Abstract
NOD2 is a member of the NLR family of proteins that participate in the activation of the innate immune response. RIP2 is a downstream kinase activated by both NOD1 and NOD2. There is scarcity of information regarding the relevance of NOD2 in periodontitis, a chronic inflammatory condition characterized by inflammatory bone resorption. We used NOD2-KO and RIP2-KO mice in a model of microbial-induced periodontitis. Heat-killed Aggregatibacter actinomycetemcomitans was injected in the gingival tissues three times/wk for 4 wk. Bone resorption was assessed by μCT analysis; osteoclasts were identified by immunohistochemical staining for TRAP and inflammation was assessed using a severity score system in H/E-stained sections. In vitro studies using primary macrophages assessed the response macrophages using qPCR-based array and multi-ligand ELISA. Bone resorption and osteoclastogenesis were significantly reduced in NOD2-KO mice. Severity of inflammation was not affected. qPCR-focused arrays and multi-ligand ELISA showed that expression of pro-inflammatory mediators was reduced in NOD2- and RIP2-deficient cells. RANKL-induced osteoclastogenesis was impaired in NOD2- and RIP2-deficient macrophages. We conclude that NOD2 is important for osteoclast differentiation and inflammatory bone resorption in vivo and also for the macrophage response to Gram-negative bacteria.
Keywords: Bone resorption, inflammation, innate immunity, macrophages, NOD2 signaling adaptor protein
Introduction
NLRs are proteins initially described as cytosolic sensors of bacterial infection or intracellular PRRs. There are 22 proteins included in the NLR family, which are further grouped into five subfamilies according to variations in their domain structures. The NLRC subfamily comprises five different proteins that are highly conserved in vertebrates: NOD1 (NLRC1), NOD2 (NLRC2) and the inflammasome-activating NLRs NLRC3, NLRC4 and NLRC5. NOD1 and NOD2 are the two most studied members and they recognize different structures in bacterial peptidoglycan: meso-diaminopimelic acid-containing peptidoglycan (meso-DAP) is the ligand for NOD1, which is present in most Gram-negative and some Gram-positive bacteria, whereas NOD2 is activated by muramyl-dipeptide (MDP), a structure found in all bacterial peptidoglycans.1
Upon recognition of their specific ligands, NOD proteins undergo a similar activation process involving self-oligomerization and interaction with a common downstream kinase called RIP2/RICK/CARDIAK that will primarily drive NF-kB and MAPK activation. Although, simplistically, the activation process is similar for NOD1 and NOD2, NOD2 activation requires two ATP hydrolysis events mediated by acidic amino acid residues, as opposed to a single initial hydrolysis event in NOD1 activation.2 This information, combined with the existence of distinct ligands, suggests that NOD1 and NOD2 may have different functional roles.
Abundant evidence of the important role of NOD2 in host response is primarily derived from studies on host–microbial interactions in the gut mucosa,3 and conditions usually associated with infection by invasive bacteria, such as enteroinvasive Escherichia coli, Pseudomonas aeruginosa, Shigella flexneri and Listeria monocytogenes. Activation of NOD proteins was initially only associated with the presence of their peptidoglycan ligands in the intracellular environment owing to bacterial invasion or by direct introduction by non-invasive bacteria presenting a type III- or type IV-secreting apparatus, such as Helicobacter pylori.4,5 However, novel functions of NOD1 and NOD2 have been described more recently, including the activation of B lymphocytes,6 modulation of dendritic cell activation and Ag presentation,7 regulation of adaptive immunity by participating in Th178 and Th1 polarization,9 particularly by cross-talk with TLR signaling. Importantly, activated NOD proteins may function as signaling scaffolds and interact with as-yet-unknown functional partners and have a significant impact on the innate and adaptive host response. Thus, it is possible that some NLRs, in addition to their PRR role, also act as signaling regulators, amplifying the activation of different pathways and modulating other immune and non-immune processes.4,10
Periodontitis is the most prevalent bone resorption-associated condition in humans.11 As a bacterial-initiated and bacterial-maintained pathology, the nature of host–microbial interactions largely dictates the extent and severity of tissue destruction. Recently, mice lacking NOD1 were shown to present reduced bone resorption in a ligature/infection model of periodontal diseases;12 moreover, both NOD1 and NOD2 were shown to mediate sensing of periodontal diseaseassociated bacteria in human embryonic kidney cells.13 Other in vivo studies report that direct injections of peptidoglycan from Gram-positive bacteria (NOD2 ligand) in the gingival tissues of mice induced significant bone loss, whereas injections of peptidoglycan from Gram-negative bacteria (NOD1 ligand, weak NOD2 activator) did not. Moreover, the association of bacterial LPS and peptidoglycan from either Gram-positive or Gram-negative bacteria synergistically enhanced bone resorption induced by LPS in this model.14 In this study, we assess the role of NOD2 and RIP2 on a microbial-induced periodontitis model, specifically on their role in inflammatory bone resorption, osteoclastogenesis and inflammation. In vitro studies using primary macrophages provide additional information on the importance of NOD2 and RIP2 for the response of macrophages to Gram-negative microorganisms and also for osteoclastogenesis.
Materials and methods
Animal use and experimental periodontitis model
We used wild-type (WT) and genetically modified [whole genome deletion—knockout (KO)—of NOD2 and RIP2: NOD2-KO and RIP2-KO, respectively] mice backcrossed to C57BL/6 background for eight generations.15,16 All animals used in these studies were between 8 and 10 wk of age. Euthanasia was always performed by cervical dislocation and the Institutional Committee on the Use of Experimental Animals approved the study protocol. Primary bone marrow-derived macrophages (BMDM) were obtained from bone marrow flushed from tibias and femurs, differentiated and expanded in the presence of M-CSF, as previously described.17 Host–microbial interactions in vivo were studied using an experimental model of microbial-induced periodontitis. In this model, heat-killed bacteria associated with periodontal disease were re-suspended in PBS at 109 CFU/ml and 3 μl of this suspension was directly injected into the gingival tissues surrounding the teeth of the mice, three times/wk for 4 wk. Euthanasia was performed by cervical dislocation 2 d after the last injection. These in vivo studies used a total of 42 mice distributed equally among three different genotypes: WT, NOD2-KO and RIP2-KO. Of the 14 mice in each genotype, six were vehicle controls and received bilateral injections of 3 μl PBS vehicle in the palatal aspect of upper first molars, whereas eight mice received injections of 3 × 106 CFU (3 μl volume) of heat-killed Gram-negative A. actinomycetemcomitans (Aa; JP2 clone/serotype b obtained from the personal collection of Dr. Mario Julio Avila-Campos, Department of Microbiology, Institute of Biological Sciences, University of Sao Paulo-USP).
