Abstract
Histologic transformation to small cell neuroendocrine prostate cancer occurs in a subset of patients with advanced prostate cancer as a mechanism of treatment resistance. Rovalpituzumab tesirine (SC16LD6.5) is an antibody-drug conjugate that targets delta-like protein 3 (DLL3) and was initially developed for small cell lung cancer. We found that DLL3 is expressed in most of the castration-resistant neuroendocrine prostate cancer (CRPC-NE) (36 of 47, 76.6%) and in a subset of castration-resistant prostate adenocarcinomas (7 of 56, 12.5%). It shows minimal to no expression in localized prostate cancer (1 of 194) and benign prostate (0 of 103). DLL3 expression correlates with neuroendocrine marker expression, RB1 loss, and aggressive clinical features. DLL3 in circulating tumor cells was concordant with matched metastatic biopsy (87%). Treatment of DLL3-expressing prostate cancer xenografts with a single dose of SC16LD6.5 resulted in complete and durable responses, whereas DLL3-negative models were insensitive. We highlight a patient with neuroendocrine prostate cancer with a meaningful clinical and radiologic response to SC16LD6.5 when treated on a phase 1 trial. Overall, our findings indicate that DLL3 is preferentially expressed in CRPC-NE and provide rationale for targeting DLL3 in patients with DLL3-positive metastatic prostate cancer.
One-sentence summary:
DLL3 is overexpressed in neuroendocrine prostate cancer and is a potential therapeutic target.
Editor’s Summary:
Lessons from one tumor help with another
Neuroendocrine prostate cancer is an aggressive tumor subtype that can arise late in the course of the disease and drive therapeutic resistance. Puca et al. demonstrate the feasibility of targeting it with an antibody-drug conjugate against delta-like protein 3 (DLL3), a recently identified therapeutic being tested in small cell lung cancer, another aggressive neuroendocrine tumor. The authors confirmed the presence of DLL3 in most neuroendocrine prostate cancers, but not in localized tumors or normal prostate, and showed its association with aggressive clinical features. They then demonstrated the effectiveness of targeting DLL3 in multiple mouse models as well as a human patient.
INTRODUCTION
A subset of castration-resistant prostate cancers evades androgen receptor (AR)–targeted therapies by losing AR expression and/or signaling dependence and acquiring neuroendocrine features (1–4). In extreme cases, histologic transformation to small cell neuroendocrine carcinoma occurs. Castration-resistant small cell neuroendocrine prostate cancer (CRPC-NE) is clinically aggressive and does not respond well to standard prostate cancer therapies. Given their similarities with small cell lung cancer (SCLC), patients are commonly treated with platinum-based chemotherapy regimens that are standardly used for SCLC. Both SCLC and CRPC-NE harbor frequent loss of the tumor suppressor genes TP53 and RB1 (2, 5, 6) and overexpress achaete-scute complex-like 1/achaete-scute homolog 1 (ASCL1), a key transcriptional regulator (7–9). ASCL1 is required for SCLC development in mice harboring loss of Tp53:Rb:Rbl2 (10). ASCL1 also regulates the expression of Notch signaling proteins including delta-like protein 3 (DLL3). DLL3 is an inhibitory ligand of the Notch receptor pathway and is expressed during brain development where it regulates somitogenesis (11). In neural tissues, DLL3 is predominantly localized in the Golgi apparatus and can inhibit Notch receptor in cis (12, 13). DLL3 is aberrantly expressed on the surface of most of the SCLC cells, and the DLL3-targeted antibody-drug conjugate rovalpituzumab tesirine (SC16LD6.5) has demonstrated antitumor activity in SCLC preclinical models and in early-phase clinical trials (14, 15).
SC16LD6.5 is a humanized antibody against DLL3 that is used for internalization of the conjugated payload consisting of a DNA-damaging pyrrolobenzodiazepine (PBD) dimer toxin. PBD dimers bind to the minor groove of DNA, causing a stop of the replication fork and arrest of tumor cells in G2-M phase with consequent induction of apoptosis (16). Preclinical studies of SC16LD6.5 in patient-derived xenograft (PDX) models of SCLC and large cell neuroendocrine carcinoma demonstrated complete and durable eradication of tumors (15). The first clinical study using SC16LD6.5 in patients with recurrent metastatic SCLC and large cell neuroendocrine lung cancers (NCT01901653) identified the recommended phase 2 dose and schedule (0.3 mg/kg every 6 weeks), dose-limiting toxicities (thrombocytopenia, skin reaction, and serosal effusions), and objective responses in 17% (11 of 65) of patients and stable disease in 54% of patients (35 of 65) (14).
On the basis of the molecular similarities with SCLC, we sought to determine whether DLL3 expression is associated with the CRPC-NE phenotype in prostate cancer and to evaluate the antitumor activity of SC16LD6.5 in DLL3-expressing prostate cancer models.
RESULTS
DLL3 expression and clinical outcomes in neuroendocrine prostate cancer
DLL3 mRNA is expressed in adult normal brain compartment tissues (fig. S1), whereas in cancer tissues, it is enriched in SCLC and other small cell carcinomas (15, 17). We evaluated mRNA and protein expression of DLL3 in a prostate cancer cohort of 423 patients (735 samples), including benign prostate (n = 173 samples), localized prostate adenocarcinoma (PCA) (n = 324 samples), CRPC adenocarcinoma (CRPC-Adeno) (n = 127 samples), and CRPC-NE (n = 111 samples). There was no detectable protein expression of DLL3 in benign prostate (0 of 103) or localized prostate cancer (1 of 194, 0.52%). In contrast, DLL3 protein expression was present in most of the CRPC-NE (36 of 47, 76.6%) and in a subset of CRPC-Adeno (7 of 56, 12.5%). The percentage of DLL3-positive cells in tumor samples expressing DLL3 protein was 63.95% in CRPC-NE and 10.37% in CRPC-Adeno (Fig. 1, A and B, and fig. S2A). The mean H-score, combining the percentage of cells expressing DLL3 and the intensity of the signal (range, 0 to 300), was 0 for benign, 0.02 for localized prostate cancer, 10.57 for CRPC-Adeno, and 216.49 for CRPC-NE (P < 0.001) (Fig. 1C). The one localized prostate cancer that demonstrated DLL3 expression harbored only 2% positive cells (fig. S2B and table S1). In DLL3-expressing CRPC-Adeno, neuroendocrine markers such as synaptophysin (SYP) were often weakly expressed (representative CRPC-Adeno case; Fig. 1D and table S1).
Fig. 1. DLL3 expression and clinical outcomes in neuroendocrine prostate cancer.
