Significance
Regulation of translational quality control during adverse growth conditions is critical to maintain cellular homeostasis. Translational quality control is maintained by proofreading by the aminoacyl-tRNA synthetases (aaRS), which ensure that cognate aminoacyl-tRNAs are provided to the ribosome for protein synthesis. Our findings now show that under oxidative stress a global conformational change in the phenylalanyl-tRNA synthetase positively regulates quality control by increasing the rate of proofreading. These results demonstrate that the translational machinery is able to rapidly and positively respond to environmental challenges to maintain the accuracy of protein synthesis.
Keywords: aminoacyl-tRNA, oxidative stress, proofreading, quality control
Abstract
Accurate translation of the genetic code is maintained in part by aminoacyl-tRNA synthetases (aaRS) proofreading mechanisms that ensure correct attachment of a cognate amino acid to a transfer RNA (tRNA). During environmental stress, such as oxidative stress, demands on aaRS proofreading are altered by changes in the availability of cytoplasmic amino acids. For example, oxidative stress increases levels of cytotoxic tyrosine isomers, noncognate amino acids normally excluded from translation by the proofreading activity of phenylalanyl-tRNA synthetase (PheRS). Here we show that oxidation of PheRS induces a conformational change, generating a partially unstructured protein. This conformational change does not affect Phe or Tyr activation or the aminoacylation activity of PheRS. However, in vitro and ex vivo analyses reveal that proofreading activity to hydrolyze Tyr-tRNAPhe is increased during oxidative stress, while the cognate Phe-tRNAPhe aminoacylation activity is unchanged. In HPX−, Escherichia coli that lack reactive oxygen-scavenging enzymes and accumulate intracellular H2O2, we found that PheRS proofreading is increased by 11%, thereby providing potential protection against hazardous cytoplasmic m-Tyr accumulation. These findings show that in response to oxidative stress, PheRS proofreading is positively regulated without negative effects on the enzyme’s housekeeping activity in translation. Our findings also illustrate that while the loss of quality control and mistranslation may be beneficial under some conditions, increased proofreading provides a mechanism for the cell to appropriately respond to environmental changes during oxidative stress.
Translation of the genetic code is an essential process that generates functional proteins in the cell. One key player in translation is the aminoacyl-tRNA synthetase, which canonically ligates an amino acid to its cognate tRNA at the 3′CCA end of the acceptor stem (1). Aminoacylated tRNA (aa-tRNA) is used to decode mRNA at the ribosome to synthesize peptides in a codon-specific manner. In the case of phenylalanyl-tRNA synthetase (PheRS), phenylalanine (Phe) is aminoacylated onto tRNAPhe to generate Phe-tRNAPhe. Many amino acids have similar chemical and physical properties. Therefore, aaRSs must be able to discriminate against noncognate amino acids to ensure accurate translation of the genetic code. To aid in this, some aaRS, including PheRS, have proofreading activity to maintain high accuracy (2, 3). Proofreading can occur before the amino acid is ligated onto a tRNA or after the ester linkage has been formed, termed pretransfer and posttransfer editing, respectively (3). For example, tyrosine (Tyr) and Phe differ in structure by one hydroxyl group; therefore, PheRS uses its proofreading activity to discriminate against ligating Tyr onto tRNAPhe. If Tyr-tRNAPhe goes to the ribosome, Tyr can be misincorporated into the growing peptide at Phe codons, causing mistranslation. Translational errors can lead to the formation of protein aggregates, which have been implicated in numerous physiological defects (4–6). Therefore, maintaining translational accuracy is an essential cellular process.
Oxidative stress, an imbalance in the levels of free oxygen and nitrogen radicals, has previously been documented to alter translational fidelity and can have adverse effects on the cell if not properly regulated (5, 7). DNA, RNA, and proteins can be affected by oxygen radicals independently; therefore, all steps of translating the genetic code are susceptible to oxidative stress (8–12). Amino acid oxidation can cause changes to proteins that allow different interactions with the environment and altered activity (13). Oxidative stress causes diverse effects on aaRSs in various organisms. For example, in the case of Escherichia coli threonyl-tRNA synthetase (ThrRS), oxidation of a critical cysteine in the editing domain leads to reversible inactivation of editing, allowing for serine (Ser) to be misacylated onto tRNAThr. Accumulation of Ser-tRNAThr in the cell has potential to cause mistranslation at Thr codons (14). Conversely, in yeast, reversible oxidation of the mitochondrial PheRS inactivates aminoacylation activity due to the formation of disulfide bonds (15). This inactivation of PheRS leads to a growth defect in yeast, revealing that the aminoacylation activity of mitochondrial PheRS is critical for yeast during oxidative stress. In the case of mammalian methionyl-tRNA synthetase (MetRS), during oxidative stress, phosphorylated MetRS is able to interact with other noncognate tRNAs. MetRS is able to aminoacylate many tRNAs in the cells during oxidative stress to increase the presence of Met in cellular proteins (16). Increasing the proportion of Met in peptides protects the rest of the proteome from oxidative damage by using the additional sulfur groups to act as sinks for oxygen radicals. These examples highlight the diverse effects of oxidative stress on enzymatic activity and substrate specificity of aaRSs during translation.
Salmonella enterica serovar Typhimurium (S. Typhimurium) is an organism that is transiently exposed to high levels of oxidative stress. It is able to adapt to the acidic environment in the gastrointestinal tract, outcompete bacteria that typically reside in the gut, and cause infection where other bacteria are often removed by the immune response (17). Salmonella survives within the host microbiome in part due to its resiliency against oxidative stress generated during the host’s immune response. One method by which Salmonella survives is through its ability to replicate within a macrophage, an immune cell that generates reactive oxygen species (ROS) (18). It has previously been shown that ROS can target free amino acids, generating imbalances to the endogenous cellular amino acid pool (7). Specifically, during oxidative stress, there is an increase in the presence of ortho, meta, and para-Tyr isomers due to Phe oxidation at various positions of the aromatic ring (7). The generation of these isomers alters the amino acid pools, and high concentrations of noncognate amino acids can lead to mistranslation, presenting a significant challenge for PheRS proofreading. Therefore, understanding how PheRS compensates for these changes in the environment will explain how translational accuracy is affected. Here we report that during oxidative stress, S. Typhimurium PheRS undergoes a conformational change that allows aminoacylation activity to be retained while simultaneously increasing proofreading activity, preventing the accumulation of m-Tyr-tRNAPhe. In vitro and ex vivo analyses reveal that oxidative stress generates a hyperaccurate PheRS, positively regulating translational quality control during oxidative stress.
