Significance
Cell surface receptors assign and display unique identities to neurons and direct proper and robust wiring of neurons to create functional neural circuits. Recent work has identified two new classes of receptors in fruit flies, called the Dpr and DIP families with 32 members, which interact in 38 pairwise combinations. These proteins are implicated in neural identity, wiring, and survival in many parts of the fly nervous system. Here, using evolutionary, biochemical, and structural analyses, we show that Dprs and DIPs are members of an ancient bilaterian family of receptors. Members of this family share functional roles relevant to wiring across species, and their expansion may have been crucial in the emergence of the bilaterian nervous systems.
Keywords: immunoglobulin superfamily, molecular evolution, nervous system, IgLON, Dpr-DIP family
Abstract
The evolution of complex nervous systems was accompanied by the expansion of numerous protein families, including cell-adhesion molecules, surface receptors, and their ligands. These proteins mediate axonal guidance, synapse targeting, and other neuronal wiring-related functions. Recently, 32 interacting cell surface proteins belonging to two newly defined families of the Ig superfamily (IgSF) in fruit flies were discovered to label different subsets of neurons in the brain and ventral nerve cord. They have been shown to be involved in synaptic targeting and morphogenesis, retrograde signaling, and neuronal survival. Here, we show that these proteins, Dprs and DIPs, are members of a widely distributed family of two- and three-Ig domain molecules with neuronal wiring functions, which we refer to as Wirins. Beginning from a single ancestral Wirin gene in the last common ancestor of Bilateria, numerous gene duplications produced the heterophilic Dprs and DIPs in protostomes, along with two other subfamilies that diversified independently across protostome phyla. In deuterostomes, the ancestral Wirin evolved into the IgLON subfamily of neuronal receptors. We show that IgLONs interact with each other and that their complexes can be broken by mutations designed using homology models based on Dpr and DIP structures. The nematode orthologs ZIG-8 and RIG-5 also form heterophilic and homophilic complexes, and crystal structures reveal numerous apparently ancestral features shared with Dpr-DIP complexes. The evolutionary, biochemical, and structural relationships we demonstrate here provide insights into neural development and the rise of the metazoan nervous system.
Ig superfamily (IgSF) proteins, which form the largest single-pass cell surface and adhesion family in humans, are crucial to animal development and have undergone large gene family expansions during metazoan evolution (1–3). They have been heavily studied in the context of development and function of the immune and nervous systems. Unlike in the immune system, neural processes, such as neurite outgrowth, guidance, and synaptic targeting, employ IgSF and other cell surface molecules that are usually conserved between vertebrates and invertebrates. As the central functionality of IgSF proteins on the cell surface is mediated through the recognition of other surface receptors and ligands, recent efforts have focused on deorphanization of these proteins in vertebrates (4) and invertebrates (5) via high-throughput interactome studies. However, genomic and interactomic data can be difficult to interpret when proteins are not annotated for function and orthologous proteins in vertebrate and invertebrate model organisms cannot be identified.
Our interactome studies on the Drosophila IgSF have revealed two protein families with distinct neural expression patterns: the Dpr family, named after the founding member defective proboscis extension response (6), and their binding partners, the Dpr-interacting proteins (DIPs) (5, 7). Dprs and DIPs form a complex network consisting of 38 interactions among 32 proteins. Most of the Dprs and DIPs that have been studied thus far are expressed exclusively in the nervous system. In the pupal optic lobe, the larval ventral nerve cord, olfactory receptor neurons, and the neuromuscular system, each Dpr and DIP is expressed in a unique subset of neurons (5, 7–9). One Dpr is also expressed in postsynaptic muscle cells (10). In the optic lobe, Dprs and DIPs are expressed in distinctive combinations in different neuronal types, and synaptic targeting defects and neuronal death have been observed in dpr11 and DIP-γ mutants (7, 8). In the neuromuscular system, dpr11 and DIP-γ mutants show synapse maturation defects, while Dpr10 and DIP-α are necessary for the formation of synapses onto specific muscle targets (10, 11). In the olfactory system, Dprs and DIPs are necessary for neuronal adhesion and glomerulus formation (9). Overall, the available data suggest that Dprs and DIPs serve neuronal wiring functions, likely by acting as “identification tags” for neurons, and physically guiding their connectivity. Since the numbers of known synaptic targeting molecules are limited, the study of Dprs and DIPs has strong potential to illuminate mechanisms involved in the development of synaptic circuits.
Dprs and DIPs have domain structures that are similar to those of many other IgSF proteins (3). Following a signal peptide, they carry two and three Ig domains, respectively. Dprs and DIPs interact via their N-terminal Ig domains, creating a pseudosymmetric Ig-Ig complex (7). The C-terminal ends of Dprs and DIPs are strongly hydrophobic, either serving as transmembrane helices or as recognition sites for glycosylphosphatidylinositol (GPI) linkages to the plasma membrane. Most Dprs and DIPs do not have intracellular domains and lack conserved features in their juxtamembrane regions.
While Dpr and DIP homologs can be identified in arthropods, it is unclear whether they exist in other animals and have also undergone large gene family expansions. Therefore, we set out to uncover Dpr and DIP homologs across major metazoan groups and establish biochemical and structural similarities among the proposed proteins. We show that Dprs and DIPs share a common ancestor with vertebrate proteins known as IgLONs, which form a family of five neuronal proteins in humans. The shared roles of these proteins in neurite outgrowth and synapse formation (12) suggest that the family consisting of these proteins, here named Wirins, is primarily involved in nervous system development across bilaterians. The Wirin family expanded independently in vertebrates, arthropods, and mollusks through multiple gene duplications. We further show that the homophilic and heterophilic interactions characteristic of Dprs and DIPs are also observed among IgLONs and the nematode orthologs. In addition, we demonstrate the molecular interfaces known to mediate Dpr-DIP interactions are used by these orthologs. Overall, we describe a family of proteins that share conserved roles in nervous system development that evolved early in bilaterians and have undergone gene family expansions in conjunction with the evolution of complex nervous systems.
