Significance
Developmental transitions during shoot development in plants are regulated by factors originating outside and within the shoot apical meristem (SAM). The best-known example of this is the vegetative-to-reproductive transition, which is initiated by a leaf-derived signal that transforms the vegetative SAM into a developmentally stable inflorescence meristem. Although the juvenile-to-adult vegetative transition (vegetative phase change) is also thought to be regulated by factors exogenous and internal to the SAM, how this process is coordinated spatially remains unknown. Here we demonstrate that the SAM specifies leaf identity early in development, but that leaves become more important determinants of shoot identity as the shoot ages. We also reveal a role for the plant aging pathway in the regulation of meristem size.
Keywords: vegetative phase change, miR156, WUSCHEL, leaf development, shoot apical meristem
Abstract
The extent to which the shoot apical meristem (SAM) controls developmental decisions, rather than interpreting them, is a longstanding issue in plant development. Previous work suggests that vegetative phase change is regulated by signals intrinsic and extrinsic to the SAM, but the relative importance of these signals for this process is unknown. We investigated this question by examining the effect of meristem-deficient mutations on vegetative phase change and on the expression of key regulators of this process, miR156 and its targets, SPL transcription factors. We found that the precocious phenotypes of meristem-deficient mutants are a consequence of reduced miR156 accumulation. Tissue-specific manipulation of miR156 levels revealed that the SAM functions as an essential pool of miR156 early in shoot development, but that its effect on leaf identity declines with age. We also found that SPL genes control meristem size by repressing WUSCHEL expression via a novel genetic pathway.
As plants mature they transition through a number of distinct developmental phases. A transition from juvenile-to-adult vegetative growth, before the onset of reproductive development, has been recognized since the late 19th century (1). Depending on the species, this transition, known as vegetative phase change (VPC), may lead to changes in, among others, leaf size and shape, plastochron length, shoot physiology, adventitious root production, disease resistance, and reproductive competence (reviewed in ref. 2). In the model plant Arabidopsis thaliana, the switch to adult growth is associated with the production of large, spatulate, and serrated leaves that produce trichomes on their abaxial surface. Leaves with a juvenile identity are small, round, smooth, and lack abaxial trichomes.
The master regulator of VPC in Arabidopsis and all other studied flowering plants is the microRNA miR156 (2). The expression of miR156 declines temporally during shoot maturation, allowing the expression of its target genes in the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) family to increase (3–5). Arabidopsis has 10 SPL genes that are targeted by miR156 and that promote adult growth to varying degrees with SPL9, SPL13, and SPL15 having the largest effect (6). miR156 and the partly redundant miR157 also exist in multigene families, with the loci MIR156A, MIR156C, MIR157A, and MIR157C being the most important for suppressing SPL activity during juvenile growth (7).
miR156 and SPL9/13/15 are all expressed throughout the shoot apex in both the shoot apical meristem (SAM) and young leaf primordia (6). While there has been recent progress in elucidating the molecular mechanisms that control the temporal decline of miR156 expression (8–13), how expression of the miR156-SPL pathway and the process of VPC are coordinated across the shoot apex remains unknown. It has long been thought that the identity of a shoot is determined by the maturation state of the SAM (14, 15). A regulatory role for the SAM is further supported by the phenotypes of plants that have reduced meristems and that immediately produce leaves with adult traits, such as wuschel (wus) and paused (psd) mutants (16, 17). However, leaf ablation studies have shown that signaling from existing juvenile leaves promotes the subsequent production of adult leaves (18–20). These results sit in apparent conflict: if juvenile leaves are required to initiate adult growth, how do wus and psd immediately produce adult leaves? We aimed to resolve this question by analyzing the effects of perturbations to the meristem on the miR156-SPL pathway. We demonstrate that the wus phenotype is a consequence of reduced miR156 expression and that expression of miR156 within the SAM, both in wus and developmentally normal meristems, affects leaf identity. We further show that expression of WUS is regulated by feedback from SPL genes.
Results
The Precocious Formation of Adult Leaves in SAM-Defective Plants Is Not Attributable to a Delay in Leaf Initiation.
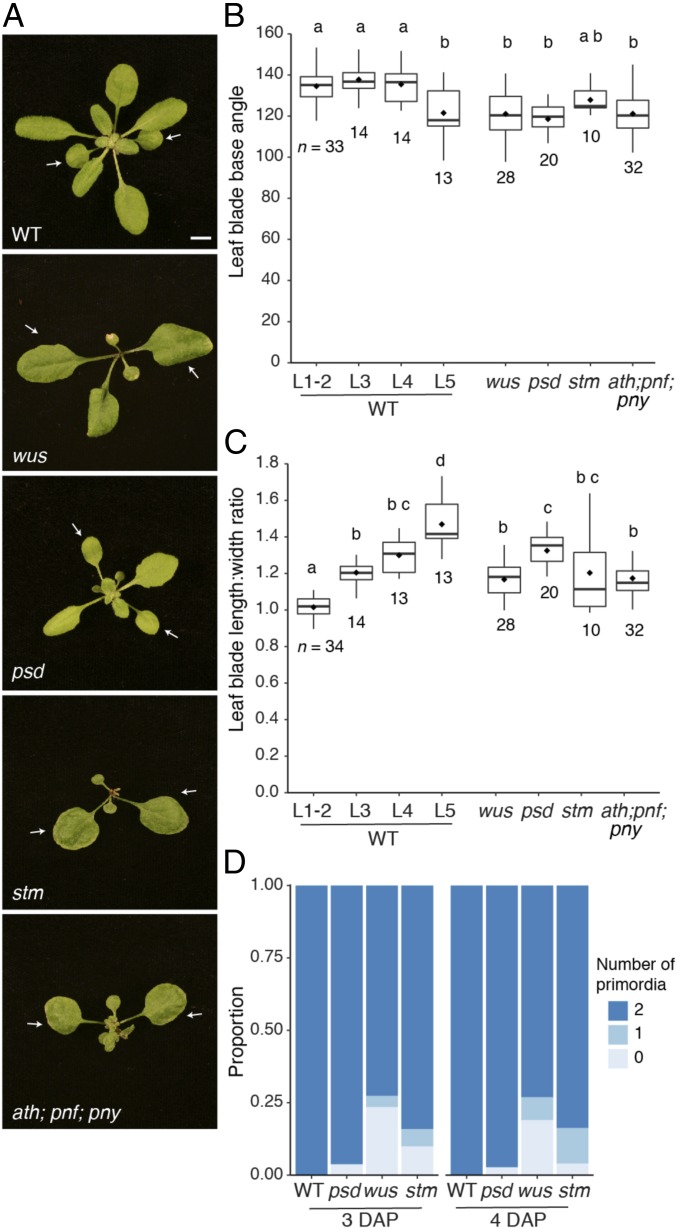
Mutations in the homeobox gene, WUS, and the tRNA export receptor, PSD, interfere with development of the SAM, delay leaf emergence, and exhibit precocious vegetative development; the first two leaves produced by these mutants prematurely display adult traits (Fig. 1 A–C) (16, 17). To determine if the effect of these mutations on leaf identity is attributable to their effect on the SAM, we examined the phenotype of two other meristem-deficient mutants: shoot meristemless (stm-1) and the triple mutant arabidopsis thaliana homeobox 1; pennywise; pound-foolish (ath; pny; pnf) (21). Like wus and psd, the first two leaves of these mutants resembled leaves produced at later plastochrons in developmentally normal plants (Fig. 1 A–C).
Fig. 1.
The precocious phenotype of meristem-defective mutants is not caused by a delay in leaf initiation. (A) The phenotype of WT Columbia and mutant plants at 23 DAP in LD conditions. White arrows point to leaves 1 and 2. (Scale bar, 5 mm.) (B and C) The angle of the leaf base (B) and the length:width ratio of the leaf blade (C) in leaves 1–5 of WT plants and in leaves 1 and 2 of meristem-defective mutants. Boxes display the interquartile range (IQR) (boxes), median (lines), and values beyond 1.5* IQR (whiskers); mean values are marked by a solid diamond (◆). Significantly distinct groups were determined by one-way ANOVA with post hoc Tukey multiple comparison test (letters indicate statistically distinct groups; P < 0.05). Sample sizes are shown on the graphs. (D) Distribution of leaf primordia number identified by staining of a LFY::GUS reporter in respective meristem-defective mutants (wus and stm were segregating 25% mutant plants; psd was homozygous for the mutation).
