Abstract
Mutations in the AHDC1 gene are associated with the Xia-Gibbs syndrome (XGS), a sporadic genetic disorder characterised by developmental delay, intellectual disability, hypotonia, obstructive sleep apnoea, dysmorphic facial features, and cerebral malformations with plagiocephaly. Here we report the case of a 13-year-old Colombian female patient with a history of developmental delay, speech delay, sleep disturbances, and dysmorphic craniofacial features. The whole exome sequencing (WES) test revealed a novel de novo heterozygous frameshift mutation in AHDC1. The present case report describes the second case of mutations in AHDC1 in a Latin American patient. A literature review showed that the clinical features were similar in all reported patients. The WES test enabled the identification of the causality of this disorder characterised by high clinical and genetic heterogeneity.
Keywords: Developmental disabilities , Whole exome sequencing , Frameshift mutations
What’s Known
In the last two decades, significant progress has been made to understand the genetic causes of intellectual disability. However, with the tests currently available, the diagnostic yield has only been possible in 40% of the cases.
Xia-Gibbs syndrome is a rare form of syndromic intellectual disability.
What’s New
Xia-Gibbs syndrome and its genotype-phenotype correlations are not fully understood. A novel pathogenic frameshift variant in AHDC1 was identified using the whole exome sequencing technique.
Inherited autosomal dominant intellectual disability pose a diagnostic challenge.
Introduction
Intellectual disability (ID) is found in 3% of paediatric patients around the world.1 Approximately 50% of IDs are considered moderate-to-severe and related to genetic disorders. De novo mutations along with autosomal dominant inheritance have recently been documented as the major cause of IDs. In the past two decades, significant progress has been made to understand the genetic causes of ID. However, with the tests currently available, the diagnostic yield has only been possible in 40% of the cases.2
Xia and colleagues (2014) performed a clinical WES test and described a de novo dominant truncating mutation in the AHDC1 gene in four patients with ID, speech delay, facial dysmorphic features, hypotonia, and sleep apnoea.3 Yang and colleagues (2015) performed the WES test on 2,157 patients with a clinical history of ID or developmental delay (DD) and identified seven patients with a de novo frameshift and nonsense mutations in the same gene.4 They also predicted a loss in protein function as a result of the mutations. They concluded that AHDC1 haploinsufficiency caused the XGS (MIM #615829), a rare syndromic form of ID with autosomal dominant inheritance. AHDC1 is located on 1p36.11 and encodes a protein of 1603 amino acids. Its function is linked to transcriptional and epigenetic regulation during the embryonic development and axonogenesis.5,6
The complete XGS phenotype and its genotype-phenotype correlations are not fully understood. Here we report the case of a 13-year-old girl with ID and dysmorphic facial features. Using the Trio-WES approach, we identified a novel pathogenic frameshift variant in AHDC1. Clinical features of the patient were compared with previously reported patients to establish clinical differences.
Case Presentation
The patient, a 13-year-old girl, was born from the second pregnancy of a 24-year-old mother and a non-consanguineous 24-year-old father. She was referred from the Neuropediatric Department to the Genetics Department for the clinical history of DD. Pregnancy was uncomplicated and the prenatal ultrasound was normal. Vaginal delivery at the 37th week of gestation was without complications and the anthropometric parameters were birth weight 2.780 kg (10th centile) and length of 47 cm (25th centile). The perinatal period was complicated by neonatal respiratory distress and the girl was monitored in the neonatal intensive care unit for 8 days. The patient was diagnosed with severe hypoxic-ischaemic encephalopathy, cephalohematoma, and neonatal jaundice. The mother reported a speech delay since the girl’s first monosyllables were spoken at the age of 18 months. At this age, she stood up by herself and began to walk independently. Anxious behaviour, poor social interaction with autism criteria (based on clinical diagnosis using the diagnostic and statistical manual of mental disorders, 5th edition), and sleep disturbances were also noted. These disturbances included snoring and difficulty in staying asleep. No family history of DD and ID was reported.
At 2 years of age, due to the suspected plagiocephaly, a computerised axial tomography of the skull with 3D reconstruction was performed. The results showed dolichocephaly and brachycephaly, a complete closure of the sutures and fontanelles, and the brain parenchyma and ventricular system without abnormalities. This coronal craniosynostosis was not surgically corrected. A G-banding karyotype was performed which showed a normal chromosomal arrangement (46, XX). At the age of 9, a neurological evaluation was performed using the Wechsler intelligence scale for children, 3rd edition (WISC-III). The patient scored 46 points on the verbal and performance scale resulting in a global intelligence quotient of 40 (<0.1 centiles). Additional tests showed a normal echocardiogram and normal hearing at 11 and 12 years of age, respectively.
