Abstract
Quercus rubra has been introduced in Europe since the end of the seventeenth century. It is widely distributed today across this continent and considered invasive in some countries.
Here, we investigated the distribution of genetic diversity of both native and introduced populations with the aim of tracing the origin of introduced populations. A large sampling of 883 individuals from 73 native and 38 European locations were genotyped at 69 SNPs. In the natural range, we found a continuous geographic gradient of variation with a predominant latitudinal component. We explored the existence of ancestral populations by performing Bayesian clustering analysis and found support for two or three ancestral genetic clusters. Approximate Bayesian Computations analyses based on these two or three clusters support recent extensive secondary contacts between them, suggesting that present-day continuous genetic variation resulted from recent admixture. In the introduced range, one main genetic cluster was not recovered in Europe, suggesting that source populations were preferentially located in the Northern part of the natural distribution. However, our results cannot refute the introduction of populations from the Southern states that did not survive in Europe.
Keywords: Quercus rubra, spatial genetic structure, genetic divergence, secondary contact, demographic inferences
Introduction
Since the sixteenth century, the development of international trade by terrestrial and maritime routes has favored deliberate and accidental introductions of species by humans far from species’ native habitats and contributed to inter-continental scale expansions (Hulme 2009, Pyšek et al. 2010). In recent decades, multiple examples of human-mediated introductions have been reported in animals (Clout and Russell 2008; Leprieur et al. 2008; Bigsby et al. 2011), plants (Reichard and White 2001; Richardson et al. 2011), fungi (Desprez-Loustau et al. 2007; Vellinga et al. 2009; Gladieux et al. 2010) and viruses (Tatem et al. 2006; Jones 2009). In some instances biological introductions has led to invasions and strong disturbances in natural ecosystems (Ehrenfeld 2010) or had concrete deleterious effects for plants, animals and human health (Pejchar and Mooney 2009; Pysek and Richardson 2010; Vilà et al. 2011).
Two key features required for a successful plant introduction are the residence time and propagule pressure (Pyšek and Jarošík 2005; Simberloff 2009; Blackburn et al. 2015). Long-standing introductions and high planting densities may trigger the shift from cultivation to naturalization of woody species (Pysek et al. 2009). Unfortunately, historical records of inter-continental movement of species are scarce and partial, which makes it difficult to accurately reconstruct the introduction history of a species. One of the most promising options to retrace introduction routes – and more broadly to decipher the evolutionary history of the introduced species – is to use analytical methods of population genetics (Cristescu 2015). Statistical analysis of genetic structure and diversity of native and introduced populations can identify sources and pathways from the native to the introduced range using a handful of DNA markers (Bossdorf et al. 2005; Miura 2007). Inferences drawn from an Approximate Bayesian Computation (ABC) framework may help to understand the demographic history of introduction (Estoup and Guillemaud 2010; Csilléry et al. 2010; Roux et al. 2011; Leroy et al. 2014).
Demographic processes are of utmost importance for investigating the success and spread of species recently introduced in new environments (Allendorf and Lundquist 2003; Facon et al. 2006; Sax et al. 2007). To adapt to novel environments, introduced species can rapidly evolve, displaying phenotypic and genetic divergence from their source populations (Bossdorf et al. 2005). Rapid adaptive evolution is quite often invoked to explain successful invasions (Maron et al. 2004; Lavergne and Molofsky 2007; Buswell et al. 2011).
Generally speaking, high genetic diversity is a prerequisite for rapid adaptation of introduced populations to new environments and ultimately for demographic success (Lavergne and Molofsky 2007). However, in the newly colonized areas, founder events and drift at the expanding front of colonization may substantially reduce genetic diversity and hamper further adaptation. Indeed, genetic bottlenecks are not unlikely, given such introduction scenarios and may generate founder effects during the early steps of introduction (Dlugosch and Parker 2008). Resulting loss of diversity will constrain adaptation of the newly introduced populations. Quite paradoxically, many empirical studies report demographic successes even in case of strong founder effects or population bottlenecks, e.g. in Phyla canescens (Xu et al. 2015), suggesting that adaptation may still occur under such circumstances (Tsutsui et al. 2000; Rollins et al. 2013; Stapley et al. 2015). There are at least two interpretations, not mutually exclusive, for this apparent paradox. First, there might be a strong publication bias because the tracking of these events is laborious and remains elusive. Thus only few instances of failed introductions have been reported. Second, simultaneous or repeated multiple introductions from different source populations in the native range can restore substantial genetic diversity, as has been observed in case studies of e.g. Alliaria petiolata, Ambrosia artemisiifolia and Phalaris arundinacea (Durka et al. 2005; Genton et al. 2005; Lavergne and Molofsky 2007). Thus, even if single populations were subjected to bottlenecks during their introduction, the introduced gene pool is a “melting pot”, restoring diversity by the later interbreeding of different sources of material (Keller and Taylor 2010; Rius and Darling 2014). Finally, surfing of advantageous mutations can also be expected during population expansion of an introduced species and contribute to rapid adaptation (Miller 2010; Lehe et al. 2012).
Native to North America, Quercus rubra L. (northern red oak) was introduced in Europe at the end of the seventeenth and beginning of the eighteenth centuries. First historical records date back to 1691, suggesting that Q. rubra has been present for more than ten generations in Europe (Goeze 1916; Bauer 1953; Magni Diaz 2004). Initially planted in parks and gardens as an ornamental, the species was progressively used in the second half of the nineteenth century for reforestation and timber production, making Q. rubra an important forest resource (Timbal et al. 1994). Quercus rubra is now planted across large areas in Europe (Magni Diaz 2004). In some European countries, it is considered an invasive alien tree species due to its successful regeneration and its large acorn production in Europe (Major et al. 2013; Woziwoda et al. 2014). The species is wind-pollinated, monecious and allogamous. Within the native range, intraspecific gene flow maintains high genetic diversity (Sork et al. 1993). Reproductive barriers and differences in acorn maturation periods prevent interspecific hybridization between Q. rubra (Quercus section Lobatae) and European white oaks (Quercus section Quercus) (Lanier et al. 1980).
Comparative studies of native and introduced populations have shown genetically based phenotypic differences for some life-history traits such as leaf phenology and growth (Daubree and Kremer 1993). Introduced populations are assumed to have undergone selection for adaptation to new local conditions encountered in Europe. Magni Diaz (2004) found similar levels of plastome diversity between native and introduced populations, suggesting that multiple sources of introductions have likely prevented strong founder effects.
In this study, we analyzed the genetic diversity of 73 native and 38 introduced populations of Q. rubra using 69 nuclear DNA markers. By comparing the genetic structure between native and introduced populations, our aim was to draw inferences about demographic processes that may have been associated with the introduction of Q. rubra in Europe. We particularly addressed the following questions: (1) What is the genetic structure of Q. rubra populations within the native range? (2) What is the distribution of genetic diversity in the European gene pool in comparison to the native gene pool? (3) What were the likely sources of origin of populations introduced in Europe?
Materials and methods
Sampling design and DNA extraction
All nuclear DNA samples were extracted from buds or leaves collected from different sampling locations in the native and introduced range, which are defined and considered as populations (Table 1). Fresh buds and leaves were harvested in a common garden experiment (combined provenance and progeny tests) comprising populations from the native and introduced ranges in 2003 by Magni Diaz (2004) and in the summer and fall 2015 by ourselves. Sixty-two native populations and 38 introduced populations were sampled. For each population, buds or leaves were harvested on one to 19 trees stemming from as many open pollinated progenies. Buds were preserved at -20°C until DNA extraction. This collection was complemented by a second sampling campaign in North America composed of 11 additional native populations. Samples were collected in naturally regenerated stands. For each population, leaves were harvested from 18 to 39 individual trees. In total, 73 native populations and 38 introduced populations were used in this study (see detailed information about populations in Appendix Table S1), each population comprising one to 39 trees. We further extracted annual mean temperatures (BIO1) for each population from the WorldClim database (http://www.worldclim.org/current).
Table 1.
Description of samples used for DNA extraction and genotyping: kind of samples, number of samples (NSAMPLES), year of sampling, sampling location, number of native (NNative) and introduced (NINTRODUCED) populations sampled.
| Samples | NSAMPLES | Year | Sampling location | NNATIVE | NINTRODUCED |
|---|---|---|---|---|---|
| Buds | 394 | 2003 | Progeny tests in France | 62 | 38 |
| Leaves | 331 | 2015 | Progeny tests in France | ||
| Leaves | 336 | 2015 | Natural population in USA | 11 | - |
| Total | 1061 | - | - | 73 | 38 |
Nuclear DNA was isolated from 1061 bud or leaf samples using the Invisorb® DNA plant HTS 96 kit (STRATEC Molecular GmbH, Berlin, Germany). DNA yield and quality were evaluated using a NanoDrop spectrophotometer (NanoDrop Technologies, Inc., Wilmington, USA). To assess the genotyping reproducibility, 77 individuals were extracted and genotyped twice.