It has been suggested that the first-formed leaves of psd and wus have an adult identity because leaf initiation does not occur in these mutants until after the shoot has transitioned to the adult phase (16, 22). This hypothesis implies that the timing of VPC is regulated independently of leaf production and is inconsistent with the evidence that leaves promote VPC. To resolve this inconsistency, we re-examined the effect of psd, wus, and stm on leaf initiation, using lines containing a LFY::GUS reporter that is expressed in leaf primordia; this reporter makes it possible to visualize young leaf primordia immediately after seed germination (SI Appendix, Fig. S1A). Three days after planting (DAP), 100% (20/20) of Col LFY::GUS plants and 96% (25/26) of psd LFY::GUS mutants had two leaf primordia, and a similar number of plants had two leaf primordia at 4 DAP (Fig. 1D). These primordia were slightly smaller in psd than in wild type (WT), consistent with previous studies indicating that psd delays leaf initiation (22). However, this result demonstrates that psd initiates leaves before VPC (which occurs between plastochron 5 and 6), implying that its effect on leaf identity cannot be attributed to this delay in leaf initiation. Because stm and wus cannot be maintained in homozygous condition, their effect on leaf initiation was determined by staining populations segregating 25% for these mutations. At 3 DAP, 84% of the seedlings segregating for stm had two leaf primordia, 6% had one primordium, and 10% had no visible primordia; by 4 DAP, the number of plants with at least one leaf primordium had increased to 96%. Given that 25% of these seedlings were homozygous for stm, this result demonstrates that a majority of stm mutants initiate at least one leaf during the juvenile phase. Similar results were obtained with wus. At 3 DAP, 72% of the seedlings segregating for wus had two leaf primordia, 4% had one leaf primordium, and 24% had no visible leaf primordia. At 4 DAP, 73% of these seedlings had two leaf primordia, 8% had one leaf primordium, and 19% had no leaf primordia. Thus, a significant number of wus seedlings initiate leaves during the juvenile phase. These results confirm previous studies indicating that psd, stm, and wus delay leaf initiation, but demonstrate that this delay does not account for the adult identity of the leaves that they produce.
To determine if the precocious phenotype of these mutants might be due to a delay in leaf expansion, we examined the relationship between the timing of leaf emergence and leaf identity in wus. We found that the first leaf of wus mutants reached a length of ≥0.5 mm at different times after planting (between 6 and 12 DAP), but that there was no qualitative difference in the final shape of leaves that emerged at different time points (SI Appendix, Fig. S1 B and C). These results suggest that the effect of psd, stm, and wus on leaf identity is not due to a delay in either leaf initiation or leaf growth.
Precocious Adult Leaves Exhibit Elevated SPL Activity.
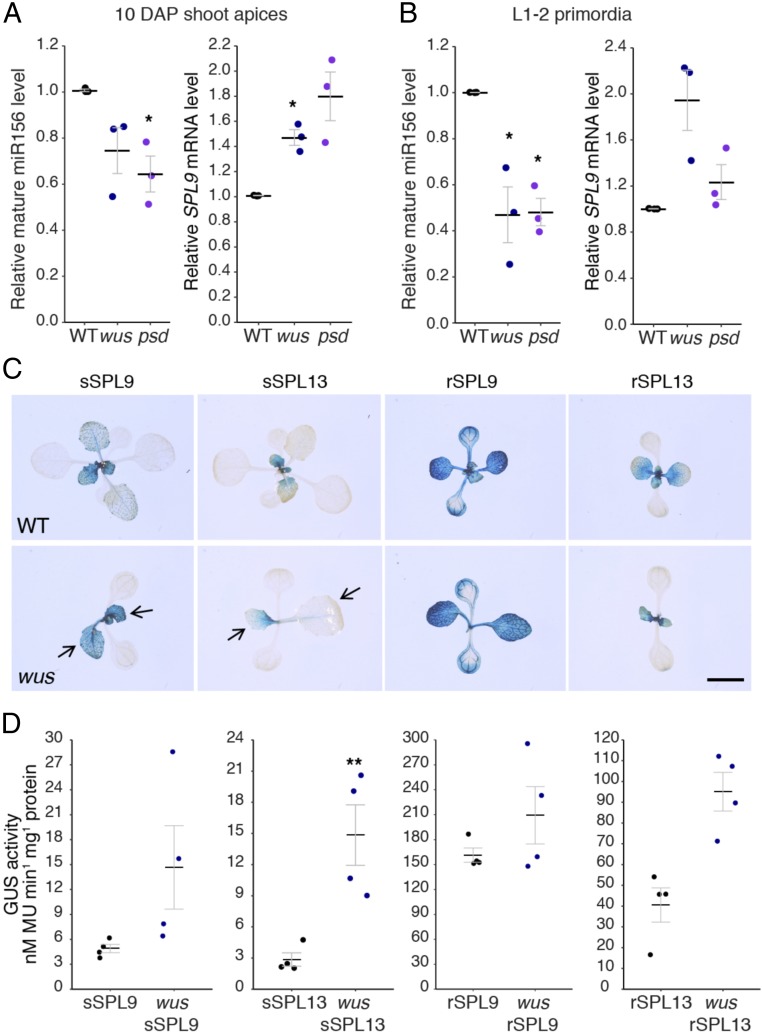
An alternative possibility is that meristem-defective mutations affect the activity of genes that regulate VPC. To test this hypothesis, we examined the effect of wus and psd on the abundance of miR156—the master regulator of VPC—and one of its direct targets, SPL9. Analyses were carried out on 10-d-old shoot apices bearing leaf primordia ≤1 mm and on 1- to 2-mm primordia of leaves 1 and 2; these samples were harvested from plants grown in short days to eliminate differences in gene expression resulting from the potential effect of these mutations on floral induction (the wus and psd mutant phenotypes persist in SD conditions). miR156 was reduced and SPL9 was elevated in both the shoot apices and leaf primordia of wus and psd compared with WT seedlings (Fig. 2 A and B). wus had no effect on miR156 expression in cotyledons (SI Appendix, Fig. S2). We then examined the effect of wus on the abundance of SPL proteins by crossing miR156-sensitive (sSPL9-GUS, sSPL13-GUS) and miR156-resistant (rSPL9-GUS, rSPL13-GUS) reporters into wus (Fig. 2C). Fluorescence assays of proteins using 4-methylumbelliferyl ß-d-glucuronide (MUG) extracted from LP1 and -2 revealed that sSPL9-GUS was expressed 3× higher and sSPL13-GUS was expressed 5.2× higher in wus than in WT primordia (Fig. 2D). rSPL9-GUS and rSPL13-GUS were also more highly expressed in wus than in WT, although to a much smaller extent than the miR156-sensitive reporters. These data suggest that the precocious vegetative phenotype of psd and wus is primarily attributable to a decrease in the abundance of miR156, although transcriptional up-regulation of SPL genes may also contribute to this phenotype.
Fig. 2.
wus and psd have reduced levels of miR156 and elevated levels of SPL9. (A and B) miR156 miRNA and SPL9 mRNA levels in 10-DAP shoot apices with leaf primordia ≥1 mm removed (A) and in isolated 1- to 2-mm primordia of leaves 1 and 2 (B) in SD conditions. Relative levels were quantified by RT-qPCR, normalized to snoR101 (for miR156) or ACT2 (for SPL9) as internal control genes and expressed as a ratio of expression to WT plants. Each data point represents a biological replicate and is the average of three technical replicates. Black bars represent the mean and gray bars the SEM. Significant differences to WT were determined by two-tailed t-test (*P < 0.05). (C) GUS staining of miRNA-sensitive and miR156-resistant SPL-GUS reporter constructs at 21 DAP in SD conditions. Black arrows point to precocious accumulation of SPL proteins in leaves 1 and 2. (Scale bar, 5 mm.) (D) MUG assays of lines shown in C. Protein was extracted from 1- to 2-mm primordia of leaves 1 and 2 in SD conditions. Each data point represents a biological replicate. Significant differences from WT were determined by two-tailed t test of log-transformed data (**P < 0.01).
