in cones, suggesting the existence of one or more common mechanisms by which diverse mutations in rods trigger nonautonomous cone degeneration (9–11). We and others have attempted to elucidate these mechanisms with the goal of developing therapies for RP that preserve cone vision regardless of the underlying mutation (12–16).
One possible contributor to nonautonomous cone degeneration in RP that has yet to be closely examined is the body’s own immune system. As they die, many cells, including photoreceptors in RP, release damage-associated molecular patterns (DAMPs) that act as endogenous danger signals and incite inflammation (17, 18). By numerous pathways, DAMPs can then stimulate proinflammatory cytokine activity or recruit immune cells, such as neutrophils and T cells, to the site of cell death (17). Even in homeostatic conditions, the retina is continuously surveyed by microglia, resident macrophages of the central nervous system (CNS) derived from myeloid progenitors in the embryonic yolk sac (19, 20). Following injury or exposure to noxious stimuli, microglia may become activated, a state characterized by acquisition of an ameboid morphology, up-regulation of cytokines, and increased phagocytosis of cell debris (21–23). Notably, activation of microglia can also be modulated by various regulatory factors from the CNS, allowing for manipulation of these cells in both experimental models and humans (24–26).
Here, we investigated the involvement of immune responses during nonautonomous cone degeneration in mouse models of RP. We found evidence of microglia activation throughout the period of cone death. We subsequently developed four AAVs to deliver genes that target retinal microglia. One of these genes, soluble CX3CL1 (sCX3CL1), also called fractalkine or neurotactin, significantly prolonged cone survival in three different mouse models of RP. Rescue of cones was accompanied by improvements in visual function, highlighting the potential of sCX3CL1 as a mutation-independent treatment for RP and other retinal diseases.
Results
Microglia Reside in the Photoreceptor Layer Throughout Cone Degeneration.
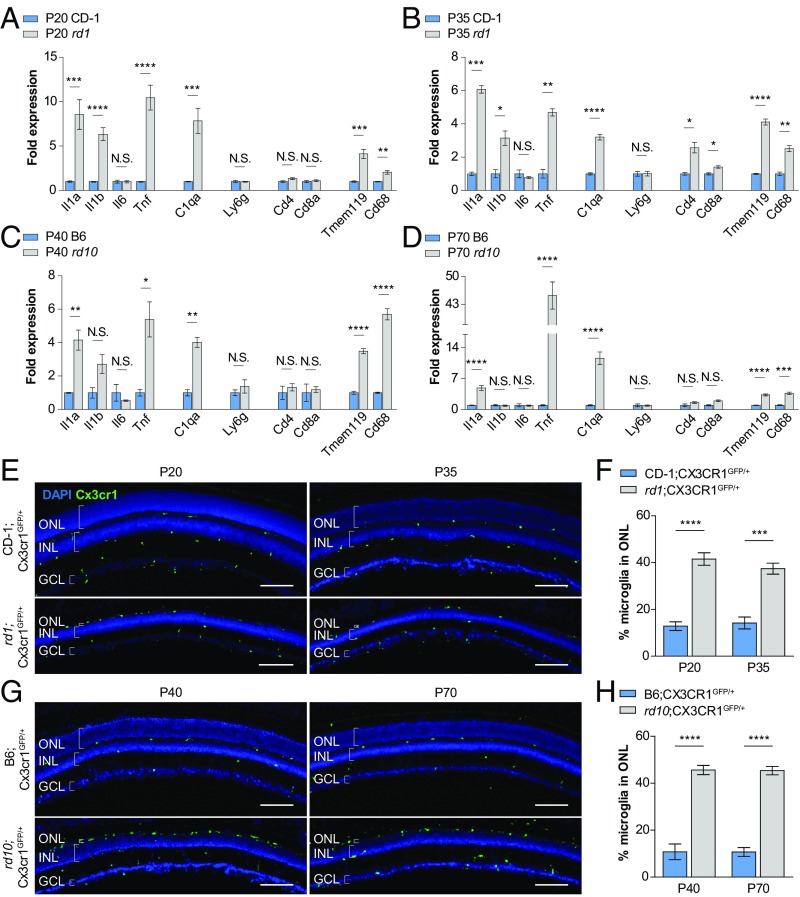
The rd1 and rd10 mouse lines are commonly used models of RP (27). Each strain harbors a different mutation in the rod-specific phosphodiesterase β-subunit, with rd1 exhibiting more rapid photoreceptor degeneration than rd10 (28). To characterize immune activity during nonautonomous cone degeneration, we first performed RT-PCR on retinas from albino rd1 and pigmented rd10 mice, as well as those from albino CD-1 and pigmented C57BL/6 (B6) mice, two strains with WT vision. Primers were designed to assay for RNAs of both innate and adaptive immune components, including inflammatory cytokines (Il1a, Il1b, Il6, and Tnf), complement (C1qa), neutrophils (Ly6g), T cells (Cd4 and Cd8a), and microglia (Tmem119 and Cd68). In each strain, two distinct time points were examined, corresponding to the onset [postnatal day 20 (P20) for rd1 and P40 for rd10] and peak (P35 for rd1 and P70 for rd10) of cone degeneration (12, 13). Compared with age-matched CD-1 and B6 retinas, rd1 and rd10 retinas demonstrated significant up-regulation of Il1a, Tnf, and C1qa at both time points, as well as Il1b specifically in rd1 mice (Fig. 1 A–D). Up-regulation of these factors also was associated with higher expression levels of Tmem119, a microglia-specific marker (29), and Cd68, a marker of lysosomal activity and microglia activation (30), but not Ly6g, Cd4, or Cd8a. As activated microglia have been previously shown to produce and secrete IL-1A, TNF, and C1Q (23), these data pointed to a possible proinflammatory role of microglia during nonautonomous cone death.
Fig. 1.
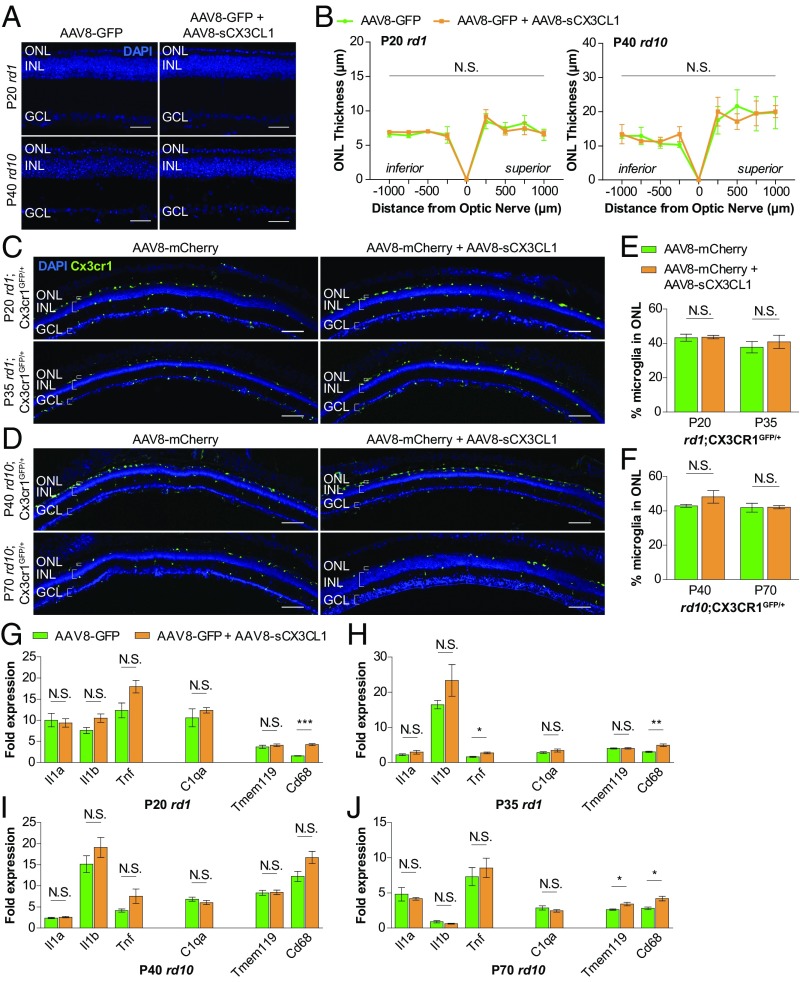
Expression of immune response genes and microglia localization during cone photoreceptor degeneration. (A–D) Whole-retina RNA expression levels of immune response genes during the onset (P20 and P40) and peak (P35 and P70) of cone degeneration in two RP mouse models (albino rd1 and pigmented rd10) versus two WT strains (albino CD-1 and pigmented B6). (E and G) Retinal cross-sections from RP and WT mice depicting Cx3cr1GFP-labeled microglia during cone degeneration. (Scale bars: 100 μm.) (F and H) Quantification of the percentage of total retinal microglia residing in the ONL during cone degeneration in RP and WT mice. Data are shown as mean ± SEM (n = 4–6 animals per condition). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by two-tailed Student’s t test with Bonferroni correction (A–D) and two-tailed Student’s t test (F and H). GCL, ganglion cell layer; INL, inner nuclear layer; N.S., not significant.