SNP marker selection and genotyping
Based on the availability of 2,394 RAD-Seq derived SNPs (Restriction-site Associated DNA Sequencing) for red oak, 1,410 bi-allelic SNPs, randomly distributed along the genome and satisfying Sequenom® selection criteria (primer design constraints), were selected, based on their polymorphism in a full sib mapping family (Konar et al. submitted). Briefly, the RAD-Seq procedure was based on genomic DNA digestions of red oak samples with two restriction enzymes (EcoRI and MseI). PCR was then performed using an iProof™ High-Fidelity DNA Polymerase (Bio-Rad, Hercules, California), followed by a purification step using AMPure XP beads (Beckman Coulter Inc., Brea, CA) before sequencing (BGI international, Cambridge, MA). Raw sequences were aligned to the de novo generated reference sequences using default parameters of BWA (BWA is a software package for mapping sequences against a reference). Picard tools (http://picard.sourceforge.net) were used to discard PCR duplicates and SAM tools were used for variant detection (SAM tools is a suite of programs for high-throughput sequencing data analysis). For this study, among the total of 1,410 bi-allelic SNPs satisfying Sequenom® selection criteria, three Sequenom® assays (W1 40 SNPs, W2 40 SNPs and W3 35 SNPs: total 115 SNPs) were designed with the MassARRAY® Assay design 3.1 software (Sequenom® 161, San Diego, USA). Multiplexes and primer sequences can be found in additional file Appendix Table S2. Genotyping was performed using a MassARRAY® System (Agena Bioscience™) and iPLEX® chemistry, according to manufacturer’s specifications. Cluster plots were visually inspected to ensure accurate genotyping calls and the data analysis was performed using MassARRAY® TYPER 4.0 genotyping software. After excluding monomorphic and unamplified loci, data analysis was performed for 80 SNPs. Individuals with more than 10 missing genotypes and loci with more than 10% of missing data were excluded from the analysis. The remaining dataset contains 883 individuals genotyped at 69 SNP loci. High reproducibility of the genotyping method (assessed on 77 replicate individuals) was found (100%). Given the discovery procedure of the SNPs, ascertainment bias was expected due to enrichment for highly polymorphic SNP. However allele frequencies were evenly distributed across the selected SNPs, indicating that SNPs with rare alleles were as abundant as SNPs with frequent alleles (data not shown). This suggests that ascertainment bias might have been lower than originally expected.
Genetic data analysis
Bayesian Clustering
TESS version 2.3, a spatially-explicit Bayesian clustering program (François et al. 2006, Chen et al. 2007), was used to determine genetic structure of populations within the native and introduced ranges. A first analysis including populations from the native range (624 individuals from 73 different geographic locations) was made to estimate the number of genetic clusters (K) in North America. A second analysis based on populations from native and introduced ranges (883 individuals from 111 locations) was carried out to identify the putative source of introductions. For both analyses, assignment of individuals to different genetic clusters (K from 2 to 8) was simulated 100 times using an admixture model (Durand et al. 2009), with 5000 sweeps of Markov Chain Monte Carlo (MCMC) and 1000 burn-in of sweeps of MCMC. The Deviance Information Criterion (DIC), a statistical measure of the model prediction capabilities, was computed by TESS for each simulation. A comparison of the best simulations based on DIC values per K was used to determine the most likely number of genetic clusters for each analysis. This procedure was complemented by a more empirical approach when the DIC criterion was not conclusive (see Results).
Multivariate analysis, population differentiation and diversity
Using the native and whole datasets, we performed Correspondence Analyses (CA) with Genetix program version 4.05 (Belkhir et al. 2004). Genetic diversity, estimated through expected heterozygosity (He), and analysis of molecular variance (AMOVA, Excoffier et al. 1992), testing for the subdivision of genetic variation among and within populations in both North America and Europe, were calculated using GenAlEx program version 6.5 (Peakall and Smouse 2012). Comparisons of He between native and introduced populations were done with adegenet package (R Core Team 2015; Jombart 2008; Jombart and Ahmed 2011) using 1000 permutations.
Approximate Bayesian Computations
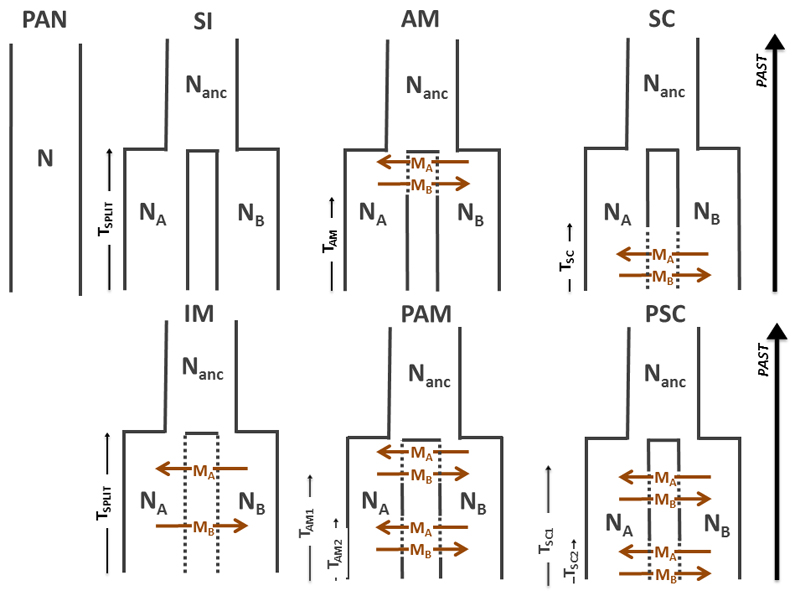
Approximate Bayesian Computation (ABC; Beaumont et al. 2000) was used to investigate the demographic history of Q. rubra. For each pair of clusters, we evaluated seven scenarios of divergence allowing gene flow at different timescales (Fig. 1). The first scenario assumed that the two investigated populations derive from a single panmictic population (PAN) with constant population size (N). The six other scenarios assumed a split of an ancestral population into two daughter populations (A and B) at time TSPLIT. All these 3 diploid populations could have different sizes (NA and NB; NANCbeing the size of the ancestral population) that have remained constant over time. Among these six scenarios, five assumed periods of gene flow since TSPLIT: ancient migration (AM), periodic ancient migration (PAM), continuous migration (IM), secondary contacts (SC) and periodic secondary contacts (PSC). The remaining scenario is a strict isolation model (SI) assuming no migration between clusters. In the AM model, migration is modelled as occurring after TSPLIT, stopping at time TAM. In the PAM model, a derivative of the previous AM model, two periods of ancient migration are assumed stopping at time TAM1 and TAM2. In the IM model, migration is assumed to have occurred continuously since TSPLIT. In the SC model, divergence without gene flow is assumed to have occurred first but at time TSC, the two populations started to exchange genes by secondary gene flow. In its derivative model (PSC), two different cycles of isolation and gene flow were assumed, with gene flow starting at time TSC1 and restarting at time TSC2 (i.e. no gene flow between TSPLIT-TSC1 and TSC1-TSC2).
Fig. 1.
Seven alternative models tested using our ABC framework: PAN: panmixia, SI: strict isolation, AM: ancient migration, IM: isolation migration, SC: secondary contact, PAM: periodic ancient migration; PSC: periodic secondary contact. N is the size of populations (NA for Pop A and NB for Pop B) for each cluster, NANC is the size of ancestral population before the division into two populations (Pop A and Pop B). MA and MB are, respectively, migration rates from population B to population A and from population A to population B. TSPLIT is the number of generations since the divergence time and TSC the number of generations since the secondary contact between both populations. TAM the number of generations since the ancient migration between both populations.
For all scenarios, four million data sets were simulated under the seven different scenarios using a pipeline composed of msnsam, priorgen and mscalc. Msnsam (Ross-Ibarra et al. 2008) is a modified version of ms (Hudson 2002) allowing variations of sample sizes between loci. Priorgen is a generator of priors developed by Ross-Ibarra et al. (2008) and modified by Roux et al. (2011, 2013) and Leroy et al. (2017) to take into account both more complex scenarios of divergence and variations in both effective population sizes and migration rates among loci, two key genomic features known to bias demographic inferences (Charlesworth et al. 1997; Castric et al. 2008; Charlesworth 2009; Roux et al. 2013; Cruickshank and Hahn 2014). For this study, we made further improvements to allow priorgen to generate random prior draws for more complex demographic scenarios with two-cycles of gene flow (PAM and PSC scenarios). Prior parameters were drawn from a large uniform distribution for times and effective population sizes as following: TSPLIT [0; 100]; NANC, NA and NB [0; 10,000,000]. For all simulations, mscalc was used to calculate 19 summary statistics (Ross-Ibara et al. 2008; Roux et al. 2011; Roux et al. 2016), including the average and standard variation for (1) the number of fixed differences, (2) the number of polymorphic sites specific to each gene pool, (3) the number of polymorphic sites existing in both gene pools, (4) Tajima's pi for each gene pool and between the two gene pools, (5) the gross divergence (Dxy), (6) the net divergence (Da), (7) Fst and (8) Pearson's correlation coefficient (see also Roux et al. 2016).
The relevance of prior-model combinations was pre-evaluated using principal component analysis to check if these combinations can produce simulated data sets reasonably similar to the observed data set. For the five models assuming gene flow, locus-specific effective migration rates MA and MB were drawn from a Beta distribution shaped by parameters a and b (see Roux et al. (2013) for details). Parameters a and b were randomly drawn from a uniform distribution: [0; 100] and [0; 500] respectively. We used a feed forward neutral network to estimate the posterior probability of the seven models (PAN, SI, AM, PAM, IM, SC, PSC). This network was performed in the R package “abc” (Csilléry et al. 2012) and used a nonlinear multivariate regression by considering the model itself as an additional parameter to be inferred under the ABC framework. We retained the 0.125% simulated values closest to the observed values for the summary statistics which were weighted by an Epanechnikov kernel (Csilléry et al. 2012). ABC analyses were performed 20 times using for each 20 trained neural networks and 8 hidden layers in the regression.
Parameter estimation was performed using the “abc” package (Csilléry et al. 2012) for the best inferred model based on the 10,000 replicated simulations (0.25% closest simulations) providing the smallest Euclidian distance. Computations were performed using 25 trained neural networks and 10 hidden layers in the regression. To check the robustness of our inferences, we computed 15,000 pseudo-observed data sets (PODS) for each scenario with priors drawn in the same distribution, as previously described. We used the same model selection procedure to obtain posterior probabilities at each of seven scenarios for each POD and then estimated robustness by using distributions of these posterior probabilities over all PODS (see Appendix Fig. S2).
