Abstract
Objective.
To evaluate the utility of amiodarone and its derivative dronedarone as novel drug repositioning candidates in EOC and to determine the potential pathways targeted by these drugs.
Methods.
Drug-predict bioinformatics platform was used to assess the utility of amiodarone as a novel drug-repurposing candidate in EOC. EOC cells were treated with amiodarone and dronedarone. Cell death was assessed by Annexin V staining. Cell viability and cell survival were assessed by MTT and clonogenics assays respectively. c-MYC and mTOR/Akt axis were evaluated as potential targets. Effect on autophagy was determined by autophagy flux flow cytometry.
Results.
“DrugPredict” bioinformatics platform ranked Class III antiarrhythmic drug amiodarone within the top 3.9% of potential EOC drug repositioning candidates which was comparable to carboplatin ranking in the top 3.7%. Amiodarone and dronedarone were the only Class III antiarrhythmic drugs that decreased the cellular survival of both cisplatin-sensitive and cisplatin-resistant primary EOC cells. Interestingly, both drugs induced degradation of c-MYC protein and decreased the expression of known transcriptional targets of c-MYC. Furthermore, stable overexpression of non-degradable c-MYC partially rescued the effects of amiodarone and dronedarone induced cell death. Dronedarone induced higher autophagy flux in EOC cells as compared to amiodarone with decreased phospho-AKT and phospho-4EBP1 protein expression, suggesting autophagy induction due to inhibition of AKT/mTOR axis with these drugs. Lastly, both drugs also inhibited the survival of EOC tumor-initiating cells (TICs).
Conclusions.
We provide the first evidence of class III antiarrhythmic agents as novel c-MYC targeting drugs and autophagy inducers in EOC. Since c-MYC is amplified in >40% ovarian tumors, our results provide the basis for repositioning amiodarone and dronedarone as novel c-MYC targeting drugs in EOC with potential extension to other cancers.
Keywords: Epithelial ovarian cancer (EOC), Drug repositioning, Class III antiarrhythmics, Amiodarone, Dronaderone, C-MYC, Tumor initiating cells (TICs)
1. Introduction
Epithelial ovarian cancer [EOC] is the most lethal gynecologic malignancy and is the fifth leading cause of cancer deaths in women [1]. This year it is estimated that >20,000 women will be newly diagnosed and >14,000 will succumb to EOC in the United States. EOC is a heterogeneous disease but the treatment outlook of EOC has remained more homogenous to date and cisplatin has still remained the first choice chemotherapy agent for EOC over recent decades [2]. This divergence of EOC heterogeneity vs. treatment dependency on cisplatin is evident by the fact that most of EOC patients who initially respond to platinum therapy relapse due to development of chemo-resistance [3]. Hence, in-spite of high initial response to platinum the outcomes are poor for advanced stage EOC with 5-year survival being <30%. Therefore, novel targeted therapeutic options are required in order to improve the lives of women diagnosed with EOC.
Traditional drug discovery approaches in oncology require billions of dollars in infrastructure and the lead compounds take more than a decade to enter clinical trial platforms at which point a majority of these drugs fail thus limiting their clinical translation potential [4,5]. Hence, economical and reliable approaches to identify novel anti-cancer are required in cancer therapeutics to enable successful clinical translation of these anti-cancer drugs. Drug repositioning offers a cost effective and safe approach in this context and several FDA approved drugs for non-oncology purposes are being evaluated as potential anti-cancer drugs in ovarian cancer [6]. For example, the anti-HIV drug Ritonavir has shown to be effective against ovarian tumor cells [7] and the anti-diabetic drug Metformin has shown promising results in ovarian cancer patient trials for its efficacy as a novel anti-cancer drug in EOC [8].
We have previously reported a novel drug repositioning platform termed “DrugPredict” that identified potential FDA approved drugs that could be repositioned in for cancer therapeutics [9]. Using DrugPredict, we have recently identified the non-steroidal anti-inflammatory drug (NSAID) Indomethacin as a novel anti-cancer drug in EOC which provides a good basis to evaluate this drug as a novel chemoadjuvant in ovarian cancer patient trials [9]. In the current study, we have evaluated Class III antiarrhythmic agents amiodarone and its derivative dronedarone as potential anti-cancer drugs in EOC. Our results provide the rationale to further characterize amiodarone and dronedarone as novel drug repositioning candidates in EOC especially given its potent inhibitory effects on the commonly activated oncogene, c-MYC.
2. Materials and methods
2.1. Cell culture and reagents
Cells were cultured in 10 cm plates in a humidified atmosphere (5% CO2) at 37 °C. At 70–90% confluence, trypsin (0.25%)/EDTA solution (Corning) was used to detach the cells from the culture plate for passaging and used for further experiments until passage 20. OV81.2 and OV81.2-CP10 primary cell line models were generated as previously described [10]. A2780 and CP70 cells were obtained from Dr. Paul Modrich (Duke University). OVCAR3 cells were purchased from ATCC. OV81.2, OV81.2-CP10, A2780, CP70, ALDHhigh CP70, ALDHlow CP70, OVCAR8 and 293 T cells were cultured in DMEM medium supplemented with 10% FBS (Gibco) and 1% penicillin-streptomycin (PS) (Gibco). OVCAR3 and OVCAR3-CP38 cells were cultured in RPMI medium (Gibco) with 10% FBS and 1%PS. Phosphate-buffered saline (PBS) was purchased from Corning. Amiodarone, dronedarone, ibutilide and sotalol were purchased from SelleckChem. Dimethyl sulfoxide (DMSO) was purchased from Fisher Chemical.
