Significance
Retrograde signaling and posttranscriptional RNA editing are important regulatory processes for chloroplast development and function in flowering plants. GUN1 is a central integrator of chloroplast retrograde signaling pathways. This study reveals an unexpected role of GUN1 in affecting plastid RNA-editing efficiency during retrograde signaling through interactions with MORF2 and demonstrates a potential role for MORF2 in retrograde signaling. In addition to increasing our knowledge of the regulation of plant organellar RNA editing under stress conditions, this research uncovers a possible link between retrograde signaling and plastid RNA editing.
Keywords: GUN1, retrograde signaling, RNA editing, MORF2/RIP2, PPR
Abstract
During development or under stress, chloroplasts generate signals that regulate the expression of a large number of nuclear genes, a process called retrograde signaling. GENOMES UNCOUPLED 1 (GUN1) is an important regulator of this pathway. In this study, we have discovered an unexpected role for GUN1 in plastid RNA editing, as gun1 mutations affect RNA-editing efficiency at multiple sites in plastids during retrograde signaling. GUN1 plays a direct role in RNA editing by physically interacting with MULTIPLE ORGANELLAR RNA EDITING FACTOR 2 (MORF2). MORF2 overexpression causes widespread RNA-editing changes and a strong genomes uncoupled (gun) molecular phenotype similar to gun1. MORF2 further interacts with RNA-editing site-specificity factors: ORGANELLE TRANSCRIPT PROCESSING 81 (OTP81), ORGANELLE TRANSCRIPT PROCESSING 84 (OTP84), and YELLOW SEEDLINGS 1 (YS1). We further show that otp81, otp84, and ys1 single mutants each exhibit a very weak gun phenotype, but combining the three mutations enhances the phenotype. Our study uncovers a role for GUN1 in the regulation of RNA-editing efficiency in damaged chloroplasts and suggests that MORF2 is involved in retrograde signaling.
Plant cells coordinately regulate the expression of nuclear and chloroplast genes that encode components of the photosynthetic apparatus. Nuclear genes that regulate chloroplast development and chloroplast gene expression provide part of this control, but information also flows in the opposite direction—from chloroplasts to the nucleus—to regulate nuclear gene expression, a process called retrograde signaling (1–4). Chloroplast-to-nucleus retrograde signaling is essential for optimizing the photoautotrophic lifestyle of plants (1–4). Previously, we performed genetic screens using the bleaching herbicide norflurazon (NF) and identified mutations in six gun (for genomes uncoupled) loci that retained nuclear gene expression after chloroplasts were damaged. GUN2, -3, -4, -5, and -6 are enzymes involved in tetrapyrrole biosynthesis or metabolism (5–9). In contrast, GUN1 encodes a chloroplast-localized P-type pentatricopeptide repeat (PPR) protein with a C-terminal small MutS-related domain (1). Typical PPR proteins are targeted to chloroplasts or mitochondria, bind organellar transcripts, and influence their expression by altering RNA sequence, turnover, processing, or translation (10). Unlike other PPR proteins, however, GUN1 does not appear to bind RNAs (11). GUN1 plays a role in multiple stress-related retrograde signaling pathways, but despite numerous studies (11–15), GUN1’s precise function in retrograde signaling remains a mystery.
In NF-treated seedlings, nuclear genes for chloroplast-destined proteins are repressed by signaling through an active intracellular retrograde signaling pathway (2, 5). Previous studies showed that, in addition to reprogramming nuclear gene expression, NF treatment also affected RNA editing in plastids (16, 17). RNA editing is a posttranscriptional modification of RNA that changes the identity of nucleotides in RNAs or that adds or deletes nucleotides so that the information in the mature RNA differs from that defined in the genome (18). In flowering plants, through RNA editing, cytidines (Cs) are converted to uridines (Us) in chloroplast and mitochondria RNAs. There are 20–60 RNA-editing sites in chloroplasts and 300 to >600 sites in mitochondria of most flowering plants (18–21). RNA editing in organelles is essential for plant growth and development (22, 23). The precise composition of the vascular plant RNA-editing complex is not definitively known, but it consists of a small RNA/protein complex (23). For example, the proteins that dictate binding specificity in chloroplasts are mainly PLS-type PPRs (10, 18, 23–26). Moreover, MULTIPLE ORGANELLAR RNA EDITING FACTORS [MORFs, also called RNA EDITING FACTOR INTERACTING PROTEINS (RIPs)] (22, 23, 27), ORGANELLE RNA RECOGNITION MOTIF proteins (28, 29), ORGANELLE ZINC-FINGER proteins (30), and PROTOPORPHYRINOGEN OXIDASE 1 (PPO1) (31) have been identified as components of the plant RNA editosome (23).
Here we show that GUN1 interacts with MORF2/RIP2 (herein only the name MORF2 will be used) to affect the efficiency of editing for multiple sites in plastid RNAs during retrograde signaling. MORF2 overexpression causes widespread plastid RNA-editing changes and regulates nuclear gene expression similarly as the gun1 mutant in retrograde signaling. MORF2 further interacts with PLS-type PPR proteins: OTP81 [also called QUINTUPLE EDITING 1 (QED1)] (24, 32), OTP84 (24), and YS1 (33). Loss of function of OTP81/QED1 (herein only the name of OTP81 will be used), OTP84, and YS1 also could disrupt retrograde signaling. Our study reveals a distinct role for GUN1 in modulating RNA-editing efficiency in damaged chloroplasts and suggests that MORF2 plays a role in retrograde signaling.
Results
Loss of GUN1 Affects the Editing Levels of Multiple Plastid RNAs During Retrograde Signaling.
