Abstract
The role of Vibrio parahaemolyticus in causing diarrhoeal disease is well known. However, phenotypic and genetic traits of this pathogen isolated from diverse sources have not been investigated in detail. In this study, we have screened samples from diarrhoeal cases (2603), brackish water fish (301) and aquatic environments (115) and identified V. parahaemolyticus in 29 (1.1%), 171 (56.8%) and 43 (37.4%) samples, respectively. Incidence of pathogenic V. parahaemolyticuswith virulence encoding thermostable-direct haemolysin gene (tdh) was detected mostly in fishes (19.3%) and waters (15.6%) than clinical samples (1.04%). The pandemic strain marker genes (toxRS and PGS-sequences) have been detected relatively more in water (6%) and fish (5%) samples than in clinical samples (0.7%). Majority of the V. parahaemolyticus isolates from clinical cases and fish samples (26.3%) belonged to classical pandemic serovars (O3:K6). In addition, several newly recognised pandemic serovars have also been identified. Pulsed field-gel electrophoresis (PFGE) analysis showed clonal relatedness (60–85%) of V. parahaemolyticus from different sources. The study observation revealed that the brackish water fishes and water bodies may act as a reservoir of pathogenic V. parahaemolyticus. Emergence of several new serovars of pandemic V. parahaemolyticussignifies the changing phenotypic characteristics of the pathogen.
Keywords: Epidemiology, Public health
1. Introduction
Vibrio parahaemolyticus is a Gram-negative, halophilic bacterium that remains one of the leading causes of diarrhoea and gastroenteritis [1, 2]. Thermostable-direct haemolysin (TDH) and TDH-relatedhaemolysin (TRH) are considered to be the major virulencefactors in this bacterium [1]. Almost all the clinical isolates expressed TDH and/or TRH encoded in the tdhand trhgenes, respectively [1, 3]. These genes are rarely found in environmental V. parahaemolyticusisolates [4, 5].
Till 1995, V. parahaemolyticushas been implicated with sporadic diarrhoea. With the emergence of unique serovar (O3:K6) in 1996, this organism has gained pandemicity and caused diarrhoeal outbreaks in several countries [6, 7, 8, 9, 10, 11, 12, 13]. The O3:K6 clone (pandemic strain) and its serovariants typically possess tdh genewith a distinctive toxRS operon that encodes a transcriptional regulator. To facilitate identification of pandemic O3:K6, group specific (GS-PCR) and ORF-8 PCRs targeting the toxRS operon and filamentous phage f237, respectively have been developed [9, 14]. Besides, PGS-PCR assay directed to detect a 930 bp sequence is used for detection of pandemic strain of this pathogen [15].
Studies conducted in India have reported the prevalence of pandemic V. parahaemolyticus strain mainly from hospitalized diarrhoeal cases [16, 17, 18, 19], sewage [20]; salt water fishes, shellfishes and coastal environment [21, 22, 23, 24]; however, no concurrent study has been made to detect the reservoirs of this pathogen. To address this lacuna, we conducted a detailed study using samples collected from different sources in and around Kolkata and also by characterizing the V. parahaemolyticus isolates.
2. Materials and methods
2.1. Sampling
Stool specimens from 2603 diarrhoeal casesadmitted in the Infectious Diseases Hospital (IDH) and outpatients attending the BC Roy Child Memorial Hospital for children, Kolkata were collected during 2008–2011. During the same period, 301 brackish and freshwater fish from different markets and 115 water samples in and around Kolkata metropolis were collected and screened for the presence of V. parahaemolyticus.
2.2. Isolation and identification of V. parahaemolyticus
All the samples were screened for V. parahaemolyticus by adopting the published cultural and biochemical methods [11, 25]. Briefly, 2–4 loopful of diarrheal stool was pre-enriched in 2 ml of alkaline peptone water (APW) containing 3% NaCl, pH 8.5. About 15 g of gill from each fish was aseptically dissected and homogenized before inoculation in 50 ml of APW. About 100 ml of water sample was filtered aseptically through 0.45μ membrane and placed in 10 ml of APW and incubated aerobically for 18 hrs at 37 °C.
A loopful of pre-enriched culture was streaked on thiosulfate citrate bile salts sucrose (TCBS) agar (Eiken, Tokyo, Japan) and incubated at 37 °C for 18–24 hrs. Presumptive identification of V. parahaemolyticus was made based on typical colony characteristics, i.e., 2–3 mm sized round non-sucrose fermenting green color colonies. Five typical colonies from each sample were selected for biochemical characterization using a multi-test medium in which V. parahaemolyticusgave acidic (yellow) butt and alkali (purple) slant (K/A) reaction [25]. V. parahaemolyticus isolates were further subjected to PCR assays targeting species-specific toxR gene [26], virulence encoding genes (tdh and trh) [27] and pandemic strain marker genes (toxRS, orf-8 and PGS-sequence [9, 14, 15].
