Abstract
Both epidemiological investigations and animal studies have linked arsenic-contaminated water to cancers, including skin, liver and lung cancers. Besides genotoxicity, arsenic exposure-related pathogenesis of disease is widely considered through epigenetic mechanisms; however, the underlying mechanism remains to be determined. Herein we explore the initial epigenetic changes via acute sodium arsenite (As) exposures of mouse embryonic fibroblast (MEF) cells and histone H3K79 methyltransferase Dot1L knockout (Dot1L−/−) MEF cells.
Our RNA-seq and Western blot data demonstrated that, in both cell lines, acute As exposure abolished histone acetyltransferase p300 at the RNA level and subsequent protein level. Consequently, p300-specific main target histone H3K27ac, a marker separating active from poised enhancers, decreased dramatically as validated by both Western blot and ChIP-qPCR/seq analyses. Concomitantly, H3K4me1 as another well-known marker for enhancers also showed significant decreases, suggesting an underappreciated crosstalk between H3K4me1 and H3K27ac involved in As exposure. Significantly, As exposure-reduced H3K27ac and H3K4me1 inhibited the expression of genes including EP300 itself and Kruppel Like Factor 4(Klf4) that both are tumor suppressor genes. Collectively, our investigations identified p300 as an internal bridging factor within cells to sense external environmental As exposure to alter chromatin, thereby changing gene transcription for disease pathogenesis.
Keywords: Sodium arsenite, Histone acetyltransferase p300, H3K27ac, Enhancers, Cancers
1. Introduction
Arsenic, as a ubiquitous element, ranks the 20th most abundant element in the earth’s crust (Mandal and Suzuki, 2002). Human exposure to environmental arsenic occurs primarily via ingestion of inorganic arsenic contaminated water. It is estimated that over 200 million individuals worldwide are exposed to drinking water containing arsenic above 10 μg/L, the safety standard set by the World Health Organization (Argos et al., 2014). Through numerous epidemiological investigations, accumulating evidence demonstrate the positive relationship between inorganic arsenic and carcinomas, including both skin and internal cancers. In Wisconsin, USA, residents who were over 35 years old and consumed arsenic-contaminated water for more than a decade suffered a higher prevalence of skin cancer than control group (Knobeloch et al., 2006). A significant dose-response relationship between arsenic concentrations and other tumors was also found in Taiwan, including bladder, kidney, and lung cancer in both sexes, and tumors of prostate and liver in males (Wu et al., 1989).
The exact pathogenic mechanisms underlying inorganic arsenic exposure remain to be determined. Inorganic arsenic shows genotoxicity through several aspects, including chromosomal aberration (Oya- Ohta et al., 1996), genomic instability (Bhattacharjee et al., 2013) and p53 dysfunction (Hamadeh et al., 1999). Besides, the role of inorganic arsenic in epigenome has also been initially explored, including DNA methylation, microRNA, and histone modification. The methylation of CpG dinucleotides at the 5′ position on the pyrimidine ring forms 5- methylcytosine (5mC). This 5mC can alter transcription by blocking the binding of transcription factors and attracting methyl-binding proteins that initiate chromatin compaction for gene silencing (Li et al., 2015; Lunnon and Mill, 2013). Therefore, hypermethylation may result in gene silencing, while de-methylation may lead to gene activation (Tellez-Plaza et al., 2014). S-adenosyl-methionine (SAM), the substrate of DNA methyltransferases, is essential for methylation of inorganic arsenic to detoxication, and arsenic exposure was associated with hypomethylation of DNA and facilitates oncogenes expression (Zhao et al., 1997). Besides, hypermethylation of p53 and p16 promoters were found in people exposed to arsenic. On the other hand, histone modification (including histone acetylation and methylation), a covalent posttranslational modification, plays an integral role in gene transcription (Allfrey and Mirsky, 1964; Dai, 2014). Intriguingly, previous report demonstrated that chronic arsenite exposure decreased H4K16ac in UROtsa cells (Jo et al., 2009). However, systematic characterizations to identify effector histone acetyltransferases and altered genomic loci with reduced histone acetylations remain to be done.
Histone acetylation is regulated by two groups of enzymes, histone acetyltransferases (HATs) and deacetylases (HDACs), with antagonizing functions (Wang et al., 2009b). Among members of HATs, p300 and CBP are homologous. Both are global transcriptional co-activators (Ogryzko et al., 1996), and play integral roles in regulation of gene transcription via at least two (not necessarily mutually exclusive) mechanisms. In scenario 1, CBP/p300 acts as associated partners of transcription factors (TFs). p300 is recruited by these TFs including p53 to acetylate nucleosomes at promoters and/or enhancers for opening chromatin, thereby facilitating the recruitment of RNA polymerase II and other general transcription factors (Guermah et al., 2006; Wang et al., 2009b). For example, we have demonstrated that the increases of histone H3K9ac and H4K16ac at transcription start sites, via inhibition of HDACs with HDAC inhibitor drugs, facilitate the RNA polymerase II recruitment (Wang et al., 2009a, b). In scenario 2, p300 functions through acetylation of TFs themselves, thereby impacting the function of TFs. For example, p300 directly acetylates exposure-relevant TF Nrf2. Acetylations of multiple lysine residues of Nrf2 increase the binding of Nrf2 to promoters (Sun et al., 2009). This increased Nrf2 binding presumably contributes to the regulation of Nrf2-regulated gene pathways.
Relevant to human health, the loss of p300 functions or simple reduction of p300 protein levels has linked to human diseases. Mutations of p300 are linked to a small percentage of patients of Rubinstein-Taybi syndrome (Roelfsema et al., 2005). These mutations lead to either truncated non-functional p300 molecules, or the loss of one copy of EP300 gene in each cell, thereby reducing the p300 amount by half. The latter suggests that the reduction of p300 amount is linked to the pathogenesis of Rubinstein-Taybi syndrome. In other words, p300 dosage is essential for normal development. Lastly, it seems that p300/CBP specifically mediate H3K18ac and H3K27ac, whereas two other homologous HATs, GCN5 and PCAF, mediate H3K9ac (Jin et al., 2011). Among multiple histone acetylation marks, H3K27ac is considered as a histone mark at active enhancers to distinguish other marks at poised enhancers (Creyghton et al., 2010).
To determine epigenetic mechanism involved in arsenic exposureassociated human disease, we herein use sodium arsenite (As) to expose mouse embryonic fibroblast cells (both wild-type and Dot1L knockout) for epigenomic profilings. Dot1L is the sole histone methyltransferase for H3K79. We choose MEF cells in our study due to following reasons: First, MEF cell line is commonly used in toxicology research. Second, wild-type and Dot1L (−/−) MEF cell lines were served as replicates, which will provide solid evidence for genes that commonly up-regulated and down-regulated after arsenic treatment, especially those involved in epigenetic regulation. We found that acute As exposure reduced the expression of p300 at both the mRNA level and the protein level. The consequence (i.e., reduction of H3K27ac) of abolished p300 was further identified. In addition, we examined the expression of two tumor suppressor genes. Collectively, our data reveal an intriguing epigenetic mechanism that arsenic exposure may impact human health through altering the functions of p300 and following histone acetylation modifications it mediated.
