Abstract
Physiological properties of tumors can be measured both in vivo and non-invasively by diffusion-weighted imaging and dynamic contrast-enhanced magnetic resonance imaging. Although these techniques have been used for more than two decades to study tumor diffusion, perfusion, and/or permeability the methods and studies on how to reduce measurement error and bias in the derived imaging metrics is still lacking in the literature. This is of paramount importance because the objective is to translate these quantitative imaging biomarkers (QIB) into clinical trials, and ultimately in clinical practice. Standardization of the image acquisition using appropriate phantoms is the first step from a technical performance standpoint. The next step is to assess whether the imaging metrics have clinical value and meet the requirements for being a QIB as defined by the Radiological Society of North America’s Quantitative Imaging Biomarkers Alliance/(QIBA®). The goal and mission of QIBA and the National Cancer Institute Quantitative Imaging Network (QIN) initiatives are to provide technical performance standards (QIBA profiles) and QIN tools for producing reliable QIBs for use in the clinical imaging community. Some of QIBA’s development of quantitative diffusion-weighted imaging and dynamic contrast-enhanced QIB profiles has been hampered by the lack of literature for repeatability and reproducibility of the derived QIBs. The available research on this topic is scant and is not in sync with improvements or upgrades in magnetic resonance imaging technology over the years. This review focuses on the need for QIBs in oncology applications and emphasizes the importance of the assessment of their reproducibility and repeatability.
Introduction
In the last decade, there have been major rapid advances in the field of magnetic resonance imaging (MRI), including advancements in hardware, acquisition pulse sequences, image reconstruction, and data analysis algorithms (1–9). These technological advances have fostered to a timely focus on quantitative MRI (qMRI), which purports the ability to derive objective metrics from images that relate to specific physical or biophysical properties of the imaged tissue. Two prime qMRI examples are diffusion-weighted imaging (DWI) and dynamic contrast enhanced (DCE)-MRI, which allow characterization of tissue cellularity inferred from water mobility and microvascular properties, derived from exogenous contrast agent (CA) kinetics, respectively. Applications of qMRI include detection of disease and its evolution in progression or response to therapies that affect the relevant biophysical property of tissue (e.g., cytotoxic therapies that reduce cellularity) (10–13). These methods have been covered in excellent reviews detailing the technical aspects and their applications (5–9,14–17). As the quantitative measurements derived from DWI (e.g., mean diffusivity) and DCE (e.g., volume transfer constant) are utilized in clinical trials of new treatment strategies, or for precision medicine and personalized cancer care, the technical confidence of these measurements in repeatability and reproducibility is ever more critical (18–20). Expert task forces of the Quantitative Imaging Biomarkers Alliance (QIBA®) have devoted significant resources to write DWI and DCE profiles and review over 1000 scientific manuscripts in this area, but such literature review efforts have yielded few original manuscripts with adequately described test-retest data. The lack of repeatability and reproducibility literature in this area creates a roadblock for clinical translation of DWI and DCE. This review focuses on the clinical and technical needs for quantitative DWI and DCE derived imaging biomarkers and provides recommendations for image acquisition, analysis, and quality control relevant to improving precision and accuracy, or reducing measurement error for the derived quantitative metrics. This review is limited to the use of DWI and DCE for evaluation of tumors in the brain, prostate, breast, liver, and head and neck, recognizing that studies of different organs imply varying technical protocols and challenges.
1. Understanding Impact of Precision and Accuracy in Quantitative Imaging
Quantitative imaging metrics reflect relevant information about a biological process by measuring biophysical parameters that could be used as biomarkers, rather than relying solely on relative differences in image signal intensity (of arbitrary scale and units), as in routine diagnostic imaging. However, the quantitative images must be standardized and optimized to generate protocols for acquisition and analysis of these biomarkers. Kessler, et al. have defined the term Quantitative Imaging Biomarker (QIB) as “An objective characteristic derived from an in vivo image measured on a ratio or interval scale as an indicator of normal biological processes, pathogenic processes, or a response to a therapeutic intervention” (21). Unlike conventional diagnostic imaging, where sensitivity and specificity are used to describe the predictive power of the qualitative test for the patient population, the technical performance of a QIB in quantitative imaging, particularly its bias (accuracy), precision (variability), and linearity, determine its inherent reliability (confidence interval) to diagnose, monitor, and predict outcome.
The objective of a test-retest study is to measure the degree to which test results are consistent over time (22–26). The internal consistency is a measure of the correlation between two sets of imaging data performed on two occasions. In a test-retest study, subjects are scanned at least twice over a short period of time to ensure that no biological change has occurred. From each image, the biomarker measurement is derived completely and independently of the results from the other scan. Sometimes a subject is scanned twice in the same study session; it is important in these instances that the subject leaves the table and is repositioned for the second scan. In other situations, particularly those involving CA administration, a subject might be scanned a second time the next day or so. For example, administering a gadolinium-based CA to a patient in a clinical trial twice on the same day is not practical due to retention of the CA in a lesion and the concern for patient safety. For this reason, it is not surprising that test-retest data are limited in sample size. Although test-retest studies are ideally performed on clinical subjects, estimates of test-retest variability can be obtained from phantom studies (27,28). It is generally recognized that these estimates are likely to underestimate the true variability seen in clinical subjects. The variability could be due to the patient movement that adds to variation in the signal intensity measurement compared with the phantom study.
2. Precision Metrology
2.1. Repeatability and Reproducibility
Repeatability represents the measurement precision, or closeness of agreement, of replicate measurements made over a short period of time. These replicate measurements are made with the same measurement procedure, operator, measuring system, operating condition, and physical location (21). Reproducibility is similar to repeatability, except that in acquiring the measurements, some aspect of the procedure, or timing, differs (e.g., different operator, different scanner, etc.) (21). For instance, systematic measurement bias between different scanners would be expected to impact reproducibility compared with more controlled (single-system) repeatability values.
Repeatability is often quantified by the within-subject standard deviation (wSD) or variance. For example, for N subjects, each with replicate measurements, one could use Table 1 to estimate the wSD. Large values of wSD indicate that confidence in any single measurement of the biomarker should be minimal because a second measurement is likely to differ considerably. Small values of the wSD boost confidence in the reliability of the measurement. Of course, “large” vs. “small” should be interpreted relative to known, or anticipated, biomarker differences between normal/abnormal tissues, across patient groups, or change with time in an individual patient/lesion undergoing treatment.
Table 1:
Steps for calculating the within-subject deviation
| Steps | Method for calculating wSD |
|---|---|
| 1 | Calculate the variance for each of N subjects from their replicate measurements. |
| 2 | Take the mean of the variances over the N subjects. This gives an estimate of the within-subject variance. |
| 3 | Take the square root of the estimated within-subject variance to get an estimate of the wSD. |
within-subject deviation (wSD)
When a patient is followed longitudinally to measure tumor treatment response or progression, there is a need to understand how to interpret observed differences in biomarker values. Small differences may be attributable to just measurement error whereas large differences exceed the expectation from simple measurement error. To reduce measurement error, standardized protocol should be implemented on pre-tested measurement device and kept consistent in the course of the study. If the wSD is known from relevant prior studies, a threshold can be calculated for when the difference between two longitudinal measurements is attributable to measurement error and when it can be confidently attributable to a true change. The “test-retest” procedure is designed to estimate wSD (22,23). From this, the repeatability coefficient (RC) is calculable (29) and represents the least significant difference between two repeated measurements taken under identical conditions, usually at a confidence level of 95%. It can be calculated as follows:
| [1] |
For example, if a test-retest of DWI is performed and the wSD is estimated at 10, then RC = 27.7. This means that if the difference between a patient’s baseline and follow-up measurements is < −27.7, or > +27.7, then real change (outside of the measurement error) has occurred with 95% confidence. If the difference is between −27.7 and +27.7, it may be due to measurement imprecision.
When the repeatability varies with the magnitude of the QIB measurements, the within-subject coefficient of variation (wCV) is sometimes used to quantify the variability (29). The wCV is commonly used with imaging biomarkers because often the wSD is small for small QIB values but increases with larger QIB values. The steps for calculating the wCV are given in Table 2. Instead of the RC, the %RC is calculated as:
| [II] |
For example, if a test-retest of DCE is performed and the wCV is estimated at 10%, then the %RC = 27.7%. This means that if the percent change (e.g., difference between a patient’s baseline and follow-up measurements divided by the baseline measurement × 100) is < −27.7% or > 27.7%, then real change has occurred with 95% confidence. If the percent change is between −27.7% and +27.7%, it may be due to measurement imprecision.
Table 2:
Steps for calculating the within-subject coefficient of variation
| Steps | Method for calculating wCV |
|---|---|
| 1 | Calculate the variance and mean for each of N subjects from their replicate measurements. |
| 2 | Calculate the wCV2 for each of the N subjects by dividing their variance by their squared mean. |
| 3 | Take the mean of the wCV2 over the N subjects. |
| 4 | Take the square root of the value in step 3 to get an estimate of the wCV. |
within-subject coefficient of variation (wCV)
Based on the current literature, %RC values for tumor apparent diffusion coefficient (ADC) region of interest (ROI) measurements derived from monoexponential modeling of DWI data in three different organs are as follows: brain = 11% (30–32), liver = 26% (33–36), and prostate = 47% (37–40). This assumes the wCV for tumors in the brain is 3.97%, 9.38% for the liver, and 16.97% for the prostate. The claim statements for the tumor ROIs in these organs can be found in the DWI QIBAprofile(https://qibawiki.rsna.org/images/7/7d/QIBADWIProfilev1.45_20170427_v5_accepted_linenumbers.pdf).
The %RC values for volume transfer constant (Ktrans) measurements in tumors, derived from pharmacokinetic modeling of DCE data, in two different organs is as follows: 21.3% for the brain and 55.7% for the prostate. This assumes that the wCV for tumors in the brain is 7.7% and 20.1% for the prostate (41,42). The claim statements for the tumor measurements in these organs can be found in the DCE QIBA profile (http://qibawiki.rsna.org/index.php/Profiles).
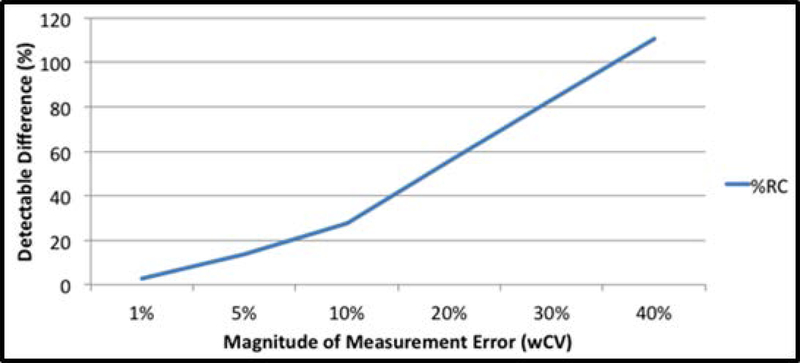
Figure 1 illustrates the impact of the wCV (or wSD) on this decision cut-point. It shows that when the imaging methods have good repeatability, earlier and more confident conclusions can be made about changes in patients’ QIB measurements. For example, when the wCV is low, e.g., 5%, a real change in Ktrans of 14% (or larger) can be detected with 95% confidence. If the wCV is moderate, e.g., 15%, a change of over 40% is needed to rule out the measurement error. A doubling of Ktrans, equivalent to 100% change, would have to occur to detect a real change with 95% confidence when the wCV is 36%.
Figure 1:
The %RC is the cut-point where a change in the biomarker measurements is considered a real change, not merely a measurement error, with 95% confidence. The graph illustrates how this cut-point increases with the within-subject CV (wCV). When the wCV is small (i.e., high precision), very small changes in the biomarker can be detected. Whereas when the wCV is large (i.e., low precision), large changes in the biomarker are needed before one can be confident that a real change has occurred.
