Significance
G protein-coupled receptors (GPCRs) form a family of signaling molecules in the membrane of cells that plays a key role in transduction of cellular responses. Little is known about how rapidly GPCRs can be activated. While the “light receptor” rhodopsin in the eye activates within 1 ms, other GPCRs are thought to activate much slower. We use two entirely different techniques with advanced time resolution to activate a dimeric metabotropic glutamate GPCR: UV light-triggered uncaging of ligand in intact cells and piezo-driven ligand application in outside-out patches. We demonstrate initial conformational rearrangements within ≈1 ms that are followed by much slower (≈20 ms) activation in the transmembrane domain. Thus, the initial activation of a nonvisual GPCR proceeds with millisecond speed.
Keywords: G protein-coupled receptors, kinetics, confocal patch-clamp fluorometry, metabotropic glutamate receptors, photouncaging
Abstract
G protein-coupled receptors (GPCRs) are key biological switches that transmit both internal and external stimuli into the cell interior. Among the GPCRs, the “light receptor” rhodopsin has been shown to activate with a rearrangement of the transmembrane (TM) helix bundle within ∼1 ms, while all other receptors are thought to become activated within ∼50 ms to seconds at saturating concentrations. Here, we investigate synchronous stimulation of a dimeric GPCR, the metabotropic glutamate receptor type 1 (mGluR1), by two entirely different methods: (i) UV light-triggered uncaging of glutamate in intact cells or (ii) piezo-driven solution exchange in outside-out patches. Submillisecond FRET recordings between labels at intracellular receptor sites were used to record conformational changes in the mGluR1. At millimolar ligand concentrations, the initial rearrangement between the mGluR1 subunits occurs at a speed of τ1 ∼ 1–2 ms and requires the occupancy of both binding sites in the mGluR1 dimer. These rapid changes were followed by significantly slower conformational changes in the TM domain (τ2 ∼ 20 ms). Receptor deactivation occurred with time constants of ∼40 and ∼900 ms for the inter- and intrasubunit conformational changes, respectively. Together, these data show that, at high glutamate concentrations, the initial intersubunit activation of mGluR1 proceeds with millisecond speed, that there is loose coupling between this initial step and activation of the TM domain, and that activation and deactivation follow a cyclic pathway, including—in addition to the inactive and active states—at least two metastable intermediate states.
G protein-coupled receptors (GPCRs) are cell surface receptors that are activated by diverse types of stimuli, including many hormones and neurotransmitters but also, taste, smell, and light (1). So far, the activation kinetics have been elucidated in detail only for one GPCR, the light receptor rhodopsin (2). This is because (i) rhodopsin can be activated synchronously by light; (ii) on activation, the covalently bound retinal undergoes spectral changes that allow precise recording with optical methods; and (iii) in rod outer segments, rhodopsin represents ∼95% of protein, allowing easy experimental access. After light activation, rhodopsin adopts its active meta-II state characterized by a rearrangement of the transmembrane helix bundle within about 1 ms via a series of short-lived intermediate conformations (3, 4).
For all other GPCRs, activation is thought to be much slower (4, 5). Functional studies measuring receptor-triggered ion currents gave activation time constants of a few seconds for the entire signaling chain, which could be reduced down to ∼200 ms on strong receptor overexpression (6). Initial studies with purified, fluorescently labeled, and reconstituted receptors reported conformational changes over many seconds (7, 8). Later, biophysical studies on nonrhodopsin GPCRs with single-molecule fluorescence, NMR, and molecular dynamics simulations identified rapid dynamic transitions within and between different off and on states in the submillisecond to hundreds of milliseconds time range (9–14). In addition, these studies indicated that the fully active state of nonrhodopsin GPCRs requires stabilization by binding to a G protein or to a (β) arrestin (13, 14). However, none of these studies provided rapid agonist-induced activation kinetics. In intact cells, agonist-induced activation of nonrhodopsin GPCRs has been studied by using FRET-based sensors combined with rapid changes in superfusion media (15, 16). For most GPCRs, these studies have yielded agonist-induced activation time constants in the 30- to 80-ms range (15–17).
When comparing the kinetics of rhodopsin and other GPCRs, the key question is whether rhodopsin is indeed a special case or whether the experimental approaches have so far limited the discovery of millisecond kinetics in nonrhodopsin GPCRs. Therefore, we set out to develop submillisecond activation and recording methods to study the activation dynamics of a nonrhodopsin GPCR in intact cells as well as in isolated outside-out membrane patches. We chose the metabotropic glutamate receptor type 1 (mGluR1) (18), a prototypical class C GPCR, because of its interesting activation mode and because earlier studies with this receptor yielded relatively fast activation time constants (19–21). Agonist binding to class C GPCRs does not occur in the heptahelical transmembrane (7-TM) domain (as in class A GPCRs) but to a “Venus Flytrap” (VFT) motif ligand binding domain in the receptors’ large N terminus. These receptors are obligate dimers, and activation involves a movement of the two protomers toward each other (19) plus an activation of the 7-TM domains, which presumably corresponds to activation of the smaller class A GPCRs (18, 20). Single-molecule FRET studies of isolated mGluR2 ligand binding domain dimers showed oscillations between active and inactive orientations in the submillisecond timescale (11), while complementary analyses of full-length solubilized receptors gave much longer dwell times of the active state of ∼80 and 180 ms for mGluR2 and mGluR3 dimers, respectively (12). Agonist-induced activation kinetics in intact cells, monitored with a series of FRET sensors, were 27 ms [or even faster (21)] for the intermolecular movement of the mGluR1 protomers toward each other and 39 ms for the 7-TM domain activation (20).
Results
Experimental Strategies.
To permit the detection of rhodopsin-like activation speeds, we developed and used synchronous activation of receptors (i) by UV light-triggered uncaging of an inactive caged glutamate derivative to rapidly release active glutamate onto intact cells and (ii) by rapid step-like piezo-controlled changes in superfusion in outside-out patches. We monitored FRET between suitably placed fluorescent labels [cyan (CFP) and yellow (YFP) variants of the GFP; see below] in the receptor by high-speed online recording, which allowed us to reach a resolution well in the submillisecond range.
