Abstract
Serum and plasma contain abundant biological information that reflect the body's physiological and pathological conditions and are therefore a valuable sample type for disease biomarkers. However, comprehensive profiling of the serological proteome is challenging due to the wide range of protein concentrations in serum.
Methods: To address this challenge, we developed a novel in-depth serum proteomics platform capable of analyzing the serum proteome across ~10 orders or magnitude by combining data obtained from Data Independent Acquisition Mass Spectrometry (DIA-MS) and customizable antibody microarrays.
Results: Using psoriasis as a proof-of-concept disease model, we screened 50 serum proteomes from healthy controls and psoriasis patients before and after treatment with traditional Chinese medicine (YinXieLing) on our in-depth serum proteomics platform. We identified 106 differentially-expressed proteins in psoriasis patients involved in psoriasis-relevant biological processes, such as blood coagulation, inflammation, apoptosis and angiogenesis signaling pathways. In addition, unbiased clustering and principle component analysis revealed 58 proteins discriminating healthy volunteers from psoriasis patients and 12 proteins distinguishing responders from non-responders to YinXieLing. To further demonstrate the clinical utility of our platform, we performed correlation analyses between serum proteomes and psoriasis activity and found a positive association between the psoriasis area and severity index (PASI) score with three serum proteins (PI3, CCL22, IL-12B).
Conclusion: Taken together, these results demonstrate the clinical utility of our in-depth serum proteomics platform to identify specific diagnostic and predictive biomarkers of psoriasis and other immune-mediated diseases.
Keywords: Proteomics, Data-Independent Acquisition Mass Spectrometry, Antibody Microarray, Biomarker, Psoriasis
Introduction
Over the past ten years, the rapid development of high throughput omics technologies has enabled us to quickly obtain a large number of physiological and pathological information of the human body 1-3. This information has begun to change the way that we understand and treat disease, from reductionism to a more holistic view 4.
Human serum and plasma are valuable in clinical and biological studies because they are a reservoir of secreted proteins from organs throughout the body, reflect the patient's physiological and pathological conditions and may contain biomarkers of disease and treatment response 4-10. Currently, there are two main strategies for analyzing the plasma/serum proteome 4. The first “triangular” approach compares protein expression differences between normal and disease conditions using proteomics technology, and then selects the proteins with significantly altered levels as biomarkers for downstream clinical validation 4, 11. The second “rectangular” method builds a knowledge base for a specific disease by quickly and accurately screening the serum/plasma proteomes from a large number of clinical samples across different time points, risk and drug treatment conditions. In general, this step employs mass spectrometry (MS) 5, 12. It is then expected that the knowledge base can be mathematically modeled to diagnose and effectively treat the disease 3, 4, 13, 14.
Geyer et al. developed a plasma proteome analysis pipeline using label-free quantitative MS, which detected 284±5 proteins containing > 40 FDA-approved biomarkers without removing high-abundance proteins 13. Using the same approach for a cohort of 43 obese people who experienced weight loss, Geyer et al. showed that the levels of 93 plasma proteins changed, including proteins associated with insulin resistance15.Their study showed that the serum proteome can be used to evaluate and monitor metabolic diseases. Liu et al. developed another approach using SWATH-MS (Sequential Windowed Acquisition of All Theoretical Fragment Ion Mass Spectra), a data independent acquisition (DIA) method with high quantitative accuracy and reproducibility. Using this method, they surveyed the changes of the plasma proteome in a longitudinal cohort containing 72 monozygotic and 44 dizygotic twins at intervals of 2-7 years. A total of 342 different proteins were quantitatively detected in 232 plasma samples. It was found that the plasma proteome was affected by genetic, environmental and longitudinal factors 5, 10.
Despite these technological advances, however, a comprehensive and in-depth determination of the serum or plasma proteome has remained a challenge due to the wide range of protein concentration 5, 8, 13, 16. Compared to MS, antibody and protein microarrays have higher sensitivity, higher throughput, and are concentration independent, thus making them suitable for detecting low-abundance proteins in serum or plasma samples 11, 17-21.
In this work, we developed an in-depth serum proteomics technology platform by combining DIA-MS and customizable antibody microarrays. We then used this two-pronged approach to study the serological protein profile of psoriasis, a common immune-mediated inflammatory skin disease characterized by skin and joint impairment and medical comorbidities 22. The disease is complex with a multifold pathogenesis affected by both hereditary and environmental factors. Moreover, psoriasis patients respond differently to therapy23. In the United States, 3.2% of the population has psoriasis with associated costs over $110 billion per year 24. Comprehensive determination of the serum proteome of psoriasis patients may help better understand the pathogenesis of psoriasis as well as identify diagnosis and treatment biomarkers. To date, only a limited number of psoriasis serum proteins have been studied 25, 26.
In this study, differentially-expressed proteins discriminating healthy volunteers from psoriasis patients were identified, as well as proteins distinguishing responders from non-responders to an oral traditional Chinese medicine. Furthermore, a positive association between the psoriasis area and severity index (PASI) score with three serum proteins (PI3, CCL22 and IL-12B) was made. Our results demonstrate the feasibility of our platform to identify diagnostic and predictive disease biomarkers in serum.
Experimental Procedures
Clinical Samples
Serum from healthy volunteers, urticaria and psoriasis patients were obtained from Guangdong Provincial Hospital of Chinese Medicine 5, 27. The detailed information pertaining to the psoriasis patients, including their demographics, co-morbidities, and medications, is provided in Tables 1 - 3. The study was approved by the ethical committees at the Beijing Proteome Research Center and Guangdong Provincial Hospital of Chinese Medicine, and performed according to the Declaration of Helsinki.
Table 1.
Clinical serum samples for the identification of psoriasis-associated proteins.
| Healthy controls (N=16) | Psoriatic patients (N=23) | |
|---|---|---|
| Age | 40.94 ± 12.61 | 47.09 ± 11.65 |
| Sex N (%) | ||
| Male | 14 (87.50) | 20 (86.96) |
| Female | 2 (12.50) | 3 (13.04) |
| PASI | / | 7.35 ± 2.72 |
Table 3.
Clinical serum samples used for validating the biomarkers of psoriasis identified in this study.
| Healthy controls (N=20) | Urticaria Patients(N=10) | Psoriatic patients (N=50) | |
|---|---|---|---|
| Age | 38.80 ± 15.20 | 41.30 ± 8.84 | 45.34 ± 12.91 |
| Sex N (%) | |||
| Male | 15 (75) | 5 (50) | 20 (78) |
| Female | 5 (25) | 5 (50) | 11 (22) |
| PASI | / | / | 12.82 ± 3.09 |
Fabrication of Psoriasis Antibody Arrays
Array design.All psoriasis-related protein targets for the psoriasis-specific antibody microarray were selected using text mining and manual curation as previously described (Figure S2)9. Briefly, the PubMed database (https://www.ncbi.nlm.nih.gov/) was searched for reported psoriasis plasma/serum proteins using “psoriasis” and “serum” or “plasma” as keywords. In parallel, the electronic PubMed abstract database was mined for psoriasis biomarkers using “psoriasis” and “biomarker” as keywords. The search resulted in a total of 544 abstracts and 364 candidate proteins. After three rounds of manual curation and de-redundancy, a total of 129 proteins associated with psoriasis remained.
Array printing.All antibodies (Bio-techne Ltd, MN, USA) were diluted to 0.2 mg/mL and then printed onto a 3D modified slide surface (Capital Biochip Corp, Beijing, China) in four replicates using an Arrayjet microarrayer (Roslin, UK). Phosphate-buffered saline (PBS) and bovine serum albumin (BSA, 100 µg/mL) (Sigma-Aldrich, MO, USA) were used as negative controls. Biotinylated BSA (100 µg/mL) and Alexa Fluor 555 goat anti-human IgG (10 μg/mL) were used as positive controls. Prepared antibody microarrays were stored at - 20 ℃ until ready to use.
