Abstract
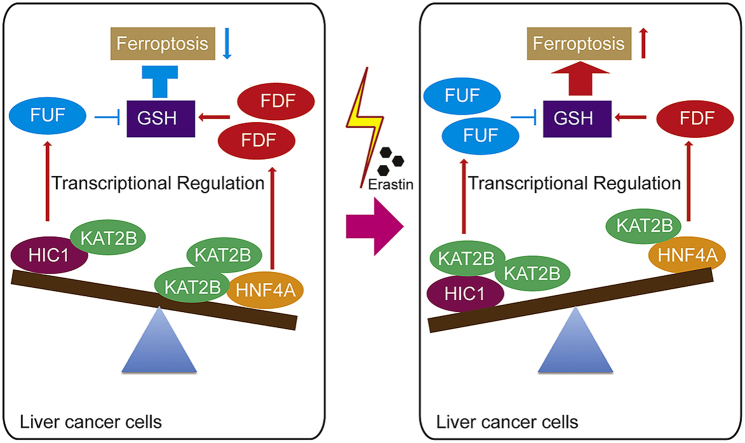
Ferroptosis is an outcome of metabolic disorders and closely linked to liver cancer. However, the mechanism underlying the fine regulation of ferroptosis in liver cancer remains unclear. Here, we have identified two categories of genes: ferroptosis up-regulated factors (FUF) and ferroptosis down-regulated factors (FDF), which stimulate and suppress ferroptosis by affecting the synthesis of GSH. Furthermore, FUF are controlled by one transcription factor HIC1, while FDF controlled by another transcription factor HNF4A. Occurrence of ferroptosis might depend on the histone acetyltransferase KAT2B. Upon stimulation of ferroptosis, dissociation of KAT2B prevents HNF4A from binding to the FDF promoter. This effect happens prior to the recruitment of KAT2B to the FUF promoter, which facilitates HIC1 binding to transcribe FUF. Clinically, HIC1 and HNF4A conversely correlate with tumor stage in liver cancer. Patients with lower HIC1 and higher HNF4A exhibit poorer prognostic outcomes. Disrupting the balance between HIC1 and HNF4A might be helpful in treating liver cancer.
Keywords: HIC1, HNF4A, GSH, Acetyltransferase, Promoter, Metabolism
Abbreviations: FUF, ferroptosis up-regulated factors; FDF, ferroptosis down-regulated factors; GSH, glutathione; HIC1, hypermethylated in cancer 1; HNF4A, hepatocyte nuclear factor 4 alpha; KAT2B, lysine acetyltransferase 2B; ROS, reactive oxygen species; VHL, von Hippel Lindau; TF, transcription factors; NRF2, nuclear factor erythroid 2-related factor 2; MT, metallothionein; DMSO, dimethylsulfoxide; DEN, diethylnitrosamine; CHX, cycloheximide; HBA1, hemoglobin subunit alpha 1; STMN1, stathmin 1; PSAT1, phosphoserine aminotransferase; IHC, immunohistochemistry; IF, immunofluorescence; WB, western blotting; qPCR, quantitative RT-PCR; co-IP, co-immunoprecipitation; ChIP, chromatin immune-precipitation; PLA, protein ligation assay; MDA, malondialdehyde; 3-PG, 3-phosphoglyceric acid; p-Pyr, phosphohydroxy pyruvate p-Ser, phosphoserine; ELISA, enzyme linked immunosorbent assay; TMT, tandem mass tags; BWA, Burrows-Wheeler Alignment; DHSs, DNase I hypersensitive sites; FDR, false discovery rate; TSS, transcription start site; STRING, Search Tool for the Retrieval of Interacting Genes; TMA, tissue microarray assay; GPX4, glutathione peroxidase 4; WT, wild-type; FDA, Food and Drug Administration
Graphical abstract
Highlights
-
•
Opposite gene expression profiles trigged by ferroptosis are identified.
-
•
Reduction of GSH is the major cause of inducing ferroptosis in liver cancer cells.
-
•
Genes related to ferroptosis are controlled by transcription factors HIC1 and HNF4A.
-
•
Stimulation of ferroptosis breaks the balance between HIC1 and HNF4A.
-
•
Lower HIC1 with higher HNF4A has poorer clinical outcomes in liver cancer patients.
1. Introduction
Liver cancer is closely related to metabolic disorders [1]. Glucose metabolism, including glycolysis and hexosamine synthesis, is aberrantly activated in liver cancer, leading to enhanced malignant phenotypes [2,3]. Moreover, dysregulation of lipid metabolism causes selective loss of intrahepatic CD4 (+) T lymphocytes and activation of c-Fos/LXRα signaling, thereby accelerating liver cancer development [[4], [5], [6]]. Therefore, targeting metabolic disorders might provide new strategies to treat liver cancer [[6], [7], [8]].
Reactive oxygen species (ROS) are chemically reactive metabolites containing oxygen that decrease in liver cancer [9,10]. Accumulation of ROS by exogenous drugs such as erastin leads to oxidative cell death, known as ferroptosis [11]. Either the suppression of glutathione (GSH) synthesis or the overload of iron induces ferroptosis [12,13]. In lung cancer, ferroptosis is inhibited because genes related to GSH biosynthesis are activated [14,15]. In renal cell carcinoma, the loss of functional von Hippel Lindau (VHL) inhibits ferroptosis, whereas restoration of functional VHL reverts renal carcinoma cells to an oxidative metabolism and renders them insensitive to the induction of ferroptosis [16]. As for liver cancer, the transcription factor (TF) nuclear factor erythroid 2-related factor 2 (NRF2) activates the expression of a series of reductases that can inhibit ferroptosis by suppressing ROS accumulation; furthermore, the elevation of proto-oncoprotein p62 prevents NRF2 from degradation and subsequently enhances transcription activity of NRF2 [17]. Additionally, metallothionein (MT)-1G has been identified as inhibiting GSH depletion and lipid peroxidation, which suppresses potential ferroptosis in liver cancer [18]. Although the molecular mechanism underlying the initiation and progression of ferroptosis is beginning to be understood, a comprehensive disclosure of the regulatory network of ferroptosis in liver cancer is not yet available.
It is well known that liver cancer cells may be able to be eliminated in a ferroptosis-dependent manner [7,19], and erastin has been reported as being capable of inducing ferroptosis and liver cancer cell death [18,20]. Improved understanding of the mechanism underlying how to regulate ferroptosis might be helpful for designing a therapeutic drug to treat liver cancer.
The aim of the present study is to comprehensively reveal the regulatory network that controls ferroptosis in liver cancer. We have uncovered that ferroptosis is tightly regulated by two categories of genes that have opposite functions. Furthermore, two TFs, HIC1 and HNF4A, have been identified to differentially and transcriptionally control these genes. Disrupting the balance between HIC1 and HNF4A is prerequisite for inducing ferroptosis upon treatment with erastin. Stimulating HIC1 while simultaneously inhibiting HNF4A might be helpful for treating liver cancer.
2. Materials and methods
2.1. Mouse experiments
For xenograft experiments, Bel-7402 cells (initial 5 × 106) under indicated treatment were subcutaneously injected into 8-week-old athymic nude mice (Bikai, Shanghai, China). Dimethylsulfoxide (DMSO) or piperazine erastin (20 mg/kg, MedChemExpress, Monmouth Junction, NJ, USA) with or without liproxstatin-1 (10 mg/kg, MedChemExpress) was subcutaneously injected once daily after xenografts were obviously formed. The tumor volume was calculated as 0.5 × L × W2, with L indicating length and W indicating width.
For liver cancer mouse model induction, the female C57BL/6J mice (with indicated proteins overexpressed or knocked out) were intraperitoneally injected with DEN (100 mg/kg) only once. Two weeks later, mice were intraperitoneally injected with CCl4 (0.5 ml/kg, dissolved in olive oil) weekly for 14 weeks until the formation of liver cancer. Next, DMSO, piperazine erastin (30 mg/kg) with or without liproxstatin-1 (12 mg/kg) was intraperitoneal injected once daily for 4 weeks.
HNF4A and HIC1 knockout mice were generated using CRISPR/Cas9 in C57BL/6J mice by Shanghai Biomodel Organism Science and Technology Development Co, Ltd. (Shanghai, China). To generate mouse models that specifically overexpress HNF4A and HIC1 in the liver, we used the pLIVE expression vector (Mirus Bio, Madison, WI, USA). Briefly, HNF4A-Myc and HIC1-FLAG were cloned into the pLIVE vector. HNF4A-Myc-pLIVE or HIC1-FLAG-pLIVE plasmids (diluted in Delivery Solution (Mirus)) were injected into the mouse's tail.
All mouse experiments were performed according to the institutional guidelines of Shanghai Tenth People's Hospital.
2.2. Tissues, cell cultures and vectors
Tumor and adjacent normal liver tissue samples were acquired at Shanghai Tenth People's Hospital under institutional approvals. Informed written consent was obtained from all the patients. The liver cancer cell lines Bel-7402, SMMC-7721, Bel-7404 and SK-Hep1 were purchased from Cell bank of Chinese Academy of Sciences (Shanghai, China). Other two types of liver cancer cell lines HepG2 and Huh7 were purchased from Cobioer (Nanjing, China). For the hepatocyte cell lines, THLE-3 was purchased from Biovector (Beijing, China) and HL-7702 from Cell bank of Chinese Academy of Sciences. Cells were cultured in DMEM and were treated with erastin (5–20 μM, St Louis, MO, USA, Sigma), ferrostatin-1 (1 μM, Sigma), or cycloheximide (CHX) (50 μg/ml, MedChemExpress). Primary mouse hepatocytes were isolated from mice using liver perfusion and digest medium (Life Technologies, Pleasanton, CA) followed by separation with 50% Percoll (Sigma) density gradient, and cultured in DMEM. The HBA1, STMN1, HIC1, HNF4A and PSAT1 expression plasmids were purchased from Origene (Beijing, China). The HBA1-sh1, STMN1-sh1, HIC1-sh1, HNF4A-sh1 and PSAT1-sh1 were purchased from Biolink (Shanghai, China). The HBA1-sh2, STMN1-sh2, HIC1-sh2, HNF4A-sh2 and PSAT1-sh2 were purchased from Open Biosystems (Huntsville, AL, USA). pTSB-CMV-GFP-EF1-PURO and pTSB-CMV-mCherry-EF1-NEO plasmids were purchased from Transheep (Shanghai, China). pHBA (HIC1)-GFP was constructed using pTSB-CMV-GFP-EF1-PURO as the backbone, and pSTMN1 (HNF4A)-mCherry was constructed using pTSB-CMV-mCherry-EF1-NEO as the backbone. Promoter regions of human HBA1, NNMT, PLIN4, STMN1, CAPG and RRM2 genes were PCR amplified from gDNA of HepG2 cells and cloned into the pGL4.21 (Promega, Madison, WI, USA) vectors. The mutant promoter plasmids were constructed using overlapping PCR.
2.3. Immunohistochemistry (IHC)
The IHC was performed using conventional protocols which are available elsewhere. The primary antibodies were anti-c-PARP (Abcam, Hong Kong, China, #ab32064), anti-HMGB1 (Abcam, #ab18256), anti-GPX4 (Abcam, #ab125066), anti-HBA1 (Abcam, #ab191183), anti-STMN1 (Abcam, #ab52630), anti-HIC1 (Abcam, #ab235037) and anti-HNF4A (R&D Systems, Wiesbaden, Germany, PP-H6939-00 and Abcam, #ab181604). The tissue microarray assay (TMA) slides used in this study were purchased from U.S. Biomax agented by Alenabio (Xi'an, China) and OUTDO Biotech Co. LTD (Shanghai, China). Immunohistochemistry staining was assessed by independent pathologists.
2.4. Immunofluorescence (IF)
Cells were firstly plated and grown in 24-well plates for 24 h. On the second day, pHBA (HIC1)-GFP (puromycin-resistant) and pSTMN1 (HNF4A)-mCherry (G418-resistant) plasmids were co-transfected into the cells. After 24 h, culture medium was replaced by the one containing puromycin (1 μg/ml) and G418 (10 mg/ml) for another 2 days. Then, cells were treated with DMSO or erastin (10 μM) for 24 h before harvest followed by being fixed and blocked. Nuclei were counterstained with DAPI. All images were collected via a confocal microscope.
2.5. Western blotting (WB)
The WB was performed using conventional protocols which are available elsewhere. The primary antibodies were anti-c-PARP (Abcam, #ab32064), anti-HMGB1 (Abcam, #ab18256), anti-GPX4 (Abcam, #ab125066), anti-GAPDH (CST, Boston, MA, USA, #5174), anti-HBA1 (Abcam, #ab191183), anti-PLIN4 (Abcam, #ab234752), anti-NNMT (Abcam, #ab223513), anti-STMN1 (Abcam, #ab52630), anti-RRM2 (Abcam, #ab209995), anti-CAPG (Abcam, #ab155688), anti-HIC1 (Abcam, #ab49326), anti-HNF4A (R&D Systems, PP-H6939-00 and Abcam, #ab181604), anti-KAT2B (Abcam, #ab12188), anti-Myc (CST, #2276) and anti-FLAG (CST, #8146).
2.6. Measurements of luciferase activity and colony formation
Luciferase activities were measured using a dual-luciferase reagent (Promega). Colony formation capacity was measured as previously described [21].
2.7. Quantitative RT-PCR (qPCR) and co-immunoprecipitation (co-IP)
Quantitative RT-PCR was performed as previously described [21]. The primers are listed in Supplementary Table S1.
The antibodies used for co-IP were anti-HIC1 (Sigma, #SAB1412231, and #SAB2103167) and anti-HNF4A (R&D Systems, PP-H6939-00), and the conventional co-IP protocols are available elsewhere.
2.8. Chromatin immune-precipitation (ChIP) and protein ligation assay (PLA)
ChIP was performed as previously described [22]. The antibodies used for ChIP were anti-H3K9Ac (Abcam, #ab4441), anti-KAT2B (Abcam, #ab12188), anti-HNF4A (R&D Systems, PP-H6939-00), anti-IgG (CST, #3900) and anti-HIC1 (Sigma, #SAB1412231). The primers are listed in Supplementary Table S1.
The PLA was performed as previously described [21]. The primary antibodies used were anti-HIC1 (Sigma, #SAB1412231 or #SAB2103167), anti-HNF4A (R&D Systems, PP-H6939-00) and anti-KAT2B (Abcam, #ab12188).
2.9. Examination of metabolites
Iron and malondialdehyde (MDA) were measured using the kits from Abcam. GSH, phospholipid, glutamate, cysteine and glycine were measured using the kits from Sigma. Cystathionine, serine, 3-phosphoglyceric acid (3-PG), phosphohydroxy pyruvate (p-Pyr) and phosphoserine (p-Ser) were measured using the enzyme linked immunosorbent assay (ELISA) kits from Lichen Biotech Ltd. (Shanghai, China).
2.10. RNA-seq and tandem mass tags (TMT) analysis
RNA-seq was performed by Biozeron Biotechnology (Shanghai, China) and analyzed as previously described [21]. TMT analysis was performed and analyzed by Luming Biotechnology (Shanghai, China).
2.11. DNase-seq data analysis
DNase-seq of HepG2 (Accession number: ENCSR149XIL) and normal liver (Accession number: ENCSR555QAY) were obtained from ENCODE database [23]. Reads were mapped to the human hg19 genome assembly by Burrows-Wheeler Alignment (BWA) [24] with default parameters, and only the read pairs with unique mapping location to the genome and with high mapping quality score (mapq≥10) were retained for further analysis. DNase I hypersensitive sites (DHSs) were called using HotSpot algorithm [25] with false discovery rate (FDR) < 0.01. To generate a high confident set of DHSs, only the sites observed both in biological replicate 1 and biological replicate 2 were used for HepG2. Sample specific DHSs were identified by comparing the DHSs of HepG2 and normal liver right lobe: the DHSs of one sample do not overlap with those of the other sample were defined as sample specific DHSs. Sample specific DHSs were annotated by Homer software [26] with the human hg19 RefSeq annotation from UCSC genome browser. The DHSs within 2 kb of a gene's transcription start site (TSS) were defined as a promoter-proximal DHS. The gene sets with only HepG2 promoter-proximal DHSs were defined as HepG2 vs. normal liver DNase-seq open gene set, and the gene sets with only normal liver promoter-proximal DHSs as normal liver vs. HepG2 DNase-seq open gene set. Normal liver vs. HepG2 DNase-seq open gene set overlapped with the genes up-regulated in erastin from TMT and RNA-seq methods to find the anti-tumorigenic genes, and HepG2 vs. Normal liver DNase-seq open gene set overlapped with the genes down-regulated in erastin from TMT and RNA-seq methods to find the pro-tumorigenic genes.
2.12. Motifs discovery of promoter-proximal DHSs
The motifs enriched in promoter-proximal DHSs of pro-ferroptosis FUF genes and anti-ferroptosis FDF genes were identified by find MotifsGenome.pl in Homer software [26].
2.13. Search tool for the retrieval of interacting genes (STRING)
HIC1, HNF4A and 10 types of histone acetyltransferases (KAT2B, EP300, KAT2A, KAT5, KAT6B, KAT7, KAT8, HAT1, NAA60) and methyltransferases (SUV39H1, SUV39H2, SETDB1, SETDB2, SETD2, EHMT1, EHMT2, MLL, MLL3, MLL4) were selected to predict their interactions in STRING database (version 10.5, https://string-db.org). We fetched all interactions which had a confidence score ≥0.27 (medium confidence).
2.14. Statistical analysis
Tests to examine the differences between groups included one-way ANOVA, log rank and the χ [2] test. ** denote p values of <0.01.
3. Results
3.1. Ferroptosis is suppressed in liver cancer and its function is to inhibit liver tumorigenesis
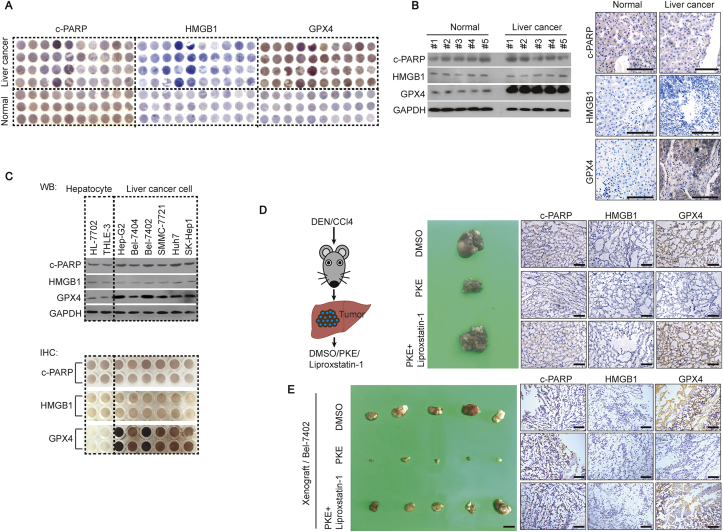
Here, we evaluated whether ferroptosis is critical in liver cancer. Through a tissue microarray assay (TMA), we noticed that glutathione peroxidase 4 (GPX4), which is negatively associated with the levels of ferroptosis [27], was highly expressed in liver cancer compared to that in normal liver tissues; however, the levels of c-PARP (biomarker of apoptosis) [27] and HMGB1 (biomarker of necrosis) [27] were similar between liver cancer and normal liver tissues (Fig. 1A). Unlike c-PARP and HMGB1, GPX4 had a higher expression level in fresh liver cancer tissues compared to that in paired normal liver tissues (Fig. 1B and Supplementary Fig. S1). By testing another 2 hepatocyte lines and 6 liver cancer cell lines, similar expression patterns of c-PARP, HMGB1 and GPX4 were also observed (Fig. 1C). These results suggested that ferroptosis might be suppressed in liver tumorigenesis. The HepG2 and Bel-7402 liver cancer cell lines, which had the lowest ferroptosis levels (Fig. 1C), were used as the main materials in the following studies.
Fig. 1.
Ferroptosis is suppressed in liver cancer. (A) TMA of c-PARP, HMGB1 and GPX4 in liver cancer and normal liver tissues, as measured by IHC. (B–C) Western blots and IHC of c-PARP, HMGB1 and GPX4 in fresh normal liver and liver cancer tissues (B), and established hepatocyte and liver cancer cell lines, as indicated (C). The IHC images in Fig. 1B are representative ones from patient #1. Scale bar, 200 μm. (D) Ferroptosis inhibited liver tumorigenesis in DEN/CCl4-treated mice. Erastin (10 mg/kg) with or without ferrostatin-1 (2 mg/kg) was treated after tumors formed in mice. The liver-bearing tumor in mice with the indicated treatment was shown. Scale bar, 1 cm, n = 5/group. The expressions of c-PARP, HMGB1 and GPX4 in the tumor were measured by IHC. Scale bar, 200 μm n = 5/group. (E) Stimulation of the ferroptosis-inhibited formation of xenografts. Xenografts in mice with indicated treatments are shown. Scale bar, 1 cm, n = 5/group. The expression of c-PARP, HMGB1 and GPX4 in xenografts were measured by IHC. Scale bar, 200 μm n = 5/group. Images of IHC and WB are representative ones of 3–5 independent experiments (except Fig. 1A).
We investigated whether ferroptosis inhibits liver tumorigenesis in DEN/CCl4-induced liver cancer and xenograft mice models. Mice were treated with piperazine erastin, the stimulus of ferroptosis which is stable in vivo, with or without simultaneous treatment of liproxstatin-1, an inhibitor of ferroptosis which is stable in vivo. We found that the size and number of tumor foci in the liver of DEN/CCl4-induced mice, the volume of xenografts formed by Bel-7402 cells, and the levels of GPX4 were noticeably reduced by piperazine erastin. However, such effects could be reversed by simultaneous treatment of liproxstatin-1 (Fig. 1D and E). The specific effects by piperazine erastin were also proved by the unchangeable levels of c-PARP and HMGB1 (Fig. 1D and E). These results demonstrated that stimulation of ferroptosis inhibits liver tumorigenesis.
3.2. Opposite gene expression profiles trigged by stimulation of ferroptosis
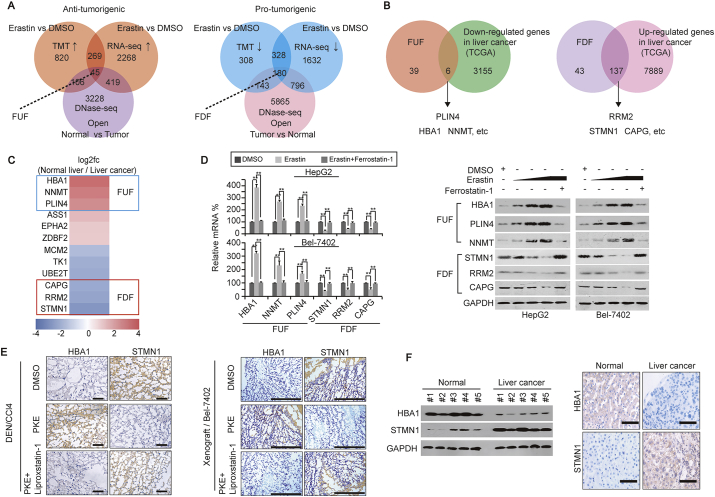
To uncover how ferroptosis is suppressed in liver cancer, we first investigated gene expression profiles altered by stimulation of ferroptosis. Through tandem mass tags (TMT) and RNA-seq, we identified 314 factors, in which both mRNA and protein were up-regulated by erastin, a well-acknowledged stimulus of ferroptosis. Through a DNase-seq, more obvious open chromatins were observed within the promoter of 45 of these 314 factors in the normal liver compared to the HepG2 cells, suggesting these 45 factors [hereafter referred to ferroptosis up-regulated factors (FUF)] might be anti-tumorigenic (Fig. 2A). In contrast, 180 possibly pro-tumorigenic factors [hereafter referred to ferroptosis down-regulated factors (FDF)] were predicted to be down-regulated by erastin, and their promoters were more open in HepG2 cells compared to the normal liver (Fig. 2A). By data mining using the TCGA database, 6/45 FUF were further identified to be down-regulated, while 137/180 FDF identified to be up-regulated in liver cancer (Fig. 2B). Additionally, via the TCGA database, HBA1, NNMT and PLIN4 were identified as the 3 most up-regulated FUF, while STMN1, RRM2 and CAPG were the 3 most down-regulated FDF in the normal liver compared to liver cancer tissues (Fig. 2C).
Fig. 2.
Differential gene expression is trigged by ferroptosis. (A) FUF and FDF were identified by TMT, RNA-seq and DNase-seq in HepG2 cells. (B) Six FUF and 137 FDF were further identified by the TCGA database in liver cancer. (C) Ranking of FUF and FDF as fold changes in the normal liver compared to liver cancer, as analyzed by the TCGA database. (D) qPCR and Western blots of FUF and FDF in the control (treated with DMSO) and HepG2 or Bel-7402 cells treated with erastin (left: 10 μM, right: 5–20 μM) with or without ferrostatin-1 (1 μM) for 24 h. (E) Representative IHC images of HBA1 and STMN1 in DEN/CCl4-induced mouse liver cancer or xenografts formed by Bel-7402 cells. Mice were treated with DMSO or erastin (for DEN/CCl4 mice: 10 mg/kg; for xenograft mice: 5 mg/kg) with or without ferrostatin-1 (for DEN/CCl4 mice: 2 mg/kg; for xenograft mice: 1 mg/kg) after liver cancer and xenografts were formed. Scale bar, 200 μm n = 5/group. (F) Expression of HBA1 and STMN1 in human liver cancer and paired normal liver tissues, as measured by Western blotting and IHC. Scale bar, 200 μm. The data are shown as the means + SD from three independent experiments (Fig. 2d). Images of IHC and WB are representative ones of 3–5 independent experiments. **, p < 0.01 indicate statistical significance. The data from Fig. 2d were analyzed by a one-way ANOVA test.
Next, we investigated whether FUF and FDF can be controlled by stimulation of ferroptosis. In cell-based in vitro experiments, we verified that mRNA and protein levels of FUF were up-regulated by erastin, while mRNA and protein levels of FDF down-regulated by erastin. However, such effects could be completely reversed by simultaneous treatment of ferrostatin-1 (a well-acknowledged inhibitor of ferroptosis) in both HepG2 and Bel-7402 cells (Fig. 2D). In DEN/CCl4-induced mouse liver cancer models, we found that injecting piperazine erastin led to a significant elevation of HBA1, while a suppression of STMN1 in liver cancer. Such effects could also be reversed by simultaneously injecting liproxstatin-1 (Fig. 2E). Moreover, similar findings could be observed in xenografts formed by Bel-7402 cells (Fig. 2E). Notably, the expression of HBA1 was suppressed in human liver cancer compared to the normal liver, while STMN1 exhibited the opposite expression pattern (Fig. 2F). These results suggested that stimulation of ferroptosis might trigger a differential expression of FUF (represented by HBA1) and FDF (represented by STMN1), which might play opposite roles in liver cancer.
3.3. Reduction of GSH leads to ferroptosis in liver cancer cells
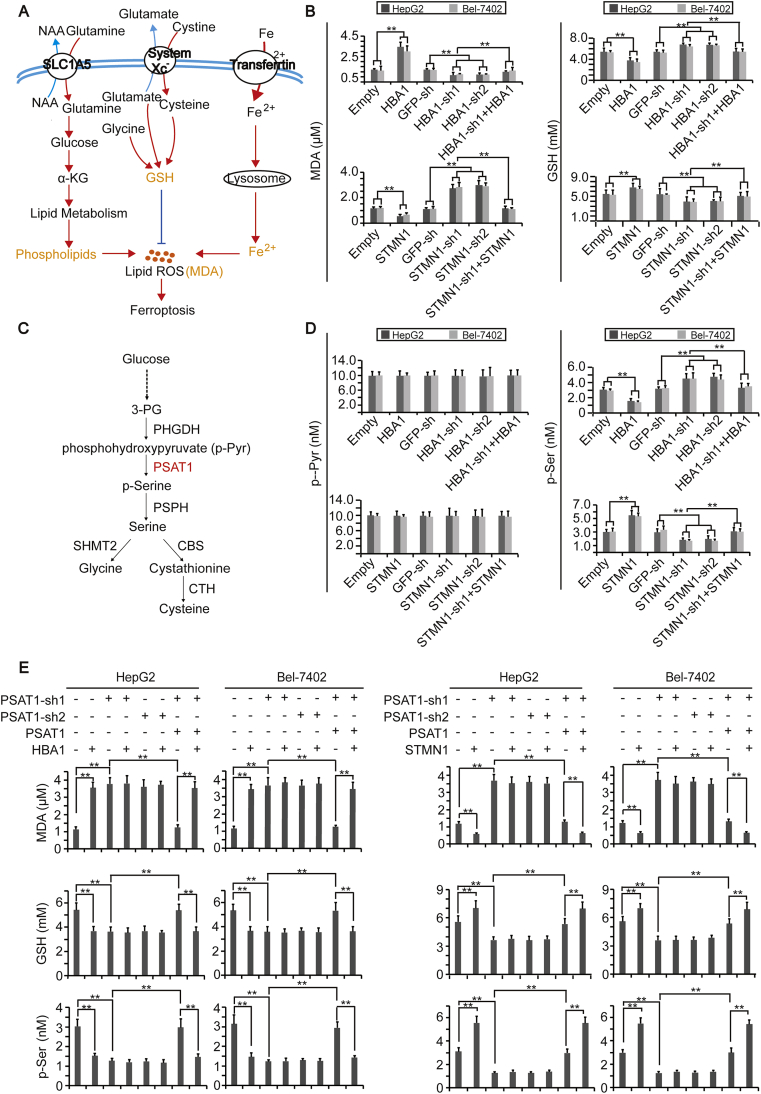
Accumulation of lipid ROS is regarded as the final step to induce ferroptosis [11,28]. At least three pathways control production of MDA, one type of lipid ROS, i.e., phospholipids and Fe2+ pathways elevate MDA levels, while the GSH pathway lowers MDA levels [29] (Fig. 3A). To further investigate the mechanism underlying how FUF and FDF influence ferroptosis, HBA1 and STMN1 were knocked down or overexpressed in HepG2 and Bel-7402 cells. We found that HBA1 and STMN1 play contrary roles in the regulation of MDA and GSH (Fig. 3B). However, neither HBA1 nor STMN1 had an effect on iron and phospholipids (Supplementary Figs. S2A–B). These results demonstrated that FUF stimulates ferroptosis, while FDF suppresses ferroptosis possibly in a GSH-dependent manner.
Fig. 3.
HBA1 and STMN1 regulate ferroptosis by affecting GSH by PSAT1. (A) Schematic presentation of signal pathways that affect accumulation of lipid ROS to cause ferroptosis. (B) Concentrations of MDA and GSH in control cells, HepG2 and Bel-7402 cells with HBA1 and STMN1 overexpressed or knocked down. (C) The metabolic axis from glucose to glycine and cysteine. (D) Concentration of p-Pyr and p-Ser in control cells, HepG2 and Bel-7402 cells with HBA1 and STMN1 overexpressed or knocked down. (E) Concentrations of MDA, GSH and p-Ser in control cells, HepG2 and Bel-7402 cells with PSAT1 knocked down in the presence or absence of simultaneous PSAT1 overexpression, with or without HBA1 or STMN1 overexpression, as indicated. The data are shown as the means + SD from three independent experiments. **, p < 0.01 indicate statistical significance. The data were analyzed by a one-way ANOVA test.
Since production of GSH can be stimulated by glutamate, cysteine and glycine, we further evaluated whether these three metabolites could be influenced by HBA1 and STMN1. Like the changes of GSH, the levels of glycine were down-regulated by HBA1, and up-regulated by STMN1 (Supplementary Figs. S2C–D), yet the level of glutamate was not altered (Supplementary Fig. S2E). Because both glycine and cysteine can be produced by the metabolic axis from glucose to serine [30,31] (Fig. 3C), we then investigated whether HBA1 and STMN1 regulate glycine and cysteine via this axis. Similar to GSH, the levels of cystathionine, serine and p-Ser were inhibited by HBA1 while being stimulated by STMN1; however, the levels of p-Pyr and 3-PG were not changed (Fig. 3D and Supplementary Figs. S2F–H), suggesting the link between p-Pyr and p-Ser is an important node targeted by HBA1 and STMN1.
Because phosphoserine aminotransferase 1 (PSAT1) has been reported to be critically important for the transition from p-Pyr to p-Ser [31], we speculated that PSAT1 might be the target for HBA1 and STMN1. To address this, we knocked down PSAT1 in control and HepG2 or Bel-7402 cells overexpressing HBA1 or STMN1. We found that knocking PSAT1 down led to an increase of MDA, whereas a decrease of GSH and p-Ser. Moreover, the opposites effects on MDA, GSH and p-Ser, which resulted from overexpressing HBA1 or STMN1, were abolished when PSAT1 was knocked down. Such effects were reversed by simultaneously overexpressing PSAT1 (Fig. 3E). However, neither HBA1 nor STMN1 was able to influence PSAT1 expression (Supplementary Fig. S2I). These results suggest that FUF and FDF might affect ferroptosis by simply influencing PSAT1 activity.
3.4. Genes related to ferroptosis are controlled by HIC1 and HNF4A
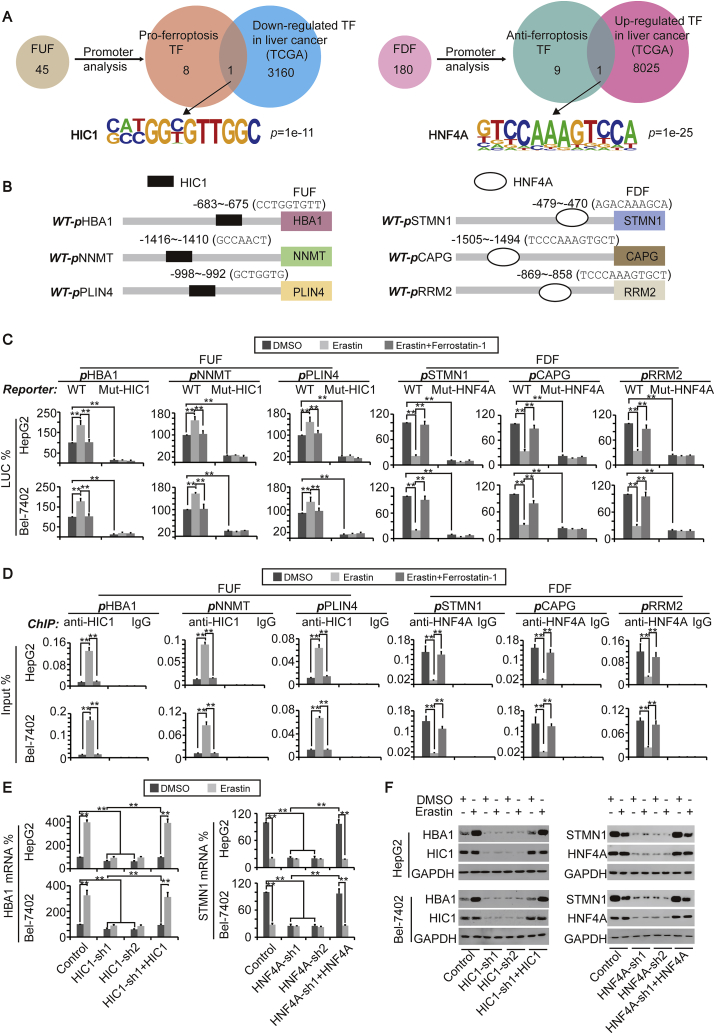
Since FUF (such as HBA1) and FDF (such as STMN1) are both important for ferroptosis, we then further investigated the mechanism underlying the regulation of FUF and FDF, to explain how ferroptosis occurs. Because both the mRNA and protein of FUF and FDF were equidirectionally changed upon erastin stimulation, we speculated that both FUF and FDF are transcriptionally controlled by stimulation of ferroptosis. To address this, we first performed a promoter analysis of 45 FUF, which is shown in Fig. 2A, and found that FUF promoters shared 9 TFs; HIC1 is the only one also predicated to be down-regulated in liver cancer by the TCGA database (Fig. 4A and Supplementary Fig. S3A). Likewise, 10 TFs were shared by the promoters of 180 FDF identified in Fig. 2A, and HNF4A was characterized as the only TF, possibly up-regulated in liver cancer (Fig. 4A and Supplementary Fig. S3A). Luciferase reporters containing FUF (HBA1, NNMT or PLIN4) or FDF (STMN1, CAPG or RRM2) promoter region with or without the HIC1 or HNF4A motif (Fig. 4B) were constructed. We found that promoter activities of FUF could be induced by treatment with erastin, and such effects could be reversed by simultaneous treatment with ferrostatin-1. Once the HIC1 motif was mutated, the basal activities of the FUF promoters were obviously reduced; furthermore, no responses to either erastin or ferrostatin-1 were observed (Fig. 4C), suggesting that HIC1 is essential for stimulation of FUF transcription upon ferroptosis induction. By contrast, stimulation of erastin led to a significant reduction of the FDF promoter activity, which could also be reversed by ferrostatin-1. Interestingly, instead of an induction of promoter activity, a reduction of basal FDF promoter activity with no response to erastin was observed after mutation of the HNF4A motif (Fig. 4C), demonstrating that HNF4A is critical for both basal transcription and suppression of FDF upon ferroptosis stimulation.
Fig. 4.
Ferroptosis-related genes are regulated by HIC1 and HNF4A. (A) A promoter analysis revealed that HIC1 and HNF4A bind to the promoters of FUF and FDF. Software Homer was used to reveal TFs binding to the promoters, and the TCGA database was used to predict factors down-regulated or up-regulated in liver cancer. (B) Location of HIC1 and HNF4A motifs within the promoters of FUF and FDF. (C) Promoter activities of FUF and FDF with (WT) or without (Mut) HIC1 or HNF4A motifs in control cells, HepG2 and Bel-7402 cells treated with erastin (10 μM) in the presence or absence of ferrostatin-1 (1 μM) for 24 h, as measured by a dual-luciferase assay. The italic “P” indicates “Promoter”. (D) HIC1 and HNF4A respond to the treatment of erastin and ferrostatin-1. Enrichment of HIC1 in the FUF promoters and HNF4A in the FDF promoters were measured by ChIP in the control cells, HepG2 and Bel-7402 cells treated with erastin (10 μM) in the presence or absence of ferrostatin-1 (1 μM) for 24 h. The non-specific IgG antibodies were parallel used as the control. The italic “P” indicates “Promoter”. (E–F) mRNA (E) and protein (F) levels of HBA1 and STMN1 in control cells, HepG2 and Bel-7402 cells with HIC1/HNF4A knocked down in the presence or absence of HIC1/HNF4A overexpression. The cells were simultaneously treated with DMSO or erastin (10 μM), as measured by qPCR (E) or WB (F). The data are shown as the means + SD from three independent experiments. **, p < 0.01 indicate statistical significance. The data were analyzed by a one-way ANOVA test. Images of WB are representative ones of 3 independent experiments.
Next, we investigated whether stimulation of ferroptosis affects HIC1 and HNF4A binding to the FUF and FDF promoter. First, binding of HIC1 and HNF4A to the promoter of FUF (including HBA1, NNMT, and PLIN4) and FDF (including STMN1, CAPG, and RRM2) were determined by a ChIP assay (Supplementary Fig. S3B). It was also found that HIC1 could be recruited to the FUF promoter, while HNF4A was released from the FDF promoter upon erastin treatment, and these outcomes could be reversed by simultaneous treating with ferrostain-1 (Fig. 4D).
We then explored the relationship between the expression of FUF/FDF and HIC1/HNF4A. We found that at the basal level, the mRNA and protein of FUF were positively associated with the levels of HIC1, and the mRNA and protein of FDF positively associated with the levels of HNF4A (Supplementary Figs. S3C–D). However, we observed that knockdown of HIC1 would lead to up-regulation of FDF, and knockdown of HNF4A would lead to up-regulation of FUF (Supplementary Fig. S3C), suggesting a loss function of either of HIC1 or HNF4A, which might facilitate the expression of genes on the opposite side.
Because binding HIC1 and HNF4A to the FUF and FDF promoters are controlled by stimulation of ferroptosis, and moreover, the expressions of FUF and FDF are regulated by HIC1 and HNF4A, we hypothesized that the stimulation of ferroptosis regulating FUF and FDF expression might also do so via HIC1 and HNF4A. To address this, erastin was treated in the control, and Bel-7402 or HepG2 cells with HIC1 or HNF4A knocked down. We found that the effects underlying erastin-induced up-regulation of HBA1 or down-regulation of STMN1 were almost not observed in control cells when HIC1 or HNF4A were knocked down; however, these outcomes could be reversed by simultaneous overexpression of HIC1 or HNF4A (Fig. 4E and F).
3.5. HIC1 and HNF4A are mutually inhibited from each other in liver tumorigenesis
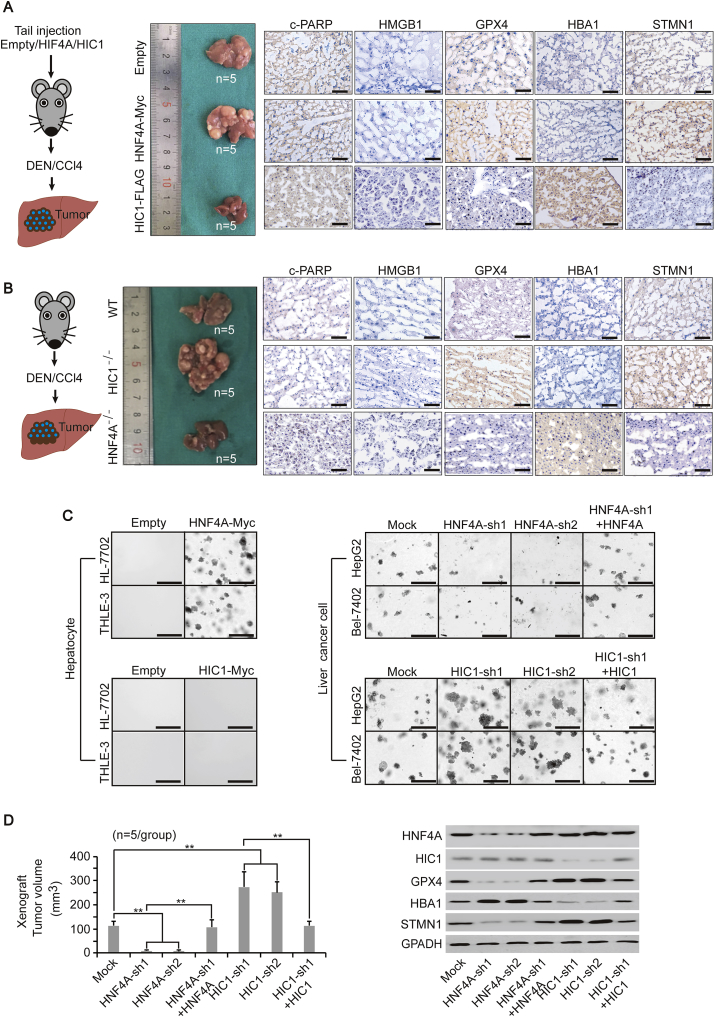
Here, we tested whether HIC1 and HNF4A also have contrary roles in liver tumorigenesis. We first specifically overexpressed HIC1 and HNF4A in the mouse liver via tail injection, and found that it is likely to form larger and more tumor foci in the liver overexpressing HNF4A after DEN/CCl4 induction, while the opposite outcome was seen in the liver overexpressing HIC1 (Fig. 5A and Supplementary Fig. S4A). At the molecular level, overexpressing HNF4A caused an obvious up-regulation of STMN1, which may have led to the elevation of GPX4. By contrary, overexpressing HIC1 increased HBA1 but decreased GPX4 (Fig. 5A). However, neither overexpressing HNF4A nor HIC1 caused changes of c-PARP and HMGB1, indicating that apoptosis and necrosis were not involved. These results suggested that changing FUF and FDF, such as HBA1 and STMN1, might affect ferroptosis and liver tumorigenesis.
Fig. 5.
HNF4A and HIC1 have the opposite function to affect liver tumorigenesis. (A) Overexpression of HNF4A and HIC1 had the opposite function in regulating liver tumorigenesis. Mice were injected with pLIVE liver-specific expressing plasmids that overexpressing HNF4A-Myc or HIC1-FLAG via the tail vein prior to the treatment of DEN/CCl4. Livers from mice under the indicated treatment are shown (n = 5/group). c-PARP, HMGB1, GPX4, HBA1 and STMN1 from the liver, as indicated, were measured by IHC. Scale bar, 200 μm. (B) Knocking out HIC1 and HNF4A led to opposite outcomes in liver tumorigenesis. Livers from WT, HIC1−/− and HNF4A−/− mice are shown (n = 5/group). The indicated proteins in the liver were measured by IHC. Scale bar, 200 μm. (C) Colony formation capacities in the hepatocyte line (HL-7702 and THLE-3) and liver cancer cell line (HepG2 and Bel-7402) under indicated treatment, as measured by the soft agar colony formation assay. Scale bar, 200 μm. (D) Volume of xenograft formed by the Bel-7402 cells was graphed (left, n = 5/group). WB of HNF4A, HIC1, GPX4, HBA1 and STMN1 in the xenograft are also shown (right). **, p < 0.01 indicate statistical significance. The data were analyzed by a one-way ANOVA test (Fig. 5D). Images of IHC, colony formation, and WB are representative ones of 3 independent experiments.
We treated DEN/CCl4 in mice with HIC1 or HNF4A knocked out, and found that lacking HIC1 stimulated liver cancer, while lacking HNF4A suppressed tumor formation (Fig. 5B). Intriguingly, although HIC1 and HNF4A directly control the expression of HBA1 and STMN1, lacking either of these two TFs enhanced the expression of protein with opposite function (Fig. 5B), suggesting HIC1 and HNF4A are critically important to keeping the balance of genes related to ferroptosis.
In cell-based experiments, hepatocyte lines HL-7702 and THLE-3 acquired colony formation capacity once HNF4A was overexpressed; however, overexpressing HIC1 had no such effects (Fig. 5C). To investigate their impact on the transformative phenotypes of liver cancer cells, we knocked down HNF4A and HIC1 in Bel-7402 and HepG2 cells. The knocking down of HNF4A impaired colony formation capacity, which could be reversed by simultaneously overexpressing HNF4A. This observation provided further evidence that HNF4A is pro-tumorigenic. By contrast, the knocking down of HIC1 enhanced colony formation capacity, which was significantly lower when HIC1 was simultaneously overexpressed (Fig. 5C).
As observed in the mice models, HNF4A significantly induced STMN1 and GPX4 expression, while HIC1 induced HBA1 but reduced GPX4 expression in both HepG2 and Bel-7402 cells (Supplementary Fig. S4B). The xenograft models also proved the opposite functions of HNF4A and HIC1 in controlling the expression of FDF and FUF, liver tumorigenesis and ferroptosis in vivo (Fig. 5D and Supplementary Fig. S4C). These data suggested that HNF4A and HIC1 play opposite roles in maintaining transformative phenotypes of liver cancer cells, possibly via their contrary functions on ferroptosis.
As described above, STMN1 and HBA1 affect GSH production to regulate ferroptosis, and moreover, HNF4A and HIC1 are upstream regulators of STMN1 and HBA1; therefore, we hypothesized that HNF4A and HIC1 also play opposites roles in regulating GSH. To address this, we tested MDA, GSH, p-Ser and p-Pyr in the livers of wild-type (WT), HNF4A−/- and HIC1−/− mice treated with or without erastin. Indeed, the basal level of MDA was remarkably elevated in HNF4A−/- mice compared to the WT mice. By contrast, the MDA was significantly suppressed when HIC1 was knocked out. Compared to the WT mice, the basal GSH and p-Ser levels were significantly decreased in HNF4A −/− mice, and significantly increased in HIC1−/− mice. Additionally, the p-Pyr was not changed regardless of changes in either HNF4A−/- or HIC1−/− mice (Supplementary Fig. S4D). In the livers of WT mice, MDA, GSH or p-Ser responded remarkably to the erastin treatment; this effect was significantly strengthened in the liver of HNF4A−/- mice. However, the induction of MDA and the reduction of GSH and p-Ser by erastin in the liver of HIC1−/− mice were not as obvious as in the WT mice (Supplementary Fig. S4D). These results suggested HNF4A antagonises HIC1, and vice versa, in the metabolism related to ferroptosis.
Then, we further investigated whether HNF4A and HIC1 mutually regulate each other. Unexpectedly, compared to the WT, knocking HNF4A out in a mouse liver had no effect on the expression of HIC1, and vice versa. Moreover, erastin also had no influence on the expression of either HNF4A or HIC1. Although the erastin-induced alteration of HBA1 was blocked when HIC1 was knocked out, the alteration of STMN1 blocked when HNF4A was knocked out, knocking HNF4A out led to an increase not only in the basal expression of HBA1, but also in the response of HBA1 to the erastin. Although knocking HIC1 out also led to an increase in the basal level of STMN1, the reduction response of STMN1 to erastin was less obvious compared to the WT one (Supplementary Fig. S4E). The promoter activities of the HBA1 and STMN1 genes were measured in primary hepatocytes from WT, HNF4A−/- or HIC1−/− mice. Expectedly, compared to the WT, the response of the HBA1 promoter to erastin was strengthened when HNF4A was knocked out. However, the response of the STMN1 promoter to erastin was weakened, which resulted from the loss of HIC1 (Supplementary Fig. S4F). These results suggested that HIC1 and HNF4A only mutually regulate function but not the expression with each other.
3.6. Stimulation of ferroptosis disrupts the balance between HIC1 and HNF4A
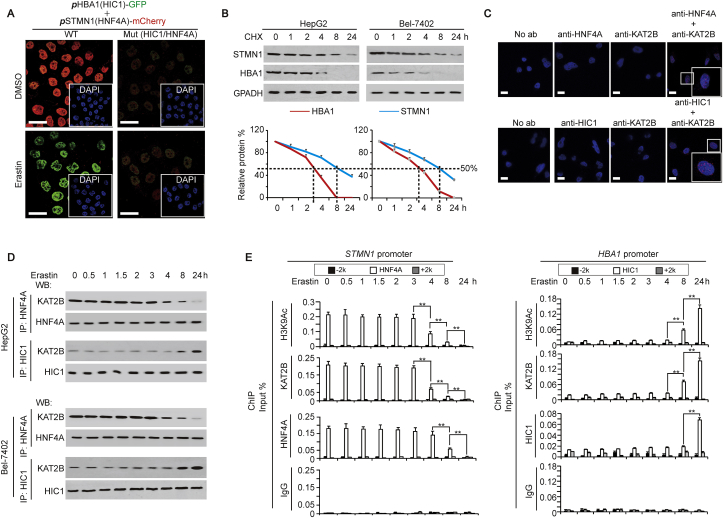
Because HIC1 and HNF4A play opposite roles in ferroptosis, and moreover, ferroptosis is suppressed in liver cancer, we speculated that the function of HNF4A is stronger than that of HIC1. To address this, the corresponding promoter regions of HBA1 genes were cloned just prior to the GFP reporters, and the corresponding promoter regions of STMN1 genes cloned just prior to the mCherry reporters. In control cells (treated with DMSO) co-transfected with WT-pHBA1-GFP (containing the HIC1 motif) and WT-pSTMN1-mCherry (containing the HNF4A motif), we found that the red fluorescence representing mCherry was much brighter than that of green fluorescence representing GFP (Fig. 6A), suggesting that the basal promoter activity of STMN1 is stronger than that of HBA1, which might be anti-ferroptosis in liver cancer cells. However, when treated with erastin, the situation was reversed, i.e., green fluorescence was brighter than red fluorescence (Fig. 6A), suggesting that ferroptosis occurs. We also observed that when HIC1 and HNF4A motifs were mutated simultaneously, the red and green fluorescence became equally weak, regardless of treating DMSO or erastin (Fig. 6A), suggesting that HIC1 and HNF4A motifs are essential for keeping the balance between FUF and FDF, and are responsible for ferroptosis stimulation. We also found that the protein half-life of FDF (including STMN1, RRM2 and CAPG) was much longer than that of FUF (including HBA1, PLIN4 and NNMT) (Fig. 6B and Supplementary Fig. S5A), suggesting that FDF are easier to maintain anti-ferroptosis transformative phenotypes of liver cancer cells.
Fig. 6.
The balance between HIC1 and HNF4A controls FUF and FDF expression. (A) Representative IF images of HepG2 cells co-transfected with WT (containing HIC1 and HNF4A motifs) or Mut (with mutated HIC1 and HNF4A motifs) promoter reporter plasmids: pHBA1-GFP and pSTMN1-mCherry. The cells were treated with same amounts of DMSO or erastin (10 μM). The nuclei are also shown via staining with DAPI. Scale bar, 50 μm. (B) CHX chase experiments of STMN1 and HBA1 in HepG2 and Bel-7402 cells. The cells were harvested at the indicated time point after adding CHX (50 μg/ml). The ratios of STMN1 or HBA1 to GAPDH were also plotted. (C) PLA showing the interaction between HNF4A and KAT2B, and between HIC1 and KAT2B in HepG2 cells. Scale bar, 50 μm. (D) Interactions between HNF4A and KAT2B, and between HIC1 and KAT2B in HepG2 and Bel-7402 cells treated with erastin (10 μM) for indicating time, as demonstrated by co-IP using anti-HNF4A or anti-HIC1 antibodies for IP, and anti-KAT2B antibodies for WB. The HNF4A or HIC1 level in each co-IP samples was adjusted to the same content. (E) Modification of H3K9Ac, and enrichment of KAT2B and HNF4A at the indicated region around the STMN1 and HBA1 promoters in HepG2 cells treated with erastin (10 μM) for the indicated time. The data are shown as the means + SD from three independent experiments. **, p < 0.01 indicate statistical significance. The data were analyzed by a one-way ANOVA test (Fig. 6B and E). Images of IF and WB are representative ones of 3 independent experiments (Fig. 6A–D).
Unexpectedly, no direct interaction between HIC1 and HNF4A was observed by protein ligation assay (PLA) (Supplementary Fig. S5B), suggesting HIC1 and HNF4A might mutually regulate ferroptosis via a third factor. To reveal this factor, we performed a STRING analysis. Because histone modification is critical for transcription [32], we focused on the interaction among HIC1, HNF4A, and enzymes critical for histone modification including methylation and acetylation. KAT2B, an acetyltransferase, was finally identified as co-interacting with both HNF4A and HIC1 (Supplementary Fig. S5C). The direct interactions between HNF4A and KAT2B, and between HIC1 and KAT2B were then confirmed by PLA (Fig. 6C). Interestingly, HIC1 and HNF4A competitively bind with KAT2B, because knocking down HNF4A increased the binding between KAT2B and HIC1, while overexpressing HNF4A led to the opposite outcome; and vice versa (Supplementary Fig. S5D).
We investigated whether stimulation of ferroptosis affects HNF4A and HIC1 binding to KAT2B. Co-IP experiments demonstrated that the interaction between HNF4A and KAT2B began to dissociate 4 h after treatment of erastin, while the interaction between HIC1 and KAT2B began to get closer 8 h after treatment of erastin in both HepG2 and Bel-7402 cells (Fig. 6D). Additionally, ChIP experiments revealed a reduced enrichment of KAT2B accompanied by a decreased level of H3K9Ac modification, a hallmark of open chromatin [33], within the region around the HNF4A motif of the STMN1 promoter, which started at 4 h after erastin treatment. This might lead to a release of HNF4A that began 8 h after erastin treatment (Fig. 6E). By contrast, we observed an induced enrichment of KAT2B with an increased level of H3K9Ac modification within the region around the HIC1 motif of the HBA1 promoter, which started 8 h after treatment with erastin. This effect might facilitate a recruitment of HIC1 onto the HBA1 promoter beginning 24 h after erastin treatment (Fig. 6E). However, signals representing the control IgG were weak and not dependent on erastin treatment for both STMN1 and HBA1 promoters, and moreover, “-2k” or “+2k” regions around the STMN1 and HBA1 promoters shown low H3K9Ac modification, KAT2B, HNF4A and HIC1 recruitment (Fig. 6E), demonstrating the specific data from the “HNF4A/HIC1” region. The above data also indicated that earlier suppressed HNF4A binding to the FDF promoter might be prerequisite for later HIC1 binding to the FUF promoter. Furthermore, a decreased expression of STMN1 was observed starting after 8 h post erastin treatment, while an increased expression of HBA1 was not seen until 24 h after erastin treatment (Supplementary Fig. S5E), further demonstrating that changes of FUF were later than changes of FDF.
3.7. Clinical significance of HNF4A and HIC1 in liver cancer
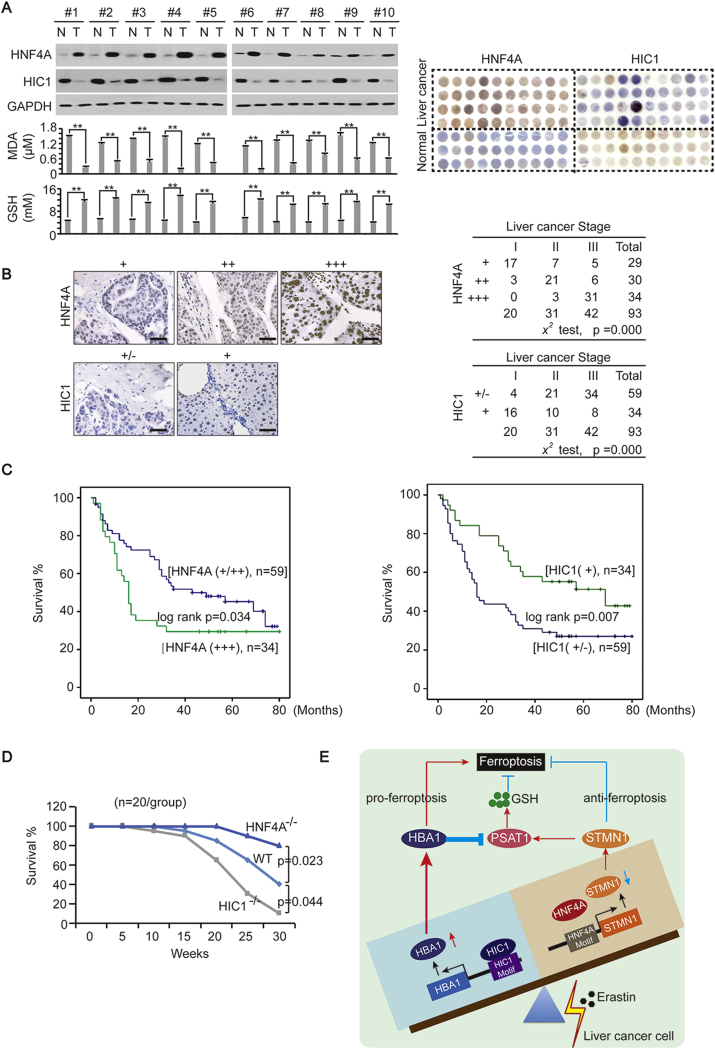
To investigate the clinical significance of HNF4A and HIC1 in liver cancer, we first evaluated their expression. In fresh tissues, we found that HNF4A was up-regulated while HIC1 was down-regulated in liver cancer compared to paired normal liver tissues. Moreover, MDA was comparably lower while GSH was higher in liver cancer than that in normal liver tissue (Fig. 7A and Supplementary Fig. S6). TMA also suggested that HNF4A was up-regulated while HIC1 was down-regulated in liver cancer compared to the normal liver (Fig. 7A). These results provided evidence that ferroptosis may be suppressed in liver cancer.
Fig. 7.
Clinical significance of HNF4A and HIC1 in liver cancer. (A) HNF4A and GSH were elevated while HIC1 and MDA were suppressed in liver cancer compared to paired normal liver tissue. WB was performed in paired liver cancer (T)-normal liver (N) from 10 patients. TMA was also performed to evaluate the HNF4A and HIC1 expression in liver cancer and in the normal liver. (B) HNF4A was positively associated and HIC1was negatively associated with the stage of liver cancer. Representative IHC images of HNF4A and HIC1 were shown. The association between the stage and the expression of HNF4A and HIC1 were analyzed by aχ [2] test. Scale bar, 200 μm. (C) Survival in patients with liver cancer after curative surgery. Patients with different expression levels of HNF4A (left) or HIC1 (right) were analyzed. (D) Survival in WT, HNF4A−/− and HIC1−/− mice with DEN/CCl4 treatment. (n = 20/group) (E) Schematic presentation of the mechanism underlying the interaction between HIC1 and HNF4A to control the balance of FUF and FDF upon stimulation of ferroptosis. The data are shown as the means + SD from three independent experiments. **, p < 0.01 indicate statistical significance. The data were analyzed by a one-way ANOVA test (Fig. 7A lower) and a log rank test (Fig. 7C and D). Images of WB and IHC are representative ones of 3 independent experiments (Fig. 7A, upper left and 7B).
Furthermore, we found that the expression of HNF4A was positively associated with the stage of liver cancer, while the level of HIC1 was negatively associated (Fig. 7B). Additionally, we found that patients with higher HNF4A or lower HIC1 expression had shorter survival times than those with lower HNF4A or higher HIC1 expression (Fig. 7C), further demonstrating that higher HNF4A with lower HIC1 is suggestive of a poorer prognostic outcome.
In mice experiments, in vivo data showed that HNF4A is linked to poor prognostic outcomes while HIC1 had the opposite effect. This conclusion was drawn by the facts that compared to the WT, HNF4A−/- mice with liver cancer induced by DEN/CCl4 had longer survival times, while HIC1−/− mice had shorter survival times (Fig. 7D).
4. Discussion
Here, one transcription factor HNF4A has been identified as suppressing ferroptosis, and another transcription factor HIC1 identified as stimulating ferroptosis in liver cancer (Fig. 7E). HNF4A is critical for liver development [34], and is up-regulated in liver cancer [35,36]. Moreover, HNF4A stimulates the EGFR-mediated proliferative response during liver cancer development [37], and induces transcription of the Hepatitis B and C virus to promote deterioration of liver cancer [38]. By contrast, HIC1 acts as a tumor suppressor, which inhibits cell growth, migration and survival [39]. Despite the function of these two TFs gradually emerges, their target genes in liver cancer cells remain largely unknown. In this study, a serial of pro-ferroptosis genes are revealed to be transcriptionally controlled by HIC1, and a serial of anti-ferroptosis genes revealed to be transcriptionally controlled by HNF4A. Interestingly, at the basal level, HNF4A-mediated anti-ferroptosis transcription is stronger than that of HIC1-mediated pro-ferroptosis transcription. However, once stimulation of ferroptosis is done, the strength of the transcription activated by these two TFs can be completely reversed; moreover, reduction of HNF4A-mediated transcription is done prior to the induction of HIC1-mediated transcription, indicating that holding the activation of HNF4A might be one of the underlying mechanisms that prevent liver cancer cells from ferroptosis. We further reveal that histone acetylation, which resulted from the binding of KAT2B, is essential for keeping the balance of the transcription competitively activated by HNF4A and HIC1 (Fig. 7E). KAT3A, a homologue of KAT2B, has already been reported to induce ferroptosis via acetylation of p53 [40], further supporting the critical roles of KAT proteins in the regulation of ferroptosis. Breaking the balance between HIC1 and HNF4A might be helpful for inducing ferroptosis in liver cancer treatment.
We also have identified STMN1 and HBA1 as the representative FDF and FUF. STMN1 is first characterized as a microtubule-destabilizing phosphoprotein [41]. Overexpression of STMN1 is observed in various tumors and is associated with poor prognosis [[42], [43], [44]]. STMN1 has been characterized as a critical metastasis stimulator in liver cancer [45,46]. Although the link between STMN1 and ferroptosis has not yet been determined, prior studies have reported that oxidative stress, a stimulus of ferroptosis, is able to increase HBA1 in HepG2 cells, indicating HBA1 is involved in ferroptosis [47,48]. However, the precise function of STMN1 and HBA1 in regulating ferroptosis has not been defined until we reveal that the two proteins have opposite functions to control GSH levels in the current study (Fig. 7E).
The links between HNF4A and STMN1, and between HIC1 and HBA1, are critically important for ferroptosis. HNF4A and HBA1 are acknowledged to regulate metabolism. HNF4A stimulates lipolysis while suppressing ER stress and lipogenesis [49,50]. The primary function of HBA1 includes transporting oxygen from the external environment to body tissues, and facilitating metabolic waste removal by assisting the transport of carbon dioxide from tissues back to the respiratory organs [51,52]. To the best of our knowledge, we are the first to reveal that HNF4A-STMN1 and HIC1-HBA1 axes oppositely regulate production of GSH via PSAT1, a key enzyme in GSH synthesis. Since the expression of PSAT1 cannot be influenced by either STMN1 or HBA1, we speculate that PSAT1 might be post-translational modified, because such modification including acetylation, glycosylation and phosphorylation, is able to regulate enzyme activity without affecting its expression [53].
Furthermore, we noticed that ferroptosis is significantly suppressed in liver cancer, and using erastin to induce ferroptosis is able to impair malignant phenotypes and further cancer development. Treating liver cancer with soferanib is approved by Food and Drug Administration (FDA) and widely accepted in clinical studies, and soferanib has been shown to induce ferroptosis in liver cancer [7,19,54]. However, the therapeutic efficacy of soferanib is transient, and almost all patients develop sorafenib resistance within a few months [55]. Interestingly, inhibition of ferroptosis can usually be detected once upon sorafenib resistance occurs [18]. Since erastin and sorafenib have similar pro-ferroptotic functions in liver cancer cells [17,18], we speculate that sorafenib equally regulates HNF4A and HIC1. Increasing the concentration of GSH by targeting HNF4A and HIC1 might improve soferanib resistance for liver cancer treatment.
Data availability
RNA-seq data have been deposited in GEO (accession number: GSE104462). TMT data have been deposited in ProteomeXchange Consortium (accession number: PXD010761). The username for reviewing TMT data is reviewer15024@ebi.ac.uk, and the password is zrLM0EBw.
Ethics approval and consent to participate
This study was approved by the Shanghai Tenth People's hospital ethics committee and written informed consent was obtained from all patients. All animal studies were approved by the Institutional Animal Care and Use Committee of the Shanghai Tenth People's Hospital.
Declaration of interests
The authors have no conflicts of interest to declare.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 81871907, 81822029, 81872288, 81774291, 81672332, 81371913, 81572330, 81472624, and 81472124), the Shanghai Municipal commission of Health and Family Planning (key developing disciplines and outstanding youth training plan no. 2017YQ024) and Shanghai Rising-Star Program (18QA1403400).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101211.
Contributor Information
Qiuhui Pan, Email: panqiuhui@263.net.
Fenyong Sun, Email: sunfenyong@126.com.
Jiayi Wang, Email: karajan2@163.com.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Bruix J., Gores G.J., Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844–855. doi: 10.1136/gutjnl-2013-306627. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut 2014; 63: 844-855. [DOI] [PMC free article] [PubMed]
- 2.Guo W., Qiu Z., Wang Z., Wang Q., Tan N., Chen T. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology. 2015;62:1132–1144. doi: 10.1002/hep.27929. [DOI] [PubMed] [Google Scholar]; Guo W, Qiu Z, Wang Z, Wang Q, Tan N, Chen T et al. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology 2015; 62: 1132-1144. [DOI] [PubMed]
- 3.Zhang X., Qiao Y., Wu Q., Chen Y., Zou S., Liu X. The essential role of YAP O-GlcNAcylation in high-glucose-stimulated liver tumorigenesis. Nat. Commun. 2017;8:15280. doi: 10.1038/ncomms15280. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang X, Qiao Y, Wu Q, Chen Y, Zou S, Liu X et al. The essential role of YAP O-GlcNAcylation in high-glucose-stimulated liver tumorigenesis. Nat Commun 2017; 8: 15280. [DOI] [PMC free article] [PubMed]
- 4.Ma C., Kesarwala A.H., Eggert T., Medina-Echeverz J., Kleiner D.E., Jin P. NAFLD causes selective CD4(+) T lymphocyte loss and promotes hepatocarcinogenesis. Nature. 2016;531:253–257. doi: 10.1038/nature16969. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ma C, Kesarwala AH, Eggert T, Medina-Echeverz J, Kleiner DE, Jin P et al. NAFLD causes selective CD4(+) T lymphocyte loss and promotes hepatocarcinogenesis. Nature 2016; 531: 253-257. [DOI] [PMC free article] [PubMed]
- 5.Bakiri L., Hamacher R., Grana O., Guio-Carrion A., Campos-Olivas R., Martinez L. Liver carcinogenesis by FOS-dependent inflammation and cholesterol dysregulation. J. Exp. Med. 2017;214:1387–1409. doi: 10.1084/jem.20160935. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bakiri L, Hamacher R, Grana O, Guio-Carrion A, Campos-Olivas R, Martinez L et al. Liver carcinogenesis by FOS-dependent inflammation and cholesterol dysregulation. J Exp Med 2017; 214: 1387-1409. [DOI] [PMC free article] [PubMed]
- 6.Nakagawa S., Wei L., Song W.M., Higashi T., Ghoshal S., Kim R.S. Molecular liver cancer prevention in cirrhosis by organ transcriptome analysis and lysophosphatidic acid pathway inhibition. Cancer Cell. 2016;30:879–890. doi: 10.1016/j.ccell.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nakagawa S, Wei L, Song WM, Higashi T, Ghoshal S, Kim RS et al. Molecular Liver Cancer Prevention in Cirrhosis by Organ Transcriptome Analysis and Lysophosphatidic Acid Pathway Inhibition. Cancer Cell 2016; 30: 879-890. [DOI] [PMC free article] [PubMed]
- 7.Wu F.Q., Fang T., Yu L.X., Lv G.S., Lv H.W., Liang D. ADRB2 signaling promotes HCC progression and sorafenib resistance by inhibiting autophagic degradation of HIF1alpha. J. Hepatol. 2016;65:314–324. doi: 10.1016/j.jhep.2016.04.019. [DOI] [PubMed] [Google Scholar]; Wu FQ, Fang T, Yu LX, Lv GS, Lv HW, Liang D et al. ADRB2 signaling promotes HCC progression and sorafenib resistance by inhibiting autophagic degradation of HIF1alpha. J Hepatol 2016; 65: 314-324. [DOI] [PubMed]
- 8.Byun J.K., Choi Y.K., Kang Y.N., Jang B.K., Kang K.J., Jeon Y.H. Retinoic acid-related orphan receptor alpha reprograms glucose metabolism in glutamine-deficient hepatoma cells. Hepatology. 2015;61:953–964. doi: 10.1002/hep.27577. [DOI] [PubMed] [Google Scholar]; Byun JK, Choi YK, Kang YN, Jang BK, Kang KJ, Jeon YH et al. Retinoic acid-related orphan receptor alpha reprograms glucose metabolism in glutamine-deficient hepatoma cells. Hepatology 2015; 61: 953-964. [DOI] [PubMed]
- 9.Yang C., Tan Y.X., Yang G.Z., Zhang J., Pan Y.F., Liu C. Gankyrin has an antioxidative role through the feedback regulation of Nrf2 in hepatocellular carcinoma. J. Exp. Med. 2016;213:859–875. doi: 10.1084/jem.20151208. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang C, Tan YX, Yang GZ, Zhang J, Pan YF, Liu C et al. Gankyrin has an antioxidative role through the feedback regulation of Nrf2 in hepatocellular carcinoma. J Exp Med 2016; 213: 859-875. [DOI] [PMC free article] [PubMed]
- 10.Piccinin E., Peres C., Bellafante E., Ducheix S., Pinto C., Villani G. Hepatic peroxisome proliferator-activated receptor gamma coactivator 1beta drives mitochondrial and anabolic signatures that contribute to hepatocellular carcinoma progression in mice. Hepatology. 2018;67:884–898. doi: 10.1002/hep.29484. [DOI] [PubMed] [Google Scholar]; Piccinin E, Peres C, Bellafante E, Ducheix S, Pinto C, Villani G et al. Hepatic peroxisome proliferator-activated receptor gamma coactivator 1beta drives mitochondrial and anabolic signatures that contribute to hepatocellular carcinoma progression in mice. Hepatology 2018; 67: 884-898. [DOI] [PubMed]
- 11.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012; 149: 1060-1072. [DOI] [PMC free article] [PubMed]
- 12.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014; 156: 317-331. [DOI] [PMC free article] [PubMed]
- 13.Wang H., An P., Xie E., Wu Q., Fang X., Gao H. Characterization of ferroptosis in murine models of hemochromatosis. Hepatology. 2017;66:449–465. doi: 10.1002/hep.29117. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang H, An P, Xie E, Wu Q, Fang X, Gao H et al. Characterization of ferroptosis in murine models of hemochromatosis. Hepatology 2017; 66: 449-465. [DOI] [PMC free article] [PubMed]
- 14.Jiang Y., Mao C., Yang R., Yan B., Shi Y., Liu X. EGLN1/c-Myc induced lymphoid-specific helicase inhibits ferroptosis through lipid metabolic gene expression changes. Theranostics. 2017;7:3293–3305. doi: 10.7150/thno.19988. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jiang Y, Mao C, Yang R, Yan B, Shi Y, Liu X et al. EGLN1/c-Myc Induced Lymphoid-Specific Helicase Inhibits Ferroptosis through Lipid Metabolic Gene Expression Changes. Theranostics 2017; 7: 3293-3305. [DOI] [PMC free article] [PubMed]
- 15.Alvarez S.W., Sviderskiy V.O., Terzi E.M., Papagiannakopoulos T., Moreira A.L., Adams S. NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature. 2017;551:639–643. doi: 10.1038/nature24637. [DOI] [PMC free article] [PubMed] [Google Scholar]; Alvarez SW, Sviderskiy VO, Terzi EM, Papagiannakopoulos T, Moreira AL, Adams S et al. NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature 2017; 551: 639-643. [DOI] [PMC free article] [PubMed]
- 16.Miess H., Dankworth B., Gouw A.M., Rosenfeldt M., Schmitz W., Jiang M. The glutathione redox system is essential to prevent ferroptosis caused by impaired lipid metabolism in clear cell renal cell carcinoma. Oncogene. 2018;37(40):5435–5450. doi: 10.1038/s41388-018-0315-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; Miess H, Dankworth B, Gouw AM, Rosenfeldt M, Schmitz W, Jiang M et al. The glutathione redox system is essential to prevent ferroptosis caused by impaired lipid metabolism in clear cell renal cell carcinoma. Oncogene 2018. [DOI] [PMC free article] [PubMed]
- 17.Sun X., Ou Z., Chen R., Niu X., Chen D., Kang R. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63:173–184. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 2016; 63: 173-184. [DOI] [PMC free article] [PubMed]
- 18.Sun X., Niu X., Chen R., He W., Chen D., Kang R. Metallothionein-1G facilitates sorafenib resistance through inhibition of ferroptosis. Hepatology. 2016;64:488–500. doi: 10.1002/hep.28574. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sun X, Niu X, Chen R, He W, Chen D, Kang R et al. Metallothionein-1G facilitates sorafenib resistance through inhibition of ferroptosis. Hepatology 2016; 64: 488-500. [DOI] [PMC free article] [PubMed]
- 19.Louandre C., Ezzoukhry Z., Godin C., Barbare J.C., Maziere J.C., Chauffert B. Iron-dependent cell death of hepatocellular carcinoma cells exposed to sorafenib. Int. J. Cancer. 2013;133:1732–1742. doi: 10.1002/ijc.28159. [DOI] [PubMed] [Google Scholar]; Louandre C, Ezzoukhry Z, Godin C, Barbare JC, Maziere JC, Chauffert B et al. Iron-dependent cell death of hepatocellular carcinoma cells exposed to sorafenib. Int J Cancer 2013; 133: 1732-1742. [DOI] [PubMed]
- 20.Ou W., Mulik R.S., Anwar A., McDonald J.G., He X., Corbin I.R. Low-density lipoprotein docosahexaenoic acid nanoparticles induce ferroptotic cell death in hepatocellular carcinoma. Free Radic. Biol. Med. 2017;112:597–607. doi: 10.1016/j.freeradbiomed.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ou W, Mulik RS, Anwar A, McDonald JG, He X, Corbin IR. Low-density lipoprotein docosahexaenoic acid nanoparticles induce ferroptotic cell death in hepatocellular carcinoma. Free Radic Biol Med 2017; 112: 597-607. [DOI] [PMC free article] [PubMed]
- 21.Zhang X., Sun F., Qiao Y., Zheng W., Liu Y., Chen Y. TFCP2 is required for YAP-dependent transcription to stimulate liver malignancy. Cell Rep. 2017;21:1227–1239. doi: 10.1016/j.celrep.2017.10.017. [DOI] [PubMed] [Google Scholar]; Zhang X, Sun F, Qiao Y, Zheng W, Liu Y, Chen Y et al. TFCP2 Is Required for YAP-Dependent Transcription to Stimulate Liver Malignancy. Cell Rep 2017; 21: 1227-1239. [DOI] [PubMed]
- 22.Wang J., Liu X., Wu H., Ni P., Gu Z., Qiao Y. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 2010; 38: 5366-5383. [DOI] [PMC free article] [PubMed]
- 23.Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]; Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489: 57-74. [DOI] [PMC free article] [PubMed]
- 24.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754-1760. [DOI] [PMC free article] [PubMed]
- 25.John S., Sabo P.J., Thurman R.E., Sung M.H., Biddie S.C., Johnson T.A. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]; John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nature genetics 2011; 43: 264-268. [DOI] [PMC free article] [PubMed]
- 26.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38: 576-589. [DOI] [PMC free article] [PubMed]
- 27.Xie Y., Zhu S., Zhong M., Yang M., Sun X., Liu J. Inhibition of aurora kinase a induces necroptosis in pancreatic carcinoma. Gastroenterology. 2017;153:1429–1443 e1425. doi: 10.1053/j.gastro.2017.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xie Y, Zhu S, Zhong M, Yang M, Sun X, Liu J et al. Inhibition of Aurora Kinase A Induces Necroptosis in Pancreatic Carcinoma. Gastroenterology 2017; 153: 1429-1443 e1425. [DOI] [PMC free article] [PubMed]
- 28.Gao M., Monian P., Quadri N., Ramasamy R., Jiang X. Glutaminolysis and transferrin regulate ferroptosis. Mol. Cell. 2015;59:298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gao M, Monian P, Quadri N, Ramasamy R, Jiang X. Glutaminolysis and Transferrin Regulate Ferroptosis. Mol Cell 2015; 59: 298-308. [DOI] [PMC free article] [PubMed]
- 29.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017; 171: 273-285. [DOI] [PMC free article] [PubMed]
- 30.DeNicola G.M., Chen P.H., Mullarky E., Sudderth J.A., Hu Z., Wu D. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 2015;47:1475–1481. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]; DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet 2015; 47: 1475-1481. [DOI] [PMC free article] [PubMed]
- 31.Mullarky E., Lucki N.C., Beheshti Zavareh R., Anglin J.L., Gomes A.P., Nicolay B.N. Identification of a small molecule inhibitor of 3-phosphoglycerate dehydrogenase to target serine biosynthesis in cancers. Proc. Natl. Acad. Sci. U. S. A. 2016;113:1778–1783. doi: 10.1073/pnas.1521548113. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mullarky E, Lucki NC, Beheshti Zavareh R, Anglin JL, Gomes AP, Nicolay BN et al. Identification of a small molecule inhibitor of 3-phosphoglycerate dehydrogenase to target serine biosynthesis in cancers. Proc Natl Acad Sci U S A 2016; 113: 1778-1783. [DOI] [PMC free article] [PubMed]
- 32.Wang S.P., Tang Z., Chen C.W., Shimada M., Koche R.P., Wang L.H. A UTX-MLL4-p300 transcriptional regulatory network coordinately shapes active enhancer landscapes for eliciting transcription. Mol. Cell. 2017;67:308–321 e306. doi: 10.1016/j.molcel.2017.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang SP, Tang Z, Chen CW, Shimada M, Koche RP, Wang LH et al. A UTX-MLL4-p300 Transcriptional Regulatory Network Coordinately Shapes Active Enhancer Landscapes for Eliciting Transcription. Mol Cell 2017; 67: 308-321 e306. [DOI] [PMC free article] [PubMed]
- 33.Jin Q., Yu L.R., Wang L., Zhang Z., Kasper L.H., Lee J.E. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J 2011; 30: 249-262. [DOI] [PMC free article] [PubMed]
- 34.Parviz F., Matullo C., Garrison W.D., Savatski L., Adamson J.W., Ning G. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat. Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]; Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G et al. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet 2003; 34: 292-296. [DOI] [PubMed]
- 35.Xu L., Hui L., Wang S., Gong J., Jin Y., Wang Y. Expression profiling suggested a regulatory role of liver-enriched transcription factors in human hepatocellular carcinoma. Cancer Res. 2001;61:3176–3181. [PubMed] [Google Scholar]; Xu L, Hui L, Wang S, Gong J, Jin Y, Wang Y et al. Expression profiling suggested a regulatory role of liver-enriched transcription factors in human hepatocellular carcinoma. Cancer Res 2001; 61: 3176-3181. [PubMed]
- 36.Dill M.T., Tornillo L., Fritzius T., Terracciano L., Semela D., Bettler B. Constitutive Notch2 signaling induces hepatic tumors in mice. Hepatology. 2013;57:1607–1619. doi: 10.1002/hep.26165. [DOI] [PubMed] [Google Scholar]; Dill MT, Tornillo L, Fritzius T, Terracciano L, Semela D, Bettler B et al. Constitutive Notch2 signaling induces hepatic tumors in mice. Hepatology 2013; 57: 1607-1619. [DOI] [PubMed]
- 37.Niehof M., Borlak J. EPS15R, TASP1, and PRPF3 are novel disease candidate genes targeted by HNF4alpha splice variants in hepatocellular carcinomas. Gastroenterology. 2008;134:1191–1202. doi: 10.1053/j.gastro.2008.01.027. [DOI] [PubMed] [Google Scholar]; Niehof M, Borlak J. EPS15R, TASP1, and PRPF3 are novel disease candidate genes targeted by HNF4alpha splice variants in hepatocellular carcinomas. Gastroenterology 2008; 134: 1191-1202. [DOI] [PubMed]
- 38.Wijetunga N.A., Pascual M., Tozour J., Delahaye F., Alani M., Adeyeye M. A pre-neoplastic epigenetic field defect in HCV-infected liver at transcription factor binding sites and polycomb targets. Oncogene. 2017;36:2030–2044. doi: 10.1038/onc.2016.340. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wijetunga NA, Pascual M, Tozour J, Delahaye F, Alani M, Adeyeye M et al. A pre-neoplastic epigenetic field defect in HCV-infected liver at transcription factor binding sites and polycomb targets. Oncogene 2017; 36: 2030-2044. [DOI] [PMC free article] [PubMed]
- 39.Ubaid U., Andrabi S.B.A., Tripathi S.K., Dirasantha O., Kanduri K., Rautio S. Transcriptional repressor HIC1 contributes to suppressive function of human induced regulatory T cells. Cell Rep. 2018;22:2094–2106. doi: 10.1016/j.celrep.2018.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ubaid U, Andrabi SBA, Tripathi SK, Dirasantha O, Kanduri K, Rautio S et al. Transcriptional Repressor HIC1 Contributes to Suppressive Function of Human Induced Regulatory T Cells. Cell Rep 2018; 22: 2094-2106. [DOI] [PMC free article] [PubMed]
- 40.Wang S.J., Li D., Ou Y., Jiang L., Chen Y., Zhao Y. Acetylation is crucial for p53-mediated ferroptosis and tumor suppression. Cell Rep. 2016;17:366–373. doi: 10.1016/j.celrep.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang SJ, Li D, Ou Y, Jiang L, Chen Y, Zhao Y et al. Acetylation Is Crucial for p53-Mediated Ferroptosis and Tumor Suppression. Cell Rep 2016; 17: 366-373. [DOI] [PMC free article] [PubMed]
- 41.Marklund U., Larsson N., Gradin H.M., Brattsand G., Gullberg M. Oncoprotein 18 is a phosphorylation-responsive regulator of microtubule dynamics. EMBO J. 1996;15:5290–5298. [PMC free article] [PubMed] [Google Scholar]; Marklund U, Larsson N, Gradin HM, Brattsand G, Gullberg M. Oncoprotein 18 is a phosphorylation-responsive regulator of microtubule dynamics. EMBO J 1996; 15: 5290-5298. [PMC free article] [PubMed]
- 42.Bai T., Yokobori T., Altan B., Ide M., Mochiki E., Yanai M. High STMN1 level is associated with chemo-resistance and poor prognosis in gastric cancer patients. Br. J. Cancer. 2017;116:1177–1185. doi: 10.1038/bjc.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bai T, Yokobori T, Altan B, Ide M, Mochiki E, Yanai M et al. High STMN1 level is associated with chemo-resistance and poor prognosis in gastric cancer patients. Br J Cancer 2017; 116: 1177-1185. [DOI] [PMC free article] [PubMed]
- 43.Cheng A.L., Huang W.G., Chen Z.C., Peng F., Zhang P.F., Li M.Y. Identification of novel nasopharyngeal carcinoma biomarkers by laser capture microdissection and proteomic analysis. Clin. Cancer Res. 2008;14:435–445. doi: 10.1158/1078-0432.CCR-07-1215. [DOI] [PubMed] [Google Scholar]; Cheng AL, Huang WG, Chen ZC, Peng F, Zhang PF, Li MY et al. Identification of novel nasopharyngeal carcinoma biomarkers by laser capture microdissection and proteomic analysis. Clin Cancer Res 2008; 14: 435-445. [DOI] [PubMed]
- 44.Zhang H.Q., Guo X., Guo S.Q., Wang Q., Chen X.Q., Li X.N. STMN1 in colon cancer: expression and prognosis in Chinese patients. Eur. Rev. Med. Pharmacol. Sci. 2016;20:2038–2044. [PubMed] [Google Scholar]; Zhang HQ, Guo X, Guo SQ, Wang Q, Chen XQ, Li XN et al. STMN1 in colon cancer: expression and prognosis in Chinese patients. Eur Rev Med Pharmacol Sci 2016; 20: 2038-2044. [PubMed]
- 45.Imura S., Yamada S., Saito Y.U., Iwahashi S., Arakawa Y., Ikemoto T. miR-223 and stathmin-1 expression in non-tumor liver tissue of patients with hepatocellular carcinoma. Anticancer Res. 2017;37:5877–5883. doi: 10.21873/anticanres.12033. [DOI] [PubMed] [Google Scholar]; Imura S, Yamada S, Saito YU, Iwahashi S, Arakawa Y, Ikemoto T et al. miR-223 and Stathmin-1 Expression in Non-tumor Liver Tissue of Patients with Hepatocellular Carcinoma. Anticancer Res 2017; 37: 5877-5883. [DOI] [PubMed]
- 46.Zheng F., Liao Y.J., Cai M.Y., Liu T.H., Chen S.P., Wu P.H. Systemic delivery of microRNA-101 potently inhibits hepatocellular carcinoma in vivo by repressing multiple targets. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zheng F, Liao YJ, Cai MY, Liu TH, Chen SP, Wu PH et al. Systemic delivery of microRNA-101 potently inhibits hepatocellular carcinoma in vivo by repressing multiple targets. PLoS Genet 2015; 11: e1004873. [DOI] [PMC free article] [PubMed]
- 47.Castro-Chavez F. Microarrays, antiobesity and the liver. Ann. Hepatol. 2004;3:137–145. [PMC free article] [PubMed] [Google Scholar]; Castro-Chavez F. Microarrays, antiobesity and the liver. Ann Hepatol 2004; 3: 137-145. [PMC free article] [PubMed]
- 48.Liu W., Baker S.S., Baker R.D., Nowak N.J., Zhu L. Upregulation of hemoglobin expression by oxidative stress in hepatocytes and its implication in nonalcoholic steatohepatitis. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024363. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu W, Baker SS, Baker RD, Nowak NJ, Zhu L. Upregulation of hemoglobin expression by oxidative stress in hepatocytes and its implication in nonalcoholic steatohepatitis. PLoS One 2011; 6: e24363. [DOI] [PMC free article] [PubMed]
- 49.Armour S.M., Remsberg J.R., Damle M., Sidoli S., Ho W.Y., Li Z. An HDAC3-PROX1 corepressor module acts on HNF4alpha to control hepatic triglycerides. Nat. Commun. 2017;8:549. doi: 10.1038/s41467-017-00772-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Armour SM, Remsberg JR, Damle M, Sidoli S, Ho WY, Li Z et al. An HDAC3-PROX1 corepressor module acts on HNF4alpha to control hepatic triglycerides. Nat Commun 2017; 8: 549. [DOI] [PMC free article] [PubMed]
- 50.Li Y., Zalzala M., Jadhav K., Xu Y., Kasumov T., Yin L. Carboxylesterase 2 prevents liver steatosis by modulating lipolysis, endoplasmic reticulum stress, and lipogenesis and is regulated by hepatocyte nuclear factor 4 alpha in mice. Hepatology. 2016;63:1860–1874. doi: 10.1002/hep.28472. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li Y, Zalzala M, Jadhav K, Xu Y, Kasumov T, Yin L et al. Carboxylesterase 2 prevents liver steatosis by modulating lipolysis, endoplasmic reticulum stress, and lipogenesis and is regulated by hepatocyte nuclear factor 4 alpha in mice. Hepatology 2016; 63: 1860-1874. [DOI] [PMC free article] [PubMed]
- 51.Buehler P.W., D'Agnillo F., Schaer D.J. Hemoglobin-based oxygen carriers: from mechanisms of toxicity and clearance to rational drug design. Trends Mol. Med. 2010;16:447–457. doi: 10.1016/j.molmed.2010.07.006. [DOI] [PubMed] [Google Scholar]; Buehler PW, D'Agnillo F, Schaer DJ. Hemoglobin-based oxygen carriers: From mechanisms of toxicity and clearance to rational drug design. Trends Mol Med 2010; 16: 447-457. [DOI] [PubMed]
- 52.Helms C., Kim-Shapiro D.B. Hemoglobin-mediated nitric oxide signaling. Free Radic. Biol. Med. 2013;61:464–472. doi: 10.1016/j.freeradbiomed.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; Helms C, Kim-Shapiro DB. Hemoglobin-mediated nitric oxide signaling. Free Radic Biol Med 2013; 61: 464-472. [DOI] [PMC free article] [PubMed]
- 53.Heiss E.H., Dirsch V.M. Regulation of eNOS enzyme activity by posttranslational modification. Curr. Pharmaceut. Des. 2014;20:3503–3513. doi: 10.2174/13816128113196660745. [DOI] [PMC free article] [PubMed] [Google Scholar]; Heiss EH, Dirsch VM. Regulation of eNOS enzyme activity by posttranslational modification. Curr Pharm Des 2014; 20: 3503-3513. [DOI] [PMC free article] [PubMed]
- 54.Tong M., Che N., Zhou L., Luk S.T., Kau P.W., Chai S. Efficacy of annexin A3 blockade in sensitizing hepatocellular carcinoma to sorafenib and regorafenib. J. Hepatol. 2018;69(4):826–839. doi: 10.1016/j.jhep.2018.05.034. [DOI] [PubMed] [Google Scholar]; Tong M, Che N, Zhou L, Luk ST, Kau PW, Chai S et al. Efficacy of annexin A3 blockade in sensitizing hepatocellular carcinoma to sorafenib and regorafenib. J Hepatol 2018. [DOI] [PubMed]
- 55.Bruix J., Qin S., Merle P., Granito A., Huang Y.H., Bodoky G. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESOURCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56–66. doi: 10.1016/S0140-6736(16)32453-9. [DOI] [PubMed] [Google Scholar]; Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017; 389: 56-66. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in GEO (accession number: GSE104462). TMT data have been deposited in ProteomeXchange Consortium (accession number: PXD010761). The username for reviewing TMT data is reviewer15024@ebi.ac.uk, and the password is zrLM0EBw.