SUMMARY
Background:
Germline mutations in telomere-related genes such as POT1 and the TERT promoter predispose to familial melanoma.
Objective:
To evaluate the prevalence of germline mutations in the POT1 gene and in the TERT promoter in a large cohort of Spanish melanoma-prone families (at least two affected individuals in first- or second-degree relatives).
Methods:
Overall, 228 CDKN2A wild-type melanoma-prone families were included in the study. Screening of POT1 was performed in one affected case of each family and TERT promoter was sequenced in one affected case from 202 families (26 families were excluded due to DNA exhaustion/degradation). Additionally, TERT promoter sequencing was extended to addition 30 CDKN2A mutated families and 70 sporadic multiple primary melanoma patients (MPM) with a family history of other cancers.
Results:
We identified four families with potentially pathogenic POT1 germline mutations: a missense variant c.233T>C (p.Ile78Thr), a nonsense variant c.1030G>T (p.Glu344*), and two variants c.255G>A (r.125_255del) and c.1792G>A (r.1791_1792insAGTA, p.Asp598Serfs*22), which we confirmed disrupted POT1 mRNA splicing. A TERT promoter variant of unknown significance (c.−125C>A) was detected in a MPM patient, but no germline mutations were detected in the TERT promoter in familial melanoma cases.
Conclusions:
Overall, 1.75% of our CDKN2A/CDK4-wild type Spanish melanoma-prone families carry probably damaging mutations in POT1. The frequency of TERT promoter germline mutations in melanoma families in our population is extremely rare.
INTRODUCTION
Around 10% of melanoma cases report a family history of melanoma. In these families, genetic variants conferring susceptibility are inherited following an autosomal dominant pattern with incomplete penetrance. To date, CDKN2A is the main high-penetrance gene involved in melanoma susceptibility and around 20% to 40% of melanoma-prone families harbor CDKN2A mutations world-wide.1, 2 Mutation screening of CDKN2A and CDK4 has been conducted in 330 Spanish melanoma-prone families from our group. Overall, CDKN2A mutations were present in 14% of families, whilst no CDK4 positive families have been identified.3
Patients with multiple primary melanomas (MPM) but without a family history of melanoma may also have an increased susceptibility to develop melanoma and CDKN2A mutations have been detected in 8–10% of sporadic MPM patients.4,5 Recent studies in melanoma-prone families using next-generation sequencing (NGS) approaches have identified other high penetrance melanoma susceptibility genes that play a role in telomere maintenance, such as TERT, POT1, TERF2IP, and ACD.6–9 Telomeres are responsible for maintaining genomic stability and chromosomal integrity.10 Germline mutations in these genes result in the deregulation of these functions conferring a high risk of developing cancer. Identifying individuals with germline mutations allows their inclusion into prevention and early diagnosis programs, which correlates with a better clinical outcome. In particular, two independent studies identified a germline mutation in the TERT promoter (c.−57T>G) in two unrelated families of Northern-European ancestry.6,11 This variant creates a new ETS transcription factor binding site in the TERT promoter and increases TERT expression.6 More recently, rare POT1 germline variants have been identified in CDKN2A wild-type melanoma-prone families from Northern- and Southern-European countries, USA, and Australia.7,8 To date, these telomere-related genes have not been extensively studied in patients of Iberian descent.
Our aim was to evaluate the prevalence of germline mutations in POT1 and the TERT promoter in a collection of Spanish patients from melanoma-prone families or a history of multiple primary melanomas.
PATIENTS AND METHODS
Families and Samples
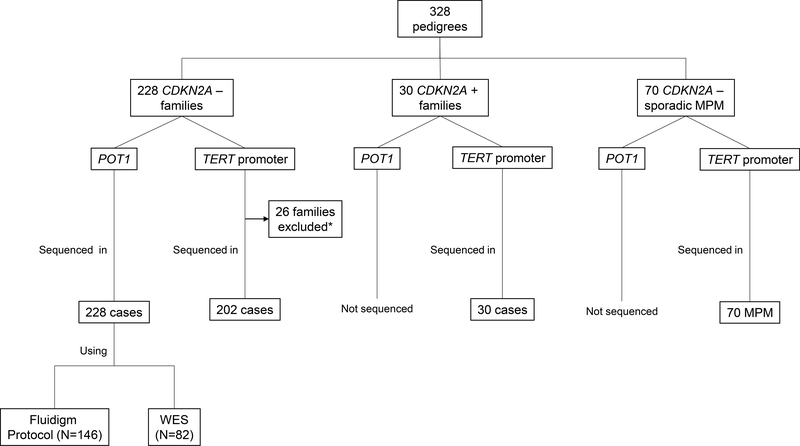
POT1 and TERT promoter molecular screening was conducted in one melanoma patient with DNA available from each of 228 CDKN2A and CDK4 wild-type families. Spain is considered a low-to-medium melanoma incidence area. The rule of two has been suggested as a genetic testing inclusion criterion.12 In fact, this inclusion criterion allows us to detect CDKN2A mutations in >10% of families with only two melanoma cases.3,5 For this reason, we included families with at least two melanoma cases in first- or second-degree relatives that were recruited at the Melanoma Unit of Hospital Clinic of Barcelona from 1994 to 2015. In addition, TERT molecular screening was performed in one melanoma patient from 30 CDKN2A mutation positive families and in 70 CDKN2A wild-type sporadic MPM patients with family history of other cancers diagnosed in first- or second-degree relatives (Fig. 1). MITF p.Glu318Lys genetic information was also available.13 All patients signed written informed consent after reading and understanding the study protocol and agreeing to participate in the study. The study was approved by the ethics committee of the Hospital Clinic of Barcelona and the National Cancer Institute, NIH.