Rods in mice, and in humans, are far more abundant than cones, representing ∼95% of photoreceptors (31, 32). In the retina, rod and cone cell bodies form a structure called the outer nuclear layer (ONL), which dramatically shrinks with rod death in RP until only a single row of cells remains, comprising the surviving cones. Pathological infiltration of microglia into the ONL has been described during the initial rod death phase of RP (33, 34). However, less is known about how microglia behave during the subsequent period of cone degeneration. To aid in visualizing microglia within the retina, RP and WT animals were bred with Cx3cr1GFP reporter mice, in which microglia are labeled with GFP (35). Only ∼10% of retinal microglia were normally located in the ONL in cross-sections from CD-1;Cx3cr1GFP/+ and B6;Cx3cr1GFP/+ WT eyes (Fig. 1 E–H). Conversely, ∼40–50% of retinal microglia could be seen in the ONL in rd1;Cx3cr1GFP/+ and rd10;Cx3cr1GFP/+ mice throughout the period of cone degeneration. Thus, even after the disappearance of rods, the period of cone degeneration had both cytokine up-regulation and continued localization of microglia to the photoreceptor layer.
Overexpression of sCX3CL1 Prolongs Cone Survival in RP Mice.
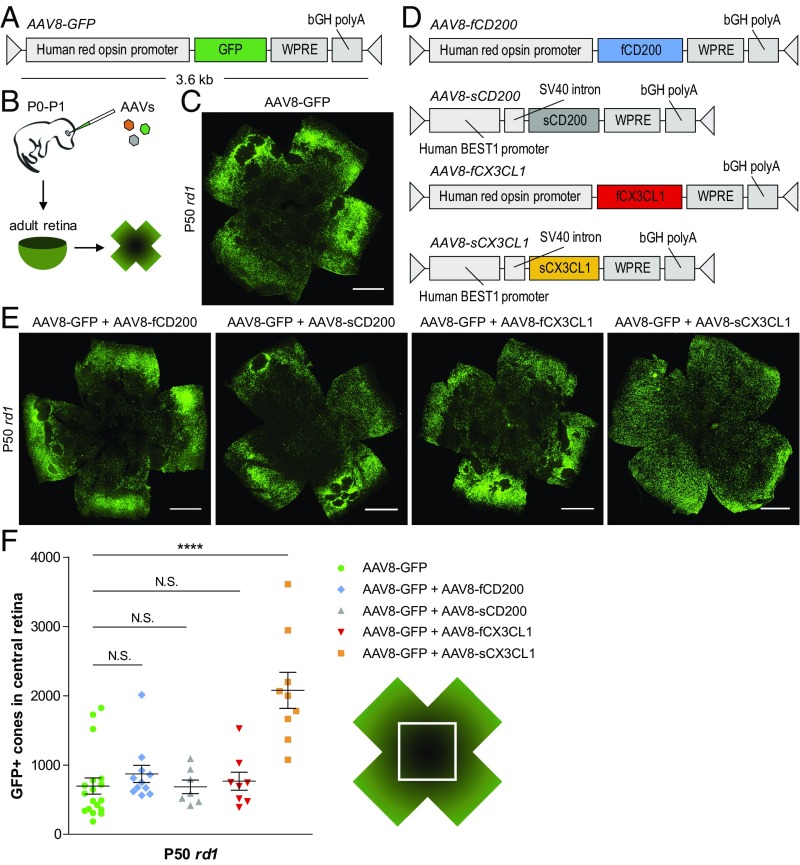
We hypothesized that during the later stages of RP, activated microglia may create a proinflammatory environment deleterious to nearby cones. We further postulated that overexpression of factors opposing microglia activation might alleviate this damage, favoring cone survival. To test this idea, a previously characterized AAV expressing GFP under the human red opsin promotor [AAV serotype 8 (AAV8)-GFP] was chosen to label cones and aid in their quantification (36) (Fig. 2A). Subretinal delivery of AAV8-GFP in neonatal mice led to brightly labeled cones throughout the entire retina, allowing for visualization of these cells in adult animals (Fig. 2B and SI Appendix, Fig. S1). AAVs were then designed to express either CD200 or CX3CL1, membrane-bound proteins reported to suppress proinflammatory activity via their respective receptors on microglia, CD200 receptor and CX3C receptor 1 (CX3CR1) (24–26). Given the hypothesized interaction between microglia and degenerating cones, full-length variants of CD200 (fCD200) and CX3CL1 (fCX3CL1) were expressed under the cone-specific human red opsin promoter (Fig. 2D). Because soluble variants of both proteins have also been described (37, 38), additional AAVs were generated for soluble CD200 (sCD200) and sCX3CL1 using the human Best1 promoter to drive expression in the retinal pigment epithelium (RPE), a cell layer adjacent to the photoreceptors (39) (SI Appendix, Fig. S2).
Fig. 2.
Effect of CD200 and CX3CL1 overexpression on cone survival. (A and B) Schematics of AAV8-GFP vector and delivery. (C) Flat-mounted P50 rd1 retina infected at P0–P1 with AAV8-GFP. (Scale bar: 1 mm.) (D) Schematics of CD200 and CX3CL1 AAV vectors. (E) Flat-mounted P50 rd1 retinas infected at P0–P1 with the indicated AAVs. (Scale bars: 1 mm.) (F) Quantification of cone survival in the central retina of P50 rd1 retinas infected with the indicated AAVs. Data are shown as mean ± SEM (n = 7–18 animals per condition). ****P < 0.0001 by two-tailed Student’s t test with Bonferroni correction. N.S., not significant.
The ability of the four AAVs (AAV8-fCD200, AAV8-sCD200, AAV8-fCX3CL1, and AAV8-sCX3CL1) to prolong cone survival was initially tested in rd1 mice, which were injected at P0–P1 and evaluated at P50. In mouse RP, cone death proceeds from the center to the periphery starting from the optic nerve head. To assay cone survival during degeneration, the central retina was therefore interrogated. Using an ImageJ module, the number of GFP-positive cones in the central retina could be reliably quantified (SI Appendix, Fig. S3). Compared with rd1 retinas infected with AAV8-GFP alone (Fig. 2C), there was no significant improvement in cone survival with the addition of AAV8-fCD200, AAV8-sCD200, or AAV8-fCX3CL1 (Fig. 2 E and F). In contrast, coinfection of AAV8-GFP with AAV8-sCX3CL1 significantly increased the number of cones remaining in the central retina (P < 0.0001), supporting a potential therapeutic effect of sCX3CL1 in RP.
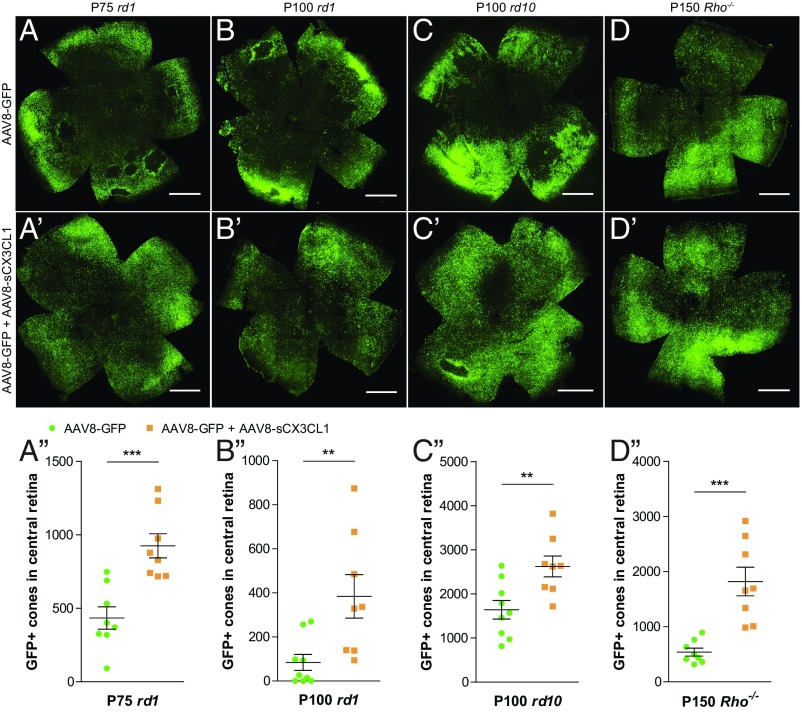
Cone survival with AAV8-sCX3CL1 was also examined in older, more degenerated rd1 mice. At P75, coinfection of AAV8-GFP with AAV8-sCX3CL1 continued to prolong cone survival compared with AAV8-GFP alone (P < 0.001) (Fig. 3 A–A′′). Even at P100, by which time the central retinas of AAV8-GFP–infected eyes were nearly devoid of cones, significantly greater cone survival with AAV8-sCX3CL1 was observed (P < 0.01) (Fig. 3 B–B′′). To determine if sCX3CL1 might provide a generic therapy for nonautonomous cone death, AAV8-sCX3CL1 was trialed in rd10 (Fig. 3 C–C′′) and Rho−/− (Fig. 3 D–D′′) mice. Rho−/− mice lack rhodopsin, the photopigment gene in rods, which is also the most commonly mutated gene in humans with autosomal dominant RP (40). Photoreceptors in the Rho−/− strain degenerate at a slower rate than in the rd1 or rd10 strain (41, 42). In rd10 and Rho−/− mice, AAV8-GFP plus AAV8-sCX3CL1 again resulted in a higher number of cones in the central retina compared with AAV8-GFP alone (P < 0.01 for rd10 and P < 0.001 for Rho−/−).