It is likewise important to know a QIB’s reproducibility when measuring the change in a patient from baseline. Often, slightly different imaging methods are used at baseline and follow-up, e.g., a different scanner, a different radiologist, or even a different facility. If the reproducibility of a QIB is known, then the minimum detectable difference can be calculated. The minimum detectable difference when different imaging methods are used is called the reproducibility coefficient and it is calculated similarly to the RC as described above (43). However, the reproducibility coefficient is often significantly larger than the RC because of the additional variance associated with the different imaging methods and their systematic biases. It is critical to recognize these sources of additional error and their effect on interpreting patients’ quantitative images.
The sample sizes of test-retest studies in the literature vary considerably, from a couple of subjects to the study by Petersen, et al. where 28 sites in Asia, Europe, and North America participated, and a total of 284 healthy volunteers were scanned (26). Usually, studies have included results from fewer subjects than that mentioned above (22–25). Obuchowski and Bullen have performed a simulation study to determine the minimum sample size needed in test-retest studies (44). They have determined that estimates of precision should be based on a sample size of at least N = 35 to provide true 95% confidence intervals for a patient’s QIB measurement and for change in the QIB over time. Note that the estimate of precision could come from a single test-retest study with N ≥ 35, or calculated as a summary measure from a metaanalysis of multiple test-retest studies (45) where the combined sample size is N ≥ 35.
2.2. Phantom-based Methodology to Improve qMRI Precision
2.2.1. DWI
Diffusion MRI assesses the Brownian motion of water molecules non-invasively. On the timescale relevant to clinical DWI, water mobility is obstructed by tissue microstructure, including extracellular tortuosity, cell membranes, organelles, and macromolecular components (46). This sensitivity to microstructure enables DWI to elucidate impediments on a micrometer-size scale to the usual random Brownian motion. The ADC value is the key quantification parameter of DWI images typically of interest for clinical decision making (3,26,30) and is derived from mono-exponential modeling of the signal intensity as a function of b-value minimizing effects of perfusion and restricted diffusion (46). The diffusion coefficient has temperature dependence in pure water of 2.4% per degree Celsius (47,48). This dependency is rarely of concern in vivo because body temperature is well regulated, and other biophysical properties have much greater influence on tissue water mobility. For instance, dense tumors typically exhibit lower values of ADC than benign tissues because of higher cell packing.
The primary use of a phantom is to standardize DWI acquisition schemes across multiple vendors, software, and hardware platforms, and certify proper calibration and performance of the systems to ensure adequate ADC measurement, accuracy, and reproducibility. The indispensable value of the phantom is in providing ground-truth parameter values fundamentally independent of measurement method both for acquisition and image analysis. Therefore, phantom measurements can be used to improve the quality of DWI images by minimizing artifacts and geometrical distortions, and ensuring a high degree of reproducibility across different sites and scanner platforms. For reproducibility evaluation, it is important that the field of view (FOV), b-values, imaging matrix, repetition time (TR), echo time (TE), parallel imaging factor, number of slices, slice positions, and slice thickness are held constant and match clinical protocols.
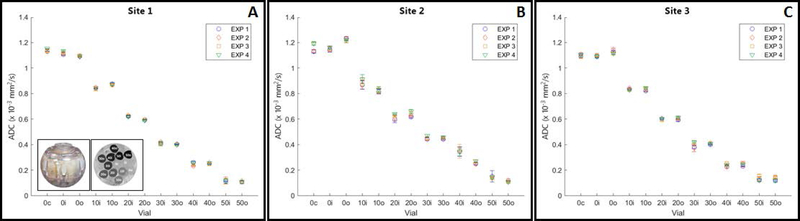
Many materials have been used in DWI phantoms such as aqueous solutions of polydimethylsiloxane, polyvinylpyrrolidone, sucrose, or polymers, liquid paraffin, alkanes, and pure water (28,49–51). The aqueous solutions and pure water are good choices for a phantom because of their non-toxicity and availability. Temperature dependence of ADC measures can be mitigated using an ice-water bath to ensure 0°C measurement across scanners (51). Moreover, the diffusion coefficient of water at 0oC is approximately 1.1 × 10–3 mm2/s, which is well within the tissue ADC range (39,52–56). However, the longitudinal relaxation time (T1) and transverse relaxation time (T2) for ice-water are much longer than most tissues. One recommended simple phantom design has been described by Chenevert, et al. (51) and consists of two cylinders of polypropylene, with the larger one containing ice and water. A smaller tube is filled with distilled water in thermal equilibrium at 0°C. Theoretically, the ADC value in this phantom should be independent of the acquisition protocol used at each site. This non-toxic and stable ice-water phantom can be readily manufactured on-site and has already been utilized for quantitative DWI quality assurance (QA) by several multi-site clinical trials (ACRIN 6698, 6701 and 6702, (57,58). Figure 2 shows representative intra-site ice-water phantom repeatability measurements acquired four times to calculate water ADC at 0° C in a multi-site setting using the same MRI imaging protocol. The CV for these 3 sites was 0.6%, 0.1%, and 1.1%. The biggest differences found between different MRI imaging protocols are in: FOV, number of slices, TR, and TE. Therefore, in practice, variability between DWI protocols influences the measured ADC value, and the differences found across measurements are about 10% from the literature value (51,59,60). The most significant differences were observed between MRI system manufactures due to distinct gradient designs leading to spatially-dependent bias in diffusion weighting b-values (28,58).
Figure 2:
Box-and-whisker plot demonstrating apparent diffusion coefficient (ADC) repeatability of water for multi-site results at 3T magnetic resonance imaging scanners using the ice water phantom. Note: Inset is the ADC map of the phantom. (Images compliments of authors from sites 1, 2, and 3: Memorial Sloan Kettering Cancer Center, Columbia University Medical Center, and University of Michigan.
The use of an application-specific phantom, such as that one developed by the National Institute of Standards and Technology (NIST) and (QIBA, to evaluate ADC measurement linearity for multi-b-value DWI studies using an array of ADC values is also recommended (61,62). The phantom is constructed using varying concentrations of polyvinylpyrrolidone (PVP, 0, 10, 20, 30, 40, and 50%) in an aqueous solution to generate physiologically relevant ADC values (63) and is available from High Precision Device, Inc (Boulder, CO). The space between the vials within the phantom can be filled with ice-water bath for temperature control. Figure 3 shows representative multi-site DWI data for 3T MRI scanners with repeatability measures. The recommended QIBA protocol for repeatability assessment with PVP phantom uses b-values of 0, 500, 900, and 2000 s/mm2 and is repeated four times, based on the guideline. QIBA provides scan protocols and software for standard analysis of quantitative DWI phantom data on the Quantitative Imaging Data Warehouse (QIBA QIDW, rsna.org/qidw). ).
Figure 3:
Repeatability results obtained using the National Institute of Standards andTechnology/Radiological Society of North America Quantitative Imaging Biomarkers Alliance diffusion-weighted magnetic resonance imaging phantom containing vials with varying concentrations of polyvinylpyrrolidone (0–50%) to generate physiologically relevant apparent diffusion coefficient (ADC) values at different vial positions (c = central; o = outer; I = inner). The phantom and ADC image are shown as inserts in the graph. Graph showing ADC (mean±sd) values for each vial in 4 experiments performed at (A) site 1, (B) site 2, and (C) at site 3. (Images compliments of author from sites 1, 2, and 3: Memorial Sloan Kettering Cancer Center; Columbia University Medical Center, and University of Michigan.
2.2.2. DCE
DCE is a noninvasive technique that measures microvascular permeability and volume fractions of the extravascular extracellular space (EES) and blood plasma space. One of the technique, involves serial acquisition of T1-weighted images before, during, and after intravenous injection of gadolinium-labeled CAs. This review will focus on the DCE T1 measurement techniques. These CAs are low-molecular weight paramagnetic complexes that extravasate to EES through vascular space. DCE analysis allows measurement of signal change on a ROI or voxel. DCE time-course data have been analyzed using heuristic approaches and quantitative kinetic analysis based on the tissue compartmental models. The heuristic, semi-quantitative measurement of the blood-normalized initial-area-under-the-gadolinium curve (IAUGCBN) has been described in the QIBA DCE profile (http://qibawiki.rsna.org/index.php/Profiles) (16,64). Areas showing rapid, high concentration uptake and fast wash-out of the CA are generally correlated with regions of malignancy within suspicious neoplastic lesions (65–67). The kinetics of the DCE time-course data depend on unique tumor vascular characteristics and thus the derived imaging metrics have found relevance in oncological applications (4,10,42,68).
With proper pharmacokinetic (PK) modeling of DCE time-course data, QIBs can be estimated. The most commonly used QIB for characterizing tumor vascular properties is the CA volume transfer constant (Ktrans), which has been detailed in the DCE profile (http://qibawiki.rsna.org/index.php/Profiles). The most common PK models used for DCE data analysis are the standard Tofts model (TM), which estimates Ktrans and volume fraction of EES (Ve) and the extended Tofts model (ETM) (14), which provides estimate of Ktrans and Ve. and an additional metric, Vp. To estimate these QIBs, such models require additional information such as input of tissue native T1 values and arterial input function (AIF).
A DCE experiment is basically a measurement of T1 changes in a tissue during the passage of CA. A static phantom has been proposed in initial work by ACRIN CQIE (69) and then by QIBA to standardize DCE acquisition protocols across hardware platforms from various vendors to verify calibration and performance of the systems. Phantom quality control may ensure adequate precision and reproducibility in T1 measurements, a prerequisite for performing quantitative DCE analysis. It also allows for the measurement of the contrast response of the employed DCE acquisition sequence across a range of T1 relaxation rates observed in in vivo DCE studies and the stability of that contrast response over time and across system upgrades (70,71).
DCE acquisition parameters can vary significantly across vendors, scanner models, station software packages, and field strengths. Often, each vendor and platform has preferred acquisition protocols. The T1 values in clinics are influenced by B1 field inhomogeneity, incomplete spoiling of transverse magnetization and MR sequence used for the range of T1 values to be measured (70). One of the QIBA recommendations is to standardize acquisition parameters to reduce sources of variability for DCE imaging, possibly at the expense of moderate protocol capabilities for some systems. Before acquiring data from subjects, it is essential that the selected pre-DCE T1 mapping protocol be performed on the static standardized phantom multiple times (or on different days) with the phantom repositioned for each experiment. This provides an estimate of true scanner variance and bias for T1 values.