2.3. Reference strains Bacterial strains
Laboratory reference strains Vp-Kx-V138 and VP230 were used as a positive control and Escherichia coli K12 strain as a negative control in the PCR assays.
2.4. Molecular characterization of isolates
2.4.1. Bacterial template DNA preparation
A loopful of overnight culture from LB agar with 3% NaCl (LBS) was taken in 1.5ml microfuge tubes containing 200μl sterile distilled water and was suspended well using a vortex mixture. This mixture was boiled for 10 min and rapidly cooled on ice. The cell suspensions were centrifuged at 4,000 rpm for 3 min and the supernatant was used as a genomic DNA template. The PCR assay was performed using the Gene Amp PCR system 2700 thermocycler (Applied Bio-systems).
2.4.2. Species-specific and virulence gene (Vp-toxR, tdh, trh) PCR
To detect the toxRgene by PCR, primers F: GTCTTCTGACGCAATCGTTG and R: ATACGAGTGGTTGCTGTCATG were used as described previously [26]. All the toxR confirmed isolates were further examined for the presence of virulence encoding tdh and trh genes using the primer pair F: CCAAATACATTTTACTTGG and R: GGTACTAAATGGCTGACATC and primer pair F: GGCTCAAAATGGTTAAGCG and R: CATTTCCGCTCTCATATGC, respectively [27].
2.4.3. Detection of pandemic marker: [toxRS new/GS-PCR; pandemic group-specificPCR (PGS-PCR) and orf-8]
V. parahaemolyticus isolated from different sources were further subjected to determine the presence of pandemic marker genes by using the published methods. GS-PCR assay was performed using GS-VP1 (TAATGAGGTAGAAACA) and GS-VP2 (ACGTAACGGGCCTACA) primer pair [9], PGS-PCR assay using F: TTCGTTTCGCGCCACAACT and R: TGCGGTGATTATTCGCGTCT [15] and orf-8gene PCR assay using F: AGTATTGCTGAAGAGTACG and R: CTCGACTTAAACGATCCCprimer pairs [14]. All the PCR amplified products were analyzed by gel electrophoresis.
2.5. Serotyping
V. parahaemolyticus isolates were serotyped using somatic (O) and capsular (K) antisera (Denka Seiken, Co. Ltd., Tokyo) [28].
2.6. Pulse field-gel electrophoresis (PFGE)
For clonal identification, the pandemic serovarswere examined by PFGE following the PulseNet protocol (CDC, 2009) [29]. Briefly, test isolates were grown on LAS and incubated overnight at 37°C. The overnight culture was suspended in a cell suspension buffer (CSB) (100 mMTris, 100mM EDTA, pH 8.0) and measured the cell density in a spectrophotometer with OD value between 1.3 and 1.5 at 600 nm. Agarose plugs were prepared by mixing equal volume of bacterial suspension with 1% low-melting agarose (Sea-Kem). After solidification, the sized plugs were treated with cell lysis buffer (50 mMTris, 50 mM EDTA, pH 8.0 and 1% Sarcosyl) followed by proteinase K (20 mg/ml) at 54 °C for 1 hour with constant shaking (150–175 rpm).
The plugs were washed twice with sterile distilled water (pre-heated to 50 °C) under vigorous shaking in a 50 °C water bath for 10–15 minand further washed 4 times with TE buffer (10 mMTris, 1 mM EDTA, pH 8.0). Agarose plugs were then equilibrated in TE buffer and were placed in 30 μl of 10X H buffer (0.1% BSA, 0.1% Triton X-100) for 45 min. After incubation, plugs were kept overnight in 150μl reaction mixture consisting 15μl 10X H buffer, 15μl 10 X BSA, 3μl NotI enzyme (45 units) [Takara, Shuzo Co. Ltd, Japan] and 117 μl sterile distilled water at 37 °C.
PFGE of the NotI digested inserts was performed by the contour clamped homogeneous electric field method on a CHEF Mapper system (Bio-Rad, Hercules, CA, USA) with 1% PFGE grade agarose in 0.5X TBE (44.5 mM Boric acid, 1.0 mM EDTA, pH 8.0) for 18 hrs using the XbaI digested DNA of Salmonella enteritidis serovar Braenderup as the standard size DNA molecular marker. A mini chiller (Bio-Rad) was used to maintain the temperature of the buffer at 14 °C. Run conditions (150 mA current, voltage-6.0V/cm, angle-120°, initial switch time-10 sec, final switch times-35 sec, linear) were generated by the auto algorithm mode of the CHEF Mapper PFGE system by using a 78–390 kb size range. After electrophoresis, the gel was stained with ethidium bromide (1 μg/ml) for 30 min and destained with water for 15 min twice. The DNA bands were visualized and photographed with the BioSpectrum AC Imaging System (USA).