Results
The Wirin Family of IgSF Proteins with Neural Wiring Functions.
Dprs and DIPs were discovered and characterized in Drosophila melanogaster. To identify their homologs in other organisms, we carried out a phylogenetic analysis. For sequence mining, we used a reciprocal BLAST strategy in which a putative Dpr or DIP homolog was defined as a protein for which the best hit in a BLAST search against D. melanogaster proteins is a Dpr or a DIP. In this way, Dpr and DIP homologs could be distinguished from dozens of distantly related IgSF proteins. All examined protostomes have both Dpr and DIP homologs. Chordates have a family of neuronal proteins called IgLONs (12, 13) that appeared as DIP homologs. Nonchordate deuterostomes and nonbilaterians lack recognizable Dpr or DIP homologs. Using human IgLONs as a reference for further reciprocal BLAST analysis, additional putative IgLON homologs were identified in protostomes: CG34353, CG7166, Klingon, Lachesin, and Wrapper. No other homologs for these proteins were found in chordates.
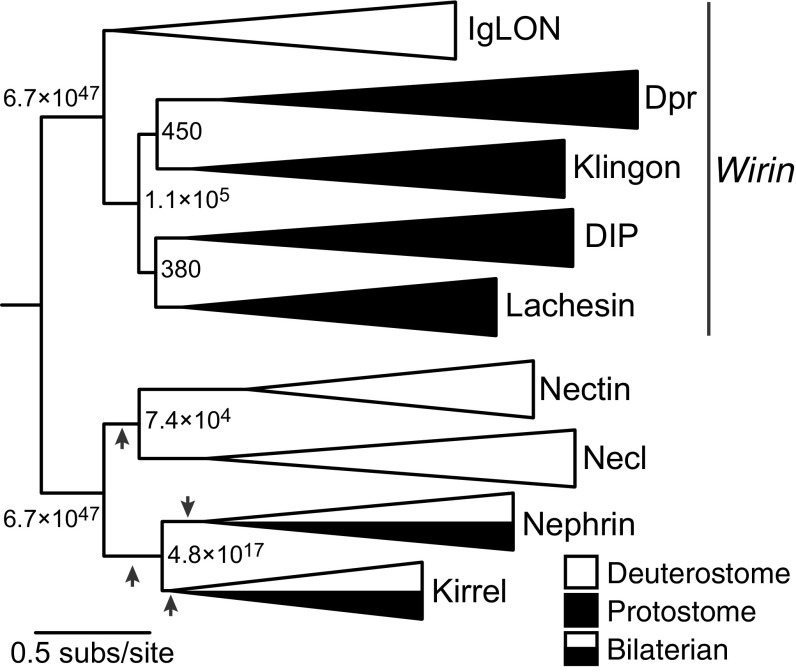
To reconstruct the history of these proteins, we inferred their maximum likelihood (ML) phylogeny (Fig. 1). Four IgSF subfamilies—Nectin, Necl, Kirrel, and Nephrin—were included as potential outgroups. DIPs, Dprs, IgLONs, Klingon, Lachesin, Wrapper, CG34353, and CG7166 form a monophyletic group that excludes all other IgSF sequences. We refer to this group as the Wirin (wiring immunoglobulin) family. There are multiple ways to place the root among the four outgroup IgSF subfamilies, all of which would imply the same number of gene gains and losses. None of these alternative rootings affect the relationships among the Wirins. Any placement of the root within the Wirins, however, entails numerous additional gene gains and losses.
Fig. 1.
The ML phylogeny of the Wirin family. Branch labels indicate approximate likelihood ratios (aLRs), defined as the likelihood of the ML topology divided by the likelihood of the next-best rearrangement of branches around the given branch. Arrows indicate alternative rootings that entail the same number of gene gains and losses as the phylogeny shown. For unreduced phylogenies, see Datasets S1–S6.
This phylogeny indicates that the Wirin family originated as a single gene in the last common ancestor of bilaterians. In deuterostomes, that progenitor evolved into the IgLON subfamily. In protostomes, a series of gene duplications gave rise to the DIP, Dpr, Klingon, and Lachesin subfamilies. IgLONs are thus coorthologous to DIPs, Dprs, Klingons, and Lachesins. Because DIPs, Dprs, and IgLONs share neuronal wiring functions (12, 13), the most parsimonious interpretation of this phylogeny is that Wirins descend from a single ancient progenitor that had similar functions in the nervous system of the bilaterian ancestor.