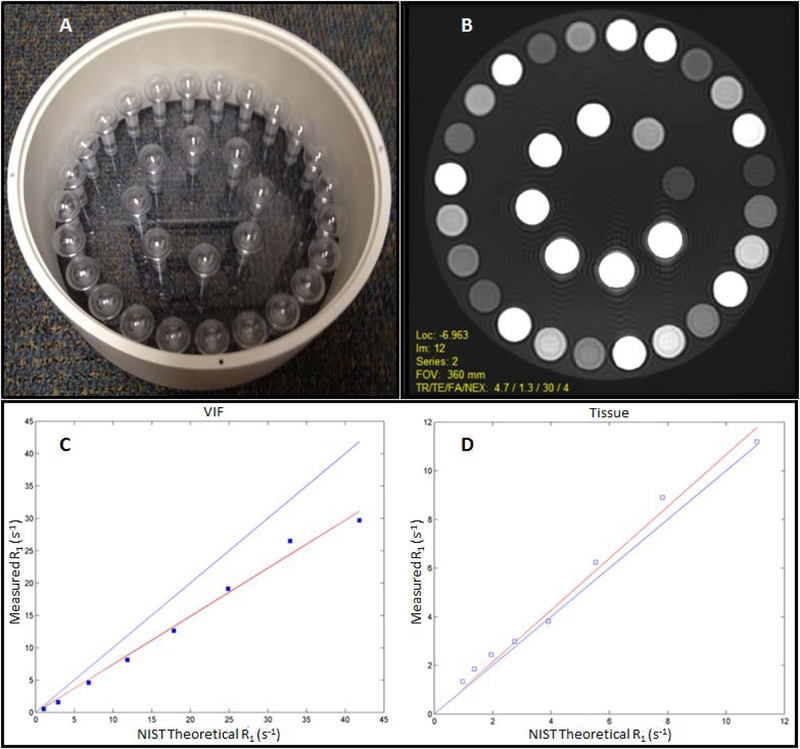
One recommended QIBA DCE T1 phantom contains vials of varying concentrations of nickel chloride solutions (70). Figure 4 shows the phantom design that consists of two sets of spherical inserts. The spheres were doped with nickel chloride to achieve T1 values spanning the range expected in vascular and tissue compartments during a DCE study. For the vascular input function spheres, the T1 values range between 0.75–41.6s−1, and for the tissue spheres, the range is 0.67–7.5s−1. To mimic the coil loading of a patient, the phantom was filled with a 30-mM sodium chloride (Sigma-Aldrich, USA) solution. The scan protocol for T1 measurement consists of acquiring coronal fast spoiled gradient echo sequences with variable flip angles (VFAs) of 30°, 25°, 20°, 15° 10°, 5°, and 2° to fully cover the range used for T1 mapping in clinical studies (70). Test-retest reliability and T1 accuracy evaluation using the QIBA DCE phantom should be considered for longitudinal studies. In addition to the DCE phantom, QIBA also provides an automated T1 quantification software application, DCETool, to analyze the data acquired from the QIBA DCE phantom (QIBA DCE-MRI WG at rsna.org/qidw ). This phantom and analysis software has been used for site qualification and requalification in support of ACRIN 6701, a DCE and DWI test-retest clinical trial in prostate cancer patients (unpublished data). Spatially-dependent B1 field inhomogeneity effects, which are more significant at higher field strengths, such as 3T, may confound VFA T1 data when acquired over large anatomic regions, necessitating B1 mapping and corrections to be included in the T1 measurement protocols. These effects will be addressed by version 2.0 of the RSNA QIBA DCE-MRI Profile that is currently under development.
Figure 4:
(A) The Quantitative Imaging Biomarkers Alliance dynamic contrast-enhanced phantom layout with 32 spheres, with different concentrations of NiCl2 solutions for varying T1 relaxation rates (R1). (B) T1-weighted magnetic resonance image of the phantom showing the 32 spheres, and (C) R1 values of the 8-vascular input function mimicking inserts compared with National Institute of Standards and Technology theoretical R1 values. (D) R1 values for the 24 tissue-mimicking inserts. (Images compliments of Edward Jackson, University of Wisconsin-Madison).
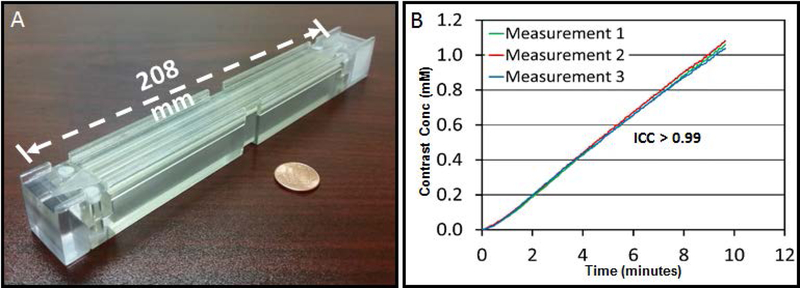
In addition to static T1 phantoms, some investigators have developed dynamic phantoms for MRI (Kim, et al. (71)), and CT (Driscoll, et al. (72)). The recent perfusion phantom was developed to correct MR scanner dependent variations in estimates of the tissue perfusion parameters in abdomen (71). The design of Kim, et al. is shown in Fig. 5A (71) is small enough to be imaged together with a patient for real-time quality assurance. Repeatability of the contrast enhancement curve of this phantom was measured using three phantoms placed at the isocenter of a 3T scanner, and the intra-class correlation coefficient was higher than 0.99 (Fig. 5B). Kim, et al. have demonstrated that this phantom significantly reduced the variation in quantifying perfusion parameters of various abdominal tissues across two different 3T scanners (71). However, the stability of this and other dynamic phantoms will need to be validated in longitudinal multi-site trials because such dynamic phantoms are difficult to produce in a manner that provides consistent results across phantoms and time.
Figure 5:
Portable perfusion phantom and its repeatability measurement. (A) Photograph of a portable perfusion phantom, and (B) contrast enhancement curves of three phantoms placed in a 3T magnetic resonance imaging scanner (temporal resolution = 2.9 s). Repeatability determined by the intraclass correlation coefficient is larger than 0.99. (Images compliments of Harrison Kim, University of Alabama at Birmingham).
2.3. Clinical data-driven Approaches to Improve qMRI Precision
2.3.1. DWI
The QIBA/Diffusion Biomarker Committee task force is dedicated to developing a DWI profile. The task force members reviewed over 1000 research articles to develop the profile claim statement based on clinical data from several organ systems, including the brain, liver, and prostate. Table 3 summarizes key scan protocol parameters for the brain (3A), prostate (3B), and liver (3D) from the QIBA profile (https://qibawiki.rsna.org/index.php/Profiles) to be relevant to a wider scientific audience. The tables have been adapted from the profile with permission from QIBA. Organs, such as breast (Table 3C) (8,73) and head and neck (Table 3E) (55,68,74), were not covered in the profile due to a lack of sufficient test-retest data. These review results point out the limited test-retest literature in various organs in the clinical oncology setting.
Table 3:
Typical Diffusion-Weighted Imaging (DWI) and Dynamic Contrast Enhanced (DCE) Acquisition Details for (A) Brain, (B) Prostate, (C) Breast, (D) Liver, and (E) Head and Neck
| (A) BRAIN | |||||
|---|---|---|---|---|---|
| Parameters | DWI | DCE | |||
| Field Strength | 1.5 T/3T | 1.5T/3T | |||
| Acquisition Sequence | SS-EPI | 3D SPGR | |||
| Receive Coil type | ≥8channel head array coil | ≥8channel head array coil | |||
| Lipid Suppression | On | On | |||
| Slice thickness (mm) | 4–5 | ≤5 | |||
| Gap thickness (mm) | 0–2 | 0–1 | |||
| FOV(mm) | 220 −240 | 220 −240 | |||
| Acquisition Matrix | 160–256 × 160–256 or 1.5–1 mm in plane resolution | 256×128–160 | |||
| Plane Orientation | Axial | Axial | |||
| Phase/frequency encode direction | AP/RL | AP/RL | |||
| Receiver bandwidth (Hz/pixel) | Max possible in freq encoding direction (acceptable > 1000) | 250 | |||
DWI Sequence Class
|
Contrast* | Pre- | Post- | ||
| # b-values | ≥ 2 (including b=0 s/mm2) | # Phases | ≥5 | 40–80 | |
| Minimum highest b-value (s/mm2) | 850–1000 | # Averages | ≥1 | 1 | |
| # Averages | ≥2 | Flip Angles (FAs) (deg) | 2–30* | 25–30 | |
| Diffusion Directions | 3 orthogonal | # Flip Angles (FAs) | 2–7 | 1 | |
| TR (ms) | 3000–5000 | TR (ms) | 3–8¥ | 3–8 | |
| TE (ms) | Minimum | TE (ms) | ≤3¥ | ≤3 | |
| In plane parallel imaging | 2 | Temporal Resolution (sec) /Total Acquisition Time (min) | <10 (ideal 5)/5–10 | ||
| Total Acquisition Time (min) | 3 |
#Contrast Dose and IV injection rate see references *Variable FAs for T10 measurement ¥Ensure TR/TE stays constant for all flip angles |
|||
| (B) PROSTATE | |||||
|---|---|---|---|---|---|
| Parameters | DWT | DCE | |||
| Field Strength | 3T | 1.5T/3T | |||
| Acquisition Sequence | SS-EPI | 3D SPGR | |||
| Receive Coil type | > 8 channel torso array coil; pelvic phase array/endorectal coils; body array coil) | > 8 channel torso array coil; pelvic phase array coil/endorectal coils; body array coil) | |||
| Lipid Suppression | On | NA | |||
| Slice thickness (mm) | 3–5 | <5 | |||
| Gap thickness (mm) | 0–1 | 0–1 | |||
| FOV(mm) | 240–260 | 260–300 | |||
| Acquisition Matrix | 224–128 × 224–128; 1–2 mm in-plane resolution | <256×l60 | |||
| Plane Orientation | Axial | Axial | |||
| Phase/frequency encode direction | AP/RL | RL/AP | |||
| Receiver bandwidth (Hz/pixel) | Max possible in freq encoding direction (acceptable > 1000) | 250 | |||
DWI Sequence Class
|
Contrast# | Pre- | Post- | ||
| # b-values | 2 (including b < 50–100 s/mm2) | # Phases | ≥5 | Sufficient to allow acquisition of at least 5 min post injection; ≥ 30 | |
| Minimum highest b-value (s/mm2) | 500–1500 | # Averages | ≥1 | 1 | |
| # Averages | 2-≥4 | Flip Angles (FAs) (deg) | 2–15* | 10–15 | |
| Diffusion Directions | ≥ 3 orthogonal |
#Flip Angles (FAs) |
3–5 | 1 | |
| TR (ms) | ≤4000 | TR (ms) | <5¥ | <5 | |
| TE (ms) | Minimum | TE (ms) | ≤2¥ | ≤2 | |
| In plane parallel imaging | 2 | Temporal Resolution (sec) /Total Acquisition Time (min) | <10/5–10 | ||
| Total Acquisition Time (min) | 3 |
#Contrast Dose and IV injection rate see references * Variable FAs for T10 measurement ¥Ensure TRITE stays constant for all flip angles |
|||
| (C) BREAST | |||||
|---|---|---|---|---|---|
| Parameters | DWI | DCE | |||
| Field Strength | 1.5T/3T | 1.5T/3T | |||
| Acquisition Sequence | SS-EPI/SE-EPI | 3D SPGR | |||
| Receive Coil type | ≥ 4 channel breast phase array coil | > 4 channel breast phase array coil | |||
| Lipid Suppression | On | On | |||
| Slice thickness (mm) | 4–5 | <2.5 | |||
| Gap thickness (mm) | 0–1 | 0 | |||
| FOV(mm) | 260–360 | To cover the entire breast whether uni- or bilateral | |||
| Acquisition Matrix | 128–192 × 128–192 | > 192×256; 1–1.5 mm in-plane resolution | |||
