Significance
Little is known about how multiple functions of a single protein are coordinated in a living cell. PARP-1 is a multidomain nuclear protein that plays a critical role in regulating developmental processes including apoptosis, DNA repair, epigenetic marking of chromatin, assembly of higher-order chromatin structures, and transcriptional activation. Using deletional isoforms of PARP-1 in in vivo and in vitro experiments, we have demonstrated that the multiple domains of PARP-1 cooperate in response to interactions with different PARP-1 targets, leading either to short-term activation of the enzyme or to prolonged and sustained activity. This sustained activity produces accumulation of pADPr in the surrounding chromatin, leading to prolonged chromatin loosening.
Keywords: PARP-1, poly(ADP-ribose), drosophila, PARP-1 regulation, protein domains
Abstract
Poly(ADP-ribose) polymerase 1 (PARP-1) is a multidomain multifunctional nuclear enzyme involved in the regulation of the chromatin structure and transcription. PARP-1 consists of three functional domains: the N-terminal DNA-binding domain (DBD) containing three zinc fingers, the automodification domain (A), and the C-terminal domain, which includes the protein interacting WGR domain (W) and the catalytic (Cat) subdomain responsible for the poly(ADP ribosyl)ating reaction. The mechanisms coordinating the functions of these domains and determining the positioning of PARP-1 in chromatin remain unknown. Using multiple deletional isoforms of PARP-1, lacking one or another of its three domains, as well as consisting of only one of those domains, we demonstrate that different functions of PARP-1 are coordinated by interactions among these domains and their targets. Interaction between the DBD and damaged DNA leads to a short-term binding and activation of PARP-1. This “hit and run” activation of PARP-1 initiates the DNA repair pathway at a specific point. The long-term chromatin loosening required to sustain transcription takes place when the C-terminal domain of PARP-1 binds to chromatin by interacting with histone H4 in the nucleosome. This long-term activation of PARP-1 results in a continuous accumulation of pADPr, which maintains chromatin in the loosened state around a certain locus so that the transcription machinery has continuous access to DNA. Cooperation between the DBD and C-terminal domain occurs in response to heat shock (HS), allowing PARP-1 to scan chromatin for specific binding sites.
The complexity and size of eukaryotic genomes require tight coordination between activation and repression of nuclear processes across tissues and organs (1, 2). Changes in the chromatin architecture of a eukaryotic nucleus coordinate gene expression in response to the extranuclear environment and are orchestrated by interactions between DNA and chromatin proteins (2, 3). The mechanisms that organize DNA into structural units are largely responsible for defining the specific functions and phenotypes of different cells (4–9).
After histones, the second most abundant protein in the eukaryotic nucleus is PARP-1, an effector protein that functions as a switch, controlling the activation and silencing of chromatin regions (10–14). PARP-1 enzymatic activity can be induced either by its interaction with nicked DNA or with histone H4 in a phosphorylated H2Av-histone-bearing nucleosome (15–17). When enzymatically active, PARP-1 covalently modifies itself and surrounding nuclear proteins by synthesizing strands of pADPr from the NAD substrate (10–13). Histones and DNA repair enzymes have been identified as PARP-1 modification targets (18–20). By shifting histones toward the more electronegative pADPr and away from the DNA molecule, PARP-1 activity initiates chromatin loosening, allowing transcription activation (21–24). The same process permits repair of damaged DNA and DNA replication (2, 6–9).
PARP-1 consists of three core domains: the N-terminal DNA binding domain (DBD), the middle automodification domain (A), and the C-terminal catalytic domain (C) (Fig. 1A) (10, 25). The DNA-binding domain contains three Zinc fingers, ZI, ZII, and ZIII, of which only ZI and ZII are capable of interacting with DNA (10, 26–28). The third ZIII represents a protein interaction subdomain and mediates interprotein interactions of other PARP-1 domains (29, 30). During the DNA damage response, ZI, ZIII, W, and Cat domains of PARP-1 form a stable active complex around a fragment of broken DNA in vitro (Fig. 1A) (29, 30). The A domain of PARP-1 is the primary target of PARP-1 activity and becomes automodified by pADPr upon PARP-1 activation (10). An automodified PARP-1 loses its ability to interact with DNA (21, 31, 32) and serves as a “shuttle” for proteins of chromatin (33). The N-terminal DBD and the C-terminal ZIII-A-W-Cat domains are responsible for PARP-1 interaction with chromatin (15, 34). Without the DBD, PARP-1 cannot bind to or be activated by DNA (15, 34). Binding of PARP-1 to histones has been shown to be regulated by its C-terminal subdomains (15, 16, 35). The presence of DBD and C-terminal domains is required for PARP-1-dependent chromatin condensation in vitro (34).
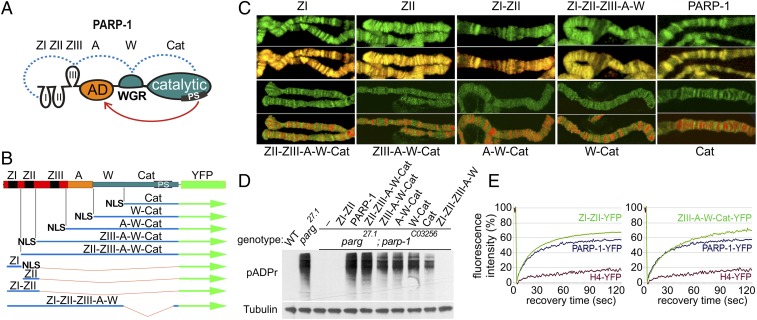
Fig. 1.