2.2. DrugPredict
DrugPredict is a novel computational drug discovery system that simultaneously performs in silico target-based and phenotypic screening on half a million compounds. It uses large amounts of publicly available data including disease genetics, chemical genetics, signaling pathways, and mouse genome-wide mutation phenotypes for the prediction candidate drugs to treat a given disease (ovarian cancer in this study). We have previously applied DrugPredict to identify repurposed candidates for ovarian cancer and experimentally validated that indomethacin can be used to kill ovarian cancer cells [9]. In this study, we used DrugPredict to predict the relevance of amiodarone in the treatment of ovarian cancer. DrugPredict was described in details in our previous study [9]. Briefly, DrugPredict applied for OC drug repositioning consists of the following steps: (1) Construct a genetic profile for OC by obtaining a list of genes that are differentially expressed in high-grade serous ovarian cancer (HGSOC) patients from TCGA (HGSOC genes); (2) correlating HGSOC genes to their homologs in mouse models and construct a mouse mutational phenotype profile for OC; (2) construct mouse mutational phenotype profile for chemicals/drugs; (3) develop algorithms to match the HGSOC-specific phenotype profile to drug profiles and prioritize FDA-approved drugs based on the phenotypic similarities; and (4) analyze repositioned drug candidates to evaluate the predictions and identify interpretable mechanisms of actions.
2.3. Generation of stable cell lines
Constitutively active c-MYCT58A plasmid was generously donated by Dr. Goutham Narla’s laboratory at Case Western Reserve University. Briefly, to generate plasmid, pMIG-cMYC (Addgene #18119) was site-direct mutated to generate a threonine - > alanine conversion at site 58. Control plasmid construct MDH1-PGK-GFP was previously donated by Dr. Chen Z. Cheng (Stanford University). c-MYCT58A stably overexpressing line was retrovirally generated in A2780. Phoenix packaging cells were seeded in a 150 mm × 150 mm cell culture plate such that the next day they would be at 50–70% confluency. 12 μg of control or cMYCT58A plasmids were transfected into cells with 6 μg pCL-ampho using Lipofectamine 2000 (Life Technologies). 8 h later media was changed and viral media was collected for 3 days post-transfection. To transduce A2780, viral media containing 5 μg/ml polybrene (Millipore) was added to cells serially for 3 days. Once cells attained confluency, cells were selected by GFP-fluorescent sorting compared to A2780 untransduced cells using FACS Aria (BD Biosciences).
2.4. Real-time PCR
Total RNA was extracted using the Total RNA Purification Plus Kit (Norgen Biotek) according to manufacturer’s instructions. For mRNA analysis, cDNA synthesis from 1 μg of total RNA was done using the Transcriptor Universal cDNA Master kit (Roche). SYBR green-based Real-time PCR was subsequently performed in triplicate using SYBR green master mix (Roche) on the Light Cycler 480 II real time PCR machine (Roche).
2.5. Immunoblotting
Whole cell protein extracts were probed with antibodies against GAPDH (1:1000) (Santa Cruz Biotechnology, CA) c-MYC 1:250 (Abcam) pAKT-Ser473 1:100(Cell Signaling), p4EBP1 1:250 (Cell Signaling) as previously described [10]. Membranes were exposed using LumiLight or LumiLightplus (Roche) method following manufacturer’s instructions.
2.6. Cell viability assay
Cells were plated in 12-well plate at 50,000 cells/well and treated the next day with the drugs. After 48 h, cells were then incubated with 3-(4,5-Dimethylthiazolyl) for 2 h and absorbance was measured at 600 nm.
2.7. Clonogenic assay
Cell survival was assessed through seeding 1000 cells/well in a 6-well plate and treated with indicated doses of the drugs the next day. On day 7, cells were fixed in a 10% acetic acid/10% methanol (in diH2O) solution and stained with 1% crystal violet (in methanol) after 7 days of growth.
2.8. Annexin V-PI staining
Cells were plated at 200,000 cells/well in a 6-well plate. After 72 h, Annexin V-PI Staining was done using the FITC or APC Annexin V Apoptosis Detection Kit II (BD Pharmingen). FACS data was acquired using BD LSR II.
2.9. Autophagy Flux flow cytometry
Autophagy Flux was determined using the CYTO-ID Autophagy detection kit (Enzo Life Sciences) as per the manufacturer’s instructions. Briefly, cells were plated at 200,000 cells/well in a 6-well plate and were treated with the drugs next day. 48 h after treatment, cells were trypsinized, washed once with the assay buffer and were stained with CYTO-ID green detection reagent in the assay buffer for 30 min at 37 °C. FACS data was acquired using BD LSRII.
2.10. ALDH sorting flow cytometry
For ALDH based sorting, ALDH assay was done using the ALDEFLUOR kit as per the protocol instructions (Stem cell Technologies) and ALDH positive and negative cells were FACS sorted from CP70 cells using BD FACS Aria II.
2.11. Tumor sphere formation assay
For tumor sphere formation assays, 1000 cells per ml were plated in 2 ml in 6-well ultra-low attachment plate (Corning) in MammoCult medium (Stem cell Technologies) in the presence of amiodarone. After 7 days, 10 × 10 stitch imaging was done at 10× (100 random images acquired) using a Retiga Aqua Blue camera (Q Imaging, Vancouver, BC) connected to a Leica DMI6000 inverted microscope. Individual images were taken and then a composite image was generated using the scan slide function in MetaMorph Imaging Software (Molecular Devices, Downingtown, PA). Subsequent integrated analysis also used MetaMorph software.
2.12. Statistical analysis
Unless otherwise noted, data are presented as mean ± SD from three-independent experiments, and Student’s t-test (two-tailed) was used to compare two groups (P < 0.05 was considered significant) for independent samples.