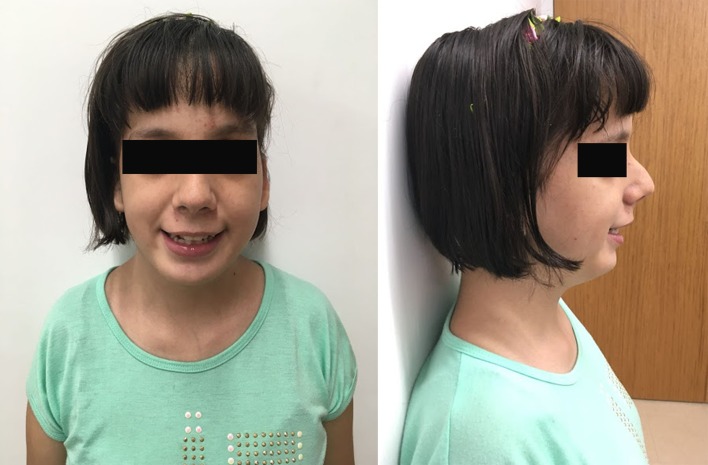
At the age of 12, the patient underwent a genetic consultation. During physical examination, macrocephaly with the cephalic perimeter of 56 cm (>97 centile) and plagiocephaly, height of 149 cm (25 centile), weight of 49.3 kg (60 centile), three trichoglyphs on the scalp, a prominent metopic ridge, asymmetric and dysmorphic facial features (figure 1) with a high palate were observed. Additionally, hypotonia of the oropharynx, hypersalivation, and multiple telangiectasia in the lower extremities were diagnosed. Neurological examination demonstrated self-harming behaviours, reduced social interactions, a tendency to seek isolation, and difficulty in performing simple tasks. No walking difficulties were observed and neither motor nor sensory deficits were detected.
Figure1.
Facial features of the patient included a broad forehead, horizontal eyebrows, small and low-set ears, mild ptosis, proptosis; upslanting palpebral fissures, mild facial hypoplasia, flat nasal bridge, nasal root depression; and an acute nasal angle, long philtrum, thin upper lip, and micrognathia.
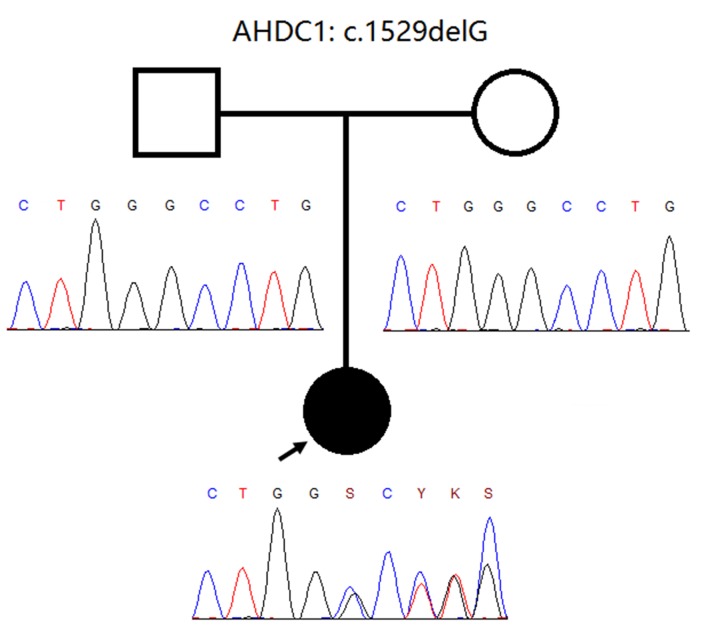
As a result of the clinical presentation, a microarray-based comparative genomic hybridisation (array-CGH; Agilent Technology, Santa Clara, CA, USA) was performed which showed standard results. Then, the Trio-WES approach was performed. Approximately 214,000 exons from all encoded sequences were enriched for consensus using genomic DNA fragments of >340,000 probes against the human genome. The sequencing was performed on the Illumina HiSeq 4000 platform until approximately 100-130X read depth was attained. Only variants in the coding region and flanking intronic regions with a minor allele frequency <1% were evaluated. Selected MAFs were compared using the following datasets: 1000 Genomes Project Consortium, dbSNP, Exome Variant Server, and Exome Aggregation Consortium (ExAc). Variants within ±20 bp of flanking intron regions were examined. A novel heterozygous de novo frameshift variant (figure 2) in AHDC1 (NM_001029882.3) was identified: c.1529delG (p.Gly510Alafs*12). This variant has not been previously reported in the literature. Variant functional prediction software tools (Mutation Taster, Condel, SIFT, and FATHMM) classified it as a damaging variant (disease-causing). Currently, it is classified as “probably pathogenic” according to the American College of Medical Genetics and Genomics (ACMG) guidelines. This variant generated a frameshift slip at codon 510. As a result, the reading frame generated a premature stop codon 12 amino acids. No other variants were identified with this methodology. The finding was confirmed by Sanger sequencing and was compatible with the diagnosis of XGS.