After the protostome and deuterostome lineages diverged, the various Wirin subfamilies expanded through subsequent gene duplications. Fig. 2A shows the ML phylogeny of IgLONs, which are unique to deuterostomes. When two poorly supported nodes are rearranged, the phylogeny becomes congruent with gene family expansion through two genome duplications thought to have occurred during early chordate evolution (14). An additional tetrapod-specific gene duplication gave rise to the total of five IgLONs in humans. In protostomes, early duplications generated one member of each major subfamily—a Dpr, a DIP, a Klingon, and a Lachesin—before the ancient split of Ecdysozoa (the superphylum containing arthropods, nematodes, and tardigrades) from Lophotrochozoa (including annelids, mollusks, and brachiopods) (Fig. 1). These subfamilies later proliferated independently in specific taxonomic groups. DIPs and Dprs expanded extensively in arthropods and mollusks (SI Appendix, Figs. S1 and S2); other protostomes included in this analysis, such as annelids, brachiopods, nematodes, and tardigrades, have just one or two Dprs and DIPs. The Klingon subfamily expanded within hexapods, resulting in the four paralogs of D. melanogaster (CG34353, CG7166, Klingon, and Wrapper; SI Appendix, Fig. S3A). Lachesins expanded specifically in lophotrochozoans, giving rise to two paralogs in mollusks and four in annelids (SI Appendix, Fig. S3B).
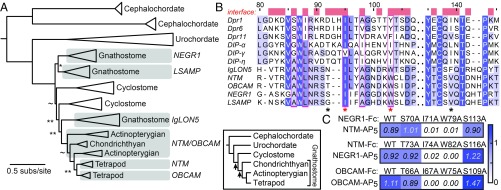
Fig. 2.
IgLONs are the vertebrate Wirins. (A) The ML phylogeny of the IgLON subfamily. aLR statistics are shown as branch supports: **<9.2 (= 2 ln100), *< 4.6 (= 2 ln10), ∼<2.2 (= 2 ln3). Unmarked branches have aLR statistics >9.2. Inset shows the established chordate phylogeny, with arrows marking two genome duplications. (B) Sequence alignment of the IG1 domains of mouse IgLONs, three Dprs, and three DIPs. Amino acids at the Dpr-DIP interface, defined by a 4-Å cutoff from the binding partner, are labeled as red blocks above the alignment. Amino acids mutated in C are labeled with an asterisk. The hydrophobic core residues of the Dpr-DIP interface observed by Cheng et al. (11) is indicated by magenta columns. Sequence numbers above the alignment are for Dpr1. (C) Mutations at the predicted interfaces of the OBCAM-OBCAM and NTM-NEGR1 complexes affect binding as tested using ECIA. To effectively compare WT to mutants, protein concentrations within each mutant series were normalized. Reported absorbance values are after subtraction of negative controls at 0.05 (±0.01) absorbance units.
IgLONs Interact with Each Other as Predicted by Homology to Dprs and DIPs.
We hypothesized that, because IgLONs are homologous to Dprs and DIPs, they may also form homophilic and heterophilic complexes. To test this, we employed the same high-throughput method, the extracellular interactome assay (ECIA), originally used in the discovery of Dpr-DIP interactions, where Fc (Fragment, crystallizable)-tagged bait are used to pull down pentamerized alkaline phosphatase-tagged prey. The coating of the bait on plates and the highly oligomerized, parallel nature of the AP5 fusions mimic cell adhesions and increase apparent affinity by 10- to 1,000-fold (5).
In the binding assay, we also included the vertebrate Nectin and Nectin-like (Necl or SynCAM, for synaptic cell adhesion molecule) proteins. Like DIPs and IgLONs, Nectins and Necls have three Ig domains (15). They are known to interact homo- and heterophilically, mediate cell-to-cell interactions in the nervous and immune systems (12, 15), and form complexes structurally similar to Dpr-DIP complexes (7). When ECIA was performed with all five mouse IgLONs and eight out of nine Nectins and Necls, we saw that Nectins and Necls interact with each other but do not produce complexes with IgLONs (SI Appendix, Fig. S4B). All IgLONs interacted with each other and could form both homo- and heterophilic complexes. This is in agreement with our prediction based on the homology of IgLONs to Dprs and DIPs and with previously reported interactions within the IgLON subfamily (16–19). While Nectins and Necls share many structural features with Dprs, DIPs, and IgLONs, their lack of any interactions with IgLONs and the monophyly of Dprs, DIPs, and IgLONs led us to conclude that they should be considered as a separate family within the IgSF (Fig. 1). The ubiquity of homo- and heterophilic interactions among Wirins and their outgroup IgSF proteins indicates that the ancestral Wirin was a homodimer.
Given the homology of Dprs, DIPs, and IgLONs, we hypothesized that structural models of IgLON complexes based on Dprs and DIPs would accurately predict interface residues in IgLON complexes. We created homology models of the IG1 domains of one homophilic (OBCAM-OBCAM) and one heterophilic (NTM-NEGR1) IgLON complex, based on the Dpr6–DIP-α structure (7). We predicted four residues, labeled by asterisks in Fig. 2B and depicted on the NTM-NEGR1 model in SI Appendix, Fig. S4C, to be at binding interfaces in IgLONs. When ECIA was repeated with single-site alanine mutants of these residues in OBCAM, NTM, and NEGR1 against WT OBCAM, NEGR1, and NTM, respectively, we observed that two out of four mutants lost all detectable binding (Fig. 2C and red asterisks in Fig. 2B). The same positions in Dpr6 (I115 and Y123) and DIP-α (I83 and I91) were previously identified to be crucial for the Dpr-DIP interaction (7). The IgLON mutations described here can be used as tools in future functional studies. Overall, our results support a close evolutionary and structural relationship between IgLONs and Dprs and DIPs.
Nematode Homologs Mimic Binding Activities of Drosophila Dprs and DIPs.