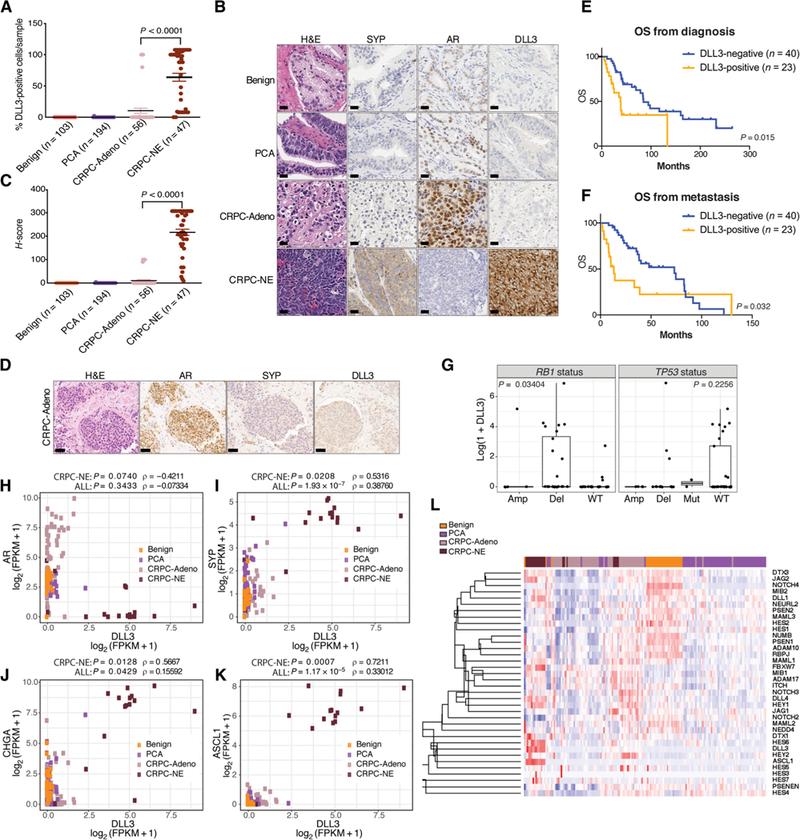
(A) Benign prostate samples (n = 103), PCA (n = 194), CRPC-Adeno (n = 56), and CRPC-NE (n = 47) were evaluated for the percentage of DLL3-positive cells per sample. Median value for CRPC-Adeno was 10.37% of DLL3-positive cells per tumor sample. Median value for CRPC-NE was 63.95% of DLL3-positive cells per tumor sample. Two-tailed t test was performed (P < 0.0001). The error bars indicate the SEM. (B) Representative IHC images of benign, PCA, CRPC-Adeno, and CRPC-NE stained by hematoxylin and eosin (H&E) and with AR, synaptophysin (SYP), and DLL3 antibodies. Scale bars, 50 μm. (C) H-score, combining the percentage of DLL3-positive cells and intensity of the signal for samples stained with DLL3 antibody (H-score: 0 to 300), two-tailed t test was used to compare CRPC-Adeno versus CRPC-NE samples (P < 0.0001). (D) Representative IHC image of a CRPC-Adeno sample stained by H&E and with AR, SYP, and DLL3 antibodies. Scale bars, 100 μm. (E) OS of patients with prostate cancer (n = 63) evaluated from the time of prostate cancer diagnosis. Patients are divided into DLL3-positive (n = 23) (orange) and DLL3-negative (n = 40) (blue) based on IHC, RNA-seq, and NanoString data. Log-rank test was used (P = 0.015). (F) OS of patients with prostate cancer (n = 63) evaluated from the time of metastasis. Patients are divided into DLL3-positive (n = 23) (orange) and DLL3-negative (n = 40) (blue). Log-rank test was used (P = 0.032). (G) Correlation of DLL3 expression and RB1 and TP53 genomic status (amplification, Amp; deletion, Del; mutation, Mut; wild-type, WT) (RB1 deletion versus no deletion: Mann-Whitney, P = 0.03404; TP53 WT versus no WT: Mann-Whitney, P = 0.2256). (H) Correlation of DLL3 and AR expression [log2 (FPKM + 1)] in benign, PCA, CRPC-Adeno, and CRPC-NE patient samples evaluated by RNA-seq (Spearman correlation within CRPC-NE samples, ρ = –0.4211, P = 0.0740; in all samples, ρ = −0.07334, P = 0.3433). (I) Correlation of DLL3 and SYP expression [log2 (FPKM + 1)] in benign, PCA, CRPC-Adeno, and CRPC-NE patient samples evaluated by RNA-seq (Spearman correlation within CRPC-NE samples, ρ = 0.5316, P = 0.0208; in all samples, ρ = 0.38760, P = 1.93E-07). FPKM, fragments per kilobase of transcript per million mapped reads. (J) Correlation of DLL3 and CHGA expression [log2 (FPKM + 1)] in benign, PCA, CRPC-Adeno, and CRPC-NE patient samples evaluated by RNA-seq (Spearman correlation within CRPC-NE samples, ρ = 0.5667, P = 0.0128; in all samples ρ = 0.15592, P = 0.0429). (K) Correlation of DLL3 and ASCL1 expression [log2 (FPKM + 1)] in benign, PCA, CRPC-Adeno, and CRPC-NE patient samples evaluated by RNA-seq (Spearman correlation within CRPC-NE samples, ρ = 0.7211, P = 0.0007; in all samples, ρ = 0.33012, P = 1.17E-05). (L) Supervised cluster analysis of Notch signaling genes in benign, PCA, CRPC-Adeno, and CRPC-NE patient samples.
We also mined published mRNA datasets (29 benign, 66 PCA, 55 CRPC-Adeno, and 19 CRPC-NE) (1,2,18) and performed targeted mRNA analysis for 166 additional cases using a custom NanoString assay described in (2) (41 benign, 64 PCA, 16 CRPC-Adeno, and 45 CRPC-NE). Similarly, DLL3 mRNA was highly expressed in 73.68 and 71.10% of CRPC-NE cases analyzed by RNA sequencing (RNA-seq) and NanoString, respectively, and in 10.91% (RNA-seq) and 12.50% (NanoString) of CRPC-Adeno samples (figs. S3 to S5). In nine cases, where mRNA data revealed DLL3 expression but immunohistochemistry (IHC) was negative, we developed probes to evaluate DLL3 mRNA expression in situ by RNA in situ hybridization (ISH); five of these nine cases demonstrated concordance between RNA ISH and IHC (fig. S6).
To understand whether DLL3 associates with prognosis in patients with prostate cancer, we examined DLL3 tumor positivity by protein expression or mRNA in relation to overall survival (OS) in patients with CRPC-Adeno or CRPC-NE, with outcomes data available (n = 63). Overall, we found that DLL3 expression was associated with worse OS compared to DLL3-negative cases with OS calculated from the time of prostate cancer diagnosis [OS, 35 months versus 81.5 months; hazard ratio (HR), 2.22; 95% confidence interval (CI) 1.04 to 4.74, P = 0.015] and from the time of metastasis (11.8 months versus 71.6 months; HR, 1.91; 95% CI 0.92 to 3.96, P = 0.032) (Fig. 1, E and F).
We evaluated the association of DLL3 expression with genomic features and found that DLL3 expression was higher in samples with RB1 genomic loss (10 of 12, 83% of evaluable patients) (RB1 deletion versus no deletion: Mann-Whitney, P = 0.03404), but there was no association between DLL3 expression and TP53 mutation or deletion (3 of 12, 25% of evaluable patients) [TP53 WT versus no WT: Mann-Whitney, P = 0.2256] (Fig. 1G). A positive correlation was observed between DLL3 and classical neuroendocrine markers SYP (Spearman correlation within CRPC-NE samples, ρ = 0.5316, P = 0.0208; in all samples, ρ = 0.38760, P = 1.9 × 10–7) and chromogranin A (CHGA) (Spearman correlation within CRPC-NE samples, ρ = 0.5667, P = 0.0128; in all samples, ρ = 0.15592, P = 0.0429); no significant anticorrelation was detected between DLL3 and AR (Spearman correlation within CRPC-NE samples, ρ = –0.4211, P = 0.0740; in all samples, ρ = –0.07334, P = 0.3433) (Fig. 1, H to J). The DLL3 promoter was hypomethylated in patients with CRPC-NE versus patients with CRPC-Adeno (fig. S7), suggesting a possible mechanism causing up-regulation of DLL3 in CRPC-NE.
DLL3 is transcriptionally regulated by ASCL1, a master regulator of neuroendocrine differentiation and classic SCLC development (10). We found that ASCL1 was also highly expressed in CRPC-NE tumors (fig. S8) and significantly correlated with DLL3 expression (Spearman correlation within CRPC-NE samples, ρ = 0.7211, P = 0.0007; in all samples, ρ = 0.33012, P = 1.17 × 10–5) (Fig. 1K). A supervised cluster analysis of Notch signaling genes [receptors, ligands, and Hes family basic helix-loop-helix (bHLH) transcription factor (HES)/Hes-related family bHLH transcription factor with YRPW motif (HEY) targets] including DLL3 and ASCL1 showed clustering of patients with CRPC-NE tumors (Fig. 1L), and ASCL1 and DLL3 were the most differentially expressed Notch signaling genes when comparing CRPC-Adeno versus CRPC-NE samples, CRPC-NE versus PCA, or CRPC-NE versus benign (fig. S9). To understand whether manipulating DLL3 expression could modify the expression of neuroendocrine-associated genes, we transiently silenced DLL3 in the DLL3-expressing CRPC-NE cell line NCI-H660 and found that this did not affect AR or CRPC-NE signaling or Notch target gene expression (fig. S10).
Temporal and intrapatient tumor heterogeneity
We assessed DLL3 expression across multiple tumor samples obtained at the time of rapid autopsy of one patient with CRPC-NE (W CM677) (19). All metastatic sites (liver, lung, and rib) and his primary tumor at autopsy showed consistent high expression of DLL3 and histological features of small cell carcinoma (Fig. 2A). When looking back to his archival primary prostate tumor biopsy at the time of initial diagnosis (3 years earlier), his primary Gleason 9 PCA was completely negative for DLL3 expression, but a metastatic CRPC-Adeno bone biopsy of his right humerus (1 year before death) was diffusely positive (Fig. 2A). These data demonstrate the acquisition of DLL3 expression with time and treatment resistance.
Fig. 2. Temporal and intrapatient tumor heterogeneity in two patients.