Figure 1. Samples and families included in the study.
The figure shows a diagram of the samples and families assessed. CDKN2A +: pedigrees with CDKN2A germline mutations; CDKN2A -: pedigrees with CDKN2A wild-type.
*Families excluded for lack of remaining DNA from cases due to DNA exhaustion or degradation.
POT1 molecular screening
Whole exome sequencing was performed on 82 samples at the National Cancer Institute. Data analysis and extraction of POT1 variants for these families was performed using the same methodology as described in Shi et al.7 The remaining 146 samples were processed at the Sanger Institute. PCR primers were designed against all annotated exons of POT1 (ENST00000357628.7, NM_015450.2). Samples were PCR-amplified and individually barcoded following the Fluidigm unidirectional sequencing protocol.14 Primers for PCR reactions were pooled up to 1151-plex per well in 9 pools. The libraries generated were sequenced, one per lane and producing 150-bp paired-end reads, on an Illumina MiSeq.14
Reads were aligned to the reference genome (GRCh37) using BWA mem. The 146 samples had at least 90% of the target bases in POT1 of high quality (base quality >= 20, read mapping quality >=50). Variants were called with the GATK HaplotypeCaller, and quality filters were set as standard (minimum number of alternate bases 2, minimum read depth 2, minimum mapping quality for SNPs 10, window size for filtering adjacent gaps 3, and exclusion of SNPs within 10bp around a gap) minus the end-distance and stand bias filters. Variant consequences were predicted with the Ensembl Variant Effect predictor release 70.
We selected all frameshift, nonsense, missense and variants predicted to affect splicing and with a frequency <0.01 in European non-Finnish ExAC samples. Specific PCR primers were designed to amplify the exons containing the selected variants (Table S1). PCR followed by Sanger sequencing was performed to validate the presence of the variants detected by NGS. PCR conditions were: denaturation at 95°C 5 min, 10 cycles (95°C 1 min, 65°C–60°C 1 min, 72°C 1 min), followed by 25 cycles (95°C 1 min, 55°C 1 min, 72°C 1 min) and extension at 72°C (10 min). Sanger Sequencing was performed using universal M13 primers by GENEWIZ (Takeley, UK). Sequences were analyzed using the SeqPilot 4.0.1 software (JSI Medical Systems, Germany). In those families with variant confirmation, the sequencing analysis was additionally extended to other relatives if samples were available for co-segregation assessment.
POT1 mRNA studies
To assess whether the two splice-site variants detected affected mRNA processing, two independent blood RNA samples from each carrier and blood RNA samples from seven healthy donors were obtained using the PAXgene Blood RNA Kit (PreAnalytiX®, Qiagen, Germany). The mRNA was reverse-transcribed to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, CA, US). Multiple PCRs were designed to amplify the mRNA region containing the coding region of exons 5 to 8, 16/17 to 19 and 17/18 to 19 of POT1 (ENST00000357628.7, NM_015450.2). These regions were amplified from the cDNA from two independent blood samples from each patient and from healthy donors. The PCR products were assessed by electrophoresis. The different fragments amplified were isolated using PureLink Quick Gel Extraction kit (Invitrogen, CA, US) and sequenced using the corresponding primers described in Table S1 by GENEWIZ (Takeley, UK).
We designed three different droplet digital PCR (ddPCR) custom assays for specific transcript quantification. This method is very sensitive and enables direct transcript comparison of alternative splicing forms,15 as well as quantification of specific exon skipping.16 To assess the amount of transcripts containing or skipping exon 7 we designed a HEX-labeled probe annealing to exon 7 and a FAM-labeled probe annealing to the intersection between exons 6 and 8, using the Bio-Rad online system for custom probe design. Both probes were combined in the same reaction. To assess the amount of transcripts containing specific mutations: r.1792G>A or r.1791_1792insAGTA, we designed two independent custom mutation assays, labeling the mutated alleles with FAM and the wild-type allele with HEX.