N- and C-terminal PARP-1 domains contribute to PARP-1 protein localization genome-wide in vivo. (A) Domains of PARP-1: The N-terminal DNA-binding domain (DBD) containing Zn fingers: ZI, ZII, ZIII (ZF domains 1, 2, and 3), the automodification domain (A), the only domain of PARP-1 known to accept pADPr, the WGR domain (W), and the C-terminal catalytic domain (C). The PARP signature (PS) is an evolutionarily conserved PARP-1 catalytic site in the Cat domain. The dotted blue line indicates known interactions between the domains induced by interactions with damaged DNA (21). The red arrow indicates automodification of PARP-1. (B) Structure of recombinant-transgenic PARP-1 constructs for in vivo experiments. (C) Localization of deletional recombinant isoforms of PARP-1 in salivary gland polytene chromosomes. Green is the fluorescence of proteins fused to YFP, red is DNA. All isoforms of PARP-1 carrying ZI and ZII demonstrate colocalization with DNA resulting in the yellow color in the overlay. All isoforms without ZI and ZII localized in active open chromatin only, resulting in the separation of red and green in the overlay. (D) PARP-1 deletional isoform activity assay in vivo. PARP-1 deletional isoforms were expressed in the parg27.1; parp-1C03256 mutant flies. All isoforms containing the Cat domain restored pADPr accumulation. (E) Both DNA- and C-terminal domains contributed to PARP-1 protein dynamic binding to chromatin in vivo. Comparative analysis of fluorescent recovery after photobleaching (FRAP) assay for recombinant protein is shown, including ZI-II-YFP, ZIII-A-W-Cat-YFP, full-length PARP-1-YFP, and H4-YFP. Data for the FRAP experiment show the average based on 10 replicates.
Because pADPr polymers are perpetually degraded by pADPr glycohydrolase (PARG) (10–13), a sustained production of pADPr by PARP-1 is required for maintaining chromatin in its loosened state and transcription activation. Transcription silencing is prompted by PARP-1 when it binds to heterochromatic regions of chromatin via its ZI, which represses repeated mobile elements in the genome (26). Therefore, PARP-1 appears to have two antagonistic functions, acting as a transcription activator and as a repressor (33, 36), depending on which specific PARP-1 domain is involved in its interaction with the nuclear targets.
To examine how interactions of individual PARP-1 domains with different targets coordinate different nuclear functions of PARP-1, we generated deletional isoforms of PARP-1 lacking each one of its domains (Fig. 1B). Because the Drosophila genome codes for a single PARP protein (37, 38), we use Drosophila as our model organism to examine the activity and interaction of PARP-1 domains in vitro and in vivo. By studying the activity and functions of deletional isoforms of PARP-1, we show how PARP-1 localization and activity are regulated by interactions between PARP-1 domains and their targets, specifically, histones and DNA.
Results
W-Cat Domains Target Active Chromatin.
It is expected that PARP-1 binds to DNA via its N-terminal domain, whereas its interaction with histones and other chromatin proteins occurs via its C-terminal domain. To examine how these interactions regulate PARP-1 positioning and function in vivo, we monitored the localization of deletional isoforms of PARP-1 fused with YFP in Drosophila (SI Appendix, Fig. S1).
In wild-type Drosophila nuclei, all PARP-1 isoforms show discrete localization in chromatin (SI Appendix, Fig. S2), confirming that each domain of PARP-1 taken on its own is sufficient for an interaction with chromatin. We tested which domains are responsible for automodification of PARP-1 and for its interaction with pADPr. Previously we reported that all proteins that have been covalently modified by pADPr (including PARP-1 itself) and proteins bound to pADPr noncovalently are subjected to turnover inside Cajal bodies (39). Because the PARG enzyme is required for clearing pADPr, all pADPr-modified proteins remain adherent to Cajal bodies in Parg null mutants (39). We compared the localization of each deletional isoform of PARP-1 in wild-type Drosophila and in Parg mutant nuclei. In the absence of the A domain, the relocation of the C-terminal catalytic domain of PARP-1 to Cajal bodies was considerably reduced. (SI Appendix, Fig. S2, Parg−/−). All PARP-1 deletional isoforms containing the A domain were enriched inside Cajal bodies (SI Appendix, Fig. S2, Parg−/−). Because the covalent automodification of PARP-1 is required for its relocation to the Cajal body, this finding indicates that these isoforms were covalently modified by pADPr.