Assessment of alveolar bone loss and inflammation
Immediately after euthanasia, tissue blocks including the upper molars and surrounding tissues were carefully dissected from the animals, rinsed in PBS and fixed in 4% paraformaldehyde for 18 h at 4°C. These samples were then rinsed in distilled water, transferred to 70% ethanol and maintained at 4°C. μCT scanning of these samples was done on a Skyscan (Aartselaar, Belgium) at a resolution of 18 μm and tridimensional images reconstructed, spacially re-oriented in a standardized orientation and analyzed using the equipment’s software (NRecon/DataViewer/CTan/CTvol; Skyscan). A standardized region of interest (ROI) of 2.5 mm3 was positioned on the tridimensional images using anatomical landmarks as reference points, and the fraction of the volume of the ROI occupied by mineralized tissue (BV%) was determined using a standard threshold for detection of mineralized tissues. Considering that the variation in the volume of similar tooth roots in different animals is negligible, a decrease in the BV% in the ROI indicates bone loss.
After scanning, the same tissue blocks used for the μCT analysis were decalcified in 0.5 M EDTA (pH 8.0) and submitted to routine processing for paraffin embedding. Five-μm-thick, semi-serial sections were obtained on the bucco-lingual (transversal) plane and stained with hematoxylin and eosin for descriptive assessment of inflammation by an experienced examiner blinded to the experimental groups, according to a severity score system (0 = no significant inflammation; 1 = mild inflammation; 2 = moderate inflammation; 3 = severe inflammation).18 We assessed a minimum of six equally spaced semi-serial sections spanning 500 μm of the mesio-distal length of each specimen. Sections from 3–4 different animals of each genotype were assessed. Scorings were performed three times with a minimum interval of 2 wk between the assessments and the most prevalent score was used.
Immunohistochemical detection of TRAP was performed using a goat polyclonal Ab (sc-30833; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and an biotin–streptavidin–DAB visualization system (LSAB2+; Dako, Carpinteria, CA, USA). A minimum of six equally spaced semi-serial sections of each experimental conditions (PBS or Aa injections) from three different animals of each genotype were stained. TRAP+ cells containing two or more nuclei present in the vicinity of the bone surface were considered osteoclasts. The number of osteoclasts in a linear extension of 400 μm from the palatal aspect of the first molar by a trained examiner blind to the experimental groups.
In vitro studies
BMDM were dissociated from the culture substrate by incubation in cold PBS (10 min, 4°C), counted and then plated (1 × 106 cells/well in 12-well plates) in RPMI1640 supplemented with penicillin/streptomycin, 10% heat-inactivated FBS and 10 ng/ml M-CSF. After incubation for 18 h to allow the attachment and recovery of the BMDM from the dissociation procedure, these cells were de-induced for 6 h in medium containing 0.2% heat-inactivated FBS and then stimulated with 1 × 106 UFC/ml heat-killed Aa (1:1 ratio bacteria:cells) for 6 (RT-qPCR arrays) and 24 (multi-ligand ELISAs) h. Negative controls were treated with the same volume of PBS vehicle used to re-suspend the bacteria. A total of six samples (unstimulated and Aa-stimulated for three genotypes) were obtained from each experiment. Three independent experiments were performed, each one using cells obtained from 2–4 mice of each genotype. For the osteoclastogenesis experiments, BMDM obtained from each genotype were seeded onto 96-well plates (5 × 104 cells/well) in 100 μl volume of RPMI supplemented with antibiotics, 10% FBS and 10 ng/ml M-CSF. Osteoclast differentiation was induced by treating these cells with 100 ng/ml murine recombinant RANKL (positive control), 1 × 106 UFC/ml of heat-killed Aa, or the combination of RANKL and 1 × 106 UFC/ml heat-killed Aa. Negative controls received only the same volume of PBS containing 0.1% BSA (vehicle) and all treatments were repeated on d 3. These osteoclastogenesis experiments were performed in duplicate and repeated independently three times. On the sixth day, medium was removed, cells were gently washed with Ca/Mg-free PBS, fixed and permeabilized with paraformaldehyde and saponin (BD Cytofix/Cytoperm; BD Biosciences, Franklin Lakes, NJ, USA), stained with phalloidin conjugated to FITC (50 μg/ml in PBS; Sigma Aldrich Co., St. Louis, MO, USA) for 40 min at room temperature (approximately 25°C), followed by extensive washing with PBS and counter-staining with Hoechst 33342 stain (0.5 μg/ml; Sigma Aldrich Co.) for 5 min. Osteoclasts were counted on a digital inverted fluorescence microscope (Evos fl; AMG, ThermoFisher Scientific, Waltham MA, USA) as large, with ring-like green fluorescent shapes containing three or more nuclei. A trained examiner counted the cells in a blind manner to the experimental groups.
RT-qPCR and qPCR arrays
Total RNA was harvested 6 h after stimulation with the bacteria using an affinity column system (RNeasy micro; Qiagen, Hilden, Germany), and six pools of RNA were prepared by combining 300 ng total RNA extracted and purified from BMDM from each individual experiment, according to the experimental group (unstimulated or Aa-stimulated) from WT, NOD2-KO and RIP2-KO mice. cDNA was synthesized from each of these six 900-ng pools of total RNA using the reagents and procedure indicated by the supplier of the PCR-based arrays (RT2 First Strand cDNA kit; SA Biosciences/Qiagen, Frederick, MD, USA). Expression of 84 genes related with innate immunity in each sample was investigated using qPCR-based arrays (RT2 ProfilerTM PCR Array Mouse Toll-Like Receptor Signaling Pathway; SA Biosciences/Qiagen) performed according to the instructions of the supplier on a StepOne Plus qPCR thermocycler (Applied Biosystems, Foster City, CA, USA) using the indicated cycling conditions (10 min/95°C, followed by 40 cycles of 15 s/95°C and 60 s/60°C). Analysis of the qPCR arrays was performed as indicated by the supplier (SA Biosciences/Qiagen, Frederick, MD, USA). Briefly, the cycle threshold (Ct) values obtained from the qPCR thermocycler software were exported into the online analysis tool provided by the supplier of the array, which yields the results of target gene regulation as a fold change relative to the indicated control sample (in this case, non-stimulated WT macrophages). Normalization was performed using the expression of GAPDH, β-actin and β-glucuronidase. These genes were automatically selected by the online analysis tool based on the panel of six housekeeping genes included in the array. The purpose of the analysis was to assess the relative regulation of the 84 target genes in comparison with the gene expression determined in unstimulated macrophages from WT mice.
Multi-ligand ELISAs
The conditioned culture media collected 24 h after stimulation of BMDM with heat-killed Aa or with the same volume of PBS diluent was aliquoted and stored at −80°C until use. Each aliquot was thawed on ice only once and immediately before its use in multi-ligand ELISAs that allow the detection of six cytokines and six chemokines associated with host-microbial interactions and chemotaxis of immune cells (SA Biosciences/Qiagen). The concentration of total protein in the conditioned media was initially determined by a Bradford assay and then the same quantities of total protein from each experimental condition was used in the ELISAs, normalizing the results and allowing for a comparison of the relative quantities of the different cytokines in each sample. Activation of intracellular signaling pathways associated with inflammatory gene expression was also determined using multi-ligand ELISAs (Cell Signaling, Danvers, MA, USA). For these experiments, stimulation of BMDM with Aa or the same volume of PBS was performed for 10, 30 and 60 min. Cell lysates from three independent experiments (using cells from 4–6 mice) were harvested and pooled according to the experimental conditions (PBS or Aa stimulation) and genotype by combining 10 μg total protein from each experiment. Data were analyzed as relative changes to unstimulated control macrophages with the same genetic background (WT, NOD2-KO and RIP2-KO) in each period. The data in these experiments were also normalized by using the same quantity of total protein and also by the expression of total p65, as recommended by the supplier of the assays.