Amplifications were carried out in a reaction volume of 20 μL on a QX100™ Droplet Digital™ PCR System (Bio-Rad, CA, US). To quantify transcripts containing or skipping exon 7, the 20 μL PCR reaction contained 10 μL of 2x ddPCR Supermix for Probes (No dUTP) (Bio-Rad, CA, US), 1 μL of FAM-labeled probe, 1 μL of HEX-labeled probe and 9 μL of cDNA. We used two independent samples from the patient with the mutation creating exon 7 skipping, to determine the percentage of each transcript. We also assessed the percentage of each transcript in three controls, for comparison. To quantify transcripts with specific mutations, the PCR reaction was performed similarly but included 1 μL of FAM/HEX-labeled probe mix and 9 μL of cDNA, instead. We used two independent samples from the patient with the point mutation affecting splicing at position c.1792, to determine the percentage of each transcript. We assessed the presence of the mutations in three controls, as negative controls for the ddPCR reaction.
Droplets were generated by the QX100™ droplet generator and PCR was performed using a C1000™ Touch thermal cycler. The thermal cycling conditions were as follows: 1 cycle of 95 °C for 10 min, 40 cycles of 94 °C for 30 s and 55 °C for 1 min, followed by 1 cycle of 98 °C for 10 min (all at a ramp rate of 2°C/sec), and optional cooling at 4 °C (ramp rate 1°C/sec). Droplets were analyzed with Quantasoft™ Software version 1.7.4 (Bio-Rad, CA, US) according to manufacturer’s instructions. Positive HEX and FAM droplet counts per well were determined for all samples and used for transcript percentage and 95% confidence interval calculation for each sample and well.
TERT promoter molecular screening
For TERT (ENST00000310581.9, NM_198253.2) promoter amplification, also covering rs2853669 (c.−245T>C, European minor allele frequency [MAF] = 0.29) polymorphism position, specific PCR primers were designed based on the primers previously reported by Horn and colleagues (Table S1).6 PCR followed by Sanger sequencing was performed as described above for POT1 molecular screening. PCR conditions were: denaturation at 95°C for 5 min, 10 cycles (95°C 1 min, 65°C 1 min, 72°C 1 min), followed by 25 cycles (95°C 1 min, 62°C 1 min, 72°C 1 min) and a terminal extension at 72°C (10 min).
RESULTS
POT1 molecular screening
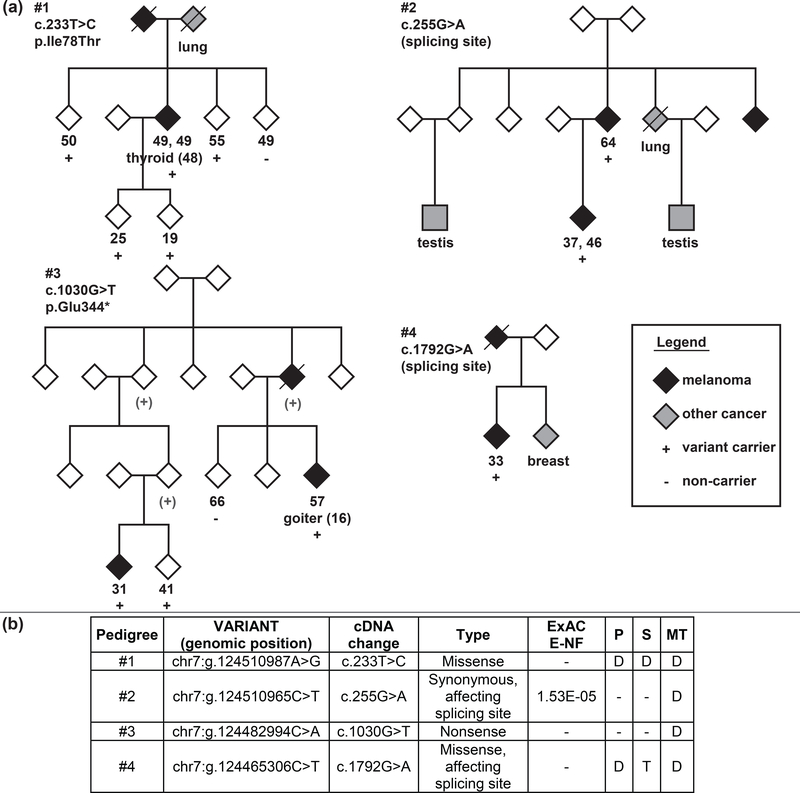
Rare POT1 variants were detected by NGS and confirmed by Sanger sequencing in 1.75% of pedigrees (4/228) (Fig. 2). Co-segregation analysis of the variants detected was extended to the rest of the family where DNA samples were available. c.233T>C (p.Ile78Thr), a missense variant predicted to be pathogenic by in silico analyses, was identified in an affected individual from a 2-case family. This individual had two melanomas and thyroid cancer. The same variant was identified in another four unaffected individuals in the family, who were 55 years old or younger. This variant is located at the DNA binding domain of POT1 and has been recently demonstrated to impair the capability to bind the telomeres.17
Figure 2. Pedigrees with POT1 variants and variant information.