Fig. 3.
Effect of AAV8-sCX3CL1 on long-term cone survival in RP mouse models. (A–D′) Flat-mounted P75 rd1 (A and A′), P100 rd1 (B and B′), P100 rd10 (C and C′), and P150 Rho−/− (D and D′) retinas infected at P0–P1 with AAV8-GFP alone or AAV8-GFP plus AAV8-sCX3CL1. (Scale bars: 1 mm.) (A′′–D′′) Quantification of cone survival in the central retina of P75 rd1 (A′′), P100 rd1 (B′′), P100 rd10 (C′′), and P150 Rho−/− (D′′) retinas. Data are shown as mean ± SEM (n = 7–9 animals per condition). **P < 0.01; ***P < 0.001 by two-tailed Student’s t test.
AAV8-sCX3CL1 Improves Cone-Mediated Visual Function.
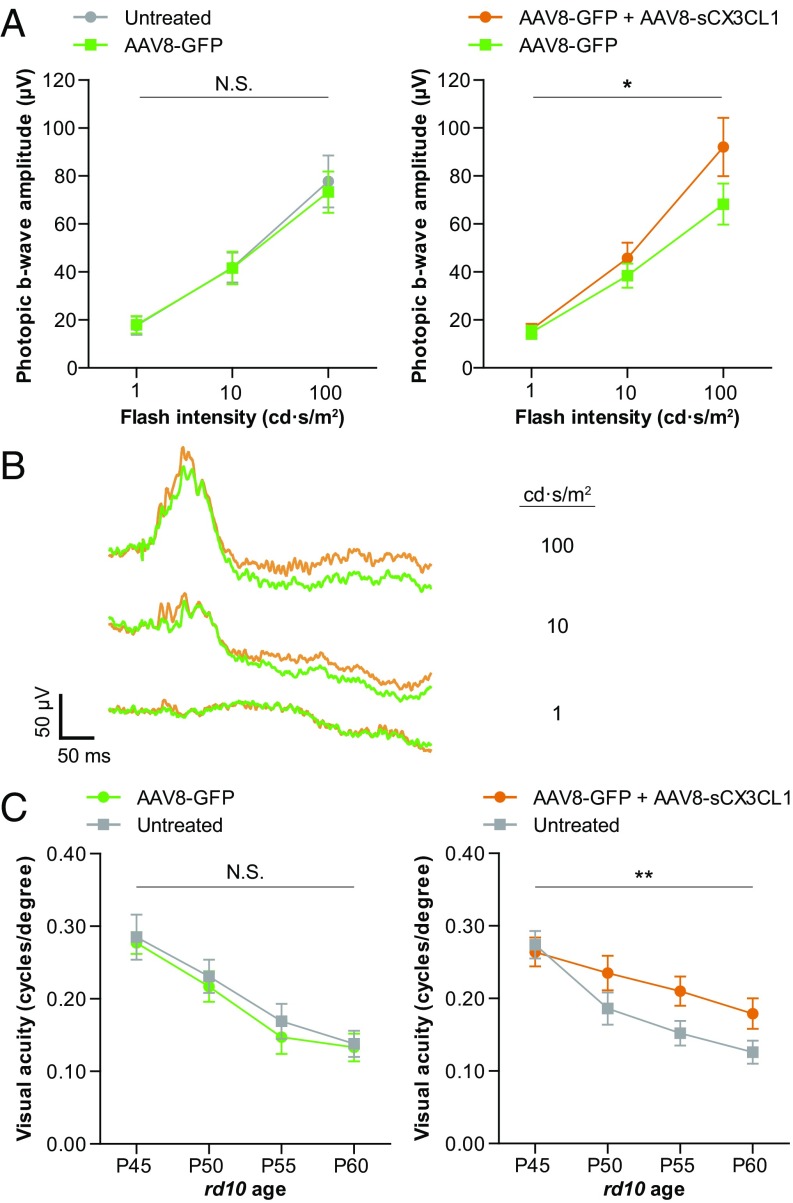
As preservation of cones by AAV8-sCX3CL1 was observed using histological assays, it was possible that vision also was rescued. Electroretinography (ERG), a physiological measure of retinal activity in response to light, can be used to reveal rod or cone activity. ERG was first used to measure photopic b-wave responses, a cone-mediated signal from the inner retina known to decline relatively early in RP in both animals and humans (1, 13). ERG recordings from P40 rd10 mice showed no difference in photopic b-waves between AAV8-GFP–infected and untreated eyes, as expected (Fig. 4A). In contrast, in animals treated with AAV8-GFP plus AAV8-sCX3CL1 in one eye and AAV8-GFP in the other, a modest but significant increase in photopic b-wave amplitudes could be seen (P < 0.05) (Fig. 4 A and B).
Fig. 4.
Effect of AAV8-sCX3CL1 on cone-mediated visual function. (A) Photopic ERG responses in P40 rd10 mice infected at P0–P1 with AAV8-GFP in one eye only (n = 12) or AAV8-GFP in one eye and AAV8-GFP plus AAV8-sCX3CL1 in the contralateral eye (n = 17). (B) Representative photopic ERG traces from a P40 rd10 animal infected with AAV8-GFP in one eye (green) and AAV8-GFP plus AAV8-sCX3CL1 in the contralateral eye (orange). (C) Optomotor assessments of visual acuity in rd10 mice at the indicated ages compared with contralateral uninjected eyes after infection with AAV8-GFP (n = 20) or AAV8-GFP plus AAV8-sCX3CL1 (n = 21). Data are shown as mean ± SEM. *P < 0.05; **P < 0.01 by two-tailed two-way ANOVA. N.S., not significant.
To evaluate vision using a behavioral test, the optomotor assay was used. This assay elicits a motor response to simulated motion, that of moving stripes. By varying the stripe width until the animal is no longer able track the stimulus, a spatial frequency threshold can be calculated, corresponding to the visual acuity in each eye (43, 44). Mice were placed under bright-light conditions to probe cone vision. In rd10 mice infected with AAV8-GFP in one eye and untreated in the other, optomotor results from P45 to P60 showed a similar drop in visual acuity between the two eyes over time (Fig. 4C). However, when animals were infected with AAV8-GFP plus AAV8-sCX3CL1 instead of AAV8-GFP alone, loss of visual acuity over the same interval was slowed in the AAV8-sCX3CL1–treated eye (P < 0.01).
AAV8-sCX3CL1 Does Not Improve Rod Survival, Microglia Localization, or Retinal Inflammation.
Absence of CX3CL1 signaling during rod degeneration in RP mice has been shown to decrease rod survival, reduce the number of microglia in the ONL, and up-regulate levels of TNF and IL-1B in the retina (33, 45). We thus investigated the effect of AAV8-sCX3CL1 on these phenotypes to uncover possible mechanisms by which sCX3CL1 gene therapy might promote cone survival. Rods normally comprise ∼95% of cells in the ONL and are thought to support cone survival through several pathways, such as secretion of trophic factors and maintenance of a normoxic environment (15, 31, 46). To examine how sCX3CL1 treatment affected rods, the thickness of the ONL in RP retinas was measured. In P20 rd1 and P40 rd10 retinas infected with AAV8-GFP, only one to two rows of nuclei remained in the ONL (Fig. 5A), consistent with near completion of rod death by the onset of cone degeneration (12, 13). Relative to these retinas, coinfection with AAV8-sCX3CL1 did not significantly alter ONL thickness (Fig. 5 A and B). This observation demonstrated not only a lack of rod preservation by AAV8-sCX3CL1 but also that cone survival was not secondary to rescue of rods.
Fig. 5.
Effect of AAV8-sCX3CL1 on microglia localization and expression of immune response genes during cone degeneration. (A) Midperipheral retinal cross-sections from rd1 and rd10 mice infected at P0–P1 with AAV8-mCherry or AAV8-mCherry plus AAV8-sCX3CL1. (Scale bars: 50 μm.) (B) Quantification of ONL thickness in rd1 and rd10 retinas during onset of cone degeneration. Cx3cr1GFP-labeled microglia in rd1 (C) and rd10 (D) retinal cross-sections infected with AAV8-mCherry or AAV8-mCherry plus AAV8-sCX3CL1 are shown. (Scale bars: 100 μm.) (E and F) Quantification of microglia residing in the ONL during cone degeneration with or without AAV8-sCX3CL1. (G–J) Whole-retina RNA expression levels of immune response genes during the onset (G and I) and peak (H and J) of cone degeneration with and without AAV8-sCX3CL1. Fold changes are relative to age-matched WT retinas. Data are shown as mean ± SEM (n = 4–6 animals per condition). *P < 0.05; **P < 0.01; ***P < 0.001 by two-tailed two-way ANOVA (B), two-tailed Student’s t test (D and F), or two-tailed Student’s t test with Bonferroni correction (G–J). GCL, ganglion cell layer; INL, inner nuclear layer; N.S., not significant.