3. Results
3.1. Class III antiarrhythmic agents have potential anti-cancer properties in EOC
DrugPredict ranked the Class III antiarrhythmic drug amiodarone within top 3.9% of potential FDA approved drugs that may be a good candidate for drug repositioning in EOC (Fig. 1A). This was comparable to the top 3.7% DrugPredict ranking of carboplatin, which is the current mainstay treatment option in EOC. Class III antiarrhythmic drugs are comprised of 6 drugs including amiodarone and its derivative dronedarone. Amiodarone has been reported to induce apoptosis in glioma cells [11] and has been found to decrease metastatic ability of breast cancer cells [12], thus suggesting that it could be a novel anti-cancer drug. However, the utility of amiodarone or its derivative dronedarone as an anti-cancer drug in EOC is yet to be explored and the mechanism driving these anti-cancer properties remains unknown.
Fig. 1.
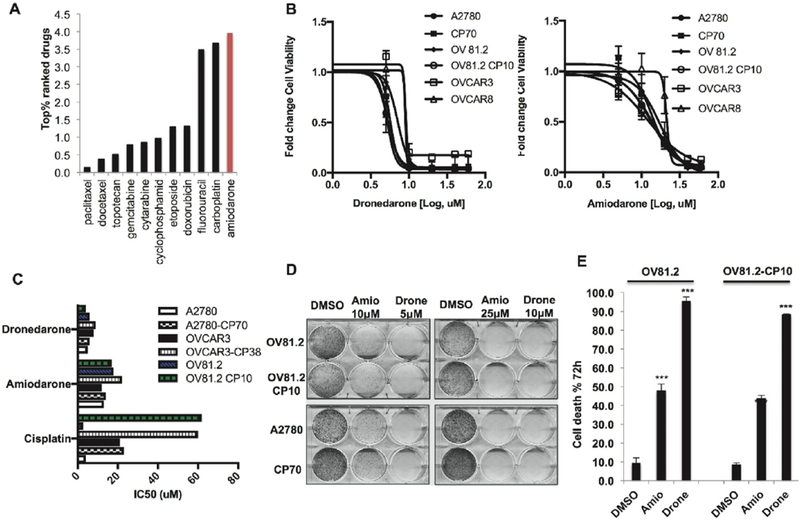
Amiodarone and dronedarone are novel drug repositioning candidates in EO. (A) DrugPredict ranking of amiodarone (Top 3.9%) showing it is comparable to the ranking of Carboplatin (Top 3.7%). (B) 48 h MTT assays with amiodarone, and dronedarone OV81.2, OV81.2-CP10, A2780, CP70, OVCAR3 and OVCAR8 cells. (C) IC50 of amiodarone and dronedarone in comparison to Cisplatin in A2780, A2780-CP70, OVCAR3, OVCAR3-CP38, OV81.2 and OV81.2-CP10 cells. (D) Clonogenics assay (Day 7) showing decreased cell survival upon treatment with amiodarone and dronedarone in OV81.2, OV81.2-CP10, A2780 and CP70 as compared to DMSO control. (E) Annexin V-PI staining flow cytometry assay (72 h) showing increased cell death in OV81.2 and OV81.2-CP10 upon treatment with amiodarone and dronedarone as compared to DMSO control. (***p < 0.0001).
In order to explore the potential anti-cancer effects of amiodarone and dronedarone in EOC, we looked at the effect of these drugs on cell viability of several ovarian cancer cell lines including primary cisplatin sensitive and resistant models that we have previously developed in the lab [10] (OV81.2 and OV81.2-CP10). Amiodarone and dronedarone decreased the viability in all cell lines with an IC50 ranging from 4 to 18 μM (Fig. 1B &1C). On the contrary, ibutilide and sotalol, which are other class III antiarrhythmic but structurally different from amiodarone [13], did not affect cell viability in any of these cells (Suppl Fig. 1). Amiodarone and dronedarone also decreased cellular survival of both cisplatin resistant (OV81.2-CP10 and CP70) and cisplatin sensitive (OV81.2 and A2780) EOC cells (Fig. 1D). Further analysis revealed that both amiodarone and dronedarone induced robust cell death in primary epithelial ovarian cancer cells as determined by Annexin-V cell death assays (Fig. 1E). Overall, through these initial studies we found that both drugs potently decrease cell survival and induce cell death in numerous EOC cells including primary high-grade serous ovarian cancer (HGSOC) cells.
3.2. Amiodarone and dronedarone are novel c-MYC targeting drugs in EOC
Next we sought to uncover the mechanisms driving amiodarone and dronedarone anti-cancer effects. Interestingly, genomic cluster and network analysis looking at the toxicity profile of amiodarone treatment in rats revealed that c-MYC regulated cellular processes were negatively affected in the hepatic tissue [14]. In addition, the DrugPredict platform found that several of the top 20 pathways associated with amiodarone (Fig. 2A) were functionally linked to c-MYC; c-MYC is reported to be an important regulator of unfolded protein response in cancer cells [15], suppression of c-MYC by glucocorticoids has been reported in cancer cells [16,17], functional roles of c-MYC is regulating several aspects of hypertrophic cardiomyopathy is well documented [18], several ATP-binding cassette (ABC) transporter genes have been identified as direct transcriptional targets of c-MYC in cancer cells [19], regulation of lipid metabolism by c-MYC is well studied in cancers [20] and lastly, functional interaction of c-MYC with mTOR signaling pathway is reported in cancers [21]. Given these functional implications of amiodarone driven regulation of c-MYC pathways and the role of c-MYC in driving EOC survival and platinum resistance [22,23], we next ascertained whether the potent effects of these drugs were due to the regulation of c-MYC.
Fig. 2.