Measurement of Psoriasis Serum Proteome using Antibody Microarrays
Biotin labeling.All serological proteins were labeled with biotin using the modified procedure as previously described 28, 29. Briefly, 10 μL serum were diluted with 90 μL filtered 1×PBS (pH 7.4) followed by 1 μL of NHS-PEG4-Biotin (20 g/L in DMSO) (Thermo Fisher Scientific, MA, USA). After incubating for 1 h at room temperature, excess biotin molecules were removed using a Bio-Spin column via centrifugation at 1000 × g for 2 min. The collected biotinylated proteins were dissolved in 500 μL of PBS containing 5% milk (w/v) and stored at 4 ℃.
Sera screening. Antibody microarrays were assembled into an incubation tray (PEPperPRINT, Heidelberg, Germany) and blocked with 600 μL 5% milk (w/v) for 1 h at room temperature. After removing the milk, the arrays were incubated with pre-labeled serum proteins at 4 °C overnight. The slides were washed three times, 10 min per wash, with PBS containing 0.05% (w/v) Tween 20 (PBST). For detection, the arrays were incubated with 2 μg/mL streptavidin-PE for 1 h at room temperature in the dark and then washed three times with PBST. After centrifuging for 2 min at 1000×g, the slide was scanned using the GenePix 4000A microarray scanner (Molecular Devices, CA, USA). The fluorescent images were analyzed and the signal intensity was extracted using the GenePix Pro image analysis software (Molecular Devices, CA, USA).
Measurement of Psoriasis Serum Proteome using DIA-MS
2 µL of serum sample were added in a 96-well plate and further diluted with lysis buffer containing 0.1 M NH4HCO3 (Sigma, MO, USA), 6 M Urea (Sigma, MO, USA), and 2 M thiourea (Sigma, MO, USA). Disulfide reduction was performed for 45 min at 33 °C, 600 rpm, with 10 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP) and then alkylated with 40 mM iodoacetamide (IAA) at 25 °C for 1 h in the dark. The protein was sequentially digested with trypsin at a ratio of 1:40 (enzyme to substrate) for 16 hrs (4 h +12 h) at 33 °C. The tryptic peptides were then acidified with 1% trifluoroacetic acid (TFA, pH 2-3) prior to C18 desalting with Sep-Pak Vac 1cc (50 mg) C18 cartridges (Waters, MA, USA) according to the manufacturer's protocol. Desalted peptides were then dried under vacuum and dissolved in 20 µL of MS buffer containing 0.1% formic acid and 2% acetonitrile in water (all HPLC grade). The peptide concentration was measured by Nanoscan (Analytik Jena AG, Jena, Germany) at an absorbance of A280 nm. 0.4 µg peptides were separated on a 30 min LC gradient using an analytical column (PepMap® RSLC, 75 µm × 250 mm, 2 µm 200 Å C18 particles) and injected into Thermo Fisher's Q Exactive HF Hybrid Quadrupole OrbitrapTM (QE-HF) mass spectrometer. The DIA acquisition scheme consisted of 24 variable windows ranging from 400 to 1000 m/z. The sequential precursor isolation window setup was as follows: 410-430, 430-450, 450-470, 470-490, 490-510, 510-530, 530-560, 560-590, 590-610, 610-630, 630-660, 660-690, 690-710, 710-730, 730-750, 750-770, 770-790, 790-820, 820-860, 860-910, 910-970 m/z (20*20, 2*40, 2*60). The resolution of MS1 was 60,000, and MS2 was 30,000. OpenSWATH (version 2.0.0 Sep 26 2017) was performed against a plasma library containing 325 proteins as described previously 30. Finally, Pyprophet limited the peptide and protein identification to a 1% false discovery rate (FDR).
Validation of biomarker candidates by ELISA
Human CCL22, PI3 and CD14 ELISA kits were obtained from R&D Systems (MN, USA). The ELISA experiments were executed according to the manufacture's protocol. Briefly, 100 µL of assay diluent, human recombinant protein standards and diluted serum samples were added to appropriate wells sequentially. The resulting plate was incubated for 2 hrs at 4 °C, followed by four times of washing with washing buffer. After removing washing buffer by aspirating, 200 µL of HRP-conjugated detection antibody was added to each well and incubated for 2 hrs at 4 °C. After washing the wells to remove unbound detection antibody, 200 µL of TMB substrate solution was added and incubated for 30 min at room temperature. Lastly, the reaction was stopped with addition of 50 µL stop solution (2 M H2SO4) and the signal was read immediately at 450 nm.
Bioinformatics analysis
Functional annotation of serological proteins employed the PANTHER database (http://pantherdb.org/) 31. The interaction network analysis of biological processes and signaling pathways used Cytoscape and ClueGO with a p-value cut-off ≤ 0.01 32. Pathway analysis employed the KEGG database (https://www.genome.jp/kegg/) 33.
Statistical analysis
Missing values in SWATH-MS data and non-signal values in microarray data were replaced with the minimum of each sample such that the signals would not be zero. Inter-sample data were normalized using quantile and log10 normalization and then subjected to Satterthwaite t-test analysis (p-value = 0.05) to identify the serological proteins related to psoriasis via the Python scipy.stats package (v1.1.0) 34.
After normalization, correlation analysis was performed with the Python scipy.stats package (v1.1.0). The Pearson correlation coefficient is used to represent correlations between continuous variables, while the Kendall rank correlation coefficient plays the same role between continuous variables and categorical variables (i.e., Drug, Sex, Smoking, Drinking and Family in the clinical data). The circos plot was made using circos (http://circos.ca/) 35. The nonbiased hierarchical clustering analysis and biased hierarchical clustering analysis were performed using R package pheatmap (v1.0.10). The values of principal components in principal components analysis (PCA) were computed using sklearn.decomposition (v0.19.1) package in Python and result charts were drawn using matplotlib (v2.2.2) and seaborn (v 0.8.1) packages.
The Psoriasis Area and Severity Index (PASI) scores is frequently used in the clinic to assess the severity of psoriasis and the treatment response 36. Therefore, the remission of psoriasis disease after therapy, or clinical improvement rate, was calculated using the equation as previously described 5, 27, 37:
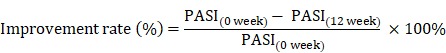 |
Data deposition
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository 77 with the dataset identifier PXD013089.
Results
Design and Fabrication of Psoriasis Antibody Microarrays
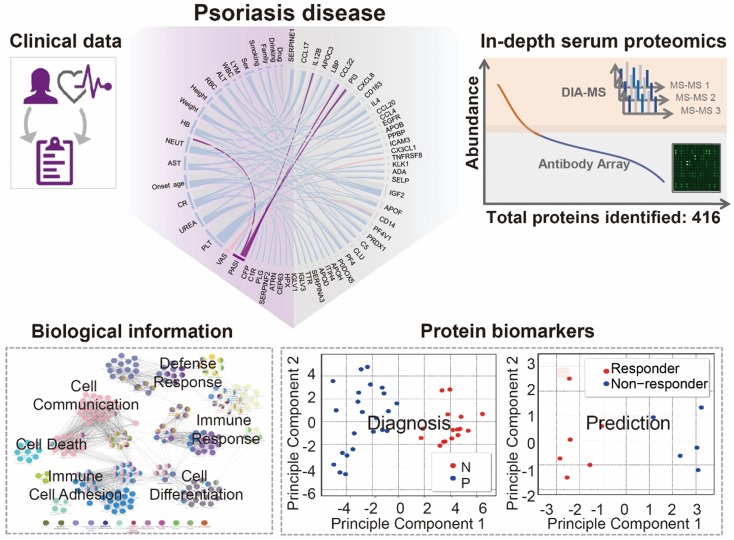
Using psoriasis as a disease model, we collected serum samples from 16 healthy controls and 23 psoriasis patients before and after 12 weeks of treatment with a well-established oral traditional Chinese medicine (i.e., YinXieLing) for psoriasis treatment (Table 1) 27, 38-41. The serum proteomes from these clinical samples were measured with our proteomics platform using DIA-MS and customizable antibody microarrays. All of the obtained data were analyzed with the following aims: 1) to elucidate the biology of psoriatic pathogenesis; 2) to find correlations between the clinical data, serum proteome and psoriasis severity index; and 3) to identity proteomic biomarkers for the diagnosis and treatment of psoriasis. The schematic illustration of this translational study using our in-depth serum proteomics approach is shown in Figure 1.
Figure 1.