The nematode Caenorhabditis elegans is an appealing candidate for the study of Dprs and DIPs, as it is a well-established model organism for neuronal wiring. Our phylogenetic analysis identified one Dpr homolog, ZIG-8, and one DIP homolog, RIG-5, in C. elegans (Fig. 3A). Similar to Dprs and DIPs, ZIG-8 and RIG-5 have two and three Ig domains, respectively. They have an N-terminal signal peptide and carry hydrophobic C-terminal sequences, indicative of membrane attachment via transmembrane helices or GPI linkages (SI Appendix, Fig. S5 A and B). ZIG-8 is a member of the zwei (two)-Ig class of proteins (20), and RIG-5 was assigned to the RIG class due to its being a neuRonal IG protein. As these protein families were defined based not on homology but on structural or functional commonalities, none of the other ZIG or RIG proteins were identified as belonging to the Wirin family.
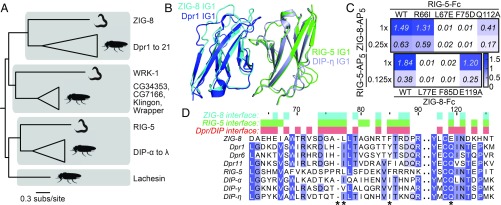
Fig. 3.
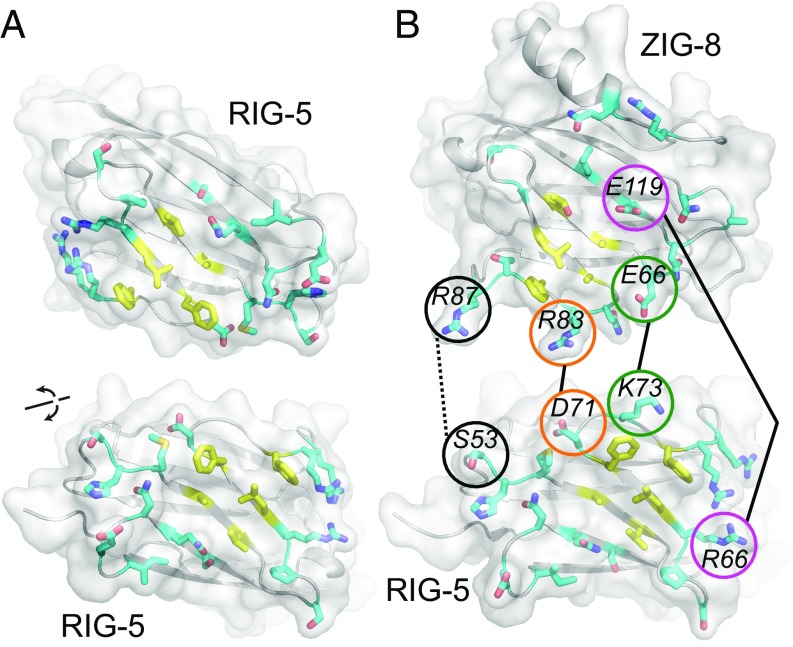
C. elegans ZIG-8 and RIG-5 bind each other using interface residues identified in Dpr-DIP complexes. (A) Phylogeny of Wirins in the two protostome model organisms, C. elegans and D. melanogaster. (B) The crystal structure of the ZIG-8–RIG-5 complex, superimposed with the Dpr1–DIP-η structure (11). The ZIG-8 N-terminal helix is removed from this view for clarity. (C) Mutations at the observed ZIG-8 and RIG-5 interface affect heterophilic binding. To effectively compare WT to mutants, protein concentrations within each mutant bait series (rows) were normalized. Each prey was tested at two concentrations to ensure that binding affinities are compared at nonsaturating concentrations. (D) Sequence alignments of IG1 domains of ZIG-8, RIG-5, Dprs, and DIPs. Asterisks indicate the four amino acid positions mutated in binding experiments.
We performed ECIA with complete ectodomains of ZIG-8 and RIG-5 and found that they form a heterophilic complex (SI Appendix, Fig. S5C). We also detected weak ZIG-8 and RIG-5 homodimers. Drosophila Dprs and DIPs form heterophilic dimers (5) and weaker homophilic dimers (11, 21), so the binding activities of ZIG-8 and RIG-5 mimic those of their fly counterparts, presumably because of their common ancestry. As is the case for Dprs and DIPs, ZIG-8–RIG-5 binding can be recapitulated with only the first Ig domains (SI Appendix, Fig. S5C).
The Structure of the ZIG-8–RIG-5 Complex Closely Resembles Fly Dpr-DIP Complex Structures.
Next, we determined the crystal structure of the ZIG-8–RIG-5 IG1-IG1 heterocomplex to 1.7-Å resolution (SI Appendix, Fig. S5F and Table S1) (PDBs: 6ON9 and 6ONB) (22, 23). ZIG-8 and RIG-5 create a pseudo-two-fold symmetric complex using their β-sheets including the GFCC′C′′ strands, similar to Dprs and DIPs. The interface area for the heterodimer is 930 Å2, which is close to the interface areas observed for Dpr and DIP complexes, ranging from 820 to 910 Å2 (excluding contributions by glycans) (7, 11, 21). The ZIG-8–RIG-5 and Dpr1–DIP-η complexes can be superimposed with a 0.92-Å rmsd for Cα atoms (145 out of 203 atoms) (Fig. 3D), indicating strong conservation of the backbone conformation across the long evolutionary interval since the common ancestor of arthropods and nematodes. Unexpectedly, ZIG-8 has an additional N-terminal helix, which is disulfide-linked to the F strand of ZIG-8 IG1 (SI Appendix, Fig. S5F).