Results
Salmonella PheRS Is Subject to Protein Oxidation.
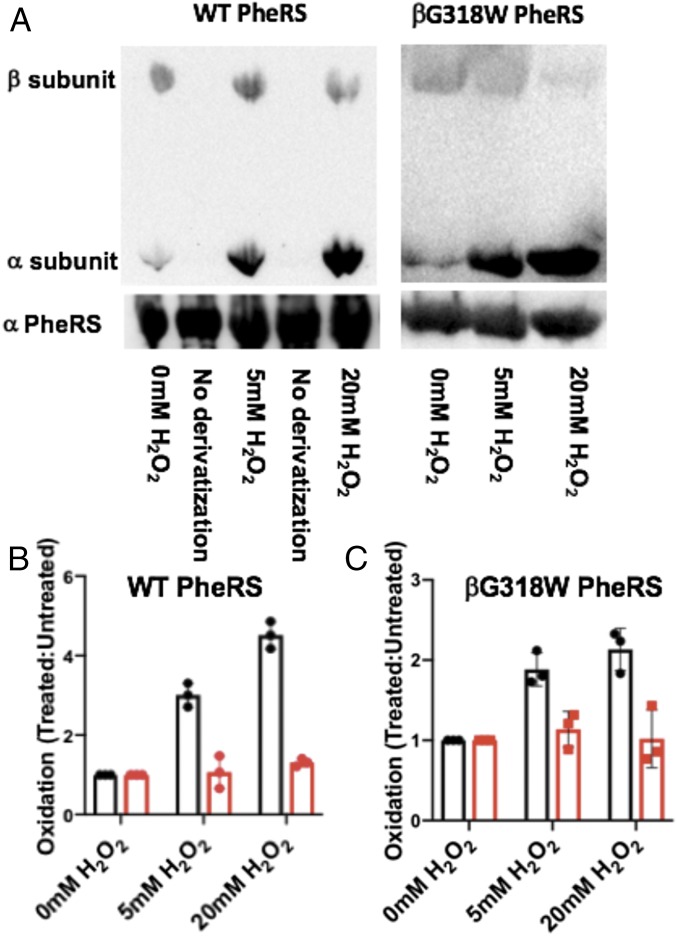
To investigate the possible role of PheRS in maintaining translational fidelity during oxidative stress, a recombinant PheRS was purified and used for subsequent in vitro analysis. S. Typhimurium and E. coli share a 95% sequence identity; therefore, a mutation in the β-subunit of PheRS (βG318W) was made in S. Typhimurium at the same position previously used to generate an editing-deficient E. coli PheRS (2). Editing-deficient S. Typhimurium PheRS was confirmed by its inability to hydrolyze Tyr-tRNAPhe (SI Appendix, Fig. S1). Using WT and editing-deficient S. Typhimurium PheRS, the role of translational fidelity during oxidative stress was further examined. Upon exposure to oxidative stress, many residues in both the WT and editing-deficient PheRS become oxidized. Immunoblot analysis, probing for 2,4-dinitrophenylhydrazine (DNPH) derivatized residues, indicated that PheRS oxidation seems to occur primarily in the α-subunit of PheRS, which includes the catalytic active site (Fig. 1). The β-subunit of PheRS, which contains the editing site, did not show significant oxidation after 5 or 20 mM H2O2 treatment (Fig. 1). This immunoblotting technique with anti-DNPH is only able to indicate ketones and aldehydes as oxidized residues; therefore, not all amino acid oxidation can be detected using these results. We used mass spectrometry and 5–5′-dithio-bis-[2-nitrobenzoic acid] (DTNB) analysis to further determine the extent of PheRS oxidation upon H2O2 treatment. Mass spectrometry analysis using oxidized recombinant proteins revealed extensive oxidation in both the WT and editing-deficient PheRS, with different residues being oxidized in each case (SI Appendix, Table S1). In addition, DTNB analysis revealed that the WT and editing-deficient PheRS have different oxidation patterns (Table 1). The WT PheRS is accessible to DTNB, while the editing-deficient PheRS does not show reactivity with DTNB, even without treatment with H2O2. Treatment of editing-deficient PheRS with 6 M guanidine hydrochloride to unfold the protein makes all cysteines accessible to the DTNB reagent. Combined, these results reveal that the oxidation of S. Typhimurium PheRS has a global effect on the structure of the protein beyond local oxidation of a single amino acid.
Fig. 1.
Quantification of oxidation of recombinant WT and editing-deficient PheRS. (A) A representative Western blot showing the oxidation of the α- and β-subunits of WT and βG318W PheRS. Lanes labeled no derivatization serve as a control to ensure the protein does not interact with the antibody unless derivatized with the aromatic structure. (B) Quantification of the relative oxidation of WT PheRS with 5 and 20 mM H2O2 relative to 0 mM H2O2. (C) Quantification of the relative oxidation of βG318W PheRS with 5 and 20 mM H2O2 relative to 0 mM H2O2. α-subunit quantified (black) and β-subunit quantified (red).
Table 1.
DTNB reactivity with cysteine
| Protein | Number of Cysteines |
| WT PheRS | 5 |
| WT PheRS + 5 mM H2O2 | 1 |
| Edt (−) PheRS | 1 |
| Edt (−) PheRS + 5 mM H2O2 | 1 |
| Edt (−) PheRS + 6M GndHCl | 4 |
Number of cysteines that react with DTNB reagent based on luminescence quantification.
Protein Oxidation Causes a Conformational Change of PheRS.