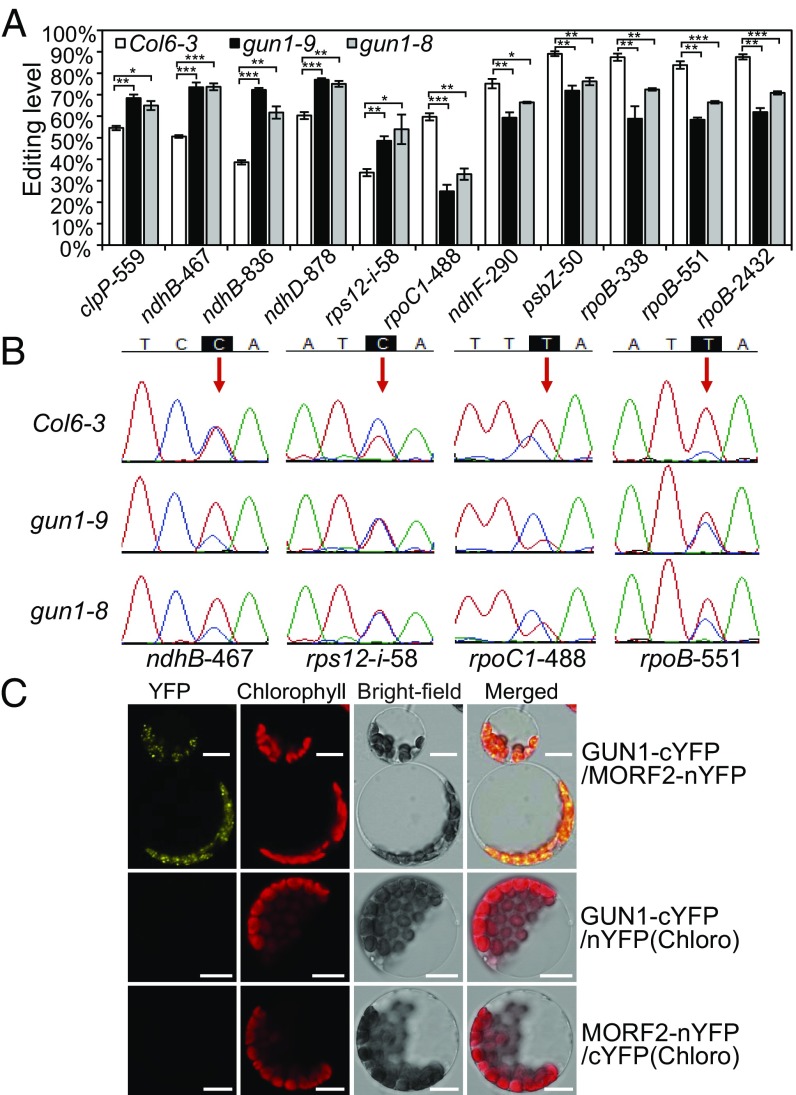
To investigate the possible link between plastid RNA editing and retrograde signaling, we compared editing levels of 34 validated plastid RNA-editing sites (34) between NF-treated and untreated Arabidopsis wild-type Col6-3 seedlings. Col6-3 is a transgenic line from Col-0 carrying a luciferase reporter under the control of the LHCB1.2 promoter and is the wild-type background of gun1 mutants (1). We found that in NF-treated wild-type seedlings, editing levels of 21 sites were affected, and 18 sites had lower editing levels, whereas the atpF-92, rpoC1-488, and rps12-i-58 sites had higher levels compared with untreated seedlings (SI Appendix, Fig. S1A). We further analyzed RNA-editing efficiency differences between wild type and two gun1 null mutant alleles, gun1-9 and gun1-8 (1). The result showed that in NF-treated gun1 mutants, RNA editing levels were increased for clpP-559, ndhB-467/836, ndhD-878, and rps12-i-58 and decreased for rpoC1-488, ndhF-290, psbZ-50, and rpoB-338/551/2432 compared with wild type (Fig. 1 A and B and SI Appendix, Fig. S1B). gun1 mutants also display retained nuclear gene expression when treated with the chloroplast translation inhibitor lincomycin (Linc) (1). We examined RNA-editing levels in Linc-treated seedlings and found that 15 sites had lower editing levels, whereas rps12-i-58 and ndhD-887 had higher editing levels in gun1-9 compared with wild type (SI Appendix, Fig. S2A). RNA-editing efficiency changes at the rps12-i-58, rpoC1-488, psbZ-50, and rpoB-551 sites had similar trends in NF- and Linc-treated gun1-9 mutants. Meanwhile, RNA editing in untreated seedlings exhibited only minor differences between wild type and gun1-9 (SI Appendix, Fig. S2B). A recent study showed that GUN1 is a short-lived protein and accumulates to detectable levels only under stress that involves retrograde signaling or during chloroplast biogenesis (15). This may explain the different effects of GUN1 on plastid RNA editing with or without NF/Linc treatment, as GUN1 protein is rapidly turned over under normal growth conditions. Therefore, our results suggest that GUN1 widely regulates the efficiency of plastid RNA editing, particularly during retrograde signaling.
Fig. 1.
GUN1 is required for the regulation of RNA-editing efficiency at multiple sites in plastids under NF treatment, and it interacts with MORF2. (A) RNA-editing levels of different sites in plastid altered by gun1 mutation under NF treatment. Col6-3 is the wild type. The x axis indicates different editing sites. The y axis represents the C to T (equal C to U in RNA) editing level. Data are mean ± SEM (three biological replicates). *P < 0.05; **P < 0.01; ***P < 0.001, two-tailed Student’s t test. (B) Sequencing chromatograms show RNA-editing profiles of representative sites indicated by red arrows. The peak for C is in blue and the peak for T is in red. (C) GUN1 interacts with MORF2 as shown by BiFC assay. nYFP: N-terminal YFP; cYFP: C-terminal YFP. nYFP(Chloro) and cYFP(Chloro) represent chloroplast-targeted nYFP and cYFP, respectively. Chlorophyll red autofluorescence indicates the localization of chloroplasts. Bright-field images correspond to the protoplast cells. Merged images show the colocalization of YFP and chloroplasts. GUN1-cYFP/nYFP(Chloro) and MORF2-nYFP/cYFP(Chloro) cotransformations are negative controls. (Scale bar, 10 μm.)
GUN1 Interacts Directly with the Plastid RNA Editosome Component MORF2.