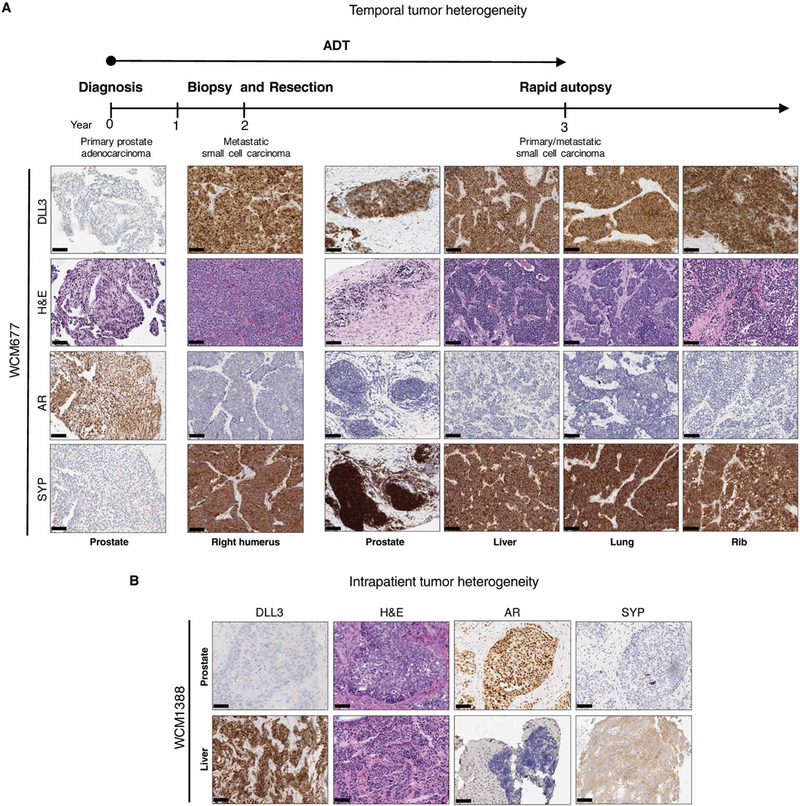
(A) Representative H&E histology images with DLL3, AR, and SYP IHC of a patient with prostate cancer, followed from initial diagnosis of prostate cancer (year 0) to autopsy (year 3). Prostate (year 0), right humerus (year 2), and autopsy (year 3). Scale bars, 100 μm. Samples were obtained from the prostate, liver, lung, and rib. Androgen deprivation therapy (ADT) treatment is indicated by the arrow. (B) Representative H&E and representative DLL3, AR, and SYP IHC images of prostate and concurrent liver biopsies derived from the same patient at the same time point. Scale bars, 100 μm.
In contrast, another patient with an atypical aggressive presentation of metastatic prostate cancer had two biopsies at the same time point with discordant DLL3 results (WCM1388). He initially presented with lytic bone, lung, and liver metastases, with a serum prostate-specific antigen (PSA) of 6.24 ng/ml. Although a prostate tumor biopsy showed PCA, a concurrently performed liver biopsy showed small cell carcinoma. Despite shared genomic features between metastatic sites, including RB1, TP53, and PTEN loss (table S2), DLL3 was not expressed in the primary tumor but was strongly expressed in the metastatic liver biopsy (Fig. 2B).
In the third patient with CRPC-NE, whose tumor tissue harbored both DLL3-positive cells (80%, ranging from weak to strong intensity) and DLL3-negative cells (20%), both cell types expressed the neuroendocrine marker SYP (fig. S11). This may reflect different stages of this plasticity phenotype in an individual tumor.
DLL3 detection in circulating tumor cells
Because CRPC-NE typically develops during later stages of prostate cancer progression and metastatic biopsies are invasive to perform, we investigated the feasibility of using circulating tumor cells (CTCs) to detect DLL3 expression in patients using the Epic Sciences platform (2, 20–22). We evaluated CTCs from 36 patients with CRPC-Adeno (n = 17) or CRPC-NE (n = 19). Both subclasses showed a similar average number of traditional cytokeratin (CK+) CTCs and CK+ CTC clusters (21) per milliliter of blood (CRPC-NE = 38.92 CTCs/ml and CRPC-Adeno = 37.26 CTCs/ml). DLL3 was detected in CTCs in 16 of 36 patients (44.4%) with a cutoff of expression defined using DLL3-positive SCLC and DLL3-negative squamous cell lung carcinoma cell lines (fig. S12). Patients with DLL3-expressing CTCs exhibited a wide range in the proportion of DLL3-positive CTCs (4 to 98%), DLL3-positive CTCs/ml (0.6 to 141.9 CTCs/ml), and DLL3 signal intensity (7 to 313). The observed median DLL3 signal intensity was 15-fold greater in DLL3-positive cells than in DLL3-negative cells (22.7 versus. 1.5). DLL3 was expressed in both CK+ and CK– CTCs (fig. S13). Overall, we observed DLL3-expressing CTCs in 11 of 19 (57.8%) patients with CRPC-NE and 5 of 17 (29.4%) patients with CRPC-Adeno (Fig. 3A). The five DLL3-expressing patients with CRPC-Adeno had aggressive clinical features including liver metastases and RB1 and TP53 genomic alterations (table S3).
Fig. 3. DLL3 detection in CTCs.
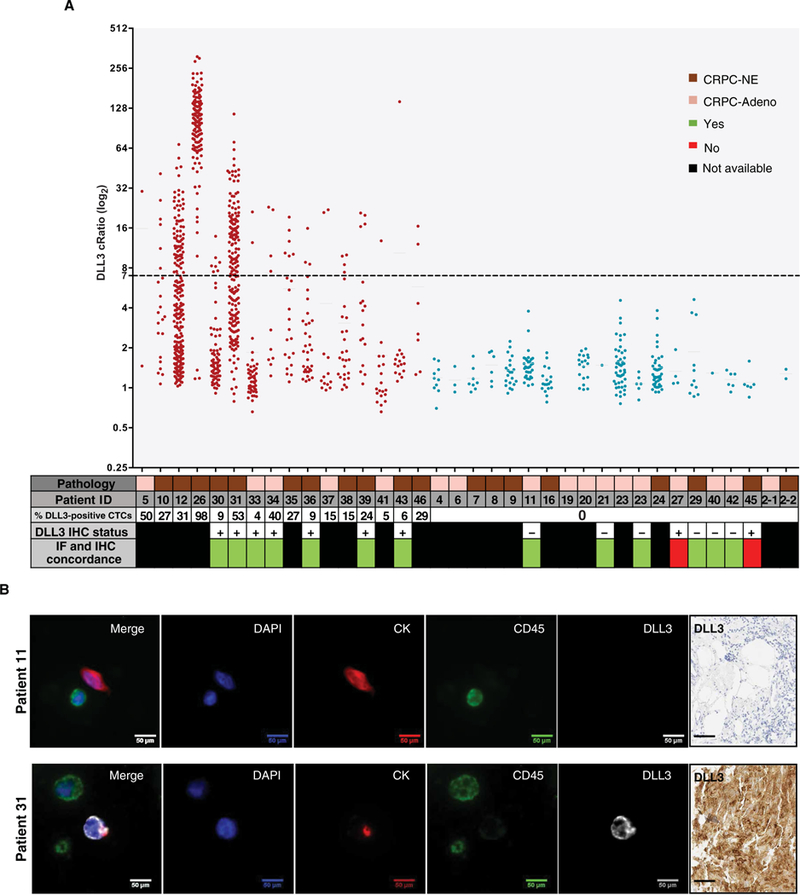
(A) Graph showing DLL3 positivity in CTCs of 36 patients with CRPC-Adeno and CRPC-NE tested with the DLL3 Epic four-color immunofluorescence (IF) assay. DLL3 cRatio (signal-to-noise ratio) is plotted along the y axis, and patient ID is plotted along the x axis. The x axis also includes a data table with additional patient-specific information including pathology [CRPC-NE (brown) or CRPC-Adeno (light brown)], percentage of DLL3-positive CTCs, DLL3 IHC status, and IF and IHC concordance. Samples expressing DLL3 are indicated in red, and samples negative for DLL3 are indicated in blue. Each dot represents a detected cell, and the dashed line at 7 along the y axis indicates the analytical threshold of positivity for DLL3. (B) Representative images of patient 11 (DLL3-negative) and patient 31 (DLL3-positive) CTCs. In the four-color composite image, blue represents 4′,6-diamidino-2-phenylindole (DAPI), and red is used for CK, green for CD45, and white for DLL3. Other images show the independent images for each channel used to create the four-color composite (DAPI, CK, CD45, and DLL3). Scale bars, 50 μm. Corresponding DLL3 IHC image is also included for both patients. Scale bars, 50 μm.
DLL3 expression in CTCs of patients was concordant with DLL3 IHC status of matched metastatic tumor biopsies in 13 of 15 patients (87%). In particular, seven of nine (78%) patients who were DLL3-positive on biopsy by IHC were also positive in CTCs, whereas six of six (100%) patients who were DLL3-negative on tumor biopsy by IHC were also negative in CTCs (Fig. 3, A and B). In patients with CRPC-NE, with both DLL3-positive and DLL3-negative CTCs, there were no significant differences in CTC morphology between cells (for example, nuclear area, nuclear speckles, nuclear major axis, cytoplasmic area, cytoplasmic speckles, cytoplasmic major axis, and nuclear/CK ratio) (fig. S14) (23).