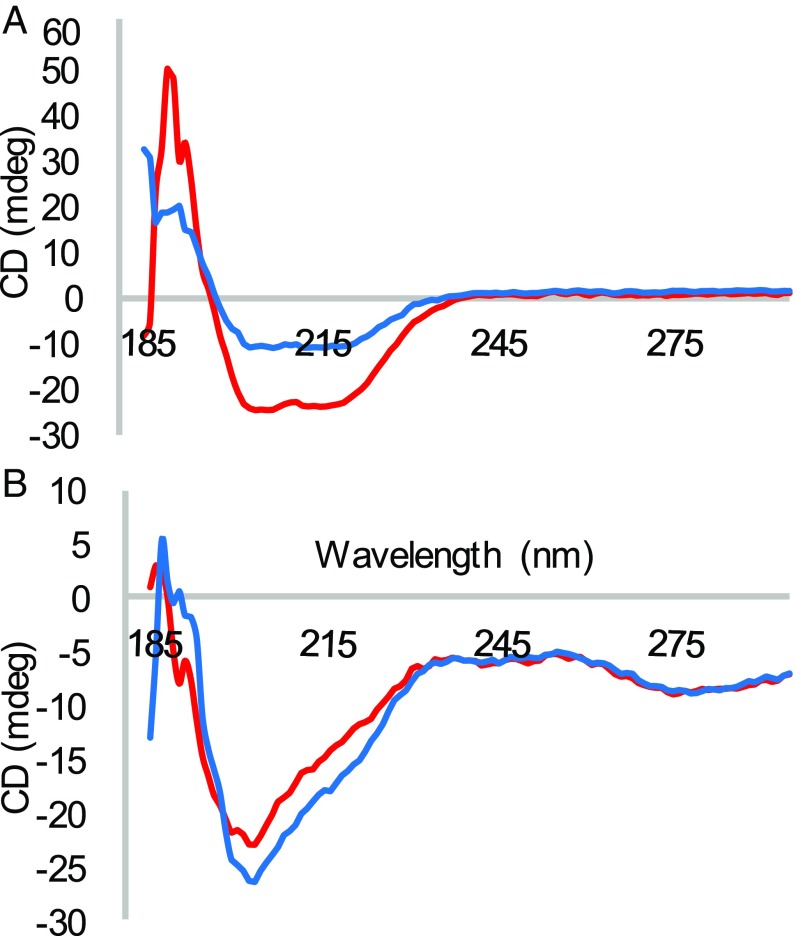
Protein oxidation can lead to the formation of disulfide bonds and other amino acid changes that induce a conformational change (15, 19). To investigate why editing-deficient and WT PheRS have different oxidation patterns, circular dichroism was used to explore the possibility of a conformational change in the editing-deficient PheRS. Circular dichroism reveals information about the secondary structure of proteins, indicating if a protein is mostly composed of β-sheets, α-helices, or is unstructured. Editing-deficient PheRS has an increase in the α-helical content compared with the WT PheRS, indicating more intrinsic structure (Fig. 2A). This increase in structure is consistent with mass spectrometry and DTNB analysis in that a more structured protein is not oxidized to the same extent as the WT PheRS. In addition, circular dichroism revealed that oxidation of both WT and editing-deficient PheRS decreases the α-helical content of the protein (Fig. 2B). Oxidation of PheRS causes a global conformational change to the protein, modifying its structure, which in turn could impact the activity of PheRS.
Fig. 2.
(A) Circular dichroism of recombinant WT (blue) and βG318W PheRS (red) between 200 and 285 nm. (B) Circular dichroism of oxidized recombinant WT (blue) and oxidized recombinant βG318W PheRS (red) between 200 and 285 nm.
Aminoacylation Activity of PheRS Is Not Affected by Oxidative Stress.
To determine if the conformational change induced by oxidative stress alters PheRS activity, in vitro analyses were performed. It has previously been reported that a change in a protein’s structure may correlate with a change in enzymatic function. For example, RfaH exists in two forms, in which the C-terminal domain is made up of α-helices or β-sheets (20). These two different conformational states are either autoinhibitory or translational activating, respectively, highlighting the potential of one protein to play two distinct roles based on its secondary structure (20). Steady-state kinetics of amino acid activation were determined by pyrophosphate exchange, revealing that oxidative stress does not impact the KM for activation of cognate Phe or the noncognate m-Tyr (Table 2) (2). In addition, tRNA aminoacylation reactions were used to determine the KM of tRNAPhe, which also revealed no significant changes during oxidative stress (Table 2). These results indicate that the conformational change caused by oxidation of PheRS does not impact PheRS activation or cognate aminoacylation of tRNAPhe.
Table 2.
Steady-state kinetics for PheRS under oxidative stress
| Phe | m-Tyr | tRNAPhe | |||||||
| Enzyme | KM (µM) | kcat (s−1) | kcat/KM (s-1/µM) | KM (µM) | kcat (s−1) | kcat/KM (s-1/µM) | KM (µM) | kcat (s−1) | kcat/KM (s-1/µM) |
| WT PheRS | 35 ± 10 | 1 ± 0.2 | 0.03 | 147 ± 17 | 0.5 ± 0.1 | 0.003 | 1.9 ± 0.7 | 6.3 ± 0.8 | 3.3 |
| WT PheRS + 5 mM H2O2 | 40 ± 14 | 0.8 ± 0.2 | 0.02 | 153 ± 31 | 0.3 ± 0.1 | 0.002 | 1.4 ± 0.7 | 7.2 ± 1 | 5.1 |
| Edit (−) PheRS | 30 ± 7 | 1.3 ± 0.3 | 0.04 | 153 ± 33 | 0.1 ± 0.01 | 0.001 | 2.7 ± 0.4 | 7.1 ± 2 | 2.6 |
| Edit (−) PheRS+ 5 mM H2O2 | 30 ± 9 | 0.6 ± 0.3 | 0.02 | 100 ± 31 | 0.2 ± 0.1 | 0.003 | 1.5 ± 0.7 | 2.4 ± 0.2 | 1.7 |
Steady-state kinetic parameters of PheRS for the activation of Phe and m-Tyr using ATP-PPi exchange and tRNAPhe using aminoacylation assays.
Oxidation of PheRS Increases Noncognate Substrate Proofreading.