Elevated miR156 Expression Suppresses the Leaf Phenotype of wus.
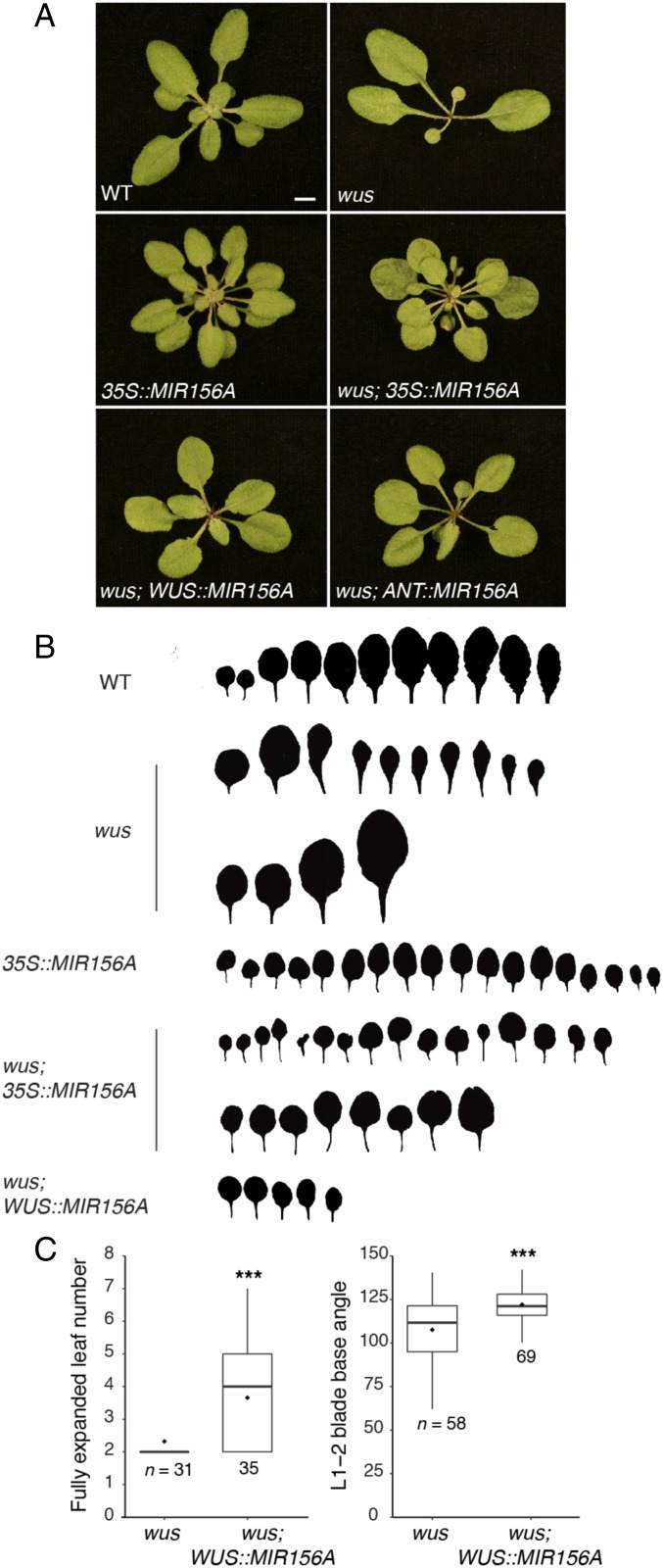
If the effect of wus on leaf identity is attributable to a decrease in the level of miR156, overexpression of miR156 should correct this phenotype. To test this prediction, we expressed miR156 constitutively and in localized regions of the shoot apex in wus mutants. Constitutive high expression of MIR156A under the regulation of the CaMV 35S promoter largely suppressed the adult leaf phenotype of wus; wus; 35S::MIR156A leaves more closely resembled the juvenilized leaves of 35S::MIR156A plants than the adult-like leaves of wus mutants (Fig. 3 A and B). 35S::MIR156A also partially suppressed the effect of wus on leaf number (Fig. 3A). Expressing MIR156A in wus using the AINTEGUMENTA (ANT) promoter—which is transcribed across the shoot apex but predominantly in incipient and young leaf primordia (23, 24) (SI Appendix, Fig. S3A)—produced a phenotype similar to that of 35S::MIR156A. Expressing miR156 in the central domain of the SAM using the WUS promoter (WUS::MIR156A) also partly rescued the effect of wus on leaf shape and leaf production (Fig. 3). Heterologous gene expression driven by the WUS promoter has previously been shown to be confined to the meristem in a wus background (25). These results therefore suggest that the precocious phenotype of wus plants is due to a reduction in the abundance of miR156 and demonstrate that expressing miR156 specifically within the SAM can partially compensate for the effect of wus on shoot development.
Fig. 3.
Enhanced miR156 expression suppresses the wus leaf phenotype. (A) Photographs were taken at 21 DAP in LD. (Scale bar, 5 mm.) (B) Heteroblasty of lines shown in A; two examples of wus and wus; 35S::MIR156A are shown to indicate the range of phenotypes observed. (C) Boxplots showing the effects of WUS::MIR156A on leaf formation at 24 DAP and the angle of blade bases in leaves 1 and 2 in a wus background. Only fully expanded leaves without polarity defects were counted. Boxes display the IQR (boxes), median (lines), and values beyond 1.5* IQR (whiskers); mean values are marked by a solid diamond (◆). Significant differences between the two lines were determined by two-tailed t test (***P < 0.001); sample sizes are indictated on the graphs.
SAM-Derived miR156 Regulates Leaf Identity.
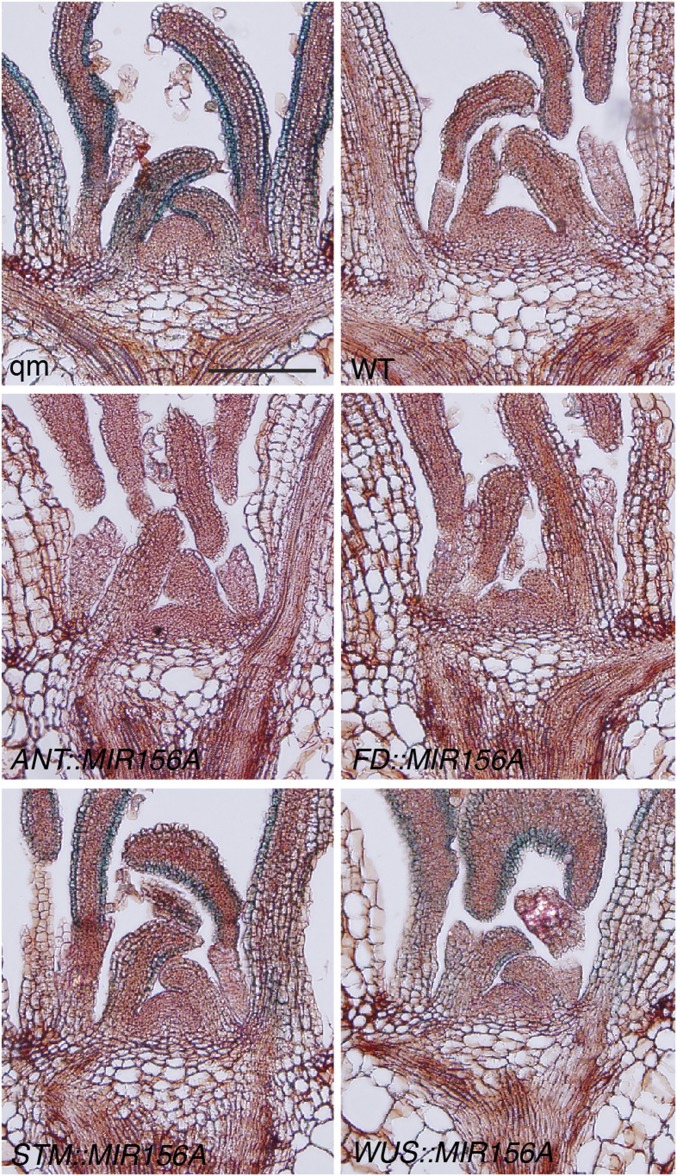
The similarity between the morphological and molecular phenotypes of plants with defective SAMs, and the ability of WUS::MIR156A to partially rescue the effect of wus on leaf shape, suggest that miR156 (or a downstream target) produced by the SAM acts noncell-autonomously to regulate leaf identity. To test this hypothesis, we manipulated the abundance of miR156 in different domains of WT and miR156-deficient shoot apices by expressing MIR156A or the miR156/157 target site mimic MIM156 [which reduces miR156/157 activity (26)] in transgenic plants using promoter sequences from ANT (27), FLOWERING LOCUS D (FD) (28, 29), STM (30), and WUS (31). Promoter::GUS fusions confirmed that the ANT sequence drives expression throughout the entire shoot apex; the FD sequence drives expression in the SAM and leaf primordia ≤250 μM in size; the STM sequence drives expression in the peripheral region of the SAM but not in leaf primordia; and the WUS sequence drives expression in the central region of the SAM (SI Appendix, Fig. S3A). Transgene expression was confirmed by RT-PCR (SI Appendix, Fig. S3B).