| Plane Orientation | Axial | Sagittal for single breast coverage; axial for bi lateral coverage | |||
| Phase/frequency encode direction | RL/AP | RL/AP for axial bilateral; HF/AP for sagittal unilateral | |||
| Receiver bandwidth (Hz/pixel) | Max possible in freq encoding direction | 250 | |||
DWI Sequence Class
|
Contrast# | Pre- | Post- | ||
| # b-values | ≥2 | # Phases | ≥2 | Sufficient to allow acquisition for ≥ 8 min post injection | |
| Minimum highest b-value (s/mm2) | 800 | # Averages | ≥1 | 1 | |
| # Averages | ≥2 | Flip Angle (FA) (deg) | 2–30* | 10–30 | |
| Diffusion Directions | 3 orthogonal | # Flip Angles (FAs) | 3–5 | 1 | |
| TR (ms) | ≥4000 | TR (ms) | <8¥ | <8 | |
| TE (ms) | Minimum | TE (ms) | ≤3¥ | ≤3 | |
| In plane parallel imaging | 2 | Temporal Resolution (sec) /Total Acquisition Time (min) | ≤ 20 /≥8 | ||
| Total Acquisition Time (min) | 4–6 |
* Contrast Dose and IV injection rate see references * Variable FAs for T10 measurement ¥ Ensure TR/TE stays constant for all flip angles |
|||
| (D) LIVER | |||||
|---|---|---|---|---|---|
| Parameters | DWI | DCE | |||
| Field Strength | 1.5T/3T | 1.5T/3T | |||
| Acquisition Sequence | SS-EPI | 3D SPGR | |||
| Receive Coil type | > 6–16 channel torso array coil | > 8–32 channel flexible or AP body array coil | |||
| Lipid Suppression | On | On | |||
| Slice thickness (mm) | 5–7 | 4–5 | |||
| Gap thickness (mm) | 0–1 | 0 | |||
| FOV(mm) | 300–450 | 280–380 | |||
| Acquisition Matrix | 160–196 × 160–192 or 2 −3 in-plane resolution | 320 × (160–192) | |||
| Plane Orientation | Axial | Oblique | |||
| Phase/frequency encode direction | AP/RL | AP/RL | |||
| Receiver bandwidth (Hz/pixel) | Max possible in freq encoding direction (acceptable > 1000) | 250 | |||
DWI Sequence Class
|
Contrast# | Pre- | Post- | ||
| # b-values | ≥2 (including one b = 50–100 s/mm2) | # Phases | ≥5 | 100 | |
| Minimum highest b-value (s/mm2) | 600–800 | # Averages | ≥2 | 1 | |
| # Averages | 2-≥4 | Flip Angles (FAs) (deg) | 2–30* | 20–30 | |
| Diffusion Directions | 3 orthogonal | # Flip Angles (FAs) | 3–5 | 1 | |
| TR (ms) | >2000 | TR (ms) | 3–7¥ | 3–7 | |
| TE (ms) | Minimum | TE (ms) | ≤5¥ | ≤5 | |
| In plane parallel imaging | 2–3 | Temporal Resolution (sec) /Total Acquisition Time (min) | < 5(ideal~3)/5 | ||
| Total Acquisition Time (min) | ~5 |
# Contrast Dose and IV injection rate see reference * Variable FAs for T10 measurement ¥Ensure TR/TE stays constant for all flip angles |
|||
| (E) HEAD AND NECK | |||||
|---|---|---|---|---|---|
| Parameters | DWI | DCE | |||
| Field Strength | 1.5T/3T | 1.5 T/3T | |||
| Acquisition Sequence | SS-EPI | 3D SPGR | |||
| Receive Coil type | Neck array or neurovascular coil | Neck array or neurovascular coil | |||
| Lipid Suppression | On | On | |||
| Slice thickness (mm) | ≥5 | >5 | |||
| Gap thickness (mm) | 0 | 0 | |||
| FOV(mm) | 220–380 | 180–220 | |||
| Acquisition Matrix | 128×128 | 256×128 | |||
| Plane Orientation | Axial | Axial | |||
| Phase/frequency encode direction | AP/RL | RL/AP | |||
| Receiver bandwidth (Hz/pixel) | Max possible in freq encoding direction | 250 | |||
DWI Sequence Class
|
Contrast# | Pre- | Post- | ||
| # b-values | ≥3 | # Phases | ≥5 | 40–80 | |
| Minimum highest b-value (s/mm2) | 1000 | # Averages | ≥ 1 | 1 | |
| # Averages | >2 | Flip Angles (FAs) (deg) | 5–30* | 15–30 | |
| Diffusion Directions | 3 orthogonal | # Flip Angles (FAs) | ≥3 | 1 | |
| TR (ms) | ≥2000 | TR (ms) | <9¥ | <9 | |
| TE (ms) | Minimum | TE (ms) | <2¥ | <2 | |
| In plane parallel imaging | 2 | Temporal Resolution (sec) /Total Acquisition Time (min) | ≤6/~5 | ||
| Total Acquisition Time (min) | 3 |
#Contrast Dose and IV injection rate see references *Variable FAs for T10 measurement ¥Ensure TR/TE stays constant for all flip angles |
|||
Clinical DWI is typically acquired using a diffusion-weighted, single-shot echo planar imaging (SS-EPI) sequence. The acquisition parameters are detailed in Table 3 for the five specific organs outlined in this review (3,8,10,74–85). Protocol optimization is a prerequisite for obtaining optimum signal-to-noise ratio for the DWI images because artifacts can be significant. Techniques that reduce the number of phase encoding steps and FOV, resulting in reduced artifacts, are preferred (86). There are newer developments in DWI building on single-shot EPI such as reduced FOV acquisition or multi-shot EPI (87,88). To reduce susceptibility artifacts and to improve spatial resolution compared to SS-EPI methods, the propeller/blade diffusion methods have also been used (89,90). Selection of optimum b-values for a specified organ is also an important parameter that should be optimized for signal-to-noise ratio (38,39). Another point in technique optimization is “landmark on the organ of interest” to confirm that organ position is close to the isocenter to minimize b-value non-uniformity across the organ (91).
Prior to the analysis, a radiologist draws a ROI on the DWI images guided by ancillary MR images, radiologic and clinical information. The ROI encompasses the entire tumor or tissue of interest. A DWI protocol includes producing an ADC map based upon a mono-exponential fit to images obtained using two or more b-values. Generally, at least three orthogonal diffusion directions are probed, with the resultant maps generated from combinations of the directional data, assuming isotropic diffusion (3,53).
| [III] |
where S and S0 are the signal intensities with and without diffusion weighting, respectively, and b is the diffusion weighting factor (b-value, s/mm2).
Most MRI scanners have capabilities for automatically producing ADC maps from the DWI images using propriety software based on mono-exponential modeling of the data. Figures 6B, 7B, 8C, 9D, and 10B show representative ADC maps derived from patients with tumors in the brain, prostate, breast, liver, and head and neck, respectively.
Figure 6:
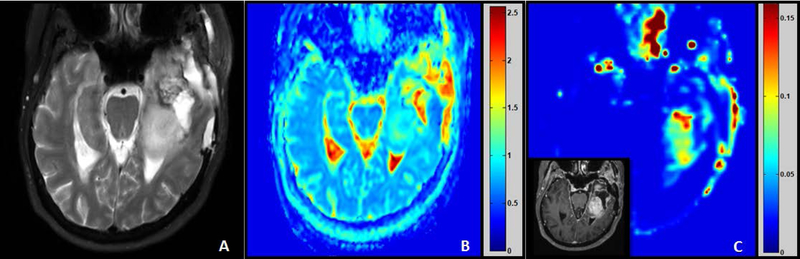
Representative pre-treatment magnetic resonance images of a patient with grade 4 brain tumor (65 years, female). (A) T2-weighted image. (B) Apparent diffusion coefficient × 10−3 (mm2/s) map generated using 3 b-values (b = 0, 100, 1000 s/mm2). (C) Ktrans (min−1) map generated from dynamic contrast enhanced data with insert of T1-weighted gadolinium contrast image. (Images compliments of Thomas Chenevert, University of Michigan).
Figure 7:
Representative pretreatment magnetic resonance images of a patient with prostate cancer—Gleason Score 4+3 (66 years, male). (A) T2-weighted image, (B) apparent diffusion coefficient × 10−3 (mm2/s) map generated using 2 b-values (i.e., b = 0, 600 s/mm2), and (C) Ktrans (min−1) map generated from dynamic contrast-enhanced data. (Images compliments of Susan M. Noworolski, University of California San Francisco).
Figure 8:
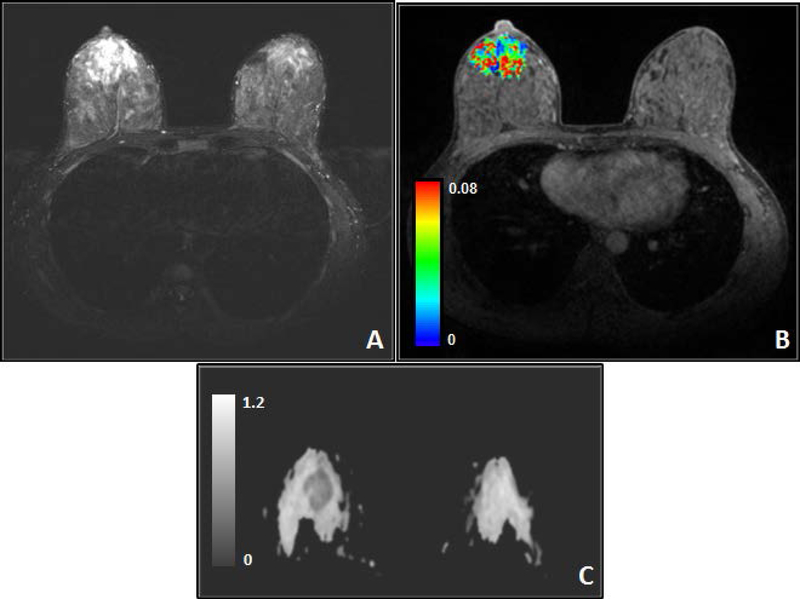
Representative magnetic resonance images from a breast cancer patient (34 years, female) with grade II invasive ductal carcinoma (IDC) in the right breast. (A) T2-weighted image with fat saturation, (B) color Ktrans (min−1) map of the tumor overlaid on T1-weighted dynamic contrast-enhanced image with fat saturation, and (C) Representative apparent diffusion coefficient × 10−3 (mm2/s) (ADC) map from a breast cancer patient (37 years, female) with grade II IDC in the right breast. Composite ADC map was generated from diffusion-weighted imaging images with b = 0 and 800 s/mm2 showing decreased ADC in tumor (arrow). (Images compliments of Wei Huang, Oregon Health & Science University).
Figure 9:
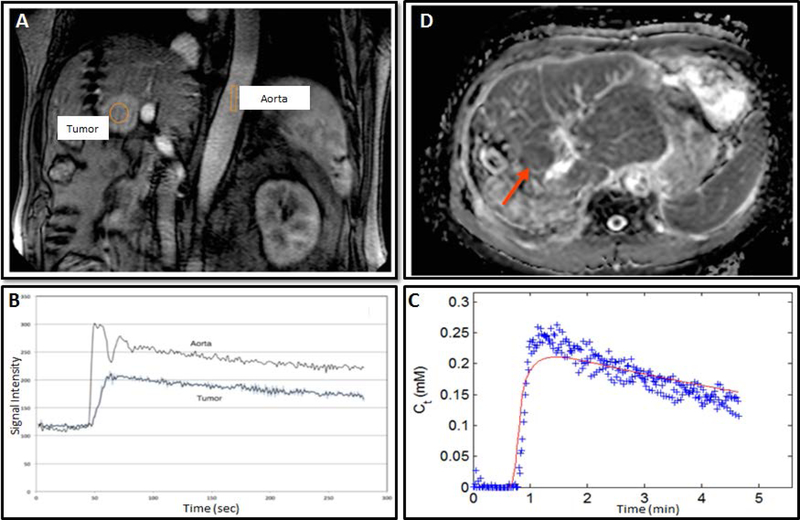
Representative magnetic resonance images from a recurrent hepatocellular carcinoma patient (57 years, male) acquired on a 3T scanner. Dynamic contrast-enhanced magnetic resonance imaging scan showing (A) enhancing tumor and (B) contrast enhancement time course. (C) The gadolinium concentration time course and extended Tofts model fit and (D) Composite apparent diffusion coefficient map generated from diffusion-weighted imaging images, with b = 0, 600 s/mm2 from same patient. (Images compliments of Sachin Jambawalikar, Columbia University Medical Center)
Figure 10:
Representative pretreatment magnetic resonance images of head and neck cancer patient (52 years, male). (A) T2-weighted image, (B) apparent diffusion coefficient ×10−3 (mm2/s) map overlaid on diffusion-weighted (b = 0 s/mm2) images generated using 10 b-values (0, 20, 50, 80, 200, 300, 500, 800, 1500, and 2000 s/mm2), (C) Ktrans (min−1) map overlaid on pre-contrast T1-weighted image. (Images compliments of Amita Shukla-Dave, Memorial Sloan Kettering Cancer Center).