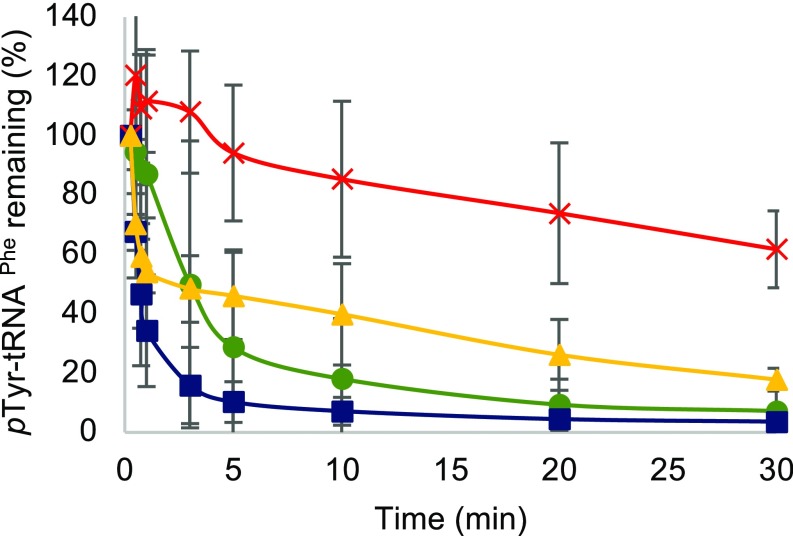
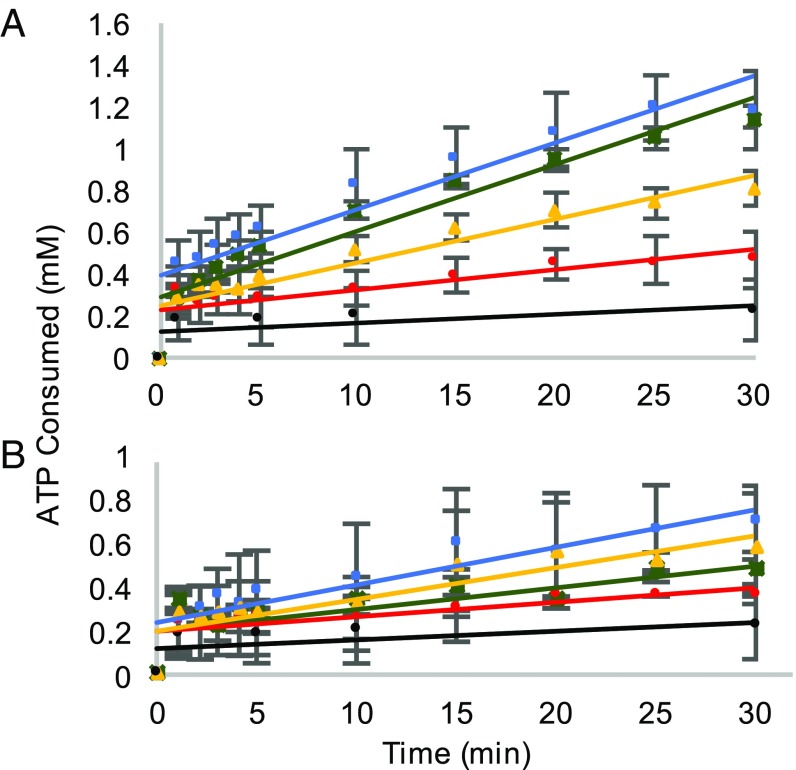
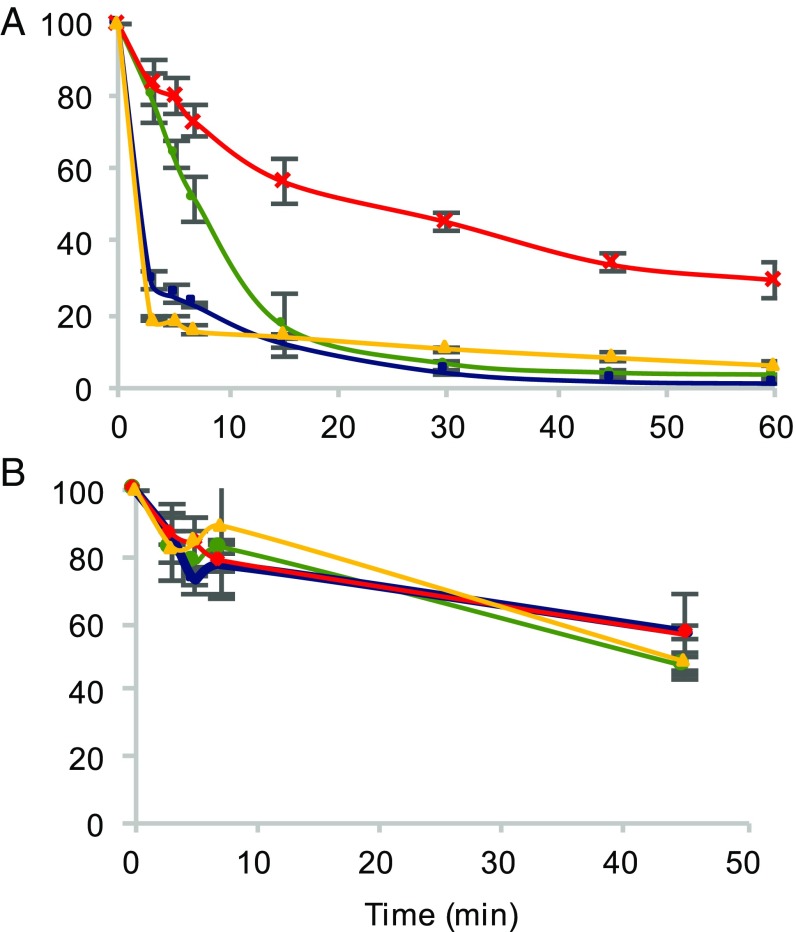
We have previously demonstrated that oxidative stress leads to elevated cytoplasmic levels of Tyr isomers, with m-Tyr accumulating to levels that lead to cytotoxic mistranslation when PheRS editing activity is impaired (7). To elucidate the effect of oxidative stress on proofreading activity of PheRS, the hydrolysis of misacylated p-Tyr-tRNAPhe was investigated (Fig. 3). In the case of the WT PheRS, treatment of PheRS with H2O2 increases the ability to hydrolyze mischarged tRNAPhe but does not impact the half-life of Tyr-tRNAPhe (Fig. 3 and SI Appendix, Fig. S2 and Table S2). Editing-deficient PheRS, upon treatment with H2O2, partially regains the ability to proofread mischarged p-Tyr-tRNAPhe by increasing hydrolysis of the ester linkage. In addition, oxidized editing-deficient PheRS decreases the half-life of Tyr-tRNAPhe by one-half of the nonoxidized editing-deficient PheRS (Fig. 3 and SI Appendix, Fig. S2 and Table S2). A lower half-life of mischarged tRNAPhe indicates more proofreading occurring. To further analyze the change in proofreading activity of oxidized PheRS, an ATP consumption analysis was performed to quantify the hydrolysis of misacylated tRNAPhe. In this analysis, ATP consumption corresponds to the combined activity of pretransfer and posttransfer editing due to the futile cycling of ATP through the reactivation of the hydrolyzed substrate. Consistent with the deacylation experiments, protein oxidation causes both WT and editing-deficient PheRS to consume more ATP after 30 min, which is indicative of an increase in pretransfer and posttransfer editing (Fig. 4A). The average rate of ATP consumed between 1 and 10 min revealed that the rate for oxidized and nonoxidized WT PheRS remained similar, with oxidized WT PheRS tending to have a higher rate of ATP consumption by 0.01 mM/min (SI Appendix, Table S3). The average rate of ATP consumed increased from 0.02 to 0.03 mM/min between 1 and 10 min when using oxidized βG318W PheRS compared with nonoxidized βG318W PheRS (SI Appendix, Table S3). This increase in proofreading activity is specific to misacylated tRNAPhe as Phe-tRNAPhe does not appear to be affected as extensively by H2O2 treatment (Fig. 4B). The slight increase in ATP consumed during Phe-tRNAPhe proofreading is not significant (SI Appendix, Table S4). Additionally, the increase in ATP consumption during oxidative stress is in a tRNA-independent manner (SI Appendix, Fig. S3). These results indicate that during oxidative stress, PheRS increases proofreading activity, while maintaining normal aminoacylation activity.
