Significance
Marine oxygen-deficient zones (ODZs) are naturally occurring midlayer oxygen-poor regions of the ocean, sandwiched between oxygenated surface and deep layers. In the absence of oxygen, microorganisms in ODZs use other compounds, such as oxidized forms of nitrogen and sulfur, as terminal electron acceptors. We identified the presence and expression of genes for both arsenic reduction and oxidation in marine ODZs, suggesting the microbial community in these waters is also cycling arsenic for respiratory gain. The existence of an arsenic respiratory cycle in pelagic waters suggests microbial arsenic metabolisms may be underestimated in the modern ocean and were likely an even more significant contributor to biogeochemical cycles in the anoxic ancient oceans when arsenic concentrations were higher.
Keywords: oxygen deficient zones, arsenic, chemoautotrophy, dissimilatory arsenate reduction, marine metagenome
Abstract
Microbial capacity to metabolize arsenic is ancient, arising in response to its pervasive presence in the environment, which was largely in the form of As(III) in the early anoxic ocean. Many biological arsenic transformations are aimed at mitigating toxicity; however, some microorganisms can respire compounds of this redox-sensitive element to reap energetic gains. In several modern anoxic marine systems concentrations of As(V) are higher relative to As(III) than what would be expected from the thermodynamic equilibrium, but the mechanism for this discrepancy has remained unknown. Here we present evidence of a complete respiratory arsenic cycle, consisting of dissimilatory As(V) reduction and chemoautotrophic As(III) oxidation, in the pelagic ocean. We identified the presence of genes encoding both subunits of the respiratory arsenite oxidase AioA and the dissimilatory arsenate reductase ArrA in the Eastern Tropical North Pacific (ETNP) oxygen-deficient zone (ODZ). The presence of the dissimilatory arsenate reductase gene arrA was enriched on large particles (>30 um), similar to the forward bacterial dsrA gene of sulfate-reducing bacteria, which is involved in the cryptic cycling of sulfur in ODZs. Arsenic respiratory genes were expressed in metatranscriptomic libraries from the ETNP and the Eastern Tropical South Pacific (ETSP) ODZ, indicating arsenotrophy is a metabolic pathway actively utilized in anoxic marine water columns. Together these results suggest arsenic-based metabolisms support organic matter production and impact nitrogen biogeochemical cycling in modern oceans. In early anoxic oceans, especially during periods of high marine arsenic concentrations, they may have played a much larger role.
For most of Earth’s history the oceans were characterized by a lack of available oxygen and as a result ancient microbes employed a variety of alternate electron acceptors (1). Proxies for these early environments are found in marine pelagic oxygen-deficient zones (ODZs) where redox gradients through the water column impact the associated microbial communities (2, 3). Naturally occurring marine ODZs are created by a combination of organic matter respiration and slow midlayer ventilation (4) and are functionally anoxic (<10 nmol L−1) (2, 5). Denitrifying metabolisms, which reduce nitrate and nitrite, are prominent (3) and ultimately result in the production of N2O and N2 gas and thus loss of N from the marine system. There is also evidence for a cryptic sulfur cycle in marine ODZs, where dissimilatory sulfate reduction produces sulfide, which is consumed by autotrophic sulfur oxidizers (6).
The oxidized inorganic arsenic compound arsenate is the thermodynamically stable form in oxygenated aqueous environments [As(V) as H2AsO4− and HAsO42−] and arsenite is the thermodynamically stable form in anoxic environments [As(III) as H3AsO30 and H2AsO3−] (7). Microbes utilizing the arsenic redox potential for energetic gains have been studied and isolated in a range of systems with moderate to high arsenic concentrations, including polluted and unpolluted soils, sediments, hot springs, lakes, wastewater, and mine drainage (8). In some arsenic-rich aquatic systems, like the alkaline Mono Lake in California (USA), arsenic supports microbial communities through a cycle of dissimilatory heterotrophic reduction of As(V) and autotrophic oxidation of As(III), analogous to the cryptic sulfur cycle in ODZs (7). A complete metabolic arsenic cycle is ancient, existing since the Archaean (9, 10), as suggested by fossilized arsenic-rich organic globules from 2.72 billion years ago (11). Arsenic concentrations have fluctuated over geologic time, with early anoxic oceans likely experiencing periods of greater arsenic abundance than today (12, 13).
While the modern ocean is not considered an arsenic-loaded system, inorganic arsenic is ubiquitous in marine waters, ranging from 15 to 20 nmol/L in the open ocean (14, 15). Measurements of arsenic in the anoxic waters of the Eastern Tropical South Pacific (16), Black Sea (17), and the anoxic fjord Saanich Inlet, British Columbia (18) show thermodynamic disequilibrium of the arsenic species in the water column which may be due, in part, to the microbial biotransformations of arsenic species. In radioisotope arsenic tracer experiments carried out on the anoxic waters from Saanich Inlet, rates of arsenite oxidation greatly diminished when antibiotics were added to the seawater (18), suggesting microbially mediated impacts on arsenic transformations. We hypothesized that the apparent redox disequilibrium of arsenic species in anoxic marine waters may be due to the presence of arsenotrophic microbes.
Results and Discussion
Full-Length Arsenic Respiratory Genes in ODZ Metagenomes.
Samples of the marine microbial community were collected on an oceanographic research cruise through the Eastern Tropical North Pacific (ETNP) ODZ in April of 2012. Filtered samples were collected at two oxygen-deficient stations (Fig. 1 and SI Appendix, Fig. S1) (19) including multiple depths through the anoxic core of the ODZ water column at station 136, and both free-living and particle-attached (>30 µm) fractions at 120 m at station BB2/141 (20). Metagenomic reads were assembled into contigs and searched for complete gene sequences for enzymes capable of arsenite oxidation (aioAB) and dissimilatory arsenate reduction (arrAB). The alpha subunits of these metabolic arsenic cycling enzymes, aioA and arrA, are found within the DMSO reductase superfamily, also known as the complex iron sulfur molybdoenzymes. This superfamily also contains other enzymes critical in respiratory redox transformations like the nitrate reductases Nap and Nar, formate dehydrogenases Fdn and Fdh, polysulfide reductase Psr, and tetrathionate reductase Ttr, among others (21). Potential arsenotrophic genes were confirmed through phylogenetic inference within the larger DMSO reductase superfamily.