To validate the crystal structure, we designed mutants of ZIG-8 and RIG-5 at the heterodimeric interface. Mutation sites are equivalent residues previously mutated in Dprs and DIPs (7, 11) and IgLONs (Fig. 2). The mutations led to weaker binding or loss of affinity between ZIG-8 and RIG-5, confirming the structure (Fig. 3C and SI Appendix, Fig. S5E). The most effective mutations for breaking the ZIG-8–RIG-5 complex were L77E (ZIG-8) and F75A (RIG-5) (see SI Appendix for sequence numbering). In our structure, both residues are buried within cavities on the interacting proteins’ surfaces (SI Appendix, Fig. S6 A and B), explaining why these residues are essential for ZIG-8–RIG-5 complex formation.
The general chemical features of the ZIG-8–RIG-5 interface are also similar to those of the Dpr and DIP interfaces, despite only moderate sequence conservation at the interface (Fig. 3D and SI Appendix, Fig. S6C). As observed before in Dpr-DIP complexes, a central hydrophobic core (labeled as yellow side chains in SI Appendix, Fig. S6 A and B) is surrounded by permissive, and usually polar, amino acids (cyan side chains) in the ZIG-8–RIG-5 complex. Finally, using surface plasmon resonance, we showed that the dissociation constant (KD) for the ZIG-8–RIG-5 complex is 10 µM (SI Appendix, Fig. S6 D and E). This is within expectations for Wirins, since Dpr-DIP complexes were shown to have KD values in the high-nanomolar to high-micromolar range, where the stronger complexes have KD values ranging from 0.4 to 10 µM (5, 7, 11, 21).
RIG-5 and ZIG-8 Homodimerize Weakly in Solution.
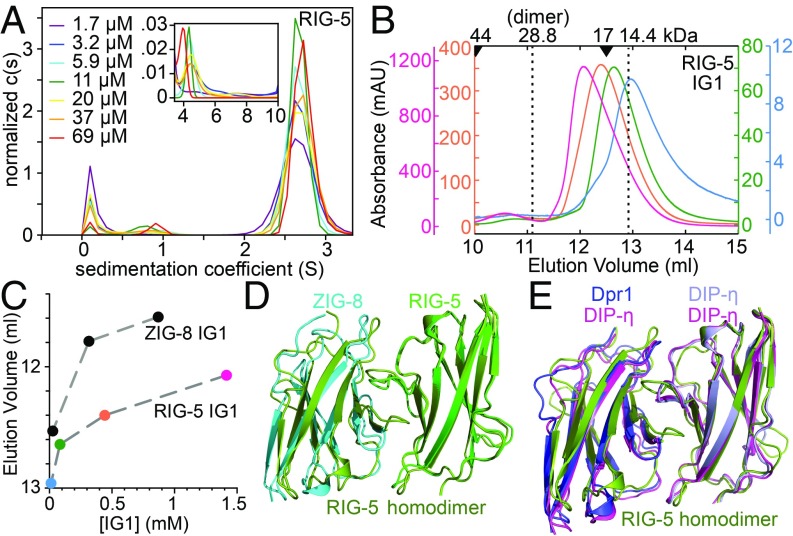
Since homodimerization appears to be a common, but not universal, feature among Wirins, we characterized the weaker homodimerization of ZIG-8 and RIG-5 in solution. Using sedimentation velocity experiments in an analytical ultracentrifuge (SV-AUC), we observed that RIG-5 and ZIG-8 exist mostly as monomers across the micromolar concentration range, with signs of homodimerization with KD values predicted in the low-millimolar range (Fig. 4A and SI Appendix, Fig. S7 A and B). We also applied RIG-5 and ZIG-8 IG1 domains on a size-exclusion chromatography column at a wider concentration range: when dilute, the elution volumes for RIG-5 and ZIG-8 were close to that expected for a monomer, but when concentrated, they behaved as larger molecules (Fig. 4 B and C). This behavior indicates a fast-exchange monomer-to-dimer equilibrium notable at 0.1 to >1 mM concentrations, in agreement with AUC results.
Fig. 4.
The IG1 domain of the C. elegans Wirins weakly homodimerize. (A) SV-AUC runs of the RIG-5 ectodomain show dimers (Inset) as a minor component in the micromolar range. For binding isotherms of RIG-5 and ZIG-8 homodimerization, see SI Appendix, Fig. S6 F and G. (B) RIG-5 IG1 elution volumes in size-exclusion chromatography decreases with increasing RIG-5 concentration, indicating a fast-exchange monomer-dimer equilibrium. Elution volumes for molecular weight standards are shown above as arrowheads. The RIG-5 IG1 construct used encodes for a ∼14.4-kDa mature glycoprotein. Absorbance was measured at a 0.2-cm path length. (C) Elution volumes plotted against loaded protein concentration indicate weak homodimerization for RIG-5 and ZIG-8, similar to what was observed for DIP-α and DIP-η (11). (D and E) Crystal structure of the RIG-5 IG1 homodimer superimposed with the ZIG-8–RIG-5 heterodimer (D) and with the Dpr1–DIP-η heterodimer and the DIP-η homodimer (E).
The Crystal Structure of the RIG-5 Homodimer.
To demonstrate that the weak RIG-5 homodimers we observed in solution are structurally similar to known DIP homodimers, we determined the crystal structure of the first Ig domain of RIG-5 to 1.42-Å resolution (SI Appendix, Table S1) (PDB: 6ON6) (24). The structure shows that RIG-5 forms symmetrical homodimers closely resembling the ZIG-8–RIG-5 heterocomplex (Fig. 4D), and Drosophila Dpr-DIP and DIP-DIP complexes (Fig. 4E). The interface area for the RIG-5 dimer is 940 Å2. The RIG-5 homodimeric interface is characterized by high shape complementarity (sc = 0.73) (25), probably to make up for lack of charge complementarity, as we previously observed for some Dpr-DIP and DIP-DIP structures (7, 11).