Fig. 3.
Hydrolysis of p-Tyr-tRNAPhe over time shown by percent of mischarged tRNAPhe remaining. Editing-deficient PheRS (red), editing-deficient PheRS + 5 mM H2O2 (yellow), WT PheRS (green), WT PheRS + 5 mM H2O2 (blue).
Fig. 4.
ATP consumption using WT and editing-deficient PheRS. (A) ATP consumption with m-Tyr-tRNAPhe. (B) ATP consumption with Phe-tRNAPhe. WT PheRS (green), WT PheRS + 5 mM H2O2 (blue), editing-deficient PheRS (red), editing-deficient PheRS + 5 mM H2O2 (yellow), and no enzyme control (black).
Misaminoacylation of tRNAPhe Decreases During Oxidative Stress.
An increase in PheRS proofreading activity during oxidative stress was further quantified by analyzing misacylation of tRNAPhe with m-Tyr using TLC. Without H2O2 treatment, editing-deficient PheRS is able to misacylate tRNAPhe with noncognate m-Tyr to a comparable extent to cognate Phe (SI Appendix, Fig. S4 A and B). Following protein oxidation, the ability of editing-deficient PheRS to mischarge tRNAPhe was decreased by ∼30% (SI Appendix, Fig. S4 A and B). Again, the aminoacylation of Phe-tRNAPhe was not affected by oxidative stress, as this editing activity is specific for noncognate misaminoacylation and does not perturb cognate aminoacylation activity (SI Appendix, Fig. S4C). These results further confirm that PheRS is able to increase its fidelity during oxidative stress, while not impacting canonical aminoacylation activity.
E. coli PheRS Proofreading Increases During Oxidative Stress in Cell Lysates.
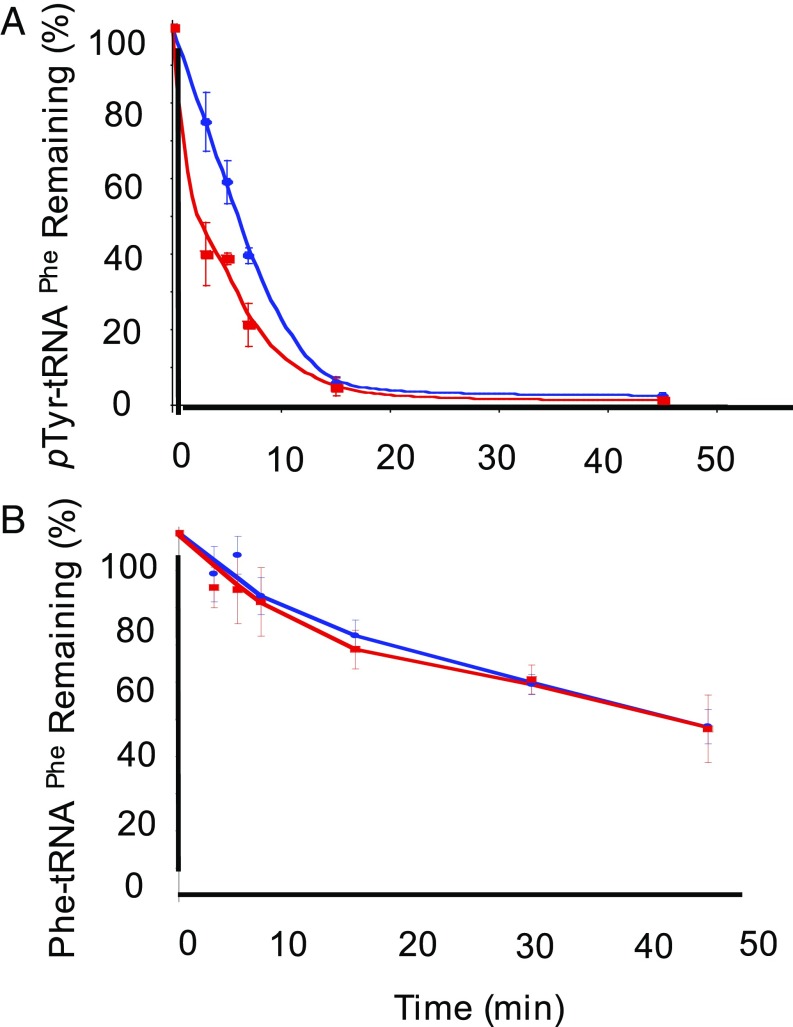
In vitro analyses revealed that treating recombinant PheRS with H2O2 leads to increased proofreading activity of PheRS. To investigate if this also happens ex vivo, E. coli cell lysates were used in deacylation assays. Midlog-phase liquid cultures of WT and editing-deficient strains of E. coli were treated with 20 mM H2O2, and cell-free lysates were prepared and assayed for aminoacylation and editing activities. The specific activity of PheRS in the oxidized cell-free lysate was significantly lower than nonoxidized cell lysates (SI Appendix, Table S5). Therefore, proofreading activity of PheRS was normalized to the specific activity of aminoacylation using the cell lysates. Using E. coli cell-free lysates in editing reactions containing misacylated p-Tyr-tRNAPhe revealed that oxidation leads to an increase in editing of misacylated tRNAPhe (Fig. 5A), without hydrolyzing cognate-aminoacylated Phe-tRNAPhe (Fig. 5B). The half-life of Tyr-tRNAPhe was also quantified to compare oxidative stress to normal conditions. The WT E. coli in the presence and absence of H2O2 have similar half-life of 11 min (SI Appendix, Fig. S5 and Table S6). Whereas the editing-deficient E. coli cell lysate under oxidative stress significantly decreases the half-life of mischarged tRNAPhe by 10 min (SI Appendix, Fig. S5 and Table S6), the half-life of Phe-tRNAPhe remains unchanged in the presence and absence of H2O2 (SI Appendix, Fig. S6 and Table S7). These results indicate that translational quality control is maintained during oxidative stress by increasing the proofreading of misacylated tRNA.