As discussed above, the QIBA/DWI Biomarker Task Force members performed an extensive literature search and found limited manuscripts with test-retest data and therefore reported the %RC for ADC in tumor ROIs derived from mono-exponential modeling of DWI data only in three different organs as follows: brain = 11% (30–32) liver = 26% (34–36,52) and prostate = 47% (37,40). The details on literature and assumptions used to inform these 95% confidence interval (CI) values are adapted from QIBA/DWI profile with permission. Specifically, meta-analysis was performed on the available test-retest study reports (e.g., 2–3 per organ) acquired with qualitatively similar acquisition protocols (detailed in the QIBA/DWI profile) to pool maximum sets of subjects (>30) sufficient to satisfy statistical significance. The estimated wCV for mean ADC in an ROI between 1–4 cm2 was 3.97% for brain (30–32), 9.38% for liver (33–35), and 16.97% for prostate (37–40). The derived CIs could likely be improved by more advanced organ-specific acquisition protocols to achieve better QIB precision.
2.3.2. DCE
Similar to Diffusion, the QIBA/Perfusion Biomarker Committee Task Force has invested significant effort in updating the original DCE profile (https://qibawiki.rsna.org/index.php/Profiles). The new version of the DCE profile, version 2.0 (under development), includes the brain (Table 3A) (75,81,92), prostate (Table 3B) (38,39,76,78), and breast (Table 3C) (8,79,93). The tables have been adapted from the working document of the profile with permission from QIBA. Specific scan parameters for head and neck (10,74,94) and liver (80,95) were not included in the profile due to limited test-retest clinical data but are reflected in Table 3D and Table 3E, respectively. It is important to emphasize that even though the DCE literature consists of many studies of tumors in various organs, important repeatability and reproducibility data are lacking. The major challenge for such DCE test-retest studies is the need to repeat the CA injection. The retest for DCE should be performed after the first CA injection has been eliminated from the patient, which typically requires about 24 hours. The half-life of common low molecular-weight gadolinium CAs is approximately 90 min, retention in some tissues and lesions can be significantly longer. This creates issues with the logistics of repeating the experiments. Additional CA injections also require institutional review board approval, which is especially pertinent given the increasing awareness of nephrogenic systemic fibrosis in patients with abnormal renal function and potential brain deposition of gadolinium in patients with normal renal function (96).
DCE is typically acquired using a T1-weighted, fast spoiled gradient recalled echo sequence, and the temporal resolution is determined by the pulse sequence acquisition parameters and the spatial resolution and anatomic coverage required for the organ under study. The rate at which the CA extravasates from the vasculature depends on the molecular size of the CA. With low molecular weight agents, the temporal resolution required to observe microvessel permeability is typically on the order of 5–20s (14,18,97).
Similar to DWI, the ROIs for data analysis are often determined by experienced radiologists. DCE data are sometimes analyzed using qualitative or semi-quantitative methods. DCE profile details the heuristic approach using the IAUGCb n parameter (http://qibawiki.rsna.org/index.php/Profiles). Accurate AIF and native tissue T1 (i.e., T10) measurements are the first necessary steps for PK modeling of DCE data with TM and ETM (14). There are several ways to determine AIF (98–101), each with its pros and cons. The AIF should be measured from the feeding vessel of the tumor. In practicality, the AIF is often measured in a nearby major blood vessel within the vicinity of the tumor. For instance, the carotid arteries are commonly used for head and neck cancer and the aorta for liver cancer. However, due to the image volume coverage, angulation, or the absence of major vessels in the scan volume, direct measurement of AIF from the acquired images may not always be possible.
The time course of the CA concentration in the tissue, Ct(t), for the TM is based on the Kety exchange equations (14):
| [IV] |
where Cp(t) is the time course of the plasma concentration of the CA, Ktrans (min−1) is the volume transfer constant (vascular space to the EES), and kep = Ktrans/Ve (min−1) is the rate constant for CA transport from the EES to vascular space.
The ETM incorporates the vascular compartment in modeling the tissue CA concentration (14). For the ETM, Ct(t), can be expressed as:
| [V] |
Figures 6C, 7C, 8B, and 10C show representative Ktrans maps derived from patients with tumors in the brain, prostate, breast, and head and neck, respectively. Figures 9A–C show CA uptake and CA time course of concentration curves for the aorta and tumors in the liver. The currently available test-retest DCE data has illustrated that the %RC for Ktrans in a tumor ROI is 21.3% for brain and 55.7% for prostate (41,42). The statistical approach used to derive this performance claim information for DCE profile is similar to the one applied in the QIBA/DWI profile.
The DCE data acquisition in clinics for organs such as breast, prostate, liver is slightly challenging when compared to brain and head and neck. In this review, we highlight a few key acquisition aspects for these organs. The typical acquisition parameter range for breast DCE is detailed in Table 3C. There is a unique difference between DCE of the breast and that of other organs, largely due to the clinical need for bilateral scanning, which require full breast coverage with high spatial resolution because breast cancer has a high incidence of contralateral and multi-focal disease (102), and clear assessment of lesion morphology is essential for cancer diagnosis (103,104). For example, the American College of Radiology breast MRI lexicon recommends that image slice thickness no more than 2 mm or thinner. There is a trade-off between spatial and temporal resolutions, when using conventional gradient-echo pulse sequences, low temporal resolution breast DCE protocols on commercial scanner systems are commonly used in clinical settings and large-scale clinical trials such as the ISPY-1 (105,106) and ISPY-2 (107) trials. Due to poor accuracy in quantitative PK analysis of DCE data (67,108,109), semi-quantitative analyses (i.e., uptake slope, percent signal change, time to peak, signal enhance ratio, etc.) of DCE time course data are generally employed under this circumstance (110). The results of a simulation study by Henderson, et al. have shown that a temporal resolution of 16 s or less is preferred for PK analysis of breast DCE data (111). Parallel imaging acceleration together with k-space under-sampling acquisition and view sharing commercially available reconstruction methods, such as TWIST (time-resolved angiography with stochastic trajectories) (112–114), DISCO (Differential Subsampling With Cartesian Ordering) (115,116), and 4D THRIVE (T1 High Resolution Isotropic Volume Examination) sequences (117) have allowed for simultaneous acquisitions of 3D breast DCE data. For breast DCE, the AIF can be determined using direct measurement from an axillary artery (118), the reference-tissue method (119,120), or the population-averaged AIF (113,114,121).
For prostate DCE, data acquisition details are given in Table 3B (122), which suggests at least a 10-stemporal resolution and 30 time points, resulting in a five-minute total scan time. Spatial resolution is approximately 1 mm in-plane with 3-mm thick sections. These values are based on the PI-RADS_v2 recommendations for clinical acquisition (123). Studies from the literature generally meet these criteria with more variation in spatial resolution (0.7 −1.9 mm in-plane resolution and 3–4 mm slices) and, for some studies, higher temporal resolution (3–10 s) (122,124). Best spatial or temporal resolutions and/or increased coverage can be obtained via the use of key-hole imaging, parallel imaging, or compressed sensing (122,125). Usually, the AIF is directly measured from iliac arteries.
For liver DCE, data acquisition details are provided in Table 3D (126–132). One of the major challenges in acquisition of liver DCE data is the respiratory motion of the abdomen. Keeping this in mind, DCE images are mostly acquired with a series of multiple breath-holds and/or shallow breathing. In order to achieve the optimum tradeoff between temporal and spatial resolution, the procedure commonly used in the clinical setting is to coach the patients to hold their breath for approximately 15 s (expiratory phase), followed by a 5s break and then repeat the acquisition multiple times for a total of 25 min (129,132). This allows to acquire high temporal resolution DCE data <5s (ideal ~3s) at the first breath hold for the accurate quantification of the AIF. Generally, 10 −12 coronal slices are acquired to bisect both the lesion and aorta ensuring that the lesion is in the center of FOV in the superior-inferior direction. In addition, on a 3T scanner, a B1-mapping sequence for correction of the T10 mapping and DCE data is recommended (126,130,131). Even though liver has dual blood supply inputs (portal vein and aorta), and tumors are highly vascular, quantitative PK analysis software applications generally use a single input TM. However, recent work for liver DCE analysis has evaluated hepatic perfusion quantification using a dual-input kinetic model (127,128).
3. Pearls and Pitfalls as qMRI Precision is Improved
The role of qMRI in clinical oncology settings has been elegantly reviewed in the past (1,10,133–138). This review is focused on the need for more test-retest studies, defining repeatability and reproducibility, and determining the extent of repeatability and reproducibility determinations that have been performed in phantoms and patients and reported in the literature with adequate technical data and details to allow a statistically robust meta-analysis. Although there are about 1000 publications for human subject DWI and DCE studies in oncology in the literature, the test-retest pool of papers is still quite limited. In the development of a QIB that can be used in clinical trials or practice, a critical step is understanding the test-retest precision for a specific acquisition and analysis protocol; hence this dearth of test-retest data is limiting both imaging biomarker discovery and clinical application of more advanced quantitative imaging methods.
4. Proposal fo r qMRI Precision Studies
QIBA (http://www.rsna.org/qiba/) seeks to improve the value and practicality of QIBs by reducing variability across devices, patients, sites, and time (43). The common platform for communicating strategies to improve technical performance for QIB applications is through QIBA Profiles. Profiles are developed using published data to generate evidence-based performance claims that inform users about what quantitative results can be achieved by following the profile guidelines (139). For example, in longitudinal claims, the Profile provides a cut-point for when a true change has occurred as well as a range of values for the true change in the biomarker (139). For each of these claims, data on the imaging procedure’s precision is critical to obtain, particularly its ability to provide repeatable measurements when there has been no biological change in the subject (21). The estimate of precision is then used to construct the cut-point to distinguish true change in the biomarker from measurement error.
The precision estimate is also essential in planning clinical trials. Whether the QIB is being used as an integrated or integral biomarker, the estimate of its precision is needed to predict the required trial size. Underestimating the wSD or wCV will lead to a trial that lacks adequate statistical power, and overestimating wSD or wCV will lead to a trial with a larger N (and higher cost) than needed. Thus, a reliable estimate of the precision is critical to clinical decision-making as well as understanding the potential role of the QIB in diagnosis, prediction, and treatment monitoring. These general guidelines are relevant both for the studies using conventional acquisition protocols and for more advanced quantitative imaging techniques (e.g. MR fingerprinting (140,141)) seeking translation in to clinical practice. Such methods hold promise for providing multiple quantitative MRI parameters from a fast (single) acquisition, although their specific implementations for quantitative DWI and DCE are currently sparse. Prior to test-retest precision studies, these advanced multi-parametric model-based methods would also need to demonstrate level of accuracy with respect to conventional acquisition and image analysis techniques (that quantify individual diffusion and perfusion parameters) using physical and digital phantoms that provide ground-truth parameter values.
There is a paucity of studies assessing the repeatability of imaging procedures for measuring QIBs. The published test-retest studies are often small (e.g., < 10), poorly designed (e.g., changing protocols, varying times between imaging), and their results are presented using metrics that are neither generalizable to other sites (e.g., intraclass correlation coefficients) nor lend themselves to meta-analyses.
In conclusion, QIBA recommends reproducibility and repeatability of DWI and DCE studies in phantoms and patients for identification of QIBs to be used in multicenter oncology trials.