Study design using in-depth serum proteomics. In this study, we developed an in-depth serum proteomics platform by combination of DIA-MS and customable antibody microarray microarrays, which allows the measurement of serum proteome in unprecedented depth (~10 orders). Furthermore, the comprehensive determination of serum proteome for psoriasis disease enable the integrative analysis of correlations between serum protein expression and clinical data as well as the identification of biological information and potential biomarkers for psoriasis disease.
To select target proteins for antibody microarray detection, we first text-mined and manually curated the PubMed database as previous described (Figure S1A) 9. Using psoriasis and plasma or serum as keywords, we identified 898 articles in which 120 protein candidates were obtained after three rounds of curation. In parallel, we identified 113 articles using psoriasis and biomarker as keywords, in which 33 candidates were obtained. After removing redundant biomarkers-of-interest, 129 plasma/serum proteins and biomarkers related to psoriasis remained (Table S1, Figure S1B). ClueGO analysis revealed that these plasma/serum proteins are indeed involved in multi-signaling pathways associated with psoriasis, including cell proliferation, cell differentiation, cell migration, programmed cell death, chemotaxis and endocytosis (Figure S2). Of note, two pivotal psoriasis pathways, the MAPK and STAT pathways, were also identified in this study using our proteomics platform (Figure S2, Table S2) 42, 43.
Based on this 129-protein dataset, we designed and prepared an antibody microarray to detect all of these psoriasis plasma/serum proteins reported in the literature (Table S1). By creating a customized antibody microarrays focused on the disease of interest, the reagents, cost and time for the serum screening and data analysis are significantly reduced compared to using high-density antibody arrays 11. Almost all (98.4%, 127/129) of the antibodies used to prepare the microarray were verified by the antibody vendors to detect the conformational or partially conformational epitopes of target proteins via ELISA, immunoprecipitation (IP), immunofluorescence (IF), immunohistochemistry (IHC) or flow cytometry. Only 1.6% (2/129) antibodies were exclusively validated by western blotting (WB), which recognizes linear epitopes (Figure S3). All 129 antibodies were printed on the surface of a 3D-modified slide with appropriate negative controls (PBS and BSA) and positive controls (biotinylated BSA and Alexa Fluor 555 goat anti-human IgG).
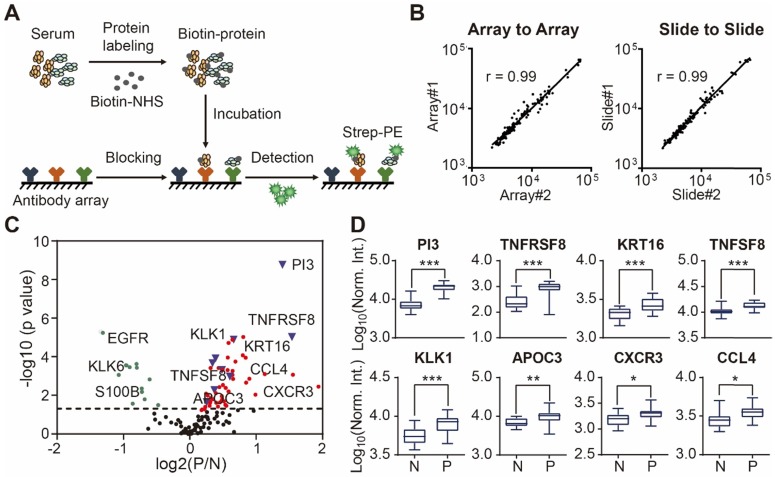
Using custom-made antibody microarrays, we screened 50 serum proteomes of healthy controls and psoriasis patients (Figure 2A, Table 1 and Table 2) 27, 38. The r correlations of array-to-array and slide-to-slide were 0.99 respectively (Figure 2B). Using serum from a healthy control, the intra- and inter-array coefficient of variation (CV) were calculated as 2.09% and 5.72%, respectively. Furthermore, the CV of antibody microarray measurements in all 50 serum samples ranged from 2.13% to 8.2%, indicating the high reproducibility of antibody microarrays in serum screening (Figure S4). The majority (86.82%; 112/129) of the proteins were detected in these serum samples (Figure S5).
Figure 2.
Design and fabrication of psoriasis-specific antibody microarrays. (A) Workflow for antibody microarray design and fabrication; (B) Reproducibility of antibody microarray detection of serum proteins; (C) Identification of psoriasis-associated proteins in serum using volcano plot analysis. Green and red dots represent down- and up-regulated proteins that were previously shown to be associated with psoriasis, respectively. Blue dots represent the novel serum proteins associated with psoriasis disease identified in this study; (D) Box plot analysis of newly identified psoriasis-associated serum proteins. The selection of differentially expressed proteins was performed using the Satterthwaite t-test analysis (p-value = 0.05). N and P represent healthy controls and psoriatic patients, respectively. *, ** and *** represent the p-value less than 0.05, 0.005 and 0.0005, respectively.
Table 2.
Clinical serum samples for the identification of biomarkers associated with psoriasis treatment.
| 0 week (n=11) | 12 weekS (n=11) | |
|---|---|---|
| Age | 44.72 ± 11.1 | 44.72 ± 11.1 |
| Sex N (%) | ||
| Male | 9 (81.82) | 9 (81.82) |
| Female | 2 (18.18) | 2 (18.18) |
| PASI | 7.73 ± 3.12 | 7.09 ± 4.89 |
To assess the relationship between proteins reported in the literature with psoriasis, we compared the protein expression between psoriasis patients (n=23) and healthy controls (n=16) using the Satterthwaite t-test analysis. In addition to the well-known psoriasis-related proteins (e.g. VEGFA, IFNG, SELE, CXCL8, IL4, APOB, CCL22, EGFR, FAS, CD40LG), we identified 9 additional proteins (i.e., PI3, TNFRSF8, PFN1, KRT16, TNFSF8, KLK1, APOC3, CXCR3, CCL4) associated with psoriasis with statistical significance (p ≤ 0.05) (Figure 2C and 2D) (Table S3).
Surprisingly, two well-known serum cytokines that play an important role in the pathogenesis of psoriasis (i.e., TNF, IL17A) were not identified after statistical analysis. We therefore tested the binding specificity of the TNF and IL17A antibodies to their target proteins with the anti-TNF and anti-IL17A antibodies, respectively, by probing a protein array containing 7 different proteins (TNF, IL2, IL17A, IL1A, CCL2, CXCL10, CXCL9 and CCL4) (Figure S6) 44. The antibodies bound to their targets in both cases, thus suggesting that the reason TNA and IL17A were not identified in this study may be due to the pathophysiologic heterogeneity of psoriasis disease in the population 45-48.
Profiling of Psoriasis Serum Proteomes using DIA-MS and Antibody Microarray Technology
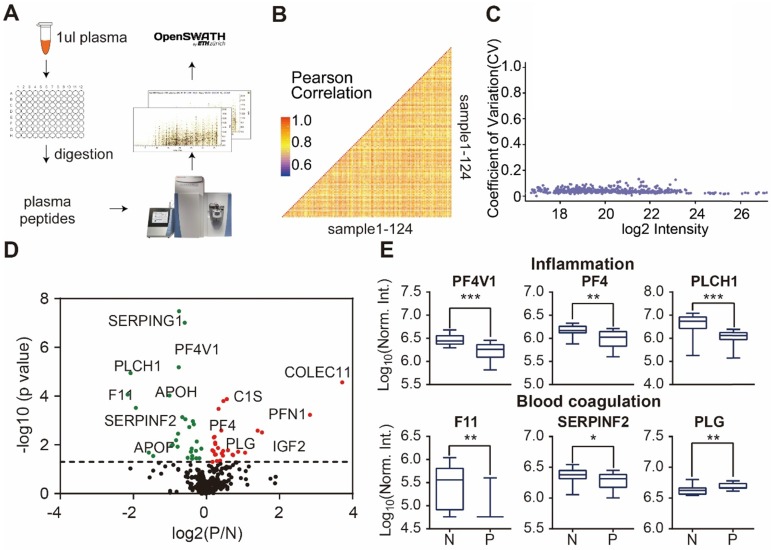
The workflow for plasma/serum screening using SWATH-MS has been described before 5, 30. In this study, we extended the SWATH methodology from TripleTOF (Sciex) to the DIA method using the QE-HF instrument (Thermo Fisher) (Figure 3A). To enable screening in high-throughput and reduce technical variation, all serum samples were added to a 96-well plate (2 μL/well), and then lysed, reduced, alkylated and digested in the same batch. DIA analysis led to the identification of 283 proteotypic peptides that uniquely belong to a single protein sequence to ensure high degree of quantitative accuracy.