To monitor the localization of PARP-1 deletional isoforms in chromatin, we used in vivo fluorescent imaging. We distinguished between inactive (condensed) and active (decondensed or loose) chromatin by applying Draq5 dye, which accumulates in areas of dense chromatin. Fluorescent imaging of salivary gland polytene chromosomes showed that all PARP-1 isoforms containing ZI and ZII were localized in a pattern similar to that of DNA stained with Draq5 (Fig. 1 B and C). Even isolated ZI and ZII were distributed along the chromatin in a similar manner. These results suggest that the first two Zn fingers display a non–site-specific affinity to DNA. In the absence of ZI and ZII, PARP-1 deletional isoforms were localized exclusively inside the open chromatin (Fig. 1C), suggesting that PARP-1 interacts with open chromatin via its C-terminal domains, regardless of whether it is bound to DNA or not. Zn fingers are not required for PARP-1 localization to active chromatin.
Using a parg−/−;parp−/− double mutant Drosophila line, we tested whether C-terminal domains of PARP-1 are sufficient for PARP-1 activation in vivo (Fig. 1D). parg−/−;parp−/− double mutant animals lack PARP-1 and cannot produce pADPr. Because these mutants also lack PARG, any modifications by poly(ADP ribosyl)ation become permanent, making their quantification straightforward. We expressed each deletional isoform individually in the parg−/−;parp−/− double mutant using ubiquitous drivers. For each transgenic stock, nuclear proteins were extracted from third-instar larvae of the same age and size. The amounts of pADPr that accumulated in each deletional isoform were assessed on Western blots using an anti-pADPr antibody. All isoforms containing the Cat domain of PARP-1 demonstrated either complete or partial restoration of pADPr accumulation (Fig. 1D). This result supports the inference made from in vivo fluorescent imaging that C-terminal domains of PARP-1 are sufficient for DNA-independent PARP-1 activation in vivo.
To examine how different domains contribute to PARP-1 distribution and activity in chromatin, we used a FRAP assay (15). We recorded two parameters for the recovery of fluorescence for a chromatin-associated protein: the speed of recovery based on the slope of the FRAP curve and the magnitude of recovery based on the plateau of the FRAP curve. Both parameters depend on the affinity of the protein to chromatin and the fraction of the protein that can dissociate without a deep remodeling of chromatin. Core histones are embedded in nucleosomes and, therefore, cannot be recovered without chromatin remodeling (Fig. 1E, H4-YFP). The full-length PARP-1 recovery curve plateaued after reaching 59% (Fig. 1E, PARP-1-YFP). Deleting either C-terminal (Fig. 1E, Left, ZI-ZII-YFP) or DNA-binding N-terminal (Fig. 1E, Right, ZIII-A-W-Cat-YFP) domains increased the proportion of PARP-1 recovered in 60 s. Thus, each domain contributes independently to the dynamics of PARP-1 localization and its interaction with chromatin.
W-Cat Domains, but Not DNA-Binding Domains Bind to the hsp70 Transcriptional Start Site.
Rapid transcriptional activation of the hsp70 gene is dependent on PARP-1 activity (14, 24, 31). To confirm that the W-Cat domain primarily contributes to the transcription activation function of PARP-1 and that DNA-binding ZI and ZII are responsible for a nonspecific binding to DNA, we examined the binding patterns of each deletional isoform at the PARP-1-dependent hsp70 locus and tested the ability of each isoform to activate the transcription of hsp70 (Fig. 2). We first compared the distribution of different PARP-1 isoforms in the hsp70 locus using a chromatin immunoprecipitation assay before and after HS treatment. To eliminate any contribution from endogenous PARP-1, we expressed YFP-fused PARP-1 isoforms in the parp −/− mutant background. Before HS, full-length PARP-1-YFP predominantly accumulated at the transcription start site (TSS) and transcription termination site (TTS) (Fig. 2A, blue bars). In addition, a significant fraction of PARP-1-YFP was bound upstream from the promoter and in the coding region of the gene (Fig. 2A, blue bars). After a 30-min HS, the full-length PARP-1 was almost completely gone from the TSS and TTS, but its binding in the coding region significantly increased (Fig. 2A, red bars). In the absence of the DNA-binding domain, PARP-1 deletional isoforms bind almost exclusively at TSS and TTS before HS (Fig. 2 B and C, blue). We did not detect any increase in their binding in the coding region after HS (Fig. 2 B and C, red). In contrast, deletional isoforms containing only ZI and ZII were widely dispersed throughout the coding region of the hsp70 locus before HS (Fig. 2D, blue). Their binding pattern did not change after HS (Fig. 2D, red).
Fig. 2.
C-terminal domains of PARP-1 are responsible for PARP-1 targeting to the TSS of the hsp70 locus, whereas DNA-binding domains target PARP-1 to areas outside of the promoter region. (A–D) The comparison of recombinant protein distribution within the hsp70 locus, ChIP assay before and after HS: PARP-1-YFP (A); ZIII-A-W-Cat-YFP (B); W-Cat-YFP (C); and ZI-ZII-YFP (D). (E) PARP-1 isoforms with C-terminal domains rescue transcription activation at the hsp70 locus following HS in parp-1C03256 mutants. The level of hsp70 mRNA was recorded before and after 30 min of HS treatment using quantitative RT PCR. (F) PARP-1 isoforms with C-terminal domains rescue histone H3 displacement from hsp70 locus, following HS in parp-1C03256 mutants. ChIP assay compares amounts of H3 histone in the promoter region of the hsp70 locus in wild-type and parp-1C03256 mutant animals expressing full-length PARP-1 (PARP-1-YFP) or PARP-1 deletional isoforms (ZI-ZII, ZIII-A-W-Cat, W-Cat) before (−) and after (+) 30 min of HS treatment. All error bars are based on the average of triplicates.