To determine whether GUN1 plays a direct or indirect role in RNA editing, we carried out a yeast two-hybrid assay and found that GUN1 interacted strongly with MORF2 (SI Appendix, Fig. S3A). MORF2 is an essential component of the RNA editosome and is required for editing at almost all sites in plastid RNAs (22). Interactions between GUN1 and MORF2 were detected in plant cells using either bimolecular fluorescence complementation (BiFC) (Fig. 1C) or firefly luciferase complementation imaging (LCI) assays (SI Appendix, Fig. S3B). For BiFC, coexpression of GUN1 fused to the C-terminal of YFP (cYFP) and MORF2 fused to the N-terminal of YFP (nYFP) reconstituted YFP fluorescence in chloroplasts. As negative controls, we first confirmed that the transit peptide of chloroplast-localized OTP81 (24) could target YFP to chloroplasts (SI Appendix, Fig. S3C). Then, GUN1-cYFP or MORF2-nYFP was coexpressed with chloroplast-localized nYFP or cYFP, respectively, and no YFP signals were observed (Fig. 1C). For LCI, coexpression of GUN1-cLUC (C-terminal luciferase fusion) and MORF2-nLUC (N-terminal luciferase fusion) led to high levels of luciferase activity, whereas negative controls showed low or no luciferase activity (SI Appendix, Fig. S3B).
We next generated several lines overexpressing MORF2 (MORF2OX) fused with 3×HA-3×FLAG tags in the wild-type Col6-3 background. We picked two lines: a strong overexpressor [MORF2OX(s)] with strong phenotypes and a relatively weaker overexpressor [MORF2OX(w)] (SI Appendix, Fig. S4 A and B) with weak phenotypes (see results below). Then we generated plants coexpressing GUN1-GFP and MORF2-3×HA-3×FLAG and performed coimmunoprecipitation of MORF2 using extracts from NF-treated seedlings followed by mass spectrometric analysis. FLAG antibodies were able to precipitate GUN1 from plants co-overexpressing GUN1-GFP and MORF2-3×HA-3×FLAG (SI Appendix, Fig. S3D), further validating the interaction between GUN1 and MORF2. However, FLAG antibodies could not precipitate native GUN1 from MORF2OX(s) plants, which might be caused by low abundance of native GUN1 (15) even in NF-treated seedlings. Together with the observation that RNA editing is abnormal in gun1 mutants, these data suggest that the interaction between GUN1 and MORF2 may regulate the editing efficiency of multiple sites within plastid RNAs during times of stress.
MORF2 Overexpression Changes the Efficiency of Plastid RNA Editing at Multiple Sites and Confers a Phenotype Similar to gun1 During Retrograde Signaling.
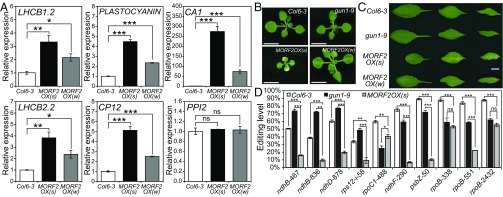
We then wanted to know if morf2 mutants or lines overexpressing MORF2 had phenotypes that resembled gun1 mutants, which would implicate a role for MORF2 in retrograde signaling. In wild type, NF treatment represses the expression of nuclear genes that encode chloroplast-destined proteins, whereas, in gun mutants, expression levels of these genes are higher than in the wild type (SI Appendix, Fig. S5A). This molecular phenotype has been defined as the gun phenotype (1, 5). Under normal conditions, morf2 homozygous plants (propagated through heterozygotes and with serious segregation distortion) are pale and die as seedlings (22). Previous studies either used morf2 homozygous plants segregated from heterozygotes (22) or transient virus-induced gene silencing (20) to study MORF2. NF bleaches seedlings, making it hard to identify morf2 homozygous plants, and transient silencing is not suitable for testing the gun phenotype. Therefore, we examined the gun phenotype of MORF2OX lines by comparing the expression differences of several nuclear-encoded photosynthesis genes (herein called retrograde signaling marker genes) (35) between NF-treated MORF2OX and wild-type seedlings. The results showed that RNA levels for all marker genes were relatively higher in MORF2OX lines than in the wild type, while the expression of the negative control gene was not affected (Fig. 2A). MORF2OX(w) had a smaller effect on marker genes expression than MORF2OX(s) (Fig. 2A). Based on the above gun phenotype criteria, both MORF2OX(s) and MORF2OX(w) have a gun phenotype under NF treatment, with MORF2OX(s) being a stronger allele than MORF2OX(w). On the other hand, Linc-treated MORF2OX(s) seedlings had lower expression levels of retrograde signaling marker genes compared with wild type (SI Appendix, Fig. S5B), which suggested that the gun phenotype of MORF2OX lines was specific to NF treatment. These results indicate that MORF2 may be involved in an NF-related retrograde signaling pathway that is different from the Linc-related pathway.
Fig. 2.
Overexpression of MORF2 confers a gun phenotype and regulates RNA editing under NF treatment. (A) The qPCR analysis of retrograde signaling marker gene expression indicates that MORF2OX lines have gun phenotypes. Total RNAs were isolated from 5 μM NF-treated seedlings grown under 24 h light (100 µmol⋅m−2⋅s−1) at 22 °C for 5 d. The x axis indicates different samples. The y axis shows the relative expression level, and expressions in Col6-3 are set to 1. The PPI2 gene is the negative control. Data are mean ± SEM (three biological replicates). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant, two-tailed Student’s t test. (B) Phenotype of MORF2OX seedlings grown on normal medium under long-day (LD) conditions (16 h light/8 h dark, 22 °C) for 14 d. (Scale bar, 0.5 cm.) (C) The leaf phenotype of 5-wk MORF2OX plants grown in soil under LD. The leaves are the rosette leaf, the first and the second cauline leaf from left to right, respectively. (Scale bar, 1 cm.) (D) The RNA-editing profile overlap between NF-treated MORF2OX(s) and gun1-9 seedlings. The x axis indicates the editing sites. The y axis shows the editing levels. Data are mean ± SEM (three biological replicates). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant, two-tailed Student’s t test.
When grown in the absence of NF, gun1-9 mutants resembled wild type (Fig. 2 B and C), possibly reflecting the limited role of GUN1 in most stages of plant development. In contrast, MORF2OX(s) plants were smaller than the wild type with pale green cotyledons at the seedling stage (Fig. 2B). At later stages of development, MORF2OX(s) plants had smaller and variegated leaves (Fig. 2C). MORF2OX(w) seedlings had variegated but not pale cotyledons (Fig. 2B). Mature MORF2OX(w) plants had normal rosette leaves and less variegation on cauline leaves (Fig. 2C). These results also suggest that MORF2 has a wider role in chloroplast development than GUN1.