We next asked how microglia responded to sCX3CL1 therapy by comparing retinas from rd1;Cx3cr1GFP/+ (Fig. 5C) and rd10;Cx3cr1GFP/+ (Fig. 5D) mice with and without AAV8-sCX3CL1. As use of AAV8-GFP in these animals complicated visualization of GFP-expressing microglia, an analogous AAV8-mCherry virus was generated for control infections. During both the onset and peak of cone degeneration in eyes receiving only AAV8-mCherry, ∼40% of retinal microglia could be found in the ONL, indicating continued localization of these cells to the vicinity of cones (Fig. 5 E and F). This percentage remained unchanged with the addition of AAV8-sCX3CL1, arguing against a role of sCX3CL1 in reducing microglial residence in the ONL.
Finally, given up-regulation of inflammatory cytokines and complement during the period of nonautonomous cone death, the effect of AAV8-sCX3CL1 on these factors was evaluated by RT-PCR. For the majority of genes tested, including Il1a, Il1b, C1qa, and Tmem119, expression levels in the retina were similar in eyes infected with AAV8-GFP or AAV8-GFP plus AAV8-sCX3CL1 (Fig. 5 G–J). The notable exception was Cd68, a microglia activation marker that was up-regulated with AAV8-sCX3CL1 throughout cone degeneration (30). Collectively, these data challenged the notion that AAV8-sCX3CL1 attenuates complement and inflammatory cytokine activity in the retina. Moreover, they showed that even with massive rod death, microglia in the ONL, and ongoing inflammation in the eye, AAV8-sCX3CL1 was still able to prolong cone survival.
AAV8-sCX3CL1 Induces Markers of Microglia Activation.
To probe for gene expression changes in microglia that might be brought about by AAV8-sCX3CL1, RNA sequencing (RNA-seq) of retinal microglia from AAV8-sCX3CL1–infected eyes was performed. Flow cytometry of RP retinas carrying the Cx3cr1GFP transgene indicated that microglia corresponded to a CD11b+ Ly6G/Ly6C− population in the retina (SI Appendix, Fig. S4), consistent with earlier studies (47, 48). Using these cell surface markers, retinal microglia from rd10 mice infected with AAV8-GFP or AAV8-GFP plus AAV8-sCX3CL1 were sorted at P70 during the peak of cone degeneration. Sorted microglia were a highly purified population, expressing microglia-specific genes, such as Fcrls, P2ry12, and Tmem119, but not markers for other retinal cell types compared with nonmicroglia (CD11b− Ly6G/Ly6C− and CD11b− Ly6G/Ly6C+) cells (49–56) (SI Appendix, Fig. S5).
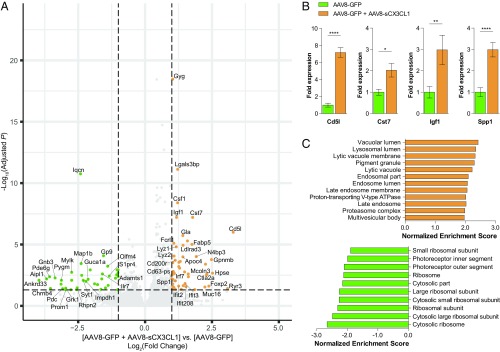
RNA-seq analysis of sorted microglia from P70 rd10 retinas infected with AAV8-sCX3CL1 demonstrated significant (adjusted P < 0.05, fold change ≥2) up-regulation and down-regulation of 50 and 40 genes, respectively (Fig. 6A and SI Appendix, Tables S2 and S3). Four of these expression changes were validated by RT-PCR on independent samples (Fig. 6B). Among the genes up-regulated with AAV8-sCX3CL1 were known markers of microglia activation during neurodegeneration, including Cst7, Spp1, Igf1, Csf1, Lyz2, Cd63-ps, and Gpnmb (57–60). In support of this, gene set enrichment analysis (GSEA) of microglia with AAV8-sCX3CL1 revealed significant enrichment of lysosome components (Fig. 6C), a prominent feature of activated microglia (30, 34, 61). Interestingly, low levels of multiple cone-specific genes, such as Aipl1, Chrnb4, and Gnb3, were observed in microglia from AAV8-GFP–treated retinas (62–64), potentially due to phagocytosis of dying cones or cone fragments. Fewer of these transcripts were detected in microglia from AAV8-sCX3CL1–treated retinas, hinting that sCX3CL1 might affect the digestion of phagocytosed materials.
Fig. 6.
Transcriptional profiling of retinal microglia during cone degeneration following sCX3CL1 overexpression. (A) Volcano plot of up-regulated and down-regulated genes from P70 rd10 retinal microglia following infection at P0–P1 with AAV8-GFP or AAV8-GFP plus AAV8-sCX3CL1. Dotted lines indicate adjusted P < 0.05 and magnitude of fold change ≥2. Full gene lists are provided in SI Appendix, Tables S2 and S3. (B) RT-PCR validation of gene expression changes in P70 rd10 retinal microglia with AAV8-sCX3CL1. (C) GSEA of P70 rd10 microglia from retinas infected with AAV8-GFP plus AAV8-sCX3CL1 (orange) compared with AAV8-GFP alone (green). Gene sets with a family-wise error rate <0.05 are displayed. Data are shown as mean ± SEM (n = 7–8 animals per condition). *P < 0.05; **P < 0.01; ****P < 0.0001 by two-tailed Student’s t test.
Normal Numbers of Microglia Are Not Necessary for Cone Rescue with AAV8-sCX3CL1.
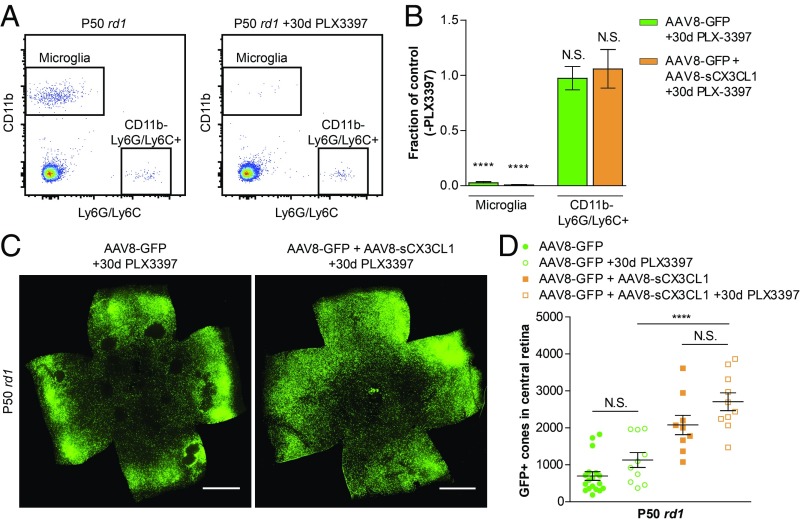
In healthy eyes, CX3CR1, the only known receptor for CX3CL1, is thought to be specifically expressed by microglia (65). This fact and the above RNA-seq data prompted us to ask what effect ablation of microglia might have on cone survival. In addition, we were curious if microglia were necessary for the rescue of cones by AAV8-sCX3CL1. It is possible to pharmacologically deplete microglia using PLX3397, a potent colony-stimulating factor 1 receptor (CSF1R) inhibitor (23, 66). To this end, rd1 mice were fed PLX3397, and depletion of retinal microglia was assessed using flow cytometry. PLX3397 treatment led to ∼99% depletion of microglia after 30 d (Fig. 7 A and B). To determine if reduction in microglia prolonged cone survival, and to test whether the activity of AAV8-sCX3CL1 in preserving cones required microglia, rd1 mice were infected with AAV8-GFP with or without AAV8-sCX3CL1 and administered PLX3397 for 30 d during the period of cone degeneration. Depletion of microglia nonsignificantly (P > 0.05) increased cone survival in both conditions (Fig. 7 C and D). Moreover, depletion of microglia did not abrogate the ability of AAV8-sCX3CL1 to rescue cones (P < 0.0001).
Fig. 7.
Effect of microglia depletion on AAV8-sCX3CL1 cone rescue. (A) Representative flow cytometry gating of microglia (CD11b+ Ly6G/Ly6C−) and CD11b− Ly6G/Ly6C+ populations in P50 rd1 retinas with or without PLX3397 treatment from P20–P49. Panels are gated on live cells (DAPI−) following doublet exclusion. (B) Fraction of microglia and CD11b− Ly6G/Ly6C+ cells remaining relative to controls in P50 rd1 retinas infected at P0–P1 with AAV8-GFP alone or AAV8-GFP plus AAV8-sCX3CL1 after 30 d of PLX3397 treatment. Retinas from littermates without PLX3397 treatment were used as controls. (C) Flat-mounted P50 rd1 retinas from PLX3397-treated mice infected with AAV8-GFP or AAV8-GFP plus AAV8-sCX3CL1. (Scale bars: 1 mm.) (D) Quantification of cone survival in the central retina of P50 rd1 retinas from PLX3397-treated mice infected with AAV8-GFP or AAV8-GFP plus AAV8-sCX3CL1. Data are shown as mean ± SEM [n = 3–4 animals per condition (A), n = 9–18 animals per condition (D)]. ****P < 0.0001 by two-tailed Student’s t test. N.S., not significant.