Antitumor activity of SC16LD6.5
To explore SC16LD6.5 as a treatment option for DLL3-expressing prostate cancer, we used DLL3-positive neuroendocrine prostate cancer models including NCI-H660 cell line xenografts and the PDX model LTL352 [described in Lin et al. (24)], and for DLL3-negative prostate cancer models, we used DU145 cell line xenografts and the patient-derived organoid xenograft (PDOX) WCM1262 PDOX [described in Puca et al. (25)] (Fig. 4A). In line with the inhibitory effect of DLL3 on Notch signaling (26), the DLL3-expressing models had lower Notch receptor expression (Fig. 4B), especially NOTCH2 (DLL3 versus NOTCH2 in patient samples, r = –0.502963, P = 3.197 × 10–12) (fig. S15). The xenografts were treated with a single dose of vehicle, IgG1LD6.5 (isotype control antibody-drug conjugate), or SC16LD6.5 administered intraperitoneally, and tumor volume was monitored after treatment. Treatment of the NCI-H660 and LTL352 xenografts with SC16LD6.5 resulted in complete responses at 35 days after treatment (Fig. 4, Cand D, and table S4), whereas DU145 and WCM1262 PDOX tumors continued growing despite the treatment, reaching the tumor volume limit allowed by our animal protocol. Although the potent PBD warhead was cytotoxic to both cell lines in vitro and presumably toxic in vivo, SC16LD6.5 preferentially inhibited cell viability of NCI-H660 cells versus DU145 (fig. S16). Together, these results indicate that SC16LD6.5 treatment in prostate cancer models expressing DLL3 markedly inhibits tumor growth.
Fig. 4. DLL3 antibody drug-conjugate activity in DLL3-expressing neuroendocrine prostate tumors in vivo.
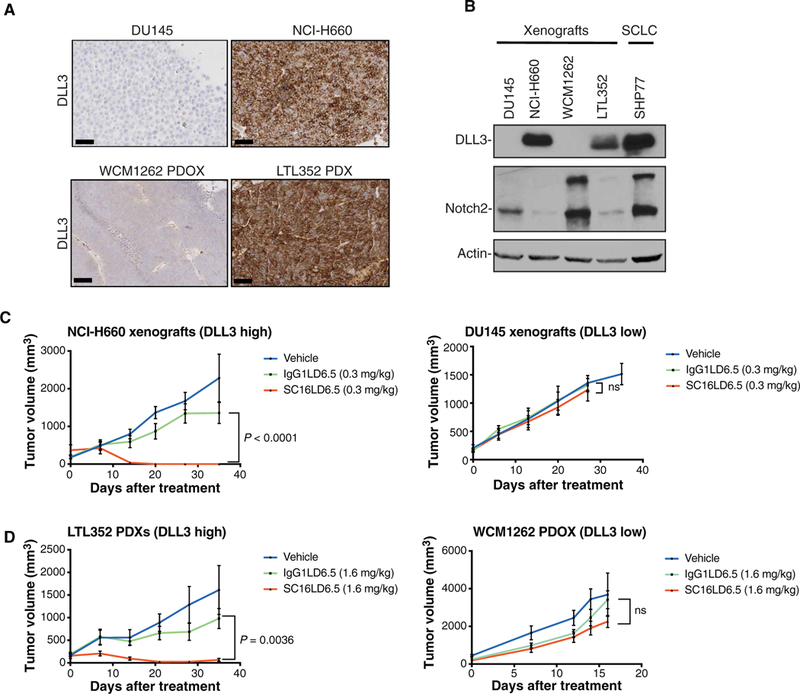
(A) Representative DLL3 IHC images of DU145 cells and WCM1262 PDOX (DLL3-negative) and NCI-H660 cells and LTL352 PDX (DLL3-positive). Scale bars, 100 μm. (B) Western blot analysis of tissue derived from DU145, NCI-H660, WCM1262, and LTL352 xenografts using DLL3 and Notch2 antibodies. Actin is used as a loading control. The SHP77 cell line is used as a control for DLL3 expression. (C) DU145 and NCI-H660 tumor volume (mm3) measurements after single-dose treatment of vehicle, IgG1LD6.5 (0.3 mg/kg), and SC16LD6.5 (0.3 mg/kg) in NU/J mice evaluated for 35 days after treatment. Two-way analysis of variance (ANOVA) test was performed (P < 0.0001). ns, not significant. (D) LTL352 PDX and WCM1262 PDOX tumor volume (mm3) measurements after single-dose treatment of vehicle, IgG1LD6.5 (1.6 mg/kg), and SC16LD6.5 (1.6 mg/kg) in NOD scid gamma mice evaluated for 35 days after treatment. Two-way ANOVA test was performed (P = 0.0036).
Clinical response in a patient enrolled on a phase 1 SC16LD6.5 basket trial
We report a patient with DLL3-expressing neuroendocrine prostate cancer treated on an ongoing phase 1 basket trial of rovalpituzumab tesirine (SC16LD6.5) (NCT02709889). This patient is 71 years old, and he underwent prostatectomy for pT3aN0, with a Gleason score 4 + 4 = 8 with tertiary pattern 5 PCA with 20% small cell carcinoma component (positive for SYP) and serum PSA of 5.9 ng/ml (Fig. 5, A and B). He received adjuvant radiotherapy and ADT. Two years later, he developed extensive nodal metastases in the setting of an undetectable PSA. A lymph node biopsy revealed metastatic small cell carcinoma (Fig. 5C) harboring TP53 mutation, RB1 deletion, and PTEN deletion and diffused positivity for DLL3 (85% cells expressed DLL3, 2+ intensity; Fig 5D). After progression on cisplatin and etoposide, he was enrolled in the SC16LD6.5 phase 1b basket trial. He was treated at a dose of 0.3 mg/kg every 6 weeks, and scans after cycle 1 showed clinical improvement including marked shrinkage of target nodal metastases (42 to 24 mm) and complete and partial responses of nontarget lesions (Fig. 5E). Scans after cycle 2 remained stable (24 mm) with no new lesions seen (Fig. 5E).
Fig. 5. Clinical response in a patient enrolled on a phase 1 SC16LD6.5 basket trial.
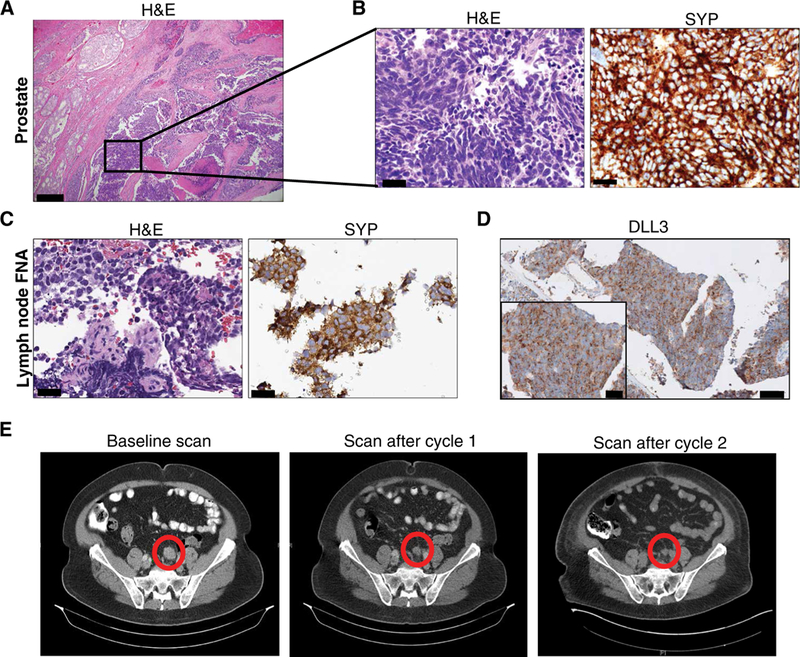
(A) Representative H&E image of radical prostatectomy specimen that shows a PCA (left) with an adjacent component of small cell neuroendocrine carcinoma (inset). Scale bar, 200 μm. (B) H&E and strong diffuse synaptophysin (SYP) IHC of the small cell carcinoma component. Scale bars, 50 μm. (C) Lymph node biopsy performed 2 years after prostatectomy. Representative images of H&E and SYP IHC are shown. Scale bars, 50 μm. Fine-needle aspiration, FNA. (D) Representative images of DLL3 IHC are shown (scale bar, 200 μm; inset scale bar, 50 μm). (E) CT scans before therapy and after cycles 1 and 2 of SC16LD6.5 (nodal metastasis highlighted by a circle).
DISCUSSION
In this study, we found that DLL3 is expressed in a subset of advanced metastatic prostate cancers and is not notably expressed in localized prostate cancer or benign tissues, making DLL3 an attractive therapeutic target. Expression of DLL3 correlated with aggressive clinical features including neuroendocrine morphology and poor survival. With the encouraging in vivo data and patient response seen with the antibody-drug conjugate rovalpituzumab tesirine (SC16LD6.5), these data provide rationale for performing DLL3-targeted testing and the clinical evaluation of DLL3-directed therapy for patients with DLL3-positive metastatic prostate cancer.
Because we found that DLL3 is mostly expressed in metastatic and treatment-resistant prostate cancer rather than localized disease, interrogating DLL3 expression for trial enrollment should be considered using metastatic tumor biopsy tissue. This approach may be challenging to perform serially. As an alternative, we showed that CTC testing for DLL3 expression is feasible and may potentially serve as a noninvasive strategy to track the emergence of DLL3 expression.