Considering MORF2’s role in the RNA editosome, we analyzed the RNA-editing profile of MORF2OX(s). morf2 homozygous plants were reported to have dramatically lower levels of editing at most editing sites in plastids (22). Our results showed that MORF2OX(s) also moderately down-regulated RNA editing at 27 sites and increased the editing of rpoC1-488 under normal growth conditions (SI Appendix, Fig. S6A). This may be an explanation for the wider role of MORF2 in chloroplast development than GUN1. NF-treated MORF2OX(s) seedlings had decreased editing levels at 28 sites (Fig. 2D and SI Appendix, Fig. S6B), including all sites affected by gun1 mutations, except clpP-559. At those 28 sites, editing levels of rpoC1-488, ndhF-290, psbZ-50, and rpoB-338/551/2432 sites decreased in both MORF2OX(s) and gun1-9 (Fig. 2D).
MORF2OX Regulates Nuclear Gene Expression Similarly to gun1-9 During Retrograde Signaling.
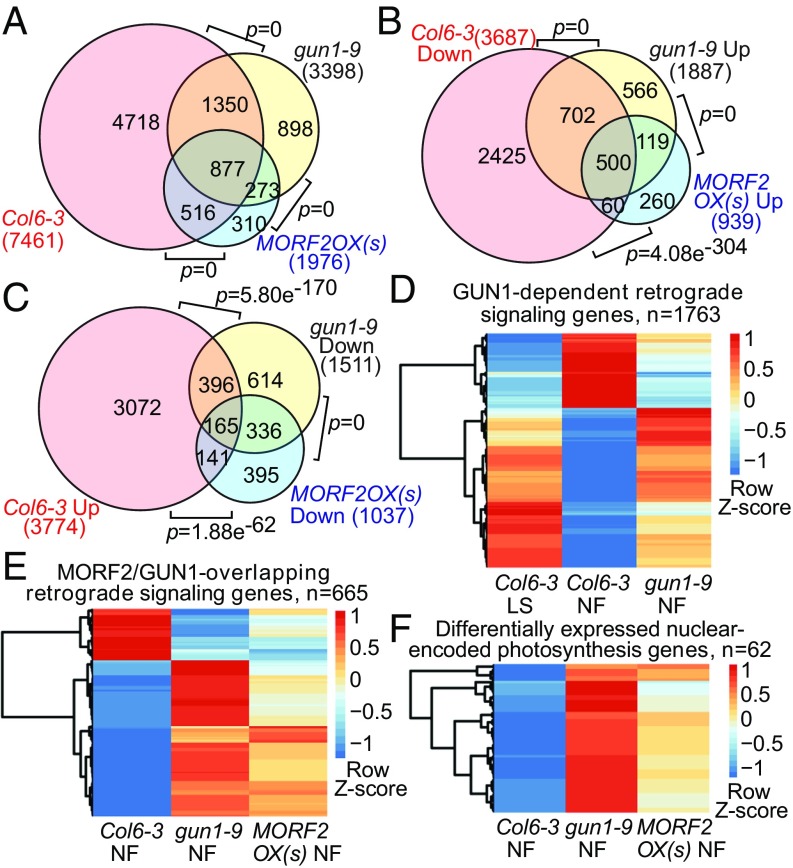
To identify a comprehensive set of genes under the control of an NF-generated signal, and to investigate the involvement of MORF2 in the GUN1-dependent retrograde signaling pathway, we performed RNA sequencing on NF-treated Col6-3, gun1-9, and MORF2OX seedlings. We identified 7,461 NF-regulated genes in Col6-3 compared with no treatment (Fig. 3A and Dataset S1A). In NF-treated seedlings, there were 3,398 differentially expressed genes (DEGs) in gun1-9, of which 2,227 were NF-regulated (Fig. 3A and Dataset S1B). Among these, expressions of 1,202 genes were repressed in wild type after NF treatment but were higher in gun1-9 than in the wild type (Fig. 3B and Dataset S2A). The expression pattern of these genes was consistent with the definition of the gun phenotype. Moreover, for another 561 genes, expressions were up-regulated in wild type after NF treatment but were lower in gun1-9 than in the wild type. These 561 genes had opposite directions of regulation between gun1-9 and wild type (Fig. 3C and Dataset S2A), which could also reflect retrograde signaling. Thus, we designated these total of 1,763 genes (Fig. 3D and Dataset S2A) as GUN1-dependent retrograde signaling genes.
Fig. 3.
Transcriptome data indicate that MORF2OX(s) regulates nuclear gene expression similarly to gun1-9 during retrograde signaling. (A) The Venn diagram shows the differentially expressed gene overlap among Col6-3 (Col6-3 NF vs. Col6-3 LS), gun1-9 (gun1-9 NF vs. Col6-3 NF), and MORF2OX(s) [MORF2OX(s) NF vs. Col6-3 NF] under NF treatment. (B) The Venn diagram shows overlapped DEGs that are down-regulated in Col6-3 (Col6-3 Down) but have a higher expression level in gun1-9 (gun1-9 Up) and MORF2OX(s) [MORF2OX(s) Up] compared with wild type under NF treatment. (C) The Venn diagram shows overlapped DEGs that were up-regulated in Col6-3 (Col6-3 Up), but had a lower expression level in gun1-9 (gun1-9 Down) and MORF2OX(s) [MORF2OX(s) Down] than in the wild type under NF treatment. P values in A–C show the statistical significance of the overlap between two groups of genes in Venn diagrams. (D–F) The hierarchical clustering of expression levels of GUN1-dependent (D), MORF2/GUN1-overlapping (E) retrograde signaling genes, and differentially expressed nuclear-encoded photosynthesis genes (F) in different samples. LS, untreated; NF, NF-treated. Heatmaps show the Z-score value of log2-transformed [(average reads per kilobase of transcript per million mapped reads) + 0.001] of each gene.