Data analysis
The statistical analysis aimed at comparing the results among different experimental conditions in each genotype (control, Aa-stimulated, etc.) and between different genotypes (WT, NOD2-KO, RIP2-KO) in instances where at least three data points from independent experiments were available. These comparisons were done using ANOVA with Bonferroni post-hoc tests and non-paired t-tests with Welch’s correction for unequal variances, respectively, assuming complete independence between the results of the three genotypes. For all these analyses, we used GraphPad Prism4 software, and statistical significance was set at 95% (P < 0.05). The qPCR-focused array data obtained with combined samples were analyzed using an online bioinformatics tool, DAVID (Database for Annotation, Visualization and Integrated Discovery; http://david.abcc.ncifcrf.gov),19,20 in an exploratory manner consistent with an hypothesis-generating study.21 The purpose of these analyses was to assess how the gene functional clusters that were up-regulated in macrophages stimulated with heat-killed bacteria were affected by deletions of NOD2 or RIP2.
Results
Role of NOD2 and RIP2 in vivo: inflammation, bone resorption and osteoclastogenesis
μCT analysis showed that NOD2-KO mice presented significantly less bone resorption, which was accompanied by a significant decrease on osteoclast numbers (Figure 1). Strikingly, reduction in bone resorption was not accompanied by a decrease in inflammation observed histologically (Figure 2). RIP2-KO mice also showed a reduction on the severity of alveolar bone resorption, but this decrease did not reach statistical significance. Similarly to NOD2-KO animals, RIP2-KO mice had no decrease on inflammation in the soft tissues adjacent to the bone. This lack of effect on the severity of the inflammatory infiltrate suggests that NOD2 and RIP2 may affect the inflammation qualitatively, affecting the phenotype of inflammatory cells and the profile of inflammatory mediators produced. This possibility was assessed in the subsequent in vitro studies, using primary BMDM, as macrophages are the prototypical Ag-presenting cell bridging innate and adaptive immunity and also osteoclast precursors. In vitro, NOD2- and RIP2-deficient BMDM were significantly less sensitive to RANKL-induced osteoclastic differentiation than BMDM from WT mice (Figure 3).
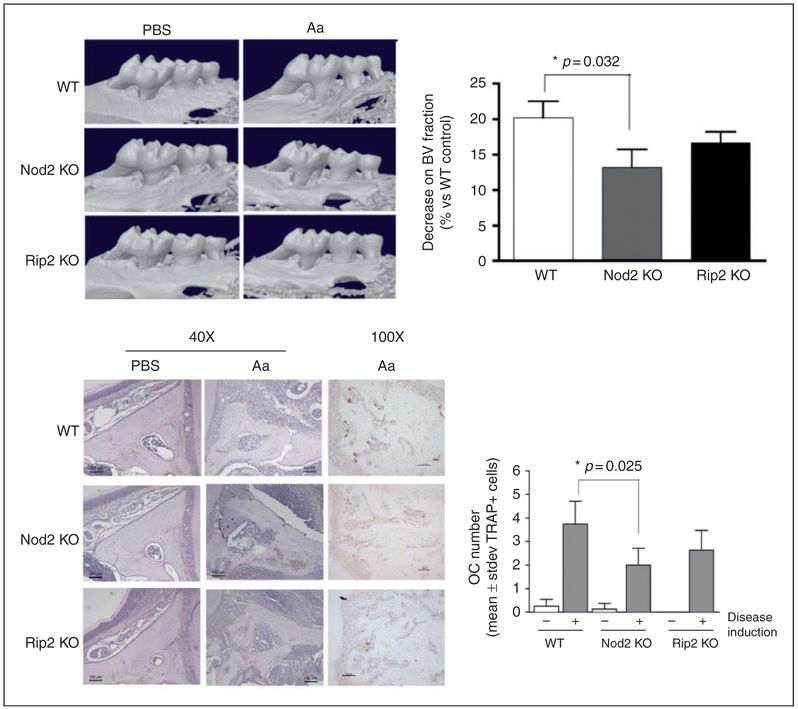
Figure 1.
Role of NOD2 and RIP2 on inflammatory bone resorption and osteoclastogenesis in vivo. Alveolar bone resorption induced in the experimental periodontitis model assessed by μCT is significantly attenuated in Nod2KO mice. (A) Representative images of tridimensional reconstructions of hemi-maxillae segments in the of mice from each genotype and experimental condition (Aa- or PBS-injected) scanned by μCT. Threshold was set to allow visualization of mineralized tissues only. The graph represents the average and SD of the relative reduction in mineralized tissue content [bone volume (BV) fraction] in the standardized ROI assessed in comparison with the BV fraction of the ROI in WT control samples (set to 100%). Samples from at least three different animals were analyzed for each group. (B) Representative images of eosin-stained sections (40× magnification) representative of each genotype and experimental condition, depicting the area in which osteoclasts were counted: from the apex of the palatal root to the area subjacent to the major palatine artery and nerve. Representative images (100× magnification) of the immunohistochemical staining for TRAP. The graph presents average and SDs of the numbers of osteoclasts in the area of interest, according to the genetic background and presence or absence of disease induction. Asterisk (*) indicates significant difference between the indicated pair of bars (Student’s t-test with Welch’s correction for unequal variances).
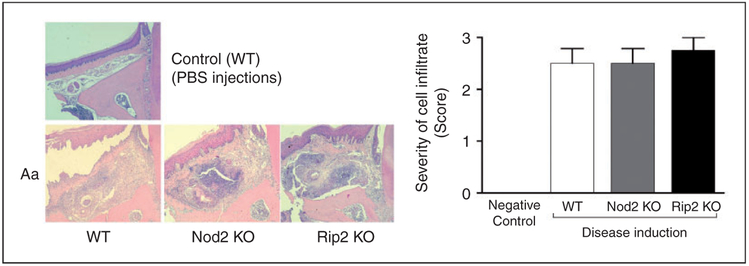
Figure 2.
Inflammatory infiltrate associated with experimental periodontitis in NOD2-KO and RIP2-KO mice. Representative images of 5 μm hematoxylin and eosin-stained sections from upper first molars (frontal or buccal–palatal plane) subjected to Aa injections, according to the genotype of the animal. A representative image of a control mouse (WT, PBS-injected) is shown for comparison purposes (no difference was noted in comparison with PBS-injected tissues of NOD2-KO and RIP2-KO mice; data not shown). The graph presents the analysis of cellular infiltrate using the severity score. At least four semi-serial sections obtained from samples from at least three different animals were analyzed for each group and these representative images were obtained at 100× magnification.