These findings support our hypothesis that DNA-independent binding contributes to specific localization of PARP-1 in TSS and TTS, whereas DNA-dependent binding is responsible for PARP-1 interaction with chromatin in a non–site-specific manner. Previously, we have reported that the TSS of the hsp70 locus is flanked by two H2Av-containing nucleosomes and that PARP-1 binding to histone H4 in these nucleosomes regulates PARP-1 localization and activation during HS (35). Taken together with this earlier finding, our data suggest that DNA-independent positioning of PARP-1 at the TSS involves PARP-1 binding with two flanking nucleosomes via its interaction with histone H4 in those nucleosomes. Without HS, full-length PARP-1 and all W-Cat-containing isoforms of PARP-1 copurified with histone H4 and H2Av in our chromatin coimmunoprecipitation experiments; DNA-binding Zn fingers of PARP-1 do not show such a specificity (SI Appendix, Fig. S3).
To test whether stable PARP-1 binding to chromatin is required for PARP-1-dependent transcription, we analyzed HS transcriptional response in a parp−/− mutation rescued with full-length PARP-1 and each of three deletional isoforms. Upon HS treatment, full-length PARP-1 rescued HS response completely. The presence of W-Cat in an isoform was sufficient to activate transcription of the hsp70 gene, albeit not at the same level as in the presence of a full-length PARP-1 (Fig. 2E). This finding is consistent with our inference that the W and Cat domains are sufficient for PARP-1 localization in promoters and its activation there. Earlier studies (17, 24) have demonstrated that PARP-1 controls nucleosome displacement at promoters during transcription (40). We found that the isolated C-terminal domain of PARP-1 (W-Cat) also has the ability to displace histones at the hsp70 locus after HS (Fig. 2F). Thus, PARP-1 localization and activation in chromatin were sufficient to maintain active chromatin during gene transcription.
Histone H4-Mediated PARP-1 Activation Is Dependent on H4 Binding to the C-Terminal W-CAT Domains of PARP-1.
To examine the roles of PARP-1 domains in binding to DNA and nucleosomal histones in vitro, we generated three deletional isoforms, including ZI-ZII (consisting of ZI-ZII only), ZIII-A-W-Cat (PARP-1 lacking ZI-ZII), and W-Cat (PARP-1 lacking ZI-ZIII as well as its A domain) (Fig. 3A). These deletional recombinant constructs and full-length PARP-1 were expressed and purified using the bacterial system. To test the affinity of different deletional isoforms of PARP-1 to components of nucleosomes, we performed two in vitro binding assays using DNA and H4 coupled to sepharose beads.
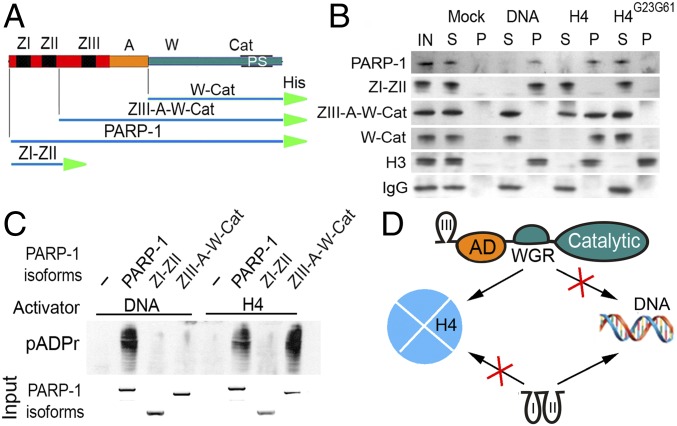
Fig. 3.
Histone H4-mediated PARP-1 activation is dependent on H4 binding to the C-terminal W-CAT domains of PARP-1. (A) Composition of deletional recombinant PARP-1 isoforms for in vitro experiments. His, 6 histidine tag. (B) In vitro binding assay (15): DNA, histone H4 and a mutant form of H4, H4G23G61 were each covalently coupled to CnBr beads and preincubated with a solution containing each deletional isoform of PARP-1. Following precipitation of beads, pellet (P) and solution (S) were subjected to PAGE and Western blot. The presence of PARP-1 isoforms in pellet and solution was detected on Western blot using anti–6XHis-tag antibody. Histone H3 was used as a positive control, which interacts with DNA and both forms of histone H4. IgG was used as a nonspecific protein-binding control. (C) In vitro activation assay. Sepharose beads with covalently attached DNA or H4 were preincubated with PARP-1 deletional isoforms and then mixed with NAD. The accumulation of pADPr was detected on Western blot using an anti-pADPr antibody. Input: the Coomassie Brilliant Blue stained PAGE gel shows the quantities of PARP-1 isoforms loaded in each reaction. (D) Diagram illustrating interactions between PARP-1 deletional isoforms and their targets (DNA and histones): ZIII-A-W-Cat (Top) interacting with H4-coupled beads only, ZI-ZII (Bottom) interacts with DNA-coupled beads but not with H4.