Receptor Activation by Agonist Uncaging.
For UV-uncaging experiments, we used a custom-built microscope setup (SI Appendix, Fig. S1) that allowed imaging and quantitative recording of CFP and YFP emissions for FRET recording. Uncaging of caged compounds was achieved via a 375-nm UV laser with a spot area of ∼35 µm (half-maximal intensity) in diameter (SI Appendix, Fig. S1C). Using caged fluorescein as a reference compound, we adjusted the laser power such that complete uncaging in this area was achieved well within a pulse duration of 300 µs (SI Appendix, Fig. S1D).
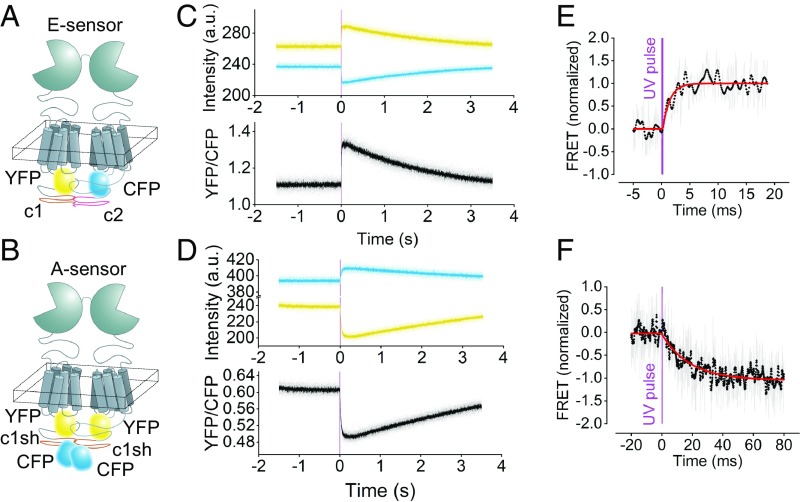
To synchronously photoactivate the mGluR1, we used a photolabile caged analog of glutamate, 4-methoxy-7-nitroindolinyl (MNI) glutamate (22). Activation of the receptors was monitored via mGluR1 FRET sensors, which were labeled with CFPs and YFPs in the second intracellular loop or at the C terminus such that they report either intermolecular (E sensor; reporting movements between the protomers) or intramolecular (A sensor; reporting movements in the transmembrane domain) conformational changes (5); these sensors were transiently expressed in HEK293T cells for live cell recording (Fig. 1). Release of glutamate from 1 mM MNI-caged glutamate by a 300-µs UV laser pulse caused a rapid change in FRET in both sensors (Fig. 1 C and D). MNI glutamate concentration and UV pulse intensity and duration were sufficient to produce full activation speed (SI Appendix, Fig. S2). Using these conditions, FRET in the E sensor increased by about 30% after the UV pulse (Fig. 1C), which is compatible with the two protomers in the mGluR1 dimer moving toward each other as seen in the inactive vs. active crystal structures of the mGluR1–VFT (23). In the case of the A sensor, FRET decreased by about 20% (Fig. 1D); a similar decrease has been seen for essentially all analogous FRET constructs reporting transmembrane conformational changes of GPCRs on activation (16, 17). This result is compatible with the notion of an outward movement of transmembrane domain VI on activation (9–11).
Fig. 1.
Millisecond activation of mGluR1 FRET sensors in living cells on UV light-triggered uncaging of MNI–l-glutamate. (A and B) Schematic of the E sensors (A) and A sensors (B) reporting intermolecular and intramolecular movements of the mGluR1, respectively. The E sensor is composed of one mGluR1 labeled with a CFP and one labeled with a YFP in the second intracellular loop; a C-terminal tail from the GABAB1 and GABAB2 receptors, respectively, assures that only dimers carrying two different labels reach the cell surface (20). The A sensor contains two mGluR1 protomers, each labeled with a YFP in the second intracellular loop and a CFP at the C terminus (20). (C and D) Fluorescence emission intensities recorded in real time in single intact cells expressing the E sensor (C) or the A sensor (D) before and after uncaging of 1 mM MNI–l-glutamate. Data collected from YFP, CFP, and the corresponding corrected FRET ratio are depicted in yellow, blue, and black, respectively. The transient nature of the signals is due to diffusion of uncaged glutamate away from the receptor. (E and F) Millisecond kinetics of the E- and A-sensor activation. Shown are the normalized corrected FRET ratios of single cells expressing the E sensor (E) and A sensor (F). The thickness of the purple line represents the duration of the UV pulse (300 µs). Unfiltered ratio traces (shown in gray) were low passed at 1.25 kHz (black dots). The red curves represent monoexponential fits with time constants τon = 1.2 ms (E) and τon = 18.4 ms (F).
The kinetics of these FRET changes, recorded with photouncaging, were remarkably fast (Fig. 1 E and F), with time constants of individual traces down to 1.2 ms (E sensor) and 18.4 ms (A sensor) and 1.9 ± 0.2 and 23.8 ± 0.7 ms for the corresponding averaged time courses, respectively (Fig. 2). This is clearly faster than observed before using superfusion systems, which generated data in the order of ∼30 ms (16, 17). The signals were transient, lasting for only a few seconds, compatible with the notion that the liberated glutamate dissipates in the solution after the UV release. Low light intensities for photouncaging of MNI glutamate resulted in slower activation kinetics, but the maximum speed was clearly reached at the higher light intensities and pulse durations used (SI Appendix, Fig. S2), suggesting that the time course shown in Fig. 1E represents the maximal speed of the system.
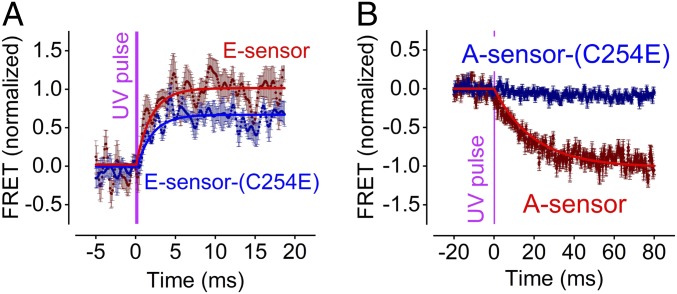
Fig. 2.