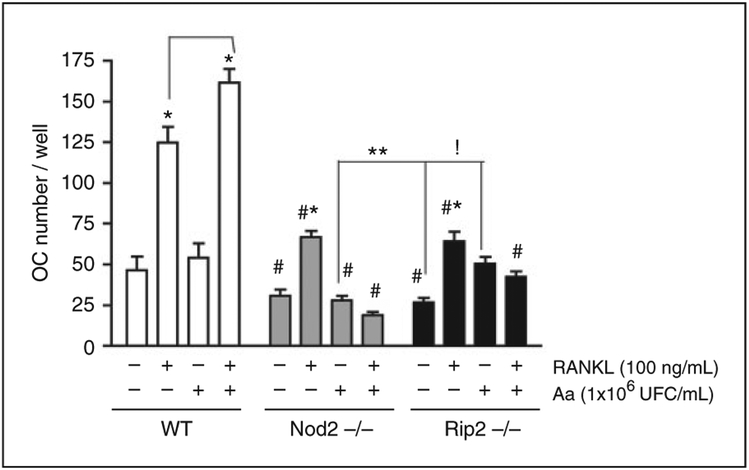
Figure 3.
Influence of NOD2 and RIP2 on osteoclastogenesis in vitro. BMDM isolated from WT, NOD2-KO or RIP2-KO mice were treated with RANKL (100 ng/ml) both independently and associated with 1 × 106 UFC/ml of heat-killed Aa in the presence of 20 ng/ml M-CSF for 6 d. RANKL and Aa treatments were repeated on d 3, and cells were fixed/permeabilized with paraformaldehyde/saponin, stained with FITC-conjugated phalloidin for the identification of actin ring formation and subsequently with Hoechst 33342 for identification of nuclei. Osteoclasts were counted on a digital inverted fluorescent microscope by a trained examiner unaware of the genotype and treatment. Data for each experimental condition within each genotype (WT, NOD2-KO, RIP2-KO) were analyzed by ANOVA followed by Bonferroni post-hoc tests. Pairwise comparisons between same experimental conditions between the different genotypes were performed with t-tests with Welch’s correction. All analyses were performed with GraphPad Prism 4 and the significance level set to 95% (P < 0.05). Asterisk (*) indicates significant difference from all other experimental conditions within each genotype; whereas exclamation mark (!) indicates significant difference between the bracketed columns within the same genotype (RIP2-deficient cells) by ANOVA. The hashtag (#) indicates significant reduction in comparison on osteoclast numbers in comparison with the same treatment in WT control cells, whereas two asterisks (**) indicates a significant difference between osteoclast numbers in the bracketed columns from different genotypes by unpaired t-tests with Welch’s correction for unequal variances.
NOD2 and RIP2 influence on the profile of inflammatory genes expressed by macrophages in response to heat-killed Aa
To assess the role of NOD2 and RIP2 on macrophage response to Gram-negative bacteria, we initially assessed the overall effect of deletion of these genes by extracting terms defining biological function of the list of genes up-regulated in stimulated macrophages using an online bioinformatics application (DAVID). To be considered ‘up-regulated’, we used a minimum of a fourfold increase over the expression level in unstimulated controls (or twofold increase on a log2 scale), which is more stringent than the guidelines for studies using treated primary cells in a microarray approach for hypothesis-generating studies.21 This stringent criterion was used owing to the use of pooled samples in the qPCR array analysis to reduce type I error, at the cost of accepting some false-negative results. In defining the functional gene clusters, DAVID parameters were also set to the highest stringency in order to generate less functional clusters, including more tightly associated genes in each cluster. The ‘score’ represents the overall enrichment score for the gene cluster, with scores > 2 considered to be of biological significance.19,20 To reduce the possibility of spurious associations, we have considered only the gene clusters associated with an enrichment score greater than 3. Table 1 indicates that the biological functions defined by clusters formed from the list of genes up-regulated in macrophages after bacterial stimulation is exactly the same in WT and NOD2-deficient macrophages (which is why the results are grouped in the same column of Table 1), whereas in RIP2-deficient macrophages the main functional annotations and their enrichment scores were different.
Table 1.
Comparison of functional annotation terms and enrichment scores for the top five gene clusters identified in the lists of genes up-regulated in macrophages by heat-killed Aa.
|
Gene cluster |
WT/NOD2−/− |
RIP2−/− |
||
|---|---|---|---|---|
| Functional annotation (top 3) | Score | Functional annotation (top 3) | Score | |
| 1 | Cytokine, cytokine activity, immune response | 10.13 | Cytokine, cytokine activity, extracellular space | 8.07 |
| 2 | Macrophage, four-helical cytokine, JAK-STAT signaling pathway | 6.79 | Disulfide bond, signal | 4.69 |
| 3 | Four-helical cytokine, JAK-STAT signaling pathway, disulfide bond | 6.40 | Defense response, regulation of cytokine production, Inflammatory response | 4.15 |
| 4 | Macrophage, lymphokine, hematopoietic cell lineage | 4.80 | Regulation of cytokine biosynthetic process,response to virus, immune response | 4.09 |
| 5 | Macrophage, hematopoietic cell lineage, myeloid leukocyte differentiation | 3.83 | Lymphokine, cytokines and inflammatory response, defense response to bacterium | 3.16 |
As both NOD2 and RIP2 did not affect the overall functional pathways modulated by heat-killed Aa in macrophages, we investigated (1) the potency of induction of gene expression in terms of the fold-change increase in comparison to unstimulated WT macrophages; and (2) the similarity of the specific genes included in the gene clusters and under the same KEGG pathway denomination term.
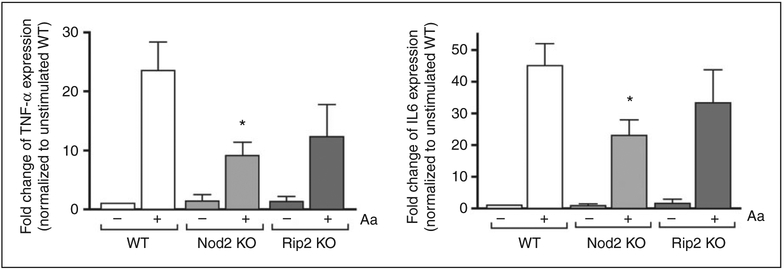
Results of the focused qPCR array indicate a clearly discernible decrease on the expression of several pro-inflammatory genes in both NOD2- and RIP2- deficient cells. Notably, macrophages lacking NOD2 stimulated with Aa presented a relative increase on the expression of anti-inflammatory IL-10 (Figure 4). These results were validated by RT-qPCR for IL-6 and TNF-α expression, performed using cDNA prepared from the RNA collected from the three independent experiments that were used to prepare the combined sample used on the arrays (Figure 5). In these confirmatory experiments, decrease on pro-inflammatory gene expression reached statistical significance only for NOD2-deficient macrophages, even though there was a discrete decrease, particularly for the expression of TNF-α in RIP2-deficient macrophages. Table 2 presents the data on the fold change up-regulation of genes according to the genotype.
Figure 4.
Overall influence of NOD2 and RIP2 on inflammation/innate immune response-associated gene expression in Aa-stimulated macrophages. Heat map of the expression of 84 genes associated with innate immunity in BMDM from WT, NOD2-KO or RIP2-KO mice stimulated with heat-killed Aa (106 UFC/ml) for 6 h. Total RNA from macrophages was harvested, quantitated and used to synthesize the cDNA used in qPCR-based arrays. These data are based on the analysis of cDNA prepared from pooled RNA samples harvested from three independent experiments (cells obtained from a minimum of three animals in each independent experiment) and the experimental groups were automatically arranged by the online data analysis tool based on the pattern of gene regulation.