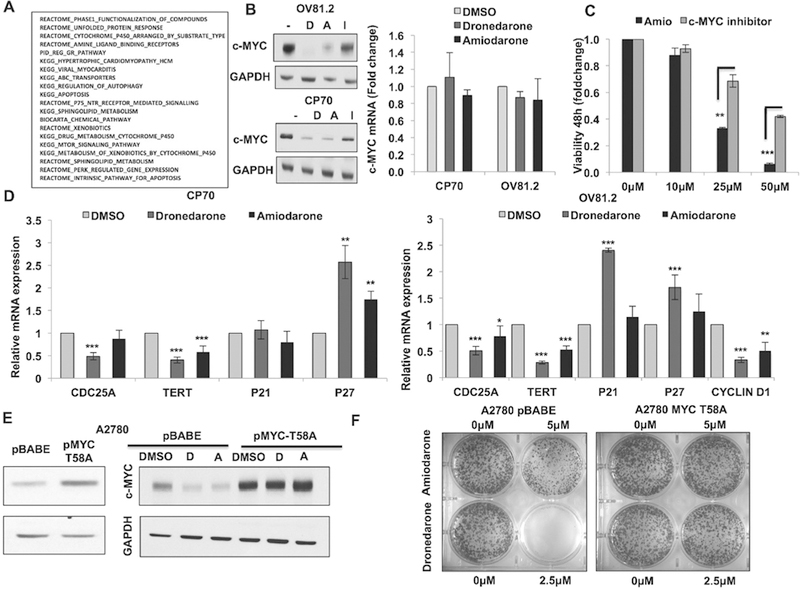
Amiodarone and dronedarone are novel c-MYC targeting drugs in EOC. (A) Top-20 predicted pathways associated with amiodarone based on DrugPredict analysis. (B) Western blots showing c-MYC protein degradation by 6h upon treatment with dronedarone (20 μM), amiodarone (20 μM) and ibutilide (20 μM) in CP70 and OV81.2 cells. (C) Real-time PCR showing changes in c-MYC mRNA by6h upon treatment with dronedarone (20 μM) and amiodarone (20 μM) in CP70 and OV81.2 cells. (D) Real-time PCR showing changes in the expression of c-MYC transcriptional targets by 6 h upon treatment with dronedarone (20 μM) and amiodarone (20 μM) in CP70 (left) and OV81.2 (right) cells. (E) Western blots (left) showing overexpression of non-degradable c-MYC (T58A) in A2780 cells and (right) effect of dronedarone (20 μM) and amiodarone (20 μM) on c-MYC protein expression in T58A cells as compared to pBABE control cells (6 h treatment). (F) Clonogenics assay (day 7) comparing the effect of dronedarone and amiodarone on viability in T58A vs. pBABE cells (** p < 0.001, *** p < 0.0001).
We found that amiodarone and dronedarone robustly decreased c-MYC protein expression in both A2780-CP70 and OV81.2 as early as 6 h (Fig. 2B). Decrease of c-MYC protein by ibutilide was less dramatic as compared to amiodarone and dronedarone, which correlated with minimal decrease in cell viability with this drug suggesting that c-MYC downregulation potentially underlies the anti-cancer drug properties of both amiodarone and dronedarone. There was minimal decrease in c-MYC mRNA levels upon treatment with amiodarone and dronedarone (~10% decrease) thus these drugs must regulate c-MYC expression by post-translational mechanisms (Fig. 2C). We further confirmed the functional inhibition of c-MYC by assessing the effects of these drugs on the expression of the transcriptional targets of c-MYC. Remarkably, mRNA expression of c-MYC activated transcriptional targets were decreased upon treatment with amiodarone and dronedarone in CP70 (CDC25A and TERT) and OV81.2 (CDC25A, TERT and Cyclin D1) (Fig. 2D). Also, expression of c-MYC inhibited transcriptional targets was increased upon treatment with these drugs in CP70 (p27) and OV81.2 (p21 and p27) (Fig. 2D). Next, to further confirm the dependency of amiodarone and dronedarone effects on c-MYC, we looked at the effects of stable overexpression of non-degradable c-MYC (T58A) on the phenotype induced by these drugs in A2780 cells which we have previously shown to express low levels of c-MYC [24] (Fig. 2E left). We found that c-MYC protein was not degraded in cells overexpressing T58A c-MYC (Fig. 2E right). Importantly, T58A c-MYC overexpression increased tolerance to both amiodarone and dronedarone showing that c-MYC overexpression rescues the effect of these drugs on cell survival (Fig. 2F). Collectively, these results further confirm that c-MYC is a functional target of amiodarone and dronedarone as is partially responsible for the anti-cancer effects of these drugs.
3.3. Amiodarone and dronedarone are novel autophagy inducing drugs in EOC
Interestingly, autophagy was one of the top 20 pathways predicted to be associated with amiodarone (Fig. 2A), consistent with a recent report showing that amiodarone induced autophagy in breast cancer cells [25]. Also, decreased c-MYC protein expression is observed during mTOR inhibition mediated autophagy induction [26]. Thus, we next looked at whether induction of autophagy could be one of the mechanisms of amiodarone and dronedarone effects in EOC. Both amiodarone and dronedarone increased Autophagy flux in EOC cells (Fig. 3A). Furthermore, mTOR and AKT pathways which have been shown to be main regulators of autophagy [27] and were predicted as one of the top 20 pathways associated with amiodarone (Fig. 2A), were decreased with amiodarone and dronedarone treatment but not with ibutilide. Both amiodarone and dronedarone decreased the expression of phospho-AKT and phospho-4EBP1 confirming inhibition of mTOR/AKT axis as compared to ibutilide, which did not decrease the expression of these proteins (Fig. 3B). These results suggest that induction of autophagy by inhibition of mTOR/AKT axis could be one of the mechanisms by which amiodarone and dronedarone decrease the survival of EOC cells and also confirm the functional reliability and utility of DrugPredict platform in identifying potential targets of novel pharmacologic agents.
Fig. 3.