The PFGE image was captured using a Gel Doc XR system (Bio-Rad). Each DNA band was normalized by aligning with the peaks of the Salmonella enterica serovar Braenderup (H9182) and analyzed using the Bio-Numerics software Version 4.0 (Applied Maths, Sint-Martens-Latem, Belgium). Degree of banding similarity was determined by comparison of the Dice coefficient, and clustering correlation coefficients were calculated by an unweighted pair-group method with arithmetic averages (UPGAMA).
3. Results
3.1. Isolation of pathogenic V. parahaemolyticus
The proportion of V. parahaemolyticus identified from diarrhoeal patients, fishes and water samples are shown in Table 1. In the species-specific PCR (Vp-toxR), all the isolates from clinical stool specimens were found to be positive for V. parahaemolyticus. Similarly, 171of 210 isolates from retail fishes and 43 of 53 isolates from water samples were identified as V. parahaemolyticus, confirming the occurrence of this pathogen in 56.8% and 37.4% of retail fish and water samples, respectively (Table 1).
Table 1.
Isolation and molecular characterization of V. parahaemolyticus isolates from clinical diarrhea, retail fish and water.
| Sample Source (no.) |
Identification (%) |
Virulence encoding gene (%) |
Pandemic strain marker (%) |
||||
|---|---|---|---|---|---|---|---|
| Culture | Vp-toxR | tdh | trh | ORF-8 | GS-PCR | PGS-PCR | |
| Clinical diarrhea (2603) | 29 (1.1) | 29 (1.1) | 27 (1.04) | 6a(0.2) | 13 (0.5) | 19 (0.7) | 19 (0.7) |
| Retail fish (301) | 210 (69.7) | 171 (56.8) | 58 (19.3) | 12 (4) | 15 (5) | ||
| Razor fish (64) (Aeoliscusstrigatus) |
53 (82.8) | 42 (65.6) | 12 (18.7) | 1 (1.5) | 2 (3.1) | ||
| Sardine (30) (Sardinops neo pilchardus) |
24 (80) | 23 (76.6) | 5 (16.7) | 2 (6.7) | 2 (6.7) | ||
| Mackerel (25) (Rastrelligerkanagurta) |
19 (76) | 19 (76) | 4 (16) | 1 (4) | 2 (8) | ||
| Conch fish (74) (Dasyatisbennettii) |
49 (66.2) | 34 (46) | 19 (25.6) | 5 (6.7) | 6 (8.1) | ||
| Phasa fish (45) (Setipinnaphasa) |
33 (73.3) | 27 (60) | 15 (33.3) | 2 (4.4) | 2 (4.4) | ||
| Rohu (45) (Labeorohita) |
17 (37.8) | 11 (24.4) | 1 (2.2) | ||||
| Hilsafish (Hilsailisha) (18) | 15 (83.3) | 15 (83.3) | 2 (11.1) | 1 (5.6) | 1 (5.6) | ||
| Water (115) | 53 (46.1) | 43 (37.4) | 18 (15.6) | 2 (1.7) | 1 (0.9) | 6 (5.2) | 7 (6) |
| River (29) | 17 | 12 (41.38) | 5 (17.2) | 1 (3.4) | 1 (3.4) | ||
| i) Ganga (21) | 9 | 4 (19) | 3 | ||||
| ii)Ischhamati (8) | 8 | 8 (100) | 2 (25) | 1 (12.5) | 1 (12.5) | ||
| Estuary (54) | 36 | 31 (57.4) | 13 (24) | 2 (3.7) | 1 (1.9) | 5 (9.2) | 6 (11.1) |
| i)Kakdweep (7) | 6 | 6 (85.7) | 5 (71.4) | 1 (14.3) | 1 (14.3) | 1 (14.3) | 2 (28.6) |
| ii)Canning (7) | 7 | 7 (100) | 1 (14.3) | ||||
| iii)Dhamakhali (34) | 23 | 18 (53) | 7 (20.6) | 1 (3) | 4 (11.8) | 4 (11.8) | |
| iv)Diamond harbour (6) | |||||||
| Fresh water (25) | |||||||
| i)Pond (16) | |||||||
| ii)Jheel (9) | |||||||
| iii)Seedling pond (7) | |||||||
One clinical isolate (tdh−/trh+) was positive in GS and PGS-PCR.