The chemical features of the RIG-5 dimerization interface are similar to those of the ZIG-8–RIG-5 and fly Dpr-DIP interfaces. Equivalent positions in the RIG-5 homodimer, ZIG-8–RIG-5 complex, Dpr-DIP complexes, and modeled IgLON complexes form a hydrophobic core (yellow in Fig. 5; SI Appendix, Figs. S7 C and D and S8). While the hydrophobic core feature is invariable among Wirins, the amino acids at the hydrophobic core are only moderately conserved. A comparison of the hydrophobic core residues in the RIG-5 and DIP-η homodimeric structures show that while every single amino acid is different at the core, their hydrophobic nature is conserved (e.g., RIG-5 L67 vs. DIP-η I84), and the main chain positions are preserved (SI Appendix, Fig. S7 C and D). The higher-affinity ZIG-8–RIG-5 complex shows high charge complementarity and more polar interactions in the periphery region compared with the weaker RIG-5 homodimer (Fig. 5). The extremely weak homophilic interactions of RIG-5 and ZIG-8 are unlikely to be physiologically significant. Since the ancestral Wirin was likely a homodimer, some of the homophilic interactions of extant Wirins might be vestiges of the ancestral Wirin’s homophilic activity. The hydrophobic core of the Wirin interfaces are not a shared feature in the outgroup IgSF families, Nectins, Necls, Kirrels, and Nephrins (SI Appendix, Fig. S8).
Fig. 5.
Nematode and arthropod Wirins interact through surfaces with similar chemical properties. (A) Open-book view of the RIG-5 homodimeric interface, with interface residues depicted in stick representation. Residues at the conserved hydrophobic core are colored yellow. (B) Open-book view of the ZIG-8–RIG-5 heterodimer, with the hydrophobic core in yellow, the periphery in cyan, and side chain-to-side chain polar interactions labeled.
Discussion
Members of the Wirin Family Perform Conserved Functions.
The evolutionary relationship we have established between Dprs and DIPs in protostomes and IgLONs in vertebrates are reflected in the biochemical and structural similarities we report here, as well as previously reported functional similarities. Dprs and DIPs are expressed in small subsets of neurons and regulate retrograde signaling, synaptic targeting, and cell death. IgLON family members have varied spatial and temporal expression patterns (reviewed in refs. 12 and 13). They are expressed both in neurons and oligodendrocytes (26) and have been observed to be localized to pre- and/or postsynaptic membranes in developing and adult brains, lending support for a synapse formation and maintenance function. NTM is localized to granule cell–Purkinje cell and mossy fiber–granule cell synapses in the cerebellum (27), while LSAMP is observed postsynaptically in granule cells of the dentate gyrus in the adult mouse hippocampus (28), and NEGR1 and OBCAM are found in postsynaptic densities in the cerebral cortex and the hippocampal CA3 region in rats (29). Synaptic localization is validated by a recent proteomic analysis of synaptic clefts, where the 199-member proteome of excitatory synapses in rat embryo cortical neurons included all five IgLONs (30). NEGR1, OBCAM, NTM, and LSAMP were also identified among proteins of the synaptic vesicle proteome in rat brains (31). Importantly, IgLONs have been shown to regulate synapse numbers in hippocampal neurons (32, 33). Support for neurite growth or axon fasciculation functions for several IgLONs have been reported (for example, see ref. 27). These data suggest that the vertebrate and protostome members of the Wirin family share functional roles in establishing connectivity in their respective nervous systems, and these shared roles are likely to derive from similar functions in the ancient common ancestor of Bilateria.
Our phylogenetic analysis identified additional Wirin subfamilies in protostomes: Klingons and Lachesins. Several previous observations connect these proteins to Dprs, DIPs, and IgLONs. Most strikingly, Drosophila Klingon interacts with a secreted leucine-rich repeat domain protein called cDIP (common Dpr- and DIP-interacting protein), which interacts with most Dprs and DIPs (5). Klingon is necessary for long-term memory formation (34) and is involved in the development of the fly photoreceptor neuron R7 (35). It is not known if Klingon and Dpr11, which is selectively expressed in one subclass of R7 neurons (7), cooperate in R7 development or connectivity. Also, similarities between Drosophila Lachesin and vertebrate IgLONs were previously recognized due to shared domain features (36, 37). Lachesin is expressed in neuronal (36), epithelial (38), and glial populations (37); its function remains poorly understood.
Finally, we identified the nematode orthologs of the Wirin family. ZIG-8, the Dpr ortholog, was first recognized as one of the candidate ZIG genes involved in the maintenance of axons within the ventral nerve cord in C. elegans (20, 39). RIG-5, the DIP ortholog, has been implicated in the navigation of axons within the ventral nerve cord (40). Among protostomes, it is intriguing to speculate that C. elegans, with a simple nervous system, has a limited Dpr/DIP repertoire (two genes), while the complex nervous systems of arthropods and mollusks utilize expanded sets of Wirin genes (37 in the fruit fly).
A Wider Neuronal Wiring Family Includes Wirins, Kirrels, Nephrins, Nectins, and Necl/SynCAMs.