a) Pedigree diagrams from families with rare POT1 germline variants. Gender has been hidden for family de-identification (except for individuals with gender-related cancer types). Below the symbol, age at diagnosis of each melanoma / age at blood sampling (for non-melanoma individuals), other cancer type / disease (age at diagnosis of other cancer/disease is indicated between parentheses) and presence “+” or absence “-” of the variant are listed. Non-tested obligated carriers are indicated as “(+)” in grey. b) Detailed information of the variant detected in each pedigree. ExAC E-NF: Variant frequency in European non-Finnish population from ExAC (http://exac.broadinstitute.org/) database. P: Polyphen2 functional prediction result (D=Probably damaging, PD=Possibly damaging, B=Benign) (http://genetics.bwh.harvard.edu/pph2/). S: SIFT functional prediction result (D=Deleterious, T=Tolerated) (http://sift.jcvi.org/). MT: Mutation Taster functional prediction result (D=Disease Causing, N=Polymorphism,) (http://www.mutationtaster.org/)
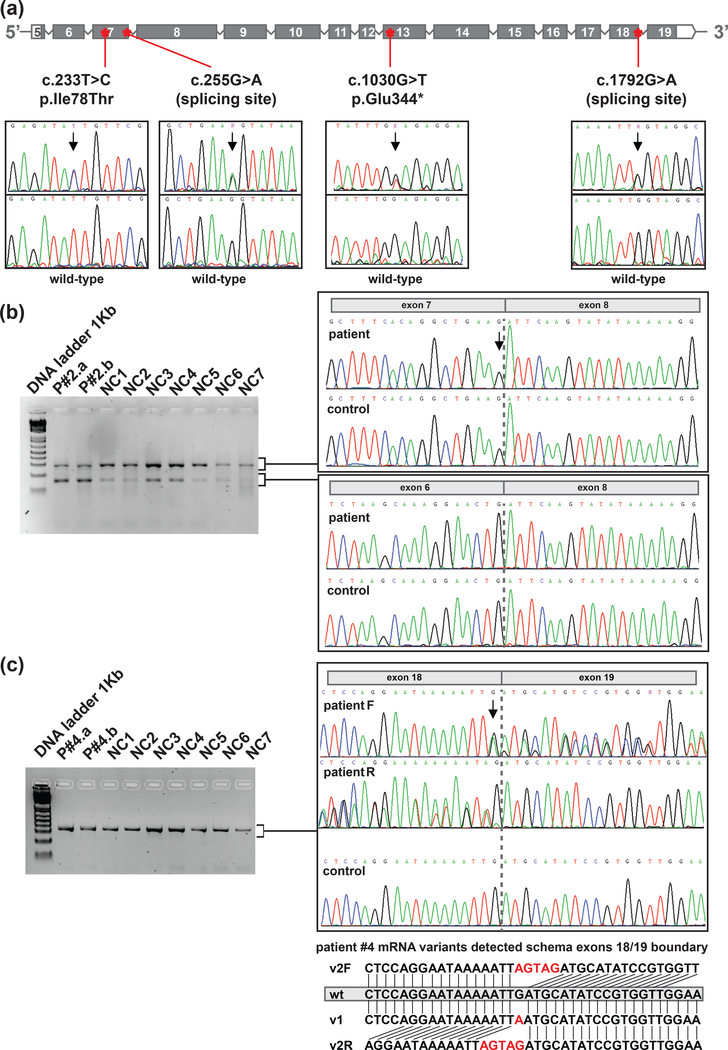
The c.255G>A (p.Lys85Lys), a synonymous variant, predicted to affect splicing, was identified in the two affected individuals available for testing from a 3-case family. One of the two affected individuals carrying the variant had two melanomas. In the family, there were also relatives with testicular cancer and lung cancer, but we were unable to test this variant in these individuals. To assess whether variant c.255G>A alters splicing, we PCR amplified and sequenced the cDNA region encompassing exons 5 to 8 and 6 to 8. Two different fragments amplified in both the carrier and 7 healthy individuals. Specific fragment sequencing confirmed exon 7 skipping of the shorter fragment, probably corresponding to an isoform that naturally excludes exon 7 (ENST00000393329.5, NM_001042594.1). We observed that the c.255G>A carrier had a higher proportion of fragments with exon 7 skipping. Furthermore, we sequenced the largest fragment, corresponding to the fragments including exon 7, using a specific internal primer that anneals exon 7. We observed only the wild-type allele, showing that the mutant allele only produces the shorter isoform (r.125_255del) (Fig. 3). We quantified the expression of each allele in blood RNA from the patient and healthy controls using ddPCR. The allele containing exon 7 skipping represented 46.85% (46.44–47.21) in the mutated patient, while the mean of transcripts with exon 7 skipping in controls represented 13.69% (11.48–15.89) of the total POT1 transcripts.