Effects of the C254E mutation on FRET signals of the mGluR1 E and A sensors. A C254E mutation was introduced in the mGluR1 E and A sensor construct s to disrupt communication between the ligand binding and transmembrane domains (24). The figures compare average FRET signals of WT- (red) and C254E-mutated (blue) sensors induced by UV light (300-µs pulse) uncaging of 1 mM MNI–l-glutamate as in Fig. 1. (A) Activation of the E sensor; fitting with a monoexponential function gave τon = 1.9 ± 0.2 ms, n = 12 (WT) and τon = 1.8 ± 0.2 ms, n = 4 (C254E). (B) Activation of the A sensor; fitting with a monoexponential function gave 23.8 ± 0.7, n = 8 (WT), and data for the C254E-mutated A sensor could not be reliably fitted to a monoexponential function due to the essentially complete suppression of the signal amplitude. Mutant data were normalized to the maximum FRET response of WT E sensor (A) and A sensor (B), respectively.
To verify that indeed these kinetics reflect inter- vs. intramolecular conformational changes in mGluR1, we made use of the C254E (20) mutant that disrupts communication from the N-terminal LB domain to the 7-TM domain in mGluR2 and five subtypes (24). When this mutation was inserted into the E and A sensors of the mGluR1, this resulted in a moderate (approximately one-third) loss of FRET signal of the E sensor (Fig. 2A) but a nearly complete loss (∼90%) of the FRET signal of the A sensor (Fig. 2B). This is compatible with the notion that the A sensor reports essentially intramolecular movements in the 7-TM domain, while the E sensor reports largely (approximately two-thirds of the FRET signal) intermolecular movements of the protomers. With a time constant from averaged traces of 1.8 ± 0.2 ms, the kinetics of the FRET signal in the C254E-mutated E sensor were indistinguishable from the WT E sensor (time constant 1.9 ± 0.2 ms). This further supports the notion that this is the speed of the intermolecular movements in the mGluR1 dimer.
Receptor Activation and Deactivation by Rapid Solution Exchange in Excised Membrane Patches.
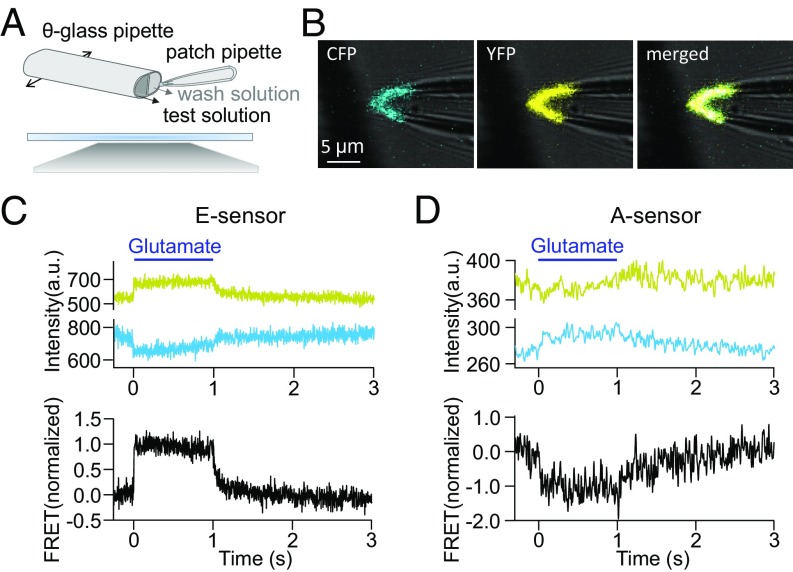
To study the fast activation kinetics obtained above with an independent approach, we performed rapid solution exchanges in small outside-out membrane patches containing the mGluR1 receptor. To this end, we used the method of confocal patch-clamp fluorometry (25–27), which has proven to be a powerful method to study rapid fluorescence changes in ligand-gated ion channels (28). For these experiments, the E and A sensors were expressed in Xenopus oocytes, and an outside-out patch at the tip of the patch pipette was positioned in front of the outlet of a theta-glass pipette that was mounted on a piezo actuator (29) (Fig. 3 A and B). This approach allowed us to jump between two solutions either containing glutamate (1 mM) or not containing glutamate in less than 220 μs (SI Appendix, Fig. S3) and to study the respective time courses of activation and deactivation for both the E and A sensors (Fig. 3 C and D). Activation of both sensors was fully reversible. When analyzing confocal CFP and YFP images (Fig. 3B), the time constants of activation were 2.4 ± 0.1 and 25 ± 7 ms for the E and A sensors, respectively (Fig. 4 A and B).
Fig. 3.
Activation of mGluR1 FRET sensors in outside-out patches in a piezo-controlled concentration jump system. (A) Scheme of the position of the patch pipette and the double-barreled theta-glass pipette for solution application. The theta-glass pipette was mounted on a piezo actuator. (B) Confocal image of an outside-out patch from a Xenopus oocyte expressing the mGluR1 E sensor. (Left) CFP (excitation at 458 nm). (Center) YFP (excitation at 514 nm). (Right) Merged. (C and D) Fluorescence signals and glutamate (1 mM)-induced FRET changes for the E sensors (C) and A sensors (D). Shown are representative photobleaching and cross-talk corrected traces of YFP (yellow) and CFP (cyan) as well as the corrected normalized FRET signal (black).
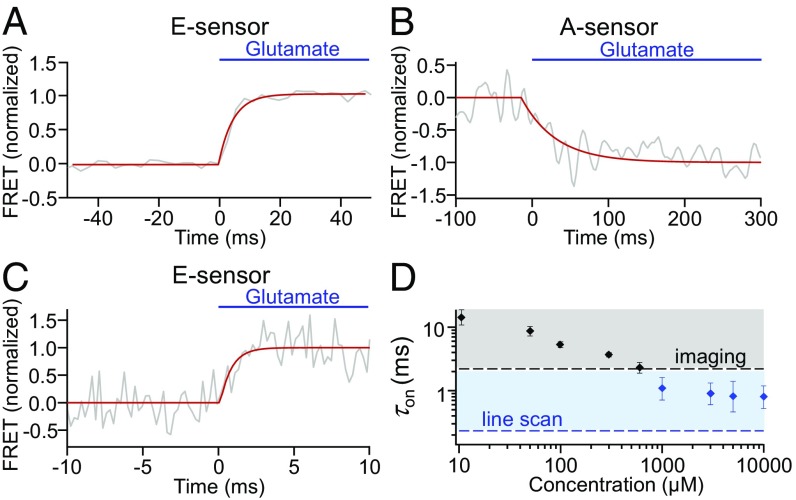
Fig. 4.