We identified 1976 DEGs in MORF2OX(s) seedlings under NF treatment (Fig. 3A and Dataset S1C), 866 of which had opposite directions of regulation between MORF2OX(s) and wild type. The expressions of 560 genes were repressed in wild type after NF treatment but were higher in MORF2OX(s) than in the wild type (Fig. 3B). For another 306 genes, RNAs accumulated to high levels in wild type after NF treatment, but were lower in MORF2OX(s) than in the wild type (Fig. 3C). Of the 866 genes, 665 were GUN1-dependent retrograde signaling genes (665 of 1,763, or 37.7%), which were designated as MORF2/GUN1-overlapping retrograde signaling genes. Compared with gun1-9, most of the MORF2/GUN1-overlapping retrograde signaling genes (Fig. 3E), including many nuclear-encoded photosynthesis genes (Fig. 3F and Dataset S2B) and retrograde signaling marker genes (SI Appendix, Fig. S7A), had similar regulation patterns but a lower fold-change in MORF2OX(s). From the transcriptome data, we concluded that MORF2OX(s) had a strong gun phenotype, although its gun phenotype was weaker than that seen in gun1-9. However, as several overlapping editing sites had lower editing levels in MORF2OX(s) compared with in gun1-9, it appeared that there was no simple correlation between RNA-editing levels and the strength of gun phenotypes. MORF2OX(w) had only 761 DEGs (SI Appendix, Fig. S7B and Dataset S1D), most of which (656 of 761, or 86%) overlapped with MORF2OX(s) (SI Appendix, Fig. S7B). A total of 217 DEGs of MORF2OX(w) were in the category of MORF2/GUN1-overlapping retrograde genes (SI Appendix, Fig. S7C). These 217 genes (including all of retrograde signaling marker genes) in MORF2OX(w) had similar expression patterns but a smaller fold-change compared with gun1-9 and MORF2OX(s) (SI Appendix, Fig. S7 A and D). This confirmed that MORF2OX(w) had an even weaker gun phenotype than MORF2OX(s). The top 50 gene ontology terms enriched in GUN1-dependent or MORF2/GUN1-overlapping retrograde signaling genes were similar. The main enriched terms were related to photosynthesis/light reaction and other chloroplast functions (SI Appendix, Fig. S8), which is consistent with the regulation pattern of retrograde signaling. From the transcriptome data, we conclude that, under NF treatment, MORF2OX lines regulate nuclear gene expression similarly as gun1-9 and that MORF2OX(s) can phenocopy about one-third of the gun phenotype of gun1-9. Together, these findings indicate that GUN1 and MORF2 have partially overlapped regulation pathways to affect nuclear gene expression during retrograde signaling.
MORF2, but Not GUN1, Interacts with Site-Specificity Factor PLS-Type PPRs.
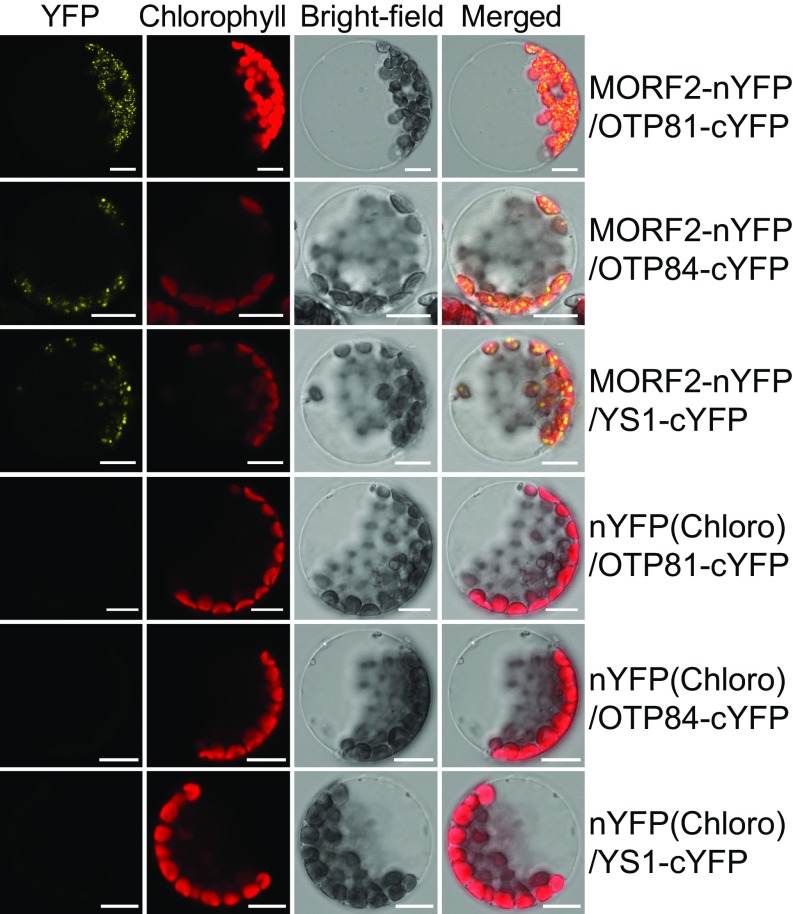
As part of the plastid RNA editosome, PLS-type PPRs bind to specific sequences near an editing site to confer specificity on an editing complex (18, 23–26). MORF2 has been reported to interact selectively with some PLS-type PPRs (22, 31). rpoB-2432, ndhF-290/psbZ-50, and rpoB-338 are four sites that had reduced editing in both gun1-9 and MORF2OX(s), and their associated site-specificity factors are OTP81/QED1 (24, 32), OTP84 (24), and YS1 (33), respectively. We showed that MORF2 interacted with OTP81, OTP84, and YS1 using a yeast two-hybrid assay (SI Appendix, Fig. S9A), which was consistent with results published recently (36). Furthermore, we confirmed these interactions in plants using BiFC (Fig. 4), LCI assays (SI Appendix, Fig. S9B), and coimmunoprecipitation of MORF2 with OTP81, OTP84, and YS1 followed by mass spectrometric analysis (SI Appendix, Fig. S3D). Moreover, we found that GUN1 did not interact with OTP81, OTP84, or YS1 directly either in yeast (SI Appendix, Fig. S9C) or in plants (SI Appendix, Fig. S9D).
Fig. 4.
MORF2 interacts with the site-specificity factors OTP81, OTP84, and YS1 as shown by BiFC assay. cYFP, C-terminal YFP; nYFP(Chloro), chloroplast-targeted nYFP; nYFP, N-terminal YFP. MORF2-nYFP interacts with OTP81-, OTP84-, and YS1-cYFP showing the YFP complementation in chloroplasts. Chlorophyll red autofluorescence indicates the localization of chloroplasts. Bright-field images correspond to protoplast cells. Merged images show the colocalization of YFP and chloroplasts. OTP81-, OTP84-, or YS1-cYFP cotransformed with nYFP(Chloro) are negative controls. (Scale bar, 10 μm).