2. Materials and methods
2.1. Cell culture and sodium arsenite exposure
MEF and Dot1L−/− cells are diploid and nontumorigenic. Both cells were cultured in high glucose Dulbecco’s Modified Eagle Medium (DMEM) with 10% (v/v) fetal bovine serum (FBS) in an incubator at 37 °C with 5% CO2. At approximately 80% confluent, cells were treated with fresh medium containing 0, 2, 5, 10 μM As for 6 h or 24 h. After exposure, cells were collected for following experiments.
2.2. Western Blot
Total protein extracts or nuclear protein extracts were prepared from cells and loaded into each lane of a 4–12% SDS-PAGE gel as described before (Wang et al., 2008). After electrophoresis, total proteins were transferred onto a nitrocellulose membrane. Membranes were blocked in TBS-T buffer (20 mM Tris base, 150 mM NaCl, 0.05% Tween 20, pH 7.4) containing 5% nonfat milk at room temperature for 2–4 h prior to the addition of primary antibody and incubation at 4 °C overnight. Nrf2 antibody (Santa Cruz, sc-13,032, Lot # C1416), p300 antibody (Santa Cruz, sc-584, Lot # C0805), H3K4me1 antibody (Abcam, ab8895, Lot # GR243233–1) and H3K27ac antibody (Abcam, ab4729, Lot # GR103379–1) were used to detect each protein. TBS-T-washed membranes were incubated with goat anti-rabbit (Santa Cruz, sc-2004, Lot # A1416) or goat anti-mouse (Santa Cruz, sc-2005, Lot #J2215) secondary antibodies in TBS-T–5% nonfat milk and washed with TBS-T, and membrane-bound antibody was visualized in a chemiluminescence assay. Beta-actin (Sigma, A5441, Lot # 030M4788) was used for normalization.
2.3. mRNA extraction and characterization
Total RNAs from exposed and control groups of two cell lines were isolated and quality controlled with protocols from Illumina. A total amount of 2 μg RNA per sample was used as input material for library construction. Ribosomal RNA (rRNA) was removed through Epicentre Ribo-zero rRNA Removal Kit (Epicentre, USA) according to the manufacturer’s instructions. Strand-specific sequencing libraries were generated through the dUTP method by using the resulting RNA from NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations. RNA-seq was performed on an Illumina Hiseq 2000 platform and 100 bp paired-end reads were generated according to Illumina’s protocol. RNA-seq data and ChIP-seq data (see below) were submitted to the GEO database (GEO number GSE118877).
2.4. Whole-genome gene expression analysis
Data analyses were performed similarly as before (Li et al., 2015). The adapter sequences were removed from the raw sequencing data and the individual libraries were converted to the FASTQ format. Sequence reads were aligned to the mouse genome (mm10) with TopHat2 (v2.0.9) and the resulting alignment files were reconstructed with Cufflinks (v2.1.1) and Scripture (beta2). For mRNA analyses, the RefSeq database (Build 37.3) was chosen as the annotation references. The read counts of each transcript were normalized to the length of the individual transcript and to the total mapped fragment counts in each sample and expressed as fragments per kilobase of exon per million fragments mapped (FPKM) of mRNAs in each sample. The mRNA differential expression analyses were applied for exposed and control groups. An adjusted P value < .05 (Student’s t-test with Benjamini- Hochberg false discovery rate (FDR) adjustment) was used as the cut-off for significantly differentially expressed genes. Differentially expressed genes (DEGs) were analyzed by enrichment analyses to detect overrepresented functional terms present in the genomic background. Gene ontology (GO) analysis was performed using the GO-seq R package, in which gene length bias was corrected.
2.5. Gene expression validation by quantitative Reverse Transcription-PCR (qRT-PCR)
Total RNAs were isolated and quality controlled using protocols from Illumina, Inc. All cDNAs were synthesized using Superscript III (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Five over- and under-expressed genes were selected for qRT-PCR assay. All qRT-PCR primers are presented in Table 1. All qPCR reactions were performed on a Roche Lightcycler 480 PCR system (Roche Applied Science, Penzberg, Germany) using Toyobo Thunderbird SYBR RT- qPCR Mix (Toyobo Life Science, Osaka, Japan), with three technical repeats. The amplification procedure was as follows: 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 20 s. Relative quantification of target genes was performed using the 2-ΔΔCT method with GAPDH as a reference gene (Livak and Schmittgen, 2001). Pearson correlation was used to calculate the association between RNA sequence and qRT-PCR (Hane et al., 1993).
Table 1.
Primers used for qRT-PCR.
| GAPDH | F: TGCACCACCAACTGCTTAGC R: GGCATGGACTGTGGTCATGAG |
| NdufaS | F: GGTGTGCTGAAGAAGACCAC R: GGTTCCGCTTTAACCATAGCC |
| Hsd17b10 | F: CGGTAATAACCGGAGGAGCC R: TGGTCATCACCCGGATACCT |
| Ndufa11 | F: GTCACACTCAATCCTCCGGG R: CGCTATGCCAAAGTACACGC |
| Cox8a | F: CGCCAAGATCCATTCGTTGC R: TCACGAAGCAGGAGGTAAGC |
| Ndufa3 | F: CCCCATTGAGCCCCTACTTC R: CGGGCACTGGGTAGTTGTAG |
| EP300 | F: CCCCAGATGGGAGGACAAAC R: ACTGGCTCCAATCTGCTGTC |
| Clock | F: GGCCACTATGTGAGAACCCC R: GACAGTCGTCCACGTTCACT |
| Postn | F: CAACGCAGCGCTATTCTGAC R: TCGGAAGCCACTTTGTCTCC |
| Notch1 | F: GAATGGCGGGAAGTGTGAAG R: CACAGCTGCAGGCATAGTCT |
| Klf4 | F: GGGAGAAGACACTGCGTCAA |
| R: GGAAGTCGCTTCATGTGGGA F: TGAGAACTTGCTGGACGGAC | |
| Crebbp | |
| R: CACTGAGGCTGGCCATGTTA | |
| UTX | F: AGCTTTTGTCGAGCCAAGGA R: GCATTGGACAAAGTGCAGGG |
| Sirt1 | F: GACTCCAAGGCCACGGATAG R: TGTTCGAGGATCTGTGCCAA |
| Hdac4 | F: GCAGATCCAGAGGCAGATCC R: TTTGGCGTCGTACATTCCCA |
2.6. Chromatin immunoprecipitation plus deep sequencing (ChIP-seq)
Chromatin immunoprecipitation (ChIP) was performed as described previously (Wang et al., 2009b; Wang et al., 2008). In brief, cells were crosslinked with 1% formaldehyde for 10 min at room temperature and the reaction was quenched with 125 mM glycine. After chromatin fragmented to 200 to 500 bp by sonication, chromatin templates from 20 million cells were used for ChIP experiment. Samples were immunoprecipitated with 2–4 μg of H3K4me1 antibody or H3K27ac antibody overnight at 4 °C followed by washing steps. After reverse crosslinking, the ChIP DNA fragments were purified and repaired using End- It™ DNA End-Repair Kit (Epicentre) followed by treatment with Taq polymerase to generate single-base 3′ overhangs used for adaptor ligation. Following ligation of a pair of Illumina adaptors to the repaired ends, the ChIP DNA was amplified using the adaptor primers. Amplified DNA fragments around 300 bp were isolated from agarose gel prior to sequencing.