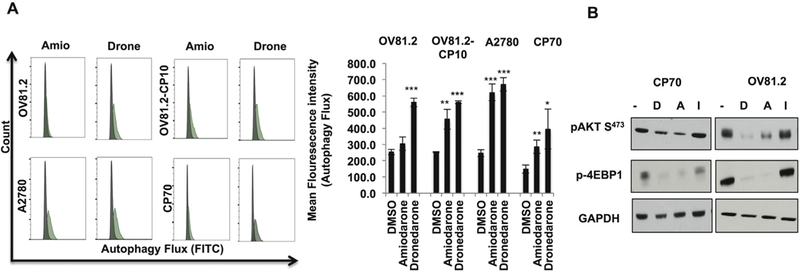
Amiodarone and dronedarone are novel autophagy inducing drugs in EOC. (A) Flow cytometry (48 h) histogram (left) and mean fluorescence intensity (right) showing increased autophagy flux upon treatment with dronedarone (20 μM) and amiodarone (20 μM) in OV81.2, OV81.2-CP10, A2780 and CP70 cells. (B) Western blots showing pAKT S473 and p-4EBP1 expression in CP70 and OV81.2 cells upon treatment with dronedarone (20 μM), amiodarone (20 μM) and ibutilide (20 μM) (*p < 0.01, **p < 0.001, ***p < 0.0001).
3.4. Amiodarone and dronedarone inhibit the survival of both tumor-initiating cells (TICs) and non-TICs in EOC
Lastly, given that tumor-initiating cells (TICs) drive tumor initiation-recurrence-metastasis and chemo-resistance in various cancers including EOC [28] and the effects of amiodarone on both TICs and non-TICs are not understood in cancers, we next looked at the effect of both amiodarone and dronedarone on a previously established TICs model ALDHhigh CP70 (TICs) and ALDHlow CP70 (non-TICs) [9,10]. Interestingly, both TICs and non-TICs exhibited similar sensitivity to these drugs (Fig. 4A and B) as determined in cell viability and clonogenic assays. Amiodarone and dronedarone induced robust cell death in both TICs and non-TICs (Fig. 4C). Ability to form tumor spheres under 3D stem-cell culture conditions is a defining trait of ovarian TICs that enables tumor initiation at low densities and also drives metastatic ability [10]. Hence, we next looked at the effects of amiodarone on ovarian TICs under 3D sphere formation culture conditions. Amiodarone induced robust decrease tumor sphere formed by ovarian TICs showing that this drug can inhibit the survival of ovarian TICs under 3D culture conditions (Fig. 4D). Interestingly, only dronedarone induced autophagy in both TICs and non-TICs whereas amiodarone did not induce autophagy in these cells (Fig. 4E). Collectively, these results reveal a novel role for both amiodarone and dronedarone in inhibiting ovarian TICs and suggest that the mechanisms driving the effects of these drugs could be different in this context.
Fig. 4.
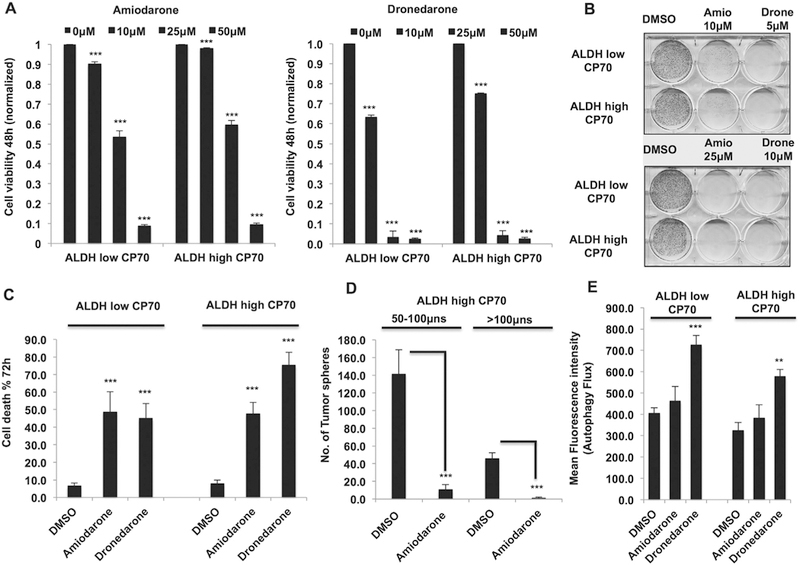
Amiodarone and dronedarone inhibit both TICs and non-TICs in EOC. (A) 48 h MTT assay showing decreased cell viability in both ALDHlow CP70 and ALDHhigh CP70 cells upon treatment with dronedarone (right) and amiodarone (left). (B) Clonogenics assay (day 7) showing decreased survival of both ALDHlow CP70 and ALDHhigh CP70 cells upon treatment with dronedarone and amiodarone. (C) 72 h Annexin-V assay showing increased cell death in both ALDHlow CP70 and ALDHhigh CP70 cells upon treatment with dronedarone and amiodarone. (D) Tumor sphere assay (7 days) showing decreased tumor spheres upon treatment with amiodarone (10 μM). (E) Flow cytometry (mean fluorescence intensity) showing increased autophagy flux upon treatment with dronedarone (20 μM) in both ALDHlow CP70 and ALDHhigh CP70 cells (**p < 0.001, ***p < 0.0001).
4. Discussion
Drug repositioning is emerging as a cost effective and reliable drug discovery approach in oncology. Identifying the “off-target” anti-cancer effects of FDA approved drugs which are already in clinical use for other diseases provides with novel drugs whose safety profile and patient tolerance data are available in detail and hence translation of these drugs into patient trials can be faster compared to traditional drug discovery approaches which often take more than a decade for the lead compounds to enter clinical trials. Also, drug repositioning offers a more economical drug discovery approach that can make afford-ability of anti-cancer drugs a reality in healthcare. Metformin has been a good example of success story in this context in ovarian cancer where-in pre-clinical studies with this drug has lead to testing it in ovarian cancer patient trials and the results have been promising. Thus, drug repositioning has the potential to improvise EOC therapeutics. Our study has identified that Class III antiarrhythmic agents amiodarone and dronedarone can be potential candidates for drug repositioning in EOC.