Figure2.
Pedigree information of the patient and Sanger sequencing electropherogram of both the patient and her parents. The pedigree is consistent with a de novo frameshift mutation.
A written informed consent was obtained from the patient’s parents for the publication of her images and clinical data for scientific purposes.
Discussion
XGS is a newly described disorder characterised by developmental delay, hypotonia, speech delay, sleep apnoea, and seizures.3 Due to the lack of reported cases, the incidence of XGS is not known. However, currently, there are 29 reported cases worldwide.3,4,7-11 It is estimated that the incidence in the general population is less than 1:1,000,000 live births.12 In all previously reported cases, including one Colombian patient,9 developmental histories have shown delayed speech and psychomotor delay. However, the diagnosis of the disorder depended on the identification of a molecular variation in AHDC1 rather than on the recognition of a distinctive set of shared facial features.7
Xia and colleagues proposed that the ID associated with mutations in AHDC1, which exists in 100% of XGS affected individuals, is caused by a dominant-negative mechanism. Clinical manifestations are a response to the lack of encoded protein, which in turn influences the response to the DNA damage and ultimately alters brain development.13
The phenotypic characteristics in our proband included a broad forehead, flat nasal bridge, and thin upper lip. These dysmorphic facial features have been consistently observed in 80% of the reported patients.3,7 To date, no patients have been clinically diagnosed based on a pattern of facial dysmorphisms. In our patient, other common features were hypotonia (reported in 90% of the patients) and sleep disturbances (reported in 50% of the patients). Independent walking is reported in 80% of the XGS patients. Our patient began walking at 2 years of age, close to the median (2.5 years) in XGS patients.7
About 60% of the patients with XGS exhibit abnormal MRI and brain malformations including thinning of the corpus callosum,7 hypomyelination, leukomalacia, and less frequently a cyst in the posterior fossa.3,4,8 Our patient exhibited dolichocephaly and brachycephaly as the main brain abnormalities and had a clinical record of craniosynostosis; previously reported in one other patient.11 Additionally, our patient exhibited auto-aggressive behaviours that are part of the less common clinical manifestations in patients with XGS; previously reported in only one patient.4 Our patient also displayed autistic features; reported in 25% of the patients.7
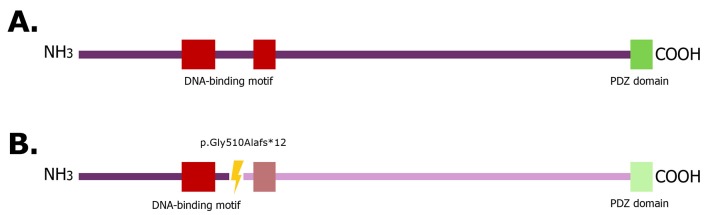
In the present case report, the described mutation introduces a premature stop codon at amino acid 522, which led to the loss of the protein DNA-binding motif as well as to a PDZ domain (figure 3). Therefore, the AHDC1 protein interactions were altered and directly linked to the neurobehavioral manifestations in XGS patients. Although no functional studies of these proteins have been reported, it is suggested that AHDC1 mutation affects the translation of essential developmental proteins. Poot and colleagues reported that an insufficient dose of this protein did not allow the development of functional protein complexes and postulated that mutations in AHDC1 affect the translation of related proteins in brain development.14
Figure3.
Wildtype AHDC1 gene structure (A) and mutated AHDC1 gene suggested structure (B). The region of frameshift mutation in ADHC1 protein is shown (yellow ray: The mutation site, light colour: The region and domain lost due to the stop codon generated by frameshift mutation).