2.7. Identification of binding regions and peaks
For ChIP-seq of H3K27ac, reads were mapped to the mm10 genome using Bowtie aligner allowing up to two mismatches (Langmead, 2010). Only the uniquely mapping reads were used for further analysis. Macs2 call peak program was used to call the peaks and the macs2 bdgdiff was used to identify the difference between MEF and Dot1L (Zhang et al., 2008).
2.8. ChIP–qPCR validation
Five genes’ promoters were selected for ChIP-qPCR assay and presented in Table 2. All qPCR reactions were performed on a Roche Lightcycler 480 PCR system (Roche Applied Science, Penzberg, Germany) using Toyobo Thunderbird SYBR RT-qPCR Mix (Toyobo Life Science, Osaka, Japan), with three technical repeats. The amplification procedure was as follows: 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 20 s. The cycle threshold (Ct) of samples was used for calculation. Each value was normalized to the percentage of Input DNA by using IP/INPUT = 2CtINPUTDNA-CtIPDNA (Alamdar et al., 2017).
Table 2.
Primers used for ChIP-qPCR validation.
| Gene | Location (5' –3') | Sequence | Size (bp) |
|---|---|---|---|
| Klf4 promoter | −1000 to − 500 | F:CTAACCGTTGGCGTGAGGAA R:CGGGTTGTTACTGCTGCAAG |
137 |
| EP300 promoter | −500 to 0 | F:TGGCACAGATTTTGGTTCACTG R: TCGCCACCATTGGTTAGTCC |
125 |
| Nqol promoter | −500 to 0 | F: GCGAGAAGAGCCCTGATTGT R: CAATATCTGGGCTCAGGCGT |
151 |
| Notch2 promoter | −1000 to −500 | F: CAGCACTGTGACAGCCCTTA R: ATAGCCTCCGTTTCGGTTGG |
221 |
| lgf2r promoter | −1500 to −1000 | F: ACCAGACACTCCGGTACTCA R: TGACGAGCCAACACAGACAG |
277 |
2.9. Statistical analysis
Results were presented in Means ± standard error of the mean (SEM). Student’s t-test was used to examine difference between two groups and ANOVA was used for multi-groups. * means P < .05, **means P < .01.
3. Results
3.1. Identification of As exposure condition
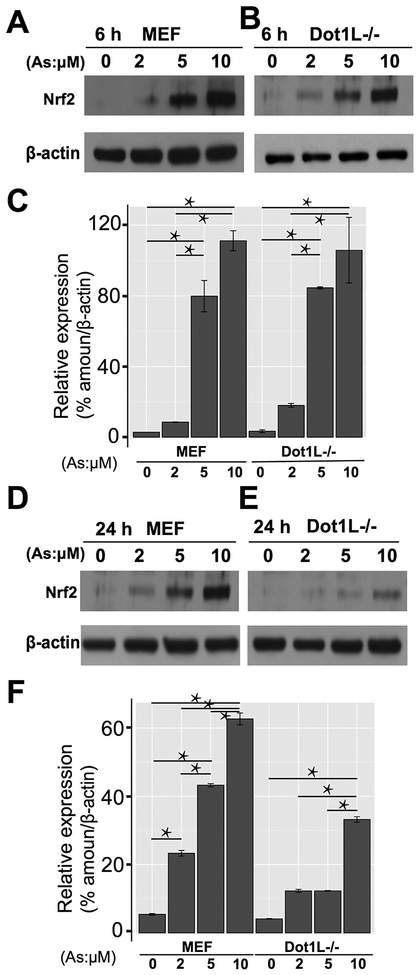
Toward the exposure condition for epigenetic insights of As exposure, we set up exposure experiments and used Nrf2 expression as a readout to characterize. Based on previous reports, we started our exposure experiments with two time points (6 h and 24 h) and four concentrations (0, 2, 5, and 10 μM). Without As exposure, there was limited amount of Nrf2 expressed in both MEF and Dot1L−/− cells (Fig. 1A and B). After 6 h treatment, the amount of Nrf2 showed dose-effect relationships with the concentrations of As in both cell lines (Fig. 1C). In addition, there was a significant difference between 2 μM and 5 μM groups. Results after 24 h exposure (Fig. 1D and E) were similar induction to 6 h treatment. However, compared with 6 h groups, which showed almost same results in both cells, 24 h groups (Fig. 1F) showed different pattern of Nrf2 concentrations in two cell lines. Intriguingly, Nrf2 protein levels were a bit less in Dot1L cell lines for unknown reason. Collectively, we consider 5 μM and 24 h appropriate for our mechanistic exploration and therefore choose this exposure condition for further experimental characterizations.
Fig. 1.
Identification of sodium arsenite exposure condition. 1A and 1B showed results of Western blot of Nrf2 protein in MEF and Dot1L−/− cells after 6 h exposure. 1C showed the quantitative results of Western blot in 1A and 1B. 1D and 1E showed results of Western blot of Nrf2 protein in MEF and Dot1L−/− cells after 24 h exposure. 1F showed the quantitative results of Western blot in 1D and 1E. * P < .05.