Plastid RNA-Editing Profile Changes Are Related to the Expression of Nuclear Genes.
To investigate more connections between RNA editing and retrograde signaling, we analyzed the RNA-editing profiles and gun phenotypes of otp81, otp84, and ys1 single and triple mutants under NF treatment. In the otp81 mutant, editing levels of seven sites were reduced compared with wild type (SI Appendix, Fig. S10). Among these, only the rpoB-2432 site exhibited reduced editing in both otp81 and gun1 mutants. The otp84 mutation affected four editing sites (SI Appendix, Fig. S10) in which editing levels of ndhF-290 and psbZ-50 were reduced in both otp84 and gun1 mutants. The rpoB-338 site was not edited in the ys1 mutant (SI Appendix, Fig. S10), while its editing level was down-regulated in gun1 mutants. Editing levels of 13 sites were affected in the otp81otp84ys1 triple mutant, of which the ndhD-878, rpoC1-488, ndhF-290, psbZ-50, and rpoB-338/2432 sites had similar regulation patterns between otp81otp84ys1 triple and gun1 mutants (SI Appendix, Fig. S10).
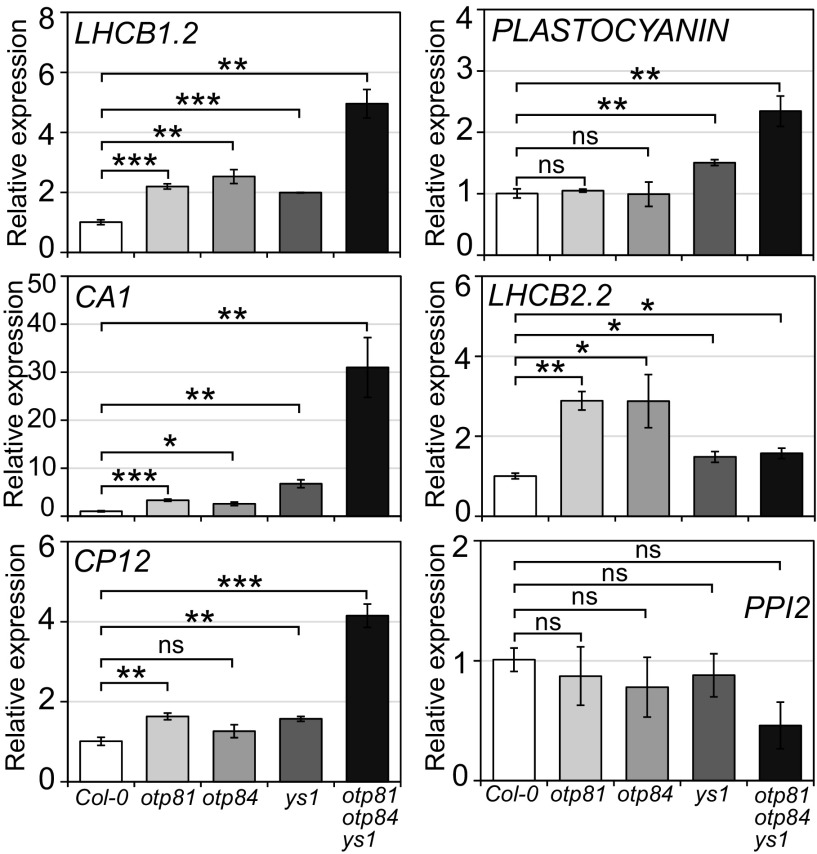
We further assayed the gun phenotypes of otp81, otp84, and ys1 single and triple mutants under NF treatment. The expressions of LHCB1.2, CA1, and LHCB2.2 were up-regulated over twofold in otp81 and otp84 single mutants compared with wild type. RNA levels of LHCB1.2 and CA1 in the ys1 single mutant also showed an over twofold up-regulation (Fig. 5). These results indicated that otp81, otp84, and ys1 single mutants had very weak gun phenotypes. In the otp81otp84ys1 triple mutant, expressions of all retrograde signaling marker genes were up-regulated over twofold compared with wild type, except for LHCB2.2, which increased only ∼1.6-fold (Fig. 5). Moreover, the expression levels of LHCB1.2, PLASTOCYANIN, CA1, and CP12 in the otp81otp84ys1 triple mutant were relatively higher than in the single mutants, indicating that the otp81otp84ys1 triple mutant had a stronger gun phenotype than the single mutants (Fig. 5). These results indicate that loss of function of OTP81, OTP84, and YS1 could partially disrupt retrograde signaling and that the editing of certain sites may be related to the expression of nuclear genes. The widespread impact on RNA editing may be important for the strong gun phenotype of gun1 mutants and MORF2OX(s) lines.
Fig. 5.
otp81, otp84, and ys1 single and triple mutants have weak gun phenotypes. The qPCR analysis of retrograde signaling marker gene expression in otp81, otp84, and ys1 single and triple mutants. Col-0 is the wild type. Total RNAs were isolated from 5 μM NF-treated whole seedlings grown under 24 h light (100 µmol⋅m−2⋅s−1) at 22 °C for 5 d. The x axis indicates the samples. The y axis shows the relative expression level of marker genes, and their expressions in Col-0 are set to one. The PPI2 gene is the negative control. Data are mean ± SEM (three biological replicates). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant, two-tailed Student’s t test.
Discussion
The chloroplast is a sensory organelle, and so signals triggered by environmental stimuli or by biochemical changes in chloroplasts can regulate nuclear gene expression via retrograde signaling pathways (2–4). However, the molecular mechanism of how signals are relayed from organelles to the nucleus in response to stress remains one of the least understood signaling pathways in plants, even though it is of fundamental importance. In this study, we reveal an unexpected function for GUN1 in regulating the efficiency of plastid RNA editing through direct interactions with MORF2 in retrograde signaling. Our studies also provide evidence that MORF2 plays a role in retrograde signaling.