Figure 3. Rare POT1 germline variants.
a) Location of the variants within the gene context (coding exons according to ENST00000357628.7 transcript) and Sanger sequencing confirmation of the variants detected. The arrows spot the position of the genomic DNA change. b) Gel electrophoresis of cDNA amplification corresponding to the coding region between exons 5 and 8 in Patient #2 (germline variant c.255G>A) and 7 non-carriers (NC). Sanger sequencing of the fragments detected. Dotted grey line marks boundary between exons. c) Gel electrophoresis of cDNA amplification corresponding to the coding region between exons 17 and 19 in Patient #4 (germline variant c.1792G>A) and 7 non-carriers (NC). Sanger sequencing of the fragments detected. A schema of the different transcript alleles detected is shown (wt: wild-type r.1792G allele, v1: r.1792G>A allele and v2 (F=Forward; R=Reverse): r.1791_1792insAGTA).Two independent blood mRNA extractions were performed for each patient with mutation. P#2.a and P#4.a stands for sample A of patient number two and four, respectively. P#2.b and P#4.b stands for sample B of each patient.
c.1030G>T (p.Glu344*), a nonsense variant, was present in all cases of a 3-case family, two tested individuals and an obligate carrier. The same variant was identified in a 41-year-old unaffected individual. This variant disrupts the ACD-binding domain.
Finally, c.1792G>A (p.Asp598Asn), a missense variant, predicted to be pathogenic and predicted to affect splicing, was identified in a melanoma patient from a 2-case family. This individual also carried the MITF p.Glu318Lys variant. Another relative in the family had breast cancer but could not be tested. To assess whether the missense variant c.1792G>A affects splicing, we PCR amplified and sequenced the cDNA region encompassing exons 16/17 to 19 and 17/18 to 19. A unique fragment was identified. Sequencing confirmed that, at the RNA level, there was an insertion of four nucleotides in the mutant allele (r.1791_1792insAGTA), which results in a reading frame alteration (p.Asp598Serfs*22). However a residual presence of the transcript containing r.1792G>A (p.Asp598Asn) was also observed. This indicates that the nucleotide change is able to activate a cryptic splicing site, but a small amount of non-spliced altered mRNA is still produced (Fig. 3). We specifically quantified the amount of transcripts with r.1791_1792insAGTA, r.1792G>A and wild-type by ddPCR. The mutant alleles were not identified in cDNA from healthy controls. In the patient, the transcripts containing r.1791_1792insAGTA represented 43.63% (41.27–45.99), r.1792G>A represented 4.05% (3.23–4.86) and wild-type transcripts 52.32% (49.89–54.76). This variant disrupts the ACD-binding domain.
TERT promoter molecular screening
We sequenced the TERT promoter in one case/family from 202 CDKN2A wild-type melanoma-prone families (26 of our 228 families analyzed for POT1 variants by NGS could not be analyzed due to exhausted or degraded DNA), 30 probands from CDKN2A mutation positive melanoma-prone families and 70 sporadic MPM with family history of other cancers were also analyzed. We detected a rare germline variant of unknown significance (c.−125C>A) in a MPM patient with a family history of breast and colon cancer. We previously reported the patient when evaluating germline mutations in a set of sporadic cases.18 No other mutations were detected in the aforementioned cases.
The distribution of the TERT promoter rs2853669 polymorphism was similar between MPM (49% TT, 47% TC, 4% CC; MAF=0.28) and familial cases (48% TT, 45% TC, 7% CC; MAF=0.30 p=0.778). The MAF in both groups was similar to that reported in 1000 Genomes in European population (0.29). There were no differences regarding age of onset (p=0.445) or the number of melanoma primaries (p=0.857) according to the rs2853669 genotype (Table S2).