Discussion
In this study, we developed a gene therapy vector, AAV8-sCX3CL1, that prolonged cone survival in three different RP mouse models and delayed the loss of cone-mediated vision. Preservation of cones with AAV8-sCX3CL1 occurred despite elevated cytokine levels in the retina, and despite the continued presence of microglia in the ONL. Depletion of up to ∼99% of microglia during cone degeneration nonsignificantly improved cone survival and did not disrupt the rescue effect of AAV8-sCX3CL1. While the mechanism by which AAV8-sCX3CL1 saves cones remains to be elucidated, our findings suggest that sCX3CL1 gene therapy could be beneficial for a wide range of patients with RP, and potentially for other patients with inflammatory processes that affect vision or other areas of the CNS.
CX3CL1 is a 373-aa protein with a single transmembrane domain that can undergo proteolytic cleavage to release sCX3CL1 into the extracellular environment (38). In the CNS, both fCX3CL1 and sCX3CL1 are primarily produced by neurons, and, by binding CX3CR1 on microglia, they are thought to regulate key aspects of microglial physiology (67, 68). One of the main responsibilities of CX3CL1 in neuron–microglia interactions is to suppress the activation of microglia (69, 70). Supporting this notion, exogenous delivery of CX3CL1 has been shown to decrease microglia activation as well as neurological deficits in animal models of Parkinson’s disease and stroke (71–73). Based on these data, we overexpressed CX3CL1 in RP mice with the hope that it would attenuate immune responses in the retina that were perpetuating nonautonomous cone death. Use of sCX3CL1 indeed prolonged cone survival during degeneration, although it did so without reducing inflammation or the number of microglia in the ONL. Interestingly, cone rescue was seen when sCX3CL1 was secreted by the RPE, but not when full-length membrane-bound CX3CL1 was expressed on cones. This result could be due to differences in the level of expression, as the RPE-specific human Best1 promoter is quite strong relative to the human red opsin promoter. Alternatively, it could be that sCX3CL1 acts on other cell types besides microglia and is better able to reach these cells when secreted. In contrast, overexpression of CD200, another repressor of microglia activation (24), failed to rescue cones whether expressed as sCD200 from the RPE or fCD200 on cones. Of note, we recently reported dose-dependent vector toxicity with certain promoters, including human Best1, resulting in dysmorphic RPE and mild photoreceptor damage (74). In this study, AAV8-sCX3CL1 containing the human Best1 promoter was nonetheless able to improve cone survival.
Activated microglia are a hallmark of early RP, given their migration into the ONL, production of inflammatory cytokines, and phagocytosis of living photoreceptors (33, 34, 45). In early RP, rods are degenerating, and these microglia activities are deleterious, as genetic ablation of microglia has been demonstrated to ameliorate rod death (34). Acute retinal detachment, another condition causing photoreceptor loss, is similarly characterized by inflammatory cytokines and phagocytic microglia (75, 76). Unlike the case with early RP, however, removal of microglia during retinal detachment was observed to accelerate photoreceptor degeneration, implying a protective role for activated microglia (76). Here, we found evidence of microglia activation during cone death in RP, as illustrated by the presence of microglia in the ONL and up-regulation of Il1a, Tnf, C1qa, and Cd68. We hypothesized that, as in early RP, these microglia might be detrimental; consequently, our goal was to develop AAVs capable of suppressing retinal microglia activation. Interestingly, drug-induced depletion of microglia in rd1 retinas provided evidence for only a slight negative effect of activated microglia on cones; only a small increase in the number of cones was seen with microglia depletion, and this change did not reach statistical significance. One explanation for this could be that while activated microglia in RP do hinder cone survival, they may also provide some benefits. We speculate that one such benefit may be increased clearance of harmful cell debris. By RNA-seq, we detected small amounts of cone-specific RNAs in microglia from AAV8-GFP–infected rd10 retinas, potentially from phagocytosis of cones or cone fragments. Because such RNAs were fewer in microglia from AAV8-sCX3CL1–infected retinas, cone debris might accumulate in control microglia if digestion of these materials cannot keep up with engulfment. Inability of microglia to complete phagocytosis may then trigger the release of factors injurious to cones, akin to the model of “frustrated phagocytosis” experienced by microglia in Alzheimer’s disease (77). As we observed up-regulation of lysosomal pathways in microglia with AAV8-sCX3CL1, these cells may more efficiently digest cone material, alleviating this frustration and favoring cone preservation.
Notably, depletion of up to ∼99% of microglia also failed to abrogate AAV8-sCX3CL1 cone rescue. Although this could indicate that sCX3CL1 prolongs cone survival independent of microglia, we cannot rule out the possibility that sCX3CL1 mediated the rescue effect via microglia before their depletion. Because AAV8-sCX3CL1 was administered before PLX3397, there may have been enough time for microglia to respond to sCX3CL1 and alter the retinal microenvironment in a manner promoting cone survival. While it normally takes around 2 mo for subretinal AAV8 expression to peak, a small amount of signal can be detected as early as 5 d postinfection (78). Consistent with this, we did see some transcriptional changes in retinal microglia 20 d after delivery of AAV8-sCX3CL1 (SI Appendix, Fig. S6), although it is difficult to interpret the relevance of such changes in delaying cone degeneration. Another scenario could be that only a few microglia are needed for AAV8-sCX3CL1 to save cones. In a recent study by Liddelow et al. (23), depletion of microglia by PLX3397 was unable to eliminate a phenotype in astrocytes induced by microglia. Specifically, the authors found that inflammatory cytokines from activated microglia caused astrocytes to damage neurons. Astrocyte neurotoxicity was absent in CSF1R-null mice, which are devoid of microglia (79). However, in other strains, neurotoxicity was still observed despite pharmacologically depleting 95% of microglia (23). Thus, it could be that the ∼1% of retinal microglia that survive PLX3397 treatment are sufficient to respond to sCX3CL1 and preserve degenerating cones. For these remaining microglia, greater sCX3CL1 signaling per cell may additionally account for the slight additivity of AAV8-sCX3CL1 and microglia depletion on cone rescue.
An alternative model, given how modest the effect of microglia depletion was on cone survival, is that nonautonomous cone death is caused by mechanisms largely independent of microglia (9, 13–15). For AAV8-sCX3CL1, the reason for cone rescue might then be due to sCX3CL1 acting on a CX3CR1-expressing cell type other than microglia. This cell type would have to be external to the retina, since none of the nonmicroglia cells in our rd1;Cx3cr1GFP/+ retinas expressed CX3CR1 when analyzed by flow cytometry. Outside of the CNS, CX3CR1 is also present on several immune cell populations, including monocytes; peripheral macrophages; and certain subsets of T cells, natural killer cells, and dendritic cells (35, 80). In these populations, one of the roles of CX3CR1 is to mediate a chemotactic response to CX3CL1 (80, 81). It is therefore plausible that sCX3CL1 secreted by the RPE might act on one of these cell types in the vascular-rich choroid, perhaps to induce migration into the subretinal space. Future work is needed to examine these possibilities and determine at a molecular level how sCX3CL1 preserves cones. Such information will aid the development of better RP treatments that more specifically target cone degeneration.
In 2017, an AAV encoding the RPE65 gene became the first gene therapy to be approved for an inherited retinal disorder (82). Despite this achievement, there are still thousands of retinal disease mutations for which no effective treatment exists (83). Addressing these lesions one by one would be cost-prohibitive and time-consuming, and specifically for RP, rods carrying the mutation have often died by the time of diagnosis (1), making gene correction therapy infeasible. Mutation-independent gene therapies are an alternative approach that, while not curative, may improve vision for a much larger number of patients, including those for whom no causal mutation has been identified. Previously, only two examples of mutation-independent gene therapies have been shown to rescue cones in animal models of RP (84). In 2015, Byrne and coworkers (85) reported better cone survival and vision in two strains of RP mice with viral-mediated expression of rod-derived cone viability factor (RdCVF), a protein normally secreted from rods that stimulates cones to take up glucose (86). That same year, our group found that cone numbers and function in three RP mouse lines could be improved with AAV-mediated delivery of antioxidant-based therapy, particularly a master antioxidant transcription factor, NRF2 (13). Here, we demonstrated that AAV8-sCX3CL1 is also a mutation-independent gene therapy capable of saving cones in different types of RP mice. Our results support further assessment of AAV8-sCX3CL1 in larger RP animal models and highlight its potential to help treat this disease in patients, regardless of their mutation.
Finally, photoreceptor death occurs in other diseases, such as age-related macular degeneration (AMD), the leading cause of blindness in the industrialized world (87). Although the pathogenesis of AMD is not fully understood, loss of rods typically precedes that of cones (88, 89), not unlike nonautonomous cone death in RP. Furthermore, genetic polymorphisms in CX3CR1, the receptor for CX3CL1, have been associated with a higher risk of AMD in patients (90). It would thus be interesting to test if sCX3CL1 can similarly alleviate retinal degeneration in AMD.
Materials and Methods
Animals.
CD-1 (catalog no. 022), rd1 (FVB/N; catalog no. 207), and B6 (catalog no. 027) mice were purchased from Charles River Laboratories. Cx3cr1GFP (catalog no. 005582) (35) and rd10 (catalog no. 004297) (27) mice on a B6 background were purchased from The Jackson Laboratory. Rhodopsin-null (Rho−/−) mice were a gift from Janis Lem, Tufts University, Boston, MA (42). Animals were subsequently bred and maintained at Harvard Medical School on a 12-h alternating light and dark cycle. All experimental procedures were approved by the Institutional Animal Care and Use Committee at Harvard University.
Plasmids.