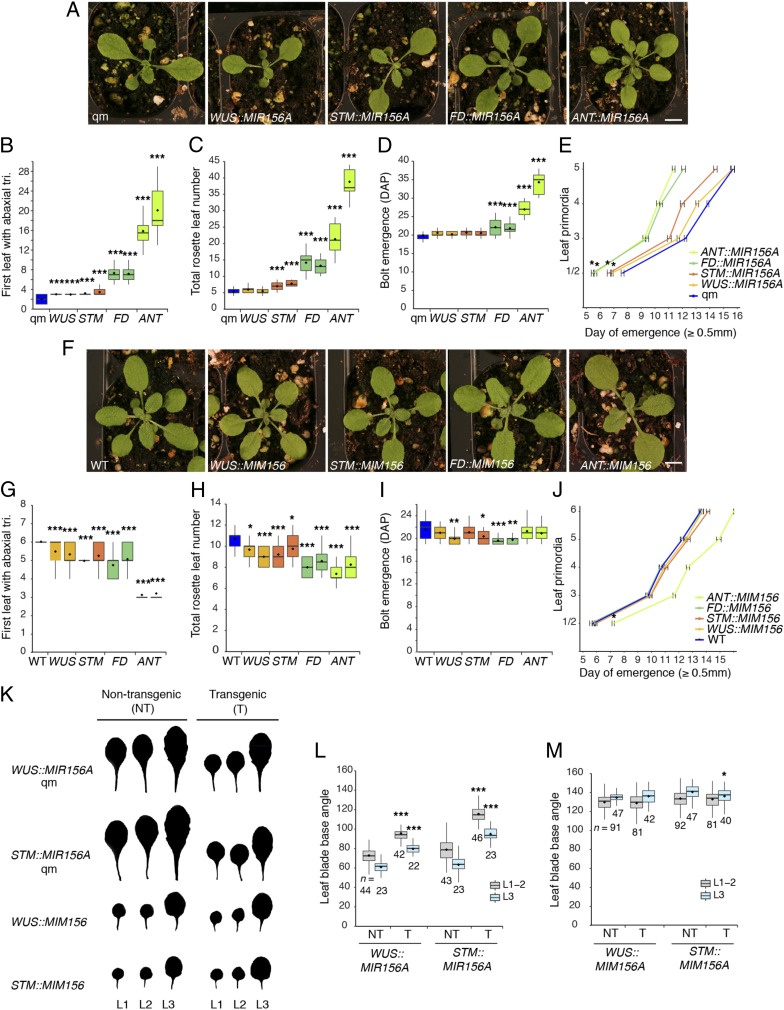
To determine if miR156 produced by the SAM is capable of rescuing the phenotype of plants deficient for miR156/157, we fused the promoter sequences mentioned above to a MIR156A genomic sequence and introduced these constructs into a mir156a mir156c mir157a mir157c quadruple mutant (mir156/157 qm) (7). WUS::MIR156A partly suppressed the leaf shape (Fig. 4 A, K, and L) and abaxial trichome phenotype (Fig. 4B) of the mir156/157 qm, and significantly accelerated the emergence of the first two leaves (Fig. 4E), but had no effect on leaf number or bolting time (Fig. 4 C and D). STM::MIR156A had a stronger effect on abaxial trichome production, leaf shape, and the rate of leaf initiation and significantly increased the number of rosette leaves, but also had no effect on bolting time. FD::MIR156A nearly completely corrected the vegetative phenotype of the mir156/157 qm (Fig. 4 A–E and SI Appendix, Fig. S3C) in long days (LD) and delayed VPC and flowering relative to WT plants in noninductive SD conditions (SI Appendix, Fig. S4). SPL expression is reduced in SD (32), which probably accounts for the stronger effect of FD::MIR156A under these conditions. The observations that meristematic expression of miR156 had the strongest effect on the morphology of early leaves (Fig. 4K and SI Appendix, Fig. S3C), was only able to delay trichome formation from leaf 2 to leaf 3 (in the case of WUS::MIR156A and STM::MIR156A) (Fig. 4B), and accelerated the emergence of leaves 1–2 but not later leaves (Fig. 4E) suggest that the regulatory capacity of SAM-derived miR156 is highest during early development—i.e., during the initiation of rosette leaves 1–3—but declines with age. In this regard, the observation that the continued expression of miR156 in leaf primordia under the ANT promoter maintained plants in the juvenile phase for a prolonged period (Fig. 4 A–C) supports the conclusion that leaves are a more important source of miR156 than the SAM throughout most of shoot development.
Fig. 4.
Localized expression of MIR156A and MIM156 in the SAM rescues loss of mir156 function and leads to precocious phase change. (A–E) Expression of MIR156A in a mir156ac mir157ac qm background. (F–J) Expression of MIM156 in a WT background. (A and F) Photographs taken at 18 DAP in LD conditions. (Scale bar, 5 mm.) (B–D and G–I) Gene names on the x axis describe the promoter used to drive expression. Two independent T3 lines are shown for each promoter. Significant differences from quadruple mutant or WT controls were determined by one-way ANOVA of log-transformed data with post hoc Dunnett’s multiple comparison test (*P < 0.05; **P < 0.01; ***P < 0.001). n = 31–48 (B and G); n = 12–24 (C, D, H, and I). (E and J) Emergence was scored when leaves were visible in the rosette without manipulating other leaves. Single representative T3 lines are shown. Error bars represent the SEM; significant differences between the timing of the emergence of leaves 1 and 2 relative to the qm or WT controls were determined by one-way ANOVA of log-transformed data with post hoc Dunnett’s multiple comparison test (*P < 0.001; n ≥ 21). (K) Silhouettes of leaves 1–3 in segregating T2 sibling plants in which the denoted transgene was either homozygous or absent. Transgene homozygosity was evaluated based on the fluorescence strength of a seed-specific red fluorescent protein marker located on the transgene. (L and M) The leaf blade base angle of the plants shown in K. Significant differences from nontransgenic sibling controls were determined by a two-tailed t test (*P < 0.05; ***P < 0.001); sample sizes are indicated on the graph. Boxes in B–D, G–I, L, and M display the IQR (boxes), median (lines), and values beyond 1.5* IQR (whiskers); mean values are marked by a solid diamond (◆). All phenotypic analyses were carried out in LD conditions.