Activation kinetics of the mGluR1 E and A sensors in an outside-out patch from a Xenopus oocyte. Shown are average responses (gray) to 1 mM glutamate (blue) applied via the piezo-controlled concentration jump system illustrated in Fig. 3. Red curves were obtained by monoexponential fits to the unfiltered data. (A and B) Activation time courses measured by confocal imaging of the E sensor (A) and A sensor (B) yielded τon = 2.4 ± 0.1 ms (n = 67) and τon = 25 ± 7 ms (n = 7), respectively. (C) Mean activation time course of the E sensor measured at improved time resolution in the line-scan mode at 4-kHz sampling rate yielded τon = 1.2 ± 0.5 ms (n = 5). (D) Concentration dependence of the E-sensor activation time constant. Data at 1,000 µM glutamate and higher concentrations (blue) were obtained from line-scan measurements, whereas at lower concentrations, they were obtained from confocal imaging. The dashed lines indicate technical limits for measuring in confocal imaging mode (black) and line-scan mode (blue) as given by the sampling rate.
Since an activation time constant of 2.4 ms is close to the limit that can be recorded by our confocal imaging setup, we proceeded to use the more rapid line-scan technique, which allows about 10 times faster recording at the expense of an increased noise level. Under these conditions, we observed time constants of 1.2 ± 0.5 ms at glutamate concentrations of 1 mM or even slightly faster at higher concentrations (Fig. 4 C and D). This is very close to the values observed with agonist uncaging (Figs. 1E and 2A). Because the time resolution of both methods is clearly sufficient to resolve submillisecond time courses (Fig. 1E and SI Appendix, Fig. S3), these results show that there are indeed conformational changes in nonvisual GPCRs that occur with rhodopsin-like speed. In contrast and again in accordance with the UV-uncaging experiments, activation of the A sensor was about 20 times slower than that of the E sensor (Fig. 4B).
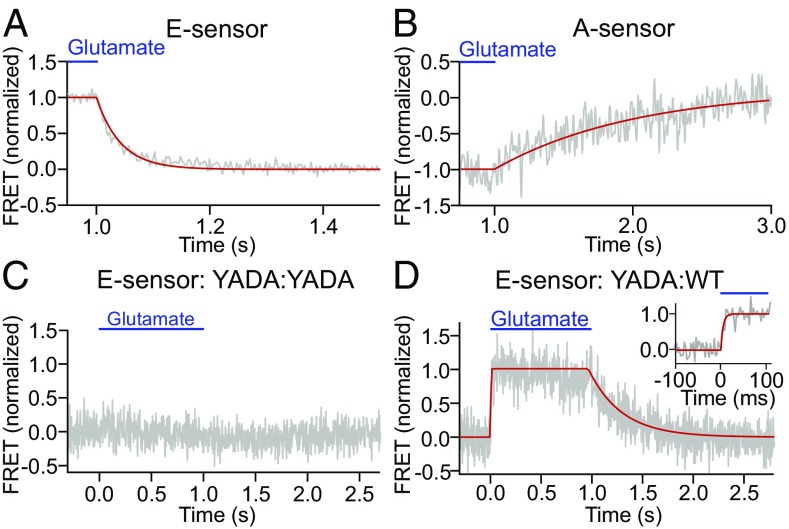
The solution exchange also allowed us to determine deactivation kinetics by jumping back to buffer-only conditions. These deactivation kinetics proved to be notably slower than the respective activation kinetics at high glutamate concentrations (Fig. 3 C and D). For the E and A sensors, the time constants were 43 ± 1 and 900 ± 60 ms, respectively (Fig. 5 A and B).
Fig. 5.
Deactivation time course in the E and A sensors and responses of activation-deficient E-sensor mutants. (A and B) Deactivation time courses in E sensors (A) and A sensors (B). Shown are averaged responses (gray) to the switch from 1 mM glutamate (blue) back to control buffer applied via the piezo-controlled concentration jump system. The time courses were fitted by a single exponential yielding the time constants τoff = 43 ± 1 ms (n = 52) for the E sensor (A) and τoff = 900 ± 60 ms (n = 6) for the A sensor (B; blue curves). (C and D) Activation and deactivation time courses in binding-deficient YADA mutant E sensors. YADA mutations were introduced in lobe II of the ligand binding domain to suppress ligand binding. (C) Binding sites in both subunits mutated. No FRET response to 1 mM glutamate was observed. (D) Response of a mixed dimer E sensor (YADA:WT) shows that one functional binding site is sufficient to permit activation. The time constants were about six times slower than observed in receptors containing two functional binding sites, yielding τon = 7 ± 2 ms (n = 6) and τoff = 295 ± 16 ms (n = 6). (Inset) Activation time course at an extended timescale.
It has been suggested that, in an activated mGluR1 dimer, only one of the 7-TM domains needs to become active but that, for full activation, the occupancy of both ligand binding sites is required (20, 30). To investigate this potential role of binding site occupancy, we generated binding-defective E-sensor protomers by introducing mutations in lobe II of the ligand binding VFT domain analogous to those described for the mGluR5 [termed YADA mutants (30)]. E-sensor dimers bearing these mutations in only one (YADA:WT) or both protomers (YADA:YADA) were then produced with the help of the dimerization domains in the C termini derived from the GABAB1 and GABAB2 receptors as described previously (20, 30). Fig. 5C shows that the YADA:YADA E sensor was completely unresponsive to glutamate, indicating that indeed ligand binding was suppressed. In contrast, the E sensor bearing only one mutation (YADA:WT) showed a clear FRET response, but the kinetics were slowed down approximately sixfold, with on- and off-time constants of 7 ± 2 and 295 ± 16 ms, respectively. This indicates that occupancy of both ligand binding sites in the mGluR1 is required for full activation speed.