XGS is an example of a disorder that illustrated the diagnostic impact of the WES test. The WES test could detect all XGS cases after several genetic tests. During the last decade, the WES test has facilitated the identification of both disease-causing genes of unknown cases and a complete characterisation of genetic variations. This was not only for genetic syndromes with unique morphological changes but also for those with more complex phenotypes and overlapping characteristics.15
Conclusion
We have reported a novel frameshift mutation in the AHDC1 gene using the WES test. The features described in the present case report were similar to those in previously reported cases except for craniosynostosis and the absence of brain malformations. The most remarkable characteristics of XGS that matched with our patient were discussed. It is recommended to conduct further, similar, experimental studies to demonstrate the pathogenicity of novel variants in AHDC1 and to better understand the full genotype-phenotype correlation.
Acknowledgement
We would like to thank the family of the patient for their participation in this study.
Conflict of Interest:None declared.
References
- 1.Michelson DJ, Shevell MI, Sherr EH, Moeschler JB, Gropman AL, Ashwal S. Evidence report: Genetic and metabolic testing on children with global developmental delay: report of the Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society. Neurology. 2011;77:1629–35. doi: 10.1212/WNL.0b013e3182345896. [DOI] [PubMed] [Google Scholar]
- 2.Rauch A, Hoyer J, Guth S, Zweier C, Kraus C, Becker C, et al. Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet A. 2006;140:2063–74. doi: 10.1002/ajmg.a.31416. [DOI] [PubMed] [Google Scholar]
- 3.Xia F, Bainbridge MN, Tan TY, Wangler MF, Scheuerle AE, Zackai EH, et al. De novo truncating mutations in AHDC1 in individuals with syndromic expressive language delay, hypotonia, and sleep apnea. Am J Hum Genet. 2014;94:784–9. doi: 10.1016/j.ajhg.2014.04.006. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang H, Douglas G, Monaghan KG, Retterer K, Cho MT, Escobar LF, et al. novo truncating variants in the AHDC1 gene encoding the AT-hook DNA-binding motif-containing protein 1 are associated with intellectual disability and developmental delay. Cold Spring Harb Mol Case Stud. 2015;1:a000562. doi: 10.1101/mcs.a000562. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45:D369–D79. doi: 10.1093/nar/gkw1102. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olah J, Vincze O, Virok D, Simon D, Bozso Z, Tokesi N, et al. Interactions of pathological hallmark proteins: tubulin polymerization promoting protein/p25, beta-amyloid, and alpha-synuclein. J Biol Chem. 2011;286:34088–100. doi: 10.1074/jbc.M111.243907. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Y, Wangler MF, McGuire AL, Lupski JR, Posey JE, Khayat MM, et al. The phenotypic spectrum of Xia-Gibbs syndrome. Am J Med Genet A. 2018;176:1315–26. doi: 10.1002/ajmg.a.38699. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosch DG, Boonstra FN, de Leeuw N, Pfundt R, Nillesen WM, de Ligt J, et al. Novel genetic causes for cerebral visual impairment. Eur J Hum Genet. 2016;24:660–5. doi: 10.1038/ejhg.2015.186. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Acero M, Acosta J. Whole-Exome Sequencing Identifies a de novo AHDC1 Mutation in a Colombian Patient with Xia-Gibbs Syndrome. Mol Syndromol. 2017;8:308–12. doi: 10.1159/000479357. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park HY, Kim M, Jang W, Jang DH. Phenotype of a Patient With a 1p36. 11-p35. 3 Interstitial Deletion Encompassing the AHDC1 Ann Lab Med 2017;37:563–5. doi: 10.3343/alm.2017.37.6.563. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller KA, Twigg SR, McGowan SJ, Phipps JM, Fenwick AL, Johnson D, et al. Diagnostic value of exome and whole genome sequencing in craniosynostosis. J Med Genet. 2017;54:260–8. doi: 10.1136/jmedgenet-2016-104215. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orphanet Consortium. AHDC1 related intellectual disability obstructive sleep apnea mild dysmorphism syndrome. [cited 2017 Dec 19] . Available from: [http://www.orpha.net/consor/cgi-bin/OC_Exp.php?lng=EN&Expert=412069. ]
- 13.Quintero-Rivera F, Xi QJ, Keppler-Noreuil KM, Lee JH, Higgins AW, Anchan RM, et al. MATR3 disruption in human and mouse associated with bicuspid aortic valve, aortic coarctation and patent ductus arteriosus. Hum Mol Genet. 2015;24:2375–89. doi: 10.1093/hmg/ddv004. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poot M, van der Smagt JJ, Brilstra EH, Bourgeron T. Disentangling the myriad genomics of complex disorders, specifically focusing on autism, epilepsy, and schizophrenia. Cytogenet Genome Res. 2011;135:228–40. doi: 10.1159/000334064. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–11. doi: 10.1056/NEJMoa1306555. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]