All datasets and programs used in this article are available from the GitHub repository: https://github.com/ThibaultLeroyFr/RedOakABC.
Results
Overall subdivision of genetic diversity
The genetic diversity calculated for native and introduced populations was similar in both gene pools. Expected heterozygosity values were not significantly different: He= 0.330 (± 0.019 SE) for the native populations and He= 0.327 (± 0.019 SE) for the introduced populations (p=0.299). The results of the AMOVA analyses indicated that the subdivision of genetic diversity between North American and European gene pools and among populations within gene pools was significant (p<0.001), albeit low (Table 2). Most of the genetic diversity was found among individuals within populations. In both North American and European areas, less than 3% of the total variation was among populations. Genetic differentiation between Europe and North America, albeit significant, was weak (0.98%). Considering both gene pools, ninety-six per cent of the total molecular variance was explained by within-population variation (Table 2).
Table 2.
Results of Analyses of Molecular Variance (AMOVA) within North America and Europe and between both gene pools of Q. rubra. df represents the degree of freedom.
| Source of variation | df. | Sum of squares |
Variance components |
Percent variation |
p-value |
|---|---|---|---|---|---|
| North America | |||||
| Among populations | 45 | 1740.4 | 0.838 | 2.84 | <0.001 |
| Within populations | 512 | 14645.7 | 28.605 | 97.16 | . |
| Europe | |||||
| Among populations | 21 | 791 | 0.847 | 2.89 | <0.001 |
| Within populations | 220 | 6253.4 | 28.424 | 97.11 | . |
| All two gene pools | |||||
| Between gene pools | 1 | 138.4 | 0.291 | 0.98 | <0.001 |
| Among populations | 66 | 2531.4 | 0.840 | 2.83 | <0.001 |
| Within populations | 732 | 20899 | 28.551 | 96.19 | . |
Native populations
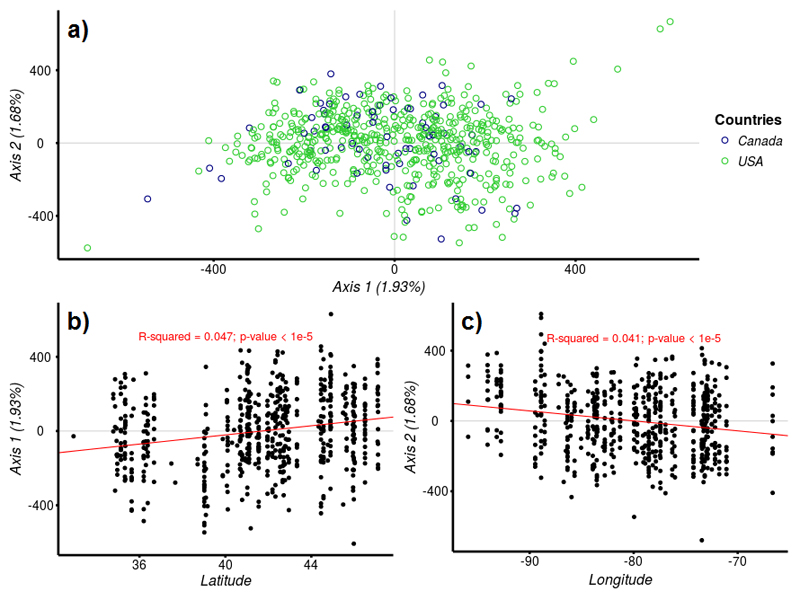
We further explored the distribution of genetic variation within the native gene pool by using a multi-locus approach. We performed a Correspondence Analysis (CA) with individuals from the native range (n=624) showing that projections of individual values along the three main axis were distributed continuously (Fig. 2a). Coordinates of individuals on the first two axes of the CA (3.61% of the variance) indicate a much greater diversity in the populations sampled in the USA compared to those sampled in Canada. We then investigated whether the continuous variation in North America follows a clinal geographic trend. Projections of individuals on the first axis are significantly correlated with the latitude of the origin of the population (Fig. 2b). Similarly, values of the projections on the second axis are significantly correlated with longitude (Fig. 2c). Finally annual mean temperatures, from observed data recorded between 1960 and 1990, were highly correlated with the first axis but not the second (see Appendix Fig. S1). Extant continuous variation across the natural distribution can be the outcome of various different evolutionary scenarios. Two contrasting scenarios illustrate this. On the one hand, one may consider a single founding population disseminating throughout the range and generating genetic clines through isolation by distance. This scenario would have resulted today in a single, large, potentially panmictic population characterized by a latitudinal genetic gradient of variation. On the other hand, we can imagine multiple ancestral populations located at the extremes of the distribution, and dissemination from these different sources would have resulted in continuous genetic variation across the natural distribution of populations as a consequence of admixture and isolation by distance. We investigated some of these scenarios by using clustering methods to explore the existence of ancestral populations and using ABC methods to reconstruct likely demographic histories.
Fig. 2.
a) Correspondence Analysis (CA) of all North American individuals. Green open circles represent individuals from USA and blue open circles represent individuals from Canada. b) Linear regression between projections on the first axis (Axis1) of the Correspondence Analysis (CA) for all North American individuals and latitude of each population origin. c) Linear regression between the projections on the second axis (Axis 2) of CA for all North American individuals and longitude of each population origin.
Ancestral populations
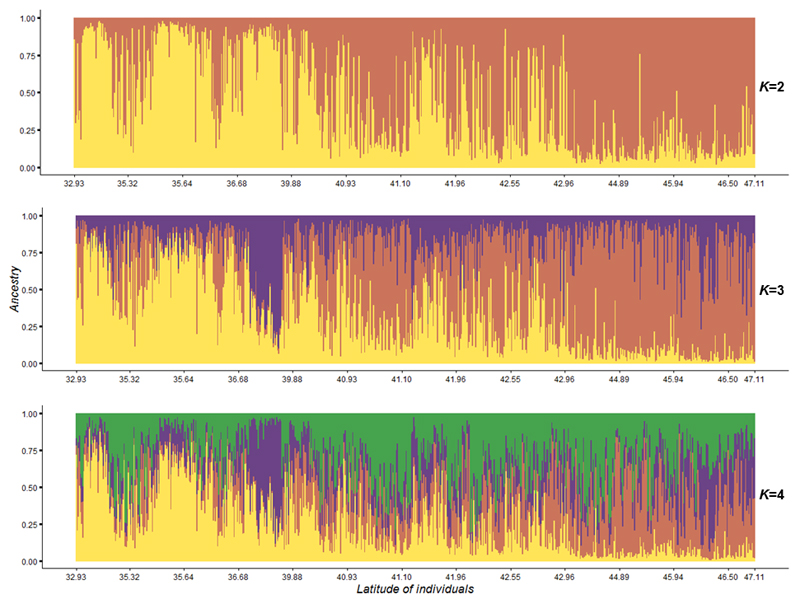
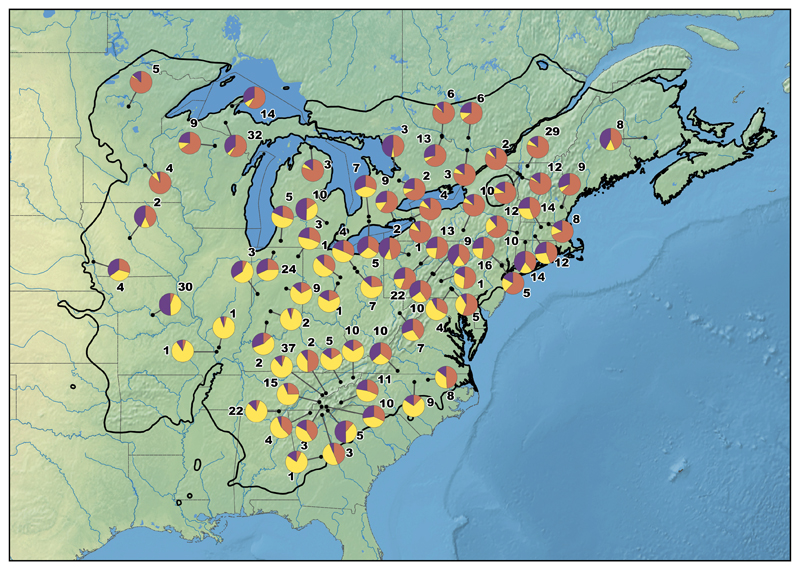
We used TESS - a spatially explicit Bayesian clustering method particularly suited to investigating discontinuities in continuous populations - to report ancestry estimates for K genetic clusters. To estimate the optimal number of clusters, i.e. the number of K that best fit the data, we used the average Deviance Information Criterion (DIC) criteria, which measures the posterior predictive error and allows comparisons of scenarios corresponding to different K values. In our case, DIC values continuously decreased without sudden variation (see Appendix Table S3), which made it impossible to identify the optimal K value using this criterion. Consequently, instead of using the DIC criterion, we checked the assignment of individuals to different genetic clusters (with different K values) and compared the outcomes by considering the number of meaningful groups of individuals. This is an empirical approach attempting to compare the consistency of assignments to clusters as the number of clusters increases. For example, we checked whether clusters at lower K values are nested in clusters when K increases. In what follows and to account for the uncertainty due to the empiricism of our approach, we considered in more detail the two cases with lower values of K (K=2 and K=3). In our study, when K=2, the Bayesian clustering analysis resulted in a latitudinal distribution of the trees from the North (G1, orange) and trees from the South (G2, yellow) and admixed trees at intermediate latitudes (Fig. 3), thus confirming the latitudinal trend observed by CA. At K=3, clustering analysis suggested that the additional cluster (G3, purple) is preferentially present in the Northcentral to Northwestern part of the distribution, although its distribution is more ubiquitous than the other two clusters. This third cluster contained a large group of individuals originating from Missouri (populations MO_2, see Appendix Table S1) and from Northwestern Michigan (populations OT_2 and MTU, see Appendix Table S1) suggesting a genetic split from Northeastern (G1, orange) and Southeastern (G2, yellow) populations (Fig. 3 and 4). At K=4, only 13 individuals (2%) were clearly assigned to one of the four cluster (Q-values above to 0.8), representing a very low proportion of the samples, whereas most individuals were admixed (n=611, 98%). In addition, the clustering appeared to be independent of geography (Fig. 3), an empirical evidence that this value of K is greater than the real number of clusters. Similar conclusions can be drawn for analyses based on more than four genetic clusters. In view of the difficulty of estimating K in our study, all our subsequent analyses have been performed assuming estimates of admixture proportions at K=2 and K=3. In what follows, Bayesian clustering should also be seen here as a pragmatic way for discretizing the continuous variation in order to ease the tracking of the source populations of the European gene pool.