Figure 3.
Detection of psoriasis serum proteins using DIA-MS. (A) High throughput screening of serum from psoriasis patients using DIA-MS; (B and C) Reproducibility of DIA-MS detection of serum proteins; (D) Identification of psoriasis-associated proteins in serum using volcano plot analysis; (E) Box plot analysis of newly identified serological psoriasis-associated proteins in inflammation and blood coagulation pathways. The selection of differentially expressed proteins was performed using the Satterthwaite t-test analysis (p-value = 0.05). N and P represent healthy controls and psoriatic patients, respectively. *, ** and *** represent the p-value less than 0.05, 0.005 and 0.0005, respectively.
Next, we analyzed the reproducibility of our DIA-MS assay, in which the Pearson correlation of 50 duplicated samples was 0.85 (0.69 - 0.95) (Figure 3B) and the average variation was 4.17% (1.75% -12.6%) (Figure 3C). The Satterthwaite t-test further identified 58 proteins differentially expressed in the serum of psoriatic patients (p ≤ 0.05) when compared to the healthy controls (Figure 3D, Table S4). Some of these proteins are known to be involved in inflammation (PF4V1, PF4, PLCH1) and blood coagulation (F11, SERPINF2 and PLG) (Figure 3E).
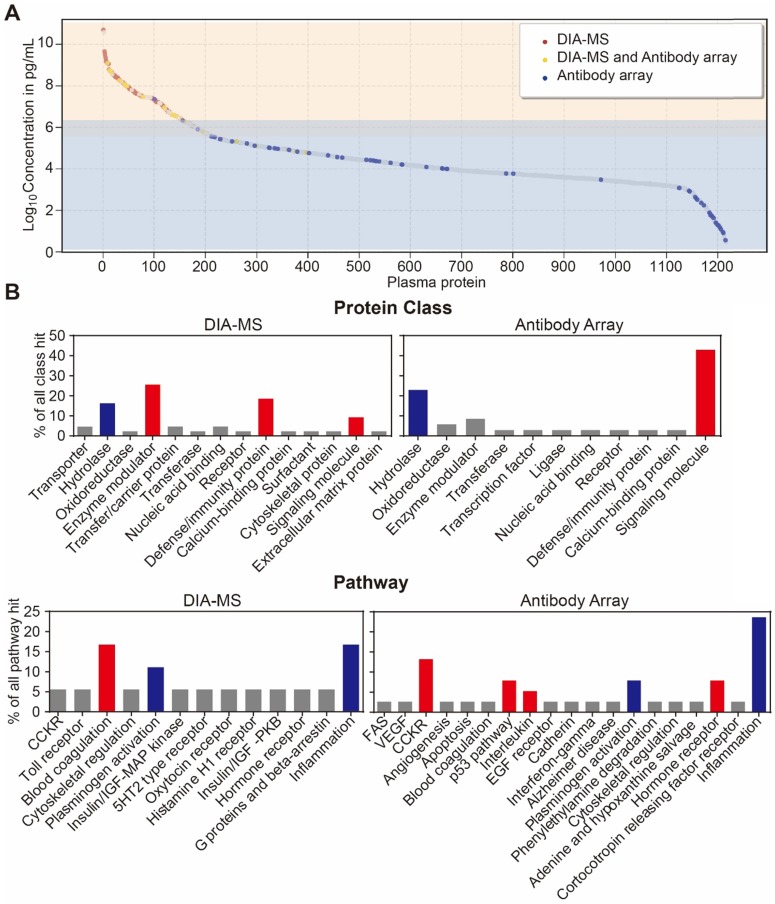
Lastly, we analyzed the distribution of serological protein concentrations detected by our DIA-MS and microarray platform using the reference concentrations from the human plasma proteome database (http://www.plasmaproteomedatabase.org/) 49 (Figure 4A, Table S5). The proteins detected by DIA-MS and antibody microarrays range from 106 to 1010 pg/mL and from 100 to 106 pg/mL, respectively, spanning a total of ~10 orders of magnitude (Figure 4A).
Figure 4.
Development of an in-depth serum proteomics platform using DIA-MS and customized antibody microarrays. (A) Distribution of serum proteins detected by our DIA-MS and antibody array two-pronged approach according to the reference concentrations in human plasma proteome database (http://www.plasmaproteomedatabase.org/). (B) are the comparison of protein classes and signaling pathways for psoriasis-associated proteins identified by DIA-MS and antibody microarrays, respectively.
Bioinformatics analysis of the serum proteins' functions identified by each proteomics method (Figure 4B) showed that most of proteins identified by DIA-MS and antibody microarrays are linked to the same cellular components and biological processes (Figure S7 and S8). However, while proteins detected by DIA-MS are enriched with enzyme modulator, defense immunity and the blood coagulation pathway, the proteins detected by antibody microarrays are enriched with signaling molecules and the CCKR, P53, interleukin and hormone receptor pathways (Figure 4B and 4C). For both DIA-MS and antibody microarrays, differentially expressed proteins in the serum of psoriasis patients were enriched in critical pathways involved in the regulation of inflammation and immune system (i.e., cytokine-chemokine, coagulation, complement) (Figure S9 and S10). Taken together, these data suggest that our in-depth proteomics analysis using DIA-MS and antibody microarrays comprehensively reveals the functional landscape of the psoriasis serum proteome, including molecular functions, protein classes, cellular components, biological processes and signaling pathways (Figure S11).
Correlation Analysis between Clinical Data, Serum Proteome and Psoriasis Severity
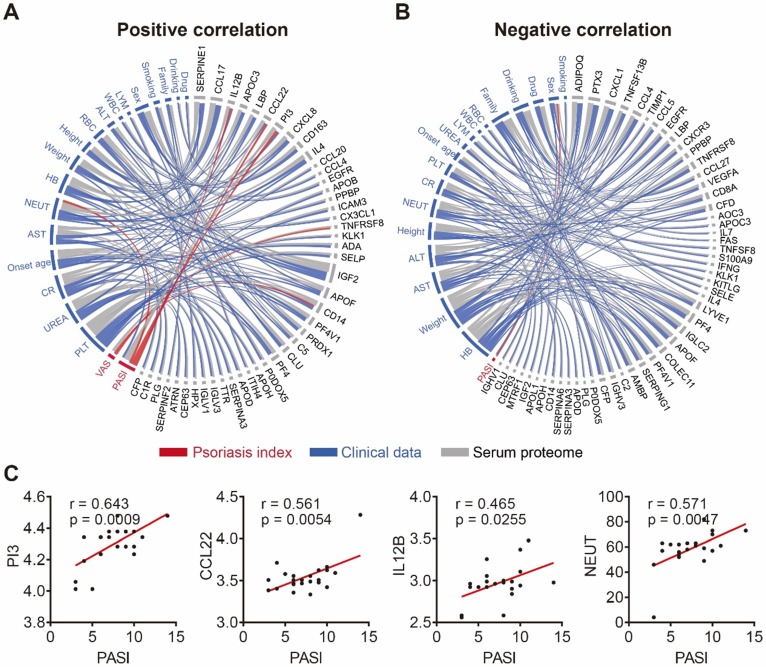
To further demonstrate the clinical utility of our in-depth serum proteomics platform, we analyzed the correlations between clinical data, serum proteome and psoriasis severity. Patients' visual analog scale (VAS) is a method sometimes used to measure itch intensity to assess psoriasis severity and treatment benefit 50, 51. Our results indicate that the number of neutrophils (NEUT) is positively correlated with the expression levels of four serum proteins (SERPINE1, PI3, IL4, CX3CL1) and negatively correlated with the expression levels of six serum proteins (CCL4, EGFR, CD8A, IFNG, IGLC2, MTRF1) (Figure S12). Notably, our results show that NEUT and sex correlated with the Psoriasis Area and Severity Index (PASI) score, which is consistent with previous reports 52, 53. In addition, we found that the PASI score is associated with three serum proteins (PI3, CCL22, IL-12B) (Figure 5C) that were uniquely different from the two serum proteins (TNFRSF8 and CD14) associated with the VAS scale (Figure S13). Among these proteins, PI3 and CCL22 were validated using ELISA (Figure S14). The positive and negative correlation data are represented by Figure 5A and Figure 5B, respectively (Table S6).