The AAV-human red opsin-GFP-woodchuck hepatitis virus posttranscriptional regulatory element (WPRE)-bovine growth hormone (bGH) polyA (AAV8-GFP) vector plasmid was a gift from Botond Roska, Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland (91), and used the promoter region originally developed by Wang et al. (92). The AAV8-mCherry vector was generated by replacing the GFP coding sequence with that of mCherry flanked by NotI and AgeI restriction sites. AAV8-fCD200 and AAV8-fCX3CL1 were then cloned by digesting AAV8-mCherry with NotI and HindIII restriction enzymes and replacing the mCherry coding sequence with the GCCGCCACC Kozak sequence, followed by the full-length mouse cDNA for CD200 (NM_010818.3) or CX3CL1 (NM_009142.3), respectively. A vector utilizing the human Best1 promoter was created by replacing the CMV promoter of the AAV-CMV-Promega intron (PI)-EGFP-WPRE-bGH plasmid, a gift from James M. Wilson, University of Pennsylvania, Philadelphia, PA, with the −540/+38-bp region of the human Best1 promoter (93). Vector plasmids for AAV8-sCD200 and AAV8-sCX3CL1 were cloned by digesting the AAV-human Best1 promoter vector with NotI and HindIII restriction enzymes and replacing the EGFP coding sequence with the GCCGCCACC Kozak sequence, followed by the first 714 bp (amino acids 1–238) of CD200 or the first 1,008 bp (amino acids 1–336) of CX3CL1, respectively, followed by a stop codon.
AAV Production and Purification.
Recombinant AAV8 vectors were generated as previously described (13, 94). Detailed procedures are provided in SI Appendix, SI Materials and Methods.
Subretinal AAV Delivery.
Subretinal injection of AAVs was performed on P0–P1 neonatal mice as previously described (13, 95). Detailed procedures are provided in SI Appendix, SI Materials and Methods.
Histology.
Enucleated eyes for retinal cross-sections were dissected in PBS. Following removal of the cornea, iris, lens, and ciliary body, the remaining eye cup was fixed in 4% paraformaldehyde for 4 h at room temperature; cryoprotected in 10%, 20%, and 30% sucrose in PBS; and embedded in a 1:1 mixture of 30% sucrose in PBS and optimal cutting temperature compound (Tissue-Tek) on dry ice. Frozen eye cups were cut on a Leica CM3050S cryostat (Leica Microsystems) into 50-μm sections for Cx3cr1GFP retinas or 20-μm sections otherwise and stained with 4′,6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific) for 5 min at room temperature before mounting with Fluoromount-G (SouthernBiotech). For flat-mounted retinas, isolated retinas were fixed in 4% paraformaldehyde for 30 min at room temperature. Four radial incisions were made to relax the retina into four leaflets, which were flattened onto a microscope slide with the ganglion cell layer facing up using a fine-haired brush. To perform antibody staining of retinal cross-sections or whole retinas, tissues were blocked with 5% goat serum or 5% BSA in PBS with 0.1% Triton X-100 for 1 h at room temperature, after which tissues were incubated with primary antibodies in block solution at 4 °C overnight followed by secondary antibodies in PBS for 2 h at room temperature. Primary antibodies included rabbit anti-CX3CL1 (ab25088, 1:500; Abcam) and rhodamine-conjugated peanut agglutinin (RL-1072, 1:1,000; Vector Laboratories). Goat anti-rabbit Alexa Fluor 594 (111-585-144, 1:1,000; Jackson ImmunoResearch) was used as a secondary antibody.
Image Acquisition and Analysis.
Images of microglia in retinal cross-sections and of flat-mounted retinas were acquired on a Keyence BZ-9000 wide-field fluorescent microscope using a 10× air objective. All other images were acquired on a Zeiss LSM710 scanning confocal microscope using a 10× air, 20× air, or 40× oil objective. Image analysis was performed using ImageJ (NIH). To calculate the percentage of microglia in the ONL, a mask was drawn around the ONL following the outlines of DAPI-labeled nuclei. Each of the microglia was determined to reside in the ONL if 50% or more of its cell body was located within the mask. To assay cone survival in flat-mounted retinas, a custom ImageJ module was created. First, to indicate the boundaries of the retina, the user marked the location of the optic nerve head and each of the four retinal leaflets as depicted in SI Appendix, Fig. S2. The module then automatically processed the image, defined the central retina, and applied a threshold to quantify the number of GFP-positive cones in this region. Detailed procedures are provided in SI Appendix, SI Materials and Methods.
ERG.
In vivo ERG recordings were performed and measured by an observer blinded to the treatment assignment using the Espion E3 System (Diagonsys) as previously described (13, 96). Detailed procedures are provided in SI Appendix, SI Materials and Methods.
Optomotor Assay.
Visual acuity was measured by an observer blinded to the treatment assignment using the OptoMotry System (CerebralMechanics) as previously described (13, 96). Detailed procedures are provided in SI Appendix, SI Materials and Methods.
RT-PCR.
For RT-PCR of whole retinas, freshly dissected retinas were homogenized using a handheld pellet pestle (Kimble Chase) in 350 μL of RLT buffer containing 1% β-mercaptoethanol. One retina was used per sample. For RT-PCR of microglia, ∼1,000 microglia per retina were sorted into 10 μL of Buffer TCL (Qiagen) to lyse cells, to which 70 μL of RLT buffer containing 1% β-mercaptoethanol was added. RNA extractions were performed using an RNeasy Micro Kit (Qiagen), followed by cDNA synthesis using the SuperScript III First-Strand Synthesis System (Invitrogen). RT-PCR reactions were conducted in triplicate using Power SYBR Green PCR Master Mix (Applied Biosystems) on a CFX96 real-time PCR detection system (BioRad) to determine cycle threshold (Ct) values. Expression levels were quantified by normalizing to the housekeeping gene Gapdh with fold changes relative to age-matched WT (CD-1 or B6) retinas. P values were calculated using ΔΔCt values. Primers for RT-PCR were designed using PrimerBank (97) with sequences available in SI Appendix, Table S1.
Flow Cytometry and Cell Sorting.
Retinal microglia were isolated using fluorescence-activated cell sorting (FACS), and data were analyzed using FlowJo 10 (TreeStar). To dissociate cells, freshly dissected retinas were incubated in 400 μL of cysteine-activated papain solution (Worthington) for 5 min at 37 °C, followed by gentle trituration with a micropipette. Dissociated cells were subsequently blocked with rat anti-mouse CD16/32 (1:100; BD Pharmingen) for 5 min on ice, followed by staining with phycoerythrin-Cy5–conjugated anti-CD11b (M1/70, 1:200; BioLegend), allophycocyanin (APC)-Cy7–conjugated anti-Ly6G (1A8, 1:200; BioLegend), and APC-Cy7–conjugated anti-Ly6C (HK1.4, 1:200; BioLegend) for 20 min on ice. After washes, cells were resuspended in FACS buffer (2% FBS, 2 mM EDTA in PBS) containing 0.5 μg/mL DAPI to label nonviable cells and passed through a 40-μm filter. Sorting was performed on a BD FACSAria II using a 70-μm nozzle according to the gating shown in SI Appendix, Fig. S4.
RNA-Seq of Microglia.
Eight biological replicates were used per experimental condition. For each retina, 1,000 microglia (CD11b+ Ly6G/Ly6C−) were sorted into 10 μL of Buffer TCL (Qiagen) containing 1% β-mercaptoethanol and immediately frozen on dry ice. For a subset of retinas, 1,000 nonmicroglia cells (CD11b−) were also sorted. Upon collection of all samples, frozen microglia and nonmicroglia lysates were thawed on wet ice and loaded into a 96-well plate for cDNA library synthesis and sequencing. A modified Smart-Seq2 protocol was performed on samples by the Broad Institute Genomics Platform (98). Libraries from 96 samples with unique barcodes were combined and sequenced on a NextSeq 500 Sequencing System (Illumina) to an expected coverage of ∼6 million reads per sample. Following demultiplexing, reads were subjected to quality control measures and mapped to the GRCm38.p6 reference genome. Reads assigned to each gene were quantified using featureCounts (99), and samples with fewer than 30% assigned reads were excluded from further analysis. Count data were analyzed using DESeq2 to identify differentially expressed genes, with an adjusted P value less than 0.05 considered significant (100). GSEA was performed using GSEA v3.0 software from the Broad Institute under default settings on the GO Cellular Component Ontology collection (580 gene sets) from the Molecular Signatures Database (MSigDB). Gene sets with a family-wise error rate less than 0.05 were considered significantly enriched.
Microglia Depletion.
Microglia were depleted using PLX3397 (SelleckChem), an orally available CSF1R inhibitor. PLX3397 was incorporated into AIN-76A rodent chow (Research Diets) at 290 mg/kg and provided ad libitum for 30 d before harvesting of the animal on P50.
Statistics.
Unpaired, two-tailed Student’s t tests were used for comparisons between two groups. For comparisons of three or more groups, a Bonferroni correction was added to the test by multiplying the resulting P value by the number of hypotheses tested. Two-tailed, two-way ANOVA was used to analyze ERG, optomotor, and ONL thickness experiments in which the same subjects are compared over a series of conditions or time points. In all cases, a P value less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Sophia Zhao and Microscopy Resources on the North Quad at Harvard Medical School for their technical assistance. This work was made possible by support from the Howard Hughes Medical Institute (S.K.W. and C.L.C.), Foundation Fighting Blindness (S.K.W.), and Fight for Sight (S.K.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901787116/-/DCSupplemental.