To determine if miR156 produced by the SAM is necessary for juvenile leaf identity, we also characterized the phenotype of WT plants transformed with constructs containing a miR156 target site mimic (MIM156) fused to the WUS, STM, FD, or ANT promoters. WUS::MIM156 and STM::MIM156 accelerated abaxial trichome production and reduced rosette leaf number but did not affect the rate of leaf initiation (Fig. 4 F–J). Expression of MIM156 from the STM promoter additionally affects the shape of leaf 3 but not leaves 1 or 2 (Fig. 4 K and M). FD::MIM156 accelerated abaxial trichome production (Fig. 4G) and the production of leaves with adult morphology (SI Appendix, Fig. S3C) and also reduced leaf number (Fig. 4H); however, like WUS::MIM156 and STM::MIM156, this construct did not affect the rate of leaf initiation (Fig. 4J). Some lines transformed with these constructs bolted slightly earlier than normal, but this effect was quite variable. These results demonstrate that miR156 produced by the SAM is both necessary and sufficient for the production of at least some aspects of juvenile leaf identity.
To determine if the ability of the WUS::MIR156A and STM::MIR156A constructs to correct the mir156/157 qm phenotype is attributable to the mobility of miR156 (or a downstream component of the miR156-SPL pathway), we tested the effects of localized miR156 activity on the accumulation of a SPL9::SPL9-GUS reporter. We crossed a SPL9::SPL9-GUS; mir156/157 qm line (7) to WUS::MIR156A; mir156/157 qm and STM::MIR156A; mir156/157 qm lines. The progeny of these crosses were thus hemizygous for SPL9::SPL9-GUS and hemizygous for either WUS::MIR156A or STM::MIR156A in a mir156/157 qm background. To ensure that the SPL9::SPL9-GUS construct was actually capable of responding to miR156, the SPL9::SPL9-GUS; mir156/157 qm line was crossed to Col and to FD::MIR156A; mir156/157 qm and ANT::MIR156A; mir156/157 qm plants. The progeny of the cross to Col were heterozygous for mir156a mir156c mir157a mir157c and hemizygous for SPL9::SPL9-GUS and were morphologically WT. Plants were harvested and stained for GUS activity 2 wk after planting when miR156/157 are relatively abundant (7).
SPL9::SPL9-GUS/+; mir156/157 qm seedlings had GUS activity in the epidermis and mesophyll of leaf primordia, but had no obvious GUS activity in the SAM (Fig. 5). In contrast, SPL9::SPL9-GUS/+ mir156/157 qm plants expressing MIR156A from either the ANT or FD promoters had no obvious GUS staining in both leaf primordia and the SAM. This result demonstrates that the SPL9::SPL9-GUS reporter is sensitive to miR156/157. Furthermore, the observation that FD::MIR156A is capable of repressing SPL9::SPL9-GUS expression outside the expression domain of the FD promoter suggests that miR156 is diffusible. This conclusion is further supported by the observation that expressing MIR156A under the regulation of the meristem-specific STM or WUS promoters strongly repressed SPL9::SPL9-GUS expression in very young leaf primordia and more weakly repressed SPL9::SPL9-GUS expression in older leaf primordia (Fig. 5 and SI Appendix, Fig. S5). This latter result suggests that miR156 diffuses from the SAM into leaf primordia. These results are consistent with the observation that WUS::MIR156A and STM::MIR156A are capable of partially rescuing the mir156/157 qm phenotype (Fig. 4), and, further, they suggest that the suppression of SPL activity during early primordia development is sufficient to determine leaf identity.
Fig. 5.
The miR156 module acts noncell-autonomously across the shoot apex. GUS-stained sections of plants hemizygous for a SPL9::SPL9-GUS reporter construct. All plants except for WT are homozygous for the qm combination mir156a miR156c mir157a miR157c; WT is heterozygous for these mutations. Plants labeled ANT::MIR156A, FD::MIR156A, STM::MIR156A, and WUS::MIR156A are hemizygous for these transgenes. (Scale bar, 100 μm.)
The miR156-SPL Pathway Represses WUS Expression.
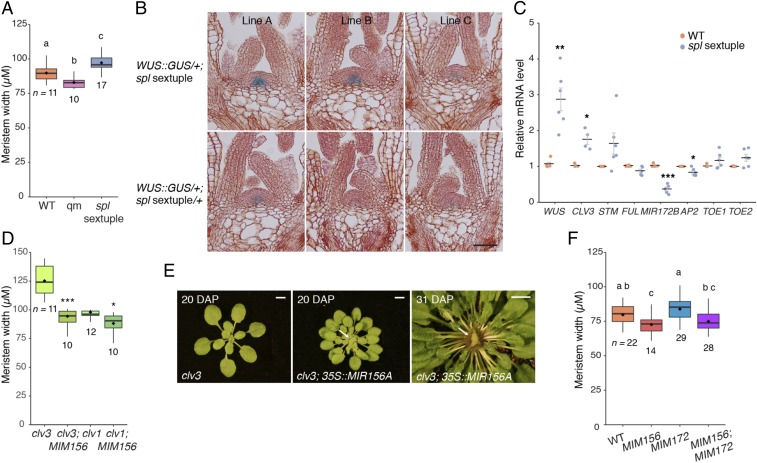
35S::MIR156A and WUS::MIR156A significantly increase the number of leaves produced by wus mutants (Fig. 3), raising the possibility that SPL genes negatively regulate the size or function of the SAM. To test this hypothesis, we measured the size of the SAM in the mir156/157 qm, in which SPL expression is strongly up-regulated (7), and in the spl2/9/10/11/13/15 sextuple mutant (spl sxm), which has very low levels of SPL activity (6). The SAM of the mir156a/157 qm was significantly narrower than WT, whereas the SAM of the spl sxm was significantly wider than WT (Fig. 6A). These results are consistent with the phenotype of plants expressing a miR156-resistant version of SPL9 (3) and suggest that SPL genes repress the growth of the SAM.
Fig. 6.
SPL genes repress WUS expression and reduce meristem size. (A, D, and F) Statistically distinct genotypes were identified by one-way ANOVA with post hoc Tukey multiple comparison test (letters indicate statistically distinct groups; P < 0.05) (A and F) or pairwise two-tailed t tests (*P < 0.05; **P < 0.01; ***P < 0.001) (D). Boxes display the IQR (boxes), median (lines), and values beyond 1.5* IQR (whiskers); mean values are marked by a solid diamond (◆). (B) Histological sections of three independent WUS::GUS lines. (Scale bar, 50 μm.) (C) mRNA levels in shoot apices with leaf primorida larger than 0.5 mm removed. Relative levels were quantified by RT-qPCR, normalized to ACT2 as an internal control gene, and expressed as a ratio to the level in WT plants. Each data point represents a biological replicate and is the average of three technical replicates. Black bars represent the mean and gray bars the SEM. Significant differences between spl sextuple and WT plants were determined by two-tailed t-test (*P < 0.05; **P < 0.01; ***P < 0.001). (E) White arrows indicate visibly fasicated mersitems (young leaves have been removed from the 31-DAP plant to more clearly show meristem size). (Scale bars, 5 mm.) All analyses were carried out on 2-wk-old plants in SD.
To determine if the expanded size of the SAM in the spl sxm is associated with enhanced WUS expression, we introduced a WUS::GUS reporter into this line. Three spl sxm; WUS::GUS lines were crossed to WT to generate plants heterozygous for these mutations and hemizygous for WUS::GUS. These phenotypically WT F1 plants were compared with spl sxm WUS::GUS/+ plants, which were identified in the T2 progeny of the original transgenic lines by the intermediate fluorescence intensity of the seed-specific OLE1::OLE1-RFP selection marker (33) on the WUS::GUS construct. Although the expression level of the WUS::GUS transgene varied between lines, GUS activity was consistently higher in spl sxm; WUS::GUS/+ than in spl sxm/+; WUS::GUS/+ plants (Fig. 6B). This result suggests that SPL transcription factors directly or indirectly repress the expression of WUS.