Acknowledgments
We acknowledge Dr. Ramesh Paudyal and Dr. David Aramburu-Nunez for their help in styling the tables and figures. We are thankful to Mr. James Keller (BS) for editing the manuscript. We acknowledge funding from NIH U01 CA211205 (ASD, LHS); U01 CA166104, P01 CA085878 and R01 CA190299 (TLC, DM); U01 CA154602, Oregon Health & Science University Circle of Giving Award (WH); SC CTSI (NIH/NCRR/NCATS) KL2TR000131 and NIH 1 L30 CA209248–01 (MSS); NIH P30CA013148 (HK); R01 CA148708 (SMN). We also acknowledge NIBIB contracts HHSN268201000050C, HHSN268201300071C, and HHSN268201500021C, which provided partial funding support for a variety of QIBA efforts.
List of Aberrations:
- AIF
arterial input function
- AP
anterior-posterior
- CA
contrast agent
- FOV
field of view
- DCE
dynamic contrast enhanced
- DWI
diffusion-weighted imaging
- ETM
extended Tofts model
- PVP
polyvinylpyrrolidone
- qMRI
quantitative magnetic resonance imaging
- QIBA
quantitative imaging biomarkers alliance
- RC
repeatability coefficient
- RL
right-left
- RSNA
radiological Society of north America
- SS-EPI
single-shot echo planar imaging
- SPGR
spoiled gradient recalled echo
- QIB
quantitative imaging biomarkers
- PK
Pharmacokinetic model
- wCV
within-subject coefficient of variation
- wSD
within-subject standard deviation
- TM
Tofts model
References
- 1.Padhani AR, Liu G, Koh DM, et al. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia 2009;11(2):102–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tofts P Quantitative MRI of the Brain Measuring Changes Caused by Disease Introduction. Quantitative Mri of the Brain: Measuring Changes Caused by Disease 2003:Xv–Xvi. [Google Scholar]
- 3.Chenevert TL, Ross BD. Diffusion imaging for therapy response assessment of brain tumor. Neuroimaging Clin N Am 2009;19(4):559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Padhani AR, Khan AA. Diffusion-weighted (DW) and dynamic contrast-enhanced (DCE) magnetic resonance imaging (MRI) for monitoring anticancer therapy. Target Oncol 2010;5(1):39–52. [DOI] [PubMed] [Google Scholar]
- 5.Barnes A, Alonzi R, Blackledge M, et al. UK quantitative WB-DWI technical workgroup: consensus meeting recommendations on optimisation, quality control, processing and analysis of quantitative whole-body diffusion-weighted imaging for cancer. Br J Radiol 2018;91(1081):20170577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beuzit L, Eliat PA, Brun V, et al. Dynamic contrast-enhanced MRI: Study of inter-software accuracy and reproducibility using simulated and clinical data. J Magn Reson Imaging 2016;43(6):1288–1300. [DOI] [PubMed] [Google Scholar]
- 7.Costa DN, Pedrosa I, Roehrborn C, Rofsky NM. Multiparametric magnetic resonance imaging of the prostate: technical aspects and role in clinical management. Top Magn Reson Imaging 2014;23(4):243–257. [DOI] [PubMed] [Google Scholar]
- 8.Partridge SC, Nissan N, Rahbar H, Kitsch AE, Sigmund EE. Diffusion-weighted breast MRI: Clinical applications and emerging techniques. J Magn Reson Imaging 2017;45(2):337–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winfield JM, Payne GS, Weller A, deSouza NM. DCE-MRI, DW-MRI, and MRS in Cancer: Challenges and Advantages of Implementing Qualitative and Quantitative Multi-parametric Imaging in the Clinic. Top Magn Reson Imaging 2016;25(5):245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jansen JF, Parra C, Lu Y, Shukla-Dave A. Evaluation of Head and Neck Tumors with Functional MR Imaging. Magn Reson Imaging Clin N Am 2016;24(1): 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li SP, Padhani AR. Tumor response assessments with diffusion and perfusion MRI. J Magn Reson Imaging 2012;35(4):745–763. [DOI] [PubMed] [Google Scholar]
- 12.Shukla-Dave A, Hricak H. Role of MRI in prostate cancer detection. NMR Biomed 2014;27(1):16–24. [DOI] [PubMed] [Google Scholar]
- 13.Galban CJ, Hoff BA, Chenevert TL, Ross BD. Diffusion MRI in early cancer therapeutic response assessment. NMR Biomed 2017;30(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tofts PS, Brix G, Buckley DL, et al. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: Standardized quantities and symbols. Journal of Magnetic Resonance Imaging 1999;10(3):223–232. [DOI] [PubMed] [Google Scholar]
- 15.Thoeny HC, Ross BD. Predicting and monitoring cancer treatment response with diffusion-weighted MRI. J Magn Reson Imaging 2010;32(1):2–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leach MO, Brindle KM, Evelhoch JL, et al. The assessment of antiangiogenic and antivascular therapies in early-stage clinical trials using magnetic resonance imaging: issues and recommendations. Br J Cancer 2005;92(9):1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koh DM, Padhani AR. Functional magnetic resonance imaging of the liver: parametric assessments beyond morphology. Magn Reson Imaging Clin N Am 2010;18(3):565–585, xii. [DOI] [PubMed] [Google Scholar]
- 18.O’Connor JP, Aboagye EO, Adams JE, et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol 2017;14(3): 169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim MM, Parolia A, Dunphy MP, Venneti S. Non-invasive metabolic imaging of brain tumours in the era of precision medicine. Nat Rev Clin Oncol 2016;13(12):725–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong KH, Panek R, Bhide SA, Nutting CM, Harrington KJ, Newbold KL. The emerging potential of magnetic resonance imaging in personalizing radiotherapy for head and neck cancer: an oncologist’s perspective. Br J Radiol 2017;90(1071):20160768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessler LG, Barnhart HX, Buckler AJ, et al. The emerging science of quantitative imaging biomarkers terminology and definitions for scientific studies and regulatory submissions. Stat Methods Med Res 2015;24(1):9–26. [DOI] [PubMed] [Google Scholar]
- 22.Ng CS, Raunig DL, Jackson EF, et al. Reproducibility of perfusion parameters in dynamic contrast-enhanced MRI of lung and liver tumors: effect on estimates of patient sample size in clinical trials and on individual patient responses. AJR Am J Roentgenol 2010;194(2):W134–140. [DOI] [PubMed] [Google Scholar]
- 23.Weller A, Papoutsaki MV, Waterton JC, et al. Diffusion-weighted (DW) MRI in lung cancers: ADC test-retest repeatability. Eur Radiol 2017;27(11):4552–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maclaren J, Han Z, Vos SB, Fischbein N, Bammer R. Reliability of brain volume measurements: a test-retest dataset. Scientific data 2014;1:140037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokoo T, Serai SD, Pirasteh A, et al. Linearity, Bias, and Precision of Hepatic Proton Density Fat Fraction Measurements by Using MR Imaging: A Meta-Analysis. Radiology 2018;286(2):486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen ET, Mouridsen K, Golay X, all named co-authors of the Qt-rs. The QUASAR reproducibility study, Part II: Results from a multi-center Arterial Spin Labeling test-retest study. NeuroImage 2010;49(1): 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hectors SJ, Wagner M, Corcuera-Solano I, et al. Comparison Between 3-Scan Trace and Diagonal Body Diffusion-Weighted Imaging Acquisitions: A Phantom and Volunteer Study. Tomography 2016;2(4):411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malyarenko D, Galban CJ, Londy FJ, et al. Multi-system repeatability and reproducibility of apparent diffusion coefficient measurement using an ice-water phantom. Journal of Magnetic Resonance Imaging 2013;37(5):1238–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raunig DL, McShane LM, Pennello G, et al. Quantitative imaging biomarkers: a review of statistical methods for technical performance assessment. Stat Methods Med Res 2015;24(1):27–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonekamp D, Nagae LM, Degaonkar M, et al. Diffusion tensor imaging in children and adolescents: reproducibility, hemispheric, and age-related differences. NeuroImage 2007;34(2):733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paldino MJ, Barboriak D, Desjardins A, Friedman HS, Vredenburgh JJ. Repeatability of quantitative parameters derived from diffusion tensor imaging in patients with glioblastoma multiforme. J Magn Reson Imaging 2009;29(5):1199–1205. [DOI] [PubMed] [Google Scholar]
- 32.Pfefferbaum A, Adalsteinsson E, Sullivan EV. Replicability of diffusion tensor imaging measurements of fractional anisotropy and trace in brain. J Magn Reson Imaging 2003;18(4):427–433. [DOI] [PubMed] [Google Scholar]
- 33.Braithwaite AC, Dale BM, Boll DT, Merkle EM. Short- and midterm reproducibility of apparent diffusion coefficient measurements at 3.0-T diffusion-weighted imaging of the abdomen. Radiology 2009;250(2):459–465. [DOI] [PubMed] [Google Scholar]
- 34.Deckers F, De Foer B, Van Mieghem F, et al. Apparent diffusion coefficient measurements as very early predictive markers of response to chemotherapy in hepatic metastasis: a preliminary investigation of reproducibility and diagnostic value. J Magn Reson Imaging 2014;40(2):448–456. [DOI] [PubMed] [Google Scholar]
- 35.Heijmen L, Ter Voert EE, Nagtegaal ID, et al. Diffusion-weighted MR imaging in liver metastases of colorectal cancer: reproducibility and biological validation. Eur Radiol 2013;23(3):748–756. [DOI] [PubMed] [Google Scholar]
- 36.Miquel ME, Scott AD, Macdougall ND, Boubertakh R, Bharwani N, Rockall AG. In vitro and in vivo repeatability of abdominal diffusion-weighted MRI. Br J Radiol 2012;85(1019): 1507–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibbs P, Pickles MD, Turnbull LW. Repeatability of echo-planar-based diffusion measurements of the human prostate at 3 T. Magn Reson Imaging 2007;25(10):1423–1429. [DOI] [PubMed] [Google Scholar]
- 38.Jambor I, Merisaari H, Aronen HJ, et al. Optimization of b-value distribution for biexponential diffusion-weighted MR imaging of normal prostate. J Magn Reson Imaging 2014;39(5):1213–1222. [DOI] [PubMed] [Google Scholar]
- 39.Jambor I, Merisaari H, Taimen P, et al. Evaluation of different mathematical models for diffusion-weighted imaging of normal prostate and prostate cancer using high b-values: a repeatability study. Magn Reson Med 2015;73(5):1988–1998. [DOI] [PubMed] [Google Scholar]
- 40.Litjens GJ, Hambrock T, Hulsbergen-van de Kaa C, Barentsz JO, Huisman HJ. Interpatient variation in normal peripheral zone apparent diffusion coefficient: effect on the prediction of prostate cancer aggressiveness. Radiology 2012;265(1):260–266. [DOI] [PubMed] [Google Scholar]
- 41.Alonzi R, Taylor NJ, Stirling JJ, et al. Reproducibility and correlation between quantitative and semiquantitative dynamic and intrinsic susceptibility-weighted MRI parameters in the benign and malignant human prostate. J Magn Reson Imaging 2010;32(1): 155–164. [DOI] [PubMed] [Google Scholar]
- 42.Jackson A, Jayson GC, Li KL, et al. Reproducibility of quantitative dynamic contrast-enhanced MRI in newly presenting glioma. Br J Radiol 2003;76(903):153–162. [DOI] [PubMed] [Google Scholar]
- 43.Sullivan DC, Obuchowski NA, Kessler LG, et al. Metrology Standards for Quantitative Imaging Biomarkers. Radiology 2015;277(3):813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Obuchowski NA, Bullen J. Quantitative imaging biomarkers: Effect of sample size and bias on confidence interval coverage. Stat Methods Med Res 2017:962280217693662. [DOI] [PubMed] [Google Scholar]
