Photosynthesis and respiration provide the chemical energy that sustains life on Earth. In eukaryotes, this essential metabolism is performed by chloroplasts and mitochondria. Although both of these organelles have small genomes that are remnants of their endosymbiotic origins, most of the proteins in these organelles are encoded by nuclear genes. One consequence of this distribution of genetic material is that many of the multisubunit protein complexes that are central to energy metabolism are composed of proteins encoded by nuclear genes and either chloroplast genes (photosynthesis) or mitochondrial genes (respiration). Thus, the coordination of nuclear and chloroplast (or mitochondrial) activities requires the anterograde flow of information from the nucleus and retrograde signaling back to the nucleus. Although we know that retrograde signals from chloroplasts (i.e., chloroplast signals) influence numerous chloroplastic and extrachloroplastic processes, we still have major gaps in our knowledge of this type of signaling (1, 2). RNA editing refers to processes that change the identities of nucleotides and processes that add or delete nucleotides from RNAs. RNA editing has been reported in viruses and diverse eukaryotes. In flowering plants, RNA editing converts cytidines to uridines in the RNAs transcribed from the chloroplast and mitochondrial genomes (3). In PNAS, Zhao et al. (4) make a strong case that RNA editing in chloroplasts contributes to a type of chloroplast-to-nucleus signaling defined by the genomes uncoupled (gun) mutants in Arabidopsis.
The gun mutant screen was the first forward genetic screen developed that specifically interrogates chloroplast-to-nucleus signaling. The mutant alleles from this screen uncouple the expression of photosynthesis-associated nuclear genes (PhANGs) from chloroplast function. Chloroplasts develop from nonphotosynthetic proplastids during the development of photosynthetic organs such as cotyledons and leaves. Light is a major positive regulator of PhANG expression. When light-grown plants are treated with inhibitors or harbor mutant alleles that block chloroplast biogenesis at a stage resembling proplastids, the expression—usually the transcription—of PhANGs is severely down-regulated. Indeed, when chloroplast biogenesis is blocked in light-grown seedlings, PhANGs are expressed at lower levels than are observed in dark-grown seedlings, which contain etioplasts—an intermediate in chloroplast biogenesis that accumulates in the absence of light-dependent gene expression and chlorophyll biosynthesis. In gun mutants, when chloroplast biogenesis is blocked, PhANG expression is significantly up-regulated relative to the wild type because of abnormal chloroplast-to-nucleus signaling. The chloroplast-to-nucleus signaling that depends on the GUN genes is also activated when the biogenesis of chloroplasts and etioplasts is not blocked but their activities are attenuated. When seedlings contain well-functioning chloroplasts, PhANG expression is indistinguishable in gun mutants and wild type. Thus, chloroplast dysfunction appears to activate this type of chloroplast-to-nucleus signaling (2).
gun mutant screens have yielded a large number of mutant alleles of genes that encode a plastidic pentatricopeptide repeat (PPR) protein named GUN1, a blue-light receptor named cryptochrome 1 (cry1), and proteins that contribute to tetrapyrrole metabolism (GUN2, GUN3, GUN4, GUN5, and GUN6), which occurs in chloroplasts and nonphotosynthetic plastids. In plants, tetrapyrroles include siroheme, heme, phytochromobilin, and chlorophyll. Increases in the levels of GUN6 (also known as ferrochelatase 1) activate chloroplast-to-nucleus signaling, probably by increasing heme biosynthesis. cry1 can accumulate in either the cytosol or the nucleus and up-regulates or down-regulates PhANG expression depending on factors that influence cellular conditions, such as chloroplast function. The specificity of the gun mutant screen indicates that chloroplast dysfunction activates a few specific mechanisms that regulate gene expression in the nucleus and that the gun phenotype is uncommon. A gun phenotype refers to the significant up-regulation of PhANG expression in seedlings containing dysfunctional chloroplasts, presumably because of abnormal chloroplast-to-nucleus signaling. The GUN genes appear to promote chloroplast function (1, 2). Consistent with this observation, all of the gun mutants that were isolated from gun mutant screens are more sensitive to the inhibitors that are typically used to block chloroplast biogenesis in these types of experiments (e.g., norflurazon and lincomycin) than wild type (5–7). Norflurazon blocks chloroplast biogenesis by inhibiting the biosynthesis of carotenoids, which are essential photosynthetic pigments. Lincomycin blocks chloroplast biogenesis by inhibiting chloroplast translation (2).
GUN1 is associated with a number of biological functions, such as chloroplast biogenesis, the regulation of nuclear gene expression, the circadian rhythm, the accumulation of anthocyanins, abiotic stress tolerance, and the development of seedlings and leaves (1, 2). Data from genetic experiments indicate that GUN1 is central to much of this type of chloroplast signaling (1, 2). However, a particular type of chloroplast signaling that is activated by lincomycin treatments was recently reported to not depend on GUN1 (8). Unfortunately, our knowledge of the biochemical function of GUN1 has been vague. In general, PPR proteins contribute to RNA metabolism in chloroplasts and mitochondria. Thus, GUN1 was suggested to participate in chloroplast-to-nucleus signaling by contributing to RNA metabolism in chloroplasts that affects the biosynthesis or transduction of a chloroplast signal (1, 2). Consistent with this idea, gun1 mutants accumulate less chloroplast RNA than wild type (9). Alternatively, based on a variety of experiments with overexpressed fusion proteins containing GUN1, such as the cross-linking of an overexpressed fusion protein containing GUN1 to nearly 300 different chloroplast proteins, including GUN6, Tadini et al. (10) proposed that GUN1 might contribute to plastid signaling by associating with some of these putative GUN1-binding proteins. Although this is an intriguing idea, at this stage, the data consistent with this idea are subject to interpretation.