Out of the total 29 isolates from clinical diarrhoeal cases, 27 were found positive for presence of tdh and 5 tdh+ isolates were also positive for trh (Table 1). Besides, one isolate was positive for trhonly. Overall, the detection rate of V. parahaemolyticus in diarrhoeal cases was very less (1.1%, 28/2603). From fish and water samples, tdh harbouring V. parahaemolyticus was detected in 19.3% (58/301) and 15.6% (18/115), respectively. Two isolates (1.7%) from water sources were positive for both tdh and trh (Table 1).
Among retail brackish water fishes, tdh positive V. parahaemolyticus was detected in 33.3% of Phasa fishes (Setipinnaphasa) followed by Conch fishes (Dasyatisbennettii) (25.6%), Razor fish (Aeoliscusstrigatus) (18.7%), Sardine (Sardinopsneopilchardus) (16.7%) and Meckerel (Rastrelligerkanagurta) (16%). Fresh water fisheslike Rohu (Labeorohita) and Hilsa (Hilsailisha) were also found to harbour tdh positiveV. parahaemolyticus in 2.2% and 11.1% of samples, respectively (Table 1). The pathogenic V. parahaemolyticuswas identified in river (17.2%) (5/29) and estuarine waters (24%) (13/54). However, no isolation of V. parahaemolyticus was made from fresh water samples viz. pond, Jheel and seedling pond (Table 1).
3.2. Occurrence of V. parahaemolyticus pandemic strain
Results of GS-PCR revealed that 0.7% of diarrhoeal cases, 4% of retail fishes and 5.2% of water samples contain edpandemic strain of V. parahaemolyticus. The PGS-PCR assay gave almost similar results (Table 1). However, orf-8 was very less, i.e., 0.5% in clinical and 0.9% in water isolates and none of fish samples (Table 1).
In the GS-PCR assay, pandemic strain of V. parahaemolyticus was recovered from 6.7% each of Conch and Sardine fishes; 5.6% of Hilsa; 4.4% of Phasa; 4% of Mackerel and 1.5% of Razor fishes. Detection of the pandemic isolates by PGS-PCR assay was slightly higher than the GS-PCR (Table 1). As detected in the GS-PCR assay, the pandemic V. parahaemolyticus was recovered in water samples from the Ischhamati river (12.5%) and estuarine waters of Kakdweep (14.3%) and Dhamakhali (11.8%) (Table 1). Like fish samples, isolation of pandemic strain from water samples by PGS-PCR was slightly more than the GS-PCR (Table 1).
3.3. Characteristics of pandemic strain
Analysis of results from clinical isolates revealed that out of a total 19 isolates (toxRS+& PGS Sequence+), 17 belonged to the established pandemicserovars (O3:K6; O1:KUT; O1:K25; O3:KUT); however, two (2) isolates belonged to O2:K4 and O8:K21 that were new in the list of pandemic serovars.
Out of 12 GS/PGS-PCR positive isolatesfrom fishes, pandemic serovars viz. O1:K33, O1:KUT, O5:KUT, O8:KUT, O10:KUT recorded in 7 isolates. Besides, serovars O1:K28; O1:K34; O3:K31; were identified as novel pandemic serovars (Table 2). From water samples, pandemic serovars viz. O4:K12; O5:KUT and OUT:KUT were identified in 4 isolates. In addition, serovar OUT:K33 was recovered as novel pandemic serovar in 2 isolates. Pandemic serovar O5:KUT and new pandemic serovar O8:K21 were found among the orf-8 positive isolatesfrom water and clinical and samples (Table 2).
Table 2.
Molecular characteristics of different serovars of V. parahaemolyticus from clinical diarrhoea, fish and water sources.