To obtain additional evidence for this conclusion, we examined WUS expression in the shoot apices of WT and spl sxm plants using RT-qPCR. Consistent with the expression of the WUS::GUS reporter, WUS transcripts were significantly more abundant in the spl sxm mutant than in WT (Fig. 6C). WUS positively regulates the expression of CLAVATA3 (CLV3), which feeds back via its receptor, CLV1, to repress WUS expression (27, 34). To determine if SPL transcription factors (TFs) repress WUS expression via this pathway, we first examined the effect of the spl sxm on CLV3 expression. We found that the spl sxm had significantly elevated levels of CLV3 transcripts (Fig. 6C), which is inconsistent with the hypothesis that SPL TFs repress WUS expression by promoting the expression of CLV3. We then tested if CLV3 and CLV1 are required for the negative effect of SPL overexpression on meristem size. If SPL TFs repress WUS expression by promoting the expression of CLV3 and/or CLV1, loss-of-function mutations in CLV3 and CLV1 should block the effect of SPL overexpression on meristem size. Specifically, the enlarged meristem phenotype of clv3 and/or clv1 should be epistatic to the small meristem phenotype of plants containing 35S::MIM156, which increases SPL expression. Instead, we found that 35S::MIM156 significantly decreased the size of the SAM in both clv1 and clv3 (Fig. 6D), whereas 35S::MIR156A dramatically enhanced the effect of these mutations on meristem size (Fig. 6E). These additive/synergistic interactions suggest that SPL TFs regulate WUS expression independently of the CLV3-CLV1 pathway. The inconsistent change in STM expression in the spl sxm (Fig. 6C) suggests that the repressive effects of SPL genes on meristem size are also independent of STM.
An alternative possibility is that SPL TFs repress WUS expression through their effect on the expression of APETALA2-like (AP2-like) TFs. Several AP2-like TFs are repressed transcriptionally by the SPL-target FRUITFULL (FUL) and posttranscriptionally by the SPL target MIR172B (4, 35, 36). Because AP2-like TFs promote the expression of WUS (35, 37), a reasonable hypothesis is that SPL TFs repress WUS expression by promoting the transcription of FUL or MIR172B. To test this hypothesis, we examined how FUL, MIR172B, and three miR172-regulated AP2 family members (AP2, TOE1, and TOE2) are expressed in the spl sxm. We saw no significant decrease in FUL expression whereas the primary transcript of MIR172B was significantly reduced in the shoot apices of spl sxm seedlings (Fig. 6C). We observed no increase in the transcript levels of AP2, TOE1, and TOE2. This latter result is consistent with previous studies showing that changes in the level of miR172 do not produce changes in the abundance of its target transcripts, possibly because miR172 regulates gene expression at a translational level (38, 39).
As a more direct test of the hypothesis SPL genes repress the growth of the SAM by repressing AP2-like gene expression, we measured the size of the SAM in MIM156, MIM172, and MIM156; MIM172 plants (Fig. 6F). Consistent with the phenotype of the mir156/157 qm mutant (Fig. 6A), MIM156 had a significantly narrower SAM than WT plants. MIM172 plants had wider meristems than WT, although this difference was not large enough to be statistically significant. The width of the SAM in MIM156; MIM172 was not significantly different from MIM156. These results support the conclusion that miR156-regulated SPL TFs repress, and miR172-regulated AP2-like TFs promote, the development of the SAM. However, the phenotype of MIM156; MIR172 is inconsistent with the hypothesis that SPL TFs regulate meristem size via the miR172-AP2 pathway. If this were true, MIM172 should be epistatic to MIM156. Rather, these results suggest that either SPL TFs and AP2-like TFs operate independently to regulate SAM size or that AP2-like TFs operate upstream of SPL TFs in this process.
Discussion
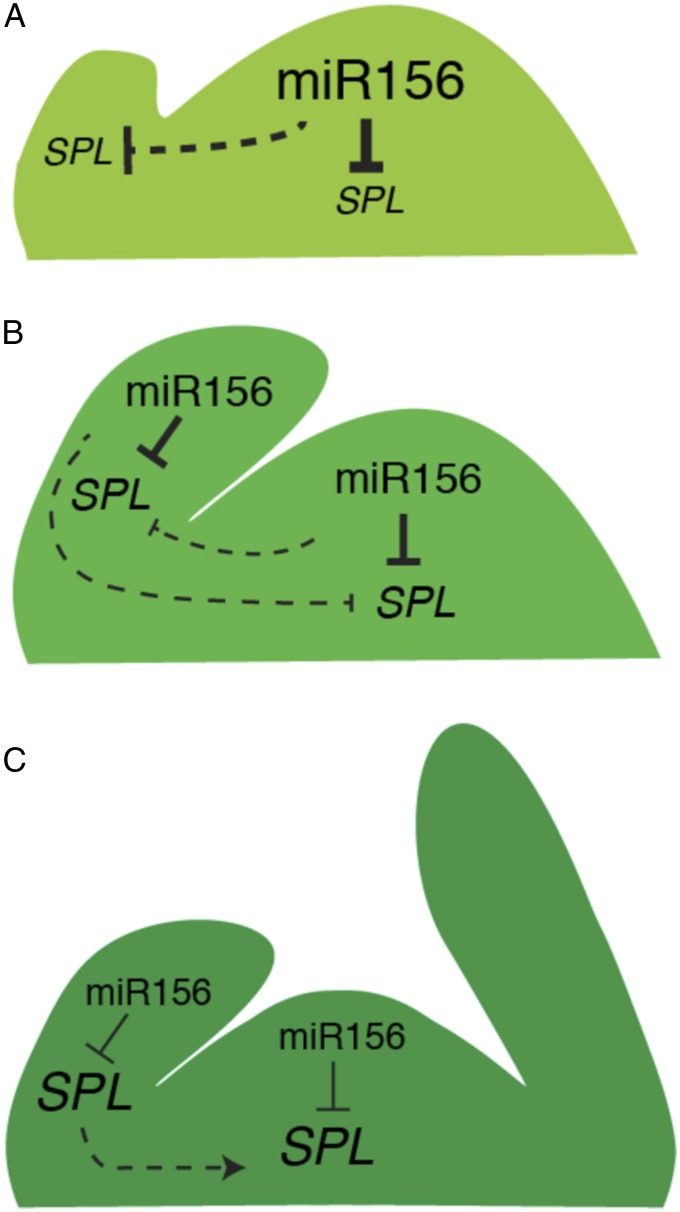
The extent to which the SAM autonomously regulates shoot development—as opposed to simply responding to regulatory factors produced by organs and tissues outside the SAM—is a classic question in plant development. Classically, vegetative phase change was thought to result from changes in the developmental identity of the SAM; indeed, the apparent stability of the juvenile and adult phases provided some of the first evidence that cells in the SAM can become determined for specific developmental fates (14, 15, 40). It is also clear that the timing of vegetative phase change can be influenced by various environmental factors and by organs/tissues external to the SAM. In particular, the discovery that preexisting leaves promote the adult identity of later-formed leaves (18–20, 41, 42) raises the possibility that vegetative identity is specified primarily by signaling from leaves to the SAM and leaf primordia, rather than by the autonomous activity of the SAM. The results presented here suggest that the identity of newly formed leaves is regulated both by SAM and by preexisting leaves, but that the relative importance of these sources of information changes during shoot development (summarized as a model in Fig. 7).
Fig. 7.
Model for the coordination of miR156 regulation across the shoot apex during vegetative development. (A) During the first few plastochrons, miR156 produced by the SAM represses SPL expression in emerging leaves. (B) During subsequent stages of juvenile growth, SPL expression in leaves is repressed predominantly by miR156 produced by leaves; SAM-derived miR156 plays a much smaller role in the regulation of SPL gene expression at these stages. (C) A temporal decline in miR156 across the shoot apex leads to the derepression of SPL genes and vegetative phase change, promoted in part by SPL protein mobility (3). Dashed lines indicate movement of either transcript (miR156) or protein (SPL) from site of expression. Lettering and line thickness reflect relative levels of expression (lettering) and repressive interactions (line thickness).