Fig. 3.
Posterior estimates of cluster membership for native individuals (n=624) at distinct values of K (from 2 to 4), obtained with TESS software. Genetic cluster of G1 is represented in orange, G2 in yellow, G3 in purple and G4 in green. Native individuals are arranged from South to North (from left to right).
Fig. 4.
Geographic distribution of the 3 genetic clusters (K=3) detected in North America. Pie diagrams represent the mean assignation of every native population to each genetic cluster and the nearby number indicates the amount of individuals contributing to the population. G1 individuals, Northeastern populations, are indicated in orange, G2, Southeastern populations, in yellow and G3, Northcentral/Northwestern populations, in purple (see main text). The black thick line delineates the natural area of Q. rubra in North America. The raster maps data are free of use and come from Natural Earth website (http://www.naturalearthdata.com/) at 1:50m scale.
Demographic scenarios
We used an Approximate Bayesian Computation (ABC) framework to elucidate occurrences and timing of contacts between ancestral populations. These methods allow explicit tests of primary vs. secondary differentiation hypotheses. Here, seven demographic models of divergence were tested within all pairs of clusters in the native range at K=2 and K=3. We considered each genetic cluster previously identified by the Bayesian clustering analysis as a population and performed our tests on the pair of clusters for each K: K2G1-K2G2, K3G1- K3G2, K3G2- K3G3 and K3G1- K3G3. For each pair of clusters we also considered the scenario of a single panmictic population, assuming the two populations of a pair were actually one single population (Fig. 1). For all pairs of clusters at K=2 and at K=3, the ABC results showed strongest support for models with at least one event of secondary contact, with posterior probabilities ranging from 0.6140 to 0.7619 (Table 3). Models with a single secondary contact (SC) clearly outperformed models assuming two cycles of secondary contacts (Table 3).
Table 3.
Average posterior probabilities for each of the seven models simulated (PAN: panmixia, SI: strict isolation, AM: ancient migration, IM: isolation migration, SC: secondary contact, PAM: periodic ancient migration; PSC: periodic secondary contact) and each pair of genetic clusters at K=2 and at K=3 in the native range. Standard deviation is given in square bracket Best models are indicated in bold.
| Range | K | Clusters (Pop A vs Pop B) |
PAN | SI | AM | IM | SC | PAM | PSC |
|---|---|---|---|---|---|---|---|---|---|
| Native | 2 | G1_vs_G2 | 0.0000 [±0.0000] | 0.0011 [±0.0018] | 0.0204 [±0.0142] | 0.0371 [±0.0058] | 0.7314 [±0.0309] | 0.0205 [±0.0140] | 0.1895 [±0.0107] |
| Native | 3 | G1_vs_G2 | 0.0001 [±0.0001] | 0.0010 [±0.0003] | 0.0779 [±0.0123] | 0.0592 [±0.0070] | 0.6140 [±0.0203] | 0.0759 [±0.0109] | 0.1719 [±0.0130] |
| Native | 3 | G1_vs_G3 | 0.0000 [±0.0000] | 0.0001 [±0.0002] | 0.0020 [±0.0014] | 0.0110 [±0.0026] | 0.7619 [±0.0093] | 0.0015 [±0.0015] | 0.2235 [±0.0084] |
| Native | 3 | G2_vs_G3 | 0.0000 [±0.0000] | 0.0000 [±0.0000] | 0.0037 [±0.0009] | 0.0176 [±0.0024] | 0.7543 [±0.0118] | 0.0039 [±0.0009] | 0.2205 [±0.0087] |
For each pair of clusters at K=2 and K=3, demographic parameters were estimated under the best-fitting model SC (see Appendix Table S4). Posterior distributions of parameters were built from 10,000 best simulations. Among all parameters, the ancestral population sizes (NANC), migration rates (MA and MB) and divergence times (TSPLIT) were found to be poorly differentiated from their prior distributions. Conversely, the timing of secondary contact (TSC) was quite well estimated and suggests that secondary gene flow occurred recently, in the last 2.08% of the divergence time between K3G1 and K3G3 (median= 0.33%), and the last 5.70% of the divergence time between K3G2 and K3G3 (median= 0.41%). The range of the posterior distribution of the divergence time between K3G1 and K3G2 is larger, suggesting that secondary contact occurred in the last 65.48% of the divergence time (median= 9.32%) (see Appendix Table S4). Using leave-one-out cross validations based on 15,000 pseudo-observed datasets (PODS) for each model, we estimated a robustness over 0.99 in support of secondary contact for all pairwise comparisons (see Appendix Fig. S2).
Introduced populations
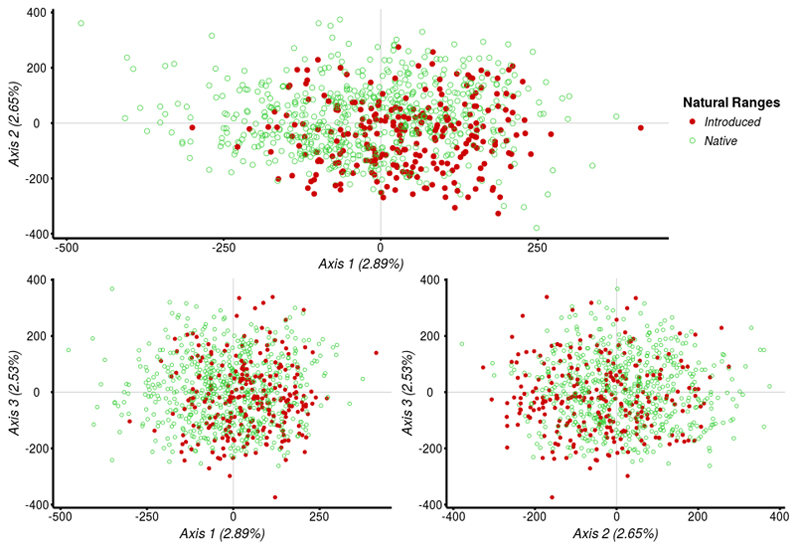
To identify the source clusters of introduced populations in Europe, we performed the same population structure analyses as conducted previously, but with the inclusion of 38 additional populations (259 introduced individuals) from the introduced range, collected from 7 European countries. The Correspondence Analysis based on all native and introduced populations showed that coordinates of European individuals are also continuously distributed on the three first axes of the CA (8.07% of the total variance, Fig. 5). Introduced populations do not show a random distribution in the space of the three CA axes.
Fig. 5.
Correspondence Analysis (CA) of all North American and European individuals. Individuals from different ranges are shown by different colors: green open circles represent individuals from the native range and red closed circles individuals from the introduced range.