Previously, we demonstrated that PARP-1 binds to a hydrophobic patch formed by Val61 and Leu23 amino acids of histone H4 (17). This patch is exposed on the surface of a H2Av histone-bearing nucleosome but hidden inside the nucleosomes that bear H2A histones (17). We confirmed that the interaction with the hydrophobic patch of histone H4 alone is sufficient for binding and activating PARP-1 in vitro (15, 17, 26). Therefore, to test binding affinities of PARP-1 domains, we used histone H4 instead of an intact nucleosome. Because an intact nucleosome also includes DNA, using an isolated H4 allowed us to target histone-binding functions of PARP-1, independent of its DNA-binding functions. The conditions of the in vitro binding assay preclude histone H4 from forming nonspecific aggregates with its binding protein partners. In this assay, H4 is involved exclusively in interactions with PARP-1 (15, 17, 26). The specificity of these interactions were confirmed in multiple reciprocal experiments (15). As a control, we used a mutant isoform of H4 where Val61 and Leu23 were replaced with glycines (H4G23G61). This mutant isoform of H4 is stable on its own and in a nucleosome (17). It interacts normally with histone H3 (Fig. 3B) but is unable to bind or activate PARP-1 (17).
Pull-down assays using DNA- and H4-coupled sepharose beads (H4-SB) (Fig. 3B) showed that ZI-ZII strongly binds to DNA but not to H4, suggesting that ZI-ZII are sufficient for DNA binding but not H4 binding. Both ZIII-A-W-Cat and W-Cat isoforms bound to H4 but not to DNA (Fig. 3B). We also found that the C-terminal but not N-terminal domains of PARP-1 recognize and bind to the H2Av-containing histone octamers with high affinity, whereas both the C- and the N-terminal domains bind to an intact mononucleosome (SI Appendix, Fig. S4).
Results reported above confirm that the two PARP-1 activators DNA and H4 bind to different domains of PARP-1 to trigger its activation. To identify which domains bind to each target, we tested the ability of DNA and H4 to activate different deletional isoforms of PARP-1. DNA could not activate the ZIII-A-W-Cat isoform, which lacks ZI and ZII because the first two Zn fingers are necessary for DNA-dependent PARP-1 activation (Fig. 3C). Despite lacking ZI-ZII, the ZIII-A-W-Cat isoform can be activated by H4, confirming that PARP-1 binding and activation mediated by histone H4 are dependent exclusively on the C-terminal domains of PARP-1 (Fig. 3D). Therefore, the C-terminal and N-terminal domains of PARP-1 have distinct functions that can be separated from one another by introducing mutations to the PARP-1 locus.
W-Cat Domain of PARP-1 Is Sufficient for H4-dependent PARP-1 Activation.
To test the role of C-terminal subdomains during PARP-1 interaction with chromatin, we used the W-Cat deletional isoform, which lacks the DBD and A domains. We found that the W-Cat isoform does not bind to DNA (Fig. 3B). We further tested whether this isoform could still interact with nucleosomal histones and become enzymatically active. To detect the enzymatic activity of the W-Cat isoform, we needed to provide an alternative target that can be modified by ADPr moieties because the W-Cat isoform lacks the A domain, which serves as the primary target for PARP-1 enzymatic activity (25). Previous studies have shown that PARP-1 also modifies histones H1, H2A, and H2B with pADPr in vitro (10, 21). Therefore, any of these histones could potentially serve as acceptors of pADPr. We compared H4-dependent activity of the W-Cat deletional isoform in the presence of these targets.
Following its activation by histone H4, full-length PARP-1 produces pADPr with and without other protein targets (Fig. 4A, Panel 1). When the W-Cat isoform was incubated with a NAD substrate and H4-coupled beads but without other protein targets, no pADPr was produced (Fig. 4A, Panel 2). This result demonstrates that PARP-1 cannot produce pADPr without its automodification domain and that histone H4 cannot serve as an acceptor of pADPr. We individually tested histones H1, H2A, H2B, H3, and H2Av to determine whether any of these could serve as an acceptor of pADPr produced by the W-Cat isoform upon its interaction with H4. Only reactions with H1 resulted in significant accumulation of pADPr in the reaction mix (Fig. 4A, Panel 3; SI Appendix, Fig. S5). Because the W-Cat isoform cannot be automodified in the absence of the A domain, all pADPrs produced by the W-Cat isoform upon its activation by H4 must have been attached to the H1 histone (Fig. 4A, Panel 3; SI Appendix, Fig. S5). This result confirms that the W-Cat domains present in the W-Cat isoform are sufficient for H4-dependent PARP-1 activation and demonstrates that H4-mediated PARP-1 activation does not lead to poly(ADP ribosyl)ation of H2A, H2B, H3, H2Av, and H4 histones (Fig. 4A, Panel 4–7) but only to the automodification of the full-length PARP-1 and modification of H1 (Fig. 4B).