Figure 5.
Validation of qPCR array data by RT-qPCR array. We used cDNA prepared from the same individual RNA samples from each of the three independent experiments performed with BMDM differentiated from WT, NOD2-KO or RIP2-KO mice. These same RNA samples were combined to prepare the pool used in the array analysis. Target gene expression was normalized to β-actin expression and fold regulation was calculated by comparison with the normalized gene expression in WT unstimulated macrophages. Asterisk (*) indicates P < 0.05 by unpaired Student’s t-tests with Welch’s correction for unequal variances.
Table 2.
Fold up-regulation of gene expression in macrophages from WT, NOD2-KO and RIP2-KO mice after 6 h-stimulation with heat-killed Aa compared with WT untreated control.
| Gene name | WT | NOD2-KO | RIP2-KO |
|---|---|---|---|
| Ccl2 | 4.05 | 6.39 | |
| Cd14 | 4.00 | ||
| Clec4e | 4.73 | 5.54 | |
| Csf2 | 6.64 | 7.78 | |
| Csf3 | 16.64 | 28.12 | 12.41 |
| Cxcl10 | 13.91 | 15.36 | 39.12 |
| Ifnb1 | 11.87 | 9.50 | 12.14 |
| Ifng | 5.69 | 7.89 | |
| Il10 | 12.36 | 26.95 | 24.99 |
| Il12a | 15.14 | 4.34 | |
| Il1a | 83.56 | 12.67 | 16.38 |
| Il1b | 45.77 | 9.48 | 13.71 |
| Il6 | 16.56 | 8.95 | 14.20 |
| Myd88 | 1243.42 | ||
| Nfkb2 | 4.94 | ||
| Nfkbia | 7.93 | 4.07 | 5.32 |
| Nfkbib | 4.31 | ||
| Ptgs2 | 17.86 | 7.64 | 7.37 |
| Tlr2 | 7.50 | ||
| Tnf | 29.52 | 15.61 | 10.79 |
| Tnfaip3 | 8.23 | 4.05 | |
| Tlr3 | 4.21 | ||
| Tlr9 | 4.25 |
As NOD2 and RIP2 affected the potency expression of inflammatory mediators induced by of microbial stimulation, we studied their influence on the production of selected proteins associated with immune response activation using multi-ligand ELISA.
Influence of NOD2 and RIP2 on the production of pro-inflammatory mediators and chemokines by bacterial-stimulated macrophages
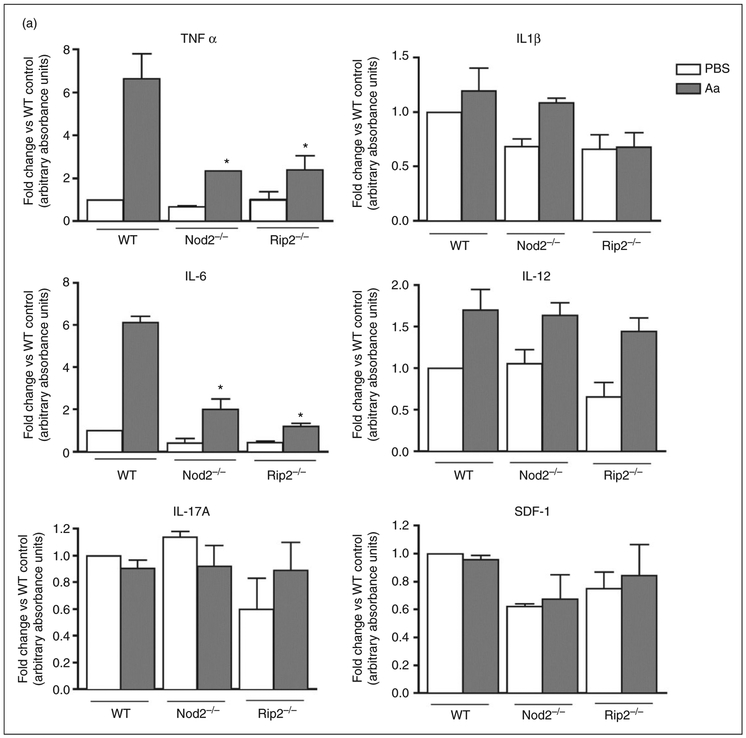
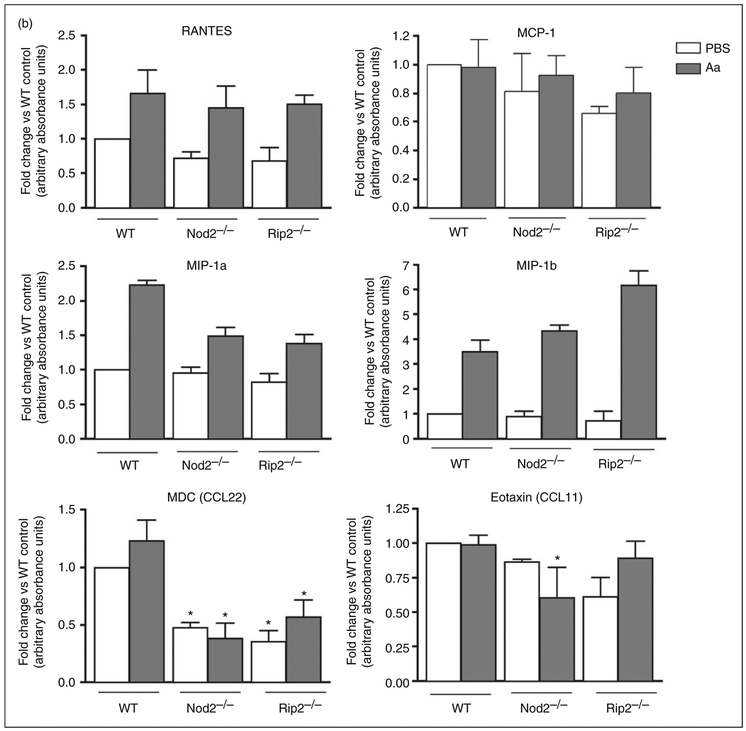
There was a statistically significant decrease on the expression of pro-inflammatory TNF-α and IL-6 in macrophages lacking NOD2 or RIP2 stimulated with heat-killed Aa (Figure 6). Notably, IL-6 protein levels were significantly decreased in RIP2-deficient macrophages, whereas the mRNA levels were not, suggesting the involvement of a post-transcriptional regulatory mechanism. The requirement of NOD2 and RIP2 for the production of TNF-α and IL-6 indicates that these genes have a profound effect on the responsiveness the macrophages to the bacterial Ags and point to the relevance of these genes in host-microbial interactions. Both NOD2 and RIP2-deficient macrophages also showed impaired production of CCL22, which is strongly chemotactic for dendritic cells and chronically activated T cells, suggesting that lack of NOD2 and RIP2 may affect adaptive immunity by indirect and direct mechanisms. Interestingly, production of CCL11, a chemotactic factor for eosinophils and primarily associated with allergic reaction and inflammation, was impaired only in NOD2-deficient macrophages. Production of IL-1β, IL-12 and IL-17A by bacterial-stimulated macrophages was not influenced by NOD2 and RIP2 (Figure 6).