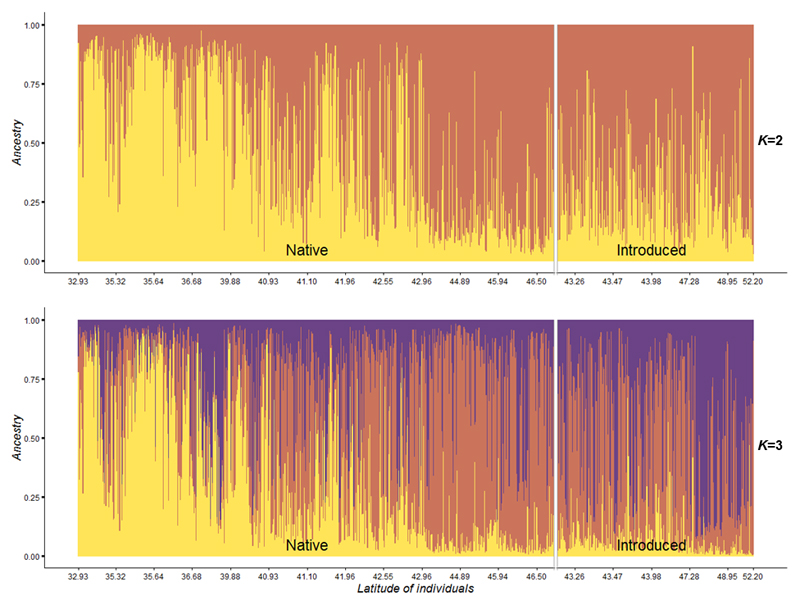
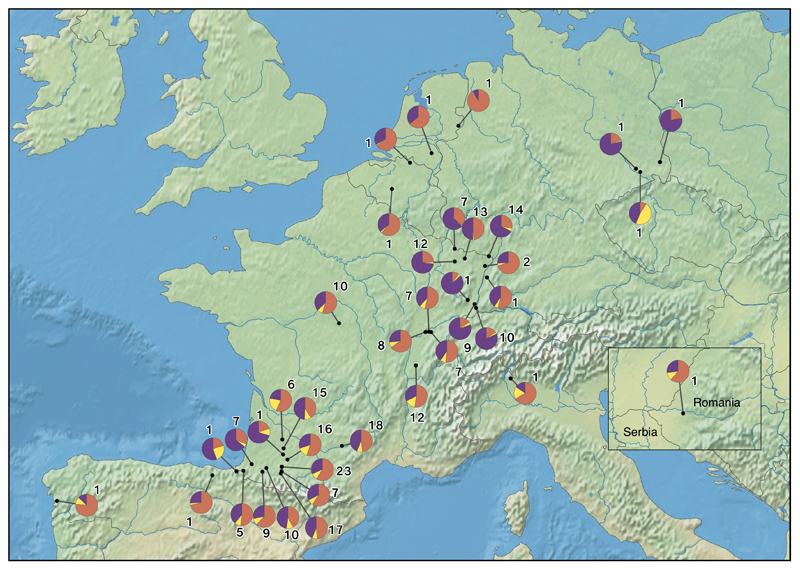
We conducted the Bayesian clustering over the whole data set comprising native and introduced individuals to explore the likely origin of the extant European gene pool. We assume in this analysis that introduced populations originated from one or more of the ancestral populations of the native range that we sampled previously. Thus the Bayesian clustering analysis were conducted at K=2 and K=3 as within the native gene pool (Fig. 6). At K=2, most European individuals (n=107, 41%) were mainly assigned (Q-values equal or above to 0.8) to the genetic cluster of Northeastern native populations (G1, orange). Only a very low proportion of samples (n=3, 1%) were strongly assigned to the other cluster (G2, yellow). The remaining (n=149, 58%) exhibited Q-values below 0.8 and were considered to be admixed. At K=3, individuals were near-exclusively assigned to two of the three native genetic clusters: Northeastern and Northcentral/Northwestern native clusters (G1, orange and G3, purple; Fig. 6). In this case, only 20.5% (n=53) were mostly assigned to the cluster G1, 13% (n=34) to the cluster G3 and none to the cluster G2. At K=3, most individuals (n=172, 66.5%) did not have a well-defined membership to any cluster (Q-values < 0.8) and were considered admixed. No geographical structure across Europe was observed (Fig. 7).
Fig. 6.
Posterior estimates of cluster membership for native and introduced individuals (respectively n=624 and n=259) at K=2 and K=3, obtained with TESS software. Genetic cluster of G1 is represented in orange, G2 in yellow and G3 in purple. Individuals are arranged according to their latitudinal coordinates from South to North (from left to right).
Fig. 7.
Geographic distribution of the 3 genetic clusters (K) found in Europe. Pie diagrams represent the mean assignation of every introduced population to each genetic cluster and the nearby number indicates the amount of individuals contributing to population. G1 individuals, Northeastern populations, are indicated in orange, G2, Southeastern populations, in yellow and G3, Northcentral/Northwestern populations, in purple. The raster maps data are free of use and come from Natural Earth website (http://www.naturalearthdata.com/) at 1:50m scale.
Discussion
In this study, our main goal was to retrace historical and demographic events that would have been associated with the introduction of Q. rubra to Europe. We proceeded stepwise by first deciphering the present genetic structure in the native distribution only. We then compared the structure in the introduced range with the extant structure in the native range and inferred putative native sources of the introduced gene pools. Overall our efforts and expectations were hampered by the low genetic differentiation of Q. rubra in its natural range (Table 2). There have been few range-wide explorations of gene diversity in Q. rubra. Earlier reports mentioned contrasting levels of differentiation (FST = 0.092 in allozymes, Sork et al. 1993; GST = 0.018 in allozymes, Daubree and Kremer 1993; FST = 0.043 in microsatellites, Borkowski et al. submitted), likely due to quite different sampling strategies. Other genetic surveys conducted on continental-wide distributed oak species report levels similar to what we found here (GST = 0.025 in Quercus petraea, Zanetto and Kremer 1995; GST = 0.020 in Quercus robur, Mariette et al. 2002). Differentiation between the introduced and native gene pool was even lower, about 1% (Table 2). Despite the overall low differentiation, we found a continuous trend of genetic variation across the natural distribution (Fig. 2). Our attempts to track the source population of introduced populations were therefore also constrained by the continuous genetic variation. To overcome these difficulties, we attempted to discretize the continuous variation by exploring the existence of ancestral populations that would have contributed through admixture to the extant continuous variation. We considered different testable scenarios that would cope to the extant distribution of genetic variation in the natural range and ease the search of source population of European populations, although they would not reproduce exactly the evolutionary history. We therefore adopted a heuristic approach by considering multiple scenarios and found that their outcomes resulted in congruent conclusions that the European populations most likely originated from the Northeastern part of the natural distribution.
Continuous geographic variation in the natural range
Our findings of continuous geographic variations are well illustrated by the correlation between the first axis of the correspondence analysis and latitude and longitude (Fig. 2b, c). These results contrast with a previous study using chloroplast DNA markers reporting only weak clinal variation in North America (Magni et al. 2005). Although chloroplast DNA markers usually exhibit very high genetic differentiation in oaks due to limited seed dispersal (Petit et al. 2002a, 2002b, 2002c), these markers provide an incomplete view of the genetic structure of populations as they are maternally inherited. In addition, chloroplast genome capture and swamping occurring during colonization dynamics may blur and hide the background nuclear genetic structure (Kremer et al. 2002; Leroy et al. 2017). Alternatively, high nuclear genetic structure may be present even with very low or no chloroplast population structure (Birchenko et al. 2009; Hoban et al. 2010; Laricchia et al. 2015).
A recent range wide genetic survey based on microsatellites also detected a latitudinal trend of variation but showed a higher divergence at the Northwestern part of the range (Borkowski et al. submitted). Apart from genetic marker data, the comparison of phenotypic traits among populations in provenance-progeny tests highlighted genetic trends across the entire native range. Phenological traits such as the timing of leaf budburst and leaf coloration exhibited strong differentiation among provenances from different geographic origins (Kriebel 1993). Timing of budburst was correlated with longitude whereas leaf coloration showed latitudinal variation (Deneke 1974; Kriebel et al. 1976; Schlarbaum and Bagley 1981). According to results obtained in range-wide provenance tests established in middle latitudes of the species range from eastern Nebraska to Northern Ohio, no latitudinal or longitudinal trend was observed for growth but populations coming from the Northern part of the range (extending from the Mississippi river to Western Maine) grew faster than other provenances (Schlarbaum and Bagley 1981; Kriebel et al. 1988; Kriebel 1993). Populations located to the West of the range limits, in Iowa, Kansas and Missouri for example, were more drought resistant and had higher survival compared to other populations of the natural range (Deneke 1974). Overall there are some congruent patterns observed between molecular genetic surveys (our results and Borkowski et al. submitted) and previous investigations conducted in common garden experiments, suggesting such potential common causes as divergent selection across large environmental gradients. However, these gradients overlap also with historical pathways of colonization (Schlarbaum et al. 1982), which may generate signatures similar to those of demographic processes. Further investigations are needed to disentangle these sources of variation.
Exploring evolutionary scenarios in the natural range
Whether the geographic trends of variation we found in the natural range bear the signatures of historical demographic events, divergent selection, or a combination of both remains an open debate. As already mentioned, demographic scenarios may entail multiple ancestral populations that disseminated across the landscape, finally generating continuous gradients of variation as a result of admixture and isolation by distance. By using Bayesian structuring analysis, we considered two scenarios (K=2 and K=3), and by explicitly exploring evolution within a single panmictic population in the ABC analysis, we also considered the case of a single ancestral population. Posterior probabilities in the ABC analysis clearly excluded the case of a single ancestral population and the two cases (K=2 and K=3) resulted in very congruent results, confirming the latitudinal trend, and suggesting two clusters preferentially distributed in the Northeast and the Southwest. When K=3, an additional more ubiquitous group was identified located more frequently in the Northcentral to the Northwestern part of the range. There might be more ancestral populations, but additional clustering will not erase the latitudinal structure. It will only refine the subdivision, but would likely require the use of more SNPs to be detected.
One question we addressed is whether divergence between these two (or three) different genetic clusters is ancient. Indeed, divergence can be the result of a recent process of isolation (“primary divergence”) or more ancient as expected under secondary contact (SC) scenarios. For all pairs of clusters, our ABC analyses found support for models assuming at least an event of secondary contact, with higher support for models assuming a single period of SC. In addition, our inferences suggested that secondary contact is very recent as compared with the period of strict isolation and showed that the high level of admixture explains the continuous pattern of genetic variation in red oak populations. More broadly, the compilation of all our ABC analyses suggests that these two (or three) different genetic groups have been separated in at least two (or three) different regions during a long part of their history, or were somehow isolated for a long period of time. One explanation for this apparent allopatric isolation is different climatic refuges, as already suspected between four species of the European white oak complex (Leroy et al. 2017). In the continental United States, the advances and retreats of Pleistocene glaciations for the last 800,000 years occurred over four major cycles, with complex patterns of retreats and advances (Balco and Rovey 2010). Long established regional populations of Q. rubra could have come into secondary contact during the migrations that must have occurred during these climate shifts.