How and when DLL3 becomes expressed during prostate cancer progression and its role in treatment resistance require further investigation. Previous data have linked members of the Notch pathway with normal prostate development and early stages of metastatic prostate cancer. A mouse model with conditional Notch1 gene deletion showed increased proliferation and tumorigenesis (27). In Pten null mice, Notch activity promoted metastasis and epithelialmesenchymal transition (28), whereas the Notch ligand Jagged 1 has been associated with prostate cancer recurrence after prostatectomy (29). Notch target genes, HEYL, HEY1, and HEY2, have been associated with prostate cancer by acting as corepressors of AR transcriptional activity by interacting with SRC1 and AR (30, 31). However, little is known about the function of Notch signaling in more advanced and non-AR-driven disease. In the current study, we found that DLL3 and ASCL1 were highly expressed in CRPC-NE and were the most differentially expressed Notch signaling genes compared to PCA. In SCLC, ASCL1 has been implicated in driving the functional loss of RB1 by cyclin-dependent kinase 5-mediated phosphorylation (32), and RB1 loss is also one of the most common alterations in CRPC-NE (2, 5, 33). Further studies focused on the functional role of ASCL1 and dysregulation of Notch activity in modulating or maintaining the neuroendocrine phenotype are warranted.
Silencing DLL3 in cell lines did not affect the expression of downstream Notch signaling genes or other molecular changes, suggesting that DLL3 is likely not a driver of the neuroendocrine phenotype in prostate cancer, at least in vitro, but may represent a potential target for the disease. In vivo studies are warranted to further explore the functional role of DLL3. Furthermore, a complex pattern of Notch signaling gene expression observed in patient tumors, with some Notch-related genes (DLL4, HES6, DTX1, and JAG2) up-regulated and others (mainly Notch 2 and 4 receptors) considerably down-regulated in CPRC-NE, highlights a potentially multifaceted role of Notch signaling in prostate cancer: It could act as an oncogene in early stages of prostate cancer by promoting metastatic progression and potentially as an oncosuppressor in later stages of the disease, as signaling is lost or inhibited along with DLL3 up-regulation. Overall, our study provides insights regarding DLL3 expression in metastatic prostate cancer and a rationale for further clinical development of DLL3-targeted therapeutics for a subset of patients with advanced prostate cancer.
MATERIALS AND METHODS
Study design
The objective of this study was to evaluate the expression of DLL3 across various stages of prostate cancer using different platforms (IHC, NanoString, and RNA-seq) and demonstrate the efficacy of rovalpituzumab tesirine (SC16LD6.5) treatment in preclinical models of advanced prostate cancer expressing DLL3. The total sample size for DLL3 expression evaluation in patient cohort was 735 (173 benign, 324 PCA, 127 CRPC-Adeno, and 111 CRPC-NE), and this was sufficient to detect statistically significant differences. For in vivo efficacy studies, the sample size was n = 5 to 9 mice per group. Animals were randomized on the basis of tumor size so that each treatment group had average tumor volumes of 150 to 450 mm3 at the time of treatment. Tumor volume was measured with a digital caliper and calculated as V (volume) = W (width)2 × L (length)/2. Data collection was stopped in the following conditions: ulcerated/necrotic tumors, skin infections with antibiotic administration, tumors reaching the protocol limit in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines, or when mice reached the end of the study. Sample size for OS analysis in the patient cohort was n = 63 and was sufficient to determine statistically significant differences using the log-rank test. Analysis of tissue microarrays (TMAs) and histological slides was performed blindly by pathologists.
Cohort description and pathology classification
Tumor biopsies were collected and approved by the Weill Cornell Medicine (WCM) Institutional Review Board (IRB) with informed consent (IRB nos. 1305013903 and 0905010441). H&E-stained slides and IHC images were reviewed by the study pathologists (J.M.M., V.S., A.V., and K.I.). Histological criteria for prostate cancer with neuroendocrine differentiation are detailed in (34).
DLL3 IHC cohort evaluation
DLL3 IHC was evaluated on a total of 400 samples from 228 patients. Samples included benign prostate tissue (n = 103, 25.75%), localized prostate adenocarcinoma (n = 194, 48.5%), castration-resistant prostate adenocarcinoma (n = 56, 14%), high-grade prostate cancer with neuroendocrine differentiation (n = 19, 4.7%), and small cell carcinoma (n = 28, 7%) (34).
Tissue microarrays
TMAs were constructed as described before (35). Both TMA and whole-section slides were stained with the anti-DLL3 antibody. Staining was evaluated in a semiquantitative manner using an H-score (36). Staining intensity (0 = no, 1 = mild, 2 = moderate, and 3 = strong staining) was multiplied by the percentage of positive cells resulting in a score ranging from 0 to 300.
Image acquisition
Microscopy images were taken using an INFINITY2 camera and the INFINITY ANALYZE software version 6.5.3 (Lumenera) with an Olympus BX43 microscope (Olympus). Some images were taken from slides scanned with the Leica Aperio AT2 digital slide scanner and the eSlide Manager software (Leica).
In vivo treatments
Organoid cells (WCM1262 PDOX) and cell lines (NCI-H660 and DU145) were subcutaneously injected with Matrigel (1:1; Corning), whereas tumor fragments from PDX LTL352 were implanted subcutaneously into NU/J mice (NCI-H660 and DU145) or nonobese diabetic mice (NOD) severe combined immunodeficient (scid) gamma (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) male mice (WCM1262 PDOX and LTL352) (the Jackson laboratory). Mice used were 6- to 8-week-old males. Tumor volume was measured every week with a digital caliper. When tumor volume reached 150 to 450 mm3, the animals were treated intraperitoneally with a single dose of vehicle (5% glucose in H2O), IgG1LD6.5, or SC16LD6.5. The doses, 0.3 mg/kg in NU/J and 1.6 mg/kg in NOD scid gamma mice, were adjusted to compensate for expected differences in clearance (37), and pharmacokinetics studies demonstrated that exposures using these doses (0.3 mg/kg in NU/J and 1.6 mg/kg in NOD scid gamma mice) were generally comparable (table S4). The number of animals used per type of xenograft was as follows: DU145 xenografts: vehicle (n = 7), IgG1LD6.5 (n = 5), and SC16LD6.5 (n = 9); NCI-H660 xenografts: vehicle (n = 8), IgG1LD6.5 (n = 8), and SC16LD6.5 (n = 8); WCM1262 PDOX xenografts: vehicle (n = 5), IgG1LD6.5 (n = 5), and SC16LD6.5 (n = 6); and LTL352 xenografts: vehicle (n = 6), IgG1LD6.5 (n = 7), and SC16LD6.5 (n = 7). The animals were sacrificed in a CO2 chamber in accordance with IACUC guidelines. Tumors were harvested and used for histology analysis or rapidly frozen.
Clinical sample RNA-seq data processing
The dataset included 55 CRPC-Adeno, 19 CRPC-NE, 66 PCA, and 29 benign samples. Of these tumor specimens, 26 CRPC-Adeno, 19 CRPC-NE, 66 PCA, and 29 benign samples were previously published (1, 2). Additional two CRPC-Adeno tumor specimens were obtained and sequenced prospectively through clinical protocols approved by the WCM IRB (IRB nos. 1305013903 and 1210013164). RNA-seq data for the remaining CRPC-Adeno cases were obtained through the Stand Up to Cancer-Prostate Cancer Foundation (SU2C-PCF) clinical trial (18), and data were accessed for this study through cBioPortal (38). Reads were mapped to the human genome reference sequence (hg19/GRC37) using STAR v2.3.0e (39). For each sample, HTSeq (40) and Cufflinks (41) were then used to generate read counts and FPKM, respectively. Gene counts and DESeq2 were used to detect differential gene expression between CRPC-NE samples and CRPC-Adeno, PCA, and benign samples (42). Boxplots, scatterplots, and heatmaps were generated using sample FPKMs in R.
Immunohistochemistry
IHC was performed on deparaffinized formalin-fixed paraffin-embedded (FFPE) sections using a Bond III automated immunostainer (Leica Microsystems). Bond Epitope Retrieval Solution 1 (ER1) (at pH 6) or ER2 was used with heat-mediated antigen retrieval. The following antibodies and conditions were used: SYP (SP11, Thermo Fisher Scientific; 1:100 dilution, ER2), AR (F39.4.1, BioGenex; 1:800 dilution with casein, ER1), and DLL3 (SP347, Spring Bioscience). Briefly, SP347 was used at 0.24 μg/ml, followed by a OptiView DAB IHC kit (Ventana) to visualize DLL3 signal. DLL3 positivity was scored at ×4 magnification for any cytoplasmic or membranous staining at any intensity in total tumor cells.