Figure 6.
Effect of NOD2 and RIP2 on the production of inflammatory cytokines and chemokines by macrophages stimulated for 24 h with heat-killed Aa. BMDM derived from WT, NOD2-KO and RIP2-KO mice were stimulated with heat-killed Aa (106 UFC/ml) for 24 h. Cell culture supernatants from three independent experiments were collected and used in multi-ligand ELISAs to detect the production of pro-inflammatory (A) cytokines and (B) chemokines. Results are presented relative to the average constitutive production observed in unstimulated macrophages from WT mice (fold change). Asterisk (*) indicates P < 0.05 in comparison with the same experimental condition in cells from WT mice by unpaired Student’s t-tests with Welch’s correction for unequal variances.
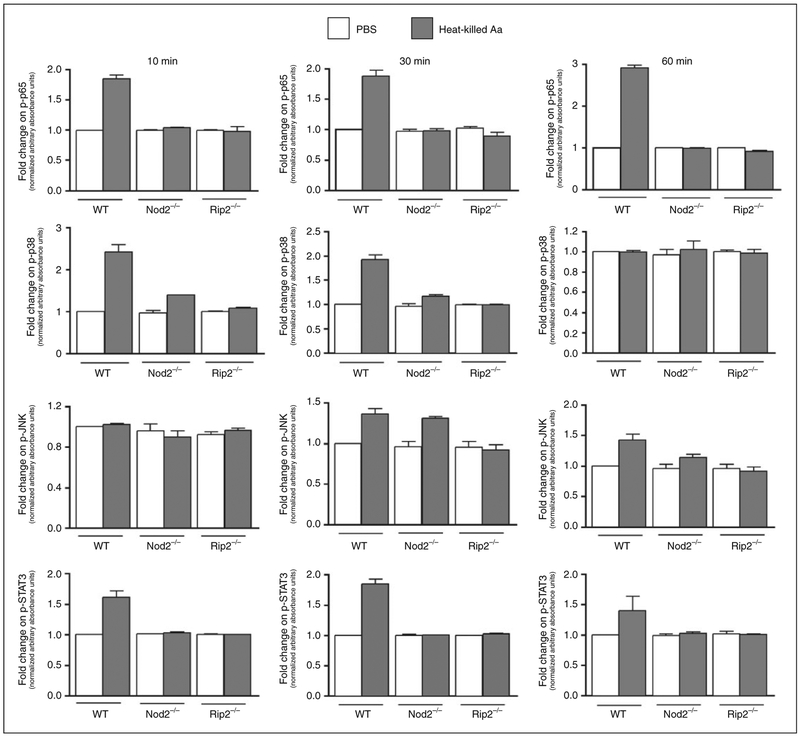
The influence of NOD2 and RIP2 on inflammatory and chemokine production may to be related with the marked attenuation of NF-κB and p38 MAPK signaling observed in NOD2- and RIP2-deficient macrophages (Figure 7). These are two major signaling pathways activated downstream of PRRs, including TLRs and NOD proteins.
Figure 7.
Relevance of NOD2 and RIP2 in the activation of signaling pathways in macrophages stimulated with heat-killed Aa. Macrophages differentiated from the bone marrow of WT, NOD2-KO or RIP2-KO mice were stimulated with heat-killed Aa for 10, 30 and 60 min. Total cell lysates were harvested and 30 μg used in multi-ligand ELISAs to detect phosphorylated forms of p65, p38, JNK and STAT3. These results were further normalized by the expression of total p65. Data were analyzed as relative change (fold change) to the normalized expression of each target protein by vehicle (PBS)-stimulated macrophages from WT mice in each experimental period (10, 30 and 60 min). Bars indicate averages and SDs of duplicate measurements. Data obtained using a pool of cell lysates from three independent experiments using cells derived from 6–8 different animals of each genotype.
Attenuation of the activation of these signaling pathways may also account for an effect on the level of gene expression, in spite of similarities on the genes and processes induced in macrophages by stimulation with Aa.
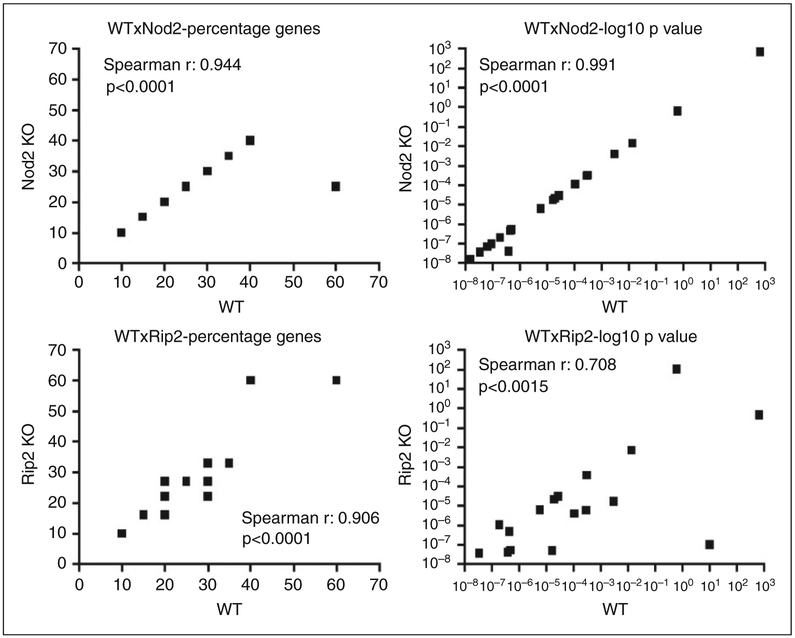
Assessment of the pathways affected using the Kyoto Encyclopedia of Genes and Genomes (KEGG) also indicated a great similarity (both on the percentage of genes and on the log10 of P-values indicating the statistical significance) in the top 20 KEGG terms defining the functions and utilities of genes that were up-regulated by heat-killed Aa in macrophages obtained from WT, NOD2-KO and RIP2-KO mice (Figure 8).
Figure 8.
Influence of NOD2 and RIP2 on the functional response (gene clusters) of BMDM to Gram-negative Aa. Correlations of percentage of genes included in each functional annotation term and of the enrichment statistical values obtained from DAVID Chart Reports. The correlation plots measure the annotation agreement between genes up-regulated by stimulation with heat-killed Aa in WT, NOD2−/− and RIP2−/− macrophages. The gene hit percentages and enrichment P-values of top enriched terms between the lists of up-regulated genes show very strong overall correlation. There is a lower correlation between the data generated from WT and RIP2−/− macrophages, suggesting that inactivation of RIP2 has a more pronounced effect on the pathways modulated by heat-killed Aa in macrophages than the inactivation of NOD2. Overall, these data indicate that heat-killed Aa induces common mechanisms/pathways in macrophages from WT, NOD2-KO and RIP2-KO mice.