Deciphering the origin of European populations
To decipher the origin of the current Q. rubra populations in Europe, we performed a second Bayesian clustering analysis based on both native and introduced populations (Fig. 6). The two cases (K=2 and K=3) came to the same conclusions. Trees of introduced populations are mostly assigned to one (when K=2) or two (K=3) ancestral populations of the natural range. To sum up, the likely geographic origin of introduced population extends in the Northern part of the natural range. Indeed European populations are more similar in terms of admixture profiles to native populations present in the Northern part (Fig. 6 and 7). However our results are not clear-cut to the point to exclude entirely origins from Southern latitudes. These results are quite consistent with historical documents reporting numerous introductions of Q. rubra in Europe from Northeastern North America during the eighteenth century, probably due to the shortest maritime route between Northern states and the ports of Western Europe, as suggested by Bauer (1954), who first raised the issue of the origin of introduced populations in Europe. Nevertheless, there are also a few records of trade exchanges with Southern states occurring at the beginning of the nineteenth century (United States. Bureau of the Census 1975). Whether acorns of red oak were introduced during that period is unknown. If it ever had been the case, then our results suggest that Southern origins have been largely extirpated since introduction either through natural selection or human interferences. Probably, Northeastern and Northcentral/Northwestern populations could have managed better to adapt to environmental conditions encountered in Europe. As mentioned by Colautti and Barrett (2013) and Hamilton et al. (2015), rapid adaptation to local climatic conditions favors survival and spread of introduced populations in new habitats.
It is worth mentioning that Q. rubra was earlier named Quercus borealis F. Michx, during which time botanists and foresters recognized Q. borealis var. maxima (Marshall) Sargent as a morphologically distinct variety based on acorn shape and higher frequency in New England (Palmer 1942). This variety was still recognized in the 1950s by European foresters (Bauer 1953; Göhre and Wagenknecht 1955), and it might well be a clue for understanding the preferential seed sourcing in Northeastern states where the pure type Q. borealis was supposedly more frequent. However, we cannot exclude the possibility that Southern native populations were never introduced in Europe. Further investigations are needed based on additional markers and phenotypic traits assessed in common garden experiments including populations of both origins.
Concluding remarks
Despite the continental presence of northern red oak in Europe, no information was available before the current study about the diversity and origins of the introduced gene pool in comparison to the native gene pool. In the current study, we investigate the likely origin of introduced populations by conducting a comparative analysis of the distribution and structure of genetic variation in the native and introduced ranges. Our task was hampered by the very weak structure in the natural range that is illustrated by the continuous pattern of genetic variation along geographic gradients. We explored ways of discretizing the continuous variation with the aim of facilitating identification of source populations of the European gene pool. This approach has shortcomings as it disrupts the underlying clinal pattern, but it results nonetheless in congruent conclusions under different scenarios, showing that the source populations of the European gene pool are likely located in the Northern part of the natural distribution. Refinements of the geographic origin may improve if the number of markers and studied populations is increased. These investigations should now be extended to explore whether evolutionary change (adaptive or neutral) has accumulated since introduction in the European gene pool, to understand the evolutionary success of the species in Europe.
Supplementary Material
Acknowledgements
We thank C. Firmat, B. Dencausse, R. Ségura, F. Bonne, T. Paul and V. Rousselet for helping during sampling collection in the field in France, R. Robichaud and T. McCleary for sampling in North America, P. Leger for helping during DNA extraction and the team of the Genome-Transcriptome platform for helping during genotyping. T. L. wants to thank C. Roux and Q. Rougemont for a long-term collaboration on demographic inferences and Approximate Bayesian Computations. We are grateful to the genotoul bioinformatics platform Toulouse Midi-Pyrenees (Bioinfo Genotoul) for providing computing resources. Part of the experiments (genotyping) were performed at the Genomic and Transcriptomic Facility of Bordeaux (grants from the Conseil Regional d’Aquitaine 20030304002FA and 20040305003FA and from the European Union, FEDER 2003227 and from Investissements d'avenir, Convention attributive d’aide N°ANR-10-EQPX-16-01). This study was carried out with financial support from the French National Research Agency (ANR) in the frame of the Investments for the future Program (ANR-10- IDEX-03-02) and from the European Research Council under the Advanced Grant TREEPEACE (#FP7-339728). BIOGECO is supported by a grant overseen by the French National Research Agency (ANR) as part of the “Investissements d’Avenir” program Labex COTE.
Contributor Information
Thibault Leroy, Email: thibault.leroy@pierroton.inra.fr.
Emilie Chancerel, Email: emilie.chancerel@inra.fr.
Jeanne Romero-Severson, Email: Jeanne.Romero-Severson.1@nd.edu.
Daniel Borkowski, Email: dborkowski@howemilitary.org.
Alexis Ducousso, Email: alexis.ducousso@pierroton.inra.fr.
Arnaud Monty, Email: arnaud.monty@ulg.ac.be.
Annabel J. Porté, Email: annabel.porte@pierroton.inra.fr.
References
- Allendorf FW, Lundquist LL. Introduction: Population Biology, Evolution, and Control of Invasive Species. Conserv Biol. 2003;17(1):24–30. doi: 10.1046/j.1523-1739.2003.02365.x. [DOI] [Google Scholar]
- Balco G, Rovey CW. Absolute chronology for major Pleistocene advances of the Laurentide ice Sheet. Geology. 2010;38(9):795–798. doi: 10.1130/G30946.1. [DOI] [Google Scholar]
- Bauer F. In: Die Roteiche. Sauerländer’s Verlag, editor. Frankfurt Am Main: 1953. [Google Scholar]
- Bauer F. Zur Rassenfrage der Roteiche. Allg. Forstzeitschrift. 1954;9:470–474. [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian Computation in Population Genetics. Genetics. 2002;162(4):2025–2035. doi: 10.1093/genetics/162.4.2025. Available from http://www.genetics.org/content/162/4/2025.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000. Université de Montpellier II; Montpellier (France): 2004. [Google Scholar]
- Bigsby KM, Tobin PC, Sills EO. Anthropogenic drivers of gypsy moth spread. Biol Invasions. 2011;13(9):2077–2090. doi: 10.1007/s10530-011-0027-6. [DOI] [Google Scholar]
- Birchenko I, Feng Y, Romero-Severson J. Biogeographical Distribution of Chloroplast Diversity in Northern Red Oak (Quercus rubra L.) Am Midl Nat. 2009;161(1):134–145. doi: 10.1674/0003-0031-161.1.134. [DOI] [Google Scholar]
- Blackburn TM, Lockwood JL, Cassey P. The influence of numbers on invasion success. Mol Ecol. 2015;24(9):1942–1953. doi: 10.1111/mec.13075. [DOI] [PubMed] [Google Scholar]
- Borkowski DS, Hoban SM, Chatwin WB, Romero-Severson J. Response to rangewide disturbances influences values of population differentiation in Quercus rubra L. 2016 [submitted] [Google Scholar]
- Bossdorf O, Auge H, Lafuma L, Rogers WE, Siemann E, Prati D. Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia. 2005;144(1):1–11. doi: 10.1007/s00442-005-0070-z. [DOI] [PubMed] [Google Scholar]
- Buswell JM, Moles AT, Hartley S. Is rapid evolution common in introduced plant species? J Ecol. 2011;99(1):214–224. doi: 10.1111/j.1365-2745.2010.01759.x. [DOI] [Google Scholar]
- Castric V, Bechsgaard J, Schierup MH, Vekemans X. Repeated Adaptive Introgression at a Gene under Multiallelic Balancing Selection. PLoS Genet. 2008;4(8):e1000168. doi: 10.1371/journal.pgen.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. Fundamental concepts in genetics: Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10(3):195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Nordborg M, Charlesworth D. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet Res. 1997;70(2):155–174. doi: 10.1017/s0016672397002954. [DOI] [PubMed] [Google Scholar]
- Chen C, Durand E, Forbes F, François O. Bayesian clustering algorithms ascertaining spatial population structure : a new computer program and a comparison study. Mol Ecol. 2007;7:747–756. doi: 10.1111/j.1471-8286.2007.01769.x. [DOI] [Google Scholar]
- Clout MN, Russell JC. The invasion ecology of mammals: A global perspective. Wildl Res. 2008;35(3):180–184. doi: 10.1071/WR07091. [DOI] [Google Scholar]
- Colautti RI, Barrett SCH. Rapid adaptation to climate facilitates range expansion of an invasive plant. Science. 2013;342(6156):364–6. doi: 10.1126/science.1242121. [DOI] [PubMed] [Google Scholar]
- Cristescu ME. Genetic reconstructions of invasion history. Mol Ecol. 2015;24(9):2212–2225. doi: 10.1111/mec.13117. [DOI] [PubMed] [Google Scholar]
- Cruickshank TE, Hahn MW. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 2014;23(13):3133–3157. doi: 10.1111/mec.12796. [DOI] [PubMed] [Google Scholar]