In our attempts to find outgroups for creating a phylogenetic tree for Wirins, we found that four other protein families, Kirrels, Nephrins, Nectins, and Necls, which have neuronal connectivity functions, are distantly related to Wirins. We previously reported strong similarities among the structures of the Dpr6–DIP-α complex, the Kirrel-Nephrin complexes, and known Nectin and Necl homo- and heterophilic complexes (7). Recent work also identified functional similarities between Kirrels and Dpr/DIPs in the organization of olfactory sensory neurons in mammals and flies (9). Wirins and these proteins interact with their homo- and heterophilic partners using the GFCC′C′′ faces of their N-terminal Ig domains (SI Appendix, Fig. S9). Furthermore, Wirins and the four distantly related families of proteins appear to adopt structures with fully extended ectodomains, unlike other neuronal IgSF proteins, such as DSCAMs, DCC, and Axonin, which adopt horseshoe-shaped structures.
No structural similarities between Wirins and the four distantly related families can be detected outside the first three Ig domains. Kirrels and Nephrins contain 5 and 10 extracellular domains, respectively, unlike the 2 and 3 domains observed in most Wirins, Nectins, and Necls. The four outgroup families have conserved intracellular regions specialized for signaling, while most Wirins do not.
A Shared Structural Architecture in Neuronal IgSF Proteins.
These connections among the Dpr, DIP, and IgLON families, Klingon, Lachesin, and their nematode orthologs help define a functional family of proteins with a shared structural architecture involved in the establishment of neuronal connectivity going back at least to the rise of bilaterians. Future studies need to investigate the evolutionary origins of the wider family of neuronal wiring molecules. It would be of interest to see if Wirins and its four related IgSF protein families arose during the period in which neurons and neuronal circuits first appeared, and if Wirin expansions correlate with increasing nervous system complexity in protostomes.
Materials and Methods
Phylogenetics.
Putative Dpr and DIP homologs were identified through a reciprocal BLASTp analysis (41). The ML phylogenies were inferred with RAxML version 8.2.12 (42). For details and unreduced phylogenies, see SI Appendix.
Crystallography of ZIG-8 and RIG-5 IG1.
Structure determination and refinement was performed using the PHENIX package (43). See SI Appendix for details on protein biochemistry and our choice for sequence numbering for RIG-5.
Supplementary Material
Acknowledgments
We thank Yeonhee J. Park and Elana Baltrusaitis for technical help and Paschalis Kratsios and J.W.T.’s laboratory for discussions and guidance. This work was supported in part by NIH Grants R01 NS097161 (to E.Ö.), R01 GM121931 (to J.W.T.), R37 NS028182 (to K.Z.), and R01 NS096509 (to K.Z.) and a Klingenstein-Simons Fellowship Award in the Neurosciences (to E.Ö.). Use of the Stanford Synchrotron Radiation Lightsource (SSRL), SLAC National Accelerator Laboratory is supported by the US Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the NIH, National Institute of General Medical Sciences (NIGMS) (including Grant P41GM103393). This work is also based upon research conducted at the Northeastern Collaborative Access Team beamlines funded by NIGMS from the NIH (Grant P30 GM124165) at the Advanced Photon Source, a US DOE Office of Science user facility operated by the Argonne National Laboratory under Contract DE-AC02-06CH11357. The Pilatus 6M detector on 24-ID-C beamline is funded by NIH Office of Research Infrastructure High-End Instrumentation Grant S10 RR029205.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.G. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6ON6, 6ON9, and 6ONB).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818631116/-/DCSupplemental.
References
- 1.Aricescu AR, Jones EY (2007) Immunoglobulin superfamily cell adhesion molecules: Zippers and signals. Curr Opin Cell Biol 19:543–550. [DOI] [PubMed] [Google Scholar]
- 2.Vogel C, Chothia C (2006) Protein family expansions and biological complexity. PLoS Comput Biol 2:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogel C, Teichmann SA, Chothia C (2003) The immunoglobulin superfamily in Drosophila melanogaster and Caenorhabditis elegans and the evolution of complexity. Development 130:6317–6328. [DOI] [PubMed] [Google Scholar]
- 4.Bushell KM, Söllner C, Schuster-Boeckler B, Bateman A, Wright GJ (2008) Large-scale screening for novel low-affinity extracellular protein interactions. Genome Res 18:622–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Özkan E, et al. (2013) An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell 154:228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura M, Baldwin D, Hannaford S, Palka J, Montell C (2002) Defective proboscis extension response (DPR), a member of the Ig superfamily required for the gustatory response to salt. J Neurosci 22:3463–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrillo RA, et al. (2015) Control of synaptic connectivity by a network of Drosophila IgSF cell surface proteins. Cell 163:1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan L, et al. (2015) Ig superfamily ligand and receptor pairs expressed in synaptic partners in Drosophila. Cell 163:1756–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barish S, et al. (2018) Combinations of DIPs and Dprs control organization of olfactory receptor neuron terminals in Drosophila. PLoS Genet 14:e1007560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashley J, et al. (2019) Transsynaptic interactions between IgSF proteins DIP-α and Dpr10 are required for motor neuron targeting specificity. eLife 8:e42690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng S, et al. (2019) Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila. eLife 8:e41028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zinn K, Özkan E (2017) Neural immunoglobulin superfamily interaction networks. Curr Opin Neurobiol 45:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan RPA, Leshchyns’ka I, Sytnyk V (2017) Glycosylphosphatidylinositol-anchored immunoglobulin superfamily cell adhesion molecules and their role in neuronal development and synapse regulation. Front Mol Neurosci 10:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dehal P, Boore JL (2005) Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3:e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samanta D, Almo SC (2015) Nectin family of cell-adhesion molecules: Structural and molecular aspects of function and specificity. Cell Mol Life Sci 72:645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gil OD, et al. (2002) Complementary expression and heterophilic interactions between IgLON family members neurotrimin and LAMP. J Neurobiol 51:190–204. [DOI] [PubMed] [Google Scholar]
- 17.Gil OD, Zanazzi G, Struyk AF, Salzer JL (1998) Neurotrimin mediates bifunctional effects on neurite outgrowth via homophilic and heterophilic interactions. J Neurosci 18:9312–9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lodge AP, McNamee CJ, Howard MR, Reed JE, Moss DJ (2001) Identification and characterization of CEPU-Se-A secreted isoform of the IgLON family protein, CEPU-1. Mol Cell Neurosci 17:746–760. [DOI] [PubMed] [Google Scholar]
- 19.Reed J, McNamee C, Rackstraw S, Jenkins J, Moss D (2004) Diglons are heterodimeric proteins composed of IgLON subunits, and Diglon-CO inhibits neurite outgrowth from cerebellar granule cells. J Cell Sci 117:3961–3973. [DOI] [PubMed] [Google Scholar]
- 20.Aurelio O, Hall DH, Hobert O (2002) Immunoglobulin-domain proteins required for maintenance of ventral nerve cord organization. Science 295:686–690. [DOI] [PubMed] [Google Scholar]
- 21.Cosmanescu F, et al. (2018) Neuron-subtype-specific expression, interaction affinities, and specificity determinants of DIP/Dpr cell recognition proteins. Neuron 100:1385–1400.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng S, Kurleto JD, Özkan E (2019) Data from “Crystal structure of the ZIG-8-RIG-5 IG1-IG1 heterodimer, tetragonal form.” Protein Data Bank. Available at https://www.rcsb.org/structure/6on9. Deposited April 20, 2019.