- 45.Huang EP, Wang XF, Choudhury KR, et al. Meta-analysis of the technical performance of an imaging procedure: guidelines and statistical methodology. Stat Methods Med Res 2015;24(1): 141–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Bihan D Apparent diffusion coefficient and beyond: what diffusion MR imaging can tell us about tissue structure. Radiology 2013;268(2):318–322. [DOI] [PubMed] [Google Scholar]
- 47.Le Bihan D, Delannoy J, Levin RL. Temperature mapping with MR imaging of molecular diffusion: application to hyperthermia. Radiology 1989; 171(3):853–857. [DOI] [PubMed] [Google Scholar]
- 48.Holz M, Heil SR, Sacco A. Temperature-dependent self-diffusion coefficients of water and six selected molecular liquids for calibration in accurate H-1 NMR PFG measurements. Phys Chem Chem Phys 2000;2(20):4740–4742. [Google Scholar]
- 49.Jerome NP, Papoutsaki MV, Orton MR, et al. Development of a temperature-controlled phantom for magnetic resonance quality assurance of diffusion, dynamic, and relaxometry measurements. Med Phys 2016;43(6):2998–3007. [DOI] [PubMed] [Google Scholar]
- 50.Newitt DC, Malyarenko D, Chenevert TL, et al. Multisite concordance of apparent diffusion coefficient measurements across the NCI Quantitative Imaging Network. J Med Imaging (Bellingham) 2018;5(1):011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chenevert TL, Galban CJ, Ivancevic MK, et al. Diffusion Coefficient Measurement Using a Temperature-Controlled Fluid for Quality Control in Multicenter Studies. Journal of Magnetic Resonance Imaging 2011;34(4):983–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bharwani N, Koh DM. Diffusion-weighted imaging of the liver: an update. Cancer Imaging 2013;13:171–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chenevert TL, Stegman LD, Taylor JM, et al. Diffusion magnetic resonance imaging: an early surrogate marker of therapeutic efficacy in brain tumors. J Natl Cancer Inst 2000;92(24):2029–2036. [DOI] [PubMed] [Google Scholar]
- 54.Jakubovic R, Zhou S, Heyn C, et al. The predictive capacity of apparent diffusion coefficient (ADC) in response assessment of brain metastases following radiation. Clin Exp Metastasis 2016;33(3):277–284. [DOI] [PubMed] [Google Scholar]
- 55.Jansen JF, Schoder H, Lee NY, et al. Tumor metabolism and perfusion in head and neck squamous cell carcinoma: pretreatment multimodality imaging with 1H magnetic resonance spectroscopy, dynamic contrast-enhanced MRI, and [18F]FDG-PET. Int J Radiat Oncol Biol Phys 2012;82(1):299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Partridge SC, Singer L, Sun R, et al. Diffusion-weighted MRI: influence of intravoxel fat signal and breast density on breast tumor conspicuity and apparent diffusion coefficient measurements. Magn Reson Imaging 2011;29(9): 1215–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mulkern RV, Ricci KI, Vajapeyam S, et al. Pediatric brain tumor consortium multisite assessment of apparent diffusion coefficient z-axis variation assessed with an ice-water phantom. Acad Radiol 2015;22(3):363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Newitt DC, Tan ET, Wilmes LJ, et al. Gradient nonlinearity correction to improve apparent diffusion coefficient accuracy and standardization in the american college of radiology imaging network 6698 breast cancer trial. J Magn Reson Imaging 2015;42(4):908–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jafar MM, Parsai A, Miquel ME. Diffusion-weighted magnetic resonance imaging in cancer: Reported apparent diffusion coefficients, in-vitro and in-vivo reproducibility. World J Radiol 2016;8(1):21–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taouli B, Beer AJ, Chenevert T, et al. Diffusion-weighted imaging outside the brain: Consensus statement from an ISMRM-sponsored workshop. J Magn Reson Imaging 2016;44(3):521–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boss MA. ‘Multicenter Study of Reproducibility of Wide Range of ADC at 0 °C” RSNA. Chicago, IL: RSNA; 2015. [Google Scholar]
- 62.Palacios EM, Martin AJ, Boss MA, et al. Toward Precision and Reproducibility of Diffusion Tensor Imaging: A Multicenter Diffusion Phantom and Traveling Volunteer Study. AJNR Am J Neuroradiol 2017;38(3):537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pierpaoli C, Joelle S, U. N, Basser PJ, Horkary F. Polyvinylpyrrolidone (PVP) Water Solutions as Isotropic Phantoms for Diffusion MRI Studies. Volume 17: Proc. Intl. Soc. Mag. Reson. Med; 2009. [Google Scholar]
- 64.NCI Recommendations for MR measurement methods at 1.5 Tesla and endpoints for use in Phase 1/2a trials of anti-cancer therapuetics affecting tumor vascular function. Dynamic contrast MRI (DCE-MRI) guidelines resulted from the NCI CIP MRI workshop on Translational Resaecrh Center in Cancer. MR Workshop on Translational Research; 2004. [Google Scholar]
- 65.Taylor JS, Tofts PS, Port R, et al. MR imaging of tumor microcirculation: promise for the new millennium. J Magn Reson Imaging 1999;10(6):903–907. [DOI] [PubMed] [Google Scholar]
- 66.Leach MO, Brindle KM, Evelhoch JL, et al. The assessment of antiangiogenic and antivascular therapies in early-stage clinical trials using magnetic resonance imaging: issues and recommendations. Brit J Cancer 2005;92(9):1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leach MO, Morgan B, Tofts PS, et al. Imaging vascular function for early stage clinical trials using dynamic contrast-enhanced magnetic resonance imaging. Eur Radiol 2012;22(7):1451–1464. [DOI] [PubMed] [Google Scholar]
- 68.Jensen LR, Garzon B, Heldahl MG, Bathen TF, Lundgren S, Gribbestad IS. Diffusion-Weighted and Dynamic Contrast-Enhanced MRI in Evaluation of Early Treatment Effects During Neoadjuvant Chemotherapy in Breast Cancer Patients. Journal of Magnetic Resonance Imaging 2011;34(5): 1099–1109. [DOI] [PubMed] [Google Scholar]
- 69.Rosen M, Kinahan PE, Gimpel JF, et al. Performance Observations of Scanner Qualification of NCI-Designated Cancer Centers: Results From the Centers of Quantitative Imaging Excellence (CQIE) Program. Acad Radiol 2017;24(2):232–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bane O, Hectors SJ, Wagner M, et al. Accuracy, repeatability, and interplatform reproducibility of T1 quantification methods used for DCE-MRI: Results from a multicenter phantom study. Magn Reson Med 2018;79(5):2564–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim H, Mousa M, Schexnailder P, et al. Portable perfusion phantom for quantitative DCE-MRI of the abdomen. Med Phys 2017;44(10):5198–5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Driscoll B, Keller H, Coolens C. Development of a dynamic flow imaging phantom for dynamic contrast-enhanced CT. Med Phys 2011;38(8):4866–4880. [DOI] [PubMed] [Google Scholar]
- 73.Sorace AG, Wu C, Barnes SL, et al. Repeatability, reproducibility, and accuracy of quantitative mri of the breast in the community radiology setting. J Magn Reson Imaging 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim S, Loevner L, Quon H, et al. Diffusion-weighted magnetic resonance imaging for predicting and detecting early response to chemoradiation therapy of squamous cell carcinomas of the head and neck. Clin Cancer Res 2009;15(3):986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bagher-Ebadian H, Jain R, Nejad-Davarani SP, et al. Model selection for DCE-T1 studies in glioblastoma. Magn Reson Med 2012;68(1):241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hambrock T, Somford DM, Huisman HJ, et al. Relationship between apparent diffusion coefficients at 3.0-T MR imaging and Gleason grade in peripheral zone prostate cancer. Radiology 2011;259(2):453–461. [DOI] [PubMed] [Google Scholar]
- 77.Kim JH, Kim JK, Park BW, Kim N, Cho KS. Apparent diffusion coefficient: prostate cancer versus noncancerous tissue according to anatomical region. J Magn Reson Imaging 2008;28(5): 1173–1179. [DOI] [PubMed] [Google Scholar]
- 78.Oto A, Yang C, Kayhan A, et al. Diffusion-weighted and dynamic contrast-enhanced MRI of prostate cancer: correlation of quantitative MR parameters with Gleason score and tumor angiogenesis. AJR Am J Roentgenol 2011;197(6): 1382–1390. [DOI] [PubMed] [Google Scholar]
- 79.Partridge SC, Stone KM, Strigel RM, DeMartini WB, Peacock S, Lehman CD. Breast DCE-MRI: influence of postcontrast timing on automated lesion kinetics assessments and discrimination of benign and malignant lesions. Acad Radiol 2014;21(9):1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taouli B, Koh DM. Diffusion-weighted MR imaging of the liver. Radiology 2010;254(1):47–66. [DOI] [PubMed] [Google Scholar]
- 81.Lin X, Lee M, Buck O, et al. Diagnostic Accuracy of T1-Weighted Dynamic Contrast-Enhanced-MRI and DWI-ADC for Differentiation of Glioblastoma and Primary CNS Lymphoma. AJNR Am J Neuroradiol 2017;38(3):485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vandecaveye V, Dirix P, De Keyzer F, et al. Diffusion-Weighted Magnetic Resonance Imaging Early after Chemoradiotherapy to Monitor Treatment Response in Head-and-Neck Squamous Cell Carcinoma. International Journal of Radiation Oncology Biology Physics 2012;82(3):1098–1107. [DOI] [PubMed] [Google Scholar]
- 83.Vandecaveye V, Dirix P, De Keyzer F, et al. Diffusion-weighted magnetic resonance imaging early after chemoradiotherapy to monitor treatment response in head-and-neck squamous cell carcinoma. Int J Radiat Oncol Biol Phys 2012;82(3):1098–1107. [DOI] [PubMed] [Google Scholar]
- 84.Donati OF, Afaq A, Vargas HA, et al. Prostate MRI: evaluating tumor volume and apparent diffusion coefficient as surrogate biomarkers for predicting tumor Gleason score. Clin Cancer Res 2014;20(14):3705–3711. [DOI] [PubMed] [Google Scholar]
- 85.Donati OF, Mazaheri Y, Afaq A, et al. Prostate cancer aggressiveness: assessment with whole-lesion histogram analysis of the apparent diffusion coefficient. Radiology 2014;271(1): 143–152. [DOI] [PubMed] [Google Scholar]
- 86.Korn N, Kurhanewicz J, Banerjee S, Starobinets O, Saritas E, Noworolski S. Reduced-FOV excitation decreases susceptibility artifact in diffusion-weighted MRI with endorectal coil for prostate cancer detection. Magn Reson Imaging 2015;33(1):56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Banerjee S, Nishimura DG, Shankaranarayanan A, Saritas EU. Reduced field-of-view DWI with robust fat suppression and unrestricted slice coverage using tilted 2D RF excitation. Magn Reson Med 2016;76(6):1668–1676. [DOI] [PubMed] [Google Scholar]
- 88.Chen NK, Guidon A, Chang HC, Song AW. A robust multi-shot scan strategy for high-resolution diffusion weighted MRI enabled by multiplexed sensitivity-encoding (MUSE). NeuroImage 2013;72:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hirokawa Y, Isoda H, Maetani YS, Arizono S, Shimada K, Togashi K. MRI artifact reduction and quality improvement in the upper abdomen with PROPELLER and prospective acquisition correction (PACE) technique. AJR Am J Roentgenol 2008;191(4): 1154–1158. [DOI] [PubMed] [Google Scholar]
- 90.Pipe JG, Farthing VG, Forbes KP. Multishot diffusion-weighted FSE using PROPELLER MRI. Magn Reson Med 2002;47(1):42–52. [DOI] [PubMed] [Google Scholar]