- Csilléry K, Blum MGB, Gaggiotti OE, François O. Approximate Bayesian Computation (ABC) in practice. Trends Ecol Evol. 2010;25(7):410–418. doi: 10.1016/j.tree.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Csilléry K, François O, Blum MGB. abc: an R package for approximate Bayesian computation (ABC) Methods Ecol Evol. 2012;3(3):475–479. doi: 10.1111/j.2041-210X.2011.00179.x. [DOI] [Google Scholar]
- Daubree J, Kremer A. Genetic and phenological differentiation between introduced and natural populations of Quercus rubra L. Ann des Sci For. 1993;50(Suppl 1):271s–280s. doi: 10.1051/forest:19930727. [DOI] [Google Scholar]
- Deneke FJ. A Red Oak Provenance Trial in Kansas. Trans Kansas Acad Sci. 1974;77(3):195. doi: 10.2307/3627319. [DOI] [Google Scholar]
- Desprez-Loustau M-L, Robin C, Buée M, Courtecuisse R, Garbaye J, Suffert F, Sache I, Rizzo DM. The fungal dimension of biological invasions. Trends Ecol Evol. 2007;22(9):472–480. doi: 10.1016/j.tree.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Dlugosch KM, Parker IM. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol. 2008;17(1):431–49. doi: 10.1111/j.1365-294X.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- Durand E, Jay F, Gaggiotti OE, Francois O. Spatial Inference of Admixture Proportions and Secondary Contact Zones. Mol Biol Evol. 2009;26(9):1963–1973. doi: 10.1093/molbev/msp106. [DOI] [PubMed] [Google Scholar]
- Durka W, Bossdorf O, Prati D, Auge H. Molecular evidence for multiple introductions of garlic mustard (Alliaria petiolata, Brassicaceae) to North America. Mol Ecol. 2005;14(6):1697–1706. doi: 10.1111/j.1365-294X.2005.02521.x. [DOI] [PubMed] [Google Scholar]
- Ehrenfeld JG. Ecosystem Consequences of Biological Invasions. Annu Rev Ecol Evol Syst. 2010;41(1):59–80. doi: 10.1146/annurev-ecolsys-102209-144650. [DOI] [Google Scholar]
- Estoup A, Guillemaud T. Reconstructing routes of invasion using genetic data: Why, how and so what? Mol Ecol. 2010;19(19):4113–4130. doi: 10.1111/j.1365-294X.2010.04773.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1007/s00424-009-0730-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facon B, Genton BJ, Shykoff J, Jarne P, Estoup A, David P. A general eco-evolutionary framework for understanding bioinvasions. Trends Ecol Evol. 2006;21(3):130–135. doi: 10.1016/j.tree.2005.10.012. [DOI] [PubMed] [Google Scholar]
- François O, Ancelet S, Guillot G. Bayesian clustering using hidden Markov random fields in spatial population genetics. Genetics. 2006;174(2):805–16. doi: 10.1534/genetics.106.059923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genton BJ, Shykoff JA, Giraud T. High genetic diversity in French invasive populations of common ragweed, Ambrosia artemisiifolia, as a result of multiple sources of introduction. Mol Ecol. 2005;14(14):4275–4285. doi: 10.1111/j.1365-294X.2005.02750.x. [DOI] [PubMed] [Google Scholar]
- Gladieux P, Zhang XG, Róldan-Ruiz I, Caffier V, Leroy T, Devaux M, Van Glabeke S, Coart E, Le Cam B. Evolution of the population structure of Venturia inaequalis, the apple scab fungus, associated with the domestication of its host. Mol Ecol. 2010;19(4):658–674. doi: 10.1111/j.1365-294X.2009.04498.x. [DOI] [PubMed] [Google Scholar]
- Goeze . Liste der seit dem 16. Jahrhundert bis auf die Gegenwart in die Gärten uns Parks Europas eingführten Bäume und Stäucher. Mitteilungen der Deutschen Dendrologischen Gesellschaft; 1916. [Google Scholar]
- Göhre K, Wagenknecht E, editors. Die Roteiche und ihr Holz. Deutscher Bauernverlag; Berlin: 1955. [Google Scholar]
- Hamilton JA, Okada M, Korves T, Schmitt J. The role of climate adaptation in colonization success in Arabidopsis thaliana . Mol Ecol. 2015;24(9):2253–2263. doi: 10.1111/mec.13099. [DOI] [PubMed] [Google Scholar]
- Hoban SM, Borkowski DS, Brosi SL, McCleary TS, Thompson LM, McLachlan JS, Pereira MA, Schlarbaum SE, Romero-Severson J. Range-wide distribution of genetic diversity in the North American tree Juglans cinerea: a product of range shifts, not ecological marginality or recent population decline. Mol Ecol. 2010;19(22):4876–4891. doi: 10.1111/j.1365-294X.2010.04834.x. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18(2):337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hulme PE. Trade, transport and trouble: Managing invasive species pathways in an era of globalization. J Appl Ecol. 2009;46(1):10–18. doi: 10.1111/j.1365-2664.2008.01600.x. [DOI] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23(14):1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24(11):1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- Jombart T, Ahmed I. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27(21):3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RAC. Plant virus emergence and evolution: Origins, new encounter scenarios, factors driving emergence, effects of changing world conditions, and prospects for control. Virus Res. 2009;141(2):113–130. doi: 10.1016/j.virusres.2008.07.028. [DOI] [PubMed] [Google Scholar]
- Keller SR, Taylor DR. Genomic admixture increases fitness during a biological invasion. J Evol Biol. 2010;23(8):1720–1731. doi: 10.1111/j.1420-9101.2010.02037.x. [DOI] [PubMed] [Google Scholar]
- Konar A, Choudhury O, Bullis R, Fiedle L, Kruser JM, Stephens MT, Gailing O, Schlarbaum SE, Coggeshall MV, Staton ME, Carlson JE, et al. The promise and performance of ddRADseq for constructing a dense genetic linkage map in the non-model tree Quercus rubra. 2016 doi: 10.1186/s12864-017-3765-8. [submitted] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour. 2015:1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer A, Kleinschmit J, Cottrell J, Cundall EP, Deans JD, Ducousso A, König AO, Lowe AJ, Munro RC, Petit RJ, Stephan BR. Is there a correlation between chloroplastic and nuclear divergence, or what are the roles of history and selection on genetic diversity in European oaks? For Ecol Manage. 2002;156(1–3):75–87. doi: 10.1016/S0378-1127(01)00635-1. [DOI] [Google Scholar]
- Kriebel H. Intraspecific variation of growth and adaptive traits in North American oak species. Ann des Sci For. 1993;50:153s–165s. doi: 10.1051/forest:19930715. [DOI] [Google Scholar]
- Kriebel HB, Bagley WT, Deneke FJ, Funsch RW, Roth P, Jokela JJ, Merritt C, Wright JW, Williams RD. Geographic variation in Quercus rubra in north central United States plantations. Silvae Genet. 1976;25(3-4):118–122. [Google Scholar]
- Kriebel HB, Merritt C, Stadt T. Genetics of Growth Rate in Quercus rubra: Provenance and Family Effects by the Early Third Decade in the North Central USA. Silvae Genet. 1988;37:193–198. [Google Scholar]
- Lanier L, Keller R, Kremer A. Le Chêne rouge (Quercus rubra L.) en France. Rev For Française. 1980;32(5):419. doi: 10.4267/2042/21428. [DOI] [Google Scholar]
- Laricchia KM, McCleary TS, Hoban SM, Borkowski D, Romero-Severson J. Chloroplast haplotypes suggest preglacial differentiation and separate postglacial migration paths for the threatened North American forest tree Juglans cinerea L. Tree Genet Genomes. 2015;11(2):30. doi: 10.1007/s11295-015-0852-3. [DOI] [Google Scholar]
- Lavergne S, Molofsky J. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc Natl Acad Sci U S A. 2007;104(10):3883–8. doi: 10.1073/pnas.0607324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehe R, Hallatschek O, Peliti L. The Rate of Beneficial Mutations Surfing on the Wave of a Range Expansion. PLoS Comput Biol. 2012;8(3):e1002447. doi: 10.1371/journal.pcbi.1002447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprieur F, Beauchard O, Blanchet S, Oberdorff T, Brosse S. Fish Invasions in the World’s River Systems: When Natural Processes Are Blurred by Human Activities. PLoS Biol. 2008;6(2):e28. doi: 10.1371/journal.pbio.0060028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy T, Le Cam B, Lemaire C. When virulence originates from non-agricultural hosts: New insights into plant breeding. Infect Genet Evol. 2014;27:521–529. doi: 10.1016/j.meegid.2013.12.022. [DOI] [PubMed] [Google Scholar]
- Leroy T, Roux C, Villate L, Bodénès C, Romiguier J, Paiva JAP, Dossat C, Aury J-M, Plomion C, Kremer A. Extensive recent secondary contacts between four European white oak species. New Phytol. 2017 doi: 10.1111/nph.14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magni CR, Ducousso A, Caron H, Petit RJ, Kremer A. Chloroplast DNA variation of Quercus rubra L. in North America and comparison with other Fagaceae. Mol Ecol. 2005;14(2):513–524. doi: 10.1111/j.1365-294X.2005.02400.x. [DOI] [PubMed] [Google Scholar]
- Magni Diaz CR. Reconstitution de l’introduction de Quercus rubra L. en Europe et conséquences génétiques dans les populations allochtones. Thèse de doctorat; Ecole Nationale du Génie Rural des Eaux et Forêts, Paris, FRANCE: 2004. [Google Scholar]
- Major KC, Nosko P, Kuehne C, Campbell D, Bauhus J. Regeneration dynamics of non-native northern red oak (Quercus rubra L.) populations as influenced by environmental factors: A case study in managed hardwood forests of southwestern Germany. For Ecol Manage. 2013;291:144–153. doi: 10.1016/j.foreco.2012.12.006. [DOI] [Google Scholar]
- Mariette S, Cottrell J, Csaikl UM, Goikoechea P, König A, Lowe AJ, Van Dam BC, Barreneche T, Bodénès C, Streiff R, Burg K, et al. Comparison of levels of genetic diversity detected with AFLP and microsatellite markers within and among mixed Q. petraea (MATT.) LIEBL. and Q. robur L. stands. Silvae Genet. 2002;51(2–3):72–79. [Google Scholar]