Discussion
In this study, using in vivo and in vitro approaches we demonstrated that NOD2 (and to a lesser extent RIP2) is relevant for inflammatory bone resorption and osteoclastogenesis in experimental periodontitis. NOD2 and RIP2 are also required for maximum response of macrophages to Gram-negative bacterium Aa. NOD2 was initially described as a cytosolic receptor, functioning as an intracellular PRR-sensing bacterial peptidoglycan in innate immune cells. The basic assumption is that the ligands have to gain access to the cytosol to activate NOD2, which may happen by active phagocytosis or by bacterial invasion of the host cells. In this study we used heat-killed Gram-negative bacteria, which included many MAMPs that can activate multiple PRRs, including NOD1 and NOD2. In our model, we think that activation of NOD2 and RIP2 may occur by three possibilities: (1) phagocytosis of heat-killed bacteria by the macrophages with internalization of the ligands; (2) indirect activation by cross-talk with TLR-mediated activation of intracellular signaling pathways; (3) indirect activation by autocrine or paracrine effects of cytokines produced by the activation of membrane-bound TLR by the bacterial Ags. We have not addressed which of these possibilities would be involved in NOD2 and RIP2 activation in macrophages, as our in vitro experiments aimed to describe the role of NOD2 and RIP2 in the response of macrophages to Gram-negative bacteria (in our study model, Aa, a Gram-negative bacterium associated with periodontitis in humans), which may activate multiple TLRs and also NOD1.
NOD2 ligand MDP has been shown to enhance osteoclastogenesis induced by LPS and inflammatory cytokines in co-cultures of osteoblasts and hematopoietic cells by increasing RANKL mRNA expression in osteoblasts.22 This could be an explanation for the decrease on alveolar bone resorption without a corresponding marked decrease on the severity of inflammation assessed histologically in this study. It is also possible that the regulation of the expression of selected genes (as indicated by the in vitro studies, such as TNF-α, IL-6, IL-10, IFN-δ, IL-12, IL-1r1 and Pglyrp in NOD2-deficient macrophages and, to a lesser extent, in RIP2-deficient macrophages) may have occurred in the absence of obvious changes in the histological aspect of cellular infiltration. Thus, it is possible that lack of NOD2 and RIP2 affected osteoclastogenesis either directly (by affecting osteoclast precursor cells) or indirectly, by modulating expression of cytokines that are important for osteoclast differentiation. Using macrophages as osteoclast precursor cells, we observed a significant decrease on RANKL-induced osteoclastogenesis in both NOD2- and RIP2-deficient cells. Interestingly, concomitant stimulation with RANKL and heat-killed Aa enhanced osteoclastogenesis in WT macrophages, but further inhibited osteoclastogenesis in NOD2- and RIP2-deficient cells, suggesting a bone-sparing role for NOD1, which is still expressed in these cells.
Importantly, the attenuation of bone resorption in our loss-of-function model using NOD2-KO mice is supported by a recent study in which NOD2 agonists were introduced in vivo (in a gain-of-function model) and caused an increase in bone resorption.14 Interestingly, a recent publication did not find any role for NOD2 in bone resorption in another model of experimental periodontal disease. Using the ligature-induced model in mice, Jiao et al.12 showed reduced alveolar bone loss in RIP2-KO mice but not in NOD2-KO mice. The authors report a decrease on neutrophil infiltration in RIP2-KO mice only, associating these effects with the increased prevalence of an endogenous Gram-negative bacterium (NI1060) with great genetic similarity to Aa. The obvious difference in the experimental model (ligature for 10 d, vs. sustained stimulation with heat-killed bacteria over 3 wk) is demonstrated by the markedly greater severity of bone loss in the ligature model, which may be representative of a more acute inflammatory response (and/or significantly greater intensity of stimulus) than in our exogenous, heat-killed bacteria injection model. Moreover, we have not assessed the potential role of the endogenous microbiota, but we assume that as we injected the microorganisms their role on our results, if any, was minimal.
RIP2 is a kinase that is a common downstream target of both NOD1 and NOD2. The differences between the outcomes assessed in the absence of NOD2 and RIP2 both in vivo and in vitro may be related with an associated impairment of NOD1 function in RIP2-deficient cells and animals. In general, attenuation of bone resorption and macrophage response was less pronounced in the absence of RIP2, which may be due to a compensatory activation of inflammation/immune response pathways due to the more severe inhibition of innate immune response. However, inhibition of RIP2 kinase decreases production of PGE2 in human monocytes by inhibiting Cox-2 mRNA gene expression,23 which agrees with our results for Ptgs2 gene expression (the murine homolog of Cox-2) in murine macrophages, and is consistent with an inhibition of bone resorption.24 However, the magnitude of Ptgs2 inhibition we observed was nearly identical in NOD2- and RIP2-deficient cells, suggesting that this decrease is not the main responsible for the differences in attenuation of bone resorption between NOD2-KO and RIP2-KO mice in vivo.
It is important to consider that the data include the assessment of gene expression at the mRNA level, which does not necessarily reflect the protein level of the cytokines and enzymatic products (such as prostaglandins). Moreover, we used a qPCR focused array approach, which assesses a limited number of genes in comparison to a microarray approach. However, this approach has the advantage of focusing on genes directly related to host–microbial interactions. The fact that we used a pooled sample from three independent experiments (each experiment including cells from 3–4 mice) is a limitation; however, we validated the array data by RT-qPCR using cDNA prepared from the same RNA samples used to prepare the pool. We consider that by pooling equal quantities of total RNA from three independent experiments (each experiment including cells obtained from multiple mice) we included the inherent experimental variability into the data and used stringent criteria (consistent with a hypothesis-generating study) to select genes considered as up- or down-regulated. IL-6 and TNF-α cytokine production by Aa-stimulated macrophages was significantly inhibited in both NOD2- and RIP2-deficient cells, whereas IL-1β and SDF-1 production was discretely inhibited in RIP2- and NOD2-deficient cells, respectively. Modulation of gene expression at the mRNA and protein level is likely to be dependent on the marked inhibition of the activation of NF-kB and p38 MAPK observed. Surprisingly, our data indicate that Aa-induced activation of STAT3 was completely blocked in NOD2- and RIP2-deficient cells, which is unexpected as JAK-STAT is not a direct target of TLR/PRR signaling and suggests a role for NOD2/RIP2 in cross-talk leading to activation of this signaling pathway.
In any case, correspondence between in vitro and in vivo data has to be considered with caution; in the in vitro studies we used only macrophages as professional Ag-presenting/phagocytosing cells and a major cell type in inflammation and innate immunity; however, the participation of multiple cell types in vivo contributing to the cytokine network may result in distinct regulation of cytokines. Interestingly, NOD2 and RIP2 are required from microbial-induced expression of CCL22, a ligand for CCR4 with specific chemotactic activity for monocytes, dendritic cells and activated T cells of Th2 phenotype,25 suggesting that NOD2 and RIP2 may influence the adaptive immune response, possibly altering the Th-type response. CCL11 is classically associated with allergic response as it is chemotactic for eosinophils but not for neutrophils or mononuclear cells.26 TLR agonists were shown to induce its expression in in vivo models of sepsis;27 we observed a significant attenuation of microbial-stimulated CCL11 only in NOD2-deficient macrophages. In fact, heat-killed Gram-negative bacteria inhibited expression of CCL11 by human airway smooth muscle cells, which may be a mechanism for the effects of bacterial immunotherapy for allergic lung conditions.28 However, these authors suggested that the inhibition of CCL11 was mediated by bacterial DNA and not by TLR4 or NOD receptors, which is contradictory to our data. NF-kB and GATA-3 are the major common transcription factors with binding sites in the promoters of both CCL22 and CCL11, indicating that NF-kB could be an important pathway affected by the deletion of NOD2 in macrophages. The possible consequences of the modulation of chemokine expression on the adaptive immunity would only be perceived in vivo, and future studies will address the role of NOD2 in the nature of the adaptive immune response.