Fig. 5.
Native E. coli PheRS is able to edit mischarged tRNAPhe using cell lysates grown under oxidative stress. (A) Hydrolysis of H3 p-Tyr-tRNAPhe over time. (B) Hydrolysis of H3 Phe-tRNAPhe over time. Editing-deficient PheRS (red), editing-deficient PheRS + 5 mM H2O2 (yellow), WT PheRS (green), WT PheRS + 5 mM H2O2 (blue).
E. coli Lacking ROS Scavenging Display Increased PheRS Proofreading.
The oxidative stress response in E. coli involves the scavenging of oxidative radicals by catalases and peroxidases to break down H2O2 into water and oxygen. Without scavenging enzymes, katG, katE, and ahpCF, oxidative stress levels in the cell increase through the accumulation of H2O2 (21). When grown anaerobically, WT E. coli accumulates low levels of oxygen radicals, while HPX− E. coli (∆katG katE ahpCF) accumulates high levels of oxygen radicals, due to the lack of scavenging activity (21). Comparing PheRS activities of WT MG1655 and HPX− E. coli cell-free lysates reveals that basal oxidative stress levels have a similar effect compared with growth in the presence of H2O2. The specific activity of PheRS in the HPX− E. coli cell-free lysate was lower than WT E. coli cell lysates (SI Appendix, Table S8). Therefore, proofreading activity was normalized to the specific activity of the cell-free lysates. Using HPX− E. coli, we demonstrate that increased H2O2 in the cell, through the lack of scavenging enzymes, generates a hyperaccurate PheRS with an increased rate of hydrolysis of pTyr-tRNAPhe (Fig. 6A and SI Appendix, Fig. S7 and Table S9). The half-life of Tyr-tRNAPhe is significantly lower in the HPX− E. coli compared with WT E. coli, indicating an increase in proofreading activity in these cells. Again, the Phe-tRNAPhe half-life does not seem to change between HPX− and WT E. coli (SI Appendix, Fig. S8 and Table S10). The extent of increased proofreading is not as high as for cells grown with H2O2, consistent with reduced accumulation of H2O2 in cells grown anaerobically. Overall, increased proofreading in E. coli cell-free lysate lacking scavenging enzymes reveals that even low levels of H2O2 produced by the cell have the potential to increase PheRS accuracy.
Fig. 6.
E. coli cell lysates lacking scavenging enzymes (HPX−) edit mischarged tRNAPhe better than WT E. coli grown anaerobically. (A) Hydrolysis of H3 p-Tyr-tRNAPhe over time. (B) Hydrolysis of H3 Phe-tRNAPhe over time. WT E. coli (blue) and HPX− E. coli (red).
Discussion
PheRS Oxidation Positively Regulates Proofreading.
To accurately decode mRNA, properly aminoacylated tRNA is required; otherwise, mistranslation can occur. During oxidative stress, there is an increase in the presence of noncognate Tyr in the amino acid pools, which poses a threat to PheRS fidelity (7). How PheRS responds to the increased presence of noncognate substrate during oxidative stress remained unknown. Mass spectrometry analysis and circular dichroism reveal that PheRS oxidation leads to a conformational change, producing a protein with less α-helices. It is not oxidation of a specific residue in PheRS, but rather a global effect on the protein’s structure. These results are in contrast to the better characterized oxidation of a cysteine residue in ThrRS that alters proofreading activity (5). However, oxidation of proteins has been shown to generally impact the structure of proteins and encourage the formation of aggregates leading to altered cellular responses (15, 19, 22). Oxidative generation of an unstructured protein causes PheRS to function in a hyperaccurate manner by more rapidly hydrolyzing the ester linkage of noncognate Tyr-tRNAPhe both in vitro and in vivo. Steady-state kinetic analyses reveal that the conformational change induced by oxidative stress does not impact cognate or noncognate amino acid activation or cognate aminoacylation activity to generate Phe-tRNAPhe. These results reveal an instance in which oxidative stress positively regulates proofreading, whereas previous reports on the effect of oxidative stress on aaRS demonstrated that proofreading activity can be inhibited by H2O2 treatment (14). This highlights the importance of using an editing-deficient PheRS in our experiments because it allows us to notice changes in proofreading activity that may be more difficult to observe for WT protein (e.g., SI Appendix, Fig. S4). Furthermore, the lack of commercially available radiolabeled m-Tyr, the preferred noncognate substrate, may also contribute to the lack of observable statistically significant differences upon oxidation of WT PheRS in some assays (e.g., SI Appendix, Table S6).
Proofreading Protects Cells from Mistranslation Under Adverse Growth Conditions.