Both amiodarone and dronedarone decreased the survival of EOC cells irrespective of their cisplatin sensitivity status. Several mechanisms could underlie function of these drugs in EOC and accordingly, pathway association for amiodarone by DrugPredict revealed several pathways that are implicated in tumorigenesis. Identification of amiodarone as an autophagy inducer has been reported in pharmacologic screening studies in cancer cells [25] which correlated with our both prediction and validation results and additionally we identified dronedarone as a more potent inducer of autophagy in EOC. Hence, induction of autophagy could be one of the mechanisms underlying the functions of both amiodarone and dronedarone in EOC. Inhibition of mTOR/AKT axis by both these drugs in primary HGSOC cells as well as prediction of amiodarone association with mTOR by DrugPredict further supports this possibility.
The most interesting observation from our study was identifying c-MYC as a functional target of both amiodarone and dronedarone, thus providing the very first evidence of c-MYC targeting by Class III antiarrhythmics in cancers including EOC. DrugPredict analysis of top associated pathways with amiodarone suggested c-MYC could be a central nodal point for amiodarone effects in cancer cells and our results validated c-MYC as a functional target of amiodarone and also dronedarone. This is of potential clinical relevance since c-MYC is the most commonly amplified gene in EOC [22] and is functionally implicated in regulating several aspects of ovarian cancer pathogenesis [23]. However, further studies assessing the efficacy of amiodarone and dronedarone in-vivo patient derived xenograft models of EOC form a critical component of evaluating these antiarrhythmic agents as novel anti-cancer drugs in EOC. Our results provide a good basis to test this possibility. Since c-MYC is amplified and also functionally implicated in several cancers, our results provide the rationale to evaluate amiodarone and dronedarone as novel c-MYC targeting drugs in other cancers too.
Several concerns need to be addressed before realizing the translational potential of amiodarone and dronedarone as novel anti-cancer drugs for the treatment of epithelial ovarian cancer. Long half-life (~60 days) and calcium/potassium channel blocking activities are the causes of the most of the side effects associated with amiodarone [29]. Repurposing amiodarone to decrease its half-life and decrease its calcium/potassium channel blocking activities would be an ideal approach to further test the translational potential of this drug as an anti-cancer drug. Dronedarone has a shorter half-life than amiodarone (24 h) and hence could be an attractive drug for re-purposing [30]. However, dronedarone also inhibits calcium/potassium channels and hence further studies are required to design re-purposed amiodarone and dronedarone derivatives that are potentially free of cardiac effects.
To summarize, our study reiterates the importance of employing Drug repositioning approaches to rapidly identify and characterize novel therapeutic modalities for the treatment of EOC. The discoveries based on such approaches offer the advantage of being potentially fast tracked into patient trials since these drugs are already FDA approved. Hence, these approaches will enable the design of pre-clinical studies with successful clinical implications.
Supplementary Material
HIGHLIGHTS.
Amiodarone and dronedarone are novel Drug-Repositioning candidates in epithelial ovarian cancer (EOC).
Amiodarone and dronedarone target c-MYC for degradation.
Amiodarone and dronedarone are inhibitors of mTOR/AKT axis in EOC.
Amiodarone and dronedarone are effective at inducing cell death in both EOC TICs and non-TICs.
Acknowledgements
We thank Norma C. and Albert I. Geller for their constant support of the Gynecological Cancer Translational Research Program and the DiFeo Laboratory (A.D.). We acknowledge the help from Cytometry & Imaging Microscopy Core Facility and the Athymic Animal and Preclinical Therapeutics Core of the Case Comprehensive Cancer Center (P30CA043703). Funding for this work was supported by grants from the National Cancer Institute, R01CA197780 (A.D.), Department of Defense, OC150553 (A.D.), The Mary Kay Foundation (A.D. & R.X.), and The Young Scientist Foundation (A.D.). QW and RX are supported by the NIH Director’s New Innovator Award under the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the National Institutes of Health (DP2HD084068, Xu), NIH National Institute of Aging (1 R01 AG057557–01, Xu), American Cancer Society Research Scholar Grant (RSG-16–049-01-MPC, Xu), and NIH Clinical and Translational Science Collaborative of Cleveland (1UL1TR002548–01, Konstan).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ygyno.2018.09.019.
Conflict of interest
None.