In summary, our data indicate an important role for both NOD2 and RIP2 on microbial-induced gene expression and RANKL-induced osteoclastogenesis in macrophages and also a relevant role for NOD2 in inflammatory-induced bone resorption and osteoclastogenesis in vivo; further exploration of the biological mechanisms may provide insight for therapeutic perspectives based on the modulation of NOD2 activation in conditions characterized by host–microbial interactions.
Acknowledgements
The authors wish to thank research technician Leandro Alves dos Santos (Department of Diagnosis and Surgery, School of Dentistry at Araraquara-Univ Estadual Paulista/UNESP) for his technical assistance in the histological processing and obtaining of the sections.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Financial support provided by Sao Paulo Research Foundation (FAPESP) grants to CRJ #2010/05783-5 and #2010/05632-7 and support from NIDCR grants R01DE017732 and R01DE021921 to DTG.
Footnotes
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Robertson SJ, Rubino SJ, Geddes K and Philpott DJ. Examining host-microbial interactions through the lens of NOD: from plants to mammals. Semin Immunol 2012; 24: 9–16. [DOI] [PubMed] [Google Scholar]
- 2.Zurek B, Proell M, Wagner RN, et al. Mutational analysis of human NOD1 and NOD2 NACHT domains reveals different modes of activation. Innate Immun 2012; 18: 100–111. [DOI] [PubMed] [Google Scholar]
- 3.Rubino SJ, Selvanantham T, Girardin SE and Philpott DJ. Nodlike receptors in the control of intestinal inflammation. Curr Opin Immunol 2012; 24: 398–404. [DOI] [PubMed] [Google Scholar]
- 4.Tigno-Aranjuez JT and Abbott DW. Ubiquitination and phosphorylation in the regulation of NOD2 signaling and NOD2-mediated disease. Biochim Biophys Acta 2012; 1823: 2022–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borzutzky A, Fried A, Chou J, et al. NOD2-associated diseases: bridging innate immunity and autoinflammation. Clin Immunol 2010; 134: 251–261. [DOI] [PubMed] [Google Scholar]
- 6.Petterson T, Jendholm J, Mansson A, et al. Effects of NOD-like receptors in human B lymphocytes and crosstalk between NOD1/NOD2 and Toll-like receptors. J Leukoc Biol 2011; 89: 177–187. [DOI] [PubMed] [Google Scholar]
- 7.Wagner CS and Cresswell P. TLR and nucleotide-binding oligomerization domain-like receptor signals differentially regulate exogenous antigen presentation. J Immunol 2012; 188: 686–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geddes K, Rubino S, Streutker C, et al. Nod1 and Nod2 regulation of inflammation in the Salmonella colitis model. Infect Immun 2010; 78: 5107–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tada H, Aiba S, Shibata K, et al. Synergistic effect of Nod1 and Nod2 agonists with toll-like receptor agonists on human dendritic cells to generate interleukin-12 and T helper type 1 cells. Infect Immun 2005; 73: 7967–7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kufer TA and Sansonetti PJ. NLR functions beyond pathogen recognition. Nat Immunol 2011; 12: 121–128. [DOI] [PubMed] [Google Scholar]
- 11.Pacios S, Kang J, Galicia J, et al. Diabetes aggravates periodontitis by limiting repair through enhanced inflammation. FASEB J 2012; 26: 1423–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiao Y, Darzi Y, Tawaratsumida K, et al. Induction of bone loss by pathobiont-mediated Nod1 signaling in the oral cavity. Cell Host Microbe 2013; 13: 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okugawa T, Kaneko T, Yoshimura A, et al. NOD1 and NOD2 mediate sensing of periodontal pathogens. J Dent Res 2010; 89: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kishimoto T, Kaneko T, Ukai T, et al. Peptidoglycan and lipopolysaccharide synergistically enhance bone resorption and osteoclastogenesis. J Periodontal Res 2012; 47: 446–454. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi KS, Chamaillard M, Ogura Y, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 2005; 307: 731–734. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi K, Inohara N, Hernandez LD, et al. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 2002; 416: 194–199. [DOI] [PubMed] [Google Scholar]
- 17.Marim FM, Silveira TN, Lima DS Jr. and Zamboni DS. A method for generation of bone marrow-derived macrophages from cryopreserved mouse bone marrow cells. PLoS One 2010; 5: e15263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu R, Bal HS, Desta T, et al. Diabetes enhances periodontal bone loss through enhanced resorption and diminished bone formation. J Dent Res 2006; 85: 510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT and Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang da W, Sherman BT and Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 21.Md Ley. Cytokine protocols, 2nd ed. Totowa, NJ, London: Humana; Springer; [distributor], 2012, p.1. [Google Scholar]
- 22.Yang S, Takahashi N, Yamashita T, et al. Muramyl dipeptide enhances osteoclast formation induced by lipopolysaccharide, IL-1 alpha, and TNF-alpha through nucleotide-binding oligomerization domain 2-mediated signaling in osteoblasts. J Immunol 2005; 175: 1956–1964. [DOI] [PubMed] [Google Scholar]
- 23.Taxman DJ, Lei Y, Zhang S, et al. ASC-dependent RIP2 kinase regulates reduced PGE2 production in chronic periodontitis. J Dent Res 2012; 91: 877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kats A, Bage T, Georgsson P, et al. Inhibition of microsomal prostaglandin E synthase-1 by aminothiazoles decreases prostaglandin E2 synthesis in vitro and ameliorates experimental periodontitis in vivo. FASEB J 2013; 27: 2328–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sebastiani S, Danelon G, Gerber B and Uguccioni M. CCL22-induced responses are powerfully enhanced by synergy inducing chemokines via CCR4: evidence for the involvement of first beta-strand of chemokine. Eur J Immunol 2005; 35: 746–756. [DOI] [PubMed] [Google Scholar]
- 26.Ponath PD, Qin S, Ringler DJ, et al. Cloning of the human eosinophil chemoattractant, eotaxin. Expression, receptor binding, and functional properties suggest a mechanism for the selective recruitment of eosinophils. J Clin Invest 1996; 97: 604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosmann M, Russkamp NF and Ward PA. Fingerprinting of the TLR4-induced acute inflammatory response. Exp Mol Pathol 2012; 93: 319–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Issa R, Sorrentino R, Sukkar MB, et al. Differential regulation of CCL-11/eotaxin-1 and CXCL-8/IL-8 by gram-positive and gramnegative bacteria in human airway smooth muscle cells. Respir Res 2008; 9: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]