Figure 5.
Correlation network of serum proteome, clinical data and psoriasis index. (A) Positive and (B) negative correlations between the serum proteome (gray), clinical data (blue) and psoriasis indices (red) using circos, respectively. The statistical methods employed are explained in the Materials and Methods section; (C) Psoriasis serum proteins associated with the PASI score.
Identification of Psoriasis Biomarkers using In-depth Serum Proteomics
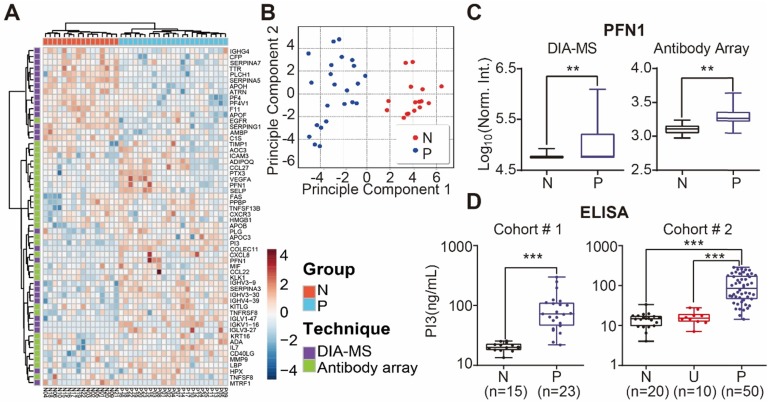
We performed unbiased hierarchical clustering on our collected serum proteome data and clinical status. The analyses showed that 87.3 % (18/23) of psoriasis patients could be distinguished from healthy controls with a specificity of 100% (16/16) (Figure S15). After using the Satterthwaite t-test analysis with a p-value of 0.01, 58 proteins in the nonbiased cluster analysis could discriminate healthy controls from psoriasis patients (Figure 6A). These results were confirmed by principle component analysis, indicating that these proteins could be used as diagnostic biomarkers for psoriasis (Figure 6B). Among these proteins, PFN1, a protein involved in actin polymerization, was cross-validated by DIA-MS and antibody microarrays (Figure 6C).
Figure 6.
Identification of serum proteins for the detection of psoriasis. (A) Classification of healthy and psoriatic patient groups based on differentially expressed proteins and unbiased clustering analysis; (B) Classification of healthy and psoriatic patient groups based on differentially expressed proteins and principle component analysis; (C) Cross-validation of PFN1 protein expression using DIA-MS and antibody microarrays. The statistical analysis was performed using the Satterthwaite t-test with a p-value of 0.01. (D) Validation of PI3 as a psoriasis biomarker using ELISA. *, ** and *** represent the p-value less than 0.05, 0.005 and 0.0005, respectively.
The potential biomarkers of psoriasis identified in our study, PI3, CCL22 and CD14, were further validated using quantitative ELISA using the same cohort of samples. We found that the expression of PI3, CCL22 and CD14 were higher in the psoriasis group (Figure 6D, Figure S14 and S16), which is consistent with our microarray results. We also validated PI3 using an independent cohort of 20 healthy controls, 10 urticaria patients, and 50 psoriasis patients (Table 3). The results show that PI3 expression is consistently higher in psoriasis patients compared to the other groups (Figure 6D).
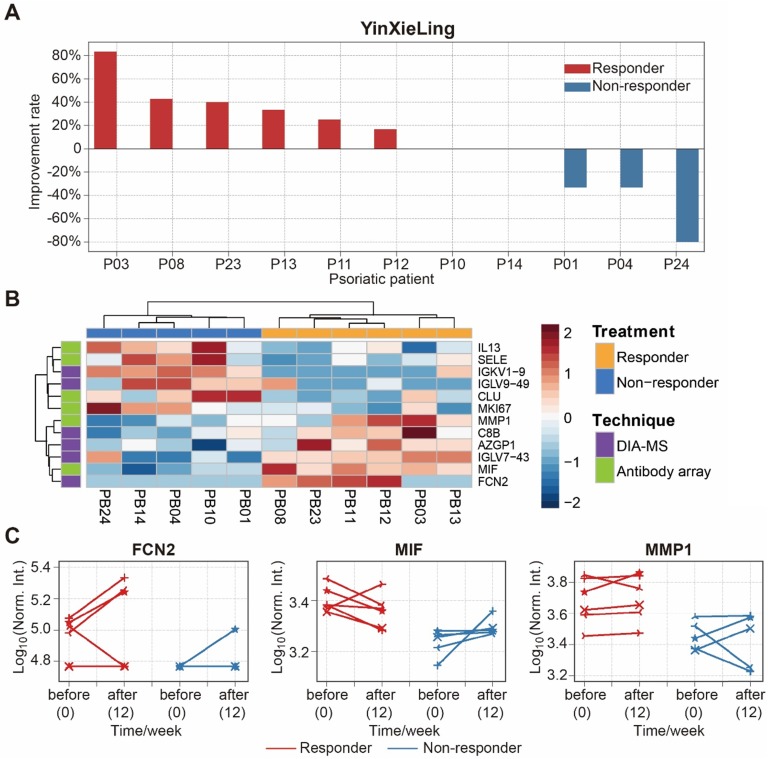
There are different drugs on the market for the treatment of psoriasis, including therapeutic monoclonal antibodies targeting specific cytokines (e.g., Adalimumab, Infliximab). Although highly effective, the use of these treatments can be limited by side effects, primary or secondary loss of efficacy, and high treatment costs 54. Therefore, finding biomarkers to predict treatment outcomes will reduce treatment costs, avoid side-effects and improve treatment effectiveness. In this study, we selected 11 psoriasis patients treated with an oral Chinese herbal medicine, YinXieLing, for 12 weeks 27, 38 (Table 2). Treatment results showed YinXieLing was effective in 6 patients and ineffective in 5 patients (Figure 7A).
Figure 7.
Identification of serum proteins to predict the efficacy of psoriasis treatment with YinXieLing. (A) Different responses of psoriasis patients to Chinese medicine treatment; (B) Classification of responders and non-responders based on differentially expressed proteins and unbiased clustering analysis; (C) Changes in protein expression levels before treatment and 12 weeks after treatment. The statistical analysis was performed using the Satterthwaite t-test with a p-value ≤ 0.05.
To identify predictive biomarkers of treatment response, we compared the serum protein expression in responders (n=6) and nonresponders (n=5) prior to the drug treatment and identified 12 differentially-expressed proteins with a p-value ≤ 0.05 (Table S7). These proteins distinguished responders and nonresponders using unbiased clustering and principle component analysis (Figure 7B and Figure S17). The proteins were then validated by measuring their expression changes after treatment with YinXieLing for 12 weeks (Figure 7C). After 12 weeks of treatment, the expression levels of FCN2, MIF and MMP1 in the serum of responders were consistently higher than nonresponders, indicating their potential as predictive biomarkers of YinXieLing effectiveness in treating psoriasis.
Discussion
In-depth proteome profiling of serum and plasma has been a major challenge due to the wide range of protein concentrations and the detection capability of current technologies. To help address this, we utilized data from DIA-MS and customizable antibody microarrays to compare the serological proteome of healthy controls and psoriasis patients. The combination of DIA-MS and antibody microarrays enabled the unprecedented in-depth analysis of the psoriasis serum proteome spanning 10 orders of magnitude in protein concentration (Figure 4A), thus allowing the exploration of protein changes during psoriasis and their relationship to clinical data (Figure 5). We found that the high proteome coverage of DIA-MS and the high sensitivity of antibody microarrays complemented each other, such that more informative data were collected on high and low abundance proteins from DIA-MS data and antibody microarrays, respectively (Figure 4B) (Figure S8-S10). This is not surprising since it is well known that the presence of highly abundant proteins can mask the detection of lower abundant proteins during mass spectrometry analysis 55.