Zhao et al. (4) make a strong case that GUN1 contributes to chloroplast-to-nucleus signaling by influencing RNA editing. They report that in norflurazon-treated seedlings, RNA editing is altered at 21 sites and that GUN1 affects RNA editing at 11 of these sites in norflurazon-treated seedlings. Importantly, they found that GUN1 has only minor effects on RNA editing in untreated seedlings containing well-functioning chloroplasts. Moreover, they show that GUN1 can interact with MULTIPLE ORGANELLAR RNA EDITING FACTOR 2 (MORF2), an essential chloroplast protein and a core component of the RNA editosome that edits many of the RNAs that are edited in chloroplasts. Although it is not possible to test whether loss-of-function alleles of morf2 influence chloroplast-to-nucleus signaling because this mutant is seedling lethal, overexpression of MORF2 induces a gun phenotype. Zhao et al. (4) also report that three RNA editing site-specificity factors named ORGANELLE TRANSCRIPT PROCESSING 81 (OTP81), ORGANELLE TRANSCRIPT PROCESSING 84 (OTP84), and YELLOW SEEDLINGS 1 (YS1) interact with MORF2 and that the otp81, otp84, and ys1 loss-of-function mutants are gun mutants, albeit subtle gun mutants. Moreover, their transcriptome analyses provide evidence that the chloroplast-to-nucleus signaling activated in gun1 mutants and MORF2 overexpression lines is similar. However, Zhao et al. (4) find no simple correlation between the levels of RNA editing and the strength of the gun phenotype.
MORF2 may help to assemble specific protein complexes at RNA editing sites. GUN1 accumulates only transiently during chloroplast biogenesis, but when chloroplasts experience dysfunction, the accumulation of GUN1 is sustained (11). Based on these observations, Zhao et al. (4) suggest that the overexpression of MORF2 and the accumulation of GUN1 may influence chloroplast-to-nucleus signaling by affecting the activity of protein complexes at RNA editing sites and that GUN1 may affect protein complexes at RNA editing sites by interacting with MORF2 (Fig. 1). They suggest two models to explain the influence of RNA editing on chloroplast-to-nucleus signaling. First, because three of the changes in RNA editing sites that were found in all of the plastid signaling-deficient lines tested were in transcripts encoding subunits of the plastid-encoded RNA polymerase and because connections between chloroplast-to-nucleus signaling and the plastid-encoded RNA polymerase were reported previously (9), they suggest that chloroplast dysfunction may influence the plastid-encoded RNA polymerase and, therefore, the transcription of a particular set of chloroplast genes, which may activate GUN1-dependent chloroplast-to-nucleus signaling. Alternatively, they suggest that chloroplast-to-nucleus signaling is influenced by complex defects in RNA editing that lead to the accumulation of misfolded chloroplast proteins that must be cleared by a chloroplast protein quality-control pathway.
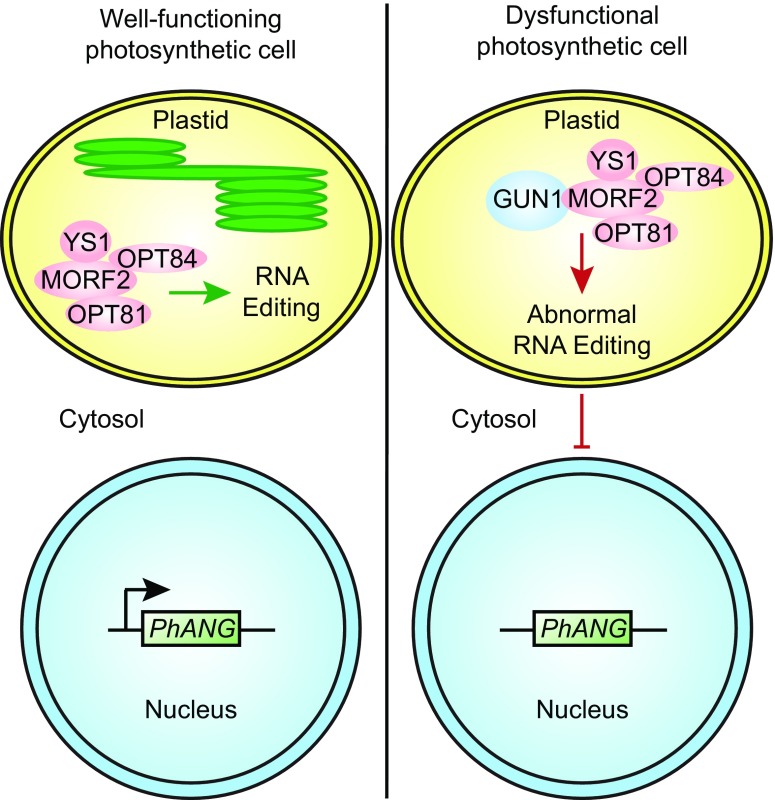
Fig. 1.