| Serovar | Source |
|||||||
|---|---|---|---|---|---|---|---|---|
| Human (n = 28) |
Fish (n = 58) |
Water (n = 18) |
||||||
| tdh/trh | toxRS&PGS-PCR | tdh | toxRS | PGS-PCR | tdh/trh | toxRS | PGS-PCR | |
| O1:K25 | 10 | 9 (6*) | ||||||
| O1:K28 | 2 | 2 | 2 | |||||
| O1:K33 | 2 | 1 | 1 | |||||
| O1:K34 | 1 | 1 | 1 | |||||
| O1:KUT | 2 | 1 (*) | 4 | 2 | 3 | |||
| O2:K4 | 1 | 1 | ||||||
| O2:K28 | 1 | |||||||
| O2:KUT | 2 | |||||||
| O3:K6 | 5 | 5 (*4) | ||||||
| O3:K31 | 4 | 2 | 1 | |||||
| O3:KUT | 4 | 2 (*1) | 3 | 1 | ||||
| O4:K8 | 1 | |||||||
| O4:K9 | 1 | |||||||
| O4:K12 | 2 | 1 | 1 | |||||
| O4:K25 | 1 | |||||||
| O4:K42 | 1 | |||||||
| O4:K63 | 1 | |||||||
| O4:KUT | 4 | 1 | ||||||
| O5:K15 | 1 | |||||||
| O5:K17 | 5 | |||||||
| O5:KUT | 8 | 2 | 3 | 7 | 2 (*1) | 2 | ||
| O6:K15 | 1 | |||||||
| O7:K19 | 1 | |||||||
| O8:K1 | 1 | |||||||
| O8:K21 | 1 | 1 (*) | ||||||
| O8:KUT | 1 | 1 | 1 | 1 | ||||
| O10:KUT | 8 | 1 | 1 | 2 | ||||
| O11:K28 | 1 | |||||||
| O11:KUT | 1 | |||||||
| OUT:K28 | 2 | |||||||
| OUT:K33 | 2 | 2 | 2 | |||||
| OUT:K55 | 1 | |||||||
| OUT:KUT | 2 | 4 | 1 | 2 | ||||
| OUT:K15 | 1 | |||||||
| Total | 27 | 19 | 58 | 12 | 15 | 18 | 6 | 7 |
Asterisk (*) in parentheses indicates the number of isolate positive for orf-8 gene. One clinical isolate with tdh−/trh+was typed as O5:KUT.
3.4. Molecular typing
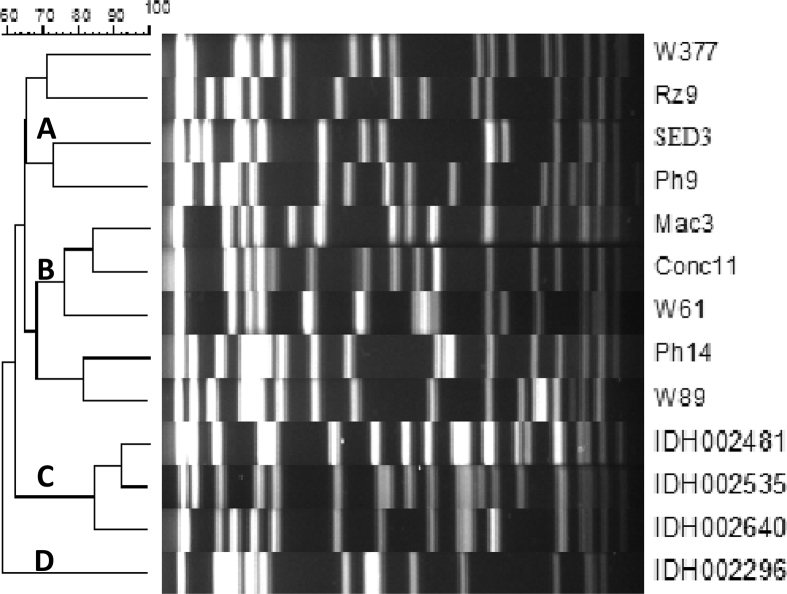
PFGE of the pandemic V. parahaemolyticus was performed to show the extent of clonal relationship between isolates from different sources. A total of 13 representative isolates (GS-PCR+ and PGS-PCR+) from human (4) fish (6) and water (3) isolates were selected and examined. Dendrogram of PFGE analysis showed three major clusters (A-C, Fig. 1). In cluster ‘C’, three clinical isolates (IDH 002481; IDH 002535 and IDH 002640) were grouped; of these, IDH 002481 and IDH 002535 belonged to a single clade with >90% similarity and also exhibited >80% relatedness with another human isolate (IDH 002640) in the same cluster. These clinical isolates had ∼62% similarity with fish and water isolates in clusters ‘A’ and ‘B’, respectively. Further, cluster ‘A’ and ‘B’ had approximately 65% similarity. The other clinical isolate (IDH002296) that discretely belonged to cluster ‘D’ showed ∼60% relatedness with other clinical (n = 3), fish (n = 6) and water (n = 3) isolates of clusters ‘A’ to ‘C’ (Fig. 1).
Fig. 1.
Phylogenetic tree (dendrogram) of Vibrio parahaemolyticus isolates from. human, fish and water using the software, Bio-Numerics (Applied Maths, Belgium). The similarity scale is on the top of figure. IDH: Human clinical isolates, RZ, SED, Ph, Mac, Conc: Isolates from fishes, W = Isolates from water, RZ- Razor; Con- Conch; Ph-Phasa; SRD- Sardine, Mac-Mackerel, W-water.