Discussion
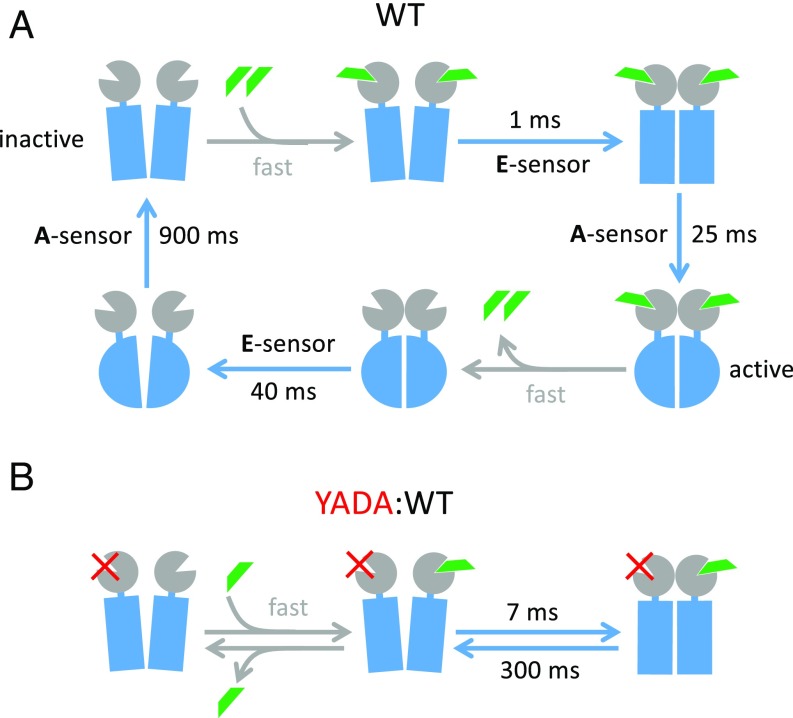
The cycle of activation and deactivation by rapid application of a saturating agonist concentration to the mGluR1 is schematically summarized in Fig. 6A. Using techniques that allow for activation in the submillisecond range, we find that the first major activation step, the rearrangement of the two protomers within the receptor dimer, occurs within 1–2 ms. This step is most likely preceded by agonist-induced closure of the ligand binding VFT domain, which mGluRs share with ionotropic receptors (23, 31) and which has been shown to switch with submillisecond kinetics both in single-molecule experiments with the isolated domains and in functional experiments (11, 32). This is faster than the dimer rearrangement that we observed. In our experiments with solution switches, the loss of concentration dependence of the activation speed at high ligand concentrations (Fig. 4D) shows that the observed timescale indeed reports the speed of the dimer rearrangement and is not limited by diffusion or binding dynamics. This apparently rate-limiting rearrangement within an mGluR, the mGluR5, has very recently been characterized structurally by cryo-EM and crystallography (33). These data suggest that closure of the VFT domain is intricately linked with a major rearrangement of the entire dimer, which also involves a repositioning of the transmembrane (7-TM) domains that rotate and come closer relative to each other. It is entirely plausible that this is what the E sensor indicates.
Fig. 6.
Schematic illustration of the activation and deactivation kinetics in mGluR1. The glutamate ligands are indicated in green. The specified times are the time constants determined by the fits. It is assumed that the ligand binding is not rate limiting [i.e., that the ligand concentration is sufficiently high and that ligand binding and closure of the VFT domain occur at submillisecond speed (“fast”)] (31, 32). (A) WT receptor. (B) YADA mutant in one protomer to eliminate ligand binding (red crosses).
The coupling between this rapid rearrangement step and activation of the transmembrane (7-TM) domain is apparently loose, resulting in ∼20-fold slower activation of the A sensor vs. the E sensor. Loose coupling between initial and later steps in the agonist-induced activation of class A GPCRs has been suggested by NMR structural studies (13) as well as molecular dynamics simulations (34), and it seems that the same is true for activation of class C GPCRs via their VFT domain. Not only activation but also, deactivation are much slower for the A sensor than the E sensor. As a consequence, the reaction scheme indicates that, in addition to the unliganded inactive (Fig. 6A, Upper Left) and the double-liganded active states (Fig. 6A, Lower Right), there are two metastable intermediate states, where either only the rearrangement (Fig. 6A, Upper Right) or the 7-TM conformation (Fig. 6A, Lower Left) corresponds to the fully active state. The roles and signaling properties of these intermediate states remain to be elucidated.
Finally, Fig. 6B shows that, for full activation and deactivation speeds of the dimer rearrangement, both ligand binding sites need to be occupied by an agonist, since knocking out one of two binding sites considerably slows the signals of the E sensor in both directions.
In recent biophysical studies on purified GPCRs, the fully active state of the receptor was only obtained in the presence of other proteins that stabilized it (i.e., a G protein, a β-arrestin, or a corresponding nanobody) (10, 13, 35, 36). This state was, therefore, not seen in the recent structural analysis of mGluR5 activation (33). Because of high intracellular levels of GTP, this fully active state is presumably only transient in intact cells, and it is possible that it may not or only in part correspond to the active state reported here.
Taken together, our data indicate that transmembrane domain activation of the mGluR1 begins with a conformational change in the receptor dimer that lasts only about 1 ms. However, in contrast to rhodopsin activation, where the active meta-II state of the transmembrane domain is achieved in 1 ms, achievement of the fully active conformation of the mGluR1 in the 7-TM domain then takes much longer (∼20 ms), indicating loose coupling between the two processes. Likewise, the return to the inactive state occurs stepwise: first, in the dimer rearrangement and second, in the 7-TM domain. The signaling competence of the resulting intermediate states remains to be identified. This stepwise process of activation and deactivation may contribute to flexibility and the receptors’ ability to trigger downstream signals.
Materials and Methods
Reagents.
Chemicals were purchased from the following suppliers: MNI-caged glutamate was from Tocris Biosciences, l-glutamine was from Pan Biotech, penicillin (100 U/mL) and streptomycin (100 µg/mL) were from Gibco, Effectene Transfection Reagent was from Qiagen, BSA was from AppliChem, and poly-d-lysine and sodium pyruvate were from Sigma-Aldrich.