Proposed interactions between RNA editing and chloroplast signaling. In well-functioning photosynthetic cells (Left), chlorophyll accumulates in the thylakoid membranes (green), and chloroplasts perform photosynthesis. The GUN1 protein does not accumulate and thus the chloroplast-to-nucleus signaling that depends on GUN1 is not active. MORF2 contributes to RNA editing (green arrow) and interacts with OPT81, OPT84, and YS1. Light signaling, tissue-specific signals, and the circadian rhythm (not shown) drive high-level expression of PhANGs (black arrow), which promotes chloroplast function. Blocking chloroplast biogenesis at a stage resembling the proplastid leads to the development of dysfunctional photosynthetic cells (Right) containing dysfunctional chloroplasts that do not accumulate chlorophyll or perform photosynthesis. In these cells, GUN1 accumulates and regulates RNA editing by interacting with MORF2 (red arrow). Abnormal RNA editing activates a chloroplast-to-nucleus signaling mechanism that down-regulates PhANG expression (red T-bar). Similarly, GUN1 accumulates during the early stages of leaf development—before chloroplast biogenesis is completed (11)—and regulates RNA editing and chloroplast-to-nucleus signaling as it does in dysfunctional photosynthetic cells.
The data from Zhao et al. (4) have a number of implications. For instance, the overexpression of MORF2 induces a gun phenotype when chloroplast biogenesis is blocked with a norflurazon treatment and not when chloroplast biogenesis is blocked with a lincomycin treatment. Only gun mutants with deficiencies in tetrapyrrole metabolism were previously reported to exhibit gun phenotypes when treated with norflurazon but not when treated with lincomycin. These data and genetic interactions between mutant alleles of GUN1 and tetrapyrrole signaling mutants are consistent with connections between tetrapyrrole-influenced chloroplast-to-nucleus signaling and RNA editing. In contrast, gun1 mutants exhibit gun phenotypes when chloroplast biogenesis is blocked with either norflurazon or lincomycin and when chloroplast dysfunction is induced with other mechanistically distinct procedures. These data and the observation that MORF2 overexpression lines have weaker gun phenotypes than gun1 mutants indicate that the interactions between GUN1 and MORF2 represent only one component of GUN1-dependent chloroplast-to-nucleus signaling. This work may also help us to advance our understanding of the integration of light and chloroplast signaling (1). Tetrapyrrole signaling was reported in untreated dark-grown seedlings but not in untreated light-grown seedlings that were deficient in one of the σ subunits of the plastid-encoded RNA polymerase (9). Thus, this σ-subunit mutant may provide a useful system for studying the influence of light-regulated development on the potential interactions between tetrapyrrole signaling and RNA editing. In addition to stimulating new research directions such as these, the breakthrough described by Zhao et al. (4) provides the clarity needed to propose any number of specific experiments that may lead to even more mechanistic insight into the chloroplast signaling defined by the GUN genes.
Acknowledgments
R.M.L.’s research is supported by the Huazhong Agricultural University Scientific & Technological Self-Innovation Foundation (Grant 2016RC009) and the National Key Research and Development Program of China, Ministry of Science and Education (Grant 2018YFD1000800).
Footnotes
The author declares no conflict of interest.
See companion article on page 10162.
References
- 1.Larkin RM. (2014) Influence of plastids on light signalling and development. Philos Trans R Soc Lond B Biol Sci 369:20130232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larkin RM. (2016) Tetrapyrrole signaling in plants. Front Plant Sci 7:1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takenaka M, Zehrmann A, Verbitskiy D, Härtel B, Brennicke A (2013) RNA editing in plants and its evolution. Annu Rev Genet 47:335–352. [DOI] [PubMed] [Google Scholar]
- 4.Zhao X, Jianyan H, Chory J (2019) GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signaling. Proc Natl Acad Sci USA 116:10162–10167.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Llamas E, Pulido P, Rodriguez-Concepcion M (2017) Interference with plastome gene expression and Clp protease activity in Arabidopsis triggers a chloroplast unfolded protein response to restore protein homeostasis. PLoS Genet 13:e1007022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song L, Chen Z, Larkin RM (2018) The genomes uncoupled mutants are more sensitive to norflurazon than wild type. Plant Physiol 178:965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao X, Huang J, Chory J (2018) genome uncoupled1 mutants are hypersensitive to norflurazon and lincomycin. Plant Physiol 178:960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ortiz-Alcaide M, et al. (2019) Chloroplasts modulate elongation responses to canopy shade by retrograde pathways involving HY5 and ABA. Plant Cell 31:384–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J (2013) Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J 73:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tadini L, et al. (2016) GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol 170:1817–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu GZ, et al. (2018) Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED1. Plant Physiol 176:2472–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]