Positively regulating proofreading provides one mechanism by which S. Typhimurium is able to maintain translational accuracy during oxidative stress. In addition, due to shifts in amino acid pools during oxidative stress generating elevated levels of noncognate substrates, the increased proofreading activity of PheRS may be important for maintaining cellular homeostasis. During oxygen-limiting growth, norvaline (Nva) accumulates and poses a threat to the accuracy of leucyl-tRNA synthetase (LeuRS) (23, 24). LeuRS uses posttransfer editing to hydrolyze Nva-tRNALeu and prevent its incorporation into growing peptide chains. Similarly, during oxidative stress, Tyr isomers accumulate, posing a threat to the accuracy of PheRS. We have demonstrated that oxidation of PheRS increases translational quality control to help maintain high fidelity of aminoacylation upon exposure to high levels of noncognate Tyr isomers. Hyperaccuracy may be especially important to PheRS because the substrate for PheRS, Phe, can directly be oxidized to cytotoxic nonproteinogenic m-Tyr. It has also been demonstrated that oxidative stress effects aaRS proofreading adversely; however, in those cases, there is not a direct correlation to higher levels of noncognate amino acid in the cell. In Candida albicans, it has been reported that mistranslation may be beneficial by generating proteome diversity (25). In addition, phosphorylated MetRS is able to recognize diverse tRNAs in the cell to increase misincorporation of Met into the proteome (16). It is proposed that mismethionylation, leading to mistranslation, may be beneficial for the cell by allowing Met to quench oxygen radicals. Here we have reported that in S. Typhimurium, mistranslation may not be beneficial during oxidative stress due to the increase in presence of o-Tyr, p-Tyr, and m-Tyr. Overall, ensuring the accuracy of translation dependent on PheRS, rather than increasing proteome diversity, may be more important for the cell to survive during oxidative stress. Similarly, it has been proposed that the loss of proofreading upon oxidation of ThrRS may primarily function in detecting oxidant levels in the environment, rather than promoting potentially beneficial mistranslation (26).
Hyperaccuracy of Oxidized PheRS and the Stringent Response.
The stringent response is a cellular stress response that is activated upon amino acid starvation. Upon accumulation of uncharged tRNA, an alarmone, guanosine pentaphosphate or tetraphosphate [(p)ppGpp], is produced to modulate transcription of amino acid biogenesis genes and suppress transcription of genes involved in translation (27). The stringent response aims to generate a balance between translation and the nutrients available. Inhibiting PheRS quality control prevents appropriate activation of the stringent response due to the accumulation of mischarged tRNAPhe (28). Without the stringent response, bacteria cannot appropriately respond to nutrient-limiting environments. In Salmonella, regulation of the stringent response is required for many cellular properties such as growth and motility, as well as Salmonella host cell invasion (29). Therefore, increasing the accuracy of translation could allow for S. Typhimurium to maintain its ability to properly activate the stringent response and invade a host when amino acid pools are perturbed. Proper activation of stress responses allows for S. Typhimurium to colonize within and infect a host. Through this work, we have expanded the knowledge of the effects oxidative stress has on translational quality control in S. Typhimurium by defining hyperaccurate PheRS. S. Typhimurium can directly benefit from hyperaccuracy of translation through accurate protein production and proteome integrity and indirectly through proper activation of the stringent response during adverse growth conditions. S. Typhimurium can use these translational stress responses to complement the well-characterized transcriptional responses that are the foundation for S. Typhimurium pathogenesis to maintain resiliency during oxidative stress and survive within the host microbiome (30–32).
Methods
Salmonella PheRS Cloning and Purification.
Genomic DNA from S. Typhimurium 14028s (from John Gunn, The Ohio State University, Columbus, OH) was used to PCR-amplify pheS and pheT, which were cloned into pET28a(+). Editing-deficient S. Typhimurium PheRS was generated by site-directed mutagenesis in pheT (G318W). Plasmids containing proteins were expressed in E. coli BL21(DE3) with 1 mmol IPTG induction for 4 h. Cells were harvested, lysed by sonication, and purified using a TALON metal affinity resin. PheRS was eluted with 250 mM imidazole, and fractions containing protein were concentrated and dialyzed overnight in 50 mM Tris pH 7.5, 100 mM KCl, 5 mM MgCl2, 3 mM 2-mercaptoethanol, 5% glycerol, and then dialyzed 4 h in similar buffer with 50% glycerol for storage.
Detection of Oxidation of PheRS.
Purified protein was incubated with 0, 5, or 20 mM H2O2 at 37 °C for 1 h. Excess H2O2 was dialyzed out using a 3-kDa filtration unit. Fifteen micrograms of protein was incubated at 25 °C for 15 min, combined with equal-volume 12% SDS and 2× volume of either dinitrophenyl hydrazine (DNPH) or control solution. Neutralization buffer was added, and samples were run on a 10% SDS/PAGE gel. Gel was transferred onto a nitrocellulose membrane. Western blot was completed using antibody dilutions as recommended by the Millipore OxyBlot Protein Oxidation Detection Kit protocol. Image intensity was quantified using imageJ. Oxidation extent was normalized to a nonoxidized protein.
Enzymatic Assays.
Detailed descriptions of methods used to analyze steady-state kinetics, proofreading of PheRS, and preparation of materials is described in the SI Appendix, Supplemental Data Methods.
Circular Dichroism.
Purified proteins (0.2 mg/mL) in 100 mM KPO4 were analyzed in a quartz absorption cuvette with 1 mm path length. Proteins were oxidized as in the MS preparation above. Absorbance values between 190 and 260 nm were recorded.
PheRS-Specific Activity of E. coli Cell Lysates.
One hundred millimoles Na2+ Hepes (pH 7.2), 30 mM KCl, 10 mM MgCl2, 2 mM ATP, 15 µM tRNAPhe, 30 µM 14C-Phe, and either 12 µg or 36 µg total protein from cell lysate were combined and incubated at 37 °C for 1 h. Time points were taken at 0 (before adding lysate), 5, 15, 30, 45, and 60 min and placed onto filter papers presoaked with 5% TCA. The filter papers were subsequently washed in 3× 5% TCA and 1× 95% EtOH. Radiation was quantified using liquid scintillation counting. The PheRS-specific activity was determined by [charged 14C-Phe-tRNAPhe (pmol)/min]/total protein (mg).
Editing of p-Tyr-tRNAPhe Using E. coli Cell Lysates.
E. coli strains were grown to an OD600 0.4, and 5 mM H2O2 was added for 1 h. Cells were harvested, lysed by sonication, and lysate was cleared by centrifugation for 1 h at 22,000 g at 4 °C. Precharged pTyr-tRNAPhe was combined with 30 mM KCl, 10 mM MgCl2, and 0.16 mg/mL of cell lysate, and reaction aliquots were taken between 0–60 min and placed onto 3-mm filter papers presoaked with 5% TCA. The filter papers were washed as above, and radiation was quantified using liquid scintillation counting.