Generation of mGluR1 FRET Sensors.
The cRNA of the protomers of the A and E sensors (20) was used as templates for the generation of mGluR1 C254E protomers. In both cases, mutant FRET sensors were generated by site-directed mutagenesis using standard molecular biology procedures and cloned into a pRK5 vector as described previously (20). Additional FRET sensors were recloned into a pGEM-HEnew vector (37) for expression in Xenopus oocytes. These constructs were used as templates to generate mGluR1 Y226A, mGluR1 D308A, and mGluR1 Y226A/D308A by site-directed mutagenesis using overlap extension PCR. For cRNA production, the constructs were linearized, and cDNA was created using the mMESSAGE mMACHINE T7 kit (Thermo Fisher).
Experiments in HEK293T Cells with Flash Photolysis.
Cell culture.
HEK293T cells were cultured in DMEM (PAN Biotech) supplemented with 10% FCS (Biochrom AG), l-glutamine, penicillin (100 U/mL), and streptomycin (100 µg/mL) at 37 °C and 7% CO2.
Transient transfection of mGluR1 FRET sensors in HEK293T cells.
For FRET experiments, cells were seeded onto poly-d-lysine–coated UV-transparent quartz coverslips (Ted Pella, Inc.) in six-well plates 12–16 h before transfection. Cells were transfected using Effectene according to the manufacturer’s instructions. cDNA amounts were 150 ng of each protomer of the WT and mutant E sensors and 150 ng of the WT and mutant A sensors per coverslip. To minimize glutamate contact with receptors, cell culture medium was exchanged for DMEM-GlutaMAX (Gibco) 24 h after transfection. Approximately 60 h after transfection, cells were washed twice with HBSS (150 mM NaCl, 2.5 mM KCl, 2 mM MgCl2, 4 mM CaCl2, 10 mM Hepes, 10 mM glucose, pH 7.4) and incubated in HBSS buffer supplemented with 1.75 U/mL glutamate-pyruvate transaminase (Roche), 4 mM sodium pyruvate, and 0.1% BSA for 1 h.
Single-cell FRET measurements in HEK293T cells.
For FRET experiments, coverslips mounted in an experimental chamber were placed on a custom-built inverted microscope (Axio Observer D1; Zeiss GmbH) kept at room temperature that was equipped with an oil-immersion objective (Fluar 100×/1.30 Oil UV; Zeiss GmbH) and a light-emitting diodes system (pE-4000; CoolLED Ltd.) as an excitation light source. On excitation with 435 ± 9 nm, fluorescence emission was simultaneously recorded at 483 ± 16 nm (CFP) and 535 ± 15 nm (YFP) before and after the addition of MNI-caged l-glutamate (1 mM final concentration). Fluorescence signals were detected by photometry systems, each of which contained a gated photomultiplier (ET Enterprises Ltd.) and photometer amplifier unit (Myotronic). Photocurrents were digitized at 10-kHz sampling frequency using an analog–digital converter Axon Digidata 1550 (Molecular Devices) and recorded with the pClamp software (Molecular Devices).
Flash photolysis in HEK293T cells.
Photouncaging of MNI-caged l-glutamate was achieved by a short 375-nm laser flash generated by a trigger-controlled UV laser source (DL 375; Rapp OptoElectronic GmbH). The laser source was coupled to the microscope via a quartz fiber optic light guide and collimated to the objective. To estimate the size of UV laser focal spot size, we used caged fluorescein. A thin layer of 5-carboxymethoxy-2-nitrobenzyl fluorescein was laid on a quartz coverslip and allowed to dry to minimize lateral diffusion of the dye. The size of the fluorescent spot was measured through the obtained image from the CMOS camera (DCC3240N; Thorlabs). Intensity image was plotted in a 3D domain, and assuming that the laser beam has an ideal Gaussian intensity profile, we approximated it with a 2D Gaussian function:
where F0 is the offset; A is the amplitude relative to the baseline; xc and yc are coordinates of the peak; w1 and w2 are x and y spreads of the blob, respectively; and θ is the orientation of the blob.
FWHM values were further calculated as follows:
Analysis of uncaging data.
Fluorescence emissions of both donor and acceptor were corrected for background, fluorophores quenching, and bleed through of donor light into the acceptor channel essentially as described previously (38). In detail, background was measured in each channel for every experiment as a fluorescence intensity of neighboring nontransfected cells. Fluorophore quenching was corrected by subtracting the corresponding exponential curves for CFP and YFP. Bleed through of CFP emission into the YFP channel was estimated as 36%. FRET ratios were further corrected for the transient inner filter effect of the nitrosoindole by-product of MNI–l-glutamate uncaging (39). Corrected photocurrents were analyzed with OriginPro software (OriginLab).
FRET values at time point t were determined as follows:
where R(t) is the observed YFP/CFP ratio, Rp is a plateau ratio value at the peak of transient signal, and Rb is a baseline ratio value before uncaging.
Experiments in Outside-Out Patches Using Confocal Patch-Clamp Fluorometry.
Oocyte preparation and cRNA injection.
Oocytes of Xenopus laevis were obtained either from Ecocyte (Castrop-Rauxel) or surgically from female adults under anesthesia (0.3% 3-aminobenzoic acid ethyl ester) as described previously (40). The procedures had approval from the authorized animal ethics committee of the Friedrich Schiller University Jena. The respective cRNAs (20–40 ng) were injected into the oocytes. The oocytes were incubated at 18 °C and used between 4 and 6 d after injection.
Piezo-controlled concentration jumps at outside-out membrane patches.
Outside-out patches from Xenopus oocytes were formed by using standard patch-clamp techniques. The patch pipettes were pulled from quartz tubing (P-2000; Sutter Instrument) with outer and inner diameters of 1.0 and 0.7 mm, respectively (VITROCOM). The corresponding pipette resistance was 0.9–2.3 MΩ. The bath and pipette solution contained 150 mM KCl, 1 mM EGTA, and 10 mM Hepes (pH 7.4 with KOH). Recording was carried out at room temperature using an Axopatch 200B amplifier (Axon Instruments). Electrophysiology was controlled by the ISO3-Software (MFK Niedernhausen).