The isolates from water (W377) and Razor fish (Rz9) belonged to the same clade in cluster ‘A’ with 70% genetic relatedness. Similarly, other two fish isolates from Serdine (SED3) and Phasa (Ph9) were grouped in the same clade with 70% similarity to each other and showed 65% similarity to Razor fish and water isolates (W377 and Rz9) belonged to other clad in cluster ‘A’. In Cluster ‘B’ isolates from Mackerel (Mac3) and Conch fish (Conc11) were grouped in the same clade with 85% similarity to each other. Further, these two isolates were related (∼75%) to one isolate (W61) from water sample. Likewise, one isolate each from fish Phasa (Ph14) and water (W89) belonged to one clade in cluster ‘B’ showed ∼80% similarity and were related (70%) to the other two types of fish isolates (Mac3 and Conc11) and one water isolate (W61).
4. Discussion
Analogous to the previous studies conducted in Indian context [7, 8, 16, 17, 18, 19, 20], we found that association of V. parahaemolyticuswith diarrhoeal cases was less. Considering environmental reservoirs of V. parahaemolyticus, ourobservation was analogous to other studies [17, 30]. Epidemiologically, this aspect is important because a larger number of coastal and inland populations are engaged in aquaculture for their livelihood and possibility of infection is high. Since V. parahaemolyticusis autochthonous to coastal waters, many of the brackish water fishes carry this organism. Occurrence of V. parahaemolyticus in fresh water fish like Rohu may be due to cross contamination with other marine fishes sold in the market. The reason for the existence of V. parahaemolyticus in Hilsa fish may be due to their anadromous migratory nature.
The performance of the GS-PCR and PGS-PCR seems to vary depending on the nature of the sample [11, 12, 24]. We found that it would be prudent to adopt both the PCR assays to detect the pandemic strain of V. parahaemolyticus, particularly from the environmental sources. However, the orf-8 PCR has identified limited number of pandemic V. parahaemolyticus isolates harbouring the filamentous phage (f237).
Majority of the clinical isolates of V. parahaemolyticus belonged to the typical pandemic serovars. However, the other new serovars such as O1:K28; O1:K34; O2:K4; O3:K31; O8:K21and OUT:K33 identified in this study had the characteristics of pandemic strain. This indicates the rapid phenotypic and genetic changes among V. parahaemolyticus. Several studies correlate El Nino event responsible for such changes in the epidemiology of V. parahaemolyticus [31, 32]. It is pertinent to mention that during 1995 and 1996, only the O3:K6 was identified as pandemic serovar; however, with the passage of time, several serovars have been recognized in this category [10, 33]. Since the overall genetic relationship was almost similara mong different isolates from human, fish and water bodies, it is likely that food animals and water bodies may act as reservoirs of pandemic strains of V. parahaemolyticus.
5. Conclusion
Pandemic strain of V. parahaemolyticus mostly detected in retail brackish water fishes and aquatic bodies with several new serovars. PFGE analysis has shown the genetic relatedness of V. parahaemolyticus isolates from different sources, signifying potential reservoirs of human infection.
Declarations
Author contribution statement
Sailen Guin, Murugan Saravanan, Prasad Anjay, Goutam Chowdhury, Pazhani Gururaja Perumal: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Thandavarayan Ramamurthy. Suresh Chandra Das: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by the Indian Council of Medical Research (ICMR), New Delhi, India under extra mural research grant (IRIS ID 2003 02200).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Nelapati S., Nelapati K., Chinnam B.K. Vibrio parahaemolyticus: an emerging foodborne pathogen - a review. Vet. World. 2012;5(1):48–62. [Google Scholar]
- 2.Newton A., Kendall M., Vugia D.J., Henao O.L., Mahon B.E. Increasing rates of Vibriosis in the United States, 1996-2010: review of surveillance data from 2 systems. Clin. Infect. Dis. 2012;54(5):391–395. doi: 10.1093/cid/cis243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honda S., Goto I., Minematsu I., Ikeda N., Asano N., Ishibashi M., Kinoshita Y., Nishibuchi N., Honda T., Miwatani T. Gastroenteritis due to Kanagawa negative Vibrio parahaemolyticus. Lancet. 1987;I:331–332. doi: 10.1016/s0140-6736(87)92062-9. [DOI] [PubMed] [Google Scholar]