- 23.Cheng S, Kurleto JD, Özkan E (2019) Data from “Crystal structure of the ZIG-8-RIG-5 IG1-IG1 heterodimer, monoclinic form.” Protein Data Bank. Available at https://www.rcsb.org/structure/6onb. Deposited April 20, 2019.
- 24.Cheng S, Kurleto JD, Özkan E (2019) Data from “Crystal structure of the RIG-5 IG1 homodimer.” Protein Data Bank. Available at https://www.rcsb.org/structure/6on6. Deposited April 20, 2019.
- 25.Lawrence MC, Colman PM (1993) Shape complementarity at protein/protein interfaces. J Mol Biol 234:946–950. [DOI] [PubMed] [Google Scholar]
- 26.Sharma K, et al. (2015) Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci 18:1819–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S, et al. (2001) Neurotrimin expression during cerebellar development suggests roles in axon fasciculation and synaptogenesis. J Neurocytol 30:927–937. [DOI] [PubMed] [Google Scholar]
- 28.Zacco A, et al. (1990) Isolation, biochemical characterization and ultrastructural analysis of the limbic system-associated membrane protein (LAMP), a protein expressed by neurons comprising functional neural circuits. J Neurosci 10:73–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyata S, et al. (2003) Biochemical and ultrastructural analyses of IgLON cell adhesion molecules, Kilon and OBCAM in the rat brain. Neuroscience 117:645–658. [DOI] [PubMed] [Google Scholar]
- 30.Loh KH, et al. (2016) Proteomic analysis of unbounded cellular compartments: Synaptic clefts. Cell 166:1295–1307.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takamori S, et al. (2006) Molecular anatomy of a trafficking organelle. Cell 127:831–846. [DOI] [PubMed] [Google Scholar]
- 32.Yamada M, et al. (2007) Synaptic adhesion molecule OBCAM; synaptogenesis and dynamic internalization. Brain Res 1165:5–14. [DOI] [PubMed] [Google Scholar]
- 33.Hashimoto T, Maekawa S, Miyata S (2009) IgLON cell adhesion molecules regulate synaptogenesis in hippocampal neurons. Cell Biochem Funct 27:496–498. [DOI] [PubMed] [Google Scholar]
- 34.Matsuno M, Horiuchi J, Tully T, Saitoe M (2009) The Drosophila cell adhesion molecule klingon is required for long-term memory formation and is regulated by Notch. Proc Natl Acad Sci USA 106:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butler SJ, Ray S, Hiromi Y (1997) klingon, a novel member of the Drosophila immunoglobulin superfamily, is required for the development of the R7 photoreceptor neuron. Development 124:781–792. [DOI] [PubMed] [Google Scholar]
- 36.Karlstrom RO, Wilder LP, Bastiani MJ (1993) Lachesin: An immunoglobulin superfamily protein whose expression correlates with neurogenesis in grasshopper embryos. Development 118:509–522. [DOI] [PubMed] [Google Scholar]
- 37.Strigini M, et al. (2006) The IgLON protein Lachesin is required for the blood-brain barrier in Drosophila. Mol Cell Neurosci 32:91–101. [DOI] [PubMed] [Google Scholar]
- 38.Llimargas M, Strigini M, Katidou M, Karagogeos D, Casanova J (2004) Lachesin is a component of a septate junction-based mechanism that controls tube size and epithelial integrity in the Drosophila tracheal system. Development 131:181–190. [DOI] [PubMed] [Google Scholar]
- 39.Bénard CY, Blanchette C, Recio J, Hobert O (2012) The secreted immunoglobulin domain proteins ZIG-5 and ZIG-8 cooperate with L1CAM/SAX-7 to maintain nervous system integrity. PLoS Genet 8:e1002819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwarz V, Pan J, Voltmer-Irsch S, Hutter H (2009) IgCAMs redundantly control axon navigation in Caenorhabditis elegans. Neural Dev 4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. [DOI] [PubMed] [Google Scholar]
- 42.Stamatakis A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams PD, et al. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.