A patch pipette with an outside-out patch was positioned in front of the outlet of a double-barreled theta glass (diameter of ∼100 µm) that was mounted on a piezo actuator (Physik Instrumente). A voltage pulse controlled the piezo and displaced the theta-glass pipette and thus, the position of the laminar stream of solution at the patch. The flow speed of the solution was set to 130 mm/s. The time course of solution exchange was measured by following the fluorescence of a 1 µM DY647 solution in the line-scan mode. Fitting of such time courses at the patch yielded a mean time constant for the solution exchange of 220 ± 30 μs (SI Appendix, Fig. S3). The solution exchange in the bulk solution was estimated to be 120 µs.
FRET experiments in membrane patches.
The relative position of the patch pipettes in front of the application pipette was carefully adjusted at a distance of 10–15 µm. Fluorescence images were recorded through an 40×/1.2 C-Apochromat water immersion objective with a confocal microscope (LSM710; Carl-Zeiss). CFP and YFP were excited with the 458- or 514-nm line of an Argon laser, and the detection channel was set to 459–508 or 517–581 nm, respectively. For fast time series, either small images (16 × 8 pixel) or line scans were acquired at 458-nm excitation. The electrophysiology setup triggered both the microscope and the piezo device.
Analysis of data in membrane patches.
Regions of interest were selected manually, and the extracted time series were analyzed with an inhouse routine written in Igor Pro-6.34 (Wavemetrics). After subtraction of cross-talk in the FRET channels, the signals of both channels were individually corrected for photobleaching. Contribution of direct excitation of YFP by 458 nm was found to be negligible. To preserve minute details of the kinetics, we calculated the corrected FRET signal by FRETCor = ChFRET − f × ChCFP. ChFRET and ChCFP are the signals in the FRET and CFP (donor) channels, respectively. The correction factor f depends on detection efficiencies and quantum yield of donor and acceptor, respectively. It was calibrated by minimizing the correlated signal fluctuations due to slight fluctuations in the pipette position while preserving the anticorrelated signal change due to FRET. With this strategy, any possible offset in the donor or FRET channel signal cannot distort the kinetics, as it is the case for signal ratios. As the absolute value of FRETCor depends on the expression level and the patch size, all signals were normalized before fitting and averaging.
To avoid any influence of possible time jitter caused, for example, by variations in flow speed, pipette distance, or LSM-trigger jitter, the time points of signal start were fitted together with the fit of the signal time courses. This avoided also blurring of the kinetics on averaging of the signals.
Fitting of mGluR1 Activation Kinetics.
Kinetics were fitted as monoexponential activation and deactivation:
Fitting individual activation or deactivation time courses or the activation/deactivation time course as a whole yielded similar results for piezo-controlled concentration jumps. Only the activation term was fitted in uncaging experiments.
Baseline (base) and amplitude (amp) of the signal were appropriately set to 0 and 1 (or −1) for normalized data. tpulse_length was set according to the experiment (typically 1 s). The time constants of activation (τon) and deactivation (τoff) were fitted. The concentration jump (t0) was fitted for piezo-controlled concentration jumps and set for uncaging experiments.
Supplementary Material
Acknowledgments
We thank Veronika Hlavackova and Klaus-Peter Hofmann for helpful discussions. This work was supported by Deutsche Forschungsgemeinschaft Grant TR166 Project C04 (to K.B. and M.J.L.) and Bundesministerium für Bildung und Forschung Grant OptiMAR (to M.J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900261116/-/DCSupplemental.
References
- 1.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Hofmann KP, et al. A G protein-coupled receptor at work: The rhodopsin model. Trends Biochem Sci. 2009;34:540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Knierim B, Hofmann KP, Ernst OP, Hubbell WL. Sequence of late molecular events in the activation of rhodopsin. Proc Natl Acad Sci USA. 2007;104:20290–20295. doi: 10.1073/pnas.0710393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lohse MJ, Hofmann KP. Spatial and temporal aspects of signaling by G-protein-coupled receptors. Mol Pharmacol. 2015;88:572–578. doi: 10.1124/mol.115.100248. [DOI] [PubMed] [Google Scholar]
- 5.Lohse MJ, Maiellaro I, Calebiro D. Kinetics and mechanism of G protein-coupled receptor activation. Curr Opin Cell Biol. 2014;27:87–93. doi: 10.1016/j.ceb.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Bünemann M, Bücheler MM, Philipp M, Lohse MJ, Hein L. Activation and deactivation kinetics of α 2A- and α 2C-adrenergic receptor-activated G protein-activated inwardly rectifying K+ channel currents. J Biol Chem. 2001;276:47512–47517. doi: 10.1074/jbc.M108652200. [DOI] [PubMed] [Google Scholar]
- 7.Yao X, et al. Coupling ligand structure to specific conformational switches in the β2-adrenoceptor. Nat Chem Biol. 2006;2:417–422. doi: 10.1038/nchembio801. [DOI] [PubMed] [Google Scholar]
- 8.Damian M, Mary S, Martin A, Pin JP, Banères JL. G protein activation by the leukotriene B4 receptor dimer. Evidence for an absence of trans-activation. J Biol Chem. 2008;283:21084–21092. doi: 10.1074/jbc.M710419200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bockenhauer S, Fürstenberg A, Yao XJ, Kobilka BK, Moerner WE. Conformational dynamics of single G protein-coupled receptors in solution. J Phys Chem B. 2011;115:13328–13338. doi: 10.1021/jp204843r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nygaard R, et al. The dynamic process of β(2)-adrenergic receptor activation. Cell. 2013;152:532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olofsson L, et al. Fine tuning of sub-millisecond conformational dynamics controls metabotropic glutamate receptors agonist efficacy. Nat Commun. 2014;5:5206. doi: 10.1038/ncomms6206. [DOI] [PubMed] [Google Scholar]