- 4.Shirai H., Ito H., Hirayama T., Nakamoto Y., Nakabayashi N., Kumagai K., Takeda Y., Nishibuchi M. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 1990;58:3568–3573. doi: 10.1128/iai.58.11.3568-3573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lovell Charles. R. Ecological fitness and virulence features of Vibrio parahaemolyticus in estuarine environments. Appl. Microbiol. Biotechnol. 2017;101:1781–1794. doi: 10.1007/s00253-017-8096-9. [DOI] [PubMed] [Google Scholar]
- 6.Okuda J., Ishibashi M., Hayakawa E., Nishino T., Takeda Y., Mukhopadhyay A.K., Garg S., Bhattacharya S.K., Nair G.B., Nishibuchi M. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 1997;35:3150–3155. doi: 10.1128/jcm.35.12.3150-3155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bag P.K., Nandi S., Bhadra R.K., Ramamurthy T., Bhattacharya S.K., Nishibuchi M., Hamabata T., Yamasaki S., Takeda Y., Nair G.B. Clonal diversity among recently emerged strains of Vibrio parahaemolyticus O3:K6 associated with pandemic spread. J. Clin. Microbiol. 1999;37:2354–2357. doi: 10.1128/jcm.37.7.2354-2357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury N.R., Chakraborty S., Ramamurthy T., Nishibuchi M., Yamasaki S., Takeda Y., Nair G.B. Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerg. Infect. Dis. 2000;6:631–636. doi: 10.3201/eid0606.000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumoto C., Okuda J., Ishibashi M., Iwanaga M., Garg P., Ramamurthy T., Wong H.C., Depaola A., Kim Y.B., Albert M.J., Nishibuchi M. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRSsequence analyses. J. Clin. Microbiol. 2000;38:578–5785. doi: 10.1128/jcm.38.2.578-585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nair G.B., Ramamurthy T., Bhattacharya S.K., Dutta B., Takeda Y., Sack David A. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 2007;20:39–48. doi: 10.1128/CMR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J., Xue F., Yang Z., Zhang X., Zeng D., Chao G., Jiang Y., Li B. Vibrio parahaemolyticus strains of pandemic serotypes identified from clinical and environmental samples from Jiangsu, China. Front. Microbiol. 2016;7:787. doi: 10.3389/fmicb.2016.00787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espejo R.T., García K., Plaza N. Insight into the origin and evolution of the Vibrio parahaemolyticus pandemic strain. Front. Microbiol. 2017;8:1397–1408. doi: 10.3389/fmicb.2017.01397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernández-Rendón C.L., Barrera-Escorcia G., Wong-Chang I., Vázquez Botello A., Gómez-Gil B., Lizárraga-Partida M.L. Toxigenic V. cholerae, V. parahaemolyticus, and V. vulnificus in oysters from the Gulf of Mexico and sold in Mexico City. Int. J. Environ. Health Res. 2018;27:1–11. doi: 10.1080/09603123.2018.1548696. [DOI] [PubMed] [Google Scholar]
- 14.Myers M.L., Panicker G., Bej A.K. PCR detection of a newly emerged pandemic Vibrio parahaemolyticus O3:K6 pathogen in pure cultures and seeded waters from the Gulf of Mexico. Appl. Environ. Microbiol. 2003;69:2194–2200. doi: 10.1128/AEM.69.4.2194-2200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okura M., Osawa R., Iguchi A., Takagi M., Arakawa E., Terajima J., Watanabe H. PCR-based identification of pandemic group Vibrio parahaemolyticus with a novel group-specific primer pair. Microbiol. Immunol. 2004;48:787–790. doi: 10.1111/j.1348-0421.2004.tb03596.x. [DOI] [PubMed] [Google Scholar]
- 16.Pal S.C., Sircar B.K., Nair G.B., Deb B.C. Epidemiology of bacterial diarrheal diseases in India with special reference to Vibrio parahaemolyticus infections. In: Takeda Y., Miwatani T., editors. Bacterial Diarrheal Diseases. KTK Scientific Publishers; Tokyo, Japan: 1985. pp. 65–73. [Google Scholar]
- 17.Kanungo S., Sur D., Ali M., You Y.A., Pal D., Manna B., Niyogi S.K., Sarkar B., Bhattacharya S.K., Clemens J.D., Nair G.B. Clinical, epidemiological, and spatial characteristics of Vibrio parahaemolyticus diarrhea and cholera in the urban slums of Kolkata, India. BMC Public Health. 2012;12:830–838. doi: 10.1186/1471-2458-12-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chowdhury G., Ghosh S., Pazhani G.P., Paul B.K., Maji D., Mukhopadhyay A.K., Ramamurthy T. Isolation and characterization of pandemic and nonpandemicstrains of Vibrio parahaemolyticus from an outbreak of diarrhea in North 24 Parganas, West Bengal, India. Foodborne Pathog. Dis. 2013;10(4):338–342. doi: 10.1089/fpd.2012.1340. [DOI] [PubMed] [Google Scholar]