Several observations suggest that the SAM promotes juvenile leaf identity during early shoot development, but plays a relatively minor role in the regulation of leaf identity at later stages of shoot development. The best evidence that the SAM is required for juvenile leaf identity early in shoot development is the observation that meristem-defective mutations, such as wus, psd, and stm, exhibit accelerated adult development. Previous studies have suggested that this phenotype is due to the effect of these mutations on leaf initiation (16, 22), but our results indicate that it is more likely attributable to their low level of miR156/157. In particular, we found that, in addition to having reduced levels of miR156, the effect of wus on leaf morphology can be suppressed by expressing miR156 in the SAM. Meristem-specific expression of miR156 also partially corrects the phenotype of the mir156/157 qm, providing additional evidence that the SAM promotes juvenile leaf identity. However, meristem-specific miR156 expression did not have a major effect on the rate of initiation or on the morphology of later-formed leaves in the mir156/157 qm. Even FD::MIR156A, which confers constitutive expression of miR156 in the SAM and young leaf primordia, did not completely correct the phenotype of the mir156/157 qm. Indeed, despite a delay in the timing of VPC for FD::MIR156A, WUS::MIR156A, and STM::MIR156A plants, all these genetic lines still progressed to the adult phase, and they did so more quickly than ANT::MIR156A plants, in which miR156 is maintained in leaf primordia. Furthermore, reducing miR156/157 in the SAM of otherwise WT plants has only a minor effect on leaf identity. Together, these results suggest that the SAM specifies the identity of the first leaves produced by the shoot, but plays a much smaller role in determining the identity of subsequent leaves. Thus, how and where a plant makes developmental decisions shifts with age and shoot status. During initial growth the SAM is fundamental to maintaining a plant in the juvenile phase. However, as a plant produces leaves the control of development becomes more spatially diffuse, and the determination of shoot identity is coordinated by peripheral organs. Such a process has been well demonstrated in the case of floral induction, where the initiation of flowering is controlled by the leaf-derived protein FLOWERING LOCUS T (FT), but the specification of floral meristem identity is largely intrinsically regulated (28, 29).
One argument against this conclusion is that wus and stm have a much larger effect on VPC than the WUS::MIM156 and STM::MIM156 transgenes. However, this is readily explained by observation that wus and stm have significantly less miR156 than plants expressing WUS::MIM156 and STM::MIM156. miR156 is broadly expressed throughout the SAM (3), so it is to be expected that mutations that delay the initiation and dramatically reduce the size of the SAM—such as wus and stm—have a more significant effect on the level of miR156 than WUS::MIM156 and STM::MIM156, which are expressed in only a small number of cells in the SAM. Indeed, it is remarkable that WUS::MIR156A and WUS::MIM156 actually affect leaf identity, given the very small number of cells in which these transgenes are expressed.
How does the SAM specify leaf identity during early development? One possibility is that cells acquire their identity in the SAM and retain this identity as they divide and differentiate into leaves. We think this is unlikely because experiments in maize have shown that the phase identity of a leaf is not determined until after leaf initiation (42). Furthermore, it would be surprising if the amount of miR156 acquired by cells in the SAM is sufficient to regulate gene expression throughout leaf development if the transcript pool of a cell decreases by 50% with each division. A more likely scenario is that miR156, or a miR156-dependent factor, diffuses from the SAM into leaves. This hypothesis is supported by our observation that the meristem-specific transgenes WUS::MIR156A and STM::MIR156A strongly repress SPL expression in young leaf primordia, but are less effective in older leaf primordia. It is also supported by previous studies indicating that miR156 acts noncell-autonomously in potato (43) and in maize (44, 45) and is consistent with a model in which early leaf development is regulated by mobile gradients of a number of small RNAs (46). In contrast, a recent study on miRNA mobility found that an artificial miRNA expressed within the WUS domain functions cell-autonomously in this domain (47). However, this study did not investigate the movement of endogenous miRNAs, and it is possible that miR156 mobility is regulated by factors specific to this miRNA. In any case, our observation that the more broadly expressed meristem-specific STM::MIR156A transgene is also capable of repressing SPL expression in leaves supports the hypothesis that the SAM promotes juvenile leaf identity because miR156 diffuses from the SAM into leaf primordia. miR156 is expressed in leaves and increases in abundance as leaves expand (7, 48). Diffusion of miR156 from preexisting leaves into the SAM and newly formed leaf primordia could explain why a reduction in the level of miR156 specifically within the SAM had relatively little effect on leaf identity. Although the movement of miR156 from the SAM into leaf primordia is the most parsimonious explanation of our results, it is also possible that a repressor of SPL activity, the expression of which is promoted by miR156, could diffuse from the SAM to leaf primordia. To unambiguously demonstrate that miR156 functions noncell-autonomously within the shoot apex, it will be necessary to develop methods for specifically sequestering this miRNA within the SAM.
SPL proteins regulate many aspects of shoot development. In addition to promoting adult leaf identity, SPL proteins repress branching (49, 50) and reduce the size of the SAM when they are overexpressed as a result of the loss of miR156 regulation (ref. 3; this report). Our results suggest that SPL proteins reduce meristem size by repressing WUS transcription independent of the CLV3-CLV1 signaling pathway. One of the ways in which SPL proteins could do this is through their effect on the expression of miR172. SPL proteins promote the expression of miR172, which in turn represses a family of AP2-like transcription factors that promote WUS expression (37). However, our results indicate that the effects of SPL proteins on meristem size are largely independent of the miR172-AP2–like pathway. SPL proteins have also been shown to restrict cytokinin signaling via interference with type-B ARABIDOPSIS RESPONSE REGULATOR (ARR) protein activity (51). Given that type-B ARRs induce WUS expression (52), another possibility is that SPL genes restrict meristem size via this mechanism. The recent finding that Arabidopsis and soybean orthologs of SPL9 and WUS physically interact (53) suggests that SPL proteins could also regulate WUS activity posttranslationally. However, we observed that wus mutants expressing miR156 under the regulation of constitutive or meristem-specific promoter produce significantly more leaves than wus. This result suggests that WUS is not absolutely required for the function of SPLs in the SAM and raises the possibility that SPL proteins regulate meristem size or activity by both WUS-dependent and WUS-independent mechanisms.
SBP/SPL genes are present in algae and all land plants (54, 55) whereas miR156/157 did not evolve until plants colonized land (56–58). In the moss Physcomitrella patens, gametophytes with reduced SBP/SPL activity are more highly branched and initiate more leafy buds (59, 60). This suggests that one of the earliest functions of miR156/157 in land plant evolution was to promote apical growth and that this function has been conserved in angiosperms. The time course of miR156/SPL evolution, and the evidence that in flowering plants loss of miR156/157 causes leaves to adopt an adult identity, suggest that the adult phase is the default state of the shoot. We suspect that miR156 and miR157 evolved in response to environmental conditions that selected for more vigorous and prolonged shoot growth and that species-specific juvenile traits evolved later, as plants expanded into a variety of distinct habitats.
Materials and Methods
Col was used as the genetic background for all lines used in this work. Detailed descriptions of the methods used to generate transgenic plants and of the molecular analyses employed are presented in SI Appendix. Details of plant growth conditions and statistical tests are included in the relevant figure legends.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center for seed; Jianfei Zhao for the MIM156; MIM172 line; Zita Ndemanu, Yui Shimokobe, and Ayanna Coleman for technical support; and members of the R.S.P. laboratory for helpful discussions. This project was funded by NIH Grant GM051893 (to R.S.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817853116/-/DCSupplemental.