CTC enumeration and biomarker analysis
The Epic Sciences platform is a slide-based, multicomponent assay designed to detect and characterize CTCs using whole blood samples. The platform can monitor up to four channels in one assay format. A base three-color assay distinguishes CTCs from white blood cells (WBCs) by using DAPI to detect the nuclei of all cells, anti-CD45 to identify the WBCs, and anti-CK to identify CTCs. Patients were divided into CRPC-Adeno and CRPC-NE on the basis of pathological assessment of their metastatic biopsy. In this study, the remaining channel was used to detect DLL3. Briefly, whole blood samples were collected in 10-ml Streck Cell-Free DNA blood collection tubes and processed. Red blood cells were lysed, and nucleated cells (~3 × 106 per slide) were deposited onto 10 to 12 microscope slides. One or two replicate slides per patient sample were stained with a cocktail of antibodies including anti-CK, anti-CD45, and DAPI. CTC enumeration was performed, and DLL3 protein expression was assessed. Slides were scanned by Epic’s rapid fluorescent scanning method, and the images of nucleated cells were analyzed using a proprietary multiparametric digital pathology algorithm (21). The algorithm used 90 cellular parameters, including marker expressions (DAPI, CK, CD45, and DLL3) and cell morphology, to differentiate candidate CTCs from surrounding WBCs. The resulting images were displayed in a web-based report and were confirmed by a trained technician.
NanoString
Tumor mRNA was extracted from FFPE tissue using the Qiagen RNeasy FFPE Kit or cells using the Qiagen RNeasy Tissue and Cell Kit. Agilent 2100 Bioanalyzer system was used for RNA quality control. For FFPE samples with more than 50% of total RNA greater than 300 nucleotide (nt), 100 ng of input RNA was used. For samples with less than 50% of total RNA greater than 300 nt, the input RNA was proportionally increased according to the extent of degradation; 200 ng was used for NCI-H660 cell line. We used a custom probe set, which included AR and CRPC-NE signaling genes (2). Raw count data were normalized using the nSolver Analysis software version 2.0, which normalizes samples according to positive and negative control probes and the geometric mean of the six housekeeping primers (ABCF1, ALAS1, B2M, CLTC, RPLP0, and TUBB). Positive cases are considered with values higher than the mean + interquartile range of the negative controls with the highest maximum value (cutoff = 6.667066).
Cell lines and transfection
The NCI-H660 cell line was purchased from American Type Culture Collection and maintained according to the manufacturers’ protocols. Cells were routinely tested for mycoplasma contamination with a negative result. Human SMARTpool ON-TARGETplus DLL3 and ON-TARGETplus Non-targeting Pool were purchased from Dharmacon (L-009506–00-0005 and D-001810–10-05). NCI-H660 cells were transfected with Lipofectamine RNAiMAX (Thermo Fisher Scientific) for 48 hours, and mRNA was extracted for NanoString.
Enhanced reduced representation bisulfite sequencing
The WCM Computational Genomics Core Facility supported enhanced reduced representation bisulfite sequencing data analysis (43). Briefly, bisulfite reads were aligned to the bisulfite-converted hg19 reference genome using Bismark (44). All samples had bisulfite conversion rates of >99.7%. To characterize the amount of methylation for each gene, we calculated the methylation of promoters by averaging the amount of methylation of all covered CpGs in each promoter (at least five CpGs), which was defined as ±2-kb windows centered on RefSeq transcription start sites.
DLL3 RNA in situ hybridization assay
The single-color chromogenic RNAscope assay (Advanced Cell Diagnostics Inc.) was applied. It uses pairs of specially designed oligonucleotide probes that, through sequence-specific hybridization, allow for target-specific single-molecule detection with a robust signal amplification system. Nonspecific off-target binding by single probes does not result in signal amplification. The probe used for this assay was RNAscope LS 2.5 Probe-Hs-DLL3 (catalog no. 411338). RNA ISH staining of slides was performed on the Leica Bond III automated stainer (Leica Biosystems Inc.). Hybridization signals were detected under bright-field microscope as brown colorimetric staining, followed by counterstaining with hematoxylin. Signals were granular and discrete brown dots that corresponded to individual RNA targets.
Controls used for each DLL3 RNA ISH experiment were (i) CRPC-NE cases with documented DLL3 overexpression by IHC and the NCI-H660 cell line, (ii) RNAscope 2.5 LS Positive Control Probe Hs-PPIB (catalog no. 313908), and (iii) benign prostate cases with negative DLL3 expression by IHC. The HALO software (Indica Labs Inc.) was used for automated image quantification of DLL3 RNA in situ expression at a single-cell resolution.
In vitro cytotoxicity of SC16LD6.5
Cell viability assays were performed on 10,000 cells/well (NCI-H660) or 4000 cells/well (DU145) treated with increasing doses of SC16LD6.5, IgG1LD6.5, SC16 (naked DLL3 antibody), or D6.5 (small-molecule PBD warhead) at the indicated concentrations for 72 hours (DU145) or 13 days (NCI-H660). Cell viability assay kit was used to measure viability according to the manufacturer’s protocol (CellTiter-Glo,Promega). Data are expressed as mean ± SEM. ANOVA test was used for multiple sample comparisons (in GraphPad Prism 8).
Serum pharmacokinetics parameters after peritoneal administration of rovalpituzumab tesirine (SC16LD6.5)
Rovalpituzumab tesirine was administered intraperitoneally between 0.2 and 1.6 mg/kg in NU/J or NOD scid naive mice (3 to 4 mice per group) respectively. Serum was collected at 5 min and 4, 24, 72, 168, and 336 hours (5 min for NOD scid only). ADC and total antibody concentrations were measured by an enzyme-linked immunosorbent assay–based approach (Meso Scale Discovery). Pharmacokinetics parameters {Cmax, Tmax, and exposure [area under the curve (0 – ∞)]} were calculated by noncompartmental analysis in WinNonlin 8.0 (Certara).
Statistical analyses
Gene expression across different subtypes (benign, PCA, CRPC-Adeno, and CRPC-NE) and genotypes (RB1 and TP53) were visualized using boxplots, with the upper and lower whiskers referring to the upper and lower quartiles and the band referring to the median. Because we could not make the assumption that gene expression data are normal, we used nonparametric tests to compare gene expression across different categories. The nonparametric two-tailed Mann-Whitney U test was used to compare gene expression between different subtypes or genotypes. The Spearman correlation coefficient and corresponding P values were used to measure the extent of correlation between two distinct genes across all subtypes and within CRPC-NE samples. All statistical analyses were performed in R using the stats package. OS was calculated from the time of prostate cancer diagnosis and from the time of metastatic disease to death. Patients still alive at the time of last follow-up were censored. OS analyses were performed using the Kaplan-Meier estimator (log-rank test). DLL3 expression in CRPC-Adeno versus CRPC-NE (percentage of positive DLL3 cells and H-score) was evaluated using a two-tailed t test with error bars representing the SD of the mean (SEM). Two-way ANOVA test using multiple comparison analysis was performed for tumor volume measurements and in vitro cytotoxic activity. Differences were considered significant when P value was less than 0.05. Original tumor measurements are provided in data file S1.
Supplementary Material
www.sciencetranslationalmedicine.org/cgi/content/full/11/484/eaav0891/DC1
Fig. S1. Genotype-Tissue Expression (GTEx) expression of DLL3 in adult tissues.
Fig. S2. Evaluation of DLL3-positive cells per tumor.
Fig. S3. DLL3 expression in patient with prostate cancer cohort by RNA-seq.
Fig. S4. DLL3 expression in prostate cancer cohort evaluated by NanoString.
Fig. S5. DLL3-positive tumors evaluated by IHC, RNA-seq, and NanoString.
Fig. S6. Representative images of DLL3 expression.
Fig. S7. DLL3 promoter methylation.
Fig. S8. ASCL1 expression evaluated by RNA-seq.
Fig. S9. Differentially expression analysis of Notch signaling genes.
Fig. S10. Silencing DLL3 in the NCI-H660 cell line.
Fig. S11. Correlation between DLL3-positive and DLL3-negative cells in a tumor biopsy.
Fig. S12. DLL3 expression cutoff in CTCs.
Fig. S13. CK expression in DLL3-positive and DLL3-negative CTCs.
Fig. S14. Correlation between DLL3-positive and DLL3-negative cells in CTCs.
Fig. S15. Expression of Notch receptors across subtypes of prostate cancer and correlation with DLL3 expression.
Fig. S16. In vitro toxicity of SC16LD6.5 in NCI-H660 and DU145.
Table S1. Clinical and pathological features of patients with CRPC-Adeno and PCA-expressing DLL3 by IHC.
Table S2. Genomic features of WCM1388 patient metastatic sites.
Table S3. Clinical features of patients with CRPC-Adeno with DLL3-positive CTCs.
Table S4. Serum pharmacokinetics parameters after peritoneal administration of rovalpituzumab tesirine (SC16LD6.5).
Data file S1. Tumor measurements for in vivo studies.
Acknowledgments:
We thank F. Khani from WCM for providing histology images of the radical prostatectomy specimen from patient illustrated in Fig. 5 and acknowledge support from the Translational Research Program at WCM Pathology and Laboratory Medicine.