DISCUSSION
The present study describes the prevalence of mutations in the telomere-related genes POT1 and the TERT promoter in a large set of Spanish melanoma-prone families. We found four probably pathogenic POT1 variants and one variant of unknown significance in TERT.
TERT was the first gene involved in telomere maintenance that was identified as a high-penetrance susceptibility gene for familial melanoma.6,11 We identified a rare variant (c.−125C>T) of unknown significance in the TERT promoter in a sporadic MPM patient with family history of breast and colon cancers. No rare TERT variants were detected in the families, irrespective of the CDKN2A mutational status. Although germline TERT promoter mutations are extremely rare in familial melanoma,6,11,19 common variants in TERT increase melanoma risk with smaller size effect.20 The TERT promoter rs2853669 polymorphism, which may affect cancer clinical outcomes,21 was not associated with age of onset or presence of multiple primaries, and its prevalence was similar among the MPM and familial cases in our study. In previous studies, the c.−57T>G variant was identified in two large melanoma pedigrees. In one of them a MPM patient with bladder cancer and basal cell carcinomas was a carrier.11 In the other study two melanoma patients also developed other cancers including ovary (both cases) and bladder, renal, breast and lung (just one case).6 Unfortunately there are not enough cases with rare germline mutations in the TERT promoter to assess whether there are specific cancer types or traits that are enriched in subjects who carry TERT promoter mutations.
Unlike TERT promoter mutations, POT1 germline mutations were present in a subset of Spanish melanoma-prone families. We have detected four probably pathogenic POT1 germline mutations in 1.75% (4/228) of CDKN2A/CDK4-wild type families: the p.Ile78Thr variant, which was previously reported in a MPM patient7 and three novel variants: a nonsense variant (p.Glu344*) and two variants affecting splicing of the POT1 main transcript (c.255G>A, r.125_255del and c.1792G>A, r.1791_1792insAGTA). The prevalence of POT1 mutations observed in this study is similar to the prevalence of the medium frequency melanoma risk variant p.Glu318Lys in the MITF gene.13 A limitation of the present study is that segregation analysis in subjects with melanoma or other cancers could not be performed in some families due to the lack of DNA availability from some affected individuals. A note of caution as regards POT1 variants pathogenic function or penetrance is necessary. In fact, in one family where we could test multiple individuals, we identified the p.Ile78Thr variant also in unaffected members, although some of them were below 50 years of age, thus still at risk of developing melanoma. Moreover, one of the variants, p.Asp598Asn, was found in an affected individual also carrying a MITF pathogenic mutation. Further studies are necessary to estimate the penetrance and effect size of POT1 variants on melanoma risk.
Clinical and phenotypic characterization of mutation carriers from families with germline alterations in melanoma susceptibility genes such as CDKN2A or BAP1 have allowed us to refine genetic counseling. CDKN2A germline mutations have been associated with the presence of atypical nevi,22 early age at diagnosis or MPM.3–5 Beyond melanoma, CDKN2A mutation carriers have increased risk of pancreatic cancer and other tobacco-related cancers, thus smoking avoidance can be recommended as a preventive strategy.3,23 Furthermore, CDKN2A carriers can be included into pancreatic cancer screening programs that allow the detection of pancreatic cancer at a resectable stage.24 BAP1 germline mutations confer risk to cutaneous and uveal melanoma, mesothelioma and renal tumors, thus ophthalmological examinations and screening for early mesothelioma or renal tumors detection can be implemented in carriers.1
Although the number of reported pedigrees with POT1 mutations is limited, MPM patients are present in multiple pedigrees,7,8,25 including the present study. Moreover, other cancer types occur in the pedigrees. A recent study has identified a POT1 germline variant in a melanoma-prone family with multiple cases of thyroid cancer and goiter.25 We identified two melanoma patients in different families carrying POT1 mutations who also develop thyroid cancer or goiter, thus supporting a role for POT1 variants in the predisposition of thyroid cancer and goiter. Robles-Espinoza and collaborators identified POT1 mutations in individuals with cutaneous melanoma and breast or lung cancer. Other tumors were developed in those families but segregation could not be confirmed.8 POT1 germline mutations have also been identified in families with Li-Fraumeni-Like syndrome with cardiac angiosarcoma,26,27 glioma,28, chronic lymphocytic leukemia (CLL)29 and Hodgkin lymphoma.30 Speedy and colleagues suggested that the penetrance of germline mutations in genes involved in shelterin complex is modest and act as moderate penetrance alleles, based on the fact that families with CLL with POT1 mutations do not segregate with glioma or melanoma and the absence of significant loss of heterozygosity in tumors of carriers.28 While the families assessed have been mostly selected for a specific tumor type, other cancers types are also reported, some of them associated with the POT1 tumor spectrum. In Robles-Espinoza and colleagues, there is an untested melanoma patient that also developed CLL in one family and an untested melanoma patient that also developed a brain tumor.8 In Calvete and colleagues there is a patient with melanoma and cardiac angiosarcoma with POT1 mutation.25 But, to date, beyond Li-Fraumeni like families (that have multiple different tumor types), most of the families assessed tend to have a predominance of a specific tumor type. There may be genetic modifiers that tend to predispose to a particular tumor type in POT1 mutation carriers. On the other hand, to our knowledge, there are no studies in which they have studied POT1 in sporadic cases of melanoma with family history of other tumors associated with POT1 germline mutations. This strategy might allow the identification of more families with POT1 mutations. Nevertheless, more studies should be performed to assess the role of POT1 in the susceptibility to other cancers.