3.2. Acute As exposure of MEF cells for mechanistic insights via RNA-seq analyses
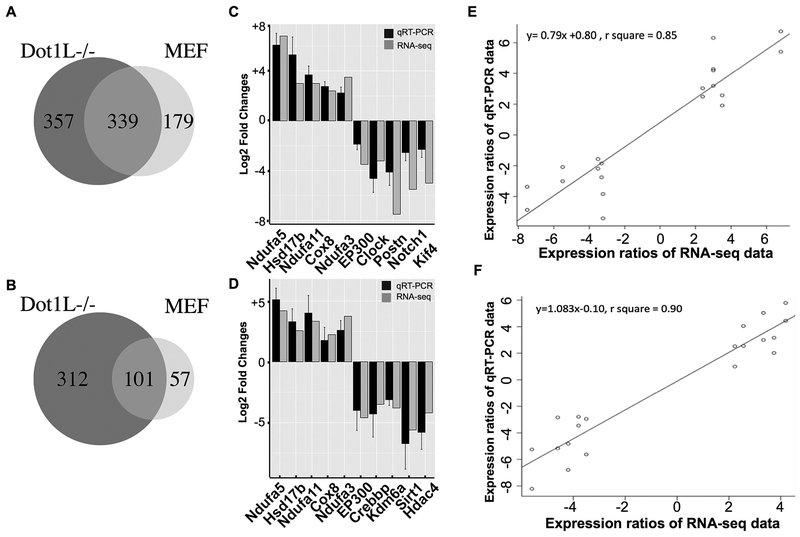
To identify an intermediate factor potentially bridging environmental exposure (external) and cellular (internal) pathway changes, we focused on transcriptomic changes. In other words, we would like to identify the bridging factor(s) based on transcript changes. To our goal, we purified mRNAs from exposed cells and followed our established protocols for RNA-seq library construction (Li et al., 2015). Sequenced reads were aligned to the mouse genome, and expression changes were analyzed. Results demonstrated that As treatment upregulated 518 genes in MEF and 696 genes in Dot1L−/− MEF cells and the overlapping genes were 339 in two cell lines (Fig. 2A). As treatment also downregulated genes, with 158 genes in MEF and 413 genes in Dot1L−/− MEF cells and the overlapping number was 101 (Fig. 2B). To further validate our RNA-seq results, we selected 10 genes for qRT-PCR analyses in both cell lines and confirmed their expression changes (Fig. 2C and D). For up-regulated genes, we choose genes with roles in response to oxidative stress; for down-regulated genes, we choose these with roles in epigenetics or tumor initiation. R squares from Pearson correlation analyses were 0.85 for MEF cell and 0.90 for Dot1L−/− cell, suggesting the results from our RNA-seq analyses reliable (Fig. 2E and F).
Fig. 2.
Sodium arsenite exposure affects the expression of genes, including tumor suppressor genes EP300 and Kif4. 2A and 2B showed up-regulated and downregulated genes after As exposure in MEF and Dot1L−/− cells. 2C and 2D showed qRT-PCR results using MEF and Dot1L−/− cells, respectively, for further validation. 2E and 2F showed Pearson correlation of results in two cell lines.
3.3. Acute As exposure inhibited histone acetyltransferase p300
We next focused on the shared genes that were repressed in both cell lines, That is, we focused on gene’s repression that was dependent on As exposure, but independent of the loss function of Dot1L. Among these 101 overlapping genes that were inhibited by As treatment in two cell lines, we noticed an intriguing candidate, EP300 (Log2 fold, −3.5 in MEF cell and − 4.6 in Dot1L−/−) (Fig. 2C and D). EP300 encodes histone acetyltransferase p300 (also known as E1A-associated protein p300), essential for regulating cell growth and differentiation and preventing the growth of tumors (Eckner et al., 1994). For the latter, many have reported the role of p300 as a tumor suppressor (Iyer et al., 2004).
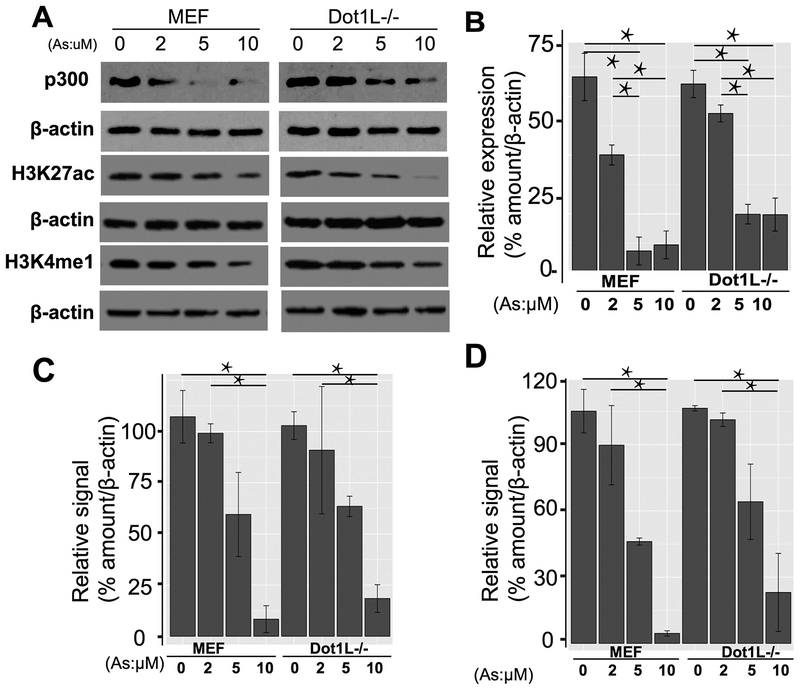
We first did qRT-PCR (Fig. 2C and D) and validated the observed changes from RNA-seq analyses. Because reduced RNA level may not reflect the actual protein level and the protein executes enzymatic activity, we next did Western blot to reveal the exact p300 protein level within cells upon As exposure. Western blot from three biological replicates demonstrated that As exposure indeed reduced p300 protein level and showed dose-effect relationships with As concentrations (Fig. 3A and B). In combination with RNA-seq and qRT-PCR data, we conclude that As exposure reduces the expression of EP300 initially at the transcription level and eventually at the protein level.
Fig. 3.
Sodium arsenite exposure inhibited histone acetyltransferase p300 and p300-mediated H3K27ac, as well as enhancer mark H3K4me1. 3A showed the expression of p300, and relative signals of H3K27ac and H3K4me1 in MEF and Dot1L−/− cells in different concentrations of As. 3B, 3C and 3D showed the quantitative results of p300, H3K27ac, and H3K4me1, respectively. *P < .05.
3.4. Altered histone acetylation H3K27ac, an enhancer marker
Next, we determined the consequence of the abolishment of p300 due to As exposure. As a HAT, p300 is expected to acetylate nucleosomal tails. Though p300 seems to acetylate many residues of histone H3 or H4 especially from in vitro assays, there are also reports suggesting H3K27ac as p300-sepcific mark (Jin et al., 2011; Tang et al., 2013). Therefore, we examined the changes of H3K27ac after As exposure. With the increasing concentration of As, our data demonstrated that the levels of H3K27ac decreased accordingly (Fig. 3A and C) from three replicates. These data demonstrate that As exposure causes the decreases of histone H3K27ac, as a consequence of diminished p300. The decrease of H3K27ac due to As exposure is quite significant, given that in previous report both CBP and p300 needed depletion for reduction of H3K27ac (Jin et al., 2011).
