Abstract
Background
The complexity of breast cancer at the clinical, morphological and genomic levels has been extensively studied in the western population. However, the mutational genomic profiles in Chinese breast cancer patients have not been explored in any detail.
Methods
We performed targeted sequencing using a panel consisting of 33 breast cancer-related genes to investigate the genomic landscape of 304 consecutive treatment-naïve Chinese breast cancer patients at Guangdong Provincial People’s Hospital (GDPH), and further compared the results to those in 453 of Caucasian breast cancer patients from The Cancer Genome Atlas (TCGA).
Results
The most frequently mutated gene was TP53 (45%), followed by PIK3CA (44%), GATA3 (18%), MAP3K1 (10%), whereas the copy-number amplifications were frequently observed in genes of ERBB2 (24%), MYC (23%), FGFR1 (13%) and CCND1 (10%). Among the 8 most frequently mutated or amplified genes, at least one driver was identifiable in 87.5% (n=267) of our GDPH cohort, revealing the significant contribution of these known driver genes in the development of Chinese breast cancer. Compared to TCGA data, the median age at diagnosis in our cohort was significantly younger (48 vs. 58 years; P<0.001), while the distribution of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER2) statuses were similar. The largest difference occurred in HR+/HER2- subtype, where 8 of the 10 driver genes compared had statistically significant differences in their frequency, while there were differences in 2 of 10 driver genes among the TNBC and HR+/HER2+ group, but none in the HR-/HER2+ patients in our cohort compared to the TCGA data. Collectively, the most significant genomic difference was a significantly higher prevalence for TP53 and AKT1 in Chinese patients. Additionally, more than half of TP53-mutation HR+/HER2- Chinese patients (~60%) are likely to harbor more severe mutations in TP53, such as nonsense, indels, and splicing mutations.
Conclusions
We elucidated the mutational landscape of cancer genes in Chinese breast cancer and further identified significant genomic differences between Asian and Caucasian patients. These results should improve our understanding of pathogenesis and/or metastatic behavior of breast cancer across races/ethnicities, including a better selection of targeted therapies.
Keywords: Genomic mutation, mutational landscape, Chinese breast cancer, next-generation sequencing, multi-gene, TP53, AKT1, GATA3, MAP3K1, PIK3CA
Introduction
Breast cancer, the leading cause of female cancer death worldwide, has been well recognized as a group of heterogeneous diseases in terms of both clinical behavior and molecular landscape (1,2). The availability of sophisticated high-throughput technology, together with well-developed bioinformatics tools, has significantly accelerated our understanding of the molecular basis of cancer. Gene expression profiles that classify breast cancers into different subtypes have yielded transcriptional signatures that are used to support therapeutic decisions (3-5). Characterizations of early breast cancer at the genomic level have cataloged the numerous genomic alterations involved in tumorigenesis and metastatic progression (6). These studies showed that breast cancer includes a large number of actionable genomic alterations, such as TP53 mutation, PIK3CA mutation, ERBB2 amplification, FGFR1 amplification, CCND1 amplification, AKT1 mutations, and GATA3 mutation (7-12).
Currently, the most authoritative, robust, and widely available tumor genomic information source is The Cancer Genome Atlas (TCGA) project, which is a comprehensive “atlas” of cancer genomic profiles (13,14). However, the TCGA breast cancer samples are largely Caucasian (69%), and the Asian ethnic group is significantly under-represented (6% or a sample size of only 65 breast cancer patients) (15). Additionally, there have been few reports of somatic mutations of breast cancer in Chinese patients using next-generation sequencing (NGS) methodology (16).
Previous large epidemiological studies have suggested that clinicopathologic features and outcomes of breast cancer vary considerably among racial and ethnic groups (17,18). Worldwide, China, which includes about one-fifth of the global population, accounts for 12.2% of all newly diagnosed breast cancers (19). Compared with some western countries, China has a relatively lower incidence of breast cancer (20). However, the incidence of breast cancer in China has increased more than twice as fast as global rates since the 1990s (19). The peak age of breast cancer onset in China is between 45 and 55 years compared to an average of between 60 and 70 years in many Western countries (21). In addition, only ~3.5% of patients are pathologically confirmed as invasive lobular breast cancer in Chinese women, the proportion of which is significantly lower than the observation in Caucasian women constituting ~10–15% of all cases (22,23). Beyond these clinicopathological factors, it is important to understand the molecular basis of breast cancer underlying these ethnic differences. There is, therefore, an urgent need to understanding genomic features in Chinese breast cancer group, compared with other ethnic groups that might be valuable in the treatment planning of Asian breast cancer patients, including Chinese.
In this study, we performed capture-based ultra-deep targeted sequencing to interrogate the mutation profiles associated with 304 treatment-naïve Chinese breast cancer patients using the customized BreastCore panel consisting of 33 breast cancer-related vital genes, spanning 140 kb of the human genome. Our objective was (I) to provide a landscape of non-synonymous genomic mutations and copy-number aberrations (CNAs) of frequently altered cancer genes in Chinese breast tumors, and (II) to identify distinctive genomic mutational patterns of Chinese breast cancer patients, compared to the TCGA data set. We hypothesized that there are significant differences in the molecular profile of Chinese breast cancer patients compared to the TCGA data.
Methods
Patients and specimens
Our Chinese cohort consisted of 305 primary treatment-naïve tumors from 304 consecutive female breast cancer patients diagnosed at Guangdong Provincial People’s Hospital (GDPH) from October 2016 to December 2017. At our center, estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) expression in each specimen with breast cancer were routinely evaluated by immunohistochemistry (IHC) staining at Department of Pathology in Guangdong Provincial People’s Hospital. ER or PR in each specimen was considered positive if more than 1% of tumor nuclei were strongly stained according to the 2010 ASCO/CAP guidelines (24). Hormonal receptor (HR) was recorded as HR-positive (HR+) in the individual specimen with either ER-positive (ER+) or PR positive, while the status was recorded as HR-negative (HR-) only when both of ER and PR were negative. Additionally, HER2 status was confirmed by IHC and/or fluorescence in situ hybridization (FISH) according to 2013 ASCO/CAP guideline (25). Primary tumor biopsies were obtained using an Institutional Review Board approved protocol, and the subsequent analysis had been approved by the Ethic Guangdong Provincial People’s Hospital (No. GDREC2014122H). All patients provided written informed consent for translational research. Sequencing assays were performed blinded to the clinical-pathological parameters in CLIA-certified Burning Rock Biotech (Guangzhou, China).
Tissue DNA extraction
DNA was extracted using QIAamp DNA FFPE tissue kit (Qiagen, California, US) according to the manufacturer’s instructions. The DNA concentration was measured by Qubit dsDNA assay (Life Technologies, California, US).
NGS library preparation and Capture-based targeted DNA sequencing
DNA was subjected to end repair, phosphorylation and adaptor ligation. Fragments of size 200–400 bp were selected by AMPure beads (Agencourt AMPure XP Kit), followed by hybridization with capture probe baits, hybrid selection with magnetic beads and PCR amplification. A bioanalyzer high-sensitivity DNA assay was subsequently performed to assess the quality and size of the fragments. Indexed samples were sequenced on Nextseq500 sequencer (Illumina, Inc., US) with pair-end reads.
Sequence data analysis
Sequence data were mapped to the human genome (hg19) using BWA aligner 0.7.10. Local alignment optimization, variant calling, and annotation were performed using GATK 3.2, MuTect, and VarScan. Variants were filtered using the VarScan filter pipeline, with loci with depth less than 100 filtered out. At least 5 supporting reads were needed for insertions or deletions (Indels); while 8 supporting reads were needed for single-nucleotide variants (SNVs) to be called. According to the ExAC, 1000 Genomes, dbSNP, ESP6500SI-V2 database, variants with population frequency over 0.1% were grouped as SNP and excluded from further analysis. Remaining variants were annotated with ANNOVAR and SnpEff v3.6. DNA translocation analysis was performed using Factera 1.4.3 as previously described. The limit of detection for SNVs is 2% for hotspots and 5% for non-hotspots. Copy number variation was detected by in-house analysis scripts based on the depth of coverage data of capture intervals. Coverage data were corrected against sequencing bias resulting from GC content and probe design. The average coverage of all captured regions was utilized to normalize the coverage of different samples to comparable scales. Copy number was calculated based on the ratio between the depth of coverage in tumor samples and average coverage of an adequate number (n>50) of samples without copy number variation as references as to each capture interval. Copy number variation is called if the coverage data of the gene region was quantitatively and statistically significantly different from its reference control. The limit of detection for CNVs is 1.5 for deletion and 2.64 for amplification. We performed capture-based targeted sequencing on 305 tumor tissue samples utilizing the BreastCore panel comprising a customized panel of 33 breast cancer-related genes, spanning 140 kb of the human genome, the majority of which have been well documented as driver genes in western patients with breast cancer (10). Collectively, we achieved with a mean coverage depth of ~1,200× across all target regions on all 305 tissues samples, and 98.6% of all target regions had coverage greater than 200×.
Statistical analysis
Data were summarized by frequency and percentage for categorical variables. Comparisons between groups were performed using Fisher’s exact or Chi-square test for these categorical variables. All statistical tests were two-sided, and differences were considered with significance when P<0.05. According to the Benjamini-Hochberg procedure, false discovery rate (FDR or q-value) was used for correcting P value for multiple hypothesis testing.
Results
Patient characteristics
In this study, our cohort included 305 primary invasive breast tumors obtained from 304 treatment-naïve consecutive patients with median age of 48 ranging from 24 to 82 years who were pathologically confirmed at Guangdong Provincial People’s Hospital (GDPH). Among them, one patient had synchronous bilateral primary breast lesions. The detailed patients’ clinicopathological features were listed in Table 1. Briefly, 57.9% of patients were pre-menopausal, and 74.0% were diagnosed at an early stage (31.9% stage I and 42.1% stage II), while 83.3% of primary tumors were HR+, 27.3% were HER2 positive (HER2+) and only 8.2% were triple negative breast cancer (TNBC; lacking expression of ER, PR, and HER2).
Table 1. Patient characteristics.
| Variables | GDPH (n=304) | TCGA (n=453) | P | |||
|---|---|---|---|---|---|---|
| No. | (%) | No. | (%) | |||
| Age [median, range] | 48 | [24–82] | 58 | [28–90] | <0.001# | |
| ≤50 years | 169 | 55.6 | 132 | 29.1 | ||
| >50 years | 135 | 44.4 | 321 | 70.9 | ||
| Sex | 0.153 | |||||
| Female | 304 | 100.0 | 449 | 99.1 | ||
| Male | 0 | 0.0 | 4 | 0.9 | ||
| Menopausal Status | <0.001# | |||||
| Pre-menopause | 176 | 57.9 | 120 | 26.5 | ||
| Post-menopause | 128 | 42.1 | 299 | 66.0 | ||
| Missing | 0 | 0.0 | 34 | 7.5 | ||
| LN status | 0.044# | |||||
| Positive | 137 | 45.1 | 229 | 50.6 | ||
| Negative | 167 | 54.9 | 219 | 48.3 | ||
| Missing | 0 | 0.0 | 5 | 1.1 | ||
| TNM stage | 0.162 | |||||
| I–II | 225 | 74.0 | 339 | 74.8 | ||
| III–IV | 79 | 26.0 | 109 | 24.1 | ||
| Missing | 0 | 0.0 | 5 | 1.1 | ||
| Pathological type | <0.001# | |||||
| IDC | 260 | 85.5 | 318 | 70.2 | ||
| ILC | 9 | 3.0 | 85 | 18.8 | ||
| Others | 35 | 11.5 | 50 | 11.0 | ||
| ER status | 0.855 | |||||
| Positive | 240 | 78.9 | 361 | 79.7 | ||
| Negative | 64 | 21.1 | 92 | 20.3 | ||
| PR status | 0.397 | |||||
| Positive | 222 | 73.0 | 310 | 68.4 | ||
| Negative | 81 | 26.6 | 141 | 31.1 | ||
| Missing | 1 | 0.3 | 2 | 0.4 | ||
| HER2 status | 0.054 | |||||
| Positive | 83 | 27.3 | 95 | 21.0 | ||
| Negative | 221 | 72.7 | 358 | 79.0 | ||
| Histologic grade | NA | |||||
| G1 | 17 | 5.6 | – | – | ||
| G2 | 116 | 38.2 | – | – | ||
| G3 | 114 | 37.5 | – | – | ||
| Missing | 7 | 2.3 | 453 | 100.0 | ||
Based on the Pearson χ2 test, the P value is calculated using Fisher’s exact test; #, P value<0.05. LN, lymph node; IDC, infiltrating ductal carcinoma; ILC, infiltrating lobular carcinoma; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
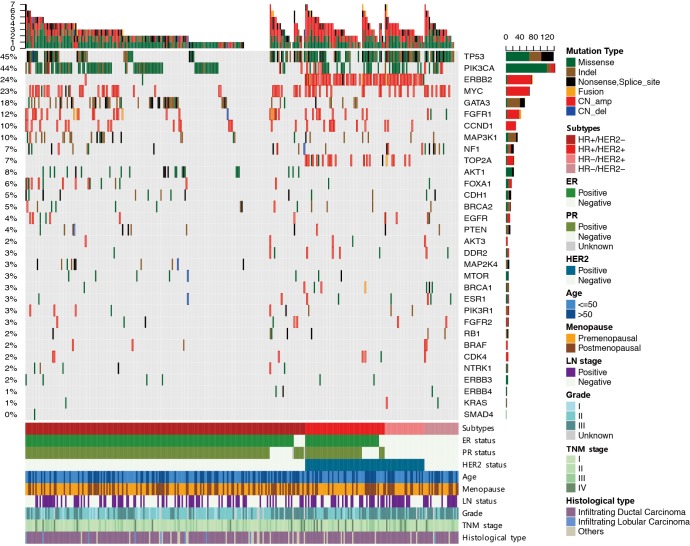
The mutational landscape of 33 cancer genes in Chinese breast tumors
At least one genomic alteration was observed in 92.1% (n=281) of tumor tissues. Altogether, there were 871 aberrant events, including 396 single-nucleotide variants (SNVs), 166 insertions or deletions (Indels), 292 copy-number amplifications, and 7 copy-number deletions. The remaining 24 samples had no mutation detected from this panel. Interestingly, there were 16.4% (n=50) of samples harbored only one altered gene, with the most frequent being PIK3CA (n=19), TP53 (n=8), GATA3 (n=4), AKT1 (n=4), ERBB2 (n=3) and CCND1 (n=3). This observation consistently suggests fundamental roles for these driver genes implicated in promoting initiation and/or progression of breast cancer, though it is relatively lower as compared to a previous whole exome-sequencing report showing that the identifiable incidence of a single driver is approximately 28% (10). The average number of altered genes in an individual specimen was 3 in our cohort (range, 0 to 7).
TP53 and PIK3CA were the most frequently mutated genes, being present in 45% and 44% of samples, respectively, while GATA3 transcription factor gene was the third most common mutation (18% of samples) (Figure 1). Two tumors harbored dual mutations in GATA3. Therefore, there was a total of 56 mutations revealing 2 hotspot mutations, including frameshift mutation occurring at proline 409 (n=15) and splicing mutation due to dinucleotide CA deletion at the exon4/5 junction (n=10). Additionally, we also identified two recurrent M294K mutations, though the type of missense mutation was relatively rare in GATA3 (n=4). All of the GATA3 mutations occurred within HR+ tumors, and 93% of them were inactivated protein-truncating mutations, suggesting the loss of function in mediating the canonical recruitment of ER transcription complex (26). In addition, we identified one conservative inframe deletion and nine missense mutations in the other ER-associated transcription factor FOXA1 gene, including 2 of recurrent D226N and 2 of F254L mutations. We also observed 8 additional HR+/HER2- tumors with FOXA1 amplification, highlighting the requirement of FOXA1 as a transcriptional pioneer factor in assisting ER aberrantly access to its genomic targets in FOXA1-mutant/amplified tumors (27).
Figure 1.
The mutational landscape of 33 cancer genes in Chinese breast tumors (n=305). Genomic alterations in 33 genes are shown in the middle panel, except the BCL2L11 gene, in which we did not identify any aberrant events. Tumor samples are grouped by clinically-defined subtypes: HR+/HER2- (n=196), HR+/HER2+ (n=56), HR-/HER2+ (n=28) and TNBC (n=25). Top bar summarizes the total number of mutations in each patient (columns); sidebar (rows) summarizes the percentage of tumors with a mutation in each gene (left-hand) and mutation composition for each gene in the entire cohort (right-hand). Clinical parameters for each patient are shown in the bottom panel. Different colors denote different types of mutations and different clinicopathological features. Indel, insertions or deletions; CN_amp, copy-number amplification; CN_del, copy-number deletion.
In our GDPH cohort, the fourth frequently mutated gene was MAP3K1 (10%), a serine/threonine protein kinase acting as an important upstream activator of mitogen-activated protein kinase (MAPK) signaling in response to stress (6). Inactivating MAP3K1 mutations, together with one of its downstream substrates encoded by MAP2K4, have been reported to be the striking features in luminal/ER+ tumors (4). In total, 40 nonsynonymous MAP3K1 mutations were identified in 28 of samples including 5 nonsense, 26 frameshift indels, 1 in-frame deletion and 8 missense mutations (Figure 1). Of note, 43% of MAP3K1-mutated tumors (12 out of 28 cases) carried dual mutations in MAP3K1, supporting the role for MAP3K1 as a potential recessive cancer gene. Furthermore, we found that 9 out of 16 tumors with single MAP3K1 mutation harbored concurrent TP53 mutations, but only one out of 12 tumors with dual MAP3K1 mutations had concurrent TP53 mutation (P=0.016). Since MAPK signaling has an important role in the stabilization and subsequent activation of p53 protein (28), dual mutations in MAP3K1 are more likely to capably drive oncogenic properties even in the absence of TP53 mutation. MAP2K4 mutations were found in 3% of HR+ samples (n=9) in a mutually exclusive manner with MAP3K1 mutation, 8 of which were predicted to be inactivating truncation mutations. KRAS (n=2) and BRAF (n=1), the upstream components of MAPK signaling, were extremely rarely mutated in breast cancer (Figure 1). But we did observe two recurrent and highly oncogenic KRAS G12V/A mutations. In contrast, NF1, a negative regulator of RAS oncogene signal transduction, was frequently mutated in Chinese breast tumors at a high frequency of 6.0% (Figure 1). A total of 6 frameshift indels, 5 nonsense, 2 splice-region, and 5 missense mutations were observed in NF1, and 70% (13 out of 18) of mutations were inactivated truncation events, implicating the tumor suppressive role for wild-type NF1 in Chinese breast cancer.
In addition to these frequently mutated genes, the other dominant genomic features were the copy-number amplifications (Figure 1), including ERBB2 (24%), MYC (23%), FGFR1 (13%) and CCND1 (10%). Incorporation of recurrent copy-number changes, together with cancer genes bearing mutations in more than ~10% of samples, generated a list of 8 driver genes, including TP53, PIK3CA, GATA3, MAP3K1, ERBB2, MYC, FGFR1, and CCND1. In line with previous reports showing the high contribution (~60%) of the 8 drivers in the western population (9,10), at least one driver was identifiable in 87.5% (n=267) of our GDPH cohort. Taken together, the 8 most frequently mutated or amplified genes dominated the genomic feature of Chinese breast cancer.
Distinctive genomic features in clinically-defined subgroups
In the hormone responsive HR+/HER2- cohort, the most frequently mutated gene was PIK3CA, occurring in 45.9% of patients followed by TP53 (28.1%) and GATA3 (24.0%) (Figure 1 and Table 2). This subgroup had the highest GATA3 mutation frequency and the lowest TP53 mutation frequency among all 4 subgroups (Figure 1). All of the AKT1 mutations (n=23) were actionable E17K mutation in our cohort and exclusively occurred in this subgroup (Table 2). In addition, very few samples (10.7%) in this group had a co-occurrence of TP53 and PIK3CA. The amplification of MYC, CCND1, and FGFR1 was also frequently observed in HR+/HER2- group (Figure 1). In the triple-positive HR+/HER2+ group (n=56), 66.1% of cases harbored TP53 mutations, and 21.4% had concurrent PIK3CA mutations (Figure 1). Almost all of TOP2A amplification was observed in ERBB2-amplified tumors, regardless of HR status (Table 2). The HR-/HER2+ breast cancer (n=28) had the highest TP53 (89.3%) mutation frequency among all 4 subgroups (Table 2), the majority of which harbored concurrent PIK3CA mutation (n=12). In the TNBC group (n=25), 80.0% of samples harbored TP53 mutation, while very few had concurrent PIK3CA mutations (n=3). Consistent with the previous report (4), aberrant MYC amplification (40%) and a BRCA1 mutation (12%) were the significant characteristics of TNBC. Intriguingly, the NF1 gene was significantly mutated in Chinese patients with TNBC at a frequency of 24.0% (P<0.001; Table 2). However, mutations in GATA3 and MAP3K1, which was frequently observed in HR+ samples, were extremely rare in HR- samples (Figure 1 and Table 2).
Table 2. Genomic features in clinically-defined subgroups from GDPH cohort.
| Variables | HR+/HER2-(n=196) | HR+/HER2+ (n=56) | HR-/HER2+ (n=28) | TNBC (n=25) | P | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | (%) | No. | (%) | No. | (%) | No. | (%) | |||||
| Age [median, range] | 48 | [24–78] | 48 | [30–67] | 48 | [27–69] | 47 | [28–82] | 0.957 | |||
| ≤50 years | 108 | 55.1 | 33 | 58.9 | 15 | 53.6 | 14 | 56.0 | ||||
| >50 years | 88 | 44.9 | 23 | 41.1 | 13 | 46.4 | 11 | 44.0 | ||||
| Menopausal status | 0.775 | |||||||||||
| Pre-menopause | 111 | 56.6 | 36 | 64.3 | 16 | 57.1 | 14 | 56.0 | ||||
| Post-menopause | 85 | 43.4 | 20 | 35.7 | 12 | 42.9 | 11 | 44.0 | ||||
| LN status | 0.265 | |||||||||||
| Positive | 80 | 40.8 | 30 | 53.6 | 15 | 53.6 | 12 | 48.0 | ||||
| Negative | 116 | 59.2 | 26 | 46.4 | 13 | 46.4 | 13 | 52.0 | ||||
| TNM stage | 0.302 | |||||||||||
| I–II | 149 | 76.0 | 40 | 71.4 | 17 | 60.7 | 20 | 80.0 | ||||
| III–IV | 47 | 24.0 | 16 | 28.6 | 11 | 39.3 | 5 | 20.0 | ||||
| Pathological type | 0.627 | |||||||||||
| IDC | 164 | 83.7 | 50 | 89.3 | 26 | 92.9 | 21 | 84.0 | ||||
| ILC | 8 | 4.1 | 0 | 0.0 | 0 | 0.0 | 1 | 4.0 | ||||
| Others | 24 | 12.2 | 6 | 10.7 | 2 | 7.1 | 3 | 12.0 | ||||
| Histologic grade | <0.001# | |||||||||||
| G1 | 21 | 10.7 | 2 | 3.6 | 0 | 0.0 | 0 | 0.0 | ||||
| G2 | 102 | 52.0 | 28 | 50.0 | 6 | 21.4 | 7 | 28.0 | ||||
| G3 | 65 | 33.2 | 26 | 46.4 | 21 | 75.0 | 15 | 60.0 | ||||
| Missing | 8 | 4.1 | 0 | 0.0 | 1 | 3.6 | 3 | 12.0 | ||||
| TP53 | <0.001# | |||||||||||
| Wild-type | 141 | 71.9 | 19 | 33.9 | 3 | 10.7 | 5 | 20.0 | ||||
| Mutant | 55 | 28.1 | 37 | 66.1 | 25 | 89.3 | 20 | 80.0 | ||||
| PIK3CA | 0.034# | |||||||||||
| Wild-type | 106 | 54.1 | 38 | 67.9 | 15 | 53.6 | 20 | 80.0 | ||||
| Mutant | 90 | 45.9 | 18 | 32.1 | 13 | 46.4 | 5 | 20.0 | ||||
| GATA3 | <0.001# | |||||||||||
| Wild-type | 149 | 76.0 | 49 | 87.5 | 28 | 100.0 | 25 | 100.0 | ||||
| Mutant | 47 | 24.0 | 7 | 12.5 | 0 | 0.0 | 0 | 0.0 | ||||
| ERBB2 | <0.001# | |||||||||||
| Wild-type | 192 | 98.0 | 7 | 12.5 | 2 | 7.1 | 25 | 100.0 | ||||
| Mutant | 4 | 2.0 | 49 | 87.5 | 26 | 92.9 | 0 | 0.0 | ||||
| AKT1 | 0.003# | |||||||||||
| Wild-type | 173 | 88.3 | 56 | 100.0 | 28 | 100.0 | 25 | 100.0 | ||||
| Mutant | 23 | 11.7 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | ||||
| TOP2A | <0.001# | |||||||||||
| Wild-type | 196 | 100.0 | 39 | 69.6 | 23 | 82.1 | 25 | 100.0 | ||||
| Mutant | 0 | 0.0 | 17 | 30.4 | 5 | 17.9 | 0 | 0.0 | ||||
| MYC | 0.001# | |||||||||||
| Wild-type | 155 | 79.1 | 43 | 76.8 | 22 | 78.6 | 15 | 60.0 | ||||
| Mutant | 41 | 20.9 | 13 | 23.2 | 6 | 21.4 | 10 | 40.0 | ||||
| NF1 | <0.001# | |||||||||||
| Wild-type | 186 | 94.9 | 55 | 98.2 | 27 | 96.4 | 19 | 76.0 | ||||
| Mutant | 10 | 5.1 | 1 | 1.8 | 1 | 3.6 | 6 | 24.0 | ||||
| BRCA1 | 0.001# | |||||||||||
| Wild-type | 195 | 99.5 | 54 | 96.4 | 28 | 100.0 | 22 | 88.0 | ||||
| Mutant | 1 | 0.5 | 2 | 3.6 | 0 | 0.0 | 3 | 12.0 | ||||
Based on the Pearson χ2 test, the P value is calculated using Fisher’s exact test; #, P value<0.05. Copy number changes of PIK3CA, NF1, and BRCA1were classified into wild-type group in this analysis. LN, lymph node; IDC, infiltrating ductal carcinoma; ILC, infiltrating lobular carcinoma.
Comparison of mutational spectrum between Chinese and TCGA breast cancer patients
To investigate the potential ethnic differences in mutation frequencies between Chinese and western breast cancer patients, we examined publicly available data from TCGA and extracted data involving 453 Caucasian samples with known clinical information (https://xenabrowser.net/; last updated on June 01 2016) (4). A detailed comparison of clinical parameters between GDPH and TCGA cohorts was summarized in Table 1. The median age at diagnosis in our GDPH cohort was 48 years, which was significantly younger than the TCGA cohort, with a median age of 58 years (P<0.001). Moreover, the distribution of histopathologic types was also statistically significantly different from TCGA cohort (P<0.001), which had more patients with infiltrating lobular carcinoma (ILC; TCGA: 18.8% vs. GDPH: 3%) and fewer with infiltrating ductal carcinoma (IDC; 70.2% vs. 85.5%). However, the distribution of ER, PR, and HER2 statuses was comparable between the two cohorts.
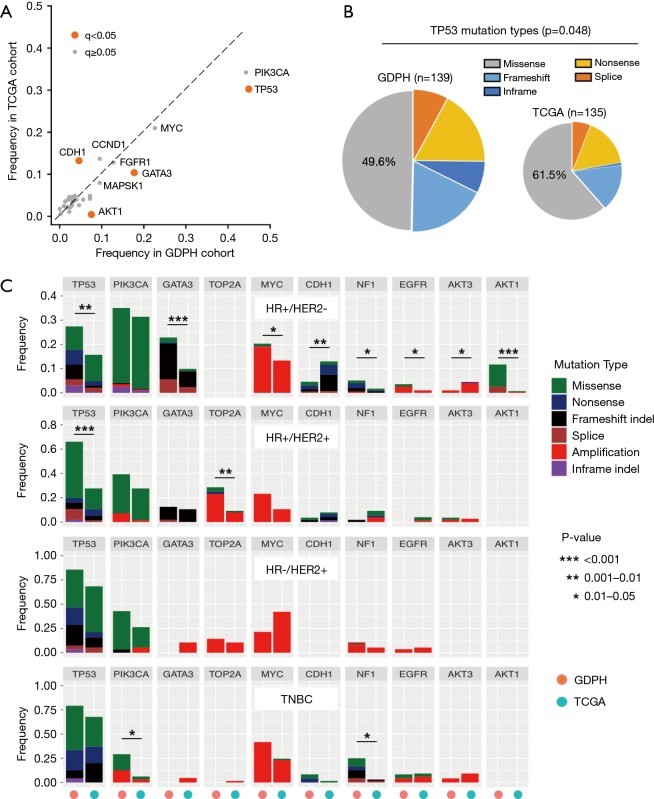
Using a stringent false discovery rate (FDR; q-value) <0.05, a total of 4 driver genes were found to be differentially mutated between the two entire cohorts (Figure 2A). TP53 (GDPH: 45% vs. TCGA: 30%; q<0.001), AKT1 (8% vs. 1%; q<0.001) and GATA3 (18% vs. 10%; q=0.036) were more frequently mutated in our cohort, whereas there was a lower prevalence for CDH1 (5% vs. 13%; q=0.036) as compared to the TCGA cohort. In addition to the observed difference in TP53 mutation frequency, we also found that the composition of TP53 mutation types also differed between the two cohorts (Figure 2B). The majority of TP53 mutations (61.5%) were missense in the TCGA cohort; however, only 49.6% of TP53 mutations were missense in our cohort (P=0.048).
Figure 2.
Comparison of mutational spectrum between Chinese and TCGA cohorts. (A) The frequency of gene alterations in GDPH and TCGA cohorts. Astringent false discovery rate (FDR; q-value) <0.05 was used, and genes in which mutations showed a significant difference in frequency between the two cohorts are labels as an orange color. (B) The composition of TP53 mutation types in GDPH and TCGA cohorts is shown, and the proportion of missense mutation is compared between the two cohorts using the Chi-square test. (C) Differences in mutation frequencies between Chinese and Caucasians (obtained from TCGA). Mutations occurring in more than 10% of patients in at least one subgroup were selected. For each gene, the left bar represents data obtained from our cohort; right bar represents TCGA data. Fisher’s exact test was used to assess differences in mutation frequencies between the two cohorts. A P value less than 0.05 and odds ratio greater than 2 or odds ratio less than 0.5 were listed. Stars (*) denote statistically significant difference between the two cohorts.
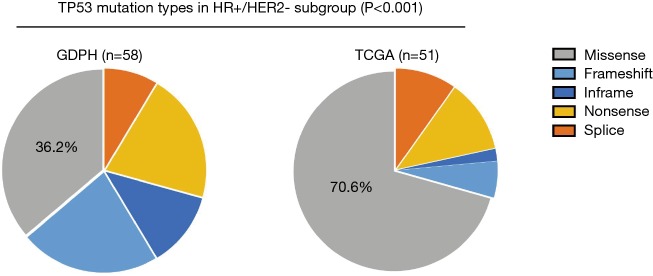
Furthermore, we compared and contrasted the mutation frequencies in driver genes occurring in >10% of cases from at least one subgroup between the two cohorts. Overall, we identified significant molecular differences in 3 of the 4 clinically-defined subtypes (Figure 2C). The largest difference was in the HR+/HER2- subtype, where 8 of the 10 driver genes compared had statistically significant differences in their frequency, while there were differences in 2 of 10 driver genes among the TNBC and HR+/HER2+, but none in the HR-/HER2+ patients in our cohort compared to the TCGA data. Thus, the HR+/HER2- cohort of Chinese breast cancer exhibited a significantly higher mutation frequency in TP53 (P=0.003), GATA3 (P<0.006), AKT1 (P<0.001), NF1 (P=0.020), MYC (P=0.044) and EGFR (P=0.031), but had a lower frequency in CDH1 (P=0.002) and AKT3 (P=0.034) as compared to the TCGA data (Figure 2C). More importantly, we found that the composition of TP53 mutation type differed most significantly in HR+/HER2- subgroup (Figure 2C and Figure S1) since the majority of TP53 mutations (70.6%) were missense in TCGA cohort, but only 36.2% in our cohort (P<0.001). This finding demonstrates that more than half of TP53-mutant HR+/HER2- Chinese patients are likely to harbor nonsense, indels or splicing mutations in TP53, the mutation types of which have been reported to result in a more significant loss of p53 protein compared to missense mutation (29). Among the HR+/HER2+ group, our cohort of Chinese breast cancer patients had a significantly higher mutation frequency in TP53 (P<0.001) and TOP2A (P=0.002) compared to the TCGA data (Figure 2C). In the HR-/HER2+ group, GDPH and TCGA cohorts showed a comparable mutation frequency across all genes listed. In the TNBC group, there was no difference in TP53 mutation frequency, but the GDPH cohort had a higher mutation frequency in NF1 (P=0.010) and PIK3CA (P=0.022) (Figure 2C). Collectively, these findings provide new insights into understanding the differences and similarities of genomic features between Chinese and western patients among the HR+, HER2+ and TNBC breast cancer.
The spectrum of TP53 mutation in Chinese breast cancer patients
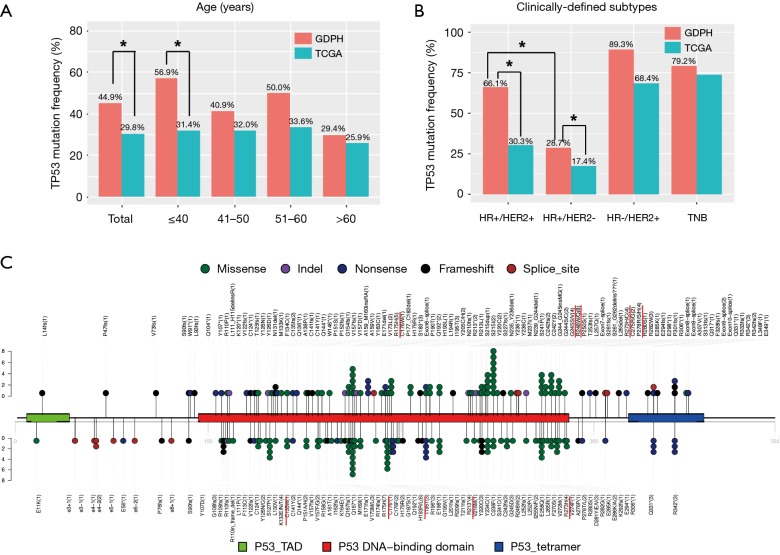
The difference in TP53 mutation frequency was the most significant differences between the Chinese and TCGA cohorts. The percentage of patients harboring TP53 mutation in each age group was shown in Figure 3A. Overall, significantly more patients in the Chinese cohort harbored TP53 mutations (P<0.001) comparing to the TCGA cohort. This difference was most prominent in patients younger than 40. When we compared the percentage of patients with TP53 mutation, the Chinese cohort had significantly higher TP53 mutation frequency among the HR+/HER2- group (GDPH 28.7% vs. TCGA 17.4%; P=0.004) and the HR+/HER2+ (GDPH 66.1% vs. TCGA 30.3%; P<0.001) group (Figure 3B). Interestingly, HR+/HER2+ patients in the Chinese cohort were significantly more likely to harbor TP53 mutation than HR+/HER2- patients (P<0.001), suggesting TP53 mutation frequency significantly varied by HER2 status. However, this phenomenon was only observed in HR+ tumors, but not in HR- tumors (Figure 3B).
Figure 3.
The spectrum of TP53 mutation in Chinese breast cancer patients. The distribution of TP53 mutations is grouped by the age of breast cancer onset (A) and clinically-defined subgroups (B). The mutation frequencies for TP53 in GDPH and TCGA cohorts are compared using Fisher’s exact test. Stars (*) denote statistically significant difference (P<0.05). (C) Lollipop diagram depicts the type and location of TP53 mutation in our cohort (top) and TCGA cohort (bottom). Different types of mutations were colored by different colored dots, and each colored dots represents one TP53 mutation. The number of patients with a specific mutation was also listed in the parenthesis.
In the GDPH cohort, we identified a total of 139 mutations in the TP53 gene. As previously reported (29,30), the distribution of the mutations was nonuniform across the gene (Figure 3C). The conserved regions of exons 5–8 harbored 78% (n=108) of the mutations, the majority of which were missense mutations (n=67) and predominantly clustered in the DNA-binding domain of the protein. Approximately 22% of mutations (n=31) resided outside exons 5–8, with 9.4% and 7.2% in exon 4 and 10, respectively. However, almost all of mutations occurring outside exons 5–8 were non-missense mutations (n=29), including 13 inframe or frameshift indels, 11 nonsense and 5 splicing mutations. We further compared the landscape of TP53 non-synonymous mutation distribution in Chinese breast cancer patients and TCGA cohort (Figure 3C). Although these distributions looked very similar, the mutational hotspots differed between the two cohorts. For example, the most frequent TP53 mutation in our cohort occurred at codon 248 (R248W/Q; n=8), whereas R175H was the most common mutation in the TCGA cohort (n=7). Additionally, the second hotspot resided at codon 193 in the TCGA cohort (H193R/L; n=6), while only one H193L mutation was observed in our GDPH cohort (Figure 3C). Collectively, we identified a total of 25 codons that were recurrently mutated in our cohort (Table 3). In addition to six of previously reported “major hotspot” codons (175, 213, 245, 248, 273 and 282) that comprise each at least 2% of all mutations (30), mutational hotspots were also frequently observed in codons 242 (n=4), 278 (n=4) and 342 (n=5). Intriguingly, only 11 of splicing mutations in TP53 were identified in our cohort, but each 3 of them were located at relatively poorly-defined codons 261, 307 and 331, respectively (Table 3). The results suggest that these specific recurrent mutations might confer selective growth advantage during the development and/or dissemination of breast cancer. Taken together, these findings provide essential information to improve our understanding of genomic differences in breast cancer across races/ethnicities.
Table 3. Recurrent TP53 mutations in GDPH cohort.
| Position (codon) | DNA/base change | Protein/amino acid change | Exon | Type of mutation | No. of altered cases |
|---|---|---|---|---|---|
| 135 | c.403dup | p.C135fs | 5 | Frameshift | 2 |
| 141 | c.420del | p.C141fs | 5 | Frameshift | 1 |
| c.422G>A | p.C141Y | 5 | Missense | 1 | |
| 157 | c.468del | p.V157fs | 5 | Frameshift | 1 |
| c.470T>A | p.V157D | 5 | Missense | 1 | |
| 173 | c.517G>T | p.V173L | 5 | Missense | 2 |
| 175# | c.524G>A | p.R175H | 5 | Missense | 5 |
| 183# | c.548C>G | p.S183* | 5 | Nonsense | 3 |
| 192 | c.574C>T | p.Q192* | 6 | Nonsense | 2 |
| 195# | c.584T>C | p.I195T | 6 | Missense | 3 |
| 205 | c.613T>C | p.Y205H | 6 | Missense | 1 |
| c.614A>G | p.Y205C | 6 | Missense | 1 | |
| 213# | c.637C>T | p.R213* | 6 | Nonsense | 2 |
| c.638G>T | p.R213L | 6 | Missense | 1 | |
| 215# | c.644G>T | p.S215I | 6 | Missense | 2 |
| c.643_645dup | p.S215dup | 6 | Inframe | 1 | |
| 220 | c.659A>G | p.Y220C | 6 | Missense | 2 |
| 236# | c.704_709del | p.N235_Y236del | 7 | Inframe | 1 |
| c.707A>G | p.Y236C | 7 | Missense | 1 | |
| c.708C>A | p.Y236* | 7 | Nonsense | 1 | |
| 242# | c.723Cdel | p.C242fs | 7 | Frameshift | 2 |
| c.725G>A | p.C242Y | 7 | Missense | 2 | |
| 244# | c.730G>A | p.G244S | 7 | Missense | 1 |
| c.730G>T | p.G244C | 7 | Missense | 1 | |
| c.727_732dup | p.G244_G245insMG | 7 | Inframe | 1 | |
| 245# | c.733G>A | p.G245S | 7 | Missense | 2 |
| c.734G>T | p.G245V | 7 | Missense | 2 | |
| 248# | c.743G>A | p.R248Q | 7 | Missense | 6 |
| c.742C>T | p.R248W | 7 | Missense | 2 | |
| 261# | c.782+1G>T | X261_splice | 7 | Splice | 1 |
| c.783-19_786delinsA | X261_splice | 8 | Splice | 1 | |
| c.783-26_795del | X261_splice | 8 | Splice | 1 | |
| 273# | c.818G>A | p.R273H | 8 | Missense | 3 |
| c.817C>T | p.R273C | 8 | Missense | 1 | |
| 275 | c.823T>C | p.C275R | 8 | Missense | 1 |
| c.823T>G | p.C275G | 8 | Missense | 1 | |
| 278# | c.832C>T | p.P278S | 8 | Missense | 2 |
| c.833C>A | p.P278H | 8 | Missense | 1 | |
| c.833C>G | p.P278R | 8 | Missense | 1 | |
| 282# | c.844C>T | p.R282W | 8 | Missense | 3 |
| 307# | c.919+1G>T | X307_splice | 8 | Splice | 1 |
| c.920-1G>C | X307_splice | 9 | Splice | 1 | |
| c.920-1_920GCdelinsTT | X307_splice | 9 | Splice | 1 | |
| 331# | c.991C>T | p.Q331* | 9 | Nonsense | 1 |
| c.993+1G>A | X331_splice | 9 | Splice | 1 | |
| c.993+2T>A | X331_splice | 9 | Splice | 1 | |
| c.994-1G>A | X331_splice | 10 | Splice | 1 | |
| 342# | c.1024C>T | p.R342* | 10 | Nonsense | 3 |
| c.1025del | p.R342fs | 10 | Frameshift | 1 | |
| c.1024del | p.R342fs | 10 | Frameshift | 1 |
#, ≥3 altered cases in our cohort; *, indicate the nonsense mutation.
Discussion
Extensive efforts have been made in comprehensive genomic sequencing of breast cancer tumors, further highlighting the genomic complexity in this heterogeneous disease. Racial diversity has been shown to be intimately associated with the pathogenesis of cancer (31). A prototypic example is the relatively high prevalence of EGFR mutation in Asian patients with lung adenocarcinoma (32). As described previously (15), only ~6% of breast tumors in TCGA are from Asian patients, suggesting that Asian patients are significantly under-represented in this publicly available database. Therefore, ethnic diversity may have a potential impact on the generalizability of the TCGA profiles to Asian breast cancer patients.
In the present study, we thus investigated the landscape of non-synonymous genomic mutations and CNAs of 33 cancer genes by using NGS methodology and established the genomic profiles of frequently altered cancer genes in Chinese breast tumors. In our GDPH cohort, the most frequently mutated gene was TP53 (45%), followed by PIK3CA (44%), GATA3 (18%), MAP3K1 (10%), whereas the copy-number amplifications were frequently observed in genes of ERBB2 (24%), MYC (23%), FGFR1 (13%) and CCND1 (10%). Among the 8 most frequently mutated or amplified genes, at least one driver was identifiable in 87.5% (n=267) of our GDPH cohort, revealing the significant contribution of these known driver genes in the development of Chinese breast cancer. This finding provides valuable genomic information for a future translational study focusing on the Chinese patients and ultimately should improve our understanding of pathogenesis and aggressive biological behavior of Chinese breast cancer.
Compared to the TCGA data, Chinese breast cancer patients had a significantly higher frequency of TP53 mutation (45% vs. 30%; q<0.001).This comparison seems valid since other studies consistently report a mutation frequency around 30% for the TP53 gene in western patients with primary breast cancer (4,9,33-35). Furthermore, we found a composition difference between the Chinese and TCGA cohorts, especially among HR+/HER2- group, in which Chinese breast cancer patients had significantly more non-missense mutations (inframe or frameshift indels, nonsense, and splicing), whereas missense mutation was the predominant mutation in TCGA cohort. To date, multiple studies have revealed a significant association between TP53 mutation and unfavorable prognosis in a number of cancer types, including breast cancer (12,29,36). It has been further reported that the type of non-missense mutation in TP53 is more strongly associated with poor survival than missense mutation in breast cancer (11,37), implicating differential clinically significance for TP53 mutation type and position (30). However, whether the relatively higher proportion of TP53 non-missense mutation, at least in part, contributes to the earlier onset of disease in Chinese patients merits further investigation.
In addition to TP53 gene, our Chinese cohort had a significantly lower mutation frequency in CDH1 (5% vs. 13%; q=0.036), but had a higher mutation prevalence for AKT1 (8% vs. 1%; q<0.001) and GATA3 (18% vs. 10%; q=0.036) when compared to the TCGA cohort. A further comparison revealed all of these differences occurred in HR+/HER2- subgroup (Figure 2C). As previously described (38,39), comprehensive comparison of the molecular portraits in different histopathologic types has demonstrated that CDH1 mutations are the best known ILC genetic hallmark (~65%), while GATA3 mutations are predominantly observed in luminal IDC than in ILC. Therefore, these differences in mutation prevalence of CDH1 and GATA3 might be attributable to the lower incidence of ILC in Chinese breast cancer (22). However, the histopathologic differences could not explain the significantly higher prevalence for AKT1 gene in our cohort, because AKT1 is more frequently mutated in ILC than in IDC (39). Of note, it has been reported that AKT1 E17K mutation occurs only in ~3–5% of western patients with primary HR+ breast cancer (4,6,9,10,34), but is significantly enriched in >10% of recurrent and metastatic samples (12,40). Recently, AKT1 mutation has been reported to be correlated with increased risk of early relapse (39), consistently implying the oncogenic potential for AKT1 E17K in driving the aggressive behavior and/or in conferring the resistance to conventional therapy of breast cancer (41). Here, we report a significantly higher prevalence for AKT1 mutation, occurring in 13% of Chinese patients with primary HR+/HER2- tumors. This finding is in agreement with ~11% of HR+/HER2- patients with AKT1 alteration in a recent report from China (42). Given the higher prevalence of AKT1 mutation in Chinese breast cancer and the success of AKT-targeted therapies (43,44), we recommend setting priorities for clinical trial development in Chinese patients with high risk of relapse or metastasis due to AKT1 mutation.
There are a few limitations associated with this study. Although we also identified several significant differences between Chinese and TCGA cohorts, such as higher prevalence for PIK3CA and NF1 alterations in Chinese TNBC group, limited sample size had hindered us from achieving a robust comparison between the two cohorts. Additionally, the prognostic values of frequently altered genes, such as TP53 mutation, AKT1 mutation, and MYC amplification, need to be confirmed using our cohort once the disease-free survival or overall survival data is more mature. Since it is a single center study using a 33-gene panel, further investigation is needed to validate our findings in a multi-center prospective trial by using a larger NGS-based panel.
Conclusions
In conclusion, we investigated the prevalence of 33 cancer genes and characterized the genomic mutational profiles of frequently altered genes in Chinese breast tumors, thus generating distinctive genomic features associated with clinically-defined subgroups in Chinese patients. More importantly, we further compared the mutational spectrum between Chinese and Caucasian patients, showing a significantly higher prevalence for TP53 and AKT1 in the Chinese population. The significant genomic differences between Asian and Caucasian patients, especially for TP53 mutations, merit further investigation. These results should improve our understanding of pathogenesis and/or metastatic behavior of breast cancer across races/ethnicities.
Figure S1.
A significantly higher non-missense mutation for TP53 in Chinese HR+/HER2- patients. The composition of TP53 mutation types in HR+/HER2- subgroup is shown, and the proportion of non-missense (frameshift, inframe, nonsense and splicing) mutation is compared between GDPH and TCGA cohorts using Fisher’s exact test.
Acknowledgements
We thank all the patients and their families for participation. We thank Dr. Kuok In Ngo for critical reading this manuscript.
Funding: This study was supported by funding from National Natural Science Foundation of China (81602645; 81071851; 81001189), Natural Science Foundation of Guangdong Province (2016A030313768; 2018A030313292) and Research Funds from Guangzhou Municipal Science and Technology Project (201707010418; 201804010430). The funding body has no roles in the design of the study and collection, analysis and interpretation of data and in writing the manuscript.
Ethical Statement: Primary tumor biopsies were obtained using an Institutional Review Board approved protocol, and this study had been approved by the Ethics Committee of Guangdong Provincial People’s Hospital (No. GDREC2014122H). All patients provided written informed consent for translational research.
Disclaimer: Availability of data and materials: The original raw sequence data have been deposited to the public databank National Omics Data Encyclopedia (NODE; http://www.biosino.org/node/index; Accession number: OEP000152).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Harbeck N, Gnant M. Breast cancer. Lancet 2017;389:1134-50. 10.1016/S0140-6736(16)31891-8 [DOI] [PubMed] [Google Scholar]
- 2.Ginsburg O, Bray F, Coleman MP, et al. The global burden of women's cancers: a grand challenge in global health. Lancet 2017;389:847-60. 10.1016/S0140-6736(16)31392-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerji S, Cibulskis K, Rangel-Escareno C, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 2012;486:405-9. 10.1038/nature11154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61-70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giltnane JM, Hutchinson KE, Stricker TP, et al. Genomic profiling of ER(+) breast cancers after short-term estrogen suppression reveals alterations associated with endocrine resistance. Sci Transl Med 2017;9. doi: . 10.1126/scitranslmed.aai7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis MJ, Ding L, Shen D, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 2012;486:353-60. 10.1038/nature11143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346-52. 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luen SJ, Asher R, Lee CK, et al. Association of Somatic Driver Alterations With Prognosis in Postmenopausal, Hormone Receptor-Positive, HER2-Negative Early Breast Cancer: A Secondary Analysis of the BIG 1-98 Randomized Clinical Trial. JAMA Oncol 2018;4:1335-43. 10.1001/jamaoncol.2018.1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nik-Zainal S, Davies H, Staaf J, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534:47-54. 10.1038/nature17676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012;486:400-4. 10.1038/nature11017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meric-Bernstam F, Frampton GM, Ferrer-Lozano J, et al. Concordance of genomic alterations between primary and recurrent breast cancer. Mol Cancer Ther 2014;13:1382-9. 10.1158/1535-7163.MCT-13-0482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meric-Bernstam F, Zheng X, Shariati M, et al. Survival Outcomes by TP53 Mutation Status in Metastatic Breast Cancer. JCO Precis Oncol 2018;2018. doi: 10.1200/PO.17.00245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network , Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113-20. 10.1038/ng.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blum A, Wang P, Zenklusen JC. SnapShot: TCGA-Analyzed Tumors. Cell 2018;173:530. 10.1016/j.cell.2018.03.059 [DOI] [PubMed] [Google Scholar]
- 15.Spratt DE, Chan T, Waldron L, et al. Racial/Ethnic Disparities in Genomic Sequencing. JAMA Oncol 2016;2:1070-4. 10.1001/jamaoncol.2016.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng L, Zhu X, Sun Y, et al. Prevalence and Prognostic Role of PIK3CA/AKT1 Mutations in Chinese Breast Cancer Patients. Cancer Res Treat 2019;51:128-40. 10.4143/crt.2017.598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iqbal J, Ginsburg O, Rochon PA, et al. Differences in breast cancer stage at diagnosis and cancer-specific survival by race and ethnicity in the United States. JAMA 2015;313:165-73. 10.1001/jama.2014.17322 [DOI] [PubMed] [Google Scholar]
- 18.Warner ET, Tamimi RM, Hughes ME, et al. Racial and Ethnic Differences in Breast Cancer Survival: Mediating Effect of Tumor Characteristics and Sociodemographic and Treatment Factors. J Clin Oncol 2015;33:2254-61. 10.1200/JCO.2014.57.1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan L, Strasser-Weippl K, Li JJ, et al. Breast cancer in China. Lancet Oncol 2014;15:e279-89. 10.1016/S1470-2045(13)70567-9 [DOI] [PubMed] [Google Scholar]
- 20.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 21.Li J, Zhang BN, Fan JH, et al. A nation-wide multicenter 10-year (1999-2008) retrospective clinical epidemiological study of female breast cancer in China. BMC Cancer 2011;11:364. 10.1186/1471-2407-11-364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng S, Bai JQ, Li J, et al. The pathologic characteristics of breast cancer in China and its shift during 1999-2008: a national-wide multicenter cross-sectional image over 10 years. Int J Cancer 2012;131:2622-31. 10.1002/ijc.27513 [DOI] [PubMed] [Google Scholar]
- 23.Christgen M, Derksen P. Lobular breast cancer: molecular basis, mouse and cellular models. Breast Cancer Res 2015;17:16. 10.1186/s13058-015-0517-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol 2010;28:2784-95. 10.1200/JCO.2009.25.6529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013;31:3997-4013. 10.1200/JCO.2013.50.9984 [DOI] [PubMed] [Google Scholar]
- 26.Takaku M, Grimm SA, Roberts JD, et al. GATA3 zinc finger 2 mutations reprogram the breast cancer transcriptional network. Nat Commun 2018;9:1059. 10.1038/s41467-018-03478-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurtado A, Holmes KA, Ross-Innes CS, et al. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 2011;43:27-33. 10.1038/ng.730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu GS. The functional interactions between the p53 and MAPK signaling pathways. Cancer Biol Ther 2004;3:156-61. 10.4161/cbt.3.2.614 [DOI] [PubMed] [Google Scholar]
- 29.Silwal-Pandit L, Vollan HK, Chin SF, et al. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin Cancer Res 2014;20:3569-80. 10.1158/1078-0432.CCR-13-2943 [DOI] [PubMed] [Google Scholar]
- 30.Hainaut P, Pfeifer GP. Somatic TP53 Mutations in the Era of Genome Sequencing. Cold Spring Harb Perspect Med 2016. doi: . 10.1101/cshperspect.a026179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calvo E, Baselga J. Ethnic differences in response to epidermal growth factor receptor tyrosine kinase inhibitors. J Clin Oncol 2006;24:2158-63. 10.1200/JCO.2006.06.5961 [DOI] [PubMed] [Google Scholar]
- 32.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 33.Basho RK, de Melo Gagliato D, Ueno NT, et al. Clinical outcomes based on multigene profiling in metastatic breast cancer patients. Oncotarget 2016;7:76362-73. 10.18632/oncotarget.12987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereira B, Chin SF, Rueda OM, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun 2016;7:11479. 10.1038/ncomms11479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lefebvre C, Bachelot T, Filleron T, et al. Mutational Profile of Metastatic Breast Cancers: A Retrospective Analysis. PLoS Med 2016;13:e1002201. 10.1371/journal.pmed.1002201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffith OL, Spies NC, Anurag M, et al. The prognostic effects of somatic mutations in ER-positive breast cancer. Nat Commun 2018;9:3476. 10.1038/s41467-018-05914-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olivier M, Langerod A, Carrieri P, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res 2006;12:1157-67. 10.1158/1078-0432.CCR-05-1029 [DOI] [PubMed] [Google Scholar]
- 38.Ciriello G, Gatza ML, Beck AH, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015;163:506-19. 10.1016/j.cell.2015.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desmedt C, Zoppoli G, Gundem G, et al. Genomic Characterization of Primary Invasive Lobular Breast Cancer. J Clin Oncol 2016;34:1872-81. 10.1200/JCO.2015.64.0334 [DOI] [PubMed] [Google Scholar]
- 40.Yates LR, Knappskog S, Wedge D, et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell 2017;32:169-84.e7. 10.1016/j.ccell.2017.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carpten JD, Faber AL, Horn C, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 2007;448:439-44. 10.1038/nature05933 [DOI] [PubMed] [Google Scholar]
- 42.Chen L, Yang L, Yao L, et al. Characterization of PIK3CA and PIK3R1 somatic mutations in Chinese breast cancer patients. Nat Commun 2018;9:1357. 10.1038/s41467-018-03867-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hyman DM, Smyth LM, Donoghue MTA, et al. AKT Inhibition in Solid Tumors With AKT1 Mutations. J Clin Oncol 2017;35:2251-9. 10.1200/JCO.2017.73.0143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim SB, Dent R, Im SA, et al. Ipatasertib plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer (LOTUS): a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol 2017;18:1360-72. 10.1016/S1470-2045(17)30450-3 [DOI] [PMC free article] [PubMed] [Google Scholar]