In conclusion, the analysis of telomere-related genes showed rare POT1 variants in a subset of Spanish melanoma-prone families, while mutations in the TERT promoter were extremely rare. If extended to additional families and cancer types, these findings may have important implications for genetic counseling.
Supplementary Material
What’s already known about this topic?
Telomere-related genes germline mutations predispose to familial melanoma.
The prevalence of germline mutations in telomere-related genes has not been widely studied in melanoma families of Iberian-descent.
What does this study add?
This study evaluates for the first time the prevalence of POT1 and TERT promoter mutations in a Hospital-based series of 228 CDKN2A-negative melanoma families from Barcelona, Spain.
We have identified POT1 mutations in 1.75% families, but no TERT promoter mutations in our series.
These results will facilitate genetic counseling and screening of melanoma families.
Acknowledgements:
Thanks to our patients and their families who are the main reason for our studies; to nurses from the Melanoma Unit of Hospital Clínic of Barcelona, Daniel Gabriel, Pablo Iglesias, Mireia Domínguez and Maria E Moliner for helping to collect patient data, and to Judit Mateu from the “Melanoma: image, genetics and immunology” group at IDIBAPS for her technical assistance.
Funding sources:
The research at the Melanoma Unit in Barcelona is partially funded by Spanish Fondo de Investigaciones Sanitarias grants PI15/00716 and PI15/00956; CIBER de Enfermedades Raras of the Instituto de Salud Carlos III, Spain, co-financed by European Development Regional Fund “A way to achieve Europe” ERDF; AGAUR 2017_SGR_1134 of the Catalan Government, Spain; European Commission under the 6th Framework Programme, Contract No. LSHC-CT-2006–018702 (GenoMEL) and by the European Commission under the 7th Framework Programme, Diagnoptics; The National Cancer Institute (NCI) of the US National Institute of Health (NIH) (CA83115); a grant from “Fundació La Marató de TV3” 201331–30, Catalonia, Spain; a grant from “Fundación Científica de la Asociación Española Contra el Cáncer” GCB15152978SOEN, Spain, and CERCA Programme / Generalitat de Catalunya. Part of the work was carried out at the Esther Koplowitz Center, Barcelona.
The whole exome sequencing analysis was in part supported by the intramural research program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics.
DJA is funded by Cancer Research UK, The Wellcome Trust and the ERC Synergy Programme.
C. D. Robles-Espinoza’s work is funded by the Wellcome Trust (Seed Award Ref. 204562/Z/16/Z) and Universidad Nacional Autónoma de México (PAPIIT award ref. IA200318).
Miriam Potrony is the recipient of a PhD Fellowship FI14/00231 (PFIS) from Instituto de Salud Carlos III, Spain.
Role of the Sponsors: The sponsors had no role in the design and conduct of the study; in the collection, analysis and interpretation of data; or in the preparation, review, or approval of the manuscript.
Footnotes
Conflict of Interest Disclosure: None Declared.