3.5. Acute As exposure decreased H3K4me1, also a well-known enhancer marker
In epigenetic community, both H3K27ac and H3K4me1 are two well-known markers for enhancers and have intriguing crosstalk for enhancer activity (Lai et al., 2017; Wang et al., 2008). With H3K27ac down, we next asked the extent to which H3K4me1 changes accordingly. To our aim, we checked the signals of H3K4me1 in the same samples. Intriguingly, our Western blot results from three replicates reveal the concomitant decreases of H3K4me1 (Fig. 3A and D). These data collectively suggest that As exposure involves underappreciated crosstalk between two enhancer markers, H3K27ac and H3K4me1.
3.6. Identification of vulnerable genomic loci with As exposure-induced loss of H3K27ac
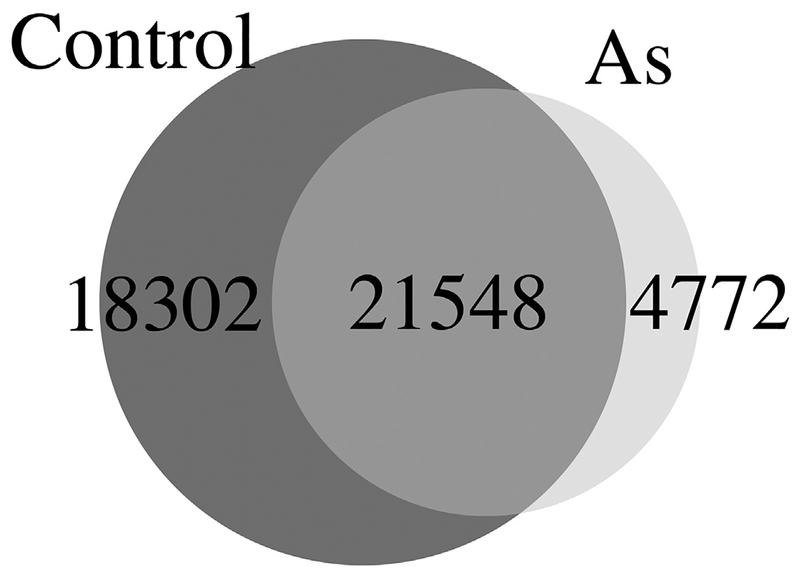
Data of our Western blotting assays reveal the changes of histone acetylation at the global level. To identify the vulnerable genomic loci with altered chromatin after As exposure, we did ChIP-seq analyses using a previously characterized anti-H3K27ac (Wang et al., 2009b; Wang et al., 2008). Following our established pipeline, we analyzed H3K27ac signals genomewide, in this case, only in MEF cells. As expected, As exposed sample has much less H3K27ac peaks compared to control samples. There are 674 peaks specific to exposed sample, whereas 7822 peaks specific to control sample. Two samples shared 18,264 peaks (Fig. 4). Among all 674 peaks specific to As exposure, there are 545 peaks located at enhancer regions. These data are consistent with our Western blotting results (Fig. 3).
Fig. 4.
Sodium arsenite exposure reduced the number of H3K27ac peaks, compared to the number in control cells. There are 674 peaks specific to As treated sample, whereas 7822 peaks specific to control sample. Two samples shared 18,264 peaks.
3.7. Association between decreased H3K27ac and inhibited gene expression
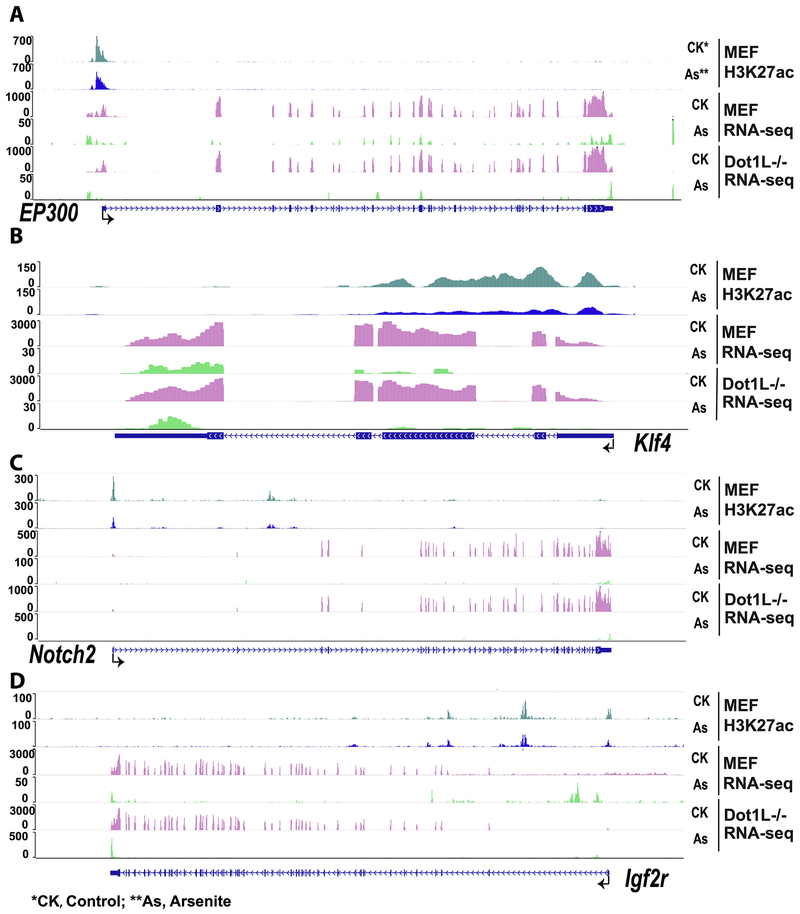
Having established that As exposure reduced H3K27ac and H3K4me1 (Figs. 3 and 4), we next determined the reductions on transcription of selected genes. Snapshots of ChIP-seq results were presented for genes including EP300, Klf4, Notch2, and Igf2r (Fig. 5). These data suggested the association between decreasing of H3K27ac and repressed transcripts of these genes. Notice the different scales used for presenting RNA-seq data.
Fig. 5.
Snapshot results from WashU EpiGenome Browser. 5A, 5B, 5C, and 5D showed ChIP-seq and mRNA-seq results of EP300, Klf4, Notch2, and Igf2r gene, respectively. As shown in this figure, transcriptional level of each gene decreased in As treated group compared with control group. The signal of H3K27ac also decreased after treatment. CK* and CK, control group; As** and As, sodium arsenite exposed group.