We found that four proteins (SERPINE1, PI3, IL4, CX3CL1) and six proteins (IFNG, CCL4, EGFR, CD8A, IGLC2, MTRF1) were positively and negatively correlated with neutrophils, respectively (Figure S12). In our previous studies, we found that the oral traditional Chinese medicine, YinXieLing, exerts its therapeutic effect on psoriasis by inhibiting of cell proliferation and pro-inflammation while increasing CD4+ Foxp3+ regulatory t cell generation 39-41. These results align well with previous studies that show that PI3 downregulates neutrophil-mediated inflammation and IFNG inhibits neutrophil accumulation 56-59.
Three serum proteins (PI3, CCL22 and IL12B) that correlated with the PASI score (i.e., disease severity) were identified in this study (Figure 6D and Figure S16). PI3 (Elafin) is an epithelial host-defense protein that is absent in normal skin but highly expressed in keratinocytes of inflamed skin, which is a hallmark of psoriasis 60. The gene expression of PI3 has been found to be up-regulated in psoriatic skin samples (n=58) compared to controls (n=63) 61. More recently, Elgharib et al. found serological PI3 was significantly elevated in 26 psoriasis patients compared to 26 healthy controls 62, which appears to also confirm our results as well. CCL22 is a chemoattractant for monocytes, dendritic cells, natural killer cells and activated T lymphocytes, and likely plays a role in trafficking these cell types to inflammatory sites. Increased CCL22 expression in psoriatic skin has been associated with the positive response to infliximab therapy, a chimeric monoclonal antibody that targets TNF alpha 23. IL12B mutation is associated with the susceptibility to psoriasis and psoriastic lesions are particularly rich in IL12B proteins 63. Our study, which employed serum samples, suggests that PI3, CCL22 and IL12B are secreted into the circulatory system. In addition, we found that two serum proteins (TNFRSF8, CD14) correlated with the VAS score (i.e., another score system to measure disease severity) (Figure S13). CD14 is a marker on monocytes and macrophages, whose expression increased in psoriasis patients, but did not correlate with the PASI score 64.
Unbiased clustering analysis and principle component analysis identified 58 proteins that distinguished healthy controls from psoriasis patients (Figure 6A and 6B). A protein that interacts with actin and regulates the structure of the cytoskeleton, PFN1, was identified as a disease biomarker of psoriasis by both DIA-MS and the antibody array (Figure 6C). Interestingly, elevated PFN1 protein levels was previously observed in the synovial fluid of psoriatic arthritis patients using liquid chromatography coupled to MS (LC-MS/MS) and multiplexed selected reaction monitoring (MRM) assays65.
Finally, this study discovered 12 proteins that appear to be related to the effectiveness of YinXieLing treatment, distinguishing responders from non-responders with a p-value ≤ 0.05 (Figure 7B). Responders had sustained high expression levels of three proteins (FCN2, MMP1, MIF) after 12 weeks of treatment (Figure 7C). FCN2 encodes a ficolin-2 proteins that activates the lectin complement signaling pathway in innate immunity 66. MMP1 (Interstitial collagenase), which cleaves I, II, III, VII and X collagens, is elevated in the serum of patients with psoriatic arthritis, rheumatoid arthritis, osteoarthritis and systemic lupus erythematosus 67. MIF (Macrophage migration inhibitory factor) is a pleiotropic pro-inflammatory factor involved in inflammation and immunological processes that is elevated in the serum and peripheral blood mononuclear cells of psoriasis patients 68, 69.
There are several limitations in this study. First, only proteins associated with psoriasis in abstracts within the PubMed database were selected as targets for antibody microarrays. A larger antibody array could be constructed based on full-length text mining. Second, the sensitivity of antibody microarrays is still limited to pg/mL to ng/mL, thus no low abundance proteins below the detection limit can be analyzed. Signal amplification technology such as near-infrared fluorescence-enhanced detection 70, proximity ligation assay 71 and single molecule array 72 may increase array sensitivity for improved protein detection. Third, proteins may be quantified differently with DIA-MS and antibody microarrays due to different assay principles and detected protein isoforms. DIA-MS is an untargeted data acquisition approach with high proteomics coverage and accurate quantification of digested peptides 73. Antibody microarrays, on the other hand, detects target proteins using pre-immobilized antibodies on the slide matrix that recognize specific epitopes 11. The accurate quantification of multiple isoforms and splice variants, in which the amino acid sequences are highly homologous, is challenging for mass spectrometry and antibody arrays since their concentrations may be dramatically different in plasma and serum 74, 75. As such, biomarkers identified by DIA-MS or antibody microarrays must be validated with a different technology, such as ELISA 76. Notably, the number of clinical samples used in this study was limited, and therefore the biomarkers will need to be validated using a large independent clinical cohort before their use in clinical practice would be feasible.
The potential biomarkers revealed by this study distinguishing healthy controls from psoriasis patients and YinXieLing responders from non-responders, may help to better diagnose and treat psoriasis patients. These data also indicate that our in-depth serum proteomics approach using DIA-MS and antibody arrays has great potential in translational studies for other diseases.
Conclusion
We developed an in-depth serum proteomics technology platform that enable the measurement of serological proteome in unprecedented depth (~10 orders). Furthermore, we demonstrated that our methodology detected a large number of serum proteins associated with psoriasis disease and validated PI3 as a diagnostic biomarker for psoriasis in an independent clinical cohort. These results demonstrate the clinical utility of our novel proteomic assays and its usefulness to further advance our understanding, diagnosis, and treatment of psoriasis. Altogether, our findings suggest that the DIA-MS combined with antibody microarrays has great potential for application in translation studies and precision medicine in the future.
Supplementary Material
Supplementary figures and table legends.
Supplementary table S1.
Supplementary table S2.
Supplementary table S3.
Supplementary table S4.
Supplementary table S5.
Supplementary table S6.
Supplementary table S7.
Acknowledgments
This project was financially supported by the National Key Basic Research Project (2018ZX09733003, 2017YFC0906703 and 2018YFA0507503), the National Natural Science Foundation of China (81673040 and 31870823), the Program on the Joint Proteomics Center for Chinese Medicine between PHOENIX Center and Guangdong Provincial Academy of Chinese Medical Sciences, the State Key Laboratory of Proteomics (SKLP-O201504, SKLP-O201703 and SKLP-K201505) and Capital's Funds for Health Improvement and Research (2018-2-4034) to X.Y. We also thank Dr. Brianne Petritis for her critical review and editing of this manuscript.
Author Contributions
D. L. M. X., H. D. and Y. L. executed the text mining and manual curation. M.X., X. Z. and J. D. executed the array experiments; T. Z., Y. S., L. Y., Q. Z., X. C. and Y. Z., executed the sample preparation and DIA-MS experiments; J. D., L. H., Y. Y., D. Y., H. D. and C. L. collected the serum samples and clinical information; K. X., D. W., Y. Z., R. T., B. D. and C. C. executed the data analysis; X. Y., T. G., D. X. and C. L. designed and supervised all experiments and wrote the paper. All authors approved the final manuscript.