References
- [1].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2017, CA Cancer J. Clin. 67 (2017) 7–30, 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- [2].Kelland L, The resurgence of platinum-based cancer chemotherapy, Nat. Rev. Cancer 7 (2007) 573–584, 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- [3].Cooke SL, Brenton JD, Evolution of platinum resistance in high-grade serous ovarian cancer, Lancet Oncol. 12 (2011) 1169–1174, 10.1016/S1470-2045(11)70123-1. [DOI] [PubMed] [Google Scholar]
- [4].Ashburn TT, Thor KB, Drug repositioning: identifying and developing new uses for existing drugs, Nat. Rev. Drug Discov. 3 (2004) 673–683, 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- [5].Bertolini F, Sukhatme VP, Bouche G, Drug repurposing in oncology-patient and health systems opportunities, Nat. Rev. Clin. Oncol. 12 (2015) 1–11, 10.1038/nrclinonc.2015.169. [DOI] [PubMed] [Google Scholar]
- [6].Banno K, Iida M, Yanokura M, Irie H, Masuda K, Kobayashi Y, Tominaga E, Aoki D, Drug repositioning for gynecologic tumors: a new therapeutic strategy for cancer, ScientificWorldJournal 2015 (2015) 341362, 10.1155/2015/341362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kumar S, Bryant CS, Chamala S, Qazi A, Seward S, Pal J, Steffes CP, Weaver DW, Morris R, Malone JM, Shammas MA, Prasad M, Batchu RB, Ritonavir blocks AKT signaling, activates apoptosis and inhibits migration and invasion in ovarian cancer cells, Mol. Cancer 8 (2009) 26, 10.1186/1476-4598-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Irie H, Banno K, Yanokura M, Iida M, Adachi M, Nakamura K, Umene K, Nogami Y, Masuda K, Kobayashi Y, Tominaga E, Aoki D, Metformin: a candidate for the treatment of gynecological tumors based on drug repositioning (Review), Oncol. Lett. 11 (2016) 1287–1293, 10.3892/ol.2016.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nagaraj AB, Wang QQ, Joseph P, Zheng C, Chen Y, Kovalenko O, Singh S, Armstrong A, Resnick K, Zanotti K, Waggoner S, Xu R, Difeo A, Using a novel computational drug-repositioning approach (DrugPredict) to rapidly identify potent drug candidates for cancer treatment, Oncogene (2017) 1–12, 10.1038/onc.2017.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nagaraj AB, Joseph P, Kovalenko O, Singh S, Armstrong A, Redline R, Resnick K, Zanotti K, Waggoner S, DiFeo A, Critical role of Wnt/β-catenin signaling in driving epithelial ovarian cancer platinum resistance, Oncotarget 6 (2015) 23720–23734, 10.18632/oncotarget.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kim IY, Kang YJ, Yoon MJ, Kim EH, Kim SU, Kwon TK, IA Kim KS Choi, Amiodarone sensitizes human glioma cells but not astrocytes to TRAIL-induced apoptosis via CHOP-mediated DR5 upregulation, Neuro-Oncology 13 (2011) 267–279, 10.1093/neuonc/noq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lee H, Su M, Lo H, Wu C, Hu J, Lo D, Chao T, Tsai H, Dai M, Cancer metastasis and EGFR signaling is suppressed by amiodarone-induced versican V2, Oncotarget (2015) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Singh BN, Antiarrhythmic drugs: a reorientation in light of recent developments in the control of disorders of rhythm [review] [75 refs], Am. J. Cardiol. 81 (1998) 3D–13D. [DOI] [PubMed] [Google Scholar]
- [14].Fukushima T, Kikkawa R, Hamada Y, Horii I, Genomic cluster and network analysis for predictive screening for hepatotoxicity, J. Toxicol. Sci. 31 (2006) 419–432, 10.2131/jts.31.419. [DOI] [PubMed] [Google Scholar]
- [15].Shajahan-Haq AN, Cook KL, Schwartz-Roberts JL, Eltayeb AE, Demas DM, Warri AM, Facey COB, Hilakivi-Clarke LA, Clarke R, MYC regulates the unfolded protein response and glucose and glutamine uptake in endocrine resistant breast cancer, Mol. Cancer 13 (2014) 10.1186/1476-4598-13-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Thulasi R, Harbour DV, Thompson EB, Suppression of c-myc is a critical step in glucocorticoid-induced human leukemic cell lysis, J. Biol. Chem. 268 (1993) 18306–18312. [PubMed] [Google Scholar]
- [17].Zhou F, Medh RD, Thompson EB, Glucocorticoid mediated transcriptional repression of c-myc in apoptotic human leukemic CEM cells, J. Steroid Biochem. Mol. Biol. 73 (2000) 195–202, 10.1016/S0960-0760(00)00080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wolfram JA, Lesnefsky EJ, Hoit BD, Smith MA, Lee HG, Therapeutic potential of c-Myc inhibition in the treatment of hypertrophic cardiomyopathy, Ther. Adv. Chronic Dis. 2 (2011) 133–144, 10.1177/2040622310393059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Porro A, Iraci N, Soverini S, Diolaiti D, Gherardi S, Terragna C, Durante S, Valli E, Kalebic T, Bernardoni R, Perrod C, Haber M, Norris MD, Baccarani M, Martinelli G, Perini G, c-MYC oncoprotein dictates transcriptional profiles of ATP-binding cassette transporter genes in chronic myelogenous leukemia CD34+ hematopoietic progenitor cells, Mol. Cancer Res. 9 (2011) 1054–1066, 10.1158/1541-7786.MCR-10-0510. [DOI] [PubMed] [Google Scholar]