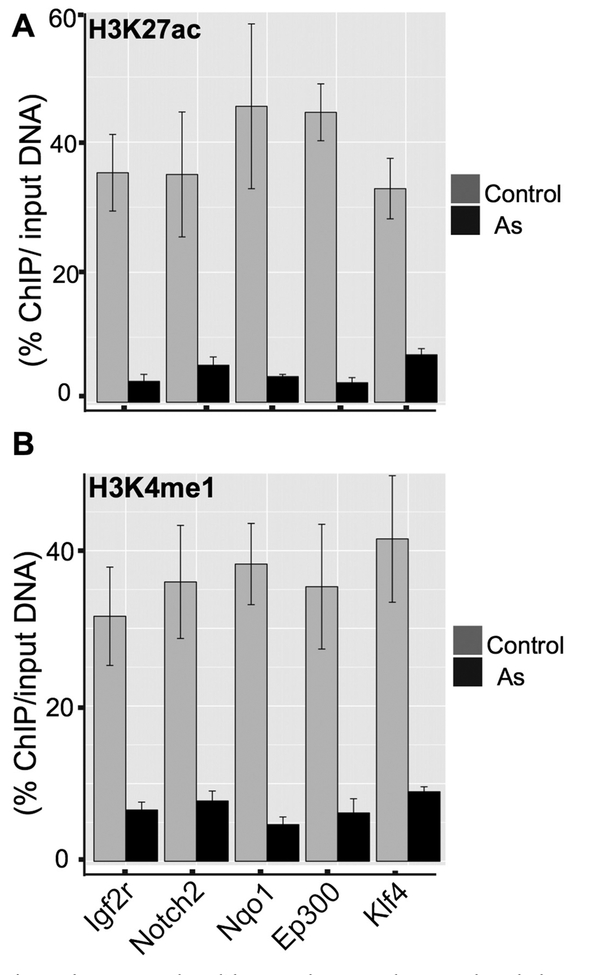
To provide independent confirmation of H3K27ac at these loci and provide additional information of H3K4me1, we did ChIP-qPCR analyses. Results from Fig. 6 showed the lower enrichment of five gene promoters after As exposure in MEF cell, which were in accordance with ChIP-seq results. Klf4, a tumor suppressor gene, showed de-acetylation of H3K27ac and down-regulation (Log2 fold, −5.0), which was validated through ChIP-qPCR (Fig. 6) after As exposure. Accompany to the decreases of H3K27ac at these loci, our ChIP-qPCR demonstrated the decreases of H3K4me1. These data convinced us that As exposure inhibited p300 protein level within cells, thereby impacting histone markers for enhancers and transcription start sites for gene transcription.
Fig. 6.
ChIP-qPCR results validate our ChIP-seq analyses. 6A showed ChIP-qPCR results using H3K27ac antibody and 6B showed ChIP-qPCR results using H3K4me1 antibody. Five genes were used for validation. Compared with control group, all five genes showed significantly lower enrichment rate.
4. Discussion
To initiate our experiments, we choose 0, 2, 5, and 10 μM As for treatment based on published papers (Chervona et al., 2012a; Eckstein et al., 2017; Shi et al., 2004; Wang et al., 2013). We use the upregulated expression of Nrf2 as a readout for initial optimization of treatment conditions. Considering the expensive nature of deep sequencing and our focus of mechanistic insights to initiate the understanding of epigenomic mechanisms during arsenic exposure, we then focus on one relatively low dose (5 μM) condition for future acute exposure. One reasonable assumption is that different levels of arsenic exposure may highly likely share similar epigenetic mechanism. Results in Fig. 3 are indeed consistent with our expectation that all three doses (2, 5, 10 μM) reduce p300 level significantly. Collectively, it seems that epigenomic profiling of cells treated with one dose works well for our purpose of identify epigenetic regulator for arsenic exposure. From our experiments, two cell lines indeed respond to arsenic exposure similarly, with minor differences in terms of the exact genes affected. However, genes such as EP300 were indeed affected in both cell lines.
We notice that previous reports of arsenic exposure are not completely consistent with each other, in terms of exposure-changed histone acetylations. In an early investigation, As exposure (7.5 μM) of HepG2 cells increased H3K9ac after 24 h, when analyzed by methods of immunofluorescence and Western blot analyses. The inhibition from As was comparable to that from a potent HDAC inhibitor trichostatin A (TSA) and from a DNMT1 inhibitor 5-azacytidine. To explain such dramatic increase of H3K9ac, investigators used the colorimetric HDAC activity assay kit (Abcam, ab1432) to report that As-inhibited HDAC activity was indistinguishable from TSA-inhibited (Ramirez et al., 2008). From the same report, histone methylation marks (H3K4me2 and me3; H3K9me2 and me3; H4K20me3; H3K27me3) were not affected as measured by immunofluorescence and Western blot. Authors also concluded the induced hyperacetylation independent of any SAM depletion that might occur due to arsenic metabolism. The aforementioned H3K9 hyperacetylation, however, is in dramatic contrast to the hypoacetylation of H3K9 in As exposed mice (Cronican et al., 2013). Prenatal exposure with 100 ppb (100 μg/L) As in drinking water significantly reduced H3K9ac genomewide in cortices and hippocampi of pups. The global hypoacetylation at H3K9 in exposed mice was consistent with the reduced H3K27ac in our exposed cell lines (. Figs 3–6), and reductions of both H3K9ac and H3K27ac might be explained by the p300 inhibition from our results (. Figs 2–3). On the other hand, the discrepancy of alteration of histone acetylations in different reports might be related to cell type and dose used, as suggested by at least one initial investigation: Two human T cell lines (Jurkat and CCRF-CEM) were exposed to As at 0.1, 1, and 100 μg/L. When checked H3K9me3, its levels in two lines were not different at 0.1 μg/L and might increase at 1 μg/L (p = .19) in Jurkat (but not in CCRF-CEM) cells; when checked H3K9ac, its levels in Jurkat were increased at both 0.1 μg/L and 1 μg/L, whereas its levels in CCRF-CEM were only increased at 1 μg/L (Pournara et al., 2016). Authors also reported similar cell typespecific global decrease of H3K9me3 in sorted CD4+ T cells (but not CD8+ T cells), from individuals with increasing urinary arsenic exposure in the Argentinean Andes. When checked individuals in Bangladesh, the total urinary arsenic was inversely correlated with H3K9ac and positively with H3K9me2 in peripheral blood mononuclear cells (PBMCs) (Chervona et al., 2012b). Note that adding one methyl group to H3K9me2 becomes H3K9me3 and both are repressive marks (Dai and Wang, 2014; Wang et al., 2009a). When examined in CD4+ T cells, it seems to us that both H3K9me2 and H3K9me3 signals correlate well (Barski et al., 2007; Wang et al., 2008). Then the different trends of H3K9me3 in individuals from Argentinean Andes and H3K9ME2 in individuals from Bangladesh in association with urinary arsenic exposure are puzzling. One obvious difference between two investigations is the cell type used: the former, CD4+ T cells; the latter, PBMCs. However, the extent to which cell types may explain the different reports remains to be done.