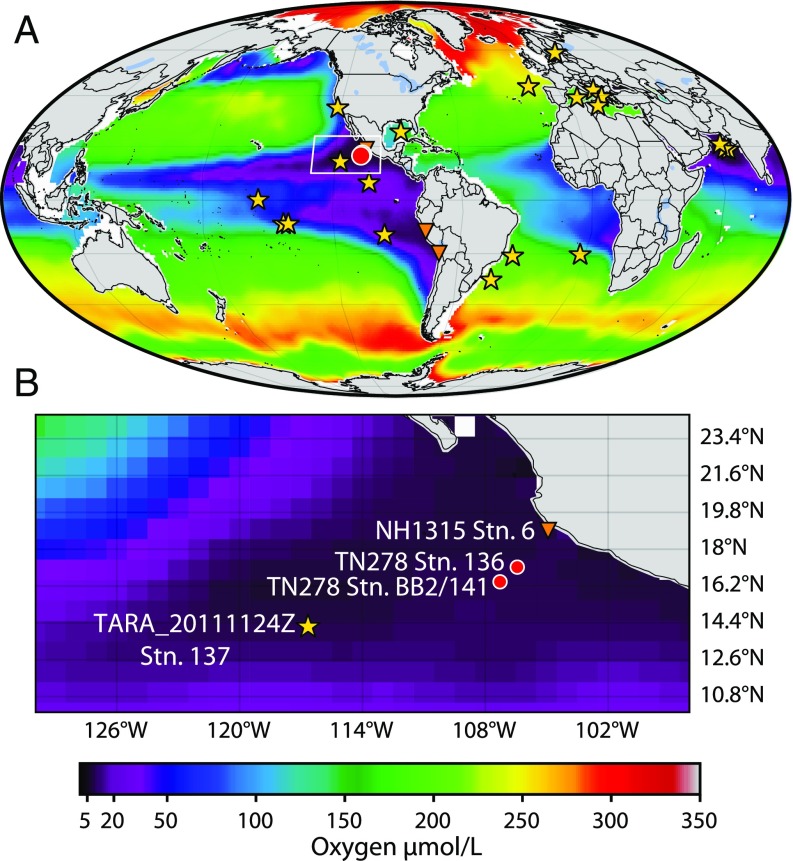
Fig. 1.
Maps displaying interpolated oxygen concentrations from 300-m depth from the World Ocean Atlas (19). (A) Global map with highlighted points. Red circles designate metagenomic sampling sites from locations described here. Orange triangles represent publicly available metatranscriptomes (34–36). Stars represent locations where arsenotrophy genes were identified in metagenomes from NCBI WGS database (SI Appendix, Table S2). The white box highlights the location of origin of metagenomes described here with a zoomed-in view of the region in B.
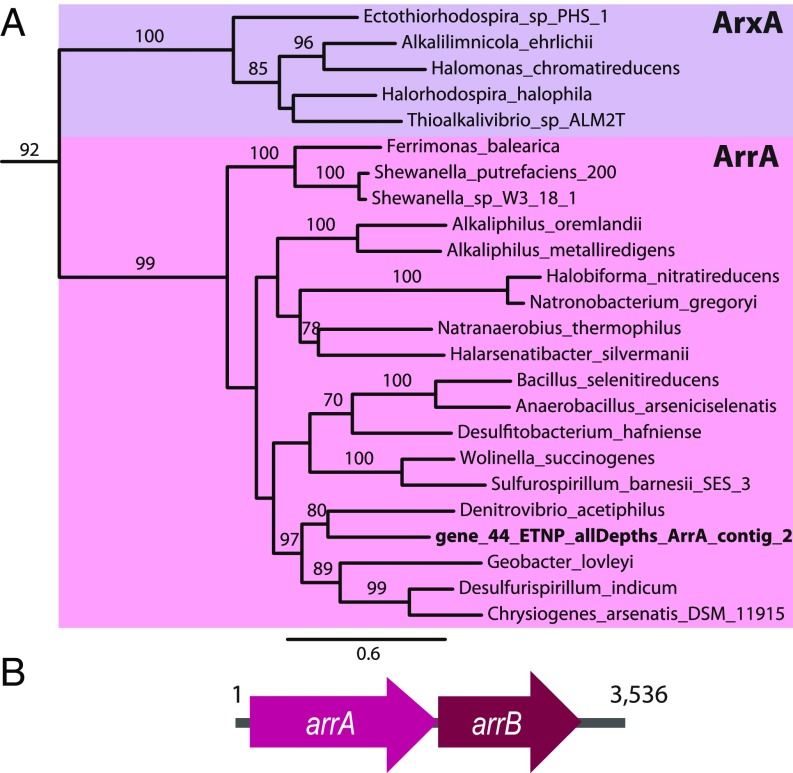
A complete dissimilatory arsenate reductase alpha subunit (arrA) sequence was identified in the ETNP ODZ metagenome assemblies (Fig. 2A); the beta subunit was also identified downstream of arrA on the same contig (Fig. 2B) and is phylogenetically closely related to the beta subunits of known ArrB sequences (SI Appendix, Fig. S2) (22). This genomic arrangement is also found in known dissimilatory arsenate-reducing prokaryotes (DARPs) (23), a phylogenetically diverse group of heterotrophs which obtain metabolic energy from the reduction of arsenate, typically using organic compounds as electron donors, although there are examples of hydrogen or reduced sulfur compounds used as electron donors (8). This ArrA sequence contains conserved residues for integral binding motifs found in the active site of functional respiratory arsenate reductase enzymes (SI Appendix, Fig. S3) (24). Notably, no reverse respiratory arsenate reductase (arxA) sequences were assembled––ArxA resembles the respiratory arsenate reductase of the DARPs (ArrA) in sequence, but functions in a reverse direction by oxidizing arsenite to arsenate (25).
Fig. 2.
Full-length dissimilatory arsenate reductase (ArrA) sequence identified in ETNP ODZ metagenomes. (A) A maximum-likelihood tree of ArrA and the closely related arsenite oxidase (ArxA) sequences including a full de novo assembled environmental ArrA sequence from the ETNP ODZ metagenome in bold. Bootstrap values ≥70 are shown. (B) The gene complement found along the contig ArrA_ETNP_allDepths_contig_2 showing the presence of a downstream arsenate reductase beta subunit arrB sequence.
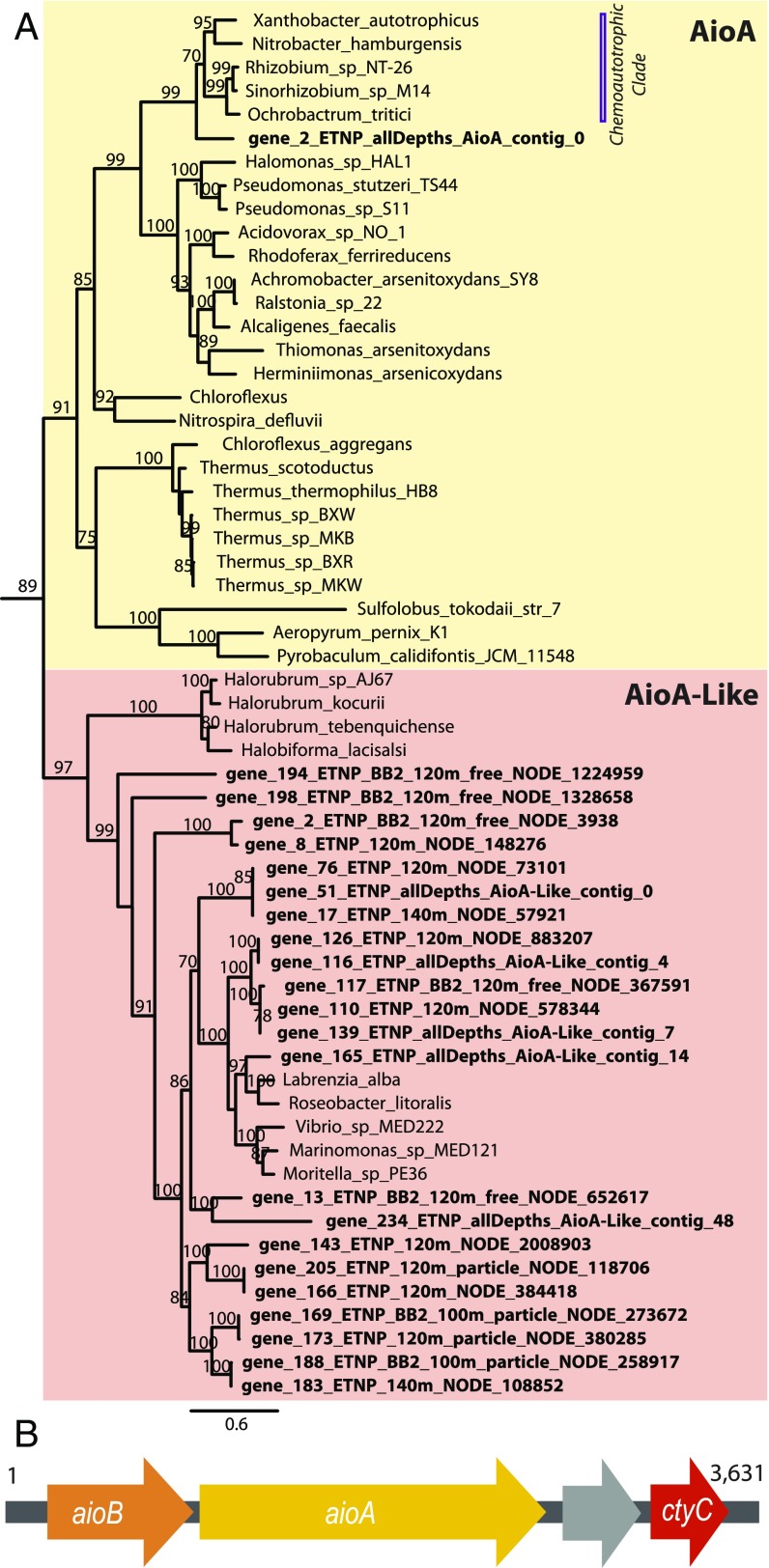
Genes for the complementary oxidation reaction in the arsenic respiratory cycle, arsenite oxidase, were also found in the ETNP ODZ. The large subunit of arsenite oxidase, AioA, is evolutionarily distinct from the ArrA/ArxA group and is found elsewhere on the DMSO reductase superfamily tree (9) (SI Appendix, Fig. S4). The Aio enzyme can be used in both dissimilatory arsenite oxidation, believed to be a detoxification strategy (8), and in chemoautotrophic arsenite oxidation, where metabolic gains are made from reduction of the respiratory chain via electron conservation during the oxidation of As(III) to As(V) (26). Most known chemoautotrophic arsenite oxidizers are strict mesophilic aerobes (8, 26); however, there are a few representatives which can utilize nitrate as the terminal electron acceptor (27–29), with some strains capable of complete denitrification by reduction of NO3 to N2 (29). Phylogenetic analysis of assembled sequences from the ETNP ODZ identified a complete aioA sequence that branched basally to a clade of sequences from Alphaproteobacteria in the Rhizobiales order with known chemoautotrophic arsenite oxidation capabilities (Fig. 3A). Genes encoding a beta subunit of arsenite oxidase (aioB) upstream and a complete cytochrome-c located downstream (Fig. 3B) were also present on the aioA-containing contig (22), a genomic organization similar to known arsenite oxidizers (26). The aioB subunit is also phylogenetically more closely related to beta subunits of chemoautotrophic arsenite oxidizers (SI Appendix, Fig. S5) than to dissimilatory arsenite oxidizers. Therefore, it is likely that the environmental assembled contig containing aioA is representative of a chemoautotrophic arsenite oxidation pathway and may be capable of denitrification.
Fig. 3.
Full-length respiratory arsenite oxidase (AioA) sequence identified in ETNP ODZ metagenomes. (A) A maximum-likelihood tree of AioA highlighted in yellow and the closely related enzyme of unknown function arsenite oxidase-like (AioA-Like) in red-orange. Full length de novo assembled environmental sequences from the ETNP are denoted by bold text. The clade which contains the environmental AioA sequence and sequences associated with chemoautotrophic activity is designated with a bar. Bootstrap values ≥70 are shown. (B) The gene complement found along the contig AioA_ETNP_allDepths_contig_0 showing the presence of the beta subunit sequence aioB and a cytochrome-C.
Numerous contigs assembled from the ETNP ODZ contained sequences closely related to AioA but branching just outside the internal node defining the AioA bacterial and archaeal clade in the DMSO reductase superfamily tree (Fig. 3A and SI Appendix, Fig. S4). This clade of sequences has been previously identified as “AioA-Like” (9) and consists of a subclade made up of a handful of Alpha- and Gammaproteobacterial sequences and another subclade of sequences recently identified in the haloarchaea (Euryarchaeota phylum) (30). The ETNP AioA-like sequences (20, 22) grouped more closely with the bacterial subclade than with the haloarchaeal group. It has been suggested that the bacterial aioA-like sequences arose via horizontal gene transfer from a Euryarchaeotal ancestor, a distinct event from the origin of aioA where the phylogenetic radiation is more congruent with the 16S rRNA phylogeny (30). An abundance of archaeal aioA-like sequences was identified in an haloarchaeal red biofilm in the arsenic-loaded Diamante Lake, Argentina and a culture of Halorubrum sp. DM2 isolated from the same location demonstrated aerobic growth on a minimal medium enriched with As(III) (31). However, the exact substrate of the AioA-Like enzyme has yet to be verified. Bacterial AioA-Like may indeed be a functional homolog to the arsenite oxidase subunit AioA. However, without further laboratory confirmation of the explicit function of this enzyme, we cannot definitively say whether this Aio-Like enzyme functions as an As(III) oxidase or whether its function has taken another evolutionary trajectory.
The longest contig containing aioA-like sequences was over 82 kilobases and provided additional insight into the metabolic capabilities of the host Proteobacterium (SI Appendix, Table S1). Assembled from a single metagenomic sample, contig ETNP_120m_NODE_73101 contains a nitrate reductase sequence (napA) which suggests the use of nitrate as an electron acceptor, as well as a gene encoding fumarate reductase, a key enzyme in the reductive TCA carbon fixation pathway (32), suggesting the potential for a chemoautotrophic lifestyle. The cooccurrence of these genes suggests that the Aio-Like enzyme may help support autotrophic carbon fixation and nitrate reduction in the ETNP ODZ.
To identify whether or not these arsenotrophy-related genes are found in other regions of the global ocean, we searched publicly available marine metagenome assemblies deposited in the National Center for Biotechnology Information (NCBI) Whole Genome Shotgun (WGS) repository (Fig. 1A and SI Appendix, Fig. S6 and Table S2). Genes for AioA, AioA-Like, and ArrA were all found on contigs assembled from oxygen-depleted waters of the Baltic Sea (33) and the aioA-like gene was also found in assemblies from the Arabian Sea ODZ. Notably, dissimilatory arsenate reductase, arrA, was only identified in assemblies from lower oxygen regions of the ocean, whereas aioA and aioA-like were also present in waters with higher oxygen concentrations such as the Eastern Tropical Pacific (aioA-like) and the oxygenated Mediterranean (aioA) (SI Appendix, Fig. S6 and Table S2) (33–35). The analysis of WGS assemblies shows the presence of aioA and aioA-like in genomes and thus the potential for microbial arsenite oxidation is not restricted only to oxygen deficient waters. The presence of these sequences in oxygenated waters may be the signature of aerobic chemoautotrophic arsenite oxidation (36), or may be associated with arsenic detoxification processes in regions of phosphate stress (8, 37).
Niche Partitioning of Arsenotrophy and Related Genes.
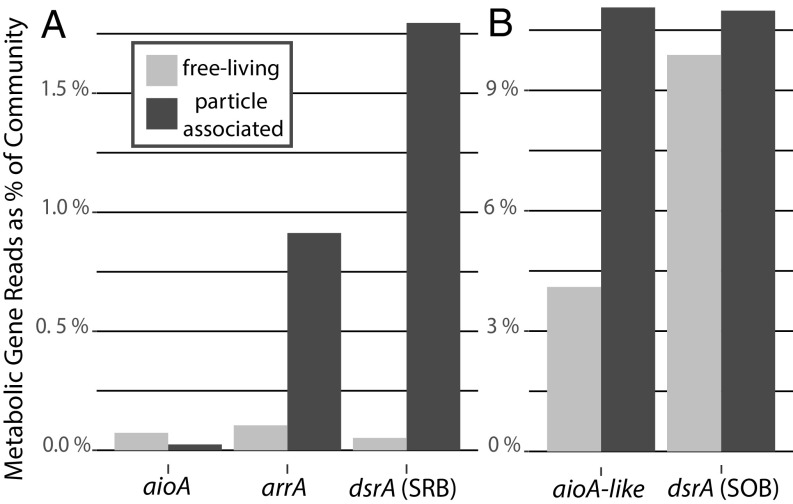
To determine the prevalence of arsenotrophic capabilities in microbial communities in the ETNP, we used a phylogenetically informed short-read placement approach to evaluate the frequency of metagenomic sequence reads associated with the genes aioA, aioA-like, and arrA compared with reads associated with the single-copy core gene RNA polymerase (rpoB) in free-living (<30 µm) or particle-associated (>30 µm) microbial communities at 120 m (Fig. 4). For comparison with a well-studied analogous metabolic cycle in ODZs, we also identified and enumerated short reads associated with the bacterial form of the sulfur-cycling gene dsrA (SI Appendix, Fig. S7), the forward form used for sulfur reduction and the reverse used for sulfur oxidation (38). The genes arrA, aioA, and forward bacterial dsrA of sulfate-reducing bacteria (SRB) were possessed by a small portion (<2%) of the microbial community, while the genes aioA-like and reverse bacterial dsrA associated with sulfur-oxidizing bacteria (SOB) were possessed by a larger fraction of the community (4–11%). Furthermore, the genes arrA, aioA-like, and forward bacterial dsrA (SRB) were more abundant in the particle-associated communities compared with the free-living fraction. It makes sense to find arrA and dsrA (SRB) inhabiting similar functional environments as they are likely both exploiting similar redox-driven niches in particles, coupling the heterotrophic breakdown of organic matter with the reduction of arsenate or oxidized sulfur compounds in the absence of more energetically favorable electron acceptors, like nitrate, which are available to the free-living microbial community (3).
Fig. 4.
Arsenic respiratory genes and other related respiratory genes show niche partitioning in genomic contribution to microbial communities among free-living (<30 μm) or particle-associated niches (>30 μm) at 120-m depth in ETNP ODZ metagenomes. Reads identified using phylogenetically informed short-read placement. Contribution of reads presented as the length-normalized number of reads associated with each target gene/the number of length-normalized RNA polymerase B, rpoB, reads in the metagenomes * 100% to give an estimate of the gene’s % contribution to the overall microbial community metagenome (53). The relative proportion of a gene’s contribution to the microbial community when the free-living and particulate fractions are compared broken up into A with a much smaller proportion of the community containing arsenotrophy genes arsenite oxidase, aioA, and dissimilatory arsenate reductase, arrA, as well as the forward version of dissimilatory sulfite reductase, dsrA, which is involved in SRB. B shows a larger proportion of the community possesses the sequences aioA-like and the reverse version of dsrA which is involved in SOB.
Active Gene Expression of Arsenotrophy and Related Genes.
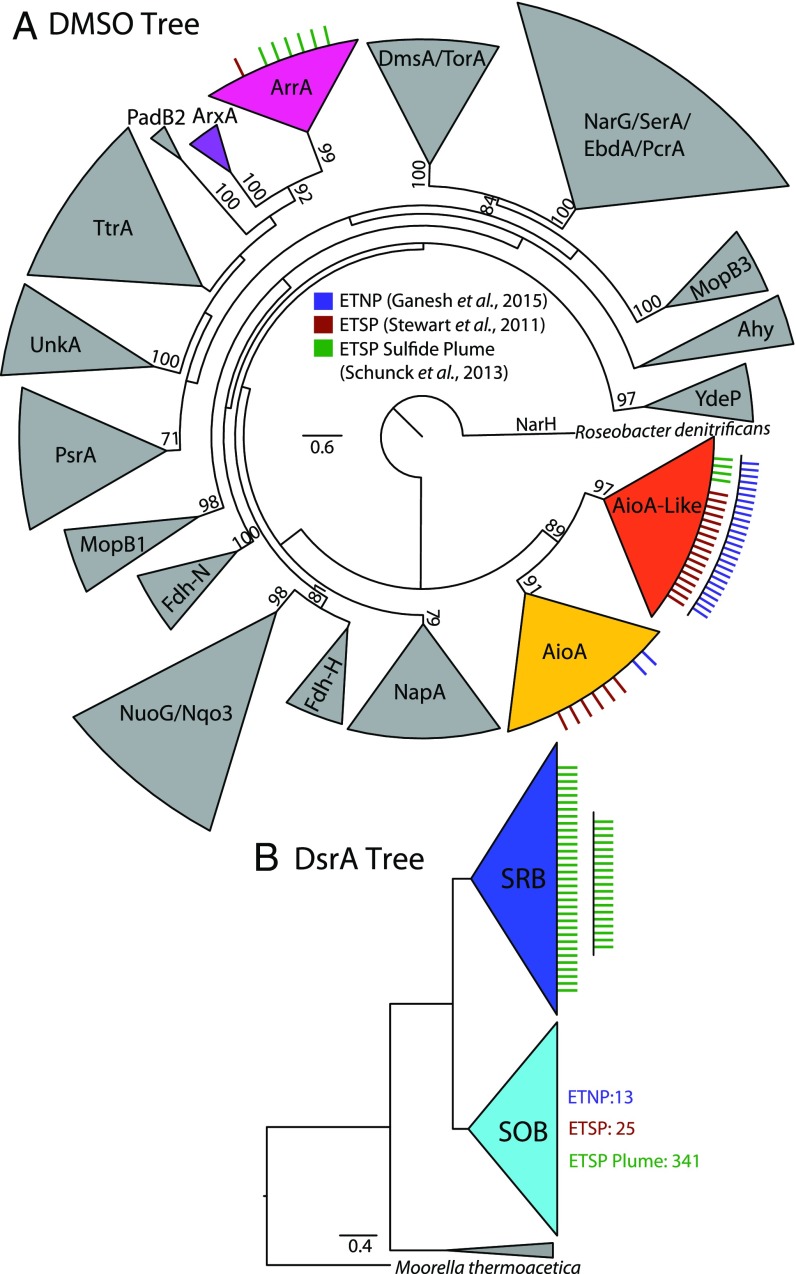
We analyzed publicly available metatranscriptomes in global ODZs (39–41) to identify whether arsenotrophic and arsenotrophy-related genes are transcriptionally active in these environments. Again using phylogenetically informed short read placement, we identified transcripts associated with arrA, aioA, and aioA-like from metatranscriptomic libraries in the ETNP (39) and two areas of the Eastern Tropical South Pacific (ETSP) (40, 41) ODZs––one ETSP library focused on the smallest free-living microbial community (0.22–1.6 µm) of a typical ODZ water column (41) and the other captured the free-living and small particle associated microbial community (0.22–10 µm) within a transient sulfide plume feature (40). Again, for comparison, we also identified transcripts associated with bacterial dsrA. Of the arsenotrophic genes, transcripts associated with aioA-like were by far the most abundant (Fig. 5A) and found in both the ETNP and the ETSP ODZs. Transcript sequences associated with aioA were also found in both the ETNP and ETSP. Dissimilatory arsenate reductase arrA and forward bacterial dsrA (SRB) transcripts were only detected in the ETSP, with the majority of sequences identified from within the sulfidic plume (Fig. 5B) (40). Thus, arsenotrophy-related genes are being expressed at a transcriptional level comparable to the forward bacterial dsrA (SRB), suggesting the active use of these metabolic pathways. The gene of unknown function aioA-like was transcriptionally active on the same order as the sulfur-oxidizing form of dsrA (SOB) in the nonsulfidic ETNP and ETSP metatranscriptomes, again suggesting that this as yet to be determined Aio-Like pathway may be of similar significance in these waters.
Fig. 5.
(A) A collapsed view of the DMSO reductase family tree (SI Appendix, Fig. S4), with associated short reads from metatranscriptomes (34–36) identified as associated with arsenotrophic enzymes through phylogenetically informed placement represented by tick marks mapped onto their respective clades; reads associated with other enzymes are not shown. (B) A phylogenetic tree of DsrA showing transcripts associated with forward bacterial DsrA associated with dissimilatory SRB and the reverse DsrA associated with SOB mapped along the respective clades for comparison with the transcripts associated with arsenotrophy in these ODZ regions captured by the publicly available transcriptomes from ODZs (34–36). Specific read identifiers mapped to these clades can be found in SI Appendix, Table S3.
Conclusion and Implications.
The presence of complete arsenotrophic gene pathways in the metagenome of the ETNP ODZ suggests microbial communities in these anoxic waters are capable of a complete bioenergetic arsenotrophic cycle. Gene expression of the arsenotrophy genes arrA and aioA indicates arsenotrophy is a viable metabolic pathway that is actively utilized in anoxic water columns. Chemoautotrophic arsenite oxidation may provide an additional fixed carbon source, contributing to the biochemical processes driving the reducing environment of ODZs. Nitrogen loss through the reduction of nitrogenous compounds to N2 has been demonstrated by some chemoautotrophic arsenite oxidizers (29); if arsenite oxidizers in the ETNP are also capable of denitrification they would impact nitrogen cycling in ODZs. The enrichment of both arsenic and sulfate reduction genes in the particulate fraction identified here is an example of redox-driven niche partitioning on sinking particles, which have zones with more reducing potential than found in the open water column (3). The cryptic arsenic cycle described here is an account of arsenotrophic cycling in an environment with nanomolar levels of available arsenic (8) suggesting these arsenic respiratory pathways may be more abundant in the environment than previously estimated.
The identification of arsenotrophic microbes living in modern marine ODZs will enable proxies for arsenic-based metabolisms in early anoxic oceans. Marine arsenic concentrations have fluctuated over geologic time, with the anoxic Precambrian ocean experiencing periods of greater arsenic load than the modern-day ocean (13) due in part to enhanced weathering of continental arsenic sulfide minerals (12). Arsenotrophic organisms may have flourished during periods of higher arsenic concentrations in an anoxic ocean, with chemoautotrophic or possibly photosynthetic arsenite oxidation supporting carbon fixation. Thus, integrating over the time and spatial scale of early anoxic oceans, arsenic-based metabolisms may have had significant implications for the biogeochemical cycling of arsenic, carbon, and nitrogen. The work described here presents a foundation to explore these metabolic pathways and their implications for biogeochemical cycling and microbial evolution in the modern-day ocean and oceans of the past.
Methods
Samples were collected during a research cruise to the ETNP in April 2012 aboard the research vessel (R/V) Thompson (cruise TN278) using 10-L Niskin bottles in a 24-bottle conductivity, temperature, and depth (CTD) water sampling rosette. The top of the ODZ was determined to be at 105 m based upon oxygen concentrations determined by switchable trace oxygen (STOX) sensor (5). Oxygen and nutrient measurements were collected as previously described (20, 42).
DNA samples were obtained from station 136 (106.543° W 17.043° N) in the anoxic zone at multiple depths (100, 110, 120, 160, 180 m). Two liters of Niskin water was vacuum filtered onto a 0.2-µm SUPOR filter. At the offshore multiday station BB2/141 (107.148° W 16.527° N), approximately four liters of water from 120 m were first filtered through a >30-µm nylon filter and the prefiltered (<30 µm) water was then filtered onto 0.2-µm Supor membrane filters (PALL Corporation). Filters were immediately frozen in a −80 °C freezer and transported on dry ice from the ship to the University of Washington where they were maintained at −80 °C.
DNA library preparation and sequencing with Illumina HiSeq 2500 were conducted in 2014 and previously described in another publication (20). DNA samples were extracted using freeze thaw followed by incubation with lysozyme and proteinase K and phenol/chloroform extraction. A Rubicon THRUPLEX kit was used for library prep using 50 ng of DNA per sample. Two libraries (ETNP_100m & ETNP_110m) were sequenced on a HiSeq 2500 in rapid mode (∼22 million 150-bp paired-end read pairs per sample) at Michigan State University. The other five libraries (ETNP_120m, ETNP_160m, ETNP_180m, ETNP_BB2_120m_free, and ETNP_BB2_120m_particle) were sequenced on a HiSeq 2500 high-output mode (∼55–150 million 125-bp paired-end read pairs per sample) at the University of Utah. Sequences were quality checked and trimmed using Trimmomatic (43).
Metagenomic reads from each depth sample were first assembled separately, processed with diginorm (44) to normalize coverage, and then de novo assembled with the VELVET assembler (45). Contigs assembled from individual depths as previously described (20) were functionally annotated using the prokka annotation pipeline (46) with reads and assembled contigs deposited at NCBI GenBank under bioproject PRJNA350692 v.1 assemblies (SI Appendix, Table S4 lists accessions for each assembly). Prokka alone was insufficient to identify DMSO reductase family-related sequences (aioA, aioA-like, arrA, and arxA). To identify these arsenotrophy-related enzymes, blast databases of all assembled contigs were created using blast version 2.2.28 (47, 48) and queried with tblastn using known reference queries. Contigs which had hits to arsenotrophy-related sequences with e values ≤ 1 × 10−50 were parsed out, and submitted to MetaGeneMark for gene prediction (49, 50). Potential genes that overlapped with the region of best blast hit were then added to the DMSO reductase tree for further identification, with full-length sequences that grouped with arsenotrophy-related clades depicted on the DMSO reductase tree (SI Appendix, Fig. S4).
To assemble the longest possible arsenotrophy-related contigs, a targeted assembly approach was used to combine metagenomes from all anoxic samples. Partial and full arsenotrophy-related sequences identified from assembled contigs from individual depths and reference regions from known arsenotrophy-related sequences (gene sequence with 10-kb regions up- and downstream) were used as queries for sequence read recruitment from each individual depth metagenome using the program FR-HIT (51). Reads recruited with FR-HIT were then assembled with the de novo assembler IDBA-UD (52) optimized for assembling data of uneven depth coverage. Arsenotrophy-related genes from these contigs were identified as above. In addition, all genes called on contigs identified as containing full-length arsenotrophy-related enzymes were annotated with InterProScan (53) with general taxonomic identity on a gene-by-gene basis being inferred at the bacterial class level through a blastn best hit to the nr/nt database. The assembled arrA amino acid sequence was then pairwise aligned with that from Shewanella ANA-3 using Jalview 2 (24, 54).
Gene trees were constructed with amino acid translated sequences from metagenomic assemblies and known reference sequences aligned with the program MUSCLE version 3.6 (55). Phylogenetic trees were then constructed with the program RAxML version 8.0.23 (56). A maximum-likelihood tree was inferred from the best of 20 trees using the amino acid substitution matrix model which resulted in the best likelihood score during a trial run (DMSO: BLOSUM62F; DsrA, AioB, and ArrB: WAGF) along with empirical character frequencies and a gamma model of rate heterogeneity using the RAxML estimated alpha value. Bootstrap analyses were conducted on all trees at n = 100. The DMSO reductase family tree and DsrA tree were used as reference trees for phylogenetic placement of metagenomic and metatranscriptomic reads.
Publicly available pelagic marine metagenomic assemblies in the NCBI WGS database were searched for the presence of arrA, aioA, and aioA-like. Multiple amino acid query sequences spanning the taxonomic range of these genes were used to identify potential genes in WGS using the SRA toolkit tool tblastn_vdb to remotely blast search assembled metagenomes annotated as marine metagenomes from seawater. Hits receiving an e value ≤ 1 × 10−80 were selected and their contig sequences obtained from WGS using Batch Entrez. Contigs were then submitted to MetaGeneMark for gene prediction (49, 50). Potential full-length arsenotrophy genes that overlapped with the region of best blast hit were added to the DMSO reductase tree for further identification.
Recruitment and placement of metagenomic reads from the samples at 120-m depth from station BB2/141 comparing the >30-µm (particulate) and <30-µm (free-living) fractions was conducted. Reads were combined into a single blast database using blast 2.2.28 (47, 48) and queried with tblastn (e-value cutoff 1 × 10−5) using full-length known reference sequences, including full-length identified environmental arsenotrophy sequences assembled in this work. Translation of blast recruited reads which were at least 33 amino acids in length after quality trimming were then identified using a phylogenetically informed placement approach by comparison with known reference sequences (20, 37, 57–59). To obtain the abundance of these reads relative to the overall prokaryotic community in the two size fractions, the length-normalized number of identified read pairs for each gene was compared with the length-normalized abundance of the single-copy core gene RNA polymerase (rpoB) in the sequenced prokaryotic community (20, 37).
A similar read recruitment and phylogenetic placement method was used to identify reads from ODZ metatranscriptomes (37, 57–59). Publicly available metatranscriptomes from oxygen deficient zones in the ETNP (39), ETSP (41), and within a sulfide plume in the ETSP (40) were downloaded locally. Reads were also recruited for identification of bacterial dsrA for comparison.
Supplementary Material
Acknowledgments
We thank John Baross and Rika Anderson for helpful discussions and feedback on this project. We also thank the chief scientists of the research cruise, Al Devol and Bess Ward, as well as the captain and crew of the R/V Thomas G. Thompson. This work was supported through a NASA Earth and Space Sciences Graduate Research Fellowship to J.K.S. and National Science Foundation Grant OCE-1138368 (to G.R.).
Footnotes
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (GenBank Bio Project PRJNA350692).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818349116/-/DCSupplemental.
References
- 1.Lyons TW, Reinhard CT, Planavsky NJ. The rise of oxygen in Earth’s early ocean and atmosphere. Nature. 2014;506:307–315. doi: 10.1038/nature13068. [DOI] [PubMed] [Google Scholar]
- 2.Ulloa O, Canfield DE, DeLong EF, Letelier RM, Stewart FJ. Microbial oceanography of anoxic oxygen minimum zones. Proc Natl Acad Sci USA. 2012;109:15996–16003. doi: 10.1073/pnas.1205009109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright JJ, Konwar KM, Hallam SJ. Microbial ecology of expanding oxygen minimum zones. Nat Rev Microbiol. 2012;10:381–394. doi: 10.1038/nrmicro2778. [DOI] [PubMed] [Google Scholar]
- 4.Wyrtki K. The oxygen minima in relation to ocean circulation. Deep-Sea Res Oceanogr Abstr. 1962;9:11–23. [Google Scholar]
- 5.Tiano L, et al. Oxygen distribution and aerobic respiration in the north and south eastern tropical Pacific oxygen minimum zones. Deep Sea Res Part I Oceanogr Res Pap. 2014;94:173–183. [Google Scholar]
- 6.Canfield DE, et al. A cryptic sulfur cycle in oxygen-minimum-zone waters off the Chilean coast. Science. 2010;330:1375–1378. doi: 10.1126/science.1196889. [DOI] [PubMed] [Google Scholar]
- 7.Oremland RS, Stolz JF. The ecology of arsenic. Science. 2003;300:939–944. doi: 10.1126/science.1081903. [DOI] [PubMed] [Google Scholar]
- 8.Amend JP, Saltikov C, Lu G-S, Hernandez J. Microbial arsenic metabolism and reaction energetics. Rev Mineral Geochem. 2014;79:391–433. [Google Scholar]
- 9.Duval S, Ducluzeau AL, Nitschke W, Schoepp-Cothenet B. Enzyme phylogenies as markers for the oxidation state of the environment: The case of respiratory arsenate reductase and related enzymes. BMC Evol Biol. 2008;8:206. doi: 10.1186/1471-2148-8-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lebrun E, et al. Arsenite oxidase, an ancient bioenergetic enzyme. Mol Biol Evol. 2003;20:686–693. doi: 10.1093/molbev/msg071. [DOI] [PubMed] [Google Scholar]
- 11.Sforna MC, et al. Evidence for arsenic metabolism and cycling by microorganisms 2.7 billion years ago. Nat Geosci. 2014;7:811–815. [Google Scholar]
- 12.Fru EC, et al. Arsenic stress after the Proterozoic glaciations. Sci Rep. 2015;5:17789. doi: 10.1038/srep17789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergman IA, Kolesov GM. Arsenic, antimony, and bismuth as indicators of the genesis of ore material in Early Precambrian ferrous quartzite formations. Geochem Int. 2012;50:816–831. [Google Scholar]
- 14.Ellwood MJ, Maher WA. An automated hydride generation-cryogenic trapping-ICP-MS system for measuring inorganic and methylated Ge, Sb and As species in marine and fresh waters. J Anal At Spectrom. 2002;17:197–203. [Google Scholar]
- 15.Cutter GA, Cutter LS. Biogeochemistry of arsenic and antimony in the North Pacific Ocean. Geochem Geophys Geosyst. 2006;7:Q05M08. [Google Scholar]
- 16.Cutter GA, Moffett JG, Nielsdóttir MC, Sanial V. Multiple oxidation state trace elements in suboxic waters off Peru: In situ redox processes and advective/diffusive horizontal transport. Mar Chem. 2018;201:77–89. [Google Scholar]
- 17.Cutter GA. Kinetic controls on metalloid speciation in seawater. Mar Chem. 1992;40:65–80. [Google Scholar]
- 18.Peterson ML, Carpenter R. Biogeochemical process affecting total arsenic and arsenic species distribution in an intermittently anoxic fjord. Mar Chem. 1983;12:295–321. [Google Scholar]
- 19.Garcia H, et al. World Ocean Atlas 2013, volume 4: Dissolved inorganic nutrients (phosphate, nitrate, silicate) NOAA Atlas NESDIS. 2014;76:25. [Google Scholar]
- 20.Fuchsman CA, Devol AH, Saunders JK, McKay C, Rocap G. Niche partitioning of the N cycling microbial community of an offshore oxygen deficient zone. Front Microbiol. 2017;8:2384. doi: 10.3389/fmicb.2017.02384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothery RA, Workun GJ, Weiner JH. The prokaryotic complex iron-sulfur molybdoenzyme family. Biochim Biophys Acta. 2008;1778:1897–1929. doi: 10.1016/j.bbamem.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Saunders JK, Fuchsman CA, McKay C, Rocap G. 2016 Arsenic respiratory pathways in the anoxic pelagic waters of the Pacific Ocean. GenBank. Available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA350692. Deposited December 22, 2016.
- 23.Andres J, Bertin PN. The microbial genomics of arsenic. FEMS Microbiol Rev. 2016;40:299–322. doi: 10.1093/femsre/fuv050. [DOI] [PubMed] [Google Scholar]
- 24.Glasser NR, Oyala PH, Osborne TH, Santini JM, Newman DK. Structural and mechanistic analysis of the arsenate respiratory reductase provides insight into environmental arsenic transformations. Proc Natl Acad Sci USA. 2018;115:E8614–E8623. doi: 10.1073/pnas.1807984115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richey C, et al. Respiratory arsenate reductase as a bidirectional enzyme. Biochem Biophys Res Commun. 2009;382:298–302. doi: 10.1016/j.bbrc.2009.03.045. [DOI] [PubMed] [Google Scholar]
- 26.van Lis R, Nitschke W, Duval S, Schoepp-Cothenet B. Arsenics as bioenergetic substrates. Biochim Biophys Acta. 2013;1827:176–188. doi: 10.1016/j.bbabio.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Oremland RS, et al. Anaerobic oxidation of arsenite in Mono Lake water and by a facultative, arsenite-oxidizing chemoautotroph, strain MLHE-1. Appl Environ Microbiol. 2002;68:4795–4802. doi: 10.1128/AEM.68.10.4795-4802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhine ED, Ní Chadhain SM, Zylstra GJ, Young LY. The arsenite oxidase genes (aroAB) in novel chemoautotrophic arsenite oxidizers. Biochem Biophys Res Commun. 2007;354:662–667. doi: 10.1016/j.bbrc.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Rhine ED, Phelps CD, Young LY. Anaerobic arsenite oxidation by novel denitrifying isolates. Environ Microbiol. 2006;8:899–908. doi: 10.1111/j.1462-2920.2005.00977.x. [DOI] [PubMed] [Google Scholar]
- 30.Rascovan N, Maldonado J, Vazquez MP, Eugenia Farías M. Metagenomic study of red biofilms from Diamante Lake reveals ancient arsenic bioenergetics in haloarchaea. ISME J. 2016;10:299–309. doi: 10.1038/ismej.2015.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ordoñez OF, Rasuk MC, Soria MN, Contreras M, Farías ME. Haloarchaea from the Andean Puna: Biological role in the energy metabolism of arsenic. Microb Ecol. 2018;76:695–705. doi: 10.1007/s00248-018-1159-3. [DOI] [PubMed] [Google Scholar]
- 32.Hügler M, Sievert SM. Beyond the Calvin cycle: Autotrophic carbon fixation in the ocean. Annu Rev Mar Sci. 2011;3:261–289. doi: 10.1146/annurev-marine-120709-142712. [DOI] [PubMed] [Google Scholar]
- 33.Dupont CL, et al. Functional tradeoffs underpin salinity-driven divergence in microbial community composition. PLoS One. 2014;9:e89549. doi: 10.1371/journal.pone.0089549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeigler Allen L, et al. Influence of nutrients and currents on the genomic composition of microbes across an upwelling mosaic. ISME J. 2012;6:1403–1414. doi: 10.1038/ismej.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sunagawa S, et al. Tara Oceans Coordinators Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 36.Santini JM, Sly LI, Schnagl RD, Macy JM. A new chemolithoautotrophic arsenite-oxidizing bacterium isolated from a gold mine: Phylogenetic, physiological, and preliminary biochemical studies. Appl Environ Microbiol. 2000;66:92–97. doi: 10.1128/aem.66.1.92-97.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saunders JK, Rocap G. Genomic potential for arsenic efflux and methylation varies among global Prochlorococcus populations. ISME J. 2016;10:197–209. doi: 10.1038/ismej.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müller AL, Kjeldsen KU, Rattei T, Pester M, Loy A. Phylogenetic and environmental diversity of DsrAB-type dissimilatory (bi)sulfite reductases. ISME J. 2015;9:1152–1165. doi: 10.1038/ismej.2014.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ganesh S, et al. Size-fraction partitioning of community gene transcription and nitrogen metabolism in a marine oxygen minimum zone. ISME J. 2015;9:2682–2696. doi: 10.1038/ismej.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schunck H, et al. Giant hydrogen sulfide plume in the oxygen minimum zone off Peru supports chemolithoautotrophy. PLoS One. 2013;8:e68661. doi: 10.1371/journal.pone.0068661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart FJ, Ulloa O, DeLong EF. Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol. 2012;14:23–40. doi: 10.1111/j.1462-2920.2010.02400.x. [DOI] [PubMed] [Google Scholar]
- 42.Devol AH. 2013. Physical, chemical and biological CTD and bottle data from R/V Thomas G. Thompson cruise TN278 in eastern tropical North Pacific Ocean (National Centers for Environmental Information, National Oceanographic and Atmospheric Administration, Asheville, NC)
- 43.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown CT, Howe A, Zhang Q, Pyrkosz AB, Brom TH. 2012. A reference-free algorithm for computational normalization of shotgun sequencing data. arXiv:1203.4802 v2. Preprint, posted May 21, 2012.
- 45.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. [Google Scholar]
- 47.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 48.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Besemer J, Borodovsky M. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 1999;27:3911–3920. doi: 10.1093/nar/27.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niu B, Zhu Z, Fu L, Wu S, Li W. FR-HIT, a very fast program to recruit metagenomic reads to homologous reference genomes. Bioinformatics. 2011;27:1704–1705. doi: 10.1093/bioinformatics/btr252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng Y, Leung HC, Yiu SM, Chin FY. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 53.Jones P, et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2–A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berger SA, Stamatakis A. Aligning short reads to reference alignments and trees. Bioinformatics. 2011;27:2068–2075. doi: 10.1093/bioinformatics/btr320. [DOI] [PubMed] [Google Scholar]
- 58.Berger SA, Krompass D, Stamatakis A. Performance, accuracy, and Web server for evolutionary placement of short sequence reads under maximum likelihood. Syst Biol. 2011;60:291–302. doi: 10.1093/sysbio/syr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berger SA, Stamatakis A. 2012 PaPaRa 2.0: A vectorized algorithm for probabilistic phylogeny-aware alignment extension. Heidelberg Institute for Theoretical Studies. Available at https://cme.h-its.org/exelixis/pubs/Exelixis-RRDR-2012-5.pdf. Accessed August 31, 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.