Funding: This study was supported by AbbVie Stemcentrx LLC, Prostate Cancer Foundation (to L.P. and H.B.), Department of Defense PC121341 (to H.B.), NIH/NCI SPORE in Prostate Cancer P50-CA211024 (to O.E., J.M.M., and H.B.), NIH grants 1R01CA194547 and 1U24CA210989 (to O.E.), and the Ann and William Bresnan Foundation (to D.M.N. and H.B.).
Footnotes
Competing interests: AbbVie Stemcentrx LLC provided research funding for this study. K.I. and L.S. are employees of AbbVie Stemcentrx LLC. L.S. is listed on patent WO 2017/201442 and related filings. L.F. R.D., and M.K. are employed by Epic Sciences Inc. D.M.N. has declared financial relationship with Roche-Genentech (served on DSMB). S.T.T. and H.B. have received advisory/consulting fees from AbbVie Stemcentrx LLC. All other authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper and/or Supplementary Materials. SC16LD6.5 is available under a material transfer agreement through submission of a proposal to the AbbVie’s Investigator Initiated Studies portal (www.abbvie.com/partnerships/additional-collaboration-opportunities/investigator-initiated-studies-iis.html).
REFERENCES AND NOTES
- 1.Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, Wang Y, Sheikh KL, Terry S, Tagawa ST, Dhir R, Nelson JB, de la Taille A, Allory Y, Gerstein MB, Perner S, Pienta KJ, Chinnaiyan AM, Wang Y, Collins CC, Gleave ME, Demichelis F, Nanus DM, Rubin MA, Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 1, 487–495 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BVSK, Varambally S, Tomlins SA, Nanus DM, Tagawa ST, van Allen EM, Elemento O, Sboner A, Garraway LA, Rubin MA, Demichelis E, Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22, 298–305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, Bianchi-Frias D, Dumpit RF, Kaipainen A, Corella AN, Yang YC, Nyquist MD, Mostaghel D, Hsieh AC, Zhang X, Corey E, Brown LG, Nguyen HM, Pienta K, Ittmann M, Schweizer M, True LD, Wise D, Rennie PS, Vessella RL, Morrissey C, Nelson PS, Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 32, 474–489.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aggarwal R, Huang J, Alumkal JJ, Zhang L, Feng FY, Thomas GV, Weinstein AS, Friedl V, Zhang C, Witte ON, Lloyd P, Gleave M, Evans CP, Youngren J, Beer TM, Rettig M, Wong CK, True L, Foye A, Playdle D, Ryan CJ, Lara P, Chi KN, Uzunangelov V, Sokolov A, Newton Y, Beltran H, Demichelis F, Rubin MA, Stuart JM, Small EJ, Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: A multi-institutional prospective study. J. Clin. Oncol. 36, 2492–2503 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, Goodrich MM, Labbé DP, Gomez EC, Wang J, Long HW, Xu B, Brown M, Loda M, Sawyers CL, Ellis L, Goodrich DW, Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355, 78–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, Leenders F, Lu X, Fernandez-Cuesta L, Bosco G, Müller C, Dahmen I, Jahchan NS, Park K-S, Yang D, Karnezis AN, Vaka D, Torres A, Wang MS, Korbel JO, Menon R, Chun S-M, Kim D, Wilkerson M, Hayes N, Engelmann D, Pützer B, Bos M, Michels S, Vlasic I, Seidel D, Pinther B, Schaub P, Becker C, Altmüller J, Yokota J, Kohno T, Iwakawa R, Tsuta K, Noguchi M, Muley T, Hoffmann H, Schnabel PA, Petersen I, Chen Y, Soltermann A, Tischler V, Choi C-M, Kim Y-H, Massion PP, Zou Y, Jovanovic D, Kontic M, Wright GM, Russell PA, Solomon B, Koch I, Lindner M, Muscarella LA, la Torre A, Field JK, Jakopovic M, Knezevic J, Castanos-Velez E, Roz L, Pastorino U, Brustugun O-T, Lund-Iversen M, Thunnissen E, Köhler J, Schuler M, Botling J, Sandelin M, Sanchez-Cespedes M, Salvesen HB, Achter V, Lang U, Bogus M, Schneider PM, Zander T, Ansén S, Hallek M, Wolf J, Vingron M, Yatabe Y, Travis WD, Nürnberg P, Reinhardt C, Perner S, Heukamp L, Büttner R, Haas SA, Brambilla E, Peifer M, Sage J, Thomas RK, Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta A, Yu X, Case T, Paul M, Shen MM, Kaestner KH, Matusik RJ, Mash1 expression is induced in neuroendocrine prostate cancer upon the loss of Foxa2. Prostate 73, 582–589 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osada H, Tatematsu Y, Yatabe Y, Horio Y, Takahashi T, ASH1 gene is a specific therapeutic target for lung cancers with neuroendocrine features. Cancer Res. 65, 10680–10685 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Augustyn A, Borromeo M, Wang T, Fujimoto J, Shao C, Dospoy PD, Lee V, Tan C, Sullivan JP, Larsen JE, Girard L, Behrens C, Wistuba II, Xie Y, Cobb MH, Gazdar AF, Johnson JE, Minna JD, ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc. Natl. Acad. Sci. U.S.A. 111, 14788–14793 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, Girard L, Minna JD, Gazdar AF, Cobb MH, Johnson JE, ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. 16, 1259–1272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunwoodie SL, Clements M, Sparrow DB, Sa X, Conlon RA, Beddington RSP, Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development 129, 1795–1806 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Chapman G, Sparrow DB, Kremmer E, Dunwoodie SL, Notch inhibition by the ligand DELTA-LIKE 3 defines the mechanism of abnormal vertebral segmentation in spondylocostal dysostosis. Hum. Mol. Genet. 20, 905–916 (2011). [DOI] [PubMed] [Google Scholar]
- 13.del Alamo D, Rouault H, Schweisguth F, Mechanism and significance of cis-inhibition in notch signalling. Curr. Biol. 21, R40–R47 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Rudin CM, Pietanza MC, Bauer TM, Ready N, Morgensztern D, Glisson BS, Byers LA, Johnson ML, Burris HA III, Robert F, Han TH, Bheddah S, Theiss N, Watson S, Mathur D, Vennapusa B, Zayed H, Lally S, Strickland DK, Govindan R, Dylla SJ, Peng SL, Spigel DR, SCRX16–001 investigators, Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: A first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol. 18, 42–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saunders LR, Bankovich AJ, Anderson WC, Aujay MA, Bheddah S, Black K, Desai R, Escarpe PA, Hampl J, Laysang A, Liu D, Lopez-Molina J, Milton M, Park A, Pysz MA, Shao H, Slingerland B, Torgov M, Williams SA, Foord O, Howard P, Jassem J, Badzio A, Czapiewski P, Harpole DH, Dowlati A, Massion PP, Travis WD, Pietanza MC, Poirier JT, Rudin CM, Stull RA, Dylla SJ, A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci. Transl. Med. 7, 302–136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartley JA, Flynn MJ, Bingham JP, Corbett S, Reinert H, Tiberghien A, Masterson LA, Antonow D, Adams L, Chowdhury S, Williams DG, Mao S, Harper J, Havenith CEG, Zammarchi F, Chivers S, van Berkel PH, Howard PW, Pre-clinical pharmacology and mechanism of action of SG3199, the pyrrolobenzodiazepine (PBD) dimer warhead component of antibody-drug conjugate (ADC) payload tesirine. Sci. Rep. 8, 10479 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koshkin VS, Elson P, Magi-Galluzzi C, McKenney J, Smith KS, Shadrach B, Emamekhoo H, Isse K, Saunders L, Kavalerchik E, Dylla S, Bheddah S, Theiss N, Watson S, Stephenson AJ, Fergany AF, Rini BI, Garcia JA, Grivas P, Prognostic value of DLL3 expression and clinicopathologic features in small cell bladder cancer (SCBC). J. Clin. Oncol. 35, 382–382 (2017). [Google Scholar]