While most investigations examined histone marks on histone H3, decreased H4K16ac in response to chronic arsenic exposure in human UROtsa cells was also reported (Jo et al., 2009). This decease of H4K16ac was caused by altered expression of one of the HATs, MYST1 (also known as MOF). MOF forms two distinct multiprotein complexes (MSL and NSL) that both acetylate H4K16ac. To summarize, three independent investigations including ours showed the decreases of investigated histone acetylation marks (H3K9ac, H3K27ac, or H4K16ac) (Cronican et al., 2013; Jo et al., 2009), whereas one report showed a contradictory increase of H3K9ac (Ramirez et al., 2008).
Through three biological replicates, we also demonstrated that acute As exposure decreased p300 at both mRNA and protein levels (Figs. 2C, D, 3A). Consequently, p300-specific H3K27ac was reduced as demonstrated by Western blot, ChIP-seq, and ChIP-qPCR analyses in our exposed mouse MEF cells. In our system, RNA-seq data did not show the altered expression of MYST1, different from the case in human UROtsa cells. Again, cell types and doses used may explain the different observation.
Our extensive preliminary data suggest that the loss of p300 (and resulting H3K27ac) is related to inorganic arsenic related diseases. Therefore, it may open a new avenue for alleviating the consequence of H3K27ac, for example, via boosting CBP enzymatic activity for at least partially compensating p300 functions (Iyer et al., 2004; Kasper et al., 2006). It is known that EP300 and its regulated gene Kif4 (silenced after arsenic exposure; Fig. 2C) are tumor suppressors (Iyer et al., 2004). For example, p300 works as a tumor suppressor in hematological tumors (Kung et al., 2000; Oike et al., 1999). Down-expression of p53 and p300 were also found in azoxymethane treated mice, which is a colon-specific carcinogen (Aizu et al., 2003). The role of Klf4 as a tumor suppressor has been validated in colorectal cancer (Zhao et al., 2004), non- Hodgkin lymphoma (Guan et al., 2010), classic Hodgkin lymphoma (Guan et al., 2010), and pancreatic ductal carcinoma (Zammarchi et al., 2011). According to our results, it is reasonable to speculate that As induced p300 downregulation, and decreased Klf4 may promote the initiation of tumors. Other researchers also reported this result (Evans et al., 2007).
In conclusion, for the first time, we reported that sodium arsenite inhibited EP300 at both mRNA and protein levels, which led to the decreased H3K27ac signals. Future experiments to confirm a similar mechanism in the long-term exposed mouse would be significant for arsenic-caused human health problems.
Acknowledgment
The laboratory of Z.W was supported by National Institutes of Health, United States (R01ES25761, U01ES026721 opportunity fund, and R21ES028351) and Johns Hopkins Catalyst Award. Q.Z thanks China Scholarship Council, China and the National Natural Science Foundation of China (81302390) for support. This work was also made possible by the ChuTian Professorship from Hubei University, China and support from GENEWIZ Suzhou. Lastly, extensive bioinformatic analyses were possible only with computational resources (and/or scientific computing services) at the Maryland Advanced Research Computing Center (MARCC). We are therefore greatly indebted to the support from MARCC.
Footnotes
Transparency document
The Transparency document associated to this article can be found, in the online version.
Conflict of Interest
ZW is a consultant of Shandong Bio-focus Gene-tech Co. Ltd. GENEWIZ Suzhou also contributes to the sequencing in this research. However, they had no influence on the experimental design and data presentation here.
References
- Aizu W, Guda K, Nambiar P, Xin T, Thibodeau M, Rosenberg DW, Giardina C, 2003. p53 and its co-activator p300 are inversely regulated in the mouse colon in response to carcinogen. Toxicol. Lett 144, 213–224. [DOI] [PubMed] [Google Scholar]
- Alamdar A, Xi G, Huang Q, Tian M, Eqani S, Shen H, 2017. Arsenic activates the expression of 3beta-HSD in mouse Leydig cells through repression of histone H3K9 methylation. Toxicol. Appl. Pharmacol 326, 7–14. [DOI] [PubMed] [Google Scholar]
- Allfrey VG, Mirsky AE, 1964. Structural modifications of histones and their possible role in the regulation of RNA synthesis. Science 144, 559. [DOI] [PubMed] [Google Scholar]
- Argos M, Parvez F, Rahman M, Rakibuz-Zaman M, Ahmed A, Hore SK, Islam T, Chen Y, Pierce BL, Slavkovich V, et al. , 2014. Arsenic and lung disease mortality in Bangladeshi adults. Epidemiology 25, 536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K, 2007. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee P, Banerjee M, Giri AK, 2013. Role of genomic instability in arsenicinduced carcinogenicity. A review. Environ. Int 53, 29–40. [DOI] [PubMed] [Google Scholar]
- Chervona Y, Arita A, Costa M, 2012a. Carcinogenic metals and the epigenome: understanding the effect of nickel, arsenic, and chromium. Metallomics 4, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervona Y, Hall MN, Arita A, Wu F, Sun H, Tseng HC, Ali E, Uddin MN, Liu X, Zoroddu MA, et al. , 2012b. Associations between arsenic exposure and global posttranslational histone modifications among adults in Bangladesh. Cancer Epidemiol. Biomark. Prev 21, 2252–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. , 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A 107, 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronican AA, Fitz NF, Carter A, Saleem M, Shiva S, Barchowsky A, Koldamova R, Schug J, Lefterov I, 2013. Genome-wide alteration of histone H3K9 acetylation pattern in mouse offspring prenatally exposed to arsenic. PLoS One 8, e53478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, 2014. Histone Modification Patterns and Their Responses to Environment. Curr. Environ. Health Rep 1, 11. [Google Scholar]
- Dai H, Wang Z, 2014. Histone modification patterns and their responses to environment. Curr. Environ. Health Rep 1, 11–21. [Google Scholar]
- Eckner R, Ewen ME, Newsome D, Gerdes M, Decaprio JA, Lawrence JB,Livingston DM, 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev 8, 869–884. [DOI] [PubMed] [Google Scholar]
- Eckstein M, Eleazer R, Rea M, Fondufe-Mittendorf Y, 2017. Epigenomic reprogramming in inorganic arsenic-mediated gene expression patterns during carcinogenesis. Rev. Environ. Health 32, 93–103. [DOI] [PubMed] [Google Scholar]
- Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C, 2007. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J. Biol. Chem 282, 33994–34002. [DOI] [PubMed] [Google Scholar]
- Guan H, Xie L, Leithauser F, Flossbach L, Moller P, Wirth T, Ushmorov A, 2010. KLF4 is a tumor suppressor in B-cell non-Hodgkin lymphoma and in classic Hodgkin lymphoma. Blood 116, 1469–1478. [DOI] [PubMed] [Google Scholar]