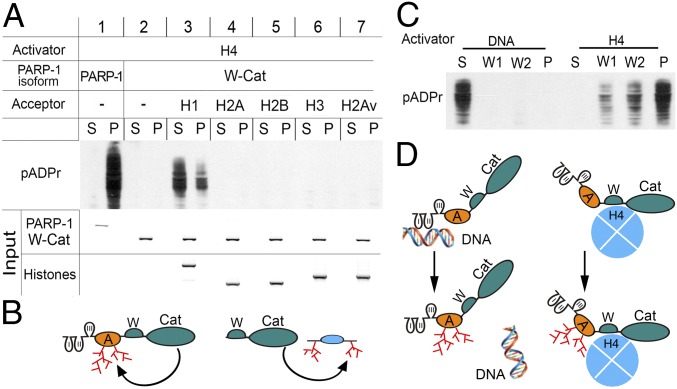
Fig. 4.
Regulation and targeting of enzymatic activity of PARP-1 by histones and DNA. (A–B) PARP-1 modifies either itself via the A domain (A) or the linker histone H1 in vitro. (A) Binding-activation assay: full-length PARP-1 and deletional isoform W-Cat were separately incubated with H4-bound sepharose beads, washed, and mixed with NAD and other histones (H1, H2A, H2B, H3, and H2Av) to induce pADPr production. The supernatant (S) was removed and the pellet (P) was washed. The absence of the A domain in the W-Cat isoform precludes automodification of PARP-1. Histones were added as potential substrates for the poly(ADP ribosyl)ation assay. Accumulation of pADPr was detected on Western blot using an anti-pADPr antibody (10H). Input: the Coomassie Brilliant Blue stained PAGE gels show the amounts of PARP-1, W-Cat, and histones loaded in each reaction. (B) Diagram illustrating the activity of full-length PARP-1 and PARP-1 deletional isoforms via the A domain and modification of H1. (C–D) Upon activation and automodification, PARP-1 loses interaction with DNA but remains bound to histone H4. (C) Interactive-activity assay of PARP-1 with sepharose beads coupled to either nicked DNA or H4. PARP-1 was incubated separately with either nicked DNA- or H4-coupled beads, washed, and mixed with NAD to trigger pADPr production. The solution was removed and the pellet was washed twice (W1, W2). The distribution of pADPr between fractions was measured after PAGE on a Western blot using an anti-pADPr antibody. (D) Diagram illustrating that automodified PARP-1 dissociates from DNA but remains bound to H4.
Unlike linker histone H1, which accumulated in the supernatant fraction upon poly(ADP ribosyl)ation (Fig. 4A, Panel 3), the PARP-1 protein remained predominantly bound to H4-coupled beads even after its automodification (Fig. 4A, Panel 1). This finding suggests that when PARP-1 is bound to and activated by H4, it remains active longer, producing a larger amount of pADPr than when it is bound to and activated by DNA. To test this inference, we compared the levels of pADPr production between PARP-1 activated by DNA and by H4 in the presence of NAD. When full-length PARP-1 was activated by H4-coupled beads, the automodified PARP-1 remained bound to H4 (Fig. 4C). This finding confirms that PARP-1 remains active and bound to H4 following PARP-1 activation. Conversely, when PARP-1 was bound to and activated by the DNA-coupled beads, the automodified PARP-1 was found predominantly in the supernatant and was detached from DNA (Fig. 4C). This assay suggests that, in a living cell, PARP-1 interaction with damaged DNA leads to a short-term hit and run activation of this protein as PARP-1 quickly detaches from DNA upon its activation.
The interaction of PARP-1 with histones and DNA, which is mediated by different domains, leads to different patterns of PARP-1 distribution through chromatin. Interaction of different PARP-1 domains with their respective substrates also results in different durations of PARP-1 activation. The function of PARP-1 in DNA repair is mediated by a short-term hit and run interaction with damaged DNA. To retain chromatin in a loosened state, prolonged binding with a histone substrate is required. A sustained interaction of PARP-1 with the core histones and sustained activity of PARP-1 may represent a mechanism responsible for maintaining open chromatin loci and an active state of transcription. PARP-1 does not require additional regulatory factors to help it bind to its substrates and to retain it there.
Discussion
PARP-1 localization and activation in chromatin is necessary for maintaining active chromatin in an open state (31, 32, 35). The poly(ADP ribosyl)ation of linker histone H1 plays a crucial role in the process (32). Our results suggest that PARP-1 domains cooperatively control its activation via DNA and histone H4 binding, which leads to pADPr accumulation. We found that DNA-binding ZI and ZII are necessary for DNA-dependent short-term hit and run activation of PARP-1, which triggers the DNA-repair pathway. The C-terminal catalytic domain of PARP-1 binds to histone H4, resulting in prolonged activation of this enzyme and sustained production of pADPr. Histone H4 binds to the PARP-1 C-terminal catalytic domain and activates PARP-1 independent of the DBD. A PARP-1 W-Cat construct is targeted to the promoter region of the hsp70 gene and can activate hsp70 transcription upon HS in the PARP-1 mutant background. Therefore, the transcription activation function of PARP-1 can be mediated independently from the DBD, although the consequent level of the transcript accumulation is considerably lower than that in the presence of full-length PARP-1. The data presented here are consistent with the previously reported finding that phosphorylation of H2Av results in exposure of key epitopes of the H4 histone, leading to PARP-1 activation (17). Therefore, it seems likely that the H4-mediated mechanism is deployed to enable PARP-1 transcriptional activation in steady-state conditions in the absence of DNA damage.