Supplementary Material
Acknowledgments
We thank James Imlay at the University of Illinois at Urbana–Champaign for generously providing us with the HPX− E. coli strain used in this study. This work was supported by the National Science Foundation (MCB-1715840), The Ohio State University Center for RNA Biology Fellowship (to R.E.S.) and Ohio State’s Second-Year Transformational Experience Program (to A.M.K.)
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901634116/-/DCSupplemental.
References
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Roy H, Ling J, Irnov M, Ibba M. Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase. EMBO J. 2004;23:4639–4648. doi: 10.1038/sj.emboj.7600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinis SA, Boniecki MT. The balance between pre- and post-transfer editing in tRNA synthetases. FEBS Lett. 2010;584:455–459. doi: 10.1016/j.febslet.2009.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, et al. Deficiencies in tRNA synthetase editing activity cause cardioproteinopathy. Proc Natl Acad Sci USA. 2014;111:17570–17575. doi: 10.1073/pnas.1420196111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ling J, Söll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc Natl Acad Sci USA. 2010;107:4028–4033. doi: 10.1073/pnas.1000315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 7.Bullwinkle TJ, et al. Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code. eLife. 2014;3:e02501. doi: 10.7554/eLife.02501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh R, Mitchell DL. Effect of oxidative DNA damage in promoter elements on transcription factor binding. Nucleic Acids Res. 1999;27:3213–3218. doi: 10.1093/nar/27.15.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willi J, et al. Oxidative stress damages rRNA inside the ribosome and differentially affects the catalytic center. Nucleic Acids Res. 2018;46:1945–1957. doi: 10.1093/nar/gkx1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogel C, Silva GM, Marcotte EM. Protein expression regulation under oxidative stress. Mol Cell Proteomics. 2011;10:M111.009217. doi: 10.1074/mcp.M111.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka M, Chock PB, Stadtman ER. Oxidized messenger RNA induces translation errors. Proc Natl Acad Sci USA. 2007;104:66–71. doi: 10.1073/pnas.0609737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nawrot B, Sochacka E, Düchler M. tRNA structural and functional changes induced by oxidative stress. Cell Mol Life Sci. 2011;68:4023–4032. doi: 10.1007/s00018-011-0773-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmad S, et al. Protein oxidation: An overview of metabolism of sulphur containing amino acid, cysteine. Front Biosci (Schol Ed) 2017;9:71–87. doi: 10.2741/s474. [DOI] [PubMed] [Google Scholar]
- 14.Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 15.Chakraborty S, Ganguli S, Chowdhury A, Ibba M, Banerjee R. Reversible inactivation of yeast mitochondrial phenylalanyl-tRNA synthetase under oxidative stress. Biochim Biophys Acta Gen Subj. 2018;1862:1801–1809. doi: 10.1016/j.bbagen.2018.04.023. [DOI] [PubMed] [Google Scholar]
- 16.Lee JY, et al. Promiscuous methionyl-tRNA synthetase mediates adaptive mistranslation to protect cells against oxidative stress. J Cell Sci. 2014;127:4234–4245. doi: 10.1242/jcs.152470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steele-Mortimer O. The Salmonella-containing vacuole: Moving with the times. Curr Opin Microbiol. 2008;11:38–45. doi: 10.1016/j.mib.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Caggiano M, et al. Oxidant-induced interprotein disulfide formation in cardiac protein DJ-1 occurs via an interaction with peroxiredoxin 2. J Biol Chem. 2016;291:10399–10410. doi: 10.1074/jbc.M115.699850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuber PK, Schweimer K, Rösch P, Artsimovitch I, Knauer SH. Reversible fold-switching controls the functional cycle of the antitermination factor RfaH. Nat Commun. 2019;10:702. doi: 10.1038/s41467-019-08567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S, You X, Imlay JA. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx- mutants of Escherichia coli. Proc Natl Acad Sci USA. 2005;102:9317–9322. doi: 10.1073/pnas.0502051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmgren A. Thioredoxin structure and mechanism: Conformational changes on oxidation of the active-site sulfhydryls to a disulfide. Structure. 1995;3:239–243. doi: 10.1016/s0969-2126(01)00153-8. [DOI] [PubMed] [Google Scholar]
- 23.Soini J, et al. Norvaline is accumulated after a down-shift of oxygen in Escherichia coli W3110. Microb Cell Fact. 2008;7:30. doi: 10.1186/1475-2859-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cvetesic N, Palencia A, Halasz I, Cusack S, Gruic-Sovulj I. The physiological target for LeuRS translational quality control is norvaline. EMBO J. 2014;33:1639–1653. doi: 10.15252/embj.201488199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miranda I, et al. Candida albicans CUG mistranslation is a mechanism to create cell surface variation. MBio. 2013;4:e00285-13. doi: 10.1128/mBio.00285-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J, Fan Y, Ling J. Mechanism of oxidant-induced mistranslation by threonyl-tRNA synthetase. Nucleic Acids Res. 2014;42:6523–6531. doi: 10.1093/nar/gku271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magnusson LU, Farewell A, Nyström T. ppGpp: A global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Bullwinkle TJ, Ibba M. Translation quality control is critical for bacterial responses to amino acid stress. Proc Natl Acad Sci USA. 2016;113:2252–2257. doi: 10.1073/pnas.1525206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azriel S, Goren A, Rahav G, Gal-Mor O. The stringent response regulator DksA is required for Salmonella enterica serovar Typhimurium growth in minimal medium, motility, biofilm formation, and intestinal colonization. Infect Immun. 2015;84:375–384. doi: 10.1128/IAI.01135-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coburn B, Grassl GA, Finlay BB. Salmonella, the host and disease: A brief review. Immunol Cell Biol. 2007;85:112–118. doi: 10.1038/sj.icb.7100007. [DOI] [PubMed] [Google Scholar]
- 31.Hannemann S, Galán JE. Salmonella enterica serovar-specific transcriptional reprogramming of infected cells. PLoS Pathog. 2017;13:e1006532. doi: 10.1371/journal.ppat.1006532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang X, Li X, Sun S, Jiang L. The transcriptional regulator VarN contributes to Salmonella Typhimurium growth in macrophages and virulence in mice. Res Microbiol. 2018;169:214–221. doi: 10.1016/j.resmic.2018.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.