REFERENCES
- 1.Potrony M, Badenas C, Aguilera P et al. Update in genetic susceptibility in melanoma. Ann Transl Med 2015; 3: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldstein AM, Chan M, Harland M et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J Med Genet 2007; 44: 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Potrony M, Puig-Butille JA, Aguilera P et al. Increased prevalence of lung, breast, and pancreatic cancers in addition to melanoma risk in families bearing the cyclin-dependent kinase inhibitor 2A mutation: implications for genetic counseling. J Am Acad Dermatol 2014; 71: 888–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puig S, Malvehy J, Badenas C et al. Role of the CDKN2A locus in patients with multiple primary melanomas. J Clin Oncol 2005; 23: 3043–51. [DOI] [PubMed] [Google Scholar]
- 5.Puig S, Potrony M, Cuellar F et al. Characterization of individuals at high risk of developing melanoma in Latin America: bases for genetic counseling in melanoma. Genet Med 2016; 18: 727–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horn S, Figl A, Rachakonda PS et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013; 339: 959–61. [DOI] [PubMed] [Google Scholar]
- 7.Shi J, Yang XR, Ballew B et al. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat Genet 2014; 46: 482–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robles-Espinoza CD, Harland M, Ramsay AJ et al. POT1 loss-of-function variants predispose to familial melanoma. Nat Genet 2014; 46: 478–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoude LG, Pritchard AL, Robles-Espinoza CD et al. Nonsense mutations in the shelterin complex genes ACD and TERF2IP in familial melanoma. J Natl Cancer Inst 2014; 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 2010; 11: 171–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harland M, Petljak M, Robles-Espinoza CD et al. Germline TERT promoter mutations are rare in familial melanoma. Fam Cancer 2016; 15: 139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leachman SA, Carucci J, Kohlmann W et al. Selection criteria for genetic assessment of patients with familial melanoma. J Am Acad Dermatol 2009; 61: 677e1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Potrony M, Puig-Butille JA, Aguilera P et al. Prevalence of MITF p.E318K in Patients With Melanoma Independent of the Presence of CDKN2A Causative Mutations. JAMA Dermatol 2016; 152: 405–12. [DOI] [PubMed] [Google Scholar]
- 14.Hunt KA, Mistry V, Bockett NA et al. Negligible impact of rare autoimmune-locus coding-region variants on missing heritability. Nature 2013; 498: 232–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun B, Tao L, Zheng YL. Simultaneous quantification of alternatively spliced transcripts in a single droplet digital PCR reaction. Biotechniques 2014; 56: 319–25. [DOI] [PubMed] [Google Scholar]
- 16.Hiller M, Spitali P, Datson N et al. Exon 51 Skipping Quantification by Digital Droplet PCR in del52hDMD/mdx Mice. Methods Mol Biol 2018; 1828: 249–62. [DOI] [PubMed] [Google Scholar]
- 17.Wong K, Robles-Espinoza CD, Rodriguez D et al. Association of the POT1 Germline Missense Variant p.I78T With Familial Melanoma. JAMA Dermatol 2019; 155: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griewank KG, Murali R, Puig-Butille JA et al. TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. J Natl Cancer Inst 2014; 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pellegrini C, Maturo MG, Martorelli C et al. Characterization of melanoma susceptibility genes in high-risk patients from Central Italy. Melanoma Res 2017; 27: 258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iles MM, Bishop DT, Taylor JC et al. The effect on melanoma risk of genes previously associated with telomere length. J Natl Cancer Inst 2014; 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rachakonda PS, Hosen I, de Verdier PJ et al. TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. Proc Natl Acad Sci U S A 2013; 110: 17426–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor NJ, Mitra N, Goldstein AM et al. Germline variation at CDKN2A and associations with nevus phenotypes among members of melanoma families. J Invest Dermatol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helgadottir H, Hoiom V, Jonsson G et al. High risk of tobacco-related cancers in CDKN2A mutation-positive melanoma families. J Med Genet 2014; 51: 545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasen H, Ibrahim I, Ponce CG et al. Benefit of Surveillance for Pancreatic Cancer in High-Risk Individuals: Outcome of Long-Term Prospective Follow-Up Studies From Three European Expert Centers. J Clin Oncol 2016; 34: 2010–9. [DOI] [PubMed] [Google Scholar]
- 25.Wilson TL, Hattangady N, Lerario AM et al. A new POT1 germline mutation-expanding the spectrum of POT1-associated cancers. Fam Cancer 2017. [DOI] [PubMed] [Google Scholar]
- 26.Calvete O, Martinez P, Garcia-Pavia P et al. A mutation in the POT1 gene is responsible for cardiac angiosarcoma in TP53-negative Li-Fraumeni-like families. Nat Commun 2015; 6: 8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calvete O, Garcia-Pavia P, Dominguez F et al. The wide spectrum of POT1 gene variants correlates with multiple cancer types. Eur J Hum Genet 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bainbridge MN, Armstrong GN, Gramatges MM et al. Germline mutations in shelterin complex genes are associated with familial glioma. J Natl Cancer Inst 2014; 107: 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Speedy HE, Kinnersley B, Chubb D et al. Germ line mutations in shelterin complex genes are associated with familial chronic lymphocytic leukemia. Blood 2016; 128: 2319–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMaster ML, Sun C, Landi MT et al. Germline mutations in Protection of Telomeres 1 in two families with Hodgkin lymphoma. Br J Haematol 2018; 181: 372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.