- Guermah M, Palhan VB, Tackett AJ, Chait BT, Roeder RG, 2006. Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell 125, 275–286. [DOI] [PubMed] [Google Scholar]
- Hamadeh HK, Vargas M, Lee E, Menzel DB, 1999. Arsenic disrupts cellular levels of p53 and mdm2: a potential mechanism of carcinogenesis. Biochem. Biophys. Res. Commun 263, 446–449. [DOI] [PubMed] [Google Scholar]
- Hane BG, Jager K, Drexler HG, 1993. The Pearson product-moment correlation coefficient is better suited for identification of DNA fingerprint profiles than band matching algorithms. Electrophoresis 14, 967–972. [DOI] [PubMed] [Google Scholar]
- Iyer NG, Ozdag H, Caldas C, 2004. p300/CBP and cancer. Oncogene 23, 4225–4231. [DOI] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K, 2011. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300- mediated H3K18/27ac in nuclear receptor transactivation. EMBO J 30, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo WJ, Ren X, Chu F, Aleshin M, Wintz H, Burlingame A, Smith MT, Vulpe CD, Zhang L, 2009. Acetylated H4K16 by MYST1 protects UROtsa cells from arsenic toxicity and is decreased following chronic arsenic exposure. Toxicol. Appl. Pharmacol 241, 294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper LH, Fukuyama T, Biesen MA, Boussouar F, Tong C, de Pauw A, Murray PJ, van Deursen JM, Brindle PK, 2006. Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol. Cell. Biol 26, 789–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobeloch LM, Zierold KM, Anderson HA, 2006. Association of arsenic-contaminated drinking-water with prevalence of skin cancer in Wisconsin’s Fox River Valley. J. Health Popul. Nutr 24, 206–213. [PubMed] [Google Scholar]
- Kung AL, Rebel VI, Bronson RT, Ch’ng LE, Sieff CA, Livingston DM, Yao TP, 2000. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev 14, 272–277. [PMC free article] [PubMed] [Google Scholar]
- Lai B, Lee JE, Jang Y, Wang L, Peng W, Ge K, 2017. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic Acids Res 45, 6388–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, 2010. Aligning short sequencing reads with Bowtie. Curr. Protocol. Bioinforma 11, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Dai H, Martos SN, Xu B, Gao Y, Li T, Zhu G, Schones DE, Wang Z, 2015. Distinct roles of DNMT1-dependent and DNMT1-independent methylation patterns in the genome of mouse embryonic stem cells. Genome Biol 16, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD, 2001. Analysis of relative gene expression data using realtime quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lunnon K, Mill J, 2013. Epigenetic studies in Alzheimer’s disease: current findings, caveats, and considerations for future studies. Am. J. Med. Genet. B 162B, 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal BK, Suzuki KT, 2002. Arsenic round the world: a review. Talanta 58,201–235. [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y, 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Oike Y, Takakura N, Hata A, Kaname T, Akizuki M, Yamaguchi Y, Yasue H, Araki K, Yamamura K, Suda T, 1999. Mice homozygous for a truncated form of CREB-binding protein exhibit defects in hematopoiesis and vasculo-angiogenesis. Blood 93, 2771–2779. [PubMed] [Google Scholar]
- Oya-Ohta Y, Kaise T, Ochi T, 1996. Induction of chromosomal aberrations in cultured human fibroblasts by inorganic and organic arsenic compounds and the different roles of glutathione in such induction. Mutat. Res 357, 123–129. [DOI] [PubMed] [Google Scholar]
- Pournara A, Kippler M, Holmlund T, Ceder R, Grafstrom R, Vahter M, Broberg K, Wallberg AE, 2016. Arsenic alters global histone modifications in lymphocytes in vitro and in vivo. Cell Biol. Toxicol 32, 275–284. [DOI] [PubMed] [Google Scholar]
- Ramirez T, Brocher J, Stopper H, Hock R, 2008. Sodium arsenite modulates histone acetylation, histone deacetylase activity and HMGN protein dynamics in human cells. Chromosoma 117, 147–157. [DOI] [PubMed] [Google Scholar]
- Roelfsema JH, White SJ, Ariyurek Y, Bartholdi D, Niedrist D, Papadia F, Bacino CA, den Dunnen JT, van Ommen GJ, Breuning MH, et al. , 2005. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am. J. Hum. Genet 76, 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Shi X, Liu KJ, 2004. Oxidative mechanism of arsenic toxicity and carcinogenesis. Mol. Cell. Biochem 255, 67–78. [DOI] [PubMed] [Google Scholar]
- Sun Z, Chin YE, Zhang DD, 2009. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol. Cell. Biol 29, 2658–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Chen WY, Shimada M, Nguyen UT, Kim J, Sun XJ, Sengoku T,McGinty RK, Fernandez JP, Muir TW, et al. , 2013. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell 154, 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez-Plaza M, Tang WY, Shang Y, Umans JG, Francesconi KA, Goessler W, Ledesma M, Leon M, Laclaustra M, Pollak J, et al. , 2014. Association of global DNA methylation and global DNA hydroxymethylation with metals and other exposures in human blood DNA samples. Environ. Health Perspect 122, 946–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. , 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet 40, 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Schones DE, Zhao K, 2009a. Characterization of human epigenomes. Curr. Opin. Genet. Dev 19, 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K, 2009b. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Humphries B, Xiao H, Jiang Y, Yang C, 2013. Epithelial to mesenchymal transition in arsenic-transformed cells promotes angiogenesis through activating beta-catenin-vascular endothelial growth factor pathway. Toxicol. Appl. Pharmacol 271, 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MM, Kuo TL, Hwang YH, Chen CJ, 1989. Dose-response relation between arsenic concentration in well water and mortality from cancers and vascular diseases. Am. J. Epidemiol 130, 1123–1132. [DOI] [PubMed] [Google Scholar]
- Zammarchi F, Morelli M, Menicagli M, Di Cristofano C, Zavaglia K, Paolucci A, Campani D, Aretini P, Boggi U, Mosca F, et al. , 2011. KLF4 is a novel candidate tumor suppressor gene in pancreatic ductal carcinoma. Am. J. Pathol 178, 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. , 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao CQ, Young MR, Diwan BA, Coogan TP, Waalkes MP, 1997. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc. Natl. Acad. Sci. U. S. A 94, 10907–10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW, 2004. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene 23, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]