- [20].Zirath H, Frenzel A, Oliynyk G, Segerstrom L, Westermark UK, Larsson K, Munksgaard Persson M, Hultenby K, Lehtio J, Einvik C, Pahlman S, Kogner P, Jakobsson P-J, Arsenian Henriksson M, MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells, Proc. Natl. Acad. Sci. 110 (2013) 10258–10263, 10.1073/pnas.1222404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pourdehnad M, Truitt ML, Siddiqi IN, Ducker GS, Shokat KM, Ruggero D, Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers, Proc. Natl. Acad. Sci. 110 (2013) 11988–11993, 10.1073/pnas.1310230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bell D, Berchuck A, Birrer M, Chien J, Cramer DW, Dao F, Dhir R, DiSaia P, Gabra H, Glenn P, Godwin AK, Gross J, Hartmann L, Huang M, Huntsman DG, Iacocca M, Imielinski M, Kalloger S, Karlan BY, Levine DA, Mills GB, Morrison C, Mutch D, Olvera N, Orsulic S, Park K, Petrelli N, Rabeno B, Rader JS, Sikic BI, Smith-McCune K, Sood AK, Bowtell D, Penny R, Testa JR, Chang K, Dinh HH, Drummond JA, Fowler G, Gunaratne P, Hawes AC, Kovar CL, Lewis LR, Morgan MB, Newsham IF, Santibanez J, Reid JG, Trevino LR, Wu Y-Q, Wang M, Muzny DM, Wheeler DA, Gibbs RA, Getz G, Lawrence MS, Cibulskis K, Sivachenko AY, Sougnez C, Voet D, Wilkinson J, Bloom T, Ardlie K, Fennell T, Baldwin J, Gabriel S, Lander ES, Ding L, Fulton RS, Koboldt DC, McLellan MD, Wylie T, Walker J, O’Laughlin M, Dooling DJ, Fulton L, Abbott R, Dees ND, Zhang Q, Kandoth C, Wendl M, Schierding W, Shen D, Harris CC, Schmidt H, Kalicki J, Delehaunty KD, Fronick CC, Demeter R, Cook L, Wallis JW, Lin L, Magrini VJ, Hodges JS, Eldred JM, Smith SM, Pohl CS, Vandin F, Raphael BJ, Weinstock GM, Mardis ER, Wilson RK, Meyerson M, Winckler W, Getz G, Verhaak RGW, Carter SL, Mermel CH, Saksena G, Nguyen H, Onofrio RC, Lawrence MS, Hubbard D, Gupta S, Crenshaw A, Ramos AH, Ardlie K, Chin L, Protopopov A, Zhang J, Kim TM, Perna I, Xiao Y, Zhang H, Ren G, Sathiamoorthy N, Park RW, Lee E, Park PJ, Kucherlapati R, Absher DM, Waite L, Sherlock G, Brooks JD, Li JZ, Xu J, Myers RM, Laird PW, Cope L, Herman JG, Shen H, Weisenberger DJ, Noushmehr H, Pan F, Triche T Jr., Berman BP, Van Den Berg DJ, Buckley J, Baylin SB, Spellman PT, Purdom E, Neuvial P, Bengtsson H, Jakkula LR, Durinck S, Han J, Dorton S, Marr H, Choi YG, Wang V, Wang NJ, Ngai J, Conboy JG, Parvin B, Feiler HS, Speed TP, Gray JW, Levine DA, Socci ND, Liang Y, Taylor BS, Schultz N, Borsu L, Lash AE, Brennan C, Viale A, Sander C, Ladanyi M, Hoadley KA, Meng S, Du Y, Shi Y, Li L, Turman YJ, Zang D, Helms EB, Balu S, Zhou X, Wu J, Topal MD, Hayes DN, Perou CM, Getz G, Voet D, Saksena G, Zhang J, Zhang H, Wu CJ, Shukla S, Cibulskis K, Lawrence MS, Sivachenko A, Jing R, Park RW, Liu Y, Park PJ, Noble M, Chin L, Carter H, Kim D, Karchin R, Spellman PT, Purdom E, Neuvial P, Bengtsson H, Durinck S, Han J, Korkola JE, Heiser LM, Cho RJ, Hu Z, Parvin B, Speed TP, Gray JW, Schultz N, Cerami E, Taylor BS, Olshen A, Reva B, Antipin Y, Shen R, Mankoo P, Sheridan R, Ciriello G, Chang WK, Bernanke JA, Borsu L, Levine DA, Ladanyi M, Sander C, Haussler D, Benz CC, Stuart JM, Benz SC, Sanborn JZ, Vaske CJ, Zhu J, Szeto C, Scott GK, Yau C, Hoadley KA, Du Y, Balu S, Hayes DN, Perou CM, Wilkerson MD, Zhang N, Akbani R, Baggerly KA, Yung WK, Mills GB, Weinstein JN, Penny R, Shelton T, Grimm D, Hatfield M, Morris S, Yena P, Rhodes P, Sherman M, Paulauskis J, Millis S, Kahn A, Greene JM, Sfeir R, Jensen MA, Chen J, Whitmore J, Alonso S, Jordan J, Chu A, Zhang J, Barker A, Compton C, Eley G, Ferguson M, Fielding P, Gerhard DS, Myles R, Schaefer C, Mills Shaw KR, Vaught J, Vockley JB, Good PJ, Guyer MS, Ozenberger B, Peterson J, Thomson E, Integrated genomic analyses of ovarian carcinoma, Nature 474 (2011) 609–615, 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Reyes-Gonzalez JM, Armaiz-Pena GN, Mangala LS, Valiyeva F, Ivan C, Pradeep S, Echevarria-Vargas IM, Rivera-Reyes A, Sood AK, Vivas-Mejia PE, Targeting c-MYC in platinum-resistant ovarian cancer, Mol. Cancer Ther. 14 (2015) 2260–2269, 10.1158/1535-7163.MCT-14-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hudson CD, Savadelis A, Nagaraj AB, Joseph P, Avril S, Difeo A, Avril N, Hudson CD, Savadelis A, Nagaraj AB, Joseph P, Avril S, DiFeo A, Avril N, Altered glutamine metabolism in platinum resistant ovarian cancer, Oncotarget 7 (2016) 41637–41649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Balgi AD, Fonseca BD, Donohue E, Tsang TCF, Lajoie P, Proud CG, Nabi IR, Roberge M, Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling, PLoS One 4 (2009) https://doi.org/10.137l/journal.pone.0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Puustinen P, Rytter A, Mortensen M, Kohonen P, Moreira JM, Jäättelä M, CIP2A oncoprotein controls cell growth and autophagy through mTORC1 activation, J. Cell Biol. 204 (2014) 713–727, 10.1083/jcb.201304012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yang ZJ, Chee CE, Huang S, Sinicrope FA, The role of autophagy in Cancer: therapeutic implications, Mol. Cancer Ther. 10 (2011) 1533–1541, 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Garson K, Vanderhyden BC, Epithelial ovarian cancer stem cells: underlying complexity of a simple paradigm, Reproduction 149 (2015) R59–R70, 10.1530/REP-14-0234. [DOI] [PubMed] [Google Scholar]
- [29].Vorperian VR, Havighurst TC, Miller S, January CT, Adverse effects of low dose amiodarone: a meta-analysis, J. Am. Coll. Cardiol. 30 (1997) 791–798, 10.1016/S0735-1097(97)00220-9. [DOI] [PubMed] [Google Scholar]
- [30].Clem JR, Farver DK, Fischer JR, Johnson TJ, Dronedarone: a safety comparison to amiodarone, Curr. Drug Saf. 5 (2010) 251–256, 10.2174/157488610791698280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