References
- 1.Goebel K. Über die Jugendzustände der Pflanzen [On the state of juvenility in plants] Flora. 1889;72:1–45. [Google Scholar]
- 2.Poethig RS. Vegetative phase change and shoot maturation in plants. Curr Top Dev Biol. 2013;105:125–152. doi: 10.1016/B978-0-12-396968-2.00005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang JW, Schwab R, Czech B, Mica E, Weigel D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell. 2008;20:1231–1243. doi: 10.1105/tpc.108.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu G, et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu M, et al. Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 2016;12:e1006263. doi: 10.1371/journal.pgen.1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He J, et al. Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana. PLoS Genet. 2018;14:e1007337. doi: 10.1371/journal.pgen.1007337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu M, Hu T, Smith MR, Poethig RS. Epigenetic regulation of vegetative phase change in Arabidopsis. Plant Cell. 2016;28:28–41. doi: 10.1105/tpc.15.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu M, Leichty AR, Hu T, Poethig RS. H2A.Z promotes the transcription of MIR156A and MIR156C in Arabidopsis by facilitating the deposition of H3K4me3. Development. 2018;145:dev152868. doi: 10.1242/dev.152868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi K, et al. Regulation of microRNA-mediated developmental changes by the SWR1 chromatin remodeling complex. Plant Physiol. 2016;171:1128–1143. doi: 10.1104/pp.16.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Y, et al. Regulation of vegetative phase change by SWI2/SNF2 chromatin remodeling ATPase BRAHMA. Plant Physiol. 2016;172:2416–2428. doi: 10.1104/pp.16.01588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Picó S, Ortiz-Marchena MI, Merini W, Calonje M. Deciphering the role of Polycomb Repressive Complex 1 (PRC1) variants in regulating the acquisition of flowering competence in Arabidopsis. Plant Physiol. 2015;168:1286–1297. doi: 10.1104/pp.15.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F, Perry SE. Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 2013;161:1251–1264. doi: 10.1104/pp.112.212282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wardlaw CW. The morphogenetic role of apical meristems: Fundamental aspects (illustrated by means of the shoot apical meristem) In: Ruhland W, editor. Differentiation and Development/Differenzierung und Entwicklung: Part 1/Teil 1. Springer; Berlin/Heidelberg: 1965. pp. 443–451. [Google Scholar]
- 15.Wareing PF. Juvenility and cell determination. In: Atherton JG, editor. Manipulation of Flowering. Butterworths; London: 1987. pp. 83–92. [Google Scholar]
- 16.Hamada S, et al. Mutations in the WUSCHEL gene of Arabidopsis thaliana result in the development of shoots without juvenile leaves. Plant J. 2000;24:91–101. doi: 10.1046/j.1365-313x.2000.00858.x. [DOI] [PubMed] [Google Scholar]
- 17.Telfer A, Bollman KM, Poethig RS. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development. 1997;124:645–654. doi: 10.1242/dev.124.3.645. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, Conway SR, Poethig RS. Vegetative phase change is mediated by a leaf-derived signal that represses the transcription of miR156. Development. 2011;138:245–249. doi: 10.1242/dev.058578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L, Xu M, Koo Y, He J, Poethig RS. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife. 2013;2:e00260. doi: 10.7554/eLife.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu S, et al. Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife. 2013;2:e00269. doi: 10.7554/eLife.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rutjens B, et al. Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J. 2009;58:641–654. doi: 10.1111/j.1365-313X.2009.03809.x. [DOI] [PubMed] [Google Scholar]
- 22.Hunter CA, Aukerman MJ, Sun H, Fokina M, Poethig RS. PAUSED encodes the Arabidopsis exportin-t ortholog. Plant Physiol. 2003;132:2135–2143. doi: 10.1104/pp.103.023309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elliott RC, et al. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi N, Jeong CW, Nole-Wilson S, Krizek BA, Wagner D. AINTEGUMENTA and AINTEGUMENTA-LIKE6/PLETHORA3 induce LEAFY expression in response to auxin to promote the onset of flower formation in Arabidopsis. Plant Physiol. 2016;170:283–293. doi: 10.1104/pp.15.00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujita H, Toyokura K, Okada K, Kawaguchi M. Reaction-diffusion pattern in shoot apical meristem of plants. PLoS One. 2011;6:e18243. doi: 10.1371/journal.pone.0018243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franco-Zorrilla JM, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 27.Schoof H, et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 28.Abe M, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- 29.Wigge PA, et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- 30.Heisler MG, et al. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 31.Bäurle I, Laux T. Regulation of WUSCHEL transcription in the stem cell niche of the Arabidopsis shoot meristem. Plant Cell. 2005;17:2271–2280. doi: 10.1105/tpc.105.032623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cardon G, et al. Molecular characterisation of the Arabidopsis SBP-box genes. Gene. 1999;237:91–104. doi: 10.1016/s0378-1119(99)00308-x. [DOI] [PubMed] [Google Scholar]
- 33.Engler C, et al. A golden gate modular cloning toolbox for plants. ACS Synth Biol. 2014;3:839–843. doi: 10.1021/sb4001504. [DOI] [PubMed] [Google Scholar]
- 34.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 35.Balanzà V, et al. Genetic control of meristem arrest and life span in Arabidopsis by a FRUITFULL-APETALA2 pathway. Nat Commun. 2018;9:565. doi: 10.1038/s41467-018-03067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 37.Würschum T, Gross-Hardt R, Laux T. APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell. 2006;18:295–307. doi: 10.1105/tpc.105.038398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brink RA. Phase change in higher plants and somatic cell heredity. Q Rev Biol. 1962;37:1–22. [Google Scholar]
- 41.Irish EE, Karlen S. Restoration of juvenility in maize shoots by meristem culture. Int J Plant Sci. 1998;159:695–701. [Google Scholar]
- 42.Orkwiszewski JA, Poethig RS. Phase identity of the maize leaf is determined after leaf initiation. Proc Natl Acad Sci USA. 2000;97:10631–10636. doi: 10.1073/pnas.180301597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhogale S, et al. microRNA156: A potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 2014;164:1011–1027. doi: 10.1104/pp.113.230714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poethig S. A non-cell-autonomous mutation regulating juvenility in maize. Nature. 1988;336:82–83. [Google Scholar]
- 45.Dudley M, Poethig RS. The heterochronic Teopod1 and Teopod2 mutations of maize are expressed non-cell-autonomously. Genetics. 1993;133:389–399. doi: 10.1093/genetics/133.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skopelitis DS, Benkovics AH, Husbands AY, Timmermans MCP. Boundary formation through a direct threshold-based readout of mobile small RNA gradients. Dev Cell. 2017;43:265–273.e6. doi: 10.1016/j.devcel.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 47.Skopelitis DS, et al. Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals. Nat Commun. 2018;9:3107. doi: 10.1038/s41467-018-05571-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie K, et al. Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice. Plant Physiol. 2012;158:1382–1394. doi: 10.1104/pp.111.190488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol. 2008;67:183–195. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiao Y, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- 51.Zhang TQ, et al. An intrinsic microRNA timer regulates progressive decline in shoot regenerative capacity in plants. Plant Cell. 2015;27:349–360. doi: 10.1105/tpc.114.135186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, et al. Cytokinin signaling activates WUSCHEL expression during axillary meristem initiation. Plant Cell. 2017;29:1373–1387. doi: 10.1105/tpc.16.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Z, et al. Genetic improvement of the shoot architecture and yield in soya bean plants via the manipulation of GmmiR156b. Plant Biotechnol J. 2018;17:50–62. doi: 10.1111/pbi.12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kropat J, et al. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci USA. 2005;102:18730–18735. doi: 10.1073/pnas.0507693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riese M, Höhmann S, Saedler H, Münster T, Huijser P. Comparative analysis of the SBP-box gene families in P. patens and seed plants. Gene. 2007;401:28–37. doi: 10.1016/j.gene.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 56.Molnár A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 57.Arazi T, et al. Cloning and characterization of micro-RNAs from moss. Plant J. 2005;43:837–848. doi: 10.1111/j.1365-313X.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- 58.Tsuzuki M, et al. Profiling and characterization of small RNAs in the liverwort, Marchantia polymorpha, belonging to the first diverged land plants. Plant Cell Physiol. 2016;57:359–372. doi: 10.1093/pcp/pcv182. [DOI] [PubMed] [Google Scholar]
- 59.Riese M, Zobell O, Saedler H, Huijser P. SBP-domain transcription factors as possible effectors of cryptochrome-mediated blue light signalling in the moss Physcomitrella patens. Planta. 2008;227:505–515. doi: 10.1007/s00425-007-0661-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cho SH, Coruh C, Axtell MJ. miR156 and miR390 regulate tasiRNA accumulation and developmental timing in Physcomitrella patens. Plant Cell. 2012;24:4837–4849. doi: 10.1105/tpc.112.103176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.