We have demonstrated that gun1 mutations affect RNA-editing levels of 11 sites in plastids (Fig. 1A), supporting a broad role for GUN1 in regulating RNA-editing efficiency. Transcriptome data showed that, in NF-treated gun1-9 seedlings, the genes for gun1 mutation-affected sites associated PLS-PPRs and broad-effect plastid RNA-editing factors were not differentially expressed, except for CRR22 and CRR28 (SI Appendix, Table S1). The only overlap between gun1 mutation-affected and CRR22- or CRR28-associated sites (25, 30) was rpoB-551, ndhB-467, and ndhD-878. Moreover, the crr22 mutation also affected the editing of ndhB-746 and ndhD-887 (25, 30), but the gun1 mutation had no effect on the editing of these two sites (SI Appendix, Fig. S1B). Based on these observations, changes in RNA-editing efficiency in gun1 mutants does not seem to be caused by secondary effects of changes in gene expression in retrograde signaling. Although we do not have data to exclude the possibility that the gun1 mutation affects protein levels of RNA-editing factors, GUN1’s role in RNA editing appears to be a direct one through its interaction with MORF2 (Fig. 1C). However, the editing efficiency of only 11 sites was affected by gun1 mutation under NF treatment, indicating the selective recruitment of GUN1 to target RNAs. Moreover, some gun1 mutation-affected editing sites had higher editing levels and others had lower editing levels compared with wild type, suggesting that additional unknown components are required to determine the editing activity at different sites. A similar observation has been reported for another RNA-editing regulator, PPO1 (31). The questions of why GUN1 is recruited to affect RNA-editing efficiency in response to stress, and how this specific recruitment is organized, deserve further studies.
MORF proteins have been shown to form both homo- and hetero-dimers among each other, and they selectively interact with PLS-type PPRs (22, 31, 36, 37), suggesting that MORFs play a key role in assembling specific protein complexes at different RNA-editing sites (36). In addition, monomers or multimers of other MORFs may be able to partially substitute for MORF2 (23). As GUN1 is a short-lived protein and accumulates under stress that involves retrograde signaling (15), it is possible that the binding of GUN1 to MORF2 affects the multimerization of MORFs or the incorporation of MORF2 into RNA editosomes to regulate RNA editing activity. GUN1 may add a level of regulation that fine-tunes plastid RNA-editing efficiency under stress conditions. MORF2 overproduction may partially mimic the loss of function of GUN1 also by affecting the multimerization of MORFs.
Our data suggest a possible link between retrograde signaling and RNA editing. GUN1 is a integrator of multiple retrograde signaling pathways and is also required for regulation of RNA-editing efficiency during retrograde signaling. Under NF treatment, MORF2OX(s) altered RNA-editing levels for multiple sites and had a strong gun1-like gun molecular phenotype (Figs. 2 and 3), suggesting that MORF2 is also involved in retrograde signaling. However, the fact that the gun phenotype of MORF2OX(s) was weaker than gun1-9 indicates that the MORF2-activated retrograde signaling explains only part of the GUN1-dependent retrograde signaling pathways. We also showed that the single and triple mutants of OTP81, OTP84, and YS1 also have very weak gun phenotypes (Fig. 5). Together, these observations further suggest a role for plastid RNA editing in affecting nuclear gene expression during retrograde signaling. These editing-level changes in plastid RNAs appear to affect nuclear gene expression.
How might a change in RNA editing trigger a retrograde signaling pathway? Among the editing sites that exhibit similar editing-level changes in gun1-9, MORF2OX(s), and otp81otp84ys1 triple mutant, three are within rpoB or rpoC1 transcripts, which encode subunits of plastid-encoded RNA polymerase (38). Previous studies have indicated that mutations in sigma factor (SIG) genes, sig2 and sig6, which encode the sigma factors of plastid RNA polymerase, also disrupt retrograde signaling (35). Therefore, it is possible that changes in RNA-editing levels for rpoB and rpoC1 transcripts ultimately affect activity of plastid-encoded RNA polymerase, resulting in abnormal transcription of a specific set of plastid genes. This may be one source of the GUN1-dependent retrograde signaling. Another possible link between retrograde signaling and RNA editing is that improper RNA editing may lead to misfolding and aggregation of corresponding proteins, which would then have to be cleared by a chloroplast protein quality control pathway. This pathway is likely essential, which would explain why we have not found retrograde signaling proteins in our genetic screens (1, 5–8). This could also explain why there is not a simple correlation between RNA-editing levels and severity of the gun phenotype. Whether a defect in RNA editing itself can act as a trigger for retrograde signaling remains to be established. Nevertheless, RNA editing is initiated in chloroplasts and leads to posttranscriptional modifications. In different treatments that activate chloroplast signaling that results in gene expression changes, defective plastid RNA-editing events have been observed (16, 17). All these cues indicate the potential of RNA editing as one of the perception signals in retrograde signaling. Continued biochemical analysis of proteins involved in RNA editing and retrograde signaling will help resolve the connections between these two fundamental processes.
Materials and Methods
Plant materials and growth conditions, analysis of RNA editing, yeast two-hybrid, BiFC, and LCI assays, plant transformation, coimmunoprecipitation, mass spectrometric analysis, gun phenotype assay, RNA-sequencing analysis, and mutant genotyping are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Jesse Woodson for useful discussions; Dr. David O’Keefe for help in text editing; and James Moresco and Jolene Diedrich for technical support. RNA-sequencing and mass spectrometric analysis were performed at the Next Generation Sequencing Core and Mass Spectrometry Core of the Salk Institute for Biological Studies supported by NIH-National Cancer Institute Grant CCSG: P30 014195; the Chapman Foundation; the Helmsley Charitable Trust; and the Helmsley Center for Genomic Medicine. The US Department of Energy (Grant DE-FG02-04ER15540 to J.C.) and the Howard Hughes Medical Institute have provided long-term support for this project.
Footnotes
The authors declare no conflict of interest.
Data deposition: The RNA-seq raw data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE110125).
See Commentary on page 9701.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820426116/-/DCSupplemental.