References
- 1.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karczewski KJ, Snyder MP. Integrative omics for health and disease. Nat Rev Genet. 2018;19:299–310. doi: 10.1038/nrg.2018.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price ND, Magis AT, Earls JC, Glusman G, Levy R, Lausted C. et al. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat Biotechnol. 2017;35:747–56. doi: 10.1038/nbt.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geyer PE, Holdt LM, Teupser D, Mann M. Revisiting biomarker discovery by plasma proteomics. Mol Syst Biol. 2017;13:942. doi: 10.15252/msb.20156297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Buil A, Collins BC, Gillet LC, Blum LC, Cheng LY. et al. Quantitative variability of 342 plasma proteins in a human twin population. Mol Syst Biol. 2015;11:786. doi: 10.15252/msb.20145728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–9. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 7.Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP. et al. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol Cell Proteomics. 2004;3:311–26. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–67. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 9.Wang D, Yang L, Zhang P, LaBaer J, Hermjakob H, Li D. et al. AAgAtlas 1.0: a human autoantigen database. Nucleic Acids Res. 2017;45:D769–D76. doi: 10.1093/nar/gkw946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J. et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–9. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu X, Schneiderhan-Marra N, Joos TO. Protein microarrays for personalized medicine. Clin Chem. 2010;56:376–87. doi: 10.1373/clinchem.2009.137158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin L, Zheng J, Yu Q, Chen W, Xing J, Chen C. et al. High throughput and accurate serum proteome profiling by integrated sample preparation technology and single-run data independent mass spectrometry analysis. J Proteomics. 2018;174:9–16. doi: 10.1016/j.jprot.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 13.Geyer PE, Kulak NA, Pichler G, Holdt LM, Teupser D, Mann M. Plasma Proteome Profiling to Assess Human Health and Disease. Cell Syst. 2016;2:185–95. doi: 10.1016/j.cels.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Chen R. et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geyer PE, Wewer Albrechtsen NJ, Tyanova S, Grassl N, Iepsen EW, Lundgren J. et al. Proteomics reveals the effects of sustained weight loss on the human plasma proteome. Mol Syst Biol. 2016;12:901. doi: 10.15252/msb.20167357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schubert OT, Rost HL, Collins BC, Rosenberger G, Aebersold R. Quantitative proteomics: challenges and opportunities in basic and applied research. Nat Protoc. 2017;12:1289–94. doi: 10.1038/nprot.2017.040. [DOI] [PubMed] [Google Scholar]
- 17.Yu X, LaBaer J. High-throughput identification of proteins with AMPylation using self-assembled human protein (NAPPA) microarrays. Nat Protoc. 2015;10:756–67. doi: 10.1038/nprot.2015.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X, Petritis B, Duan H, Xu D, LaBaer J. Advances in cell-free protein array methods. Expert Rev Proteomics. 2018;15:1–11. doi: 10.1080/14789450.2018.1415146. [DOI] [PubMed] [Google Scholar]
- 19.Lin YY, Lu JY, Zhang J, Walter W, Dang W, Wan J. et al. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136:1073–84. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ayoglu B, Mitsios N, Kockum I, Khademi M, Zandian A, Sjoberg R. et al. Anoctamin 2 identified as an autoimmune target in multiple sclerosis. Proc Natl Acad Sci U S A. 2016;113:2188–93. doi: 10.1073/pnas.1518553113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Cao Z, Duan H, Yu X. Glycosylation Profiling of Tumor Marker in Plasma Using Bead-Based Immunoassay. Methods Mol Biol. 2019;1871:413–20. doi: 10.1007/978-1-4939-8814-3_23. [DOI] [PubMed] [Google Scholar]
- 22.Farahnik B, Beroukhim K, Abrouk M, Nakamura M, Zhu TH, Singh R. et al. Brodalumab for the Treatment of Psoriasis: A Review of Phase III Trials. Dermatol Ther (Heidelb) 2016;6:111–24. doi: 10.1007/s13555-016-0121-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusumoto S, Kajihara I, Nagamoto E, Makino K, Ichihara A, Aoi J. et al. Increased CCL22 expression in psoriatic skin predicts a good response to infliximab therapy. Br J Dermatol. 2014;171:1259–61. doi: 10.1111/bjd.13091. [DOI] [PubMed] [Google Scholar]
- 24.Brezinski EA, Dhillon JS, Armstrong AW. Economic Burden of Psoriasis in the United States: A Systematic Review. JAMA Dermatol. 2015;151:651–8. doi: 10.1001/jamadermatol.2014.3593. [DOI] [PubMed] [Google Scholar]
- 25.Hong X, Jiang S, Marmolejo N, Vangipuram R, Ramos-Rojas E, Yuan Y. et al. Serum vascular endothelial growth factor receptor 3 as a potential biomarker in psoriasis. Exp Dermatol. 2018;27:1053–1057. doi: 10.1111/exd.13727. [DOI] [PubMed] [Google Scholar]
- 26.Reindl J, Pesek J, Kruger T, Wendler S, Nemitz S, Muckova P. et al. Proteomic biomarkers for psoriasis and psoriasis arthritis. J Proteomics. 2016;140:55–61. doi: 10.1016/j.jprot.2016.03.040. [DOI] [PubMed] [Google Scholar]
- 27.Deng J, Yao D, Lu C, Wen Z, Yan Y, He Z. et al. Oral Chinese herbal medicine for psoriasis vulgaris: protocol for a randomised, double-blind, double-dummy, multicentre clinical trial. BMJ open. 2017;7:e014475. doi: 10.1136/bmjopen-2016-014475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang R, Jiang W, Yang J, Mao YQ, Zhang Y, Yang W. et al. A biotin label-based antibody array for high-content profiling of protein expression. Cancer Genomics Proteomics. 2010;7:129–41. [PubMed] [Google Scholar]
- 29.Ayoglu B, Chaouch A, Lochmuller H, Politano L, Bertini E, Spitali P. et al. Affinity proteomics within rare diseases: a BIO-NMD study for blood biomarkers of muscular dystrophies. EMBO Mol Med. 2014;6:918–36. doi: 10.15252/emmm.201303724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo T, Kouvonen P, Koh CC, Gillet LC, Wolski WE, Rost HL. et al. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nat Med. 2015;21:407–13. doi: 10.1038/nm.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–86. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D61. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 35.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt J, Wozel G. The psoriasis area and severity index is the adequate criterion to define severity in chronic plaque-type psoriasis. Dermatology. 2005;210:194–9. doi: 10.1159/000083509. [DOI] [PubMed] [Google Scholar]
- 37.Papp KA, Langley RG, Lebwohl M, Krueger GG, Szapary P, Yeilding N. et al. Efficacy and safety of ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with psoriasis: 52-week results from a randomised, double-blind, placebo-controlled trial (PHOENIX 2) Lancet. 2008;371:1675–84. doi: 10.1016/S0140-6736(08)60726-6. [DOI] [PubMed] [Google Scholar]
- 38.Yao DN, Lu CJ, Wen ZH, Yan YH, Xuan ML, Li XY. et al. Oral PSORI-CM01, a Chinese herbal formula, plus topical sequential therapy for moderate-to-severe psoriasis vulgaris: pilot study for a double-blind, randomized, placebo-controlled trial. Trials. 2016;17:140. doi: 10.1186/s13063-016-1272-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han L, Sun J, Lu CJ, Zhao RZ, Lu Y, Lin HJ. et al. Formula PSORI-CM01 inhibits the inflammatory cytokine and chemokine release in keratinocytes via NF-kappaB expression. Int Immunopharmacol. 2017;44:226–33. doi: 10.1016/j.intimp.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 40.Chen H, Liu H, Lu C, Wang M, Li X, Zhao H. et al. PSORI-CM02 Formula Increases CD4+ Foxp3+ Regulatory T Cell Frequency and Ameliorates Imiquimod-Induced Psoriasis in Mice. Front Immunol. 2017;8:1767. doi: 10.3389/fimmu.2017.01767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei JA, Han L, Lu CJ, Zhao RZ, Sun J, Lu Y. et al. Formula PSORI-CM01 eliminates psoriasis by inhibiting the expression of keratinocyte cyclin B2. BMC Complement Altern Med. 2016;16:255. doi: 10.1186/s12906-016-1234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raychaudhuri SK, Abria C, Raychaudhuri SP. Regulatory role of the JAK STAT kinase signalling system on the IL-23/IL-17 cytokine axis in psoriatic arthritis. Ann Rheum Dis. 2017;76:e36. doi: 10.1136/annrheumdis-2016-211046. [DOI] [PubMed] [Google Scholar]