References
- 1.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 2.Daiger SP, Sullivan LS, Bowne SJ. Genes and mutations causing retinitis pigmentosa. Clin Genet. 2013;84:132–141. doi: 10.1111/cge.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berson EL. Retinitis pigmentosa: Unfolding its mystery. Proc Natl Acad Sci USA. 1996;93:4526–4528. doi: 10.1073/pnas.93.10.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haim M. Epidemiology of retinitis pigmentosa in Denmark. Acta Ophthalmol Scand Suppl. 2002;80:1–34. doi: 10.1046/j.1395-3907.2002.00001.x. [DOI] [PubMed] [Google Scholar]
- 5.Ali RR, et al. Gene transfer into the mouse retina mediated by an adeno-associated viral vector. Hum Mol Genet. 1996;5:591–594. doi: 10.1093/hmg/5.5.591. [DOI] [PubMed] [Google Scholar]
- 6.Murata T, et al. Ocular gene therapy: Experimental studies and clinical possibilities. Ophthalmic Res. 1997;29:242–251. doi: 10.1159/000268022. [DOI] [PubMed] [Google Scholar]
- 7.Maguire AM, et al. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sacchetti M, Mantelli F, Merlo D, Lambiase A. Systematic review of randomized clinical trials on safety and efficacy of pharmacological and nonpharmacological treatments for retinitis pigmentosa. J Ophthalmol. 2015;2015:737053. doi: 10.1155/2015/737053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narayan DS, Wood JPM, Chidlow G, Casson RJ. A review of the mechanisms of cone degeneration in retinitis pigmentosa. Acta Ophthalmol. 2016;94:748–754. doi: 10.1111/aos.13141. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, et al. Two-step reactivation of dormant cones in retinitis pigmentosa. Cell Reports. 2016;15:372–385. doi: 10.1016/j.celrep.2016.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komeima K, Rogers BS, Lu L, Campochiaro PA. Antioxidants reduce cone cell death in a model of retinitis pigmentosa. Proc Natl Acad Sci USA. 2006;103:11300–11305. doi: 10.1073/pnas.0604056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Punzo C, Kornacker K, Cepko CL. Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat Neurosci. 2009;12:44–52. doi: 10.1038/nn.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong W, MacColl Garfinkel AE, Li Y, Benowitz LI, Cepko CL. NRF2 promotes neuronal survival in neurodegeneration and acute nerve damage. J Clin Invest. 2015;125:1433–1445. doi: 10.1172/JCI79735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkatesh A, et al. Activated mTORC1 promotes long-term cone survival in retinitis pigmentosa mice. J Clin Invest. 2015;125:1446–1458. doi: 10.1172/JCI79766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aït-Ali N, et al. Rod-derived cone viability factor promotes cone survival by stimulating aerobic glycolysis. Cell. 2015;161:817–832. doi: 10.1016/j.cell.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Murakami Y, et al. Receptor interacting protein kinase mediates necrotic cone but not rod cell death in a mouse model of inherited degeneration. Proc Natl Acad Sci USA. 2012;109:14598–14603. doi: 10.1073/pnas.1206937109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zitvogel L, Kepp O, Kroemer G. Decoding cell death signals in inflammation and immunity. Cell. 2010;140:798–804. doi: 10.1016/j.cell.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 18.Murakami Y, et al. Necrotic cone photoreceptor cell death in retinitis pigmentosa. Cell Death Dis. 2015;6:e2038. doi: 10.1038/cddis.2015.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman SM, Wong WT. Microglia in the retina: Roles in development, maturity, and disease. Annu Rev Vis Sci. 2018;4:45–77. doi: 10.1146/annurev-vision-091517-034425. [DOI] [PubMed] [Google Scholar]
- 20.Salter MW, Stevens B. Microglia emerge as central players in brain disease. Nat Med. 2017;23:1018–1027. doi: 10.1038/nm.4397. [DOI] [PubMed] [Google Scholar]
- 21.Block ML, Hong J-S. Microglia and inflammation-mediated neurodegeneration: Multiple triggers with a common mechanism. Prog Neurobiol. 2005;76:77–98. doi: 10.1016/j.pneurobio.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Lynch MA. The multifaceted profile of activated microglia. Mol Neurobiol. 2009;40:139–156. doi: 10.1007/s12035-009-8077-9. [DOI] [PubMed] [Google Scholar]
- 23.Liddelow SA, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoek RM, et al. Down-regulation of the macrophage lineage through interaction with OX2 (CD200) Science. 2000;290:1768–1771. doi: 10.1126/science.290.5497.1768. [DOI] [PubMed] [Google Scholar]
- 25.Biber K, Neumann H, Inoue K, Boddeke HWGM. Neuronal ‘on’ and ‘off’ signals control microglia. Trends Neurosci. 2007;30:596–602. doi: 10.1016/j.tins.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Cardona AE, et al. Control of microglial neurotoxicity by the fractalkine receptor. Nat Neurosci. 2006;9:917–924. doi: 10.1038/nn1715. [DOI] [PubMed] [Google Scholar]
- 27.Chang B, et al. Retinal degeneration mutants in the mouse. Vision Res. 2002;42:517–525. doi: 10.1016/s0042-6989(01)00146-8. [DOI] [PubMed] [Google Scholar]
- 28.Pennesi ME, et al. Long-term characterization of retinal degeneration in rd1 and rd10 mice using spectral domain optical coherence tomography. Invest Ophthalmol Vis Sci. 2012;53:4644–4656. doi: 10.1167/iovs.12-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett ML, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci USA. 2016;113:E1738–E1746. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bodea L-G, et al. Neurodegeneration by activation of the microglial complement-phagosome pathway. J Neurosci. 2014;34:8546–8556. doi: 10.1523/JNEUROSCI.5002-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. I. Structural analysis using light and electron microscopy. J Comp Neurol. 1979;188:245–262. doi: 10.1002/cne.901880204. [DOI] [PubMed] [Google Scholar]
- 32.Curcio CA, Sloan KR, Kalina RE, Hendrickson AE. Human photoreceptor topography. J Comp Neurol. 1990;292:497–523. doi: 10.1002/cne.902920402. [DOI] [PubMed] [Google Scholar]
- 33.Peng B, et al. Suppression of microglial activation is neuroprotective in a mouse model of human retinitis pigmentosa. J Neurosci. 2014;34:8139–8150. doi: 10.1523/JNEUROSCI.5200-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao L, et al. Microglial phagocytosis of living photoreceptors contributes to inherited retinal degeneration. EMBO Mol Med. 2015;7:1179–1197. doi: 10.15252/emmm.201505298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung S, et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q, Timmers AM, Guy J, Pang J, Hauswirth WW. Cone-specific expression using a human red opsin promoter in recombinant AAV. Vision Res. 2008;48:332–338. doi: 10.1016/j.visres.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 37.Wong KK, et al. Characterization of CD200 ectodomain shedding. PLoS One. 2016;11:e0152073. doi: 10.1371/journal.pone.0152073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bazan JF, et al. A new class of membrane-bound chemokine with a CX3C motif. Nature. 1997;385:640–644. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- 39.Young RW. The renewal of photoreceptor cell outer segments. J Cell Biol. 1967;33:61–72. doi: 10.1083/jcb.33.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sung CH, et al. Rhodopsin mutations in autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci USA. 1991;88:6481–6485. doi: 10.1073/pnas.88.15.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gargini C, Terzibasi E, Mazzoni F, Strettoi E. Retinal organization in the retinal degeneration 10 (rd10) mutant mouse: A morphological and ERG study. J Comp Neurol. 2007;500:222–238. doi: 10.1002/cne.21144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lem J, et al. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc Natl Acad Sci USA. 1999;96:736–741. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prusky GT, Alam NM, Beekman S, Douglas RM. Rapid quantification of adult and developing mouse spatial vision using a virtual optomotor system. Invest Ophthalmol Vis Sci. 2004;45:4611–4616. doi: 10.1167/iovs.04-0541. [DOI] [PubMed] [Google Scholar]
- 44.Douglas RM, et al. Independent visual threshold measurements in the two eyes of freely moving rats and mice using a virtual-reality optokinetic system. Vis Neurosci. 2005;22:677–684. doi: 10.1017/S0952523805225166. [DOI] [PubMed] [Google Scholar]
- 45.Zabel MK, et al. Microglial phagocytosis and activation underlying photoreceptor degeneration is regulated by CX3CL1-CX3CR1 signaling in a mouse model of retinitis pigmentosa. Glia. 2016;64:1479–1491. doi: 10.1002/glia.23016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu DY, Cringle SJ, Su EN, Yu PK. Intraretinal oxygen levels before and after photoreceptor loss in the RCS rat. Invest Ophthalmol Vis Sci. 2000;41:3999–4006. [PubMed] [Google Scholar]