References
- 1.Koussevitzky S, et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- 2.Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet. 2008;9:383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ. Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu Rev Plant Biol. 2016;67:25–53. doi: 10.1146/annurev-arplant-043015-111854. [DOI] [PubMed] [Google Scholar]
- 4.de Souza A, Wang J-Z, Dehesh K. Retrograde signals: Integrators of interorganellar communication and orchestrators of plant development. Annu Rev Plant Biol. 2017;68:85–108. doi: 10.1146/annurev-arplant-042916-041007. [DOI] [PubMed] [Google Scholar]
- 5.Susek RE, Ausubel FM, Chory J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- 6.Larkin RM, Alonso JM, Ecker JR, Chory J. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science. 2003;299:902–906. doi: 10.1126/science.1079978. [DOI] [PubMed] [Google Scholar]
- 7.Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodson JD, Perez-Ruiz JM, Chory J. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol. 2011;21:897–903. doi: 10.1016/j.cub.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larkin RM. Tetrapyrrole signaling in plants. Front Plant Sci. 2016;7:1586. doi: 10.3389/fpls.2016.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barkan A, Small I. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol. 2014;65:415–442. doi: 10.1146/annurev-arplant-050213-040159. [DOI] [PubMed] [Google Scholar]
- 11.Tadini L, et al. GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol. 2016;170:1817–1830. doi: 10.1104/pp.15.02033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruckle ME, Larkin RM. Plastid signals that affect photomorphogenesis in Arabidopsis thaliana are dependent on GENOMES UNCOUPLED 1 and cryptochrome 1. New Phytol. 2009;182:367–379. doi: 10.1111/j.1469-8137.2008.02729.x. [DOI] [PubMed] [Google Scholar]
- 13.Cottage A, Mott EK, Kempster JA, Gray JC. The Arabidopsis plastid-signalling mutant gun1 (genomes uncoupled1) shows altered sensitivity to sucrose and abscisic acid and alterations in early seedling development. J Exp Bot. 2010;61:3773–3786. doi: 10.1093/jxb/erq186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paieri F, et al. The DEAD-box RNA helicase RH50 is a 23S-4.5S rRNA maturation factor that functionally overlaps with the plastid signaling factor GUN1. Plant Physiol. 2018;176:634–648. doi: 10.1104/pp.17.01545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu G-Z, et al. Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED1. Plant Physiol. 2018;176:2472–2495. doi: 10.1104/pp.18.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kakizaki T, Yazu F, Nakayama K, Ito-Inaba Y, Inaba T. Plastid signalling under multiple conditions is accompanied by a common defect in RNA editing in plastids. J Exp Bot. 2012;63:251–260. doi: 10.1093/jxb/err257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tseng C-C, Lee C-J, Chung Y-T, Sung T-Y, Hsieh M-H. Differential regulation of Arabidopsis plastid gene expression and RNA editing in non-photosynthetic tissues. Plant Mol Biol. 2013;82:375–392. doi: 10.1007/s11103-013-0069-5. [DOI] [PubMed] [Google Scholar]
- 18.Takenaka M, Zehrmann A, Verbitskiy D, Härtel B, Brennicke A. RNA editing in plants and its evolution. Annu Rev Genet. 2013;47:335–352. doi: 10.1146/annurev-genet-111212-133519. [DOI] [PubMed] [Google Scholar]
- 19.Chateigner-Boutin A-L, Small I. A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res. 2007;35:e114. doi: 10.1093/nar/gkm640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bentolila S, Oh J, Hanson MR, Bukowski R. Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 2013;9:e1003584. doi: 10.1371/journal.pgen.1003584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichinose M, Sugita M. RNA editing and its molecular mechanism in plant organelles. Genes (Basel) 2016;8:5. doi: 10.3390/genes8010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takenaka M, et al. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc Natl Acad Sci USA. 2012;109:5104–5109. doi: 10.1073/pnas.1202452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun T, Bentolila S, Hanson MR. The unexpected diversity of plant organelle RNA editosomes. Trends Plant Sci. 2016;21:962–973. doi: 10.1016/j.tplants.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Hammani K, et al. A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell. 2009;21:3686–3699. doi: 10.1105/tpc.109.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okuda K, et al. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell. 2009;21:146–156. doi: 10.1105/tpc.108.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stern DB, Goldschmidt-Clermont M, Hanson MR. Chloroplast RNA metabolism. Annu Rev Plant Biol. 2010;61:125–155. doi: 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- 27.Bentolila S, et al. RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc Natl Acad Sci USA. 2012;109:E1453–E1461. doi: 10.1073/pnas.1121465109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun T, et al. An RNA recognition motif-containing protein is required for plastid RNA editing in Arabidopsis and maize. Proc Natl Acad Sci USA. 2013;110:E1169–E1178. doi: 10.1073/pnas.1220162110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi X, Bentolila S, Hanson MR. Organelle RNA recognition motif-containing (ORRM) proteins are plastid and mitochondrial editing factors in Arabidopsis. Plant Signal Behav. 2016;11:e1167299. doi: 10.1080/15592324.2016.1167299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun T, et al. A zinc finger motif-containing protein is essential for chloroplast RNA editing. PLoS Genet. 2015;11:e1005028. doi: 10.1371/journal.pgen.1005028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang F, et al. Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing. Proc Natl Acad Sci USA. 2014;111:2023–2028. doi: 10.1073/pnas.1316183111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagoner JA, Sun T, Lin L, Hanson MR. Cytidine deaminase motifs within the DYW domain of two pentatricopeptide repeat-containing proteins are required for site-specific chloroplast RNA editing. J Biol Chem. 2015;290:2957–2968. doi: 10.1074/jbc.M114.622084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou W, et al. The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J. 2009;58:82–96. doi: 10.1111/j.1365-313X.2008.03766.x. [DOI] [PubMed] [Google Scholar]
- 34.Ruwe H, Castandet B, Schmitz-Linneweber C, Stern DB. Arabidopsis chloroplast quantitative editotype. FEBS Lett. 2013;587:1429–1433. doi: 10.1016/j.febslet.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 35.Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J. Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J. 2013;73:1–13. doi: 10.1111/tpj.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bayer-Császár E, et al. The conserved domain in MORF proteins has distinct affinities to the PPR and E elements in PPR RNA editing factors. Biochim Biophys Acta Gene Regul Mech. 2017;1860:813–828. doi: 10.1016/j.bbagrm.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 37.Zehrmann A, et al. Selective homo- and heteromer interactions between the multiple organellar RNA editing factor (MORF) proteins in Arabidopsis thaliana. J Biol Chem. 2015;290:6445–6456. doi: 10.1074/jbc.M114.602086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steiner S, Schröter Y, Pfalz J, Pfannschmidt T. Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol. 2011;157:1043–1055. doi: 10.1104/pp.111.184515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.