- 19.Pazhani G.P., Bhowmik S.K., Ghosh S., Guin S., Dutta S., Rajendran K., Saha D.R., Nandy R.K., Bhattacharya M.K., Mukhopadhyay A.K., Ramamurthy T. Trends in the epidemiology of pandemic and non-pandemic strains of Vibrio parahaemolyticus isolated from diarrheal patients in Kolkata, India. PLoS Negl. Trop. Dis. 2014;8(5):2815. doi: 10.1371/journal.pntd.0002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkar B., Chowdhury N.R., Nair G.B., Nishibuchi M., Yamasaki S., Takeda Y., Gupta S.K., Bhattacharya S.K., Ramamurthy T. Molecular characterization of Vibrio parahaemolyticus of similar serovars isolated from sewage and clinical cases of diarrhea in Calcutta, India. World J. Microbiol. Biotechnol. 2003;19:771–776. [Google Scholar]
- 21.Dileep V., Kumar H.S., Kumar Y., Nishibuchi M., Karunasagar I., Karunasagar I. Application of polymerase chain reaction for detection of Vibrio parahaemolyticus associated with tropical seafoods and coastal environment. Lett. Appl. Microbiol. 2003;36:423–427. doi: 10.1046/j.1472-765x.2003.01333.x. [DOI] [PubMed] [Google Scholar]
- 22.Anjay, Das S.C., Kumar A., Kaushik P., Kurmi B. Detection of Vibrio parahaemolyticus from saltwater fish samples by Vp-toxR PCR. Indian J. Fish. 2013;60(1):141–143. [Google Scholar]
- 23.Anjay, Das S.C., Kumar A., Kaushik P., Kurmi B. Occurrence of Vibrio parahaemolyticus in marine fish and shellfish. Indian J. Geo-Marine Sci. 2014;43(5):887–990. [Google Scholar]
- 24.Parthasarathy S., Das S.C., Kumar A. Occurrence of pathogenic Vibrio parahaemolyticus in crustacean shellfishes in coastal parts of Eastern India. Vet. World. 2016;9(3):330–336. doi: 10.14202/vetworld.2016.330-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaper J.B., Remmers E.F., Colwell R.R. A medium for the presumptive identification of Vibrio parahaemolyticus. J. Food Prot. 1980;43:936–938. doi: 10.4315/0362-028X-43.12.936. [DOI] [PubMed] [Google Scholar]
- 26.Kim Y.B., Okuda J., Matsumoto C., Takahashi N., Hashimoto S., Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the tox-R gene. J. Clin. Microbiol. 1999;37:1173–1177. doi: 10.1128/jcm.37.4.1173-1177.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tada J., Ohashi T., Nishimura N., Shirasaki Y., Ozaki H., Fukushima S., Takano J., Nishibuchi M., Takeda Y. Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol. Cell. Probes. 1992;6:477–487. doi: 10.1016/0890-8508(92)90044-x. [DOI] [PubMed] [Google Scholar]
- 28.Elliot E.L., Kaysner C.A., Jackson L., Tamplin M.L. USFDA Bacterial Analytical Manual. eighth ed. AOAC International; Arlengton, Va: 1995. Vibrio cholerae, Vibrio parahaemolyticus, Vibrio vulnifllus and other Virbiospp; pp. 9.01–9.27. [Google Scholar]
- 29.CDC 2009. www.cdc.gov/pulsenet/.../PulseNetAsia_Pacific
- 30.Cabrera-Garcia M.E., Vazquez-salinas C., Quinones-Ramirez E.I. Serologic and molecular characterization of Vibrio parahaemolyticus strains isolated from seawater and fish products of the Gulf of Mexico. Appl. Environ. Microbiol. 2004;70:6401–6406. doi: 10.1128/AEM.70.11.6401-6406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Urtaza J., Huapaya B., Gavilan R.G., Blanco-Abad V., Ansede-Bermejo J., Cadarso-Suarez C., Figueiras A., Trinanes J. Emergence of asiatic Vibrio diseases in south America in phase with El niño. Epidemiology. 2008;19(6):829–837. doi: 10.1097/EDE.0b013e3181883d43. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez-Escalona N., Gavilan R.G., Toro M., Zamudio M.L., Martinez-Urtaza J. Outbreak of Vibrio parahaemolyticus sequence type 120, Peru, 2009. Emerg. Infect. Dis. 2016;22(7):1235–1237. doi: 10.3201/eid2207.151896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhuiyan N.A., Ansaruzzaman M., Kamruzzaman M., Alam K., Chowdhury N.R., Nishibuchi M., Faruque S.M., Sack D.A., Takeda Y., Nair G.B. Prevalence of the pandemic genotype of Vibrio parahaemolyticus in Dhaka, Bangladesh, and significance of its distribution across different serotypes. J. Clin. Microbiol. 2002;40:284–286. doi: 10.1128/JCM.40.1.284-286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]