Significance
The Hippo signaling pathway regulates cell proliferation in response to cell contact and a variety of other extracellular stimuli. It controls the activity and nuclear localization of cotranscriptional activator YAP, which interacts with DNA binding transcription factor TEAD, for the expression of target genes involved in cell proliferation. We show here that the expression level and transcriptional activity of TEAD are actively controlled by cell density through the modulation of its palmitoylation status. TEAD palmitoylation is controlled via fatty acid synthase and depalmitoylases in response to cell density. Our study indicates that the regulation of palmitoylation status is a potential target for controlling TEAD-dependent processes, perhaps including cancer growth.
Keywords: Hippo signaling, TEAD, palmitoylation, fatty acid synthase, depalmitoylase
Abstract
The Hippo pathway is involved in regulating contact inhibition of proliferation and organ size control and responds to various physical and biochemical stimuli. It is a kinase cascade that negatively regulates the activity of cotranscription factors YAP and TAZ, which interact with DNA binding transcription factors including TEAD and activate the expression of target genes. In this study, we show that the palmitoylation of TEAD, which controls the activity and stability of TEAD proteins, is actively regulated by cell density independent of Lats, the key kinase of the Hippo pathway. The expression of fatty acid synthase and acetyl-CoA carboxylase involved in de novo biosynthesis of palmitate is reduced by cell density in an Nf2/Merlin-dependent manner. Depalmitoylation of TEAD is mediated by depalmitoylases including APT2 and ABHD17A. Palmitoylation-deficient TEAD4 mutant is unstable and degraded by proteasome through the activity of the E3 ubiquitin ligase CHIP. These findings show that TEAD activity is tightly controlled through the regulation of palmitoylation and stability via the orchestration of FASN, depalmitoylases, and E3 ubiquitin ligase in response to cell contact.
The Hippo signaling pathway contributes to the regulation of contact inhibition of proliferation, cell differentiation, and organ size control (1). The pathway is comprised of a highly conserved kinase cascade that leads to the activation of Lats1/2 (Large tumor suppressor kinase 1/2), which regulates the activity and subcellular localization of transcriptional coactivators YAP (Yes-associated protein) and its paralogue TAZ (transcriptional coactivator with PDZ-binding motif). Mst1/2 (Mammalian Ste20-like kinases 1/2) and MAP4Ks (Mitogen-activated protein kinase kinase kinase kinases) phosphorylate and activate the Lats1/2 kinases in response to a number of upstream signals (2–5). In a growth permissive state, YAP/TAZ are translocated into the nucleus where they interact with various DNA binding transcription factors, especially TEADs (TEA domain family members), but also p73, Runx2, and ERBB4 (6). Because YAP/TAZ lack the DNA binding domain, the transcription of their downstream target genes is determined by DNA binding transcription factors.
TEAD is required for YAP/TAZ to transcribe the target genes involved in cell proliferation, oncogenic transformation, and epithelial–mesenchymal transition (7). Mammals express four TEAD genes (TEAD1–4) with similar domain architectures: the N-terminal DNA binding domain, and the C-terminal protein interaction domain, which binds to various transcriptional coactivators such as YAP/TAZ and the Vestigial-like (VGLL) (8). Although they have identical DNA binding domains and recognize the same DNA sequence, each TEAD shows tissue-specific and developmental stage-specific expression patterns (9, 10). This suggests a unique biological function for each TEAD protein. In addition, the activity and subcellular localization of TEAD is regulated by biological conditions and during development (8, 11–13).
Recent studies suggest that palmitoylation of TEAD is important for its stability and activity (14, 15). However, it is not known whether the state of TEAD palmitoylation is regulated in the cell or controlled by upstream signals that affect the pathway. S-palmitoylation is a posttranslational modification of proteins in which a saturated fatty acid, palmitate, is attached via thioester linkage to cysteine residue and, less frequently, to serine and threonine residues of protein (16, 17). Palmitate is supplied exogenously by diet or de novo biosynthesized by fatty acid synthase (FASN) (18), the only mammalian enzyme that catalyzes palmitate from acetyl CoA, malonyl-CoA, and NADPH. Unlike other lipid modifications, such as prenylation and myristoylation, the palmitoylation process is reversible and can be dynamically modulated in response to cell stimulation (16). Some proteins, including TEAD, undergo nonenzymatic autopalmitoylation by direct binding to palmitoyl-CoA (14). The protein palmitoylation process can also be mediated by the palmitoyl-acyl-transferase (PAT) enzymes, which transfer palmitoyl group to -SH group on a target protein (17). The palmitate turnover of palmitoylated proteins is regulated by depalmitoylases, such as acyl-protein thioesterase (APT), which remove palmitoyl group from target proteins (17).
Protein palmitoylation contributes to membrane association and, by enhancing hydrophobicity, exerts pleotropic effects on protein–protein interaction, protein activity, aggregation, clustering, and stability (16, 17). Approximately 2,000 proteins including EGFR, H/N-Ras, Cdc42, and PSD-95 are known to be palmitoylated in mammals (19). Recent studies have shown that the activity of TEAD1 and stability of TEAD2 protein are regulated by palmitoylation, the mutation of palmitoylated cysteine residues preventing its binding to YAP and decreasing its stability (14, 15). However, the regulatory mechanism that dynamically controls TEAD palmitoylation and its biological roles has not yet been identified. In this study, we show that the cell contact or Nf2/merlin-dependent regulation of TEAD palmitoylation is controlled by the expression of FASN, depalmitoylases, and E3 ubiquitin ligase.
Results
Density and Nf2-Dependent Regulation of TEAD Activity Is Controlled in a Lats-Independent Manner.