Our data suggest that the DBD of PARP-1 is not strictly required for histone H4-dependent PARP-1 activation. This domain is, however, strictly necessary for DNA-mediated PARP-1 activation. Even though both ZI and ZII of the DBD have high binding affinity to DNA, it has been shown that only ZI is absolutely necessary for PARP-1 activation induced by DNA damage (29). This finding raises an interesting issue concerning additional functions of the DBD in the absence of DNA damage. We found that YFP-tagged PARP-1 isoforms with ZI and ZII colocalized with DNA in chromatin in a nonspecific genome-wide manner. A ChIP assay showed that the ZI- and ZII-bearing isoform was absent from the hsp70 TSS region but enriched outside this TSS (Fig. 2D). In addition, despite possessing the histone-binding and Cat domains, the isoforms which lacked the DBD (ZIII-A-W-Cat and W-Cat) could not fully restore the transcription activation function in the hsp70 gene, suggesting that the DBD interaction with DNA is also required for full transcription activation (Fig. 2E). Similar to other DNA-binding transcription factors, such as the pioneer factor FoxA (41), the pattern of PARP-1 binding has both specific and nonspecific properties in chromatin. We propose that the DNA-binding domain assists with targeting PARP-1 to the TSS region by scanning chromatin for binding sites (Fig. 5). Our findings suggest that the DBD and the C-terminal catalytic domain (W-Cat) of PARP-1 represent a cooperative mechanism that determines where and how PARP-1 induces transcription.
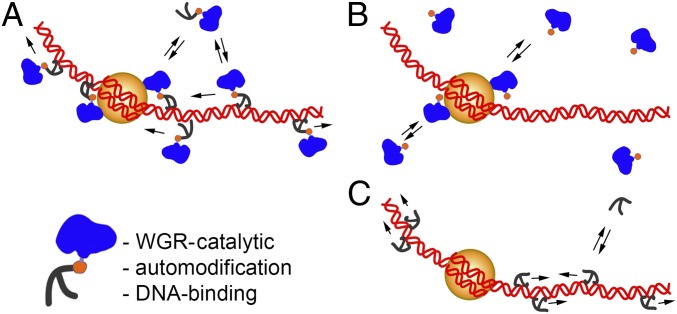
Fig. 5.
Model of PARP-1 dual binding to chromatin. (A) Full-length PARP-1 has two chromatin-binding regions: N-terminal ZI and ZII bind to DNA nonspecifically, allowing PARP-1 to scan chromatin by “walking” along DNA and the C-terminal domain which recognizes and binds to the H2Av-bearing nucleosome specifically. (B) The C-terminal domain recognizes specific epitopes of the H2Av-bearing nucleosome. (C) The ZI-ZII deletional isoform binds to DNA nonspecifically and does not interact with the nucleosome.
Experimental Procedures
Flies were cultured on standard cornmeal–molasses-agar media at 22 °C unless otherwise indicated. The fly stocks were generated by the standard genetic methods or obtained from the Bloomington Drosophila Stock Center and the Exelixis Collection at the Harvard Medical School. To make transgenic UAS::YFP constructs containing deletional isoforms of PARP-1, we generated fragments of PARP-1 cDNA corresponding to these deletions using PCR. The chromatin and PARP protein complexes were immunoprecipitated using an anti-GFP rabbit polyclonal antibody (Torrey Pines Biolabs). DNA from the elutes was measured by real-time (RT) PCR. Polytene chromosomes prepared from the salivary glands of wandering third-instar larvae were stained as described. RNA was reverse transcribed and RT PCR assays were performed using the StepOnePlus RT PCR System (Applied Biosystems). Detailed information is provided in SI Appendix, Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
Drs. Jeff Peterson and Sergei Nechaev contributed valuable comments on the paper. We also thank Elena Kotova and Mikael Garabedian for assistance with protein purification and interaction experiments, creating PARP-1 recombinant constructs and in vivo imaging. Research was supported by a grant from the National Science Foundation MCB-1616740 (to A.V.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901183116/-/DCSupplemental.
References
- 1.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 2.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 3.Blais A, Dynlacht BD. Constructing transcriptional regulatory networks. Genes Dev. 2005;19:1499–1511. doi: 10.1101/gad.1325605. [DOI] [PubMed] [Google Scholar]
- 4.Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 5.Benabdallah NS, Bickmore WA. Regulatory domains and their mechanisms. Cold Spring Harb Symp Quant Biol. 2015;80:45–51. doi: 10.1101/sqb.2015.80.027268. [DOI] [PubMed] [Google Scholar]
- 6.Wolffe AP. Transcriptional regulation in the context of chromatin structure. Essays Biochem. 2001;37:45–57. doi: 10.1042/bse0370045. [DOI] [PubMed] [Google Scholar]
- 7.Bustin M, Catez F, Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 8.Izzo A, Schneider R. The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics. Biochim Biophys Acta. 2016;1859:486–495. doi: 10.1016/j.bbagrm.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Flanagan TW, Brown DT. Molecular dynamics of histone H1. Biochim Biophys Acta. 2016;1859:468–475. doi: 10.1016/j.bbagrm.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 10.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 11.Luo X, Kraus WL. On PAR with PARP: Cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraus WL, Hottiger MO. PARP-1 and gene regulation: Progress and puzzles. Mol Aspects Med. 2013;34:1109–1123. doi: 10.1016/j.mam.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Thomas C, Tulin AV. Poly-ADP-ribose polymerase: Machinery for nuclear processes. Mol Aspects Med. 2013;34:1124–1137. doi: 10.1016/j.mam.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimoto M, et al. The HSF1-PARP13-PARP1 complex facilitates DNA repair and promotes mammary tumorigenesis. Nat Commun. 2017;8:1638. doi: 10.1038/s41467-017-01807-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinnola A, Naumova N, Shah M, Tulin AV. Nucleosomal core histones mediate dynamic regulation of poly(ADP-ribose) polymerase 1 protein binding to chromatin and induction of its enzymatic activity. J Biol Chem. 2007;282:32511–32519. doi: 10.1074/jbc.M705989200. [DOI] [PubMed] [Google Scholar]
- 16.Clark NJ, Kramer M, Muthurajan UM, Luger K. Alternative modes of binding of poly(ADP-ribose) polymerase 1 to free DNA and nucleosomes. J Biol Chem. 2012;287:32430–32439. doi: 10.1074/jbc.M112.397067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas CJ, et al. Kinase-mediated changes in nucleosome conformation trigger chromatin decondensation via poly(ADP-ribosyl)ation. Mol Cell. 2014;53:831–842. doi: 10.1016/j.molcel.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Messner S, et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38:6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Zamudio R, Ha HC. Histone ADP-ribosylation facilitates gene transcription by directly remodeling nucleosomes. Mol Cell Biol. 2012;32:2490–2502. doi: 10.1128/MCB.06667-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell. 2010;39:736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci USA. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krupitza G, Cerutti P. Poly(ADP-ribosylation) of histones in intact human keratinocytes. Biochemistry. 1989;28:4054–4060. doi: 10.1021/bi00435a063. [DOI] [PubMed] [Google Scholar]
- 23.Althaus FR, et al. Interactions of poly(ADP-ribose) with nuclear proteins. Biochimie. 1995;77:423–432. doi: 10.1016/0300-9084(96)88155-7. [DOI] [PubMed] [Google Scholar]
- 24.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kameshita I, Matsuda M, Nishikimi M, Ushiro H, Shizuta Y. Reconstitution and poly(ADP-ribosyl)ation of proteolytically fragmented poly(ADP-ribose) synthetase. J Biol Chem. 1986;261:3863–3868. [PubMed] [Google Scholar]
- 26.Kotova E, Jarnik M, Tulin AV. Uncoupling of the transactivation and transrepression functions of PARP1 protein. Proc Natl Acad Sci USA. 2010;107:6406–6411. doi: 10.1073/pnas.0914152107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buki KG, Bauer PI, Hakam A, Kun E. Identification of domains of poly(ADP-ribose) polymerase for protein binding and self-association. J Biol Chem. 1995;270:3370–3377. doi: 10.1074/jbc.270.7.3370. [DOI] [PubMed] [Google Scholar]
- 28.Kirsanov KI, et al. Minor grove binding ligands disrupt PARP-1 activation pathways. Oncotarget. 2014;5:428–437. doi: 10.18632/oncotarget.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langelier MF, Planck JL, Roy S, Pascal JM. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: Structural and functional insights into DNA-dependent PARP-1 activity. J Biol Chem. 2011;286:10690–10701. doi: 10.1074/jbc.M110.202507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336:728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 32.Kim MY, Mauro S, Gévry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Muthurajan UM, et al. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc Natl Acad Sci USA. 2014;111:12752–12757. doi: 10.1073/pnas.1405005111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wacker DA, et al. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol Cell Biol. 2007;27:7475–7485. doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotova E, et al. Drosophila histone H2A variant (H2Av) controls poly(ADP-ribose) polymerase 1 (PARP1) activation in chromatin. Proc Natl Acad Sci USA. 2011;108:6205–6210. doi: 10.1073/pnas.1019644108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji Y, Tulin AV. The roles of PARP1 in gene control and cell differentiation. Curr Opin Genet Dev. 2010;20:512–518. doi: 10.1016/j.gde.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miwa M, Hanai S, Poltronieri P, Uchida M, Uchida K. Functional analysis of poly(ADP-ribose) polymerase in Drosophila melanogaster. Mol Cell Biochem. 1999;193:103–107. [PubMed] [Google Scholar]
- 38.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 39.Kotova E, Jarnik M, Tulin AV. Poly (ADP-ribose) polymerase 1 is required for protein localization to Cajal body. PLoS Genet. 2009;5:e1000387. doi: 10.1371/journal.pgen.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 41.Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009;23:804–809. doi: 10.1101/gad.1775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.