- 91.Malkyarenko DI, Chenevert TL. Practical estimate of gradient nonlinearity for implementation of apparent diffusion coefficient bias correction. J Magn Reson Imaging 2014;40(6):1487–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mills SJ, Soh C, Rose CJ, et al. Candidate biomarkers of extravascular extracellular space: a direct comparison of apparent diffusion coefficient and dynamic contrast-enhanced MR imaging--derived measurement of the volume of the extravascular extracellular space in glioblastoma multiforme. AJNR Am J Neuroradiol 2010;31(3):549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yankeelov TE, Lepage M, Chakravarthy A, et al. Integration of quantitative DCE-MRI and ADC mapping to monitor treatment response in human breast cancer: initial results. Magn Reson Imaging 2007;25(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vandecaveye V, De Keyzer F, Nuyts S, et al. Detection of head and neck squamous cell carcinoma with diffusion weighted MRI after (chemo)radiotherapy: Correlation between radiologic and histopathologic findings. International Journal of Radiation Oncology Biology Physics 2007;67(4):960–971. [DOI] [PubMed] [Google Scholar]
- 95.Chandarana H, Taouli B. Diffusion and perfusion imaging of the liver. Eur J Radiol 2010;76(3):348–358. [DOI] [PubMed] [Google Scholar]
- 96.Kanda T, Oba H, Toyoda K, Kitajima K, Furui S. Brain gadolinium deposition after administration of gadolinium-based contrast agents. Jpn J Radiol 2016;34(1):3–9. [DOI] [PubMed] [Google Scholar]
- 97.Tofts PS. Modeling tracer kinetics in dynamic Gd-DTPA MR imaging. J Magn Reson Imaging 1997;7(1):91–101. [DOI] [PubMed] [Google Scholar]
- 98.Chen J, Yao J, Thomasson D. Automatic determination of arterial input function for dynamic contrast enhanced MRI in tumor assessment. Med Image Comput Comput Assist Interv 2008;11(Pt 1):594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li X, Welch EB, Arlinghaus LR, et al. A novel AIF tracking method and comparison of DCE-MRI parameters using individual and population-based AIFs in human breast cancer. Phys Med Biol 2011;56(17):5753–5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Parker GJ, Roberts C, Macdonald A, et al. Experimentally-derived functional form for a population-averaged high-temporal-resolution arterial input function for dynamic contrast-enhanced MRI. Magn Reson Med 2006;56(5):993–1000. [DOI] [PubMed] [Google Scholar]
- 101.Wang S, Fan X, Medved M, et al. Arterial input functions (AIFs) measured directly from arteries with low and standard doses of contrast agent, and AIFs derived from reference tissues. Magn Reson Imaging 2016;34(2):197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lehman CD, Gatsonis C, Kuhl CK, et al. MRI evaluation of the contralateral breast in women with recently diagnosed breast cancer. New Engl J Med 2007;356(13):1295–1303. [DOI] [PubMed] [Google Scholar]
- 103.Schnall MD, Blume J, Bluemke DA, et al. Diagnostic architectural and dynamic features at breast MR imaging: Multicenter study. Radiology 2006;238(1):42–53. [DOI] [PubMed] [Google Scholar]
- 104.D’Orsi CJ SE, Mendelson EB, Morris EA, et al. ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System. Reston, VA: American College of Radiology; 2013. [Google Scholar]
- 105.Hylton NM, Blume JD, Bernreuter WK, et al. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy--results from ACRIN 6657/I-SPY TRIAL. Radiology 2012;263(3):663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hylton NM, Gatsonis CA, Rosen MA, et al. Neoadjuvant Chemotherapy for Breast Cancer: Functional Tumor Volume by MR Imaging Predicts Recurrence-free Survival-Results from the ACRIN 6657/CALGB 150007 I-SPY 1 TRIAL. Radiology 2016;279(1):44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Newitt DC, Aliu SO, Witcomb N, et al. Real-Time Measurement of Functional Tumor Volume by MRI to Assess Treatment Response in Breast Cancer Neoadjuvant Clinical Trials: Validation of the Aegis SER Software Platform. Transl Oncol 2014;7(1):94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heisen M, Fan XB, Buurman J, van Riel NAW, Karczmar GS, Romeny BMT. The Influence of Temporal Resolution in Determining Pharmacokinetic Parameters From DCE-MRI Data. Magnet Reson Med 2010;63(3):811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Di Giovanni P, Azlan CA, Ahearn TS, Semple SI, Gilbert FJ, Redpath TW. The accuracy of pharmacokinetic parameter measurement in DCE-MRI of the breast at 3 T. Phys Med Biol 2010;55(1): 121132. [DOI] [PubMed] [Google Scholar]
- 110.Roberts C, Issa B, Stone A, Jackson A, Waterton JC, Parker GJ. Comparative study into the robustness of compartmental modeling and model-free analysis in DCE-MRI studies. J Magn Reson Imaging 2006;23(4):554–563. [DOI] [PubMed] [Google Scholar]
- 111.Henderson E, Rutt BK, Lee TY. Temporal sampling requirements for the tracer kinetics modeling of breast disease. Magn Reson Imaging 1998;16(9): 1057–1073. [DOI] [PubMed] [Google Scholar]
- 112.Tudorica LA, Oh KY, Roy N, et al. A feasible high spatiotemporal resolution breast DCE-MRI protocol for clinical settings. Magn Reson Imaging 2012;30(9):1257–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huang W, Li X, Chen Y, et al. Variations of dynamic contrast-enhanced magnetic resonance imaging in evaluation of breast cancer therapy response: a multicenter data analysis challenge. Transl Oncol 2014;7(1): 153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tudorica A, Oh KY, Chui SY, et al. Early Prediction and Evaluation of Breast Cancer Response to Neoadjuvant Chemotherapy Using Quantitative DCE-MRI. Transl Oncol 2016;9(1):8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saranathan M, Rettmann DW, Hargreaves BA, Clarke SE, Vasanawala SS. DIfferential Subsampling with Cartesian Ordering (DISCO): a high spatio-temporal resolution Dixon imaging sequence for multiphasic contrast enhanced abdominal imaging. J Magn Reson Imaging 2012;35(6):1484–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Morrison CK, Henze Bancroft LC, DeMartini WB, et al. Novel High Spatiotemporal Resolution Versus Standard-of-Care Dynamic Contrast-Enhanced Breast MRI: Comparison of Image Quality. Invest Radiol 2017;52(4):198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Beck GM, De Becker J, Jones AC, von Falkenhausen M, Willinek WA, Gieseke J. Contrast-enhanced timing robust acquisition order with a preparation of the longitudinal signal component (CENTRA plus) for 3D contrast-enhanced abdominal imaging. J Magn Reson Imaging 2008;27(6):1461–1467. [DOI] [PubMed] [Google Scholar]
- 118.Huang W, Tudorica LA, Li X, et al. Discrimination of Benign and Malignant Breast Lesions by Using Shutter-Speed Dynamic Contrast-enhanced MR Imaging. Radiology 2011;261(2):394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li X, Arlinghaus LR, Ayers GD, et al. DCE-MRI analysis methods for predicting the response of breast cancer to neoadjuvant chemotherapy: pilot study findings. Magn Reson Med 2014;71(4):1592–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li X, Kang H, Arlinghaus LR, et al. Analyzing Spatial Heterogeneity in DCE- and DW-MRI Parametric Maps to Optimize Prediction of Pathologic Response to Neoadjuvant Chemotherapy in Breast Cancer. Transl Oncol 2014;7(1):14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schabel MC, Morrell GR, Oh KY, Walczak CA, Barlow RB, Neumayer LA. Pharmacokinetic Mapping for Lesion Classification in Dynamic Breast MRI. Journal of Magnetic Resonance Imaging 2010;31(6): 13711378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Verma S, Turkbey B, Muradyan N, et al. Overview of dynamic contrast-enhanced MRI in prostate cancer diagnosis and management. AJR Am J Roentgenol 2012;198(6):1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Weinreb JC, Barentsz JO, Choyke PL, et al. PI-RADS Prostate Imaging - Reporting and Data System: 2015, Version 2. Eur Urol 2016;69(1):16–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Starobinets O, Korn N, Iqbal S, et al. Practical aspects of prostate MRI: hardware and software considerations, protocols, and patient preparation. Abdom Radiol (NY) 2016;41(5):817–830. [DOI] [PubMed] [Google Scholar]
- 125.Rosenkrantz AB, Geppert C, Grimm R, et al. Dynamic Contrast-Enhanced MRI of the Prostate With High Spatiotemporal Resolution Using Compressed Sensing, Parallel Imaging, and Continuous Golden-Angle Radial Sampling: Preliminary Experience. Journal of Magnetic Resonance Imaging 2015;41(5): 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chung S, Kim D, Breton E, Axel L. Rapid B1+ mapping using a preconditioning RF pulse with TurboFLASH readout. Magn Reson Med 2010;64(2):439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jajamovich GH, Calcagno C, Dyvorne HA, Rusinek H, Taouli B. DCE-MRI of the liver: reconstruction ofthe arterial input function using a low dose pre-bolus contrast injection. PLoS One 2014;9(12):e115667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Koh TS, Thng CH, Hartono S, et al. Dynamic contrast-enhanced MRI of neuroendocrine hepatic metastases: A feasibility study using a dual-input two-compartment model. Magn Reson Med 2011;65(1):250–260. [DOI] [PubMed] [Google Scholar]
- 129.Le Y, Dale B, Akisik F, Koons K, Lin C. Improved T1, contrast concentration, and pharmacokinetic parameter quantification in the presence of fat with two-point Dixon for dynamic contrast-enhanced magnetic resonance imaging. Magn Reson Med 2016;75(4):1677–1684. [DOI] [PubMed] [Google Scholar]
- 130.Sacolick LI, Wiesinger F, Hancu I, Vogel MW. B1 mapping by Bloch-Siegert shift. Magn Reson Med 2010;63(5): 1315–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Voigt T, Nehrke K, Doessel O, Katscher U. T1 corrected B1 mapping using multi-TR gradient echo sequences. Magn Reson Med 2010;64(3):725–733. [DOI] [PubMed] [Google Scholar]
- 132.Wang H, Cao Y. Correction of arterial input function in dynamic contrast-enhanced MRI of the liver. J Magn Reson Imaging 2012;36(2):411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Griffith B, Jain R. Perfusion Imaging in Neuro-Oncology: Basic Techniques and Clinical Applications. Magn Reson Imaging Clin N Am 2016;24(4):765–779. [DOI] [PubMed] [Google Scholar]
- 134.Mazaheri Y, Akin O, Hricak H. Dynamic contrast-enhanced magnetic resonance imaging of prostate cancer: A review of current methods and applications. World J Radiol 2017;9(12):416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pinker K, Helbich TH, Morris EA. The potential of multiparametric MRI of the breast. Br J Radiol 2017;90(1069):20160715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wibmer AG, Sala E, Hricak H, Vargas HA. The expanding landscape of diffusion-weighted MRI in prostate cancer. Abdom Radiol (NY) 2016;41(5):854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yang X, Knopp MV. Quantifying tumor vascular heterogeneity with dynamic contrast-enhanced magnetic resonance imaging: a review. J Biomed Biotechnol 2011;2011:732848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang X, Liang J, Heverhagen JT, et al. Improving the pharmacokinetic parameter measurement in dynamic contrast-enhanced MRI by use of the arterial input function: theory and clinical application. Magn Reson Med 2008;59(6):1448–1456. [DOI] [PubMed] [Google Scholar]
- 139.Obuchowski NA, Buckler A, Kinahan P, et al. Statistical Issues in Testing Conformance with the Quantitative Imaging Biomarker Alliance (QIBA) Profile Claims. Acad Radiol 2016;23(4):496–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature 2013;495(7440):187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen Y, Jiang Y, Pahwa S, et al. MR Fingerprinting for Rapid Quantitative Abdominal Imaging. Radiology 2016;279(1):278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]