- 43.Mavropoulos A, Rigopoulou EI, Liaskos C, Bogdanos DP, Sakkas LI. The role of p38 MAPK in the aetiopathogenesis of psoriasis and psoriatic arthritis. Clin Dev Immunol. 2013;2013:569751. doi: 10.1155/2013/569751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venkataraman A, Yang K, Irizarry J, Mackiewicz M, Mita P, Kuang Z. et al. A toolbox of immunoprecipitation-grade monoclonal antibodies to human transcription factors. Nat Methods. 2018;15:330–8. doi: 10.1038/nmeth.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McInnes IB. Psoriatic arthritis: embracing pathogenetic and clinical heterogeneity? Clin Exp Rheumatol. 2016;34:9–11. [PubMed] [Google Scholar]
- 46.Wallstrom G, Anderson KS, LaBaer J. Biomarker discovery for heterogeneous diseases. Cancer Epidemiol Biomarkers Prev. 2013;22:747–55. doi: 10.1158/1055-9965.EPI-12-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bai F, Zheng W, Dong Y, Wang J, Garstka MA, Li R. et al. Serum levels of adipokines and cytokines in psoriasis patients: a systematic review and meta-analysis. Oncotarget. 2018;9:1266–78. doi: 10.18632/oncotarget.22260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kyriakou A, Patsatsi A, Vyzantiadis TA, Sotiriadis D. Serum levels of TNF-alpha, IL-12/23p40, and IL-17 in plaque psoriasis and their correlation with disease severity. J Immunol Res. 2014;2014:467541. doi: 10.1155/2014/467541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nanjappa V, Thomas JK, Marimuthu A, Muthusamy B, Radhakrishnan A, Sharma R. et al. Plasma Proteome Database as a resource for proteomics research: 2014 update. Nucleic Acids Res. 2014;42:D959–65. doi: 10.1093/nar/gkt1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pedersen CB, McHorney CA, Larsen LS, Lophaven KW, Moeller AH, Reaney M. Reliability and validity of the Psoriasis Itch Visual Analog Scale in psoriasis vulgaris. J Dermatolog Treat. 2017;28:213–20. doi: 10.1080/09546634.2016.1215405. [DOI] [PubMed] [Google Scholar]
- 51.Flytstrom I, Stenberg B, Svensson A, Bergbrant IM. Patients' visual analogue scale: a useful method for assessing psoriasis severity. Acta Derm Venereol. 2012;92:347–8. doi: 10.2340/00015555-1237. [DOI] [PubMed] [Google Scholar]
- 52.Merola JF, Wu S, Han J, Choi HK, Qureshi AA. Psoriasis, psoriatic arthritis and risk of gout in US men and women. Ann Rheum Dis. 2015;74:1495–500. doi: 10.1136/annrheumdis-2014-205212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schon MP, Broekaert SM, Erpenbeck L. Sexy again: the renaissance of neutrophils in psoriasis. Exp Dermatol. 2017;26:305–11. doi: 10.1111/exd.13067. [DOI] [PubMed] [Google Scholar]
- 54.Raut AS, Prabhu RH, Patravale VB. Psoriasis clinical implications and treatment: a review. Crit Rev Ther Drug Carrier Syst. 2013;30:183–216. doi: 10.1615/critrevtherdrugcarriersyst.2013005268. [DOI] [PubMed] [Google Scholar]
- 55.Kim B, Araujo R, Howard M, Magni R, Liotta LA, Luchini A. Affinity enrichment for mass spectrometry: improving the yield of low abundance biomarkers. Expert Rev Proteomics. 2018;15:353–66. doi: 10.1080/14789450.2018.1450631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ollague JE, Nousari CH. Expression of Elafin in Dermatitis Herpetiformis. Am J Dermatopathol. 2018;40:1–6. doi: 10.1097/DAD.0000000000000915. [DOI] [PubMed] [Google Scholar]
- 57.Stalberg C, Noda N, Polettini J, Jacobsson B, Menon R. Anti-inflammatory Elafin in human fetal membranes. J Perinat Med. 2017;45:237–44. doi: 10.1515/jpm-2016-0139. [DOI] [PubMed] [Google Scholar]
- 58.Small DM, Zani ML, Quinn DJ, Dallet-Choisy S, Glasgow AM, O'Kane C. et al. A functional variant of elafin with improved anti-inflammatory activity for pulmonary inflammation. Mol Ther. 2015;23:24–31. doi: 10.1038/mt.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nandi B, Behar SM. Regulation of neutrophils by interferon-gamma limits lung inflammation during tuberculosis infection. J Exp Med. 2011;208:2251–62. doi: 10.1084/jem.20110919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pol A, Pfundt R, Zeeuwen P, Molhuizen H, Schalkwijk J. Transcriptional regulation of the elafin gene in human keratinocytes. J Invest Dermatol. 2003;120:301–7. doi: 10.1046/j.1523-1747.2003.12043.x. [DOI] [PubMed] [Google Scholar]
- 61.Gudjonsson JE, Ding J, Johnston A, Tejasvi T, Guzman AM, Nair RP. et al. Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J Invest Dermatol. 2010;130:1829–40. doi: 10.1038/jid.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elgharib I, Khashaba SA, Elsaid HH, Sharaf MM. Serum elafin as a potential inflammatory marker in psoriasis. Int Immunopharmacol. 2019;58:205–9. doi: 10.1111/ijd.14217. [DOI] [PubMed] [Google Scholar]
- 63.Cargill M, Schrodi SJ, Chang M, Garcia VE, Brandon R, Callis KP. et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–90. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schopf RE, Dobmeyer J, Dobmeyer T, Morsches B. Soluble CD14 monocyte antigen in suction blister fluid and serum of patients with psoriasis. Dermatology. 1993;186:45–9. doi: 10.1159/000247301. [DOI] [PubMed] [Google Scholar]
- 65.Cretu D, Prassas I, Saraon P, Batruch I, Gandhi R, Diamandis EP. et al. Identification of psoriatic arthritis mediators in synovial fluid by quantitative mass spectrometry. Clin Proteomics. 2014;11:27. doi: 10.1186/1559-0275-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kilpatrick DC, St Swierzko A, Matsushita M, Domzalska-Popadiuk I, Borkowska-Klos M, Szczapa J. et al. The relationship between FCN2 genotypes and serum ficolin-2 (L-ficolin) protein concentrations from a large cohort of neonates. Hum Immunol. 2013;74:867–71. doi: 10.1016/j.humimm.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 67.Keyszer G, Lambiri I, Nagel R, Keysser C, Keysser M, Gromnica-Ihle E. et al. Circulating levels of matrix metalloproteinases MMP-3 and MMP-1, tissue inhibitor of metalloproteinases 1 (TIMP-1), and MMP-1/TIMP-1 complex in rheumatic disease. Correlation with clinical activity of rheumatoid arthritis versus other surrogate markers. J Rheumatol. 1999;26:251–8. [PubMed] [Google Scholar]
- 68.Gilliver SC, Emmerson E, Bernhagen J, Hardman MJ. MIF: a key player in cutaneous biology and wound healing. Exp Dermatol. 2011;20:1–6. doi: 10.1111/j.1600-0625.2010.01194.x. [DOI] [PubMed] [Google Scholar]
- 69.Shimizu T. Role of macrophage migration inhibitory factor (MIF) in the skin. J Dermatol Sci. 2005;37:65–73. doi: 10.1016/j.jdermsci.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 70.Zhang B, Kumar RB, Dai H, Feldman BJ. A plasmonic chip for biomarker discovery and diagnosis of type 1 diabetes. Nat Med. 2014;20:948–53. doi: 10.1038/nm.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J. et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 72.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L. et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28:595–9. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martins-de-Souza D, Faca VM, Gozzo FC. DIA is not a new mass spectrometry acquisition method. Proteomics. 2017;17:1700017. doi: 10.1002/pmic.201700017. [DOI] [PubMed] [Google Scholar]
- 74.Stastna M, Van Eyk JE. Analysis of protein isoforms: can we do it better? Proteomics. 2012;12:2937–48. doi: 10.1002/pmic.201200161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tipton JD, Tran JC, Catherman AD, Ahlf DR, Durbin KR, Kelleher NL. Analysis of intact protein isoforms by mass spectrometry. J Biol Chem. 2011;286:25451–8. doi: 10.1074/jbc.R111.239442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu X, Song L, Petritis B, Bian X, Wang H, Viloria J. et al. Multiplexed Nucleic Acid Programmable Protein Arrays. Theranostics. 2017;7:4057–70. doi: 10.7150/thno.20151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma J. et al. iProX: an integrated proteome resource. Nucleic Acids Res. 2019;47:D1211–D1217. doi: 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and table legends.
Supplementary table S1.
Supplementary table S2.
Supplementary table S3.
Supplementary table S4.
Supplementary table S5.
Supplementary table S6.
Supplementary table S7.