- Maron JL, Vilà M, Bommarco R, Elmendorf S, Beardsley P. Rapid evolution of an invasive plant. Ecol Monogr. 2004;74(2):261–280. doi: 10.1890/03-4027. [DOI] [Google Scholar]
- Miller JR. Survival of mutations arising during invasions. Evol Appl. 2010;3(2):109–121. doi: 10.1111/j.1752-4571.2010.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura O. Molecular genetic approaches to elucidate the ecological and evolutionary issues associated with biological invasions. Ecol Res. 2007;22(6):876–883. doi: 10.1007/s11284-007-0389-5. [DOI] [Google Scholar]
- Palmer EJ. The Red Oak Complex in the United States. Am Midl Nat. 1942;27(3):732–740. doi: 10.2307/2420922. [DOI] [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research - an update. Bioinformatics. 2012;28(19):2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchar L, Mooney HA. Invasive species, ecosystem services and human well-being. Trends Ecol Evol. 2009;24(9):497–504. doi: 10.1016/j.tree.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Brewer S, Bordács S, Burg K, Cheddadi R, Coart E, Cottrell J, Csaikl UM, van Dam B, Deans JD, Espinel S, et al. Identification of refugia and post-glacial colonisation routes of European white oaks based on chloroplast DNA and fossil pollen evidence. For Ecol Manage. 2002a;156(1–3):49–74. doi: 10.1016/S0378-1127(01)00634-X. [DOI] [Google Scholar]
- Petit RJ, Csaikl UM, Bordács S, Burg K, Coart E, Cottrell J, van Dam B, Deans JD, Dumolin-Lapègue S, Fineschi S, Finkeldey R, et al. Chloroplast DNA variation in European white oaks Phylogeography and patterns of diversity based on data from over 2600 populations. For Ecol Manage. 2002b;156:5–26. doi: 10.1016/S0378-1127(01)00645-4. [DOI] [Google Scholar]
- Petit RJ, Latouche-Hallé C, Pemonge M-H, Kremer A. Chloroplast DNA variation of oaks in France and the influence of forest fragmentation on genetic diversity. For Ecol Manage. 2002c;156(1–3):115–129. doi: 10.1016/S0378-1127(01)00638-7. [DOI] [Google Scholar]
- Pyšek P, Jarošík V. Invasive Plants: Ecological and Agricultural Aspects. Birkhäuser-Verlag; Basel: 2005. Residence time determines the distribution of alien plants; pp. 77–96. [DOI] [Google Scholar]
- Pyšek P, Jarosik V, Hulme PE, Kühn I, Wild J, Arianoutsou M, Bacher S, Chiron F, Didziulis V, Essl F, Genovesi P, et al. Disentangling the role of environmental and human pressures on biological invasions across Europe. Proc Natl Acad Sci. 2010;107(27):12157–12162. doi: 10.1073/pnas.1002314107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyšek P, Křivánek M, Jarošik V. Planting intensity, residence time, and species traits determine invasion success of alien woody species. Ecology. 2009;90(10):2734–2744. doi: 10.1890/08-0857.1. [DOI] [PubMed] [Google Scholar]
- Pyšek P, Richardson DM. Invasive species, environmental change, and health. Annu Rev Environ Resour. 2010;35:25–55. doi: 10.1146/annurev-environ-033009-095548. [DOI] [Google Scholar]
- Reichard SH, White P. Horticulture as a Pathway of Invasive Plant Introductions in the United States. Bioscience. 2001;51(2):103. doi: 10.1641/0006-3568(2001)051[0103:HAAPOI]2.0.CO;2. [DOI] [Google Scholar]
- Richardson DM, Carruthers J, Hui C, Impson FAC, Miller JT, Robertson MP, Rouget M, Le Roux JJ, Wilson JRU. Human-mediated introductions of Australian acacias - a global experiment in biogeography. Divers. Distrib. 2011;17(5):771–787. doi: 10.1111/j.1472-4642.2011.00824.x. [DOI] [Google Scholar]
- Rius M, Darling JA. How important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol Evol. 2014;29(4):233–242. doi: 10.1016/j.tree.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Rollins LA, Moles AT, Lam S, Buitenwerf R, Buswell JM, Brandenburger CR, Flores-Moreno H, Nielsen KB, Couchman E, Brown GS, Thomson FJ, et al. High genetic diversity is not essential for successful introduction. Ecol Evol. 2013;3(13):4501–4517. doi: 10.1002/ece3.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. DISTRUCT: A program for the graphical display of population structure. Mol Ecol Notes. 2004;4(1):137–138. doi: 10.1046/j.1471-8286.2003.00566.x. [DOI] [Google Scholar]
- Ross-Ibarra J, Wright SI, Foxe JP, Kawabe A, DeRose-Wilson L, Gos G, Charlesworth D, Gaut BS. Patterns of Polymorphism and Demographic History in Natural Populations of Arabidopsis lyrata. PLoS One. 2008;3(6):e2411. doi: 10.1371/journal.pone.0002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C, Castric V, Pauwels M, Wright SI, Saumitou-Laprade P, Vekemans X. Does Speciation between Arabidopsis halleri and Arabidopsis lyrata Coincide with Major Changes in a Molecular Target of Adaptation? PLoS One. 2011;6(11):e26872. doi: 10.1371/journal.pone.0026872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C, Tsagkogeorga G, Bierne N, Galtier N. Crossing the Species Barrier: Genomic Hotspots of Introgression between Two Highly Divergent Ciona intestinalis Species. Mol Biol Evol. 2013;30(7):1574–1587. doi: 10.1093/molbev/mst066. [DOI] [PubMed] [Google Scholar]
- Roux C, Fraïsse C, Romiguier J, Anciaux Y, Galtier N, Bierne N. Shedding Light on the Grey Zone of Speciation along a Continuum of Genomic Divergence. PLOS Biol. 2016;14(12):e2000234. doi: 10.1371/journal.pbio.2000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sax DF, Stachowicz JJ, Brown JH, Bruno JF, Dawson MN, Gaines SD, Grosberg RK, Hastings A, Holt RD, Mayfield MM, O’Connor MI, et al. Ecological and evolutionary insights from species invasions. Trends Ecol Evol. 2007;22(9):465–71. doi: 10.1016/j.tree.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Schlarbaum SE, Adams RP, Bagley WT, Wayne WJ. Postglacial migration pathways of Quercus rubra L., northern red oak, as indicated by regional genetic variation patterns. Silvae Genet. 1982;31:150–158. [Google Scholar]
- Schlarbaum SE, Bagley WT. Intraspecific Genetic Variation of Quercus rubra L., Northern Red Oak. Silvae Genet. 1981;30(2–3):50–56. [Google Scholar]
- Simberloff D. The role of propagule pressure in biological invasions. Annu Rev Ecol Evol Syst. 2009;40(1):81–102. doi: 10.1146/annurev.ecolsys.110308.120304. [DOI] [Google Scholar]
- Sork V, Huang S, Wiener E. Macrogeographic and fine-scale genetic structure in a North American oak species, Quercus rubra L. Ann des Sci For. 1993;50(1):261s–270s. doi: 10.1051/forest:19930726. [DOI] [Google Scholar]
- Stapley J, Santure AW, Dennis SR. Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species. Mol Ecol. 2015;24(9):2241–2252. doi: 10.1111/mec.13089. [DOI] [PubMed] [Google Scholar]
- Steiner KC. Autumn predation of northern red oak seed crops. In: Gottschalk KW, Fosbroke SLC, editors. 10th Central Hardwood Forest Conference; Morgantown, West Virginia. 1995. pp. 489–494. http://nrs.fs.fed.us/pubs/gtr/gtr_ne197/gtr_ne197_489.pdf. [Google Scholar]
- Steiner KC, Abrams MD, Bowersox TW. Advance reproduction and other stand characteristics in Pennsylvania and French stands of Northern Red Oak. In: Gillespie AR, Parker GR, Pope PE, Rink G, editors. 9th Central Hardwood Forest Conference; West Lafayette, Indiana. 1993. pp. 473–483. [Google Scholar]
- Tatem AJ, Hay SI, Rogers DJ. Global traffic and disease vector dispersal. Proc Natl Acad Sci. 2006;103(16):6242–6247. doi: 10.1073/pnas.0508391103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timbal J, Kremer A, Le Goff N, Nepveu G. Le chêne rouge d’Amérique. Institut National de la Recherche Agronomique; Paris: 1994. [Google Scholar]
- Tsutsui ND, Suarez AV, Holway DA, Case TJ. Reduced genetic variation and the success of an invasive species. Proc Natl Acad Sci U S A. 2000;97(11):5948–53. doi: 10.1073/pnas.100110397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States. Bureau of the Census. Water transportation. Historical Statistics of the United States : Colonial Times to 1970. 1975;2:742–766. https://fraser.stlouisfed.org/scribd/?item_id=5808&filepath=/docs/publications/histstatus/hstat1970_cen_1975_v2.pdf. [Google Scholar]
- Vellinga EC, Wolfe BE, Pringle A. Global patterns of ectomycorrhizal introductions. New Phytol. 2009;181(4):960–973. doi: 10.1111/j.1469-8137.2008.02728.x. [DOI] [PubMed] [Google Scholar]
- Vilà M, Espinar JL, Hejda M, Hulme PE, Jarošík V, Maron JL, Pergl J, Schaffner U, Sun Y, Pyšek P. Ecological impacts of invasive alien plants: a meta-analysis of their effects on species, communities and ecosystems. Ecol Lett. 2011;14(7):702–708. doi: 10.1111/j.1461-0248.2011.01628.x. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-Statistics for the Analysis of Population Structure. Evolution (N. Y) 1984;38(6):1358–1370. doi: 10.2307/2408641. [DOI] [PubMed] [Google Scholar]
- Woziwoda B, Kopeć D, Witkowski J. The negative impact of intentionally introduced Quercus rubra L. on a forest community. Acta Soc Bot Pol. 2014;83(1):39–49. doi: 10.5586/asbp.2013.035. [DOI] [Google Scholar]
- Xu C-Y, Tang S, Fatemi M, Gross CL, Julien MH, Curtis C, van Klinken RD. Population structure and genetic diversity of invasive Phyla canescens: implications for the evolutionary potential. Ecosphere. 2015;6(9):1–21. doi: 10.1890/ES14-00374.1. [DOI] [Google Scholar]
- Zanetto A, Kremer A. Geographical structure of gene diversity in Quercus petraea (Matt.) Liebl. I. Monolocus patterns of variation. Heredity. 1995;75(5):506–517. doi: 10.1038/hdy.1995.167. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.