- 18.Robinson D, van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera J-M, Montgomery B, Taplin M-E, Pritchard CC, Attard G, Beltran H, Abida WM, Bradley RK, Vinson J, Cao X, Vats P, Kunju LP, Hussain M, Feng FY, Tomlins SA, Cooney KA, Smith DC, Brennan C, Siddiqui J, Mehra R, Chen Y, Rathkopf DE, Morris MJ, Solomon SB, Durack JC, Reuter VE, Gopalan A, Gao J, Loda M, Lis RT, Bowden M, Balk SP, Gaviola G, Sougnez C, Gupta M, Yu EY, Mostaghel A, Cheng HH, Mulcahy H, True LD, Plymate SR, Dvinge H, Ferraldeschi R, Flohr P, Miranda S, Zafeiriou Z, Tunariu N, Mateo J, Perez-Lopez R, Demichelis E, Robinson BD, Schiffman MA, Nanus DM, Tagawa ST, Sigaras A, Eng KW, Elemento O, Sboner A, Heath EI, Scher HI, Pienta KJ, Kantoff P, de Bono JS, Rubin MA, Nelson PS, Garraway LA, Sawyers CL, Chinnaiyan AM, Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pisapia DJ, Salvatore S, Pauli C, Hissong E, Eng K, Prandi D, Sailer V-W, Robinson BD, Park K, Cyrta J, Tagawa ST, Kossai M, Fontugne J, Kim R, Sigaras A, Rao R, Pancirer D, Faltas B, Bareja R, Molina AM, Nanus DM, Rajappa P, Souweidane MM, Greenfield J, Emde A-K, Robine N, Elemento O, Sboner A, Demichelis F, Beltran H, Rubin MA, Mosquera JM, Next-generation rapid autopsies enable tumor evolution tracking and generation of preclinical models. JCO Precis. Oncol. 1, 1–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beltran H, Jendrisak A, Landers M, Mosquera JM, Kossai M, Louw J, Krupa R, Graf RP, Schreiber NA, Nanus DM, Tagawa ST, Marrinucci D, Dittamore R, Scher HI, The initial detection and partial characterization of circulating tumor cells in neuroendocrine prostate cancer. Clin. Cancer Res. 22, 1510–1519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Werner SL, Graf RP, Landers M, Valenta DT, Schroeder M, Greene SB, Bales N, Dittamore R, Marrinucci D, Analytical validation and capabilities of the epic CTC platform: Enrichment-free circulating tumour cell detection and characterization. J. Circ. Biomarkers 4, 3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorente D, Olmos D, Mateo J, Dolling D, Bianchini D, Seed G, Flohr P, Crespo M, Figueiredo I, Miranda S, Scher HI, Terstappen LWMM, de Bono JS, Circulating tumour cell increase as a biomarker of disease progression in metastatic castration-resistant prostate cancer patients with low baseline CTC counts. Ann. Oncol. 29, 1554–1560 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Scher HI, Graf RP, Schreiber NA, McLaughlin B, Jendrisak A, Wang Y, Lee J, Greene S, Krupa R, Lu D, Bamford P, Louw JE, Dugan L, Vargas HA, Fleisher M, Landers M, Heller G, Dittamore R, Phenotypic heterogeneity of circulating tumor cells informs clinical decisions between AR signaling inhibitors and taxanes in metastatic prostate cancer. Cancer Res. 77, 5687–5698 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin D, Wyatt AW, Xue H, Wang Y, Dong X, Haegert A, Wu R, Brahmbhatt S, Mo F, Jong L, Bell RH, Anderson S, Hurtado-Coll A, Fazli L, Sharma M, Beltran H, Rubin M, Cox M, Gout PW, Morris J, Goldenberg L, Volik SV, Gleave ME, Collins CC, Wang Y, High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res. 74, 1272–1283 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Puca L, Bareja R, Prandi D, Shaw R, Benelli M, Karthaus WR, Hess J, Sigouros M, Donoghue A, Kossai M, Gao D, Cyrta J, Sailer V, Vosoughi A, Pauli C, Churakova Y, Cheung B, Deonarine LD, McNary TJ, Rosati R, Tagawa ST, Nanus DM, Mosquera JM, Sawyers CL, Chen Y, Inghirami G, Rao RA, Grandori C, Elemento O, Sboner A, Demichelis F, Rubin MA, Beltran H, Patient derived organoids to model rare prostate cancer phenotypes. Nat. Commun. 9, 2404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ladi E, Nichols JT, Ge W, Miyamoto A, Yao C, Yang L-T, Boulter J, Sun YE, Kintner C,Weinmaster E, The divergent DSL ligand Dll3 does not activate notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J. Cell Biol. 170, 983–992 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang XD, Leow CC, Zha J, Tang Z, Modrusan Z, Radtke F, Aguet M, de Sauvage FJ, Gao WQ, Notch signaling is required for normal prostatic epithelial cell proliferation and differentiation. Dev. Biol. 290, 66–80 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Kwon O-J, Zhang L, Wang J, Su Q, Feng Q, Zhang XHF, Mani SA, Paulter R, Creighton CJ, Ittmann MM, Xin L, Notch promotes tumor metastasis in a prostate-specific Pten-null mouse model. J. Clin. Invest. 126, 2626–2641 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santagata S, Demichelis F, Riva A, Varambally S, Hofer MD, Kutok JL, Kim R, Tang J, Montie JE, Chinnaiyan AM, Rubin MA, Aster JC, JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 64, 6854–6857 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Belandia B, Powell SM, García-Pedrero JM, Walker MM, Bevan CL, Parker MG, Hey1, a mediator of notch signaling, is an androgen receptor corepressor. Mol. Cell. Biol. 25, 1425–1436 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lavery DN, Villaronga MA, Walker MM, Patel A, Belandia B, Bevan CL, Repression of androgen receptor activity by HEYL, a third member of the hairy/enhancer-of-split-related family of notch effectors. J. Biol. Chem. 286, 17796–17808 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meder L, König K, Ozretić L, Schultheis AM, Ueckeroth F, Ade CP, Albus K, Boehm D, Rommerscheidt-Fuss U, Florin A, Buhl T, Hartmann W, Wolf J, Merkelbach-Bruse S, Eilers M, Perner S, Heukamp LC, Buettner R, NOTCH, ASCL1, p53 and RB alterations define an alternative pathway driving neuroendocrine and small cell lung carcinomas. Int. J. Cancer 138, 927–938 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen C-C, Wongvipat J, Ku S-Y, Gao D, Cao Z, Shah N, Adams EJ, Abida W, Watson PA, Prandi D, Huang C-H, de Stanchina E, Lowe SW, Ellis L, Beltran H, Rubin MA, Goodrich DW, Demichelis F, Sawyers CL, SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1 -deficient prostate cancer. Science 355, 84–88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Epstein JI, Amin MB, Beltran H, Lotan TL, Mosquera J-M, Reuter VE, Robinson BD, Troncoso P, Rubin MA, Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am. J. Surg. Pathol. 38, 756–767 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mucci NR, Akdas G, Manely S, Rubin MA, Neuroendocrine expression in metastatic prostate cancer: Evaluation of high throughput tissue microarrays to detect heterogeneous protein expression. Hum. Pathol. 31, 406–414 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Hirsch FR, Varella-Garcia M, Bunn PA Jr., di Maria MV, Veve R, Bremnes RM, Barón AE, Zeng C, Franklin WA, Epidermal growth factor receptor in non-small-cell lung carcinomas: Correlation between gene copy number and protein expression and impact on prognosis. J. Clin. Oncol. 21, 3798–3807 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Sharma SK, Chow A, Monette S, Vivier D, Pourat J, Edwards KJ, Dilling TR, Abdel-Atti D, Zeglis BM, Poirier JT, Lewis JS, Fc-mediated anomalous biodistribution of therapeutic antibodies in immunodeficient mouse models. Cancer Res. 78, 1820–1832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N, The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. CancerDiscov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders S, Pyl PT, Huber W, HTSeq—A python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akalin A, Garrett-Bakelman FE, Kormaksson M, Busuttil J, Zhang L, Khrebtukova I, Milne TA, Huang Y, Biswas D, Hess JL, Allis CD, Roeder RG, Valk PJM, Löwenberg B, Delwel R, Fernandez HF, Paietta E, Tallman MS, Schroth GP, Mason CE, Melnick A, Figueroa ME, Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLOS Genet. 8, e1002781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krueger F, Andrews SR, Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
www.sciencetranslationalmedicine.org/cgi/content/full/11/484/eaav0891/DC1
Fig. S1. Genotype-Tissue Expression (GTEx) expression of DLL3 in adult tissues.
Fig. S2. Evaluation of DLL3-positive cells per tumor.
Fig. S3. DLL3 expression in patient with prostate cancer cohort by RNA-seq.
Fig. S4. DLL3 expression in prostate cancer cohort evaluated by NanoString.
Fig. S5. DLL3-positive tumors evaluated by IHC, RNA-seq, and NanoString.
Fig. S6. Representative images of DLL3 expression.
Fig. S7. DLL3 promoter methylation.
Fig. S8. ASCL1 expression evaluated by RNA-seq.
Fig. S9. Differentially expression analysis of Notch signaling genes.
Fig. S10. Silencing DLL3 in the NCI-H660 cell line.
Fig. S11. Correlation between DLL3-positive and DLL3-negative cells in a tumor biopsy.
Fig. S12. DLL3 expression cutoff in CTCs.
Fig. S13. CK expression in DLL3-positive and DLL3-negative CTCs.
Fig. S14. Correlation between DLL3-positive and DLL3-negative cells in CTCs.
Fig. S15. Expression of Notch receptors across subtypes of prostate cancer and correlation with DLL3 expression.
Fig. S16. In vitro toxicity of SC16LD6.5 in NCI-H660 and DU145.
Table S1. Clinical and pathological features of patients with CRPC-Adeno and PCA-expressing DLL3 by IHC.
Table S2. Genomic features of WCM1388 patient metastatic sites.
Table S3. Clinical features of patients with CRPC-Adeno with DLL3-positive CTCs.
Table S4. Serum pharmacokinetics parameters after peritoneal administration of rovalpituzumab tesirine (SC16LD6.5).
Data file S1. Tumor measurements for in vivo studies.