- 47.Liyanage SE, et al. Flow cytometric analysis of inflammatory and resident myeloid populations in mouse ocular inflammatory models. Exp Eye Res. 2016;151:160–170. doi: 10.1016/j.exer.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murinello S, et al. Assessing retinal microglial phagocytic function in vivo using a flow cytometry-based assay. J Vis Exp. 2016;116:e54677. doi: 10.3791/54677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butovsky O, et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hickman SE, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akimoto M, et al. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc Natl Acad Sci USA. 2006;103:3890–3895. doi: 10.1073/pnas.0508214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shekhar K, et al. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell. 2016;166:1308–1323.e30. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roesch K, et al. The transcriptome of retinal Müller glial cells. J Comp Neurol. 2008;509:225–238. doi: 10.1002/cne.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chintalapudi SR, et al. Isolation of primary murine retinal ganglion cells (RGCs) by flow cytometry. J Vis Exp. 2017;125:e55785. doi: 10.3791/55785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cherry TJ, Trimarchi JM, Stadler MB, Cepko CL. Development and diversification of retinal amacrine interneurons at single cell resolution. Proc Natl Acad Sci USA. 2009;106:9495–9500. doi: 10.1073/pnas.0903264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 2003;34:53–58. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- 57.Keren-Shaul H, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169:1276–1290.e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 58.Song WM, et al. Humanized TREM2 mice reveal microglia-intrinsic and -extrinsic effects of R47H polymorphism. J Exp Med. 2018;215:745–760. doi: 10.1084/jem.20171529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chiu IM, et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 2013;4:385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iaccarino HF, et al. Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature. 2016;540:230–235. doi: 10.1038/nature20587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neumann H, Kotter MR, Franklin RJM. Debris clearance by microglia: An essential link between degeneration and regeneration. Brain. 2009;132:288–295. doi: 10.1093/brain/awn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iribarne M, et al. Aipl1 is required for cone photoreceptor function and survival through the stability of Pde6c and Gc3 in zebrafish. Sci Rep. 2017;7:45962. doi: 10.1038/srep45962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Decembrini S, et al. Cone genesis tracing by the Chrnb4-EGFP mouse line: Evidences of cellular material fusion after cone precursor transplantation. Mol Ther. 2017;25:634–653. doi: 10.1016/j.ymthe.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tummala H, et al. Mutation in the guanine nucleotide-binding protein β-3 causes retinal degeneration and embryonic mortality in chickens. Invest Ophthalmol Vis Sci. 2006;47:4714–4718. doi: 10.1167/iovs.06-0292. [DOI] [PubMed] [Google Scholar]
- 65.Combadière C, et al. CX3CR1-dependent subretinal microglia cell accumulation is associated with cardinal features of age-related macular degeneration. J Clin Invest. 2007;117:2920–2928. doi: 10.1172/JCI31692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elmore MRP, et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82:380–397. doi: 10.1016/j.neuron.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paolicelli RC, Bisht K, Tremblay MÈ. Fractalkine regulation of microglial physiology and consequences on the brain and behavior. Front Cell Neurosci. 2014;8:129. doi: 10.3389/fncel.2014.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lauro C, Catalano M, Trettel F, Limatola C. Fractalkine in the nervous system: Neuroprotective or neurotoxic molecule? Ann N Y Acad Sci. 2015;1351:141–148. doi: 10.1111/nyas.12805. [DOI] [PubMed] [Google Scholar]
- 69.Zujovic V, Benavides J, Vigé X, Carter C, Taupin V. Fractalkine modulates TNF-alpha secretion and neurotoxicity induced by microglial activation. Glia. 2000;29:305–315. [PubMed] [Google Scholar]
- 70.Mizuno T, Kawanokuchi J, Numata K, Suzumura A. Production and neuroprotective functions of fractalkine in the central nervous system. Brain Res. 2003;979:65–70. doi: 10.1016/s0006-8993(03)02867-1. [DOI] [PubMed] [Google Scholar]
- 71.Nash KR, et al. Fractalkine over expression suppresses α-synuclein-mediated neurodegeneration. Mol Ther. 2015;23:17–23. doi: 10.1038/mt.2014.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pabon MM, Bachstetter AD, Hudson CE, Gemma C, Bickford PC. CX3CL1 reduces neurotoxicity and microglial activation in a rat model of Parkinson’s disease. J Neuroinflammation. 2011;8:9. doi: 10.1186/1742-2094-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cipriani R, et al. CX3CL1 is neuroprotective in permanent focal cerebral ischemia in rodents. J Neurosci. 2011;31:16327–16335. doi: 10.1523/JNEUROSCI.3611-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiong W, et al. AAV cis-regulatory sequences are correlated with ocular toxicity. Proc Natl Acad Sci USA. 2019;116:5785–5794. doi: 10.1073/pnas.1821000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakazawa T, et al. Characterization of cytokine responses to retinal detachment in rats. Mol Vis. 2006;12:867–878. [PubMed] [Google Scholar]
- 76.Okunuki Y, et al. Microglia inhibit photoreceptor cell death and regulate immune cell infiltration in response to retinal detachment. Proc Natl Acad Sci USA. 2018;115:E6264–E6273. doi: 10.1073/pnas.1719601115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sokolowski JD, Mandell JW. Phagocytic clearance in neurodegeneration. Am J Pathol. 2011;178:1416–1428. doi: 10.1016/j.ajpath.2010.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Natkunarajah M, et al. Assessment of ocular transduction using single-stranded and self-complementary recombinant adeno-associated virus serotype 2/8. Gene Ther. 2008;15:463–467. doi: 10.1038/sj.gt.3303074. [DOI] [PubMed] [Google Scholar]
- 79.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Imai T, et al. Identification and molecular characterization of fractalkine receptor CX3CR1, which mediates both leukocyte migration and adhesion. Cell. 1997;91:521–530. doi: 10.1016/s0092-8674(00)80438-9. [DOI] [PubMed] [Google Scholar]
- 81.Haskell CA, Cleary MD, Charo IF. Unique role of the chemokine domain of fractalkine in cell capture. Kinetics of receptor dissociation correlate with cell adhesion. J Biol Chem. 2000;275:34183–34189. doi: 10.1074/jbc.M005731200. [DOI] [PubMed] [Google Scholar]
- 82.Russell S, et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: A randomised, controlled, open-label, phase 3 trial. Lancet. 2017;390:849–860. doi: 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ran X, et al. ‘RetinoGenetics’: A comprehensive mutation database for genes related to inherited retinal degeneration. Database (Oxford) 2014;2014:bau047. doi: 10.1093/database/bau047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fortuny C, Flannery JG. Mutation-independent gene therapies for rod-cone dystrophies. Adv Exp Med Biol. 2018;1074:75–81. doi: 10.1007/978-3-319-75402-4_10. [DOI] [PubMed] [Google Scholar]
- 85.Byrne LC, et al. Viral-mediated RdCVF and RdCVFL expression protects cone and rod photoreceptors in retinal degeneration. J Clin Invest. 2015;125:105–116. doi: 10.1172/JCI65654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Léveillard T, et al. Identification and characterization of rod-derived cone viability factor. Nat Genet. 2004;36:755–759. doi: 10.1038/ng1386. [DOI] [PubMed] [Google Scholar]
- 87.Pennington KL, DeAngelis MM. Epidemiology of age-related macular degeneration (AMD): Associations with cardiovascular disease phenotypes and lipid factors. Eye Vis (Lond) 2016;3:34. doi: 10.1186/s40662-016-0063-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Curcio CA, Medeiros NE, Millican CL. Photoreceptor loss in age-related macular degeneration. Invest Ophthalmol Vis Sci. 1996;37:1236–1249. [PubMed] [Google Scholar]
- 89.Shelley EJ, Madigan MC, Natoli R, Penfold PL, Provis JM. Cone degeneration in aging and age-related macular degeneration. Arch Ophthalmol. 2009;127:483–492. doi: 10.1001/archophthalmol.2008.622. [DOI] [PubMed] [Google Scholar]
- 90.Chan C-C, Tuo J, Bojanowski CM, Csaky KG, Green WR. Detection of CX3CR1 single nucleotide polymorphism and expression on archived eyes with age-related macular degeneration. Histol Histopathol. 2005;20:857–863. doi: 10.14670/hh-20.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Busskamp V, et al. Genetic reactivation of cone photoreceptors restores visual responses in retinitis pigmentosa. Science. 2010;329:413–417. doi: 10.1126/science.1190897. [DOI] [PubMed] [Google Scholar]
- 92.Wang Y, et al. A locus control region adjacent to the human red and green visual pigment genes. Neuron. 1992;9:429–440. doi: 10.1016/0896-6273(92)90181-c. [DOI] [PubMed] [Google Scholar]
- 93.Esumi N, Oshima Y, Li Y, Campochiaro PA, Zack DJ. Analysis of the VMD2 promoter and implication of E-box binding factors in its regulation. J Biol Chem. 2004;279:19064–19073. doi: 10.1074/jbc.M309881200. [DOI] [PubMed] [Google Scholar]
- 94.Grieger JC, Choi VW, Samulski RJ. Production and characterization of adeno-associated viral vectors. Nat Protoc. 2006;1:1412–1428. doi: 10.1038/nprot.2006.207. [DOI] [PubMed] [Google Scholar]
- 95.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xue Y, et al. CRALBP supports the mammalian retinal visual cycle and cone vision. J Clin Invest. 2015;125:727–738. doi: 10.1172/JCI79651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang X, Spandidos A, Wang H, Seed B. PrimerBank: A PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40:D1144–D1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 99.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 100.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.