- 12.Vafabakhsh R, Levitz J, Isacoff EY. Conformational dynamics of a class C G-protein-coupled receptor. Nature. 2015;524:497–501. doi: 10.1038/nature14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manglik A, et al. Structural insights into the dynamic process of β2-adrenergic receptor signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregorio GG, et al. Single-molecule analysis of ligand efficacy in β2AR-G-protein activation. Nature. 2017;547:68–73. doi: 10.1038/nature22354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vilardaga JP, Bünemann M, Krasel C, Castro M, Lohse MJ. Measurement of the millisecond activation switch of G protein-coupled receptors in living cells. Nat Biotechnol. 2003;21:807–812. doi: 10.1038/nbt838. [DOI] [PubMed] [Google Scholar]
- 16.Lohse MJ, Nuber S, Hoffmann C. Fluorescence/bioluminescence resonance energy transfer techniques to study G-protein-coupled receptor activation and signaling. Pharmacol Rev. 2012;64:299–336. doi: 10.1124/pr.110.004309. [DOI] [PubMed] [Google Scholar]
- 17.Lohse MJ, et al. Kinetics of G-protein-coupled receptor signals in intact cells. Br J Pharmacol. 2008;153:S125–S132. doi: 10.1038/sj.bjp.0707656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pin JP, Bettler B. Organization and functions of mGlu and GABAB receptor complexes. Nature. 2016;540:60–68. doi: 10.1038/nature20566. [DOI] [PubMed] [Google Scholar]
- 19.Tateyama M, Abe H, Nakata H, Saito O, Kubo Y. Ligand-induced rearrangement of the dimeric metabotropic glutamate receptor 1α. Nat Struct Mol Biol. 2004;11:637–642. doi: 10.1038/nsmb770. [DOI] [PubMed] [Google Scholar]
- 20.Hlavackova V, et al. Sequential inter- and intrasubunit rearrangements during activation of dimeric metabotropic glutamate receptor 1. Sci Signal. 2012;5:ra59. doi: 10.1126/scisignal.2002720. [DOI] [PubMed] [Google Scholar]
- 21.Marcaggi P, Mutoh H, Dimitrov D, Beato M, Knöpfel T. Optical measurement of mGluR1 conformational changes reveals fast activation, slow deactivation, and sensitization. Proc Natl Acad Sci USA. 2009;106:11388–11393. doi: 10.1073/pnas.0901290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canepari M, Nelson L, Papageorgiou G, Corrie JET, Ogden D. Photochemical and pharmacological evaluation of 7-nitroindolinyl-and 4-methoxy-7-nitroindolinyl-amino acids as novel, fast caged neurotransmitters. J Neurosci Methods. 2001;112:29–42. doi: 10.1016/s0165-0270(01)00451-4. [DOI] [PubMed] [Google Scholar]
- 23.Kunishima N, et al. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 24.Rondard P, et al. Coupling of agonist binding to effector domain activation in metabotropic glutamate-like receptors. J Biol Chem. 2006;281:24653–24661. doi: 10.1074/jbc.M602277200. [DOI] [PubMed] [Google Scholar]
- 25.Zheng J, Zagotta WN. Patch-clamp fluorometry recording of conformational rearrangements of ion channels. Sci STKE. 2003;2003:PL7. doi: 10.1126/stke.2003.176.pl7. [DOI] [PubMed] [Google Scholar]
- 26.Biskup C, et al. Relating ligand binding to activation gating in CNGA2 channels. Nature. 2007;446:440–443. doi: 10.1038/nature05596. [DOI] [PubMed] [Google Scholar]
- 27.Kusch J, et al. How subunits cooperate in cAMP-induced activation of homotetrameric HCN2 channels. Nat Chem Biol. 2011;8:162–169. doi: 10.1038/nchembio.747. [DOI] [PubMed] [Google Scholar]
- 28.Nache V, Eick T, Schulz E, Schmauder R, Benndorf K. Hysteresis of ligand binding in CNGA2 ion channels. Nat Commun. 2013;4:2866. doi: 10.1038/ncomms3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jonas P. Fast application of agonists to isolated membrane patches. In: Sakmann B, Neher E, editors. Single-Channel Recording. Plenum Press; New York: 1995. pp. 231–243. [Google Scholar]
- 30.Kniazeff J, et al. Closed state of both binding domains of homodimeric mGlu receptors is required for full activity. Nat Struct Mol Biol. 2004;11:706–713. doi: 10.1038/nsmb794. [DOI] [PubMed] [Google Scholar]
- 31.O’Hara PJ, et al. The ligand-binding domain in metabotropic glutamate receptors is related to bacterial periplasmic binding proteins. Neuron. 1993;11:41–52. doi: 10.1016/0896-6273(93)90269-w. [DOI] [PubMed] [Google Scholar]
- 32.Popescu G, Robert A, Howe JR, Auerbach A. Reaction mechanism determines NMDA receptor response to repetitive stimulation. Nature. 2004;430:790–793. doi: 10.1038/nature02775. [DOI] [PubMed] [Google Scholar]
- 33.Koehl A, et al. Structural insights into the activation of metabotropic glutamate receptors. Nature. 2019;566:79–84. doi: 10.1038/s41586-019-0881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dror RO, et al. Activation mechanism of the β2-adrenergic receptor. Proc Natl Acad Sci USA. 2011;108:18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye L, Van Eps N, Zimmer M, Ernst OP, Prosser RS. Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature. 2016;533:265–268. doi: 10.1038/nature17668. [DOI] [PubMed] [Google Scholar]
- 36.Staus DP, et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature. 2016;535:448–452. doi: 10.1038/nature18636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 38.Börner S, et al. FRET measurements of intracellular cAMP concentrations and cAMP analog permeability in intact cells. Nat Protoc. 2011;6:427–438. doi: 10.1038/nprot.2010.198. [DOI] [PubMed] [Google Scholar]
- 39.Trigo FF, Corrie JET, Ogden D. Laser photolysis of caged compounds at 405 nm: Photochemical advantages, localisation, phototoxicity and methods for calibration. J Neurosci Methods. 2009;180:9–21. doi: 10.1016/j.jneumeth.2009.01.032. [DOI] [PubMed] [Google Scholar]
- 40.Thon S, Schmauder R, Benndorf K. Elementary functional properties of single HCN2 channels. Biophys J. 2013;105:1581–1589. doi: 10.1016/j.bpj.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.