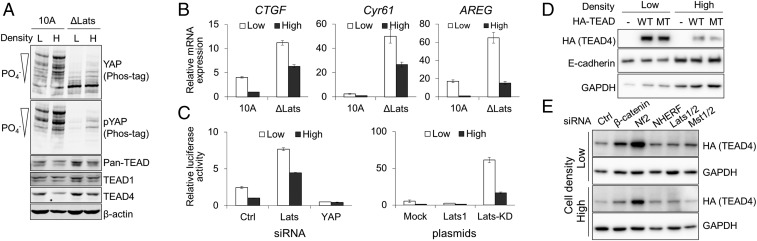
The phosphorylation and localization of YAP are controlled by cell density via the Hippo pathway. CRISPR-mediated knockout (KO) of Lats1/2, the core kinase of the Hippo pathway, in an immortalized human mammary epithelial cell line, MCF-10A, decreased the phosphorylation of YAP in sparse and dense cell cultures (Fig. 1A). However, the expression of YAP target gene mRNAs was reduced by increases in cell density even in Lats KO MCF-10A cells (Fig. 1B). Additionally, the depletion of Lats1/2 or overexpression of dominant negative Lats1 strongly increased the YAP-TEAD transcriptional reporter activity in cells at low density; however, the reporter activity was significantly reduced in cells at high density (Fig. 1C). This implies the presence of another regulatory pathway, in addition to the Lats-dependent regulation of YAP activity, which inhibits the transcriptional activation of YAP-TEAD target genes in cells at high density.
Fig. 1.
Density-dependent regulation of YAP-TEAD activity. (A) The effects of cell density on YAP and TEAD protein expression. Parental (10A) and Lats1/2 KO (ΔLats) MCF-10A cells were grown at low (L) or high (H) cell densities. Cells were lysed, and cell lysates were separated on regular or 25 μM phos-tag conjugated SDS/PAGE and subjected to Western blot analysis with the indicated antibodies. The phosphorylation status of YAP was evaluated by using phos-tag analysis with anti-YAP or anti–phospho-YAP Ser127 antibodies. (B) Quantitative analysis of YAP-TEAD target genes’ mRNA expression. The expression of known YAP-TEAD target genes CTGF, Cyr61, and AREG in parental and Lats1/2 KO MCF-10A cells grown at low and high densities were analyzed using qPCR. (C) YAP-TEAD Reporter assay. 293T cells were transfected with control, Lats1/2, or YAP siRNA and HOP-flash reporter, a YAP/TAZ-TEAD activity reporter (41). Cells were harvested and reseeded at different densities, and luciferase activity was measured. Error bars represent SD. 293T cells were transfected with pcDNA3, p2xFlag-Lats1, or p2xFlag-Lats1-KD (K734M), a dominant negative mutant of Lats1, together with HOP-flash reporter and cultured at different cell densities before the luciferase assay. (D) The expression of exogenous TEAD4. Parental MCF-10A cells and MCF-10A cells stably expressing HA-tagged wild-type (WT) or YAP binding-deficient mutant (MT) TEAD4 were grown at different cell densities. E-cadherin and GAPDH were used as internal controls. (E) Regulation of exogenous TEAD4 protein expression by depletion of Hippo signaling components. MCF-10A cells stably expressing HA-tagged TEAD4 were transfected with indicated siRNAs and cultured at low or high cell densities.
Because TEAD is a major DNA-binding transcription factor of YAP/TAZ that controls proliferation and apoptosis, we investigated whether the expression of TEAD is controlled by cell density. TEAD4 protein was down-regulated when the density of MCF-10A cells increased, while TEAD1 protein level was not changed (Fig. 1A). The reduced level of TEAD4 and YAP-TEAD reporter activity in high-density cells was also observed in MDCK and 293A cells (SI Appendix, Fig. S1). The overall TEAD level, which was detected by the Pan-TEAD antibody, was partially reduced in MCF-10A cells at high density. The cell density-dependent regulation of TEAD4 protein expression was Lats-independent because Lats1/2 KO did not prevent the reduction of TEAD4 protein in MCF-10A cells at high density (Fig. 1A). The expression of TEAD4 mRNAs was not affected by the change in cell density, which suggests that posttranscriptional regulation controls TEAD4 levels (SI Appendix, Fig. S2). This was further validated by the stable expression of TEAD4 in MCF-10A cells. Expression of exogenous wild-type TEAD4 or YAP binding-deficient TEAD4 showed density-dependent reductions of TEAD4 protein (Fig. 1D). This implies that the density-dependent regulation of TEAD4 protein level is independent from YAP binding. To identify the Hippo signaling component that contributes to the posttranslational regulation of TEAD4 protein level, we depleted Hippo signaling components in HA-tagged TEAD4 expressing MCF-10A cells through siRNA transfection and examined the exogenous TEAD4 protein expression. Depletion of Nf2/Merlin increased the level of HA-tagged TEAD4 in MCF-10A cells at low and high densities (Fig. 1E), while depletion of Lats or Mst kinases did not influence the exogenous TEAD4 protein expression level. Nf2 is a membrane associated protein that mediates contact inhibition of growth, in part through the regulation of the Hippo pathway (20). These results suggest that cell density and Nf2 play important roles in the regulation of TEAD4 levels in a Lats-independent manner.
Palmitoylation of TEAD Is Decreased in Cells at High Density.
TEAD undergoes autopalmitoylation at evolutionarily conserved cysteine residues (14). This posttranslational modification plays a key role in the regulation of YAP/TAZ binding affinity (14) and protein stability (15). Based on the decreased activity and expression level of TEAD4 protein in MCF-10A cells at high density, we hypothesized that TEAD4 activity and stability are regulated by cell density through the modulation of its palmitoylation status. To detect the palmitoylation status in intact cells, we adopted the acyl-resin–assisted capture (Acyl-RAC) technique (21), which captures S-acylated (typically palmitoylated) proteins by the cleavage of thioester bonds with the nucleophile hydroxylamine and subsequent binding to thiopropyl Sepharose beads. To validate the specificity of Acyl-RAC, 293A cells were transfected with wild-type or palmitoylation-deficient mutant TEAD4 expression vectors, and purified wild-type or mutant TEAD proteins were subjected to the Acyl-RAC procedure. As shown in SI Appendix, Fig. S3, Acyl-RAC detected the palmitoylation of wild-type TEAD4 in a hydroxylamine-dependent manner (lane 3), whereas palmitoylation-deficient mutant abrogated its detection by Acyl-RAC (lane 8).
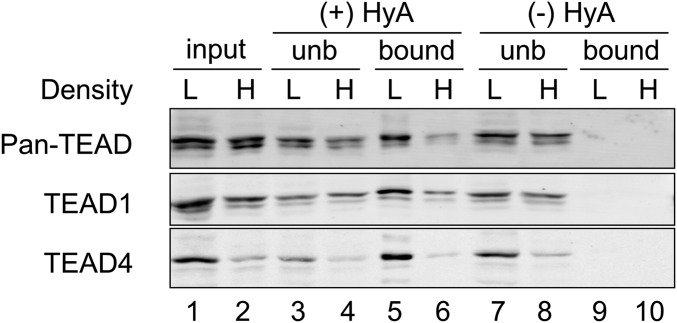
We then examined protein lysates from sparse and dense cell density MCF-10A cells using the Acyl-RAC procedure. In this assay, hydroxylamine untreated samples (lanes 9, 10) and unbound to thiopropyl Sepharose beads (lanes 3, 4, 7, 8) were used as negative controls (Fig. 2). Using the Acyl-RAC technique, we discovered that the palmitoylation of TEAD4 was greatly decreased in MCF-10A cells at high density (lanes 5, 6; Fig. 2). Intriguingly, the palmitoylation status of endogenous TEADs detected with anti–pan-TEAD and anti-TEAD1 were also greatly reduced by the increase of cell density (lanes 5, 6), while their protein levels showed a small reduction in MCF-10A cells at high density (lanes 1, 2). The palmitoylation of pan-TEAD, TEAD1, and TEAD4 were reduced at similar levels in response to increased cell density, suggesting that the palmitoylation of individual TEAD family members is regulated by the same mechanism. This also indicates that the decreased levels of TEAD4 and/or activity of multiple TEADs in MCF-10A cells at high density is resulting from the reduction of palmitoylation on TEAD.
Fig. 2.
Density-dependent palmitoylation of TEAD. Acyl-RAC of TEAD in MCF-10A cells grown at low or high cell densities. The palmitoylation status of TEAD was analyzed using the Acyl-RAC technique. H, high cell density; HyA, hydroxylamine; L, low cell density; unb, unbound.
FASN Is Important for the Activity of TEAD.
The unique reversible nature of protein palmitoylation, like phosphorylation, is controlled by multiple mechanisms (17, 22). First, the cellular level of palmitate is regulated by the uptake of dietary palmitate and the de novo palmitate synthesis via acetyl-CoA carboxylase (ACC) and FASN, enzymes responsible for the synthesis of palmitate. Second, the protein palmitoylation process is mediated either by the PAT enzyme or by nonenzymatic autopalmitoylation. Third, depalmitoylating enzymes catalyze the hydrolysis of S-acylated cysteine residues.
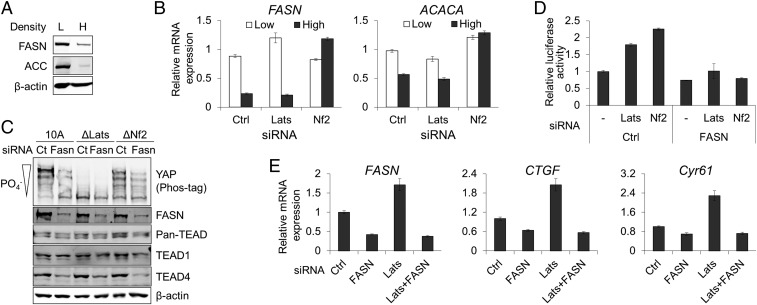
As the palmitoylation status of TEAD is changed by cell density in MCF-10A cells, we investigated the regulatory mechanism of TEAD palmitoylation. We first examined the expression of ACC and FASN in cells grown in different cell densities. Previous studies have demonstrated that the up-regulation of FASN is induced by diverse biological stimuli such as growth factors (23) and steroid hormone (24). Importantly, loss of Nf2 increases the expression of lipogenesis-related genes including FASN and acetyl-CoA carboxylase 1 (ACC1, encoded by ACACA) (25). Following the growth of cells to a postconfluent state, we observed density-dependent decreases of FASN and ACC proteins in MCF-10A, MDCK, and 293A cells (Fig. 3A and SI Appendix, Fig. S1A). The expression of FASN and ACC1 mRNAs was decreased in both parental and Lats1/2-depleted MCF-10A cells at high cell density, suggesting the cell density-dependent transcriptional regulation of ACC and FASN mRNA expression (Fig. 3B). In contrast, depletion or KO of Nf2 prevented the density-dependent decrease in FASN (Fig. 3B and SI Appendix, Fig. S4A) and ACACA (Fig. 3B) mRNAs expression. Depletion of Nf2 in Lats1/2 KO MCF-10A cells prevented the density-dependent decrease of FASN mRNA (SI Appendix, Fig. S4B) further confirmed the role of Nf2 in the regulation of FASN expression. This suggests that the expression of FASN and ACC1 is decreased in MCF-10A cells at high density in a Nf2-dependent, but Lats-independent, manner. This also implies that the reduction of TEAD palmitoylation in MCF-10A cells at high cell density is due to the down-regulation of FASN and ACC1 and, subsequently, the reduction of cellular palmitate level. Addition of exogenous palmitate to MCF-10A cells at high density increased the level of TEAD4 protein in a dose-dependent manner (SI Appendix, Fig. S10A).
Fig. 3.
FASN-dependent regulation of YAP-TEAD activity. (A) Analysis of FASN and ACC expression. The expression of FASN and ACC proteins in MCF-10A at low and high cell densities was analyzed by Western blotting. H, high cell density; L, low cell density. (B) Quantitative analysis of FASN and ACACA mRNA expression. The expression of FASN and ACACA mRNAs in parental, and Lats1/2 or Nf2-depleted MCF-10A at low or high cell densities was analyzed using qPCR. (C) Biochemical effects of FASN depletion. Endogenous FASN was depleted by siRNA transfection in parental (10A), Lats1/2 (ΔLats), or Nf2 KO (ΔNf2) MCF-10A cells. Phosphorylation of YAP was monitored by phos-tag SDS/PAGE and Western blotting. (D) HOP-flash reporter. YAP-TEAD reporter assay was carried out in 293A cells transfected with control, Lats1/2, or Nf2 siRNAs in the presence/absence of FASN siRNA. (E) Quantitative analysis of YAP-TEAD target genes’ mRNA expression. The expressions of CTGF and Cyr61 mRNA in Lats1/2 and/or FASN-depleted MCF-10A cells were analyzed using qPCR.
We next investigated whether the expression of FASN controls the activity of TEAD. The depletion of FASN showed dramatic effects on TEAD. Similar to the density-dependent regulation of TEAD, FASN siRNA transfection significantly reduced the level of TEAD4 in a Lats-independent manner while TEAD1 level was not changed (Fig. 3C). The depletion of FASN also decreased the level of nuclear TEAD4 in a Lats-independent manner, but the localization of YAP was not changed by FASN siRNA transfection (SI Appendix, Fig. S5). The depletion of FASN in MCF-10A cells by siRNA transfection did not increase the phosphorylation of YAP in parental and Nf2 KO MCF-10A cells (Fig. 3C). YAP protein remained unphosphorylated in FASN-depleted Lats KO MCF-10A cells (Fig. 3C).
The depletion of FASN also interfered with YAP-TEAD transcriptional activity. Transfection of FASN siRNA into 293A cells abolished the induced expression of YAP-TEAD reporter activity by Lats1/2 or Nf2 depletion (Fig. 3D). Furthermore, RNAi-mediated depletion of FASN prevented the increased expression of YAP-TEAD downstream target genes by Lats1/2 depletion (Fig. 3E). Taken together, these data indicate that FASN controls the level and activity of TEAD, but not YAP. Since TEAD has been shown to be autopalmitoylated (14), this suggests that synthesis of palmitate, rather than activities of PAT, controls the palmitoylation of TEAD.
Identification of TEAD4 Depalmitoylating Enzymes.
Because palmitoylation of TEAD is reduced in MCF-10A cells at high cell density, we hypothesized that depalmitoylating enzymes may be involved in depalmitoylation of TEAD. Two acyl-protein thioesterases, APT1 and APT2, have been presumed to be solely responsible for the depalmitoylation of all intracellular S-palmitoylated proteins. However, recent studies have identified novel depalmitoylating enzymes including APTL1, ABHD17A, ABHD17B, and ABHD17B (26, 27).
To identify candidate depalmitoylating enzymes, we performed BioID, a technique developed for the identification of protein–protein interaction in living cells (28). To determine the specificity of the BioID method, flag-tagged YAP was coexpressed with BirA*, which was fused to TEAD4 in 293A cells. As negative controls, BirA* fusions to YAP binding-deficient TEAD4 (29) or palmitoylation-deficient TEAD4 were used. These mutants are known from previous studies to not interact with YAP (14, 29). As expected, BirA* fused to wild-type but not mutant TEAD4 biotinylated YAP, confirming the specificity of BioID methods (SI Appendix, Fig. S6).
To identify the depalmitoylating enzyme for TEAD4, we screened the depalmitoylases using the BioID method. Twenty-five myc-tagged serine hydrolases were cotransfected with TEAD4-BirA* into 293A cells and biotin was treated for 14 h. Depalmitoylases were immunoprecipitated with Myc-agarose beads, followed by the detection of biotinylation. We found that ABHD14B, ABHD17A, ABHD17C, APT2, RBBP9, PPME1, SERHL, and MGLL are biotinylated by TEAD4-BirA* (SI Appendix, Fig. S7A).
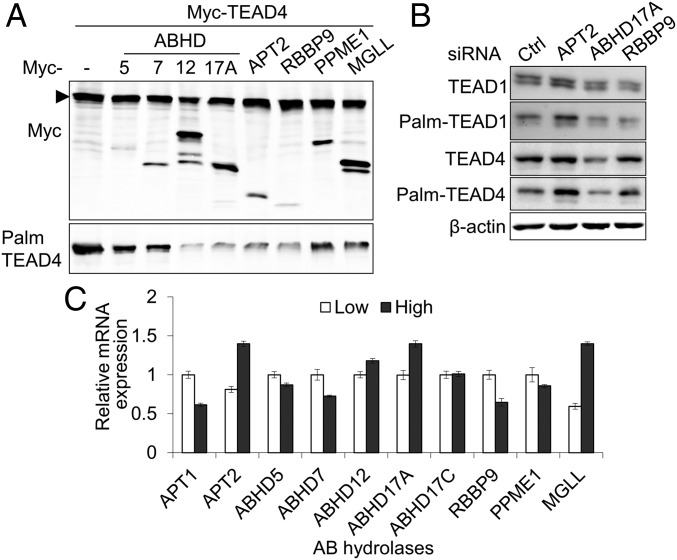
To determine whether these enzymes can depalmitoylate TEAD4, we cotransfected selected serine hydrolases with myc-tagged TEAD4 and detected palmitoylation status via the Acyl-RAC technique. As a control, serine hydrolases (ABHD7, ABHD12) that were not observed to be biotinylated by TEAD4-BirA* (SI Appendix, Fig. S7A) were included. We found that ABHD17A, APT2, RBBP9 robustly depalmitoylated TEAD4 (Fig. 4A). PPME1 and MGLL mildly reduced the palmitoylation of TEAD4. Reduction of TEAD4 protein by the overexpression of ABHD12 is likely to result from the reduction of TEAD4 palmitoylation (lane 4). We next examined whether inhibiting the expression of depalmitoylases might increase the palmitoylation of TEAD proteins. Depletion of APT2 by siRNA transfection led to a significant increase in the palmitoylation of TEAD1 and TEAD4 (Fig. 4B and SI Appendix, Fig. S8). Depletion of ABHD17A or RBBP9 showed minimal effect on the palmitoylation of TEAD proteins. This suggests that APT2 is the main depalmitoylase for TEAD1 and TEAD4.
Fig. 4.
Identification of TEAD4 depalmitoylating enzymes. (A) Analysis of palmitoylation by Acyl-RAC analysis. Depalmitoylation of TEAD4 by the overexpression of serine hydrolases was determined by Acyl-RAC. (B) Increase of TEAD palmitoylation by the depletion of APT2. Endogenous serine hydrolases in MCF-10A cells were depleted by siRNA transfection. The palmitoylation status of TEAD1 and TEAD4 was determined by Acyl-RAC. (C) Quantitative analysis of serine hydrolases mRNA expression. The expression of serine hydrolases in MCF-10A cells at low or high densities was determined by qPCR.
Next, we assessed the mRNA expression of selected serine hydrolases in sparse and dense cell density MCF-10A cells. Interestingly, the mRNA levels of APT2, ABHD17A, and MGLL, but not RBBP9, were substantially increased in MCF-10A cells at high density (Fig. 4C). Taken together, these results suggest that reduced palmitoylation of TEAD4 in MCF-10A cells at high density may be due to an increase in the expression of depalmitoylases including APT2.
E3 Ubiquitin Ligase CHIP Regulates the Stability of TEAD4 Protein.
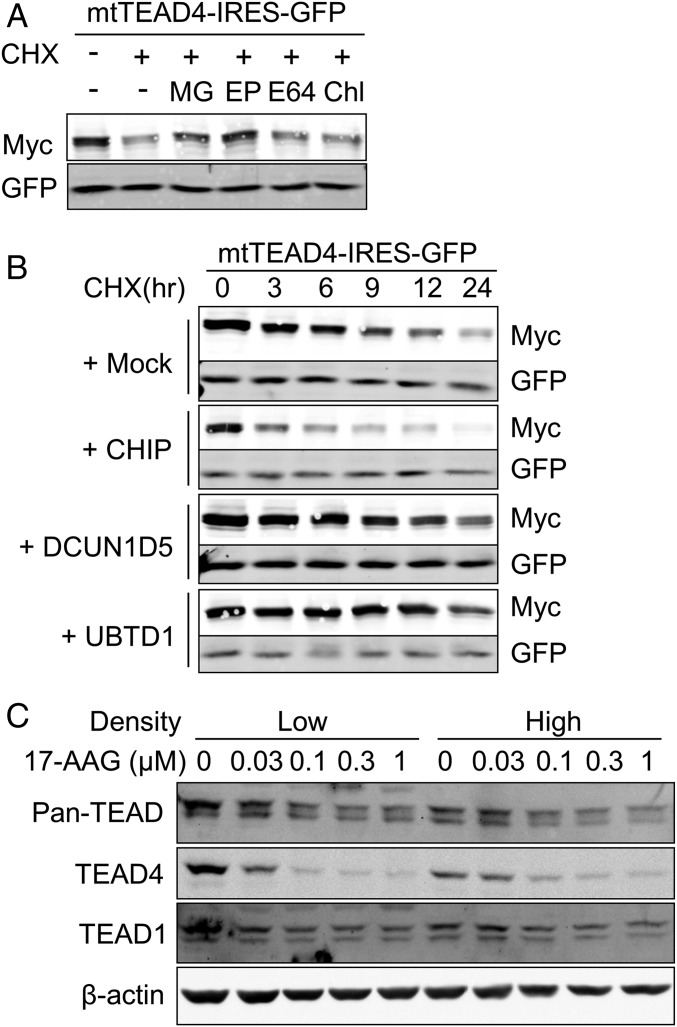
Mutation of palmitoylation sites reduces the TEAD2 protein levels relative to wild-type TEAD2 (15). To assess the protein half-life of wild-type or palmitoylation-deficient mutant TEAD4 proteins, protein synthesis was inhibited with cycloheximide (CHX). Compared with relatively stable wild-type TEAD4, with a half-life of >24 h, the palmitoylation-deficient TEAD4 mutant was relatively unstable, with a half-life of 9 h (SI Appendix, Fig. S9). In contrast, proteasome inhibition by MG132 or epoxomicin led to an increase in mutant TEAD4 protein (Fig. 5A). Inhibition of lysosomal degradation by chloroquine, however, did not increase the level of mutant TEAD4 (Fig. 5A), suggesting that the palmitoylation-deficient mutant TEAD4 protein is degraded through the proteasome pathway. These results suggest that the depalmitoylation of the TEAD protein triggers the degradation of protein by the ubiquitin-proteasome pathway.
Fig. 5.
E3 ubiquitin ligase CHIP promotes the degradation of unpalmitoylated TEAD4. (A) Inhibition of palmitoylation-deficient TEAD4 degradation by proteasome inhibitors. 293A cells transfected with palmitoylation-deficient TEAD4-IRES-GFP (mtTEAD4-IRES-GFP) were treated with 10 μM CHX in the presence of MG (10 μM MG-132, proteasome inhibitor), EP (1 μM Epoxomicin, proteasome inhibitor), E64 (10 μM E-64, cysteine proteases inhibitor), or Chl (2.5 μM Chloroquine, autophagy inhibitor) for 9 h. The expression levels of mutant-TEAD4 and GFP were analyzed by Western blot. (B) Promotion of the degradation of unpalmitoylated TEAD4 by E3 ubiquitin ligase CHIP. 293A cells transfected with mtTEAD4-IRES-GFP and indicated E3-ubiquitin ligases. After 24 h of transfection, cells were treated with 10 μM CHX for the indicated time, followed by Western blot analysis. (C) Degradation of TEAD stimulated by the Hsp90 inhibitor 17-AAG. MCF-10A cells at low or high densities were treated with 17-AAG for 24 h at the indicated concentrations. MCF-10A cells at low or high densities were treated for 24 h with 17-AAG at the indicated concentrations. TEAD protein level was observed by Western blot.
To identify the E3 ubiquitin ligase that is involved in the proteasomal degradation of unpalmitoylated TEAD4, we performed BioID. E3 ubiquitin ligases, which are described as TEAD binding proteins in the BioGRID, a curated database of protein–protein interactions (30), were cotransfected with palmitoylation deficient-TEAD4-BirA*. Using the BioID technique, we found that CHIP (Carboxyl terminus of Hsp70-interacting protein), DCUN1D5 (DCN1-like protein 5), and UBTD1 (Ubiquitin domain-containing protein 1) interact with the palmitoylation-deficient TEAD4 mutant (SI Appendix, Fig. S7B).
For further validation, CHIP, DCUN1D5, and UBTD1 were cotransfected with the palmitoylation-deficient TEAD4-IRES-GFP construct and the half-life of mutant TEAD4 was determined by treatment with CHX. Expression of DCUN1D5 and UBTD1 did not destabilize the mutant TEAD4; however, overexpression of CHIP greatly destabilized mutant TEAD4 (Fig. 5B). Compared with palmitoylation-deficient TEAD4 with a half-life of 9 h, overexpression of CHIP reduced the half-life of mutant TEAD to less than 3 h (Fig. 5B). Depletion of endogenous CHIP by siRNA transfection prevented FASN depletion-induced destabilization of TEAD4 (SI Appendix, Fig. S10B).
CHIP directly interacts with molecular chaperones Hsp70/Hsp90 and functions as a protein quality control system, which degrades the misfolded- or nonnative-Hsp70/Hsp90 clients. We therefore checked whether the inhibition of Hsp90 triggers the degradation of TEAD using treatment with 17-AAG, an Hsp90 inhibitor. Inhibition of Hsp90 by 17-AAG is known to facilitate the CHIP-mediated degradation of Hsp90 clients, such as XRCC1 (31). Treatment with 17-AAG reduced the level of endogenous TEAD4 in MCF-10A cells at low and high densities (Fig. 5C). The endogenous level of TEAD detected with pan-TEAD and TEAD1 antibodies were also decreased by the treatment of 17-AAG (Fig. 5C). This implies that TEAD is a Hsp90 client protein and sensitive to 17-AAG–induced protein degradation. Taken together, these findings indicate that E3 ubiquitin ligase CHIP regulates the stability of TEAD4 and the inhibition of Hsp90 destabilizes TEAD.
Discussion
In this study, we demonstrate that TEAD activity is controlled by cell density and Nf2 through the regulation of its palmitoylation status. The expression level, transcriptional activity, and palmitoylation of TEAD are reduced in MCF-10A cells at high density in a Lats-independent manner. FASN, which plays a key role in de novo fatty acid biosynthesis, is critical for the activity of TEAD. Inhibition of FASN reduces the TEAD4 protein level and prevents the activation of YAP-TEAD target gene expression. Addition of exogenous palmitate increased the level of TEAD4 in high cell density MCF-10A cells. The expression of FASN and ACC decreases in MCF-10A cells at high density while the mRNA levels of several depalmitoylases, including APT2 and ABHD17A, increases. Furthermore, depalmitoylated TEAD4 is degraded by the E3 ubiquitin-protein ligase, CHIP, which targets misfolded clients of Hsp70/Hsc70/Hsp90 chaperones. Our results indicate that TEAD activity can be modulated by the regulation of palmitoylation status (SI Appendix, Fig. S11). Therefore, palmitoylation status is a potential target for controlling TEAD-dependent processes, perhaps including cancer growth.
The palmitoylation of TEAD family members is reduced by an increase in cell density, which affects either stability or activity. The stabilities of individual TEAD protein in MCF-10A cells, however, are differentially regulated by cell density. In contrast to the significant reduction in the TEAD4 protein level, the TEAD1 protein shows minor reductions in MCF-10A cells at high density (Fig. 1A). This is consistent with other studies that have also provided evidence for differential effects of palmitoylation on distinct TEAD members (14, 15, 32). The mutation of the conserved palmitoylation sites in TEAD1 does not change its expression level, but completely prevents its binding to YAP (14). In contrast, the mutation of conserved cysteine residue in TEAD2 substantially reduces the protein level (15). Similarly, Mesrouze et al. (32) showed that the palmitoylation of TEAD4 is dispensable for YAP/TAZ binding but is important for its stability. This implies that the increase in cell density results in the loss of YAP-TEAD transcriptional activity by the decrease of palmitoylation in all TEAD members; however, the effect of depalmitoylation on individual TEAD members varies. This explains the context-dependent regulation of TEAD in the binding of different transcriptional coactivators and activation of downstream gene expression. Because the palmitoylation of TEAD1 is dispensable for binding to Vgll4 (14), alteration of the TEAD1 palmitoylation status can lead to switching of binding coactivator and target gene expressions (33).
We found that the expression of FASN is controlled by cell density and Nf2 (Fig. 3 A and B). In normal human tissues, FASN is expressed at minimal levels and the high expression of FASN is limited to lipogenic tissues such as liver, fetal lung, lactating breast, and adipose tissue. However, in rapidly growing cancer cells, whose hallmark is the constitutive activation of the metabolic pathway (34), the expression of FASN is commonly up-regulated to enhance the production of lipids for membrane biosynthesis (22). Moreover, its expression in cancer cells increases in a stage-dependent manner and is associated with poor survival rates of cancer patients (18). Due to its metabolo-oncogenic nature, FASN is regarded as a high priority therapeutic target for cancer therapy (18). For example, the inhibition of FASN in ovarian and breast cancer cells yields antitumor effects that reduce cell proliferation while inducing apoptosis and chemosensitivity (18, 35).
Expression of FASN is mediated by upstream signals including growth factor receptors, such as EGFR and ERBB2, and steroid hormones (22). In our study, we found that the FASN and ACC levels in MCF-10A cells decrease when the cell density of the culture increases (Fig. 3A). More importantly, the loss of Nf2 increases the transcription of FASN and ACC1 mRNAs in MCF-10A cells at high cell density (Fig. 3B). Nf2 directly associates with α-catenin at the adherens junction (36) and plays an important role in contact inhibition of proliferation by modulating receptor kinase signaling (37) and activating the Hippo pathway (38). Nf2 has been reported to regulate the Hippo pathway at multiple levels. It negatively regulates YAP/TAZ by the activation of the Hippo core kinase cascade at the plasma membrane and cell junctions, as well as by the direct inhibition of CRL4DCAF1 E3 ubiquitin ligase, which suppress Lats kinases in the nucleus (38). Our results suggest another important role of Nf2 in the regulation of contact inhibition of proliferation through the inactivation of TEAD by limiting cellular level of palmitate.
We show that TEAD palmitoylation is also controlled by the density-dependent expression of depalmitoylases such as APT2 (Fig. 4). Depalmitoylation is a highly regulated process that responds to a wide range of upstream signals. For instance, APT activity is increased in serum starvation condition, while the treatment of EGF rapidly inhibits the depalmitoylation activity of APT (39). These imply that the expression of APT2 is increased in restrained growth conditions, such as serum starvation and high cell density.
Unlike the cysteine protease-dependent degradation of TEAD1 by Vgll4 (40), unpalmitoylated TEAD4 is degraded by the proteasome pathway (Fig. 5A). E3 ubiquitin ligase CHIP mediates the degradation of unpalmitoylated TEAD4 in an Hsp90-dependent manner (Fig. 5 B and C). Inhibition of Hsp90 by 17-AAG promotes the degradation of TEAD (Fig. 5C).
In summary, we have shown that FASN, depalmitoylases, Hsp90, and E3 ubiquitin ligase mediate the regulation of TEAD activity. This suggests that the modulation of these mediators might be a relevant strategy for therapeutic intervention of cancers with hyperactivated YAP/TAZ-TEAD.
Methods
MCF-10A cells (a gift from Joan S. Brugge, Harvard Medical School, Boston) were cultured in DMEM/F12 medium supplemented with 5% horse serum, 0.5 μg/mL hydrocortisone, 100 ng/mL cholera toxin, 10 μg/mL insulin, and 20 ng/mL recombinant human EGF. 293T (American Type Culture Collection; ATCC), 293A (Thermo Fisher Scientific), and MDCK cells were maintained in DMEM with 10% FBS. Mycoplasma infection was monitored by PCR-based assay (ATCC). Lats1/2 or Nf2 KO MCF-10A cells were generated by CRISPR-Cas9 technology.
Supplementary Material
Acknowledgments
This work has been supported by National Institute of General Medical Science at the National Institutes of Health Grants R01GM106659 and R35GM122467 (to B.M.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819400116/-/DCSupplemental.
References
- 1.Gumbiner BM, Kim NG (2014) The Hippo-YAP signaling pathway and contact inhibition of growth. J Cell Sci 127:709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan EH, et al. (2005) The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24:2076–2086. [DOI] [PubMed] [Google Scholar]
- 3.Li Q, et al. (2014) The conserved misshapen-warts-Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in Drosophila. Dev Cell 31:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng Z, et al. (2015) MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat Commun 6:8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng Y, et al. (2015) Identification of happyhour/MAP4K as alternative hpo/Mst-like kinases in the Hippo kinase cascade. Dev Cell 34:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim MK, Jang JW, Bae SC (2018) DNA binding partners of YAP/TAZ. BMB Rep 51:126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao B, et al. (2008) TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22:1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin KC, Park HW, Guan KL (2017) Regulation of the Hippo pathway transcription factor TEAD. Trends Biochem Sci 42:862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacquemin P, et al. (1998) Differential expression of the TEF family of transcription factors in the murine placenta and during differentiation of primary human trophoblasts in vitro. Dev Dyn 212:423–436. [DOI] [PubMed] [Google Scholar]
- 10.Kaneko KJ, Cullinan EB, Latham KE, DePamphilis ML (1997) Transcription factor mTEAD-2 is selectively expressed at the beginning of zygotic gene expression in the mouse. Development 124:1963–1973. [DOI] [PubMed] [Google Scholar]
- 11.Home P, et al. (2012) Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc Natl Acad Sci USA 109:7362–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin KC, et al. (2017) Regulation of Hippo pathway transcription factor TEAD by p38 MAPK-induced cytoplasmic translocation. Nat Cell Biol 19:996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ota M, Sasaki H (2008) Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development 135:4059–4069. [DOI] [PubMed] [Google Scholar]
- 14.Chan P, et al. (2016) Autopalmitoylation of TEAD proteins regulates transcriptional output of the Hippo pathway. Nat Chem Biol 12:282–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noland CL, et al. (2016) Palmitoylation of TEAD transcription factors is required for their stability and function in Hippo pathway signaling. Structure 24:179–186. [DOI] [PubMed] [Google Scholar]
- 16.Hentschel A, Zahedi RP, Ahrends R (2016) Protein lipid modifications–More than just a greasy ballast. Proteomics 16:759–782. [DOI] [PubMed] [Google Scholar]
- 17.Tabaczar S, Czogalla A, Podkalicka J, Biernatowska A, Sikorski AF (2017) Protein palmitoylation: Palmitoyltransferases and their specificity. Exp Biol Med (Maywood) 242:1150–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menendez JA, Lupu R (2017) Fatty acid synthase (FASN) as a therapeutic target in breast cancer. Expert Opin Ther Targets 21:1001–1016. [DOI] [PubMed] [Google Scholar]
- 19.Sanders SS, et al. (2015) Curation of the mammalian palmitoylome indicates a pivotal role for palmitoylation in diseases and disorders of the nervous system and cancers. PLoS Comput Biol 11:e1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harvey KF, Zhang X, Thomas DM (2013) The Hippo pathway and human cancer. Nat Rev Cancer 13:246–257. [DOI] [PubMed] [Google Scholar]
- 21.Forrester MT, et al. (2011) Site-specific analysis of protein S-acylation by resin-assisted capture. J Lipid Res 52:393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menendez JA, Lupu R (2007) Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer 7:763–777. [DOI] [PubMed] [Google Scholar]
- 23.Bian Y, Yu Y, Wang S, Li L (2015) Up-regulation of fatty acid synthase induced by EGFR/ERK activation promotes tumor growth in pancreatic cancer. Biochem Biophys Res Commun 463:612–617. [DOI] [PubMed] [Google Scholar]
- 24.Santolla MF, et al. (2012) G protein-coupled estrogen receptor mediates the up-regulation of fatty acid synthase induced by 17β-estradiol in cancer cells and cancer-associated fibroblasts. J Biol Chem 287:43234–43245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stepanova DS, et al. (2017) An essential role for the tumor-suppressor Merlin in regulating fatty acid synthesis. Cancer Res 77:5026–5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin DT, Conibear E (2015) ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. eLife 4:e11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokoi N, et al. (2016) Identification of PSD-95 depalmitoylating enzymes. J Neurosci 36:6431–6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim DI, et al. (2014) Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci USA 111:E2453–E2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, et al. (2010) Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev 24:290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oughtred R, et al. (2019) The BioGRID interaction database: 2019 update. Nucleic Acids Res 47:D529–D541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang Q, et al. (2014) HSP90 regulates DNA repair via the interaction between XRCC1 and DNA polymerase β. Nat Commun 5:5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mesrouze Y, et al. (2017) Effect of the acylation of TEAD4 on its interaction with co-activators YAP and TAZ. Protein Sci 26:2399–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pobbati AV, Hong W (2013) Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol Ther 14:390–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanahan D, Weinberg RA (2011) Hallmarks of cancer: The next generation. Cell 144:646–674. [DOI] [PubMed] [Google Scholar]
- 35.Bauerschlag DO, et al. (2015) Fatty acid synthase overexpression: Target for therapy and reversal of chemoresistance in ovarian cancer. J Transl Med 13:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gladden AB, Hebert AM, Schneeberger EE, McClatchey AI (2010) The NF2 tumor suppressor, Merlin, regulates epidermal development through the establishment of a junctional polarity complex. Dev Cell 19:727–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McClatchey AI, Fehon RG (2009) Merlin and the ERM proteins–Regulators of receptor distribution and signaling at the cell cortex. Trends Cell Biol 19:198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooper J, Giancotti FG (2014) Molecular insights into NF2/Merlin tumor suppressor function. FEBS Lett 588:2743–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kathayat RS, Elvira PD, Dickinson BC (2017) A fluorescent probe for cysteine depalmitoylation reveals dynamic APT signaling. Nat Chem Biol 13:150–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin Z, et al. (2016) Acetylation of VGLL4 regulates Hippo-YAP signaling and postnatal cardiac growth. Dev Cell 39:466–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim NG, Gumbiner BM (2015) Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. J Cell Biol 210:503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.