Significance
NFAT:AP-1 cooperation is pivotal for the productive immune response, but when this response is exacerbated, it is implicated in autoimmune and inflammatory diseases. Current immunosuppressive drugs used in clinic, such as cyclosporin A, completely inhibit the NFAT activation and have strong side effects to patients by inhibiting other calcineurin substrates, highlighting the importance of identifying new immunomodulatory drugs. Here, we performed a high-throughput screen and identified Compound 10, a small drug-like molecule that inhibits the NFAT:AP-1:DNA interaction on the ARRE-2 element. Compound 10 does not affect all the NFAT:AP-1 composite sites, but it demonstrates that it is possible to target transcription factor protein complexes on DNA with small molecules, and encourages the study of such molecules for purposes of immunomodulation.
Keywords: NFAT, Fos, Jun, FRET assay, cyclosporin A
Abstract
The transcription factor nuclear factor of activated T cells (NFAT) has a key role in both T cell activation and tolerance and has emerged as an important target of immune modulation. NFAT directs the effector arm of the immune response in the presence of activator protein-1 (AP-1), and T cell anergy/exhaustion in the absence of AP-1. Envisioning a strategy for selective modulation of the immune response, we designed a FRET-based high-throughput screen to identify compounds that disrupt the NFAT:AP-1:DNA complex. We screened ∼202,000 small organic compounds and identified 337 candidate inhibitors. We focus here on one compound, N-(3-acetamidophenyl)-2-[5-(1H-benzimidazol-2-yl)pyridin-2-yl]sulfanylacetamide (Compound 10), which disrupts the NFAT:AP-1 interaction at the composite antigen-receptor response element-2 site without affecting the binding of NFAT or AP-1 alone to DNA. Compound 10 binds to DNA in a sequence-selective manner and inhibits the transcription of the Il2 gene and several other cyclosporin A-sensitive cytokine genes important for the effector immune response. This study provides proof-of-concept that small molecules can inhibit the assembly of specific DNA–protein complexes, and opens a potential new approach to treat human diseases where known transcription factors are deregulated.
The transcription factor NFAT (nuclear factor of activated T cells) is a well-known regulator of gene expression during T cell activation and differentiation (1–3). The NFAT family comprises five proteins, NFAT1–NFAT5; at least one NFAT family member is expressed in almost every cell type (3). NFAT is involved in the regulation of many pivotal cell functions, such as the cell cycle, apoptosis, and angiogenesis (4–7). All NFAT proteins share a conserved DNA-binding domain (DBD) that specifies binding to the DNA core sequence (A/T)GGAAA (1–3, 6). Four of the NFAT proteins, NFAT1–NFAT4 (also known as NFATc1–NFATc4), are regulated by Ca2+ and the Ca2+-dependent phosphatase calcineurin through a second conserved domain, the NFAT homology region, which is heavily phosphorylated in the inactive, cytoplasmic form of NFAT (8). Upon cell stimulation, Ca2+ influx activates calcineurin, which dephosphorylates NFAT and induces NFAT nuclear translocation (1, 2, 9). In the nucleus, NFAT regulates gene transcription, either alone or in collaboration with nuclear protein partners that are activated by other signaling pathways (1–3, 6).
The Ca2+-calcineurin-NFAT pathway has proved to be an important target of immune modulation. Primary examples are the immunosuppressive drugs cyclosporin A (CsA) and FK506, which inhibit NFAT activation by inhibiting the phosphatase activity of calcineurin, thus preventing all cellular functions mediated by either calcineurin or NFAT (1, 2, 9). CsA and FK506 have several toxic side effects, such as nephrotoxicity, which arises from their ability to inhibit calcineurin in cells outside the immune system (1, 2, 10). In earlier work, we argued that blocking the protein–protein interface between calcineurin and NFAT would be a more selective way of blocking the Ca2+-calcineurin-NFAT pathway, compared with blocking calcineurin activity directly. To test this hypothesis, we defined the interface between NFAT and calcineurin, showed that a peptide from the interface could block NFAT activity, and used peptide selection to generate an optimized high-affinity binding peptide (VIVIT) that was a potent blocker of the calcineurin–NFAT interaction and of NFAT dephosphorylation and NFAT-dependent cytokine gene induction in T cells (11–14). The peptide inhibitor showed a degree of selectivity, because it did not interfere with calcineurin–NF-κB signaling in T cells (12). We also used a fluorescence polarization screen to identify small organic molecules (termed INCA compounds) that inhibited binding of the VIVIT peptide to recombinant calcineurin and blocked calcineurin–NFAT signaling in cells (15). However, we and others later demonstrated that the calcineurin–VIVIT interface was used not only by NFAT, but also by many other calcineurin substrates (14, 16, 17). Cell-permeant linear (18, 19) and cyclic (20) derivatives of the VIVIT peptide have been identified.
In this study, we explore a strategy for selective modulation of the immune response. The strategy is based on our data showing that NFAT has a key role in T cell activation as well as T cell tolerance. We have demonstrated that NFAT induces different programs of gene expression, depending on what signaling pathways and transcription factors are active at the same time (21–23). In T cells, a major NFAT partner is activator protein-1 (AP-1), formed by dimers of Fos and Jun family proteins (24, 25). T cell receptor (TCR) stimulation induces prolonged Ca2+ influx so that NFAT remains in the nucleus for long periods of time (26, 27). In contrast, Fos and Jun are transcriptionally induced by TCR stimulation and by costimulatory pathways that activate PKC-θ, but this activation is transient (25, 28, 29). Thus, in the early phase of T cell activation, NFAT forms complexes with AP-1 proteins and is involved in the productive immune response, regulating the expression of cytokines, including IL-2, IL-4, IL-5, IL-13, IFN-γ, and GM-CSF (23–25, 27, 30). In contrast, under conditions of prolonged antigen exposure in the absence of costimulation, AP-1 activation dies away (31, 32) and NFAT drives a different transcriptional program of T cell anergy and exhaustion, characterized by the expression of inhibitory receptors, such as PD-1, LAG-3, TIM-3, and CTLA-4 (22, 33). The genes regulated by NFAT in these two different programs were recently elucidated by our group through genome-wide analyses (22), using T cells expressing a mutant NFAT protein (NFAT-RIT) that does not interact with AP-1 (34), and hence recapitulates the program of anergy/exhaustion induced by NFAT in the absence of costimulation (22). Together, these results indicate that disruption of the NFAT:AP-1 interaction might redirect the transcriptional profile of T cells from a productive to a tolerogenic immune response, whereas stabilizing the NFAT:AP-1 interaction would be expected to have the reverse effect of promoting T cell activation.
Our goal in this study was to identify small organic molecules that disrupt the NFAT:AP-1:DNA complex. We designed a FRET-based high-throughput screen using the best-characterized example of cooperative binding of NFAT and AP-1 to DNA, which occurs at the composite antigen-receptor response element-2 (ARRE-2) site of the Il2 promoter (35). We screened a library of ∼202,000 small-molecule compounds using this FRET assay and identified 960 compounds as candidate inhibitors. Here we focus on follow-up studies with one of these compounds, N-(3-acetamidophenyl)-2-[5-(1H-benzimidazol-2-yl)pyridin-2-yl]sulfanylacetamide (PubChem ID: 1432799), which we refer to as “Compound 10.” We show that Compound 10 disrupts the NFAT:AP-1 interaction at several but not all composite NFAT:AP-1 sites, without inhibiting the binding of NFAT or Fos-Jun dimers to DNA; and that it exerts these inhibitory effects by binding in a sequence-selective manner to DNA. Transcriptional profiling showed that Compound 10 inhibited the production of IL-2 and several other CsA-sensitive NFAT target cytokine genes, including Il4, Il5, Il13, Il17a, Il17f, and Il21. Overall, our study provides proof-of-concept that transcription factor protein complexes on DNA can be targeted by small organic molecules, and provides an approach for treating the many classes of human diseases that are triggered by deregulated expression or activation of specific transcription factors.
Results
A FRET Assay to Quantify Assembly of the Cooperative NFAT:AP-1 Complex on DNA.
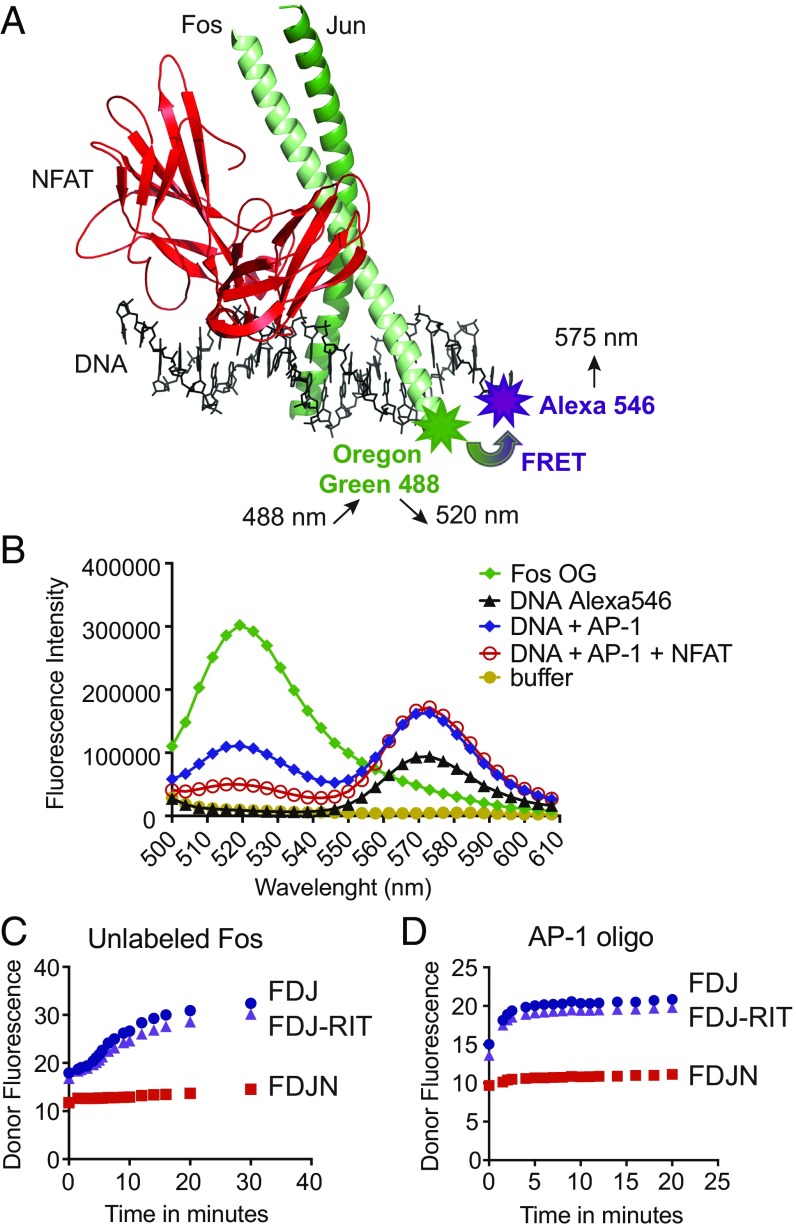
To identify compounds that disrupt the cooperative complex of NFAT and AP-1 on DNA, we devised a FRET assay (Fig. 1 A and B) using an oligonucleotide spanning the distal ARRE-2 from the murine Il2 promoter and three recombinant proteins expressed in Escherichia coli: the DBD of NFAT1 and the basic leucine zipper (bZIP) regions of c-Fos and c-Jun (SI Appendix, Fig. S1A). The ARRE-2 site contains a consensus binding site for NFAT and a weak binding site for AP-1 (24, 36). A single cysteine residue introduced near the N terminus of the c-Fos bZIP region was labeled with the donor fluorophore, Oregon green (OG) maleimide-488; the acceptor fluorophore was Alexa-546, introduced at the 3′ end of the sense ARRE-2 DNA strand (Fig. 1A). Protein–DNA mixtures were excited at 488 nm, a wavelength that excites the donor Fos-OG fluorophore with minimal background signal from direct excitation of the Alexa-546 acceptor; and fluorescence emission was scanned from 500 to 610 nm (Fig. 1B). There was a clear decrease in donor Fos-OG fluorescence (emission peak at 520 nm) and an increase in acceptor Alexa-546 fluorescence (emission peak at 575 nm) when DNA and AP-1 were incubated together, and a further decrease in donor Fos-OG fluorescence when NFAT was added (Fig. 1B). The drop in donor fluorescence was not observed when the proteins were incubated with unlabeled ARRE-2 DNA, indicating that it reflected bona fide energy transfer between donor and acceptor fluorophores.
Fig. 1.
Design of the FRET assay. (A) Structure of the NFAT:AP-1 complex bound to the ARRE-2 element of the murine IL-2 promoter (PDB ID code 1A02). The DBD of NFAT (red) is in contact with the AP-1 dimer of Fos (light green) and Jun (dark green). The NFAT site in the ARRE-2 oligonucleotide is a consensus binding site for NFAT, whereas the AP-1 site is a nonconsensus site that differs appreciably from the consensus AP-1 site TGAC/GTCA. Modified from figure 2 in ref. 35. Fos was labeled with the donor fluorophore Oregon Green 488 maleimide. The 3′ end of the sense strand of the ARRE-2 DNA oligonucleotide was labeled with the acceptor fluorophore Alexa-546. (B) Fluorescence emission scan of the donor Fos-OG alone (emission peak at 520 nm, green curve), the acceptor DNA-Alexa 546 alone (emission peak at 575 nm, black curve), and the indicated protein–DNA complexes (blue and red curves). The assay was excited at 488 nm; concentrations used were 20 nM Alexa-546–labeled ARRE-2–DNA, 20 nM Fos-OG, 20 nM Jun, and 40 nM NFAT1 DBD. (C and D) Dissociation kinetics of the loosely bound Fos:Jun:DNA (FJD) and Fos:Jun:DNA:NFAT-RIT (FJD-RIT) complexes versus the cooperatively bound Fos:Jun:DNA:NFAT (FJDN) complex upon the addition of 200 nM unlabeled Fos (C) or 20 nM consensus AP-1 oligonucleotide (D). The initial complexes had been assembled from 20 nM Fos-OG, 20 nM ARRE-2–Alexa546 DNA, and 20 nM Jun, with the inclusion of 40 nM wild-type NFAT DBD or 40 nM NFAT-RIT DBD where indicated. Donor fluorescence is plotted. The assay was read using a Synergy 2 (Biotek) plate reader with 485/20-nm and 528/20-nm filters for excitation and emission, respectively.
The quaternary NFAT:Fos:Jun:ARRE-2 DNA complex is considerably more stable than either the ternary Fos:Jun:ARRE-2 DNA complex or the mutant quaternary NFAT-RIT:Fos:Jun:ARRE-2 DNA complex, where NFAT-RIT is a mutant NFAT1 that binds DNA comparably to wild-type NFAT1 but is unable to interact with AP-1 (34). This is shown in the context of the FRET assay by the prompt increase in donor Fos-OG fluorescence upon addition of either unlabeled Fos or unlabeled AP-1 oligonucleotide, which leads to dissociation of the preformed Fos:Jun:ARRE-2 and NFAT-RIT:Fos:Jun:ARRE-2 complexes, but spares the cooperative NFAT:Fos:Jun:ARRE-2 complex (Fig. 1 C and D).
In the final automated FRET assay format, we mixed together Alexa-546–labeled ARRE-2 DNA with peptides comprising the Jun bZIP region and the OG-labeled Fos bZIP region, then added wild-type NFAT DBD or NFAT-RIT DBD. After incubation for 2 h at room temperature, we plotted the ratio between the raw fluorescence of the acceptor and donor fluorophores. As expected, addition of wild-type NFAT DBD, which supports formation of the quaternary complex, increased the acceptor/donor FRET ratio relative to no addition, by increasing the acceptor fluorescence while decreasing the donor fluorescence, whereas addition of NFAT-RIT DBD did not (SI Appendix, Fig. S1B). This method of evaluating the FRET signal gave very stable results, and the plates could be stored and read without introducing any variations for a period of 5 h to overnight. The Z′-factor of the assay performed in 384-well plates was 0.73 (SI Appendix, Fig. S1C). In conclusion, the FRET assay that we developed efficiently quantified the formation of the stable NFAT:Fos:Jun:ARRE-2 DNA complex.
A High-Throughput Screen to Identify Inhibitors of the NFAT:AP-1:DNA Complex.
The FRET assay described in the previous section was used to screen more than 202,000 compounds at the University of California, Los Angeles, Molecular Screening Shared Resource facility (Materials and Methods and SI Appendix, Fig. S1D). All compounds were tested at a final nominal concentration of 10 μM. Compounds that interfered with the formation or stability of the NFAT:Fos:Jun:DNA complex were expected to decrease the acceptor fluorescence and increase the donor fluorescence, thus decreasing the acceptor/donor FRET ratio. The assay was performed in ∼650 plates; each plate included 32 wells of positive controls with NFAT (columns 1 and 2), and 32 negative control wells without NFAT (columns 23 and 24) (SI Appendix, Fig. S1E). Collaborative Drug Discovery software was used to calculate the Z-factor for each plate using raw data obtained at both acceptor and donor wavelengths, without removing outliers; then plate averages, plate SDs, and individual-compound z-scores were recalculated after removing outliers (Materials and Methods).
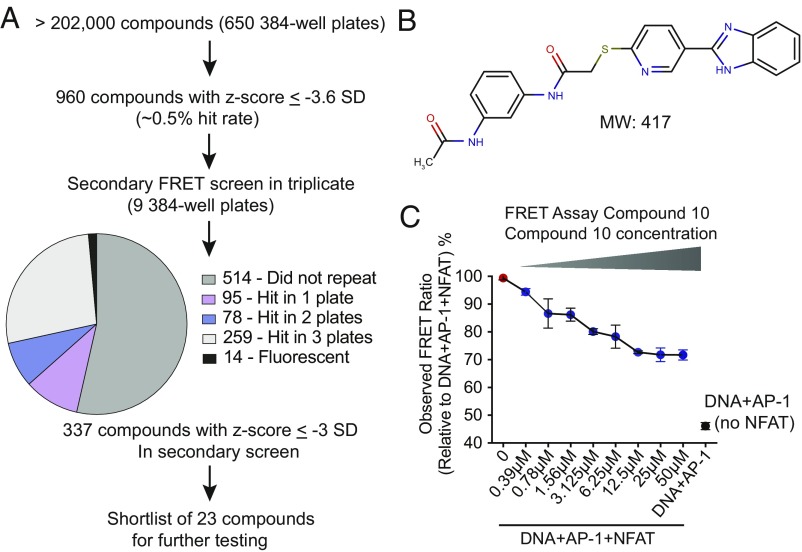
At the conclusion of the screen, we identified 960 compounds with a z-score ≤ −3.6 SD (hit rate 0.5%). These hits were retested in triplicate in a secondary screen, using the same FRET assay and workflow. The results of the secondary screen are summarized in Fig. 2A. Of the 960 retested compounds, 337 passed the secondary screen; we excluded 14 compounds that interfered with the FRET assay because of their high intrinsic fluorescence (Materials and Methods), and three known DNA intercalators/binders (aurintricarboxylic acid, mitoxantrone, and pyridostatin). From the list of 337 compounds, we selected 23 high-scoring compounds and purchased stocks for more detailed testing. Compounds that showed a dose-dependent inhibition response in the FRET assay were tested in an EMSA assay, and then further evaluated for their capacity to inhibit cytokine production. The remainder of this study is focused on a single compound, referred to here as Compound 10 (for structure see Fig. 2B), which was the only compound that inhibited both protein–DNA complex assembly in the EMSA and the production of IL-2 by T cells. Compound 10 displayed an IC50 in the FRET assay of ∼2 μM (Fig. 2C).
Fig. 2.
Results of the high-throughput FRET assay, selection and structure of Compound 10. (A) Summary of the high-throughput NFAT-AP1 inhibitor screen and flowchart of compound selection. (B) Structure of Compound 10 (PubChem ID: 1432799). (C) Concentration-dependent inhibition of the FRET signal by Compound 10. The results are plotted as observed FRET ratio (acceptor fluorescence/donor fluorescence) relative to the FRET ratio of wells containing the complete assay mix without Compound 10. Red symbol, control wells containing the complete assay mix (10 nM ARRE-2–Alexa-546, 10 nM Fos-OG, 10 nM Jun, and 20 nM NFAT); black symbol, control wells with NFAT omitted; blue symbols, wells with complete assay mix and the indicated concentrations of Compound 10. IC50 = ∼2 μM.
Compound 10 Inhibits IL-2 Production More Effectively than TNF Production.
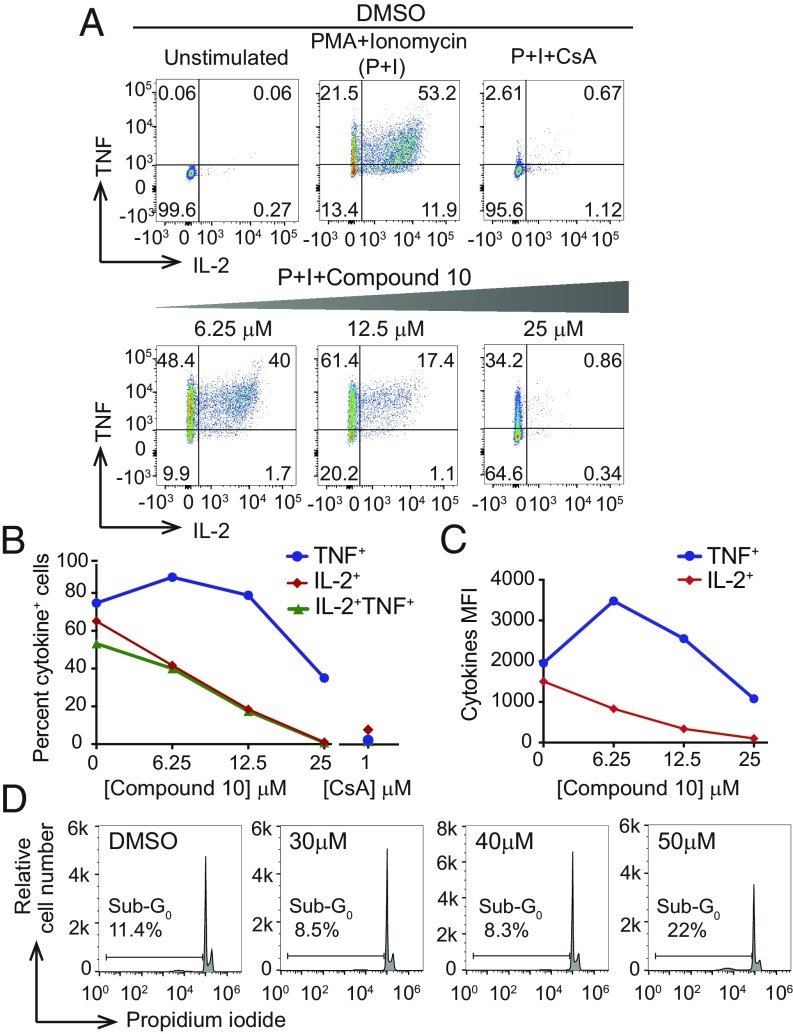
To study Compound 10 further, we preincubated freshly isolated primary murine CD4+ T cells for 1 h with different concentrations of Compound 10 (6.25, 12.5, and 25 μM) or with DMSO vehicle, and then stimulated the cells with PMA (10 nM) and ionomycin (500 nM) for 5 h. The cells were fixed, permeabilized, and stained for the cytokines IL-2 and TNF (Fig. 3); production of both these cytokines is dependent on activation of the calcineurin/NFAT pathway as judged by the nearly complete loss of IL-2 and TNF production in cells pretreated with 1 μM of the calcineurin inhibitor CsA (Fig. 3A). Under these conditions, concentrations of up to 40 μM of Compound 10 were not toxic to the freshly isolated CD4+ T cells (Fig. 3D).
Fig. 3.
Compound 10 inhibits production of the cytokines IL-2 and TNF by total CD4+ murine T cells under conditions where it is not toxic to the cells. Primary murine CD4+ T cells were incubated in the absence (DMSO control) or presence of the indicated concentrations of Compound 10 for 1 h, then either left unstimulated or stimulated with 10 nM PMA and 500 nM ionomycin for an additional 4 h. Where indicated, 1 μM of CsA was added 15 min before stimulation. (A–C) After stimulation, the cells were fixed, washed, permeabilized, and assessed for cytokine production by intracellular staining and flow cytometry. (B) Percentage of cells in A expressing IL-2, TNF, or both cytokines. (C) Mean fluorescent intensities (MFI) for the individual cytokines in cells scored as positive in A. (D) Toxicity was assessed by propidium iodide staining and flow cytometry. Cells were treated with Compound 10 as above, then stimulated with PMA and ionomycin for 4 h. The sub-G0 population is indicated in each panel. Under these conditions, cells treated with 30 or 40 μM of Compound 10 showed no increase in the sub-G0 population, whereas cells treated with 50 μM Compound 10 showed a twofold increase compared with untreated cells. The data are representative of at least two independent experiments.
Notably, treatment with 6.25 μM or 12.5 μM Compound 10 resulted in a dose-dependent decrease of the frequency of freshly isolated primary murine CD4+ T cells that produced IL-2, but no change or even a slight increase in TNF-producing cells. We observed a reproducible increase in the mean fluorescence intensity for TNF with 6.25 μM Compound 10, raising the possibility that limiting amounts of nuclear NFAT are displaced from some NFAT:AP-1 sites to other sites, like the κ3 site (Fig. 3 A–C). While both early and late IL-2 production are dependent on NFAT:AP-1 cooperation (21, 22), the early phase of TNF production involves NFAT dimer formation at the κ3 element of the TNF promoter (2, 37, 38). We interpret the data of Fig. 3 to indicate that Compound 10 has a major inhibitory effect on early IL-2 production by CD4+ T cells, with less effect on early TNF production. This in turn suggested that Compound 10 at low concentrations inhibits formation of the quaternary NFAT:Fos:Jun:DNA complex on the IL-2 promoter ARRE-2 site (as already shown in the FRET assay), but has little or no effect on formation of the NFAT dimer on the TNF promoter κ3 site.
In addition, we tested the ability of Compound 10 to inhibit IL-2 and TNF production by murine and human Th1 cells (SI Appendix, Fig. S2) under conditions where it was not toxic to the cells. All mouse Th1 cells produced IL-2 and TNF, and the production of both cytokines was blocked by Compound 10 at concentrations above 10 μM (SI Appendix, Fig. S2, Upper). Human Th1 cells required higher concentrations of Compound 10 than were required for mouse Th1 cells to achieve an equivalent inhibition of IL-2 production (SI Appendix, Fig. S2, Lower).
Compound 10 Inhibits Formation of the Quaternary NFAT:Fos:Jun:DNA Complex on Some but Not All Composite NFAT:AP-1 Sites.
Compound 10 might, in principle, inhibit IL-2 production in several ways, starting with inhibiting NFAT binding to the ARRE-2 site. It might also disrupt AP-1 binding to DNA; block formation of the quaternary complex of NFAT, Fos, and Jun on DNA; or bind directly to or intercalate into DNA. To answer this point, we performed EMSA.
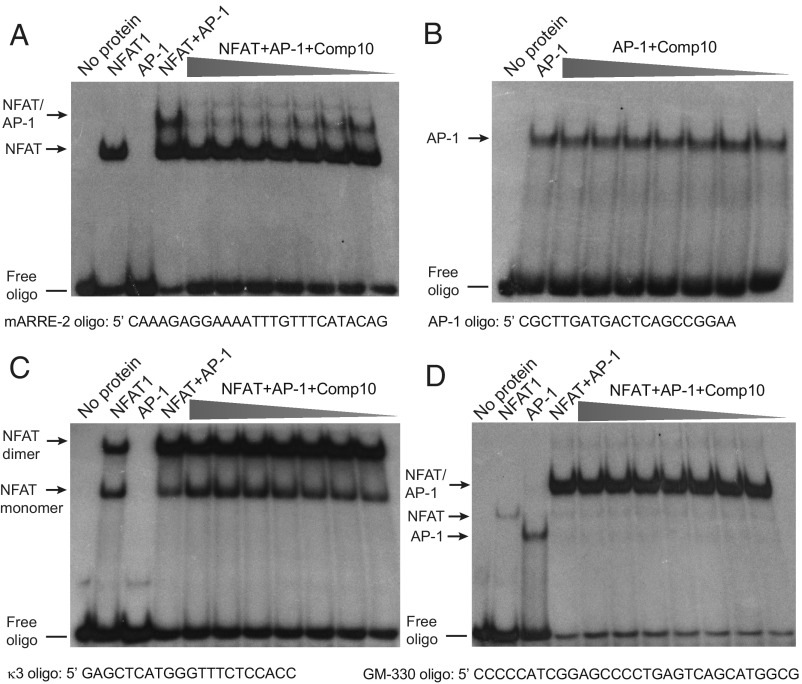
At concentrations ranging from 3.9 μM to nominally 250 μM, Compound 10 had no effect on binding of NFAT wild-type (Fig. 4A) or NFAT-RIT DBD to the murine ARRE-2 site (SI Appendix, Fig. S3A), although it inhibited assembly of the composite NFAT:AP-1:DNA complex in a dose-dependent way with an IC50 of ∼5 μM (Fig. 4A). Consistent with our previous finding that Compound 10 was more effective at inhibiting IL-2 production by mouse compared with human Th1 cells (SI Appendix, Fig. S2), Compound 10 was less effective at inhibiting cooperative NFAT:AP-1 binding to the human ARRE-2 site, which differs slightly in sequence, than to the mouse site (compare Fig. 4A and SI Appendix, Fig. S3B).
Fig. 4.
Compound 10 inhibits formation of the quaternary NFAT:Fos:Jun:DNA complex on the ARRE-2 site, but not on the GM-330 element, without inhibiting the direct binding of NFAT or AP-1 to DNA. 500 nM of Fos and 500 nM of Jun proteins were incubated with 20,000 CPMs of the indicated [γ32P] ATP-labeled oligos in the absence or in the presence of Compound 10 (concentrations: 250, 125, 62.5, 31.3, 15.6, 7.8, and 3.9 μM, respectively) for 10 min, then 10 nM of wild-type NFAT DBD or NFAT-RIT DBD was added for a further 20 min. DNA–protein complexes were analyzed by EMSA and are indicated by the arrows. Results are representative of at least two independent experiments. (A) Compound 10 inhibits formation of the quaternary NFAT:Fos:Jun:DNA complex on the murine ARRE-2 element. (B) Compound 10 does not inhibit binding of AP-1 (Fos:Jun heterodimers) to the consensus AP-1 binding site. (C) Compound 10 does not inhibit binding of NFAT monomers or dimers to the TNF κ3 element. (D) Compound 10 does not inhibit formation of the quaternary NFAT:Fos:Jun:DNA complex on the GM-330 GM-CSF enhancer element.
Compound 10 did not inhibit Fos:Jun DNA binding in an EMSA that used a consensus AP-1 site oligonucleotide and the bZIP regions of Fos and Jun from the high-throughput screen (Fig. 4B), demonstrating that it does not block AP-1 binding to DNA. Besides complexing with AP-1, NFAT proteins can also form complexes with other protein partners and, in some sites, NFAT can bind as dimers, such as to the κ3 element of TNF promoter (37). Compound 10 did not block the binding of NFAT monomers or dimers to the κ3 element of the TNF promoter at concentrations up to 15.6 μM (Fig. 4C), consistent with its inability to block TNF protein production by freshly isolated CD4+ T cells at these concentrations (Fig. 3) and confirming that it did not directly block NFAT DNA binding.
Finally, we tested the well-established NFAT:AP-1 composite site from the −330 GM-CSF enhancer (30) (Fig. 4D). We found that increasing concentrations of Compound 10 did not inhibit the formation of the composite NFAT:Fos:Jun:DNA complex on the GM-CSF enhancer element as it did on the ARRE-2 oligonucleotide (compare Fig. 4 A and D).
Together, these data suggested that Compound 10 did not act by inhibiting the protein–protein interactions between NFAT and AP-1 proteins bound to DNA, because these interactions should not differ substantially between the two composite sites. Rather, the ability of Compound 10 to inhibit formation of the cooperative NFAT:Fos:Jun:DNA complex was sensitive to the sequence of the composite NFAT:AP-1 DNA oligonucleotide used in the EMSA assay. To test this hypothesis, we devised a fluorescence-based assay to measure the direct binding of Compound 10 to DNA.
Compound 10 Binds in a Sequence-Selective Manner Directly to DNA.
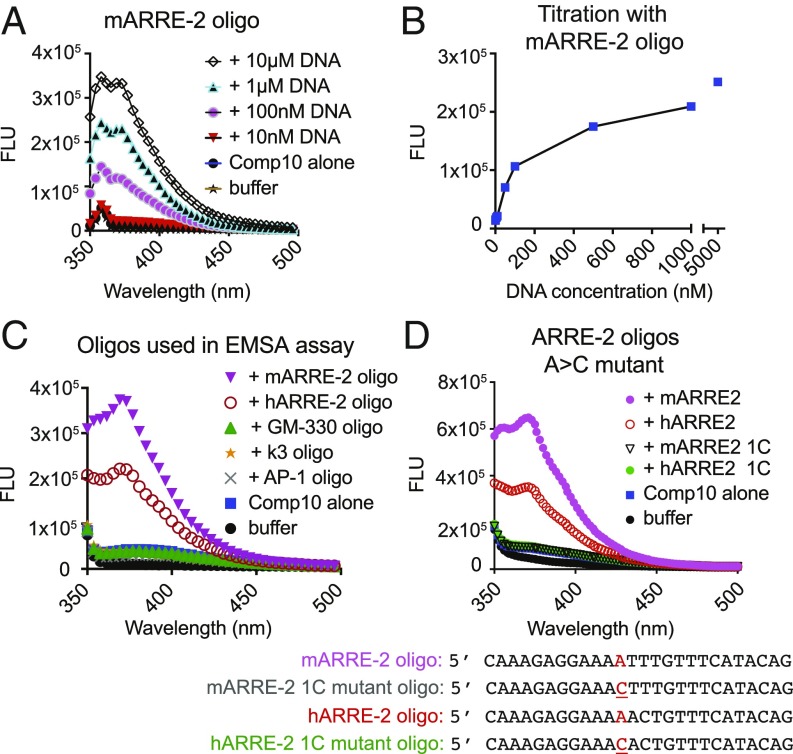
Compound 10 was weakly fluorescent when excited at 310 or 320 nm, with an emission peak ∼380 nm (Fig. 5A). Upon incubation with increasing concentrations of ARRE-2 DNA (range 10 nM to 5 μM), the fluorescence of Compound 10 increased in a dose-dependent manner (Fig. 5 A and B) and the peak emission was blue-shifted, indicating that the compound binds directly to DNA. A plot of fluorescence emission against increasing concentration of the murine ARRE-2 oligonucleotide is shown in Fig. 5B. Consistent with the EMSA data (Fig. 4 and SI Appendix, Fig. S3B), the murine ARRE-2 oligonucleotide at 500 nM caused a greater increase in the fluorescence of Compound 10 than the human ARRE-2 oligonucleotide at the same concentration; and there was no increase in fluorescence when Compound 10 was incubated with oligonucleotides containing the consensus AP-1 binding site, the κ3 element from the TNF promoter, or the GM-330 element of the GM-CSF enhancer (Fig. 5C). Together, these results indicate that Compound 10 binds in a sequence-selective manner to DNA rather than just intercalating into DNA in a nonsequence-specific manner.
Fig. 5.
Compound 10 binds to DNA. Fluorescence emission of Compound 10 (50 nM) alone or in the presence of the indicated oligonucleotides. The emission peak of Compound 10 bound to DNA is at ∼370 nm. Excitation wavelength, 310 nm or 320 nm. FLU, fluorescence units. Results are representative of at least two independent experiments. (A) Compound 10 binds to the murine ARRE-2 oligonucleotide in a concentration-dependent manner. Excitation was at 320 nm for this experiment, and the sharp early peak at 360 nm in these scans is the expected Raman peak due to scattering from water. In the remaining figure panels, excitation was at 310 nm, and therefore the Raman peak was observed just below 350 nm. (B) Titration of Compound 10 (50 nM) with increasing concentrations of the murine ARRE-2 oligo. DNA concentrations: 0, 10, 50, 100, 500 nM; 1 and 5 μM, respectively. Fluorescence emission was read at 377 nm. (C) Compound 10 binds to the murine and human ARRE-2 oligonucleotides, but not to the AP-1 consensus, TNF κ3 element and GM-330 site from GM-CSF enhancer, used for EMSA assays. All oligonucleotides were present at 500 nM. (D) A single A > C substitution that interrupts a stretch of adenines and thymines abrogates binding of Compound 10 to the mouse and human ARRE-2 oligonucleotides (500 nM).
To investigate these sequence preferences, we first measured the binding of Compound 10 to variants of the core sequence of the murine ARRE-2 oligonucleotide (AGGAAAATTTGTTTCA) in which the consensus NFAT site (GGAAAA) was eliminated by a GG > CC mutation. There was a clear decrease in Compound 10 binding as judged by increase in fluorescence intensity but the binding was not abrogated (SI Appendix, Fig. S4A). Moreover, substituting the core NFAT-binding sequence GGAAA into the weak NFAT site in the GM-330 oligonucleotide (SI Appendix, Fig. S4B), or changing the weak nonconsensus AP-1 site (TGTTTCA) in the mARRE-2 oligo to a consensus site (TGACTCA) (SI Appendix, Fig. S4C, mutant 1) did not alter Compound 10 binding appreciably. However, changing the internal sequence of the mARRE2 site (GGAAAATTT to GGAATTGAT) abrogated Compound 10 binding (SI Appendix, Fig. S4C, mutant 2). A molecule with the dimensions of Compound 10, binding as a monomer, could make specific contacts with only a few nucleotides, implying that its binding sites in DNA would be relatively common. Incubation with murine genomic DNA led to a strong dose-dependent change in the fluorescence of Compound 10 (SI Appendix, Fig. S4D); comparison with binding of Compound 10 to ARRE-2 sets an upper limit of about one binding site of affinity comparable to ARRE-2 per 100 bp in genomic DNA. (A large number of potential binding sites would be consistent with the limited effects on basal gene expression described below, if the bulk of the sites do not coincide with functionally critical DNA sequences, or if Compound 10 is readily displaced by most transcription factors at their physiological concentrations.) Finally, we made a single A > C substitution in both the human and murine ARRE-2 oligonucleotides, which in each case interrupted a short AT-rich stretch in the core NFAT:AP-1 composite site; Compound 10 binding was abrogated in each case (Fig. 5D). A table summarizing the effects of the different sequence substitutions on Compound 10 binding is presented in SI Appendix, Fig. S4E.
Compound 10 Alters the Gene Expression Pattern of Stimulated T Cells and Inhibits the Expression of Many NFAT-Dependent Genes.
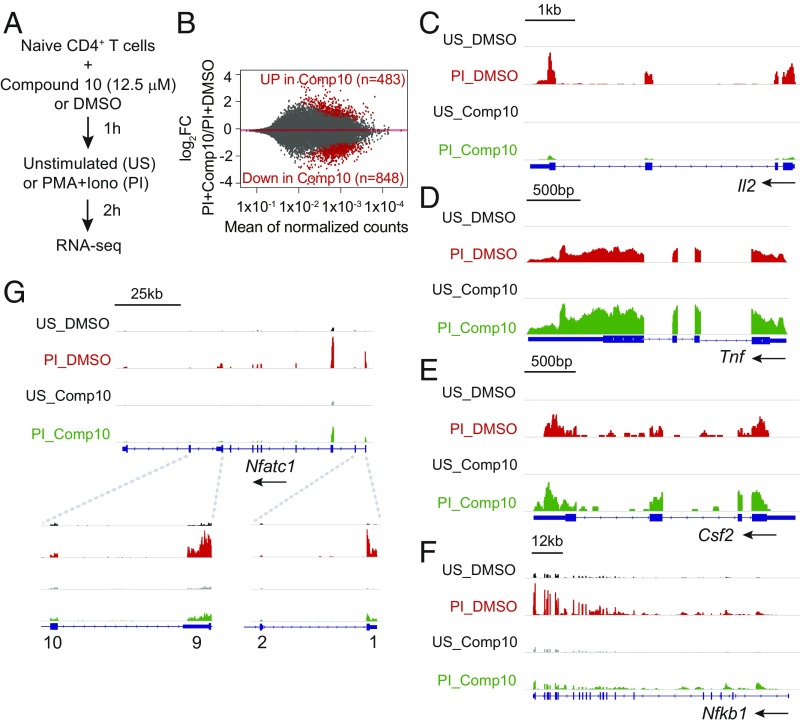
To evaluate the effects of Compound 10 on gene transcription by activated T cells, we performed RNA sequencing (RNA-seq) analysis with total freshly isolated CD4+ T cells, purified (CD62LhighCD69low) naïve CD4+ T cells, or Th1 cells prepared from total CD4+ T cells by prior activation with anti-CD3/anti-CD28 under Th1 polarizing conditions (39). The cells were preincubated for 1 h with 12.5 μM Compound 10 or DMSO (vehicle control), and then left unstimulated or stimulated with PMA and ionomycin (P+I) for a short time (2 h) (Fig. 6A). Analysis of the RNA-seq data for naïve CD4+ T cells showed that out of a total of 1,603 genes induced by PMA + ionomycin [log2 fold-change (log2FC) > 0.4, Padj ≤ 0.05], Compound 10 suppressed the induction of 848 genes (52.9%) (Fig. 6B and SI Appendix, Table S1). Many but not all of these activation-induced genes were known NFAT target genes: they included Il2 and Nfatc1 (encoding NFAT2) short isoform (40) (Fig. 6 C and G), as well as Cd160, Erg1, and Xcl1 (22) (Dataset S1). Nfkb1 (encoding NF-κB p50) is shown in Fig. 6F as an example of a gene not reported to be regulated by NFAT whose up-regulation is sensitive to Compound 10. In agreement with its effect on TNF protein expression (Fig. 3A), Compound 10 also appeared to increase Tnf mRNA slightly relative to the levels observed in DMSO-treated control cells at these early times (Fig. 6D). Consistent with its lack of effect in the GM-330 EMSA assay (Fig. 4D), Compound 10 did not inhibit Csf2 (encoding GM-CSF) mRNA expression (Fig. 6E). Compound 10 also decreased the basal expression of 120 genes in unstimulated T cells (Dataset S1).
Fig. 6.
Transcription profile regulated by Compound 10 in naïve CD4+ T cells. (A) Schematic representation of the RNA-seq assay. Sorted naïve CD4+ T cells were preincubated with Compound 10 or DMSO for 1 h, then left unstimulated (US) or stimulated with PMA (10 nM) and ionomycin (500 nM) (PI) for 2 h. (B) MA plots indicating the number of genes up-regulated (483) or down-regulated (848) by Compound 10 in stimulated naïve CD4+ T cells (PI+Compound 10) versus DMSO control (PI+DMSO). The y axis indicates the log2FC in expression of individual mRNAs in the PI+Compound 10 condition compared with the PI+DMSO condition. Genes scored as significantly up-regulated or down-regulated, with FDR threshold = 0.05, are represented by red symbols. (C–G) Genome browser views of RNA-seq signal at the Il2, Tnf, Csf2, Nfkb1, and Nfatc1 loci in unstimulated (US) and stimulated (PI) conditions, in the absence (DMSO) or in the presence (Comp10) of Compound 10, as indicated. Blue boxes correspond to exons, and arrows indicate the direction of transcription.
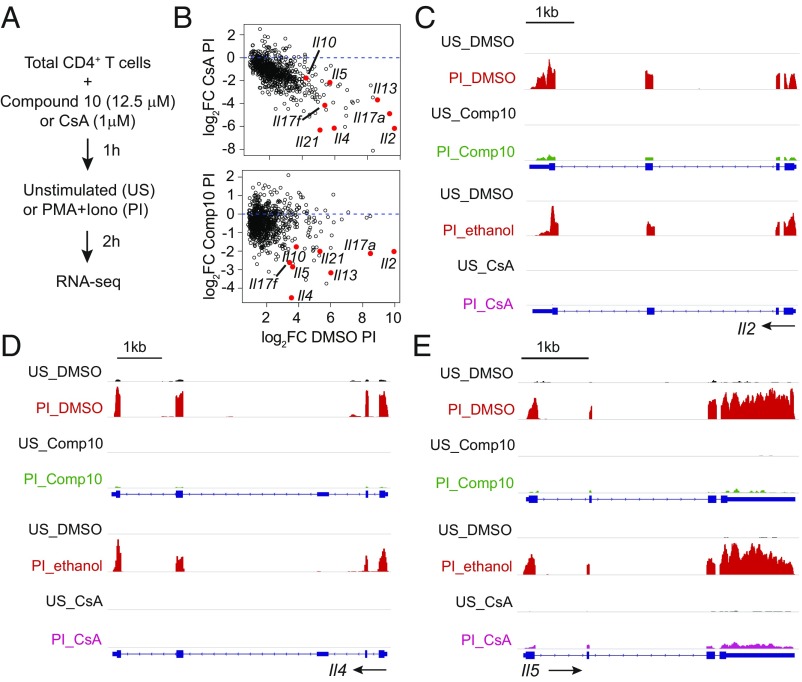
To assess the effect of Compound 10 on genes that were induced by the calcium-calcineurin-NFAT pathway, we performed RNA sequencing on total CD4+ T cells treated in separate experiments with Compound 10 (Dataset S2) or with the calcineurin inhibitor CsA (Dataset S3). There was an overlap of 801 genes induced by PMA and ionomycin in the two experiments; of these, CsA blocked 400 genes with a false-discovery rate (FDR) < 0.05, and Compound 10 blocked 136 genes. Eighty-four genes in the latter subset were inhibited by CsA as well as by Compound 10 (Fig. 7 and SI Appendix, Fig. S5 and Table S2). Notable among these were many cytokine genes, including Il2, Il4, Il5, Il10, Il13, Il17a, Il17f, and Il21, which are well-established targets of the calcium-calcineurin-NFAT pathway.
Fig. 7.
Comparing the effects of Compound 10 and CsA on T cell transcription. (A) Schematic representation of the RNA-seq assay. Total CD4+ T cells were preincubated with Compound 10 (12.5 μM) or DMSO for 1 h, or, in an independent experiment, with ethanol or CsA (1 μM) for 15 min, then left unstimulated (US) or stimulated with PMA (10 nM) and ionomycin (500 nM) (PI) for 2 h. (B) MA plots showing the effect of either CsA or Compound 10 on the genes that were induced by PMA and Ionomycin in both experiments. (C–E) Genome browser views of RNA-seq signals at the Il2, Il4, and Il5 loci in unstimulated (US) and stimulated (PI) conditions. The three genes were up-regulated by PMA+ionomycin in the vehicle control samples (DMSO or ethanol, red), and their up-regulation was inhibited in the presence of Compound 10 (green) or CsA (pink). Blue boxes correspond to exons, and arrows indicate the direction of transcription.
In conclusion, our data with Compound 10 provide proof-of-concept that it is possible to target the assembly of transcriptional complexes that regulate gene expression. Compound 10 disrupts the formation of the NFAT:AP-1:DNA complex at some but not all composite NFAT:AP-1 sites, with an effect dependent on the local DNA sequence. Predominant among the affected sites is the ARRE-2 site in the Il2 promoter. In addition, Compound 10 interferes with the expression of many NFAT target genes, particularly cytokine genes, which play an important role in T cell activation and in the effector immune response.
Discussion
One objective of our research is to devise ways to redirect the transcriptional programs of T cell activation and tolerance with small drug-like compounds. Over the last several years, we obtained evidence that modulating the formation or stability of the NFAT:AP-1 complex is an attractive way to influence the T cell immune response. In the presence and absence of AP-1, NFAT turns on T cell transcriptional programs of activation versus hyporesponsiveness, respectively (22, 25, 33); thus, small drug-like compounds that stabilize or disrupt the formation of the quaternary NFAT:Fos:Jun:DNA complex without affecting NFAT binding per se could potentially redirect T cell transcription from an “effector” to a “tolerance” program and vice versa.
To identify compounds that modulate the stability of the NFAT:Fos:Jun:DNA complex, we developed a FRET assay using the structurally best-characterized example of cooperative binding of NFAT and AP-1 on DNA, which occurs at the distal ARRE-2 of the murine IL-2 promoter (36). The ARRE-2 site contains a consensus binding site for NFAT and a weak binding site for AP-1 (formed by dimers of Fos and Jun), and is sustained by contacts between the NFAT DBD and the leucine zipper regions of Fos and Jun (35, 41) (Fig. 1A). We and others previously showed that, individually, these proteins bind weakly to the ARRE-2 site, but together they bind synergistically (1, 35, 42). In addition to ARRE-2, a number of composite sites at which NFAT alone or AP-1 alone bind with varying affinities have been identified, but at which the quaternary complex forms synergistically with much stronger affinity (1, 25, 30).
The FRET-based assay described here was designed to identify compounds that interfere with the formation of the quaternary NFAT:Fos:Jun:DNA complex. In principle, such interfering compounds could have different modes of action: by disrupting the DNA-binding of NFAT, of Fos:Jun dimers, or both; by disrupting the protein–protein interactions among NFAT, Fos, and Jun; by intercalating nonspecifically into DNA; or by binding specifically to DNA sequences within the ARRE-2 composite site. We have used several types of assays to show that Compound 10, the candidate compound that we focused on here, belongs in the last category.
We deduced the mechanism of action of Compound 10 through a series of experiments that were largely independent of one another. First, we showed by EMSA that Compound 10 exhibited varying efficacy in blocking NFAT:Fos:Jun complex formation on three tested NFAT:AP-1 composite sites: it was highly effective on the ARRE-2 site of the mouse IL-2 promoter (Fig. 4A), less effective on the human IL-2 promoter ARRE-2 site (SI Appendix, Fig. S3), and completely ineffective on the GM-330 site of the GM-CSF enhancer (Fig. 4D). These are all composite NFAT:AP-1 elements with synergistic binding of NFAT and AP-1, suggesting strongly that Compound 10 binding depends on the DNA sequence of the composite site. A direct binding assay confirmed this hypothesis (Fig. 5 and SI Appendix, Fig. S4). Notably, Compound 10 did not affect binding of AP-1 to a consensus AP-1 element, or binding of NFAT to the κ3 site of the TNF promoter; this property is important because NFAT is the core DNA binding component required to toggle between T cell activation and T cell hyporesponsiveness (“anergy” or “exhaustion”) in vivo.
Finally, we showed by transcriptional profiling (RNA-seq) that Compound 10 interfered with the expression of several categories of genes in T cells stimulated with pharmacological agents—the phorbol ester PMA and the calcium ionophore ionomycin, which operate by activating protein kinase C and store-operated calcium entry, respectively—to mimic physiological activation (Fig. 6 and SI Appendix, Tables S1 and S2). In PMA-ionomycin–activated T cells, Compound 10 interfered with the expression of many NFAT-related genes, particularly a number of cytokine genes whose expression was also inhibited by CsA (e.g., Il2, Il4, Il5, Il10, Il13, Il17a, Il17f, and Il21) (Fig. 7 and SI Appendix, Fig. S5). Compound 10 also diminished the expression of other genes shown to be direct NFAT targets in chromatin immunoprecipitation-sequencing experiments, including Cd160, Erg1, Nfatc1 short isoform (encoding NFAT2), and Xcl1 (22). These observations suggest that, despite the ability of Compound 10 to bind many sites in genomic DNA (SI Appendix, Fig. S4D) and the likelihood that it has additional nonspecific effects, at least some of its prominent effects in cells are tied to its ability to inhibit NFAT binding at physiologically relevant NFAT:AP-1 composite sites in chromatin.
The aryl tetrasaccharide of calicheamicin γ1I was shown to displace NFAT from the human ARRE-2 site (43). This effect seems to parallel the effect of Compound 10, but an NMR solution structure of the aryl tetrasaccharide in complex with a different oligonucleotide uncovered an important difference: the calicheamicin-derived inhibitor binds in the DNA minor groove at –AGGA– and distorts the local DNA geometry (44). Distortion of the core NFAT recognition site would be expected to affect all NFAT transcriptional complexes indiscriminately, and not only NFAT:AP-1 complexes. Our data for Compound 10 establish that it is feasible to target the assembly of an NFAT:AP-1 complex without interfering with the DNA binding of either partner individually. This is a promising step toward selectively blocking the assembly of NFAT:AP-1 complexes, but the efficacy of Compound 10 depends on its binding to nonconserved DNA sequences at individual composite NFAT:AP-1 sites, and so some sites are spared.
We have not defined precisely how Compound 10 binds to ARRE-2 DNA, but the nucleotide sequence –AATT– is a critical determinant (Fig. 5D and SI Appendix, Fig. S4). Hoechst 33258, a benzimidazole compound loosely similar to Compound 10, inserts edgewise into the DNA minor groove along an –AATT– sequence in an otherwise unrelated oligonucleotide (PDB ID code 1DNH) (45). A superposition of structures 1DNH and 1A02 suggests that similar insertion of Compound 10 into the minor groove of the ARRE-2 oligonucleotide would be incompatible with the narrowed minor groove of the NFAT:AP-1:DNA complex (SI Appendix, Fig. S6). Narrowing of the minor groove is coupled to the local bending of DNA that allows formation of an extended protein–protein interface between NFAT and AP-1 (35), so the inhibitory effect of Compound 10 on complex formation could be explained if Compound 10 acts as a wedge that prevents narrowing of the minor groove and increases local rigidity of the DNA. If our goal were only to target ARRE-2, we could follow established principles to design compounds with higher specificity and higher affinity for the minor groove in this region of ARRE-2 (46–48). However, our purpose is to destabilize the broad class of complexes at NFAT:AP-1 composite sites relative to NFAT:partner complexes at other sites, and thus the appropriate target is the protein–protein interface.
There is considerable clinical potential for compounds targeting NFAT:AP-1 complexes, because they would be expected to switch the T cell transcriptional program from activation to the hyporesponsive (anergic/exhausted) state. As such, they would be effective in treating graft-versus-host disease, which can be a serious complication following organ transplants or following bone marrow transplants used to treat certain cancer patients. Moreover, NFAT:AP-1–dependent transcriptional signaling is essential in the development and function of osteoclasts (2, 49), the cells that are overactive in osteoporosis. Chemical derivatives of an NFAT:AP-1 inhibitor designed to direct it specifically to bone could offer a new treatment option for this debilitating disease. Such inhibitors could also have other clinical uses, for example in treating autoimmune disease.
Of the 960 compounds identified in the primary screen, 337 compounds had significant effects in the secondary screen. The genome-wide effects of selected additional inhibitors can be tested exactly as we have done here for Compound 10, focusing specifically on blockers that act at the NFAT:AP-1 interface. Complementary studies might also develop assays for compounds that have the converse effect: that is, stabilize formation of the quaternary NFAT:AP-1:DNA complex. These compounds would potentially be useful in cancer immunotherapy because they would have the opposite effect of switching the T cell transcriptional program away from anergy/exhaustion and toward full activation.
Materials and Methods
Purification of Recombinant Proteins.
E. coli strain BL21 (DE3) were transformed with the plasmids described in this section and grown at 37 °C with the appropriate antibiotic. When the OD600 reached 0.5–0.8, protein expression was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside, American Bio) for 5 h, at 30 °C. The NFAT1 DBD containing a 6xHis-tag was previously cloned into the pQE vector (50), and used as template to construct the RIT NFAT DBD by site-directed mutagenesis of the residues R468A/I469A/T535G (RIT NFAT, described in ref. 34). The DBD proteins were purified using TALON superflow chromatography (GE). The proteins were eluted in 50 mM Tris⋅Cl pH 7.5, 300 mM NaCl, 300 mM imidazole. After gel filtration, the proteins were kept in 20 mM Tris⋅Cl pH 7.5, 150 mM NaCl, 2 mM TCEP. Fos and Jun bZIP domains (63 amino acids each) were cloned into pET11a vector. Redox-sensitive Cys-154 in Fos and Cys-272 in Jun have been previously mutated to Ser (35). A unique Cys residue proximal to the Fos N terminal was introduced via site-directed mutagenesis in the Fos bZIP construct (I142C). Jun was purified by cation-exchange chromatography using an SP Sepharose Fastflow column (GE) following the manufacturer’s instructions, eluted with a salt gradient up to 2 M NaCl, 50 mM Hepes pH 7.2, 2 mM β-mercaptoethanol. Fos was subcloned into pET28a vector, which has a 6xHis-tag, using the restriction sites NdeI and BamHI, and purified with TALON superflow chromatography (GE). Elution buffer was: 50 mM Tris⋅Cl pH 7.5, 300 mM NaCl, 300 mM imidazole. After gel filtration, the proteins were kept in 50 mM Tris⋅Cl pH 7.5, 300 mM NaCl, 2 mM TCEP.
FRET Assay.
For the FRET assay, 20 nM of Fos-OG 488, 20 nM Jun, and 20 nM murine ARRE-2–Alexa-546 oligo diluted in FRET buffer (20 mM Tris⋅Cl pH 7.5, 50 mM NaCl, 5% Glycerol, 0.45 mg/mL BSA, 2 mM TCEP) were mixed in cuvettes or in 384-well plate. After 15 min of incubation, 40 nM of wild-type or RIT NFAT DBD was added on top of the well. To read the assay in cuvettes, a spectrofluorometer (Photon Technology International) was used. Excitation wavelength was 480 nm; the emission fluorescence was scanned from 500 to 600 nm. Plates were read in plate reader (Envision, Perkin-Elmer). Excitation filter was FITC485; emission filters were FITC535 to read the donor fluorescence and Rhodamine590 to read the acceptor fluorescence. All of the filters have a bandwidth of 16–20 nm.
High-Throughput Screening.
Assay conditions.
Each plate included 32 “low-control” wells (columns 1 and 2) containing NFAT, Fos-OG, Jun, and DNA:Alexa-546; and 32 “high-control” wells (columns 23 and 24) containing only Fos-OG, Jun, and DNA:Alexa-546. Columns 3–22 had the complete reaction mix and test compounds. Automated liquid handling was performed using ELX406 (Biotec Instruments) dispenser. Next, 10 nM of Fos-OG 488, 10 nM Jun, and 10 nM murine ARRE-2:Alexa-546 oligo diluted in 15 μL of FRET buffer (20 mM Tris⋅Cl pH 7.5, 50 mM NaCl, 5% Glycerol, 0.45 mg/mL BSA, 2 mM TCEP) were dispensed in a 384-well plate. Next, 250 nL of DMSO or test compounds (202,880 in total) were pin-dispensed using a Biomek FX liquid handler. After 1-h incubation at room temperature, 10 μL of FRET buffer containing 20 nM of NFAT DBD (or FRET buffer only for the high-control weels) were dispensed using the ELX406 (Biotec Instruments) automatic dispenser. After 2 h of incubation at room temperature, the plates were read on a Flex Station (Molecular Devices) or an Acquest (Molecular Devices) plate reader. Excitation filter was FITC485; emission filters were FITC535 and Rhodamine590.
Defining the hits.
The FRET ratio for each well, calculated by the ratio between the acceptor and the donor raw fluorescence, was annotated into the Collaborative Drug Discovery software. A Z′-factor (51, 52) was calculated for each plate based on FRET ratios measured in the low-control and high-control wells, and only plates with Z′ ≥ 0.7 were considered. Test wells with an absolute Alexa-546 signal more than 3 SD above the mean of all wells with compounds were excluded as containing fluorescent compounds. After this step, the mean Alexa-546 signal and its SD were recalculated, followed by a second round of exclusion of wells containing fluorescent compounds. Wells having an absolute OG488 signal more than twice the mean of the signal in high-control wells were also considered to contain fluorescent compounds and excluded. Then, using the FRET ratio, a z-score was calculated for each test well with the formula: z-score = (x − μPlate)/σPlate, where x is the observed FRET ratio, and μPlate and σPlate are the mean and SD of the observed FRET ratios of all nonexcluded wells in the plate.
Secondary screen and subsequent tests.
The 960 initial hits with z-scores ≤ −3.6 SD identified in the high-throughput screen (Dataset S4) were retested in triplicate, using the same assay and workflow. Wells containing fluorescent compounds were defined as above and excluded. The FRET ratio was then calculated for each well, and the z-score was calculated for each test well using the formula: z-score = (x − μlow-control)/σlow-control, where x is the observed FRET ratio, and μlow-control and σlow-control are the mean and SD of the low-control wells (columns 1–2) in the plate (Dataset S5). The threshold for confirming a hit in the secondary assay was set at a z-score ≤ −3 in at least two of three replicates. From the 337 compounds that reached this criterion, we purchased stocks of 23 high-scoring compounds. Three compounds were insoluble under conditions of the assay, and one was itself fluorescent. Six of the remaining 19 compounds showed dose-dependent inhibition in the FRET assay and were further tested in an EMSA assay. Four of them inhibited the binding of NFAT:AP-1 complex to the ARRE-2 DNA site. These four compounds were tested in T cells for their capacity to inhibit cytokine production. Compound 10 was the only compound that inhibited IL-2 production.
Reconstitution and working with Compound 10.
Compound 10 was purchased from MolPort or Asinex, reconstituted in DMSO (Millipore) in a final concentration of 10 mM, vortexed for 30 min, and sonicated for 45 min in a bath 1510 Branson ultrasonic cleaner. For all of the experiments, Compound 10 was diluted to 1 mM in DMSO, and further dilutions were made in T cell media or in the indicated buffer. When different final concentrations of Compound 10 were tested in the same experiment, all wells had the same final concentration of DMSO.
Gel-shift assays.
Oligonucleotide duplexes were ordered from IDT. Murine ARRE-2: 5′ CAA AGA GGA AAA TTT GTT TCA TAC AG; human ARRE-2: 5′ CAA AGA GGA AAA ACT GTT TCA TAC AG; AP-1: 5′ CGC TTG ATG ACT CAG CCG GAA; κ3: 5′ GAG CTC ATG GGT TTC TCC ACC and GM-330: 5′ CCC CCA TCG GAG CCC CTG AGT CAG CAT GGC G. Fifty nanograms of oligonucleotides were labeled for 1 h at 37C with T4 polynucleotide kinase (10 U; New England Biolabs) and 50 μCi [γ32] dATP (Perkin-Elmer). Probes were then purified with MicroSpin G-25 columns (GE). 500 nM of Fos and 500 nM of Jun were incubated with the indicated labeled oligonucleotides (15,000–20,000 counts per minute) and 0.5 μg per reaction of poly(dI:dC) (Sigma) in binding buffer (10 mM Hepes pH 7.0, 125 mM NaCl, 10% glycerol, 0.25 mM DTT, 0.8 mg/mL BSA). When indicated, increasing concentrations of Compound 10 were also added to the reaction. After 10 min, 10 nM of NFAT DBD were added and the reaction was incubated for other 20 min at room temperature. DNA–protein complexes were separated by electrophoresis under nondenaturating conditions on a 4% polyacrylamide gel in 1× TBE buffer. The gel was prerun for 1 h at 100 V and samples were run for an additional 90 min at 200 V. The gel was dried onto Whatman filter paper and analyzed by autoradiography.
Fluorescence assay.
The indicated concentrations of DNA in Fig. 5 and SI Appendix, Fig. S4 (1 nM–10 μM) were mixed with 50 nM of Compound 10 in 130 μL final volume of binding buffer (20 mM Tris⋅Cl pH 7.5, 50 mM NaCl) and transferred to a cuvette. The readings were performed using a spectrofluorometer (Photon Technology International). Excitation wavelength was 310 nm; emission scan was 350–500 nm. DNA was diluted in water. The oligonucleotides used are listed as follows: mARRE-2 oligo: 5′ CAA AGA GGA AAA TTT GTT TCA TAC AG; hARRE-2 oligo: 5′ CAA AGA GGA AAA ACT GTT TCA TAC AG; AP-1 oligo: 5′ CGC TTG ATG ACT CAG CCG GAA; κ3 oligo: 5′ GAG CTC ATG GGT TTC TCC ACC; GM-330 oligo: 5′ CCC CCA TCG GAG CCC CTG AGT CAG CAT GGC G; mARRE-2 1C mutant oligo: 5′ CAA AGA GGA AAC TTT GTT TCA TACA G; hARRE-2 1C mutant oligo: 5′ CAA AGA GGA AAC ACT GTT TCA TAC AG; mARRE-2 GG > CC oligo: 5′ CAA AGA CCA AAA TTT GTT TCA TAC AG; GM-330 GC > AA oligo: 5′ CCC CCA TCG GAA ACC CTG AGT CAG CAT GGC G; Variant mARRE2 oligo: 5′ CAA AGA GGA AAA TTT GTT TCA GCC GGA A; mARRE-2 Mutant 1 oligo: 5′ CAA AGA GGA AAA TTT GAC TCA GCC GGA A; mARRE-2 Mutant 2 oligo: 5′ CAA AGA GGA ATT GAT GAC TCA GCC GGA A; Short NFAT oligo: 5′ TTG CTG GAA AAA TAG; mARRE-2 Mutant 3 oligo: 5′ CAA AGA GGA AAC CCT GAC TCA GCC GGA A. Underlining indicates nucleotide alterations relative to the canonical ARRE-2 or GM-330 oligonucleotide.
RNA sequencing.
Naïve or total CD4+ T cells were left unstimulated or were stimulated with PMA (10 nM) and ionomycin (500 nM) for 2 h. Total RNA was extracted from using the RNeasy kit. RNA quality was evaluated with Bioanalyzer RNA pico kit (Agilent Technologies Inc). Poly(A)-selected RNA was amplified using the SMARTseq2 protocol (53). Briefly, purified RNA was reverse-transcribed using SuperScript II, Oligo dT30 VN primers and template switching primers. A preamplification step of nine PCR cycles was performed using the Kapa HiFI Hoststart kit (Kapa Biosystems). The PCR product was purified using AmpureXP beads (Beckman Coulter) and 1 ng was further used for library preparation using the Nextera XT LibraryPrep kit (Illumina). Tagmented DNA was amplified with a 12-cycle PCR and again purified with AmpureXP beads. Library size distribution and yield were evaluated using the Bioanalyzer high-sensitivity DNA kit. Libraries were pooled at equimolar ratio and sequenced with the rapid run protocol on the Illumina HiSeq 2500 with 50 single end cycles.
RNA-seq analysis.
Trimmed reads were mapped to the mouse transcriptome (mm10) based on GENCODE annotations using Kallisto (https://www.nature.com/articles/nbt.3519). Gene-level counts imported using tximport (https://f1000research.com/articles/4-1521/v2) were analyzed using the R package DESeq2 (https://genomebiology.biomedcentral.com/articles/10.1186/s13059-014-0550-8). Differentially expressed genes were identified as those passing a test (FDR threshold = 0.05) where the alternate hypothesis was that the absolute log2FC was greater than 0.4. This strategy is more stringent and statistically more accurate than thresholding on log2FC after a test whose alternate hypothesis merely states that log2FC is different from zero. Data have been deposited in the GEO database (54).
Supplementary Material
Acknowledgments
We are grateful to Victor Wong for helping with Th1 differentiation experiments; the Flow Cytometry Core Facility at the La Jolla Institute; Jason Greenbaum and Zheng “Alex” Fu of the Bioinformatics Core and Jeremy Day of the Sequencing Core for help with RNA sequencing; and Babak Tofig and Bryan France for assistance in performing the high-throughput screen at the University of California, Los Angeles. This work was supported by NIH Grants AI109842 and AI040127. The HiSeq 2500 was funded by NIH S10OD016262 and the FACSAria Cell Sorter by the S10RR027366. G.P.M. was supported by a Conselho Nacional de Desenvolvimento Cientifico e Tecnologico fellowship, Brazil. E.G.-A. was supported by a Consejo Nacional de Ciencia y Technología/University of California Institute for Mexico and the United States fellowship from Mexico/United States.
Footnotes
Conflict of interest statement: A.R. and P.G.H. are founders of CalciMedica, Inc., and members of its scientific advisory board.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE125446).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820604116/-/DCSupplemental.
References
- 1.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: Regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 2.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 3.Macian F. NFAT proteins: Key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 4.Baksh S, DeCaprio JA, Burakoff SJ. Calcineurin regulation of the mammalian G0/G1 checkpoint element, cyclin dependent kinase 4. Oncogene. 2000;19:2820–2827. doi: 10.1038/sj.onc.1203585. [DOI] [PubMed] [Google Scholar]
- 5.Jauliac S, et al. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat Cell Biol. 2002;4:540–544. doi: 10.1038/ncb816. [DOI] [PubMed] [Google Scholar]
- 6.Mognol GP, Carneiro FR, Robbs BK, Faget DV, Viola JP. Cell cycle and apoptosis regulation by NFAT transcription factors: New roles for an old player. Cell Death Dis. 2016;7:e2199. doi: 10.1038/cddis.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernández GL, et al. Selective inhibition of vascular endothelial growth factor-mediated angiogenesis by cyclosporin A: Roles of the nuclear factor of activated T cells and cyclooxygenase 2. J Exp Med. 2001;193:607–620. doi: 10.1084/jem.193.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamura H, et al. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell. 2000;6:539–550. doi: 10.1016/s1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- 9.Shaw KT, et al. Immunosuppressive drugs prevent a rapid dephosphorylation of transcription factor NFAT1 in stimulated immune cells. Proc Natl Acad Sci USA. 1995;92:11205–11209. doi: 10.1073/pnas.92.24.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sigal NH, et al. Is cyclophilin involved in the immunosuppressive and nephrotoxic mechanism of action of cyclosporin A? J Exp Med. 1991;173:619–628. doi: 10.1084/jem.173.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aramburu J, et al. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol Cell. 1998;1:627–637. doi: 10.1016/s1097-2765(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 12.Aramburu J, et al. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science. 1999;285:2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Rao A, Hogan PG. Structural delineation of the calcineurin-NFAT interaction and its parallels to PP1 targeting interactions. J Mol Biol. 2004;342:1659–1674. doi: 10.1016/j.jmb.2004.07.068. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Zhang L, Rao A, Harrison SC, Hogan PG. Structure of calcineurin in complex with PVIVIT peptide: Portrait of a low-affinity signalling interaction. J Mol Biol. 2007;369:1296–1306. doi: 10.1016/j.jmb.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 15.Roehrl MH, et al. Selective inhibition of calcineurin-NFAT signaling by blocking protein-protein interaction with small organic molecules. Proc Natl Acad Sci USA. 2004;101:7554–7559. doi: 10.1073/pnas.0401835101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heath VL, Shaw SL, Roy S, Cyert MS. Hph1p and Hph2p, novel components of calcineurin-mediated stress responses in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3:695–704. doi: 10.1128/EC.3.3.695-704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Rao A, Hogan PG. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 2011;21:91–103. doi: 10.1016/j.tcb.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noguchi H, et al. A new cell-permeable peptide allows successful allogeneic islet transplantation in mice. Nat Med. 2004;10:305–309. doi: 10.1038/nm994. [DOI] [PubMed] [Google Scholar]
- 19.Qian Z, et al. Structure-based optimization of a peptidyl inhibitor against calcineurin-nuclear factor of activated T cell (NFAT) interaction. J Med Chem. 2014;57:7792–7797. doi: 10.1021/jm500743t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu T, Qian Z, Xiao Q, Pei D. High-throughput screening of one-bead-one-compound libraries: Identification of cyclic peptidyl inhibitors against calcineurin/NFAT interaction. ACS Comb Sci. 2011;13:537–546. doi: 10.1021/co200101w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macián F, et al. Transcriptional mechanisms underlying lymphocyte tolerance. Cell. 2002;109:719–731. doi: 10.1016/s0092-8674(02)00767-5. [DOI] [PubMed] [Google Scholar]
- 22.Martinez GJ, et al. Cutting edge: NFAT transcription factors promote the generation of follicular helper T cells in response to acute viral infection. J Immunol. 2016;196:2015–2019. doi: 10.4049/jimmunol.1501841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mognol GP, et al. Exhaustion-associated regulatory regions in CD8+ tumor-infiltrating T cells. Proc Natl Acad Sci USA. 2017;114:E2776–E2785. doi: 10.1073/pnas.1620498114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain J, et al. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature. 1993;365:352–355. doi: 10.1038/365352a0. [DOI] [PubMed] [Google Scholar]
- 25.Macián F, López-Rodríguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- 26.Oh-Hora M, et al. Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat Immunol. 2008;9:432–443. doi: 10.1038/ni1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feske S, Draeger R, Peter HH, Eichmann K, Rao A. The duration of nuclear residence of NFAT determines the pattern of cytokine expression in human SCID T cells. J Immunol. 2000;165:297–305. doi: 10.4049/jimmunol.165.1.297. [DOI] [PubMed] [Google Scholar]
- 28.Jain J, Valge-Archer VE, Rao A. Analysis of the AP-1 sites in the IL-2 promoter. J Immunol. 1992;148:1240–1250. [PubMed] [Google Scholar]
- 29.Altman A, Isakov N, Baier G. Protein kinase Ctheta: A new essential superstar on the T-cell stage. Immunol Today. 2000;21:567–573. doi: 10.1016/s0167-5699(00)01749-7. [DOI] [PubMed] [Google Scholar]
- 30.Cockerill PN, et al. Human granulocyte-macrophage colony-stimulating factor enhancer function is associated with cooperative interactions between AP-1 and NFATp/c. Mol Cell Biol. 1995;15:2071–2079. doi: 10.1128/mcb.15.4.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Whaley CD, Mondino A, Mueller DL. Blocked signal transduction to the ERK and JNK protein kinases in anergic CD4+ T cells. Science. 1996;271:1272–1276. doi: 10.1126/science.271.5253.1272. [DOI] [PubMed] [Google Scholar]
- 32.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Pereira RM, Hogan PG, Rao A, Martinez GJ. Transcriptional and epigenetic regulation of T cell hyporesponsiveness. J Leukoc Biol. 2017;102:601–615. doi: 10.1189/jlb.2RI0317-097R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macián F, García-Rodríguez C, Rao A. Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. EMBO J. 2000;19:4783–4795. doi: 10.1093/emboj/19.17.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 36.Jain J, Miner Z, Rao A. Analysis of the preexisting and nuclear forms of nuclear factor of activated T cells. J Immunol. 1993;151:837–848. [PubMed] [Google Scholar]
- 37.Goldfeld AE, McCaffrey PG, Strominger JL, Rao A. Identification of a novel cyclosporin-sensitive element in the human tumor necrosis factor alpha gene promoter. J Exp Med. 1993;178:1365–1379. doi: 10.1084/jem.178.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCaffrey PG, Goldfeld AE, Rao A. The role of NFATp in cyclosporin A-sensitive tumor necrosis factor-alpha gene transcription. J Biol Chem. 1994;269:30445–30450. [PubMed] [Google Scholar]
- 39.Avni O, et al. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 40.Chuvpilo S, et al. Autoregulation of NFATc1/A expression facilitates effector T cells to escape from rapid apoptosis. Immunity. 2002;16:881–895. doi: 10.1016/s1074-7613(02)00329-1. [DOI] [PubMed] [Google Scholar]
- 41.Diebold RJ, Rajaram N, Leonard DA, Kerppola TK. Molecular basis of cooperative DNA bending and oriented heterodimer binding in the NFAT1-Fos-Jun-ARRE2 complex. Proc Natl Acad Sci USA. 1998;95:7915–7920. doi: 10.1073/pnas.95.14.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterson BR, Sun LJ, Verdine GL. A critical arginine residue mediates cooperativity in the contact interface between transcription factors NFAT and AP-1. Proc Natl Acad Sci USA. 1996;93:13671–13676. doi: 10.1073/pnas.93.24.13671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho SN, Boyer SH, Schreiber SL, Danishefsky SJ, Crabtree GR. Specific inhibition of formation of transcription complexes by a calicheamicin oligosaccharide: A paradigm for the development of transcriptional antagonists. Proc Natl Acad Sci USA. 1994;91:9203–9207. doi: 10.1073/pnas.91.20.9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikemoto N, et al. Calicheamicin-DNA complexes: Warhead alignment and saccharide recognition of the minor groove. Proc Natl Acad Sci USA. 1995;92:10506–10510. doi: 10.1073/pnas.92.23.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teng MK, Usman N, Frederick CA, Wang AH. The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG) Nucleic Acids Res. 1988;16:2671–2690. doi: 10.1093/nar/16.6.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr Opin Struct Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 47.Kawamoto Y, Bando T, Sugiyama H. Sequence-specific DNA binding Pyrrole-imidazole polyamides and their applications. Bioorg Med Chem. 2018;26:1393–1411. doi: 10.1016/j.bmc.2018.01.026. [DOI] [PubMed] [Google Scholar]
- 48.Bhaduri S, Ranjan N, Arya DP. An overview of recent advances in duplex DNA recognition by small molecules. Beilstein J Org Chem. 2018;14:1051–1086. doi: 10.3762/bjoc.14.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim K, et al. Nuclear factor of activated T cells c1 induces osteoclast-associated receptor gene expression during tumor necrosis factor-related activation-induced cytokine-mediated osteoclastogenesis. J Biol Chem. 2005;280:35209–35216. doi: 10.1074/jbc.M505815200. [DOI] [PubMed] [Google Scholar]
- 50.Jain J, Burgeon E, Badalian TM, Hogan PG, Rao A. A similar DNA-binding motif in NFAT family proteins and the Rel homology region. J Biol Chem. 1995;270:4138–4145. [PubMed] [Google Scholar]
- 51.Birmingham A, et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 2009;6:569–575. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 53.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 54.Mognol GP, et al. 2019 Targeting the NFAT:AP-1 transcriptional complex on DNA with a small-molecule inhibitor